Abstract
The ErbB2 (Her2/neu epidermal growth receptor family) oncogene is overexpressed in 30% to 40% of human breast cancers. Cyclin D1 is the regulatory subunit of the holoenzyme that phosphorylates and inactivates the retinoblastoma (pRb) tumor suppressor and is an essential downstream target of ErbB2‐induced tumor growth. Herein, we demonstrate that ErbB2 induces the activity of the Notch signaling pathway. ErbB2 induction of DNA synthesis, contact‐independent growth, and mammosphere induction required Notch1. ErbB2‐induced cyclin D1 and cyclin D1 expression was suficient to induce Notch1 activity, and conversely, genetic deletion of Notch1 in mammary epithelial cells using foxed Notch (Notchfl/fl) mice demonstrated that cyclin D1 is induced by Notch1. Genetic deletion of cyclin D1 or small interfering RNA (siRNA) to cyclin D1‐reduced Notch1 activity and reintroduction of cyclin D1 into cyclin D1‐deficient cells restored Notch1 activity through the inhibition of Numb, an endogenous inhibitor of Notch1 activity. Thus, cyclin D1 functions downstream as a genetic target of Notch1, amplifies Notch1 activity by repressing Numb, and identifies a novel pathway by which ErbB2 induces Notch1 activity via the induction of cyclin D1.
Keywords: cancer biology, oncogenes, signal transduction
Introduction
Cancer arises as a result of the accumulation of multiple genetic lesions that ultimately result in unregulated cell cycle entry and loss of the apoptotic response. 1 , 2 Although a malignant transformation of different cell types may exhibit different combinations of cellular dysfunction, three dominant hallmarks of cancer cells are unregulated proliferation, evasion of apoptosis, and invasion. 2 One cell fate determinant in undifferentiated, proliferative cell populations, such as stem and progenitor cells, is the evolutionarily conserved Notch transmembrane receptor. 3
Four Notch homologs have been identified in the mammalian cells, 4 and in several malignancies, and all four are capable of inducing fibroblast transformation. 4 The full‐length Notch protein is processed to an approximately 120 kDa intracellular fragment and a 190 kDa extracellular segment before it reaches the plasma membrane. 5 Cleavage of the Notch intracellular domain (NICD) occurs after the ligand has bound to the extracellular portion of the Notch1 receptor and through a series of proteolytic reactions mediated by tumor necrosis factor‐alpha‐converting enzyme (TACE), presenilin/γ‐secretase, and Kuzbanian. 6 , 7 Presenilin1 is a multipass transmembrane protein that mediates the intramembrane proteolysis of the Notch receptor, liberating the intracellular domain, after ligand binding. The Notch ligands Jagged 1, Jagged 2, and Delta 1 8 have been identified in the mammalian cells, and interact with Notch to induce rapid cleavage, nuclear translocation, and phosphorylation of Notch after ligand binding. 8 The consequent release of the NICD leads to nuclear translocation, where it interacts with CSL (CBF‐1, suppressor of Hairless, LAG1; also known as RBP‐κJ) 1 , 2 , 9 , 10 and lymphoid enhancer‐binding factor 1 (LEF‐1). CSL functions as a transcriptional repressor binding co‐repressors and histone deacetylases. 5 , 6 , 7
A growing body of evidence supports a role for each of the four Notch genes in tumorigenesis. The elevated expression of Notch1 and Notch2 are associated with cervical, colon, and lung carcinomas. 11 , 12 In addition, a subset of T‐cell leukemias exhibits translocation of the NICD t(7:9)(q34;q34) to the enhancer and promoter elements of the T‐cell receptor (TCRB), leading to T‐cell transformation. 13 Simian vacuolating virus 40 T antigen (SV40T)‐mediated transformation increases Notch1 expression and cell cycle progression, 14 and overexpression of constitutively active Notch (caN) transforms early adenovirus oncoprotein 1 (E1A)‐immortalized rat kidney epithelial (RKE) cells. 15 The activation of the Notch locus is involved in mammary tumorigenesis. 16 , 17 Notch1 is highly expressed in primary breast carcinomas 18 and is rearranged by mouse mammary tumor virus (MMTV) proviral insertion that results in caN. 19
Our recent studies demonstrated that Notch1 expression in RKE cells induced contact‐independent growth in a cyclin D1‐dependent manner. 20 Cyclin D1 abundance correlated with Notch1 activity during embryogenesis and Notch1‐induced cyclin D1 expression and transcription, requiring a CBF‐binding site in the cyclin D1 promoter. 20 Notch signaling is activated in human breast cancer, 21 whereas a negative regulator of Notch1, Numb, is reduced in this disease. 22 Since the Her2/neu epidermal growth receptor family (ErbB2) oncogene is overexpressed or amplified in approximately 30% of human breast cancer, the current studies were conducted to determine whether ErbB2 or other oncogenes implicated in human breast cancer govern Notch1 activity.
Methods
Cell lines and cell culture
Breast cancer cell lines (MCF‐7, SKBR3, MDA‐MB231, MDA‐MB435, MDA‐MB436, BS‐184, BT‐474 and T47D) were routinely cultured in Dulbecco's modified eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). The parental MCF‐10A cell line was cultured as previously described. 23 The MCF‐10A sublines were derived by transducing cells with oncogene‐expressing retrovirus expression vectors, retroviral vector control (pBABE), pBABE/c‐Myc, pBABE/H‐Ras G12V, pBABE/ErbB2 (the ErBb2 construct contains a constitutively active rat homolog, also known as NeuT 24 ), pBABE/Ras‐ErbB2, and pBABE/v‐Src. These sublines were cultured in DMEM supplemented with 10% FBS. All cells were grown in a humidified atmosphere with 5% CO2 at 37°C.
Transformation
The retroviral transduction was performed as described. 25 The pBABE/vector and its oncogenic derivatives were previously co‐transfected with pSV‐Y‐A‐MLV amphotropic helper vector into 293T cells. Parental MCF‐10A cells were infected with a virus‐containing medium and selected in 2 μg/mL puromycin for 2 weeks. The expression of the oncogenes was confirmed by Western blotting and the neoplastic transformation was assessed by anchorage‐independent growth.
Plasmid construction and transgenic mice
The caN1 construct was a generous gift from Dr. Igor Prudovsky, Maine Medical Centre Research Institute. 26 The constitutively active ErbB2 (NeuT) construct was obtained by cloning the complete coding region from the pSV2/ErbB2 (a kind gift from Dr. William J. Muller, McGill University, Canada) into the pCMV5 vector using the HindII and SalI restriction sites to produce the plasmid pCMV5/ErbB2. The ErbB2 complementary deoxynucleic acid (cDNA) from the pCMV5/ErbB2 was subcloned into the pBABE/puro vector using the EcoRI and SalI restriction sites to produce the plasmid pBABE/ErbB2. pBABE/c‐Myc and pBABE/Ras‐ErbB2 have been previously described. 27 The pBABE/H‐Ras G12V and pBABE/v‐Src were a kind gift from Dr. Michael Lisanti (Tomas Jefferson University, PA, USA). The expression vector for Musashi, 28 the reporter plasmid TCF reporter luciferase (TOP‐luc), 29 and the Cp‐binding factor 1(8)‐luciferase (CBF(8)‐luc) reporter constructs 30 were previously described. The expression plasmids encoding adenovirus‐directing Cre expression (Ad‐Cre) or control virus (Ad‐Null) 31 and transgenic animals carrying foxed Notch1 alleles (Notchfl/fl) 20 were previously described. Notchfl/fl mammary epithelial cells (MECs) were generated from Notchfl/fl transgenic mice and cultured as previously described. 32 MECs were treated with either Ad‐Cre or Ad‐Null at a concentration of 2 × 10 7 plaque forming units/mL for 5 days. Cyclin D1 KO and cyclin D1 “rescued” mouse embryonic fibroblasts (MEFs) were prepared as previously described. 33 The Tomas Jefferson University ethics committee approved all laboratory animal procedures.
Western blot analysis
Western blot analysis was carried out as previously described. 34 The antibodies were obtained from the following sources: rabbit polyclonal antibody to the NICD (07‐220; Upstate Biotechnology, Lake Placid, NY, USA), rabbit monoclonal antibody to Musashi (ab21268; Abcam, Cambridge, MA, USA), rabbit polyclonal antibody to Numb (ab14140; Abcam), rabbit polyclonal antibody to guanine dissociation inhibitor (GDI, 4809; RTG, Gaithersburg, MA, USA), Cyclin D1 (DCS‐6; Santa Cruz Biotechnology, California, CA, USA) and c‐Neu (PC04‐100UG; Oncogene Science, Boston, MA, USA). The appropriate horseradish peroxidase‐conjugated secondary antibodies were subsequently applied, and immunodetection was visualized by chemiluminescence. Densitometry was performed using an Alpha Imager software (Alpha Innotech, San Leandra, CA, USA).
Tissue microarrays and immunohistochemistry
Human breast tissue microarrays were constructed from paraffin‐embedded tissues using the cutting‐edge matrix assembly (CEMA) technique 35 and consisted of 80 invasive ductal carcinomas and 20 normal breast tissues. Antigen retrieval was conducted for 30 minutes using Citra (Biogenex, San Ramon, CA, USA) in a steamer. Slides were blocked with peroxidase for 15 minutes and 10% normal goat serum for 30 minutes at room temperature. Subsequently, the arrays were incubated for 1 hour, with an antibody specific for the NICD (100‐401‐405; Rockland, Gilbertsville, PA, USA) or ErbB2 (A0485; Dako, Carpinteria, CA, USA) at dilutions 1:500 and 1:100, respectively. The slides were washed in TBS‐Tween and incubated with a polymer labeled with secondary antibody (Alexa Flour 488 goat anti‐rabbit; Invitrogen, Carlsbad, CA, USA), for 30 minutes at room temperature, then washed with phosphate buffer saline (PBS). The arrays were stored overnight at 4°C. Twentyfour hours later, peroxidase and protein blocks were performed again as described above, then the arrays were incubated with a cytokeratin antibody (M3515, mouse anti‐human, clone AE1/AE3; DakoCytomation, Carpinteria, CA, USA), diluted 1:50, for 1 hour at room temperature. The slides were washed in TBS‐Tween and incubated with a polymer labeled with secondary antibody (T‐862, Texas Red goat anti‐mouse; Molecular Probes, Carlsbad, CA, USA), for 30 minutes at room temperature, then washed with PBS. The slides were then stained with 4′‐6‐diamidino‐2‐phenylindole (DAPI). Immunofluorescent images were taken using a PM2000 microscope (HistoRX, New Haven, CT, USA; magnification ×60). These images were then analyzed for the intensity of the NICD and ErbB2 expression in human breast epithelial cells using the AQUA/PM2000 platform (HistoRX). The study of unidentified, archival breast tissues were performed under guidelines approved by the Tomas Jefferson University Institutional Review Board.
γ‐secretase inhibition and Heregulin assays
SKBR3, MCF‐10A, and MCF‐10A/ErbB2 cells were grown on six well plates (100,000 cells/well). The cells were treated for 24 hours with combinations of 2 nM Heregulin (recombinant Human β1/; R&D systems, Minneapolis, MN, USA) and either 1 μM or 5 μM γ‐secretase inhibitor X (GSI, 565771; EMD Biosciences, San Diego, CA, USA).
Luciferase reporter assays and transfection
The CBF(8)‐luc plasmid, pBABE/ErbB2 expression vector, Musashi, and caN1 constructs were transfected using Easy Transgater (America Pharma Source, Gaithersburg, MD, USA) into MCF‐7 or SKBR3 cells, according to the manufacturers' guidelines. The appropriate control vector constructs were used for normalization. At 36 hours post transfection, a luciferase assay was conducted as previously described. 36 The small interfering RNA (siRNA)‐coupled luciferase assay used Easy Transgater‐si (America Pharma Source) to transfect MCF7 cells with 150 nM cyclin D1 and Allstar control siRNA (Qiagen, Valencia, CA, USA), for 72 hours. At 36 hours post siRNA transfection, the CBF(8)‐luc and caN1 constructs were transfected. At 72 hours post transfection, a luciferase assay was conducted as previously described. 36 The relative luciferase activities were calculated by normalizing transfection efficiency according to the Renilla luciferase activities and compared to the pGL2, control vector for the CBF(8)‐luc plasmid. The ErbB2 inhibitor CP‐654577 was provided by Dr. James Moyer, Pfizer, and has been previously described. 37
Soft agar assay for anchorage‐independent growth
The details concerning the soft agar assay have been previously described. 38 SKBR3 and MCF‐10A/ErbB2 cells were used in the soft agar assay. The cells were treated for 24 hours with 2 nM Heregulin and either 1 μM or 5 μM GSI.
Mammosphere production
Normal breast tissue from mastectomies was dissociated mechanically and enzymatically, as previously described. 39 Mammospheres were cultured as previously described. 40 The mammospheres were treated for 5 days with 2 nM DSL (Notch ligand‐Delta, Serrate and Lag; Genescript Piscataway, NJ, USA), 5 nM Heregulin, and 5 μM GSI. In total, 100 μL of the media containing mammospheres was added to the wells in a 96‐well plate and counted using a Zeiss Axiovert 200 microscope (magnification ×10). Data are expressed as mammospheres/1,000 cells. This study was approved by the Institutional Review Board, Thomas Jefferson University.
Cell‐cycle analysis and siRNA
In total, 1 × 105 exponentially growing SKBR3 cells were transfected, to a final concentration, with either 150 nM Notch1 (Ambion, Austin, TX, USA) or Allstar control siRNA, using oligofectamine (Invitrogen) according to the manufacturers' protocol. The cells were starved in serum‐depleted medium (0.3% FBS) for a period of 72 hours post siRNA transfection and further stimulated with 2 nM Heregulin for 24 hours. At the 96‐hour time‐point, the cells were collected by trypsinization, fixed in 10% methanol, and resuspended in PBS containing 20 mg/mL propidium iodide (PI) and 5 U/mL ribonuclease (RNAse). The cells were analyzed by an flourescence‐activated cell sorting (FACS) analysis to determine cell cycle status.
Results
ErbB2 correlation with activated Notch1 in human breast cancer
The activation of Notch signaling results in a sequential cleavage by TACE, presenilin/γ‐secretase, and Kuzbanian, leading to a subsequent release of the NICD. In order to examine the activation of Notch signaling in human MECs, Western blotting was conducted using an antibody specific to the NICD. A comparison was made with a series of MCF‐10A clonal derivatives transduced with retroviral expression vectors including H‐Ras G12V, as Ras is known to activate Notch signaling in fibroblasts. 18 Compared to retroviral vector control, H‐Ras G12V transduction of MCF‐10A cells did not induce NICD abundance ( Figure 1A ). c‐Myc and v‐Src transduction of MCF‐10A also resulted in no significant change. In contrast, transduction of cells with the constitutively active ErbB2 construct increased the abundance of the NICD 6‐fold ( Figure 1B ). Collectively, these studies indicate that ErbB2 substantially enhances Notch signaling in human breast cancer cells.
Figure 1.
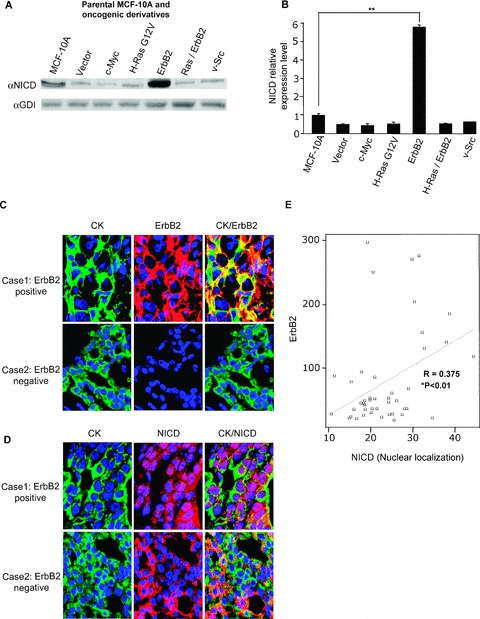
Oncogene regulation of Notch activity. (A) Notch1 expression determined in parental MCF‐10A cells and MCF‐10A oncogenic derivatives by Western blot with an antibody specific to the NICD. GDI was used as a protein loading control. (B) Densitometry of the NICD from Western blots (N= 6, **p < 0.001). (C) ErbB2 abundance assessed by immunohistochemical staining of invasive ductal carcinomas. Case 1 is an ErbB2‐positive invasive ductal carcinoma and case 2 is an ErbB2‐negative invasive ductal carcinoma (magnification ×60). (D) Nuclear localization of NICD assessed by immunohistochemical staining of invasive ductal carcinomas (case 1 and case 2 as above; magnification ×60). (E) Scatter plot showing a positive correlation between nuclear NICD staining and ErbB2 staining in invasive ductal carcinomas (N= 45, *p < 0.01, ErbB2 staining intensity and Notch1 staining intensity compared by the Pearson correlation; 2‐tailed). CK is the cytokeratin antibody (M3515, mouse antihuman, clone AE1/AE3; DakoCytomation).
To determine whether the activation of the NICD occurred in human breast cancer cells and if it was induced by ErbB2, a tissue array of human breast cancers and normal tissues were analyzed with an antibody directed to the NICD and ErbB2. A quantitative immunohistochemical staining, using the AQUA system, demonstrated evidence for cytoplasmic staining of NICD in normal and ErbB2‐negative breast tissue. There was a positive correlation of nuclear localization of the NICD in ErbB2‐positive invasive ductal carcinomas in comparison to ErbB2‐negative tissue ( Figure 1C–E ).
ErbB2 induction of Notch1 signaling
In order to examine further the mechanism by which the activation of ErbB2 induces Notch1 signaling, MCF‐10A cells were treated with Heregulin to induce, via ErbB3/ErbB2 heterodimerization, the activation of ErbB2 signaling. The addition of 2 nM Heregulin increased NICD expression 3‐fold. Notch signaling is known to require γ‐secretase activity. To determine the requirement of γ‐secretase in ErbB2 activation of Notch1, the GSI was added to the Heregulin‐treated cells (parental MCF‐10A, MCF‐10A/ErbB2, and SKBR3 cells). The addition of GSI had no significant effect on the basal Notch activity in parental MCF‐10A cells. However, the Heregulin‐induced activation of Notch signaling was abrogated by the addition of GSI at either 1 μM or 5 μM ( Figure 2A and B ). In addition, the Heregulin‐mediated induction of Notch1 activity was reduced by the GSI in MCF‐10A/ErbB2 and SKBR3 cells.
Figure 2.
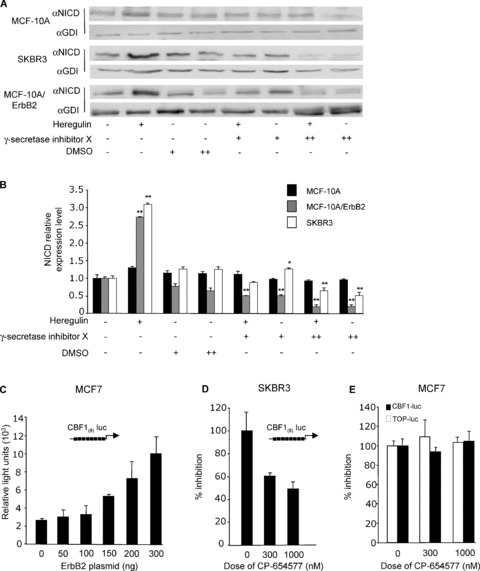
Heregulin induction of Notch activation isγ‐secretase‐dependent. (A) MCF‐10A, MCF‐10A/ErbB2, and SKBR3 cells were treated with Heregulin (2 nM) and the γ‐secretase X inhibitor at 1 mM/mL (+) and 5 mM/mL (++) for 24 hours. Western blots were probed with an antibody specific to the NICD. Membranes were stripped and reprobed for the loading control GDI. (B) densitometry of Western blots (N= 6, *p < 0.05, **p < 0.001 compared by paired t‐test to Heregulin and DMSO controls). (C) MCF‐7 cells were cotransfected with the reporter plasmid CBF(8)‐luc and an expression vector encoding ErbB2. Luciferase activity is shown as mean ± standard error of mean (SEM) for N > 5 separate transfections. (D) SKBR3 cells were transfected with CBF(8)‐luc, and an ErbB2 inhibitor, CP‐654577, was added at dual concentrations. (E) MCF‐7 cells were transfected with CBF(8)‐luc and TCF‐luciferase (TOP‐luc) and treated with CP‐654577. Luciferase activity was determined 36 hours post transfection, normalized for protein and expressed as a percentage of control.
To determine whether the activation of ErbB2 was sufficient for the induction of CBF reporter activity, the multimeric CBF(8)‐luc reporter, which is a heterologous reporter encoding multimeric copies of the HEY‐CBF binding site, was transfected into MCF‐7 cells in conjunction with an ErbB2 expression vector. ErbB2 induced CBF reporter activity ( Figure 2C ). To determine the role of endogenous ErbB2 in mediating Notch1 activity, SKBR3 cells, which overexpress ErbB2, were transfected with the CBF(8)‐luc reporter plasmid. The addition of the ErbB2 inhibitor (CP‐654577) reduced CBF reporter activity by 50% ( Figure 2D ). In breast cancer cells with low levels of ErbB2 expression, such as MCF‐7 cells, endogenous CBF activity was unaffected by the ErbB2 inhibitor ( Figure 2E ). These studies suggest ErbB2 can activate CBF transcriptional activity, and that endogenous CBF activity correlates with the relative activity of ErbB2 in human breast cancer cells.
ErbB2 induction of DNA synthesis, colony formation, and mammospheres involves Notch1
Previous studies have demonstrated that the transduction of MCF‐10A cells with cDNAs encoding the NICD and RBP‐KJ/VP16 fusion protein is sufficient for the induction of colony formation. 21 In order to determine the relative contribution of ErbB2 to the activation of Notch1 and colony formation, MCF‐10A/ErbB2 cells were examined in colony formation assays. The addition of Heregulin increased the number of colonies 2‐fold ( Figure 3A ). The addition of the GSI abrogated the induction of colonies by Heregulin, consistent with the ability of the GSI to reduce the Heregulin‐induced NICD ( Figure 3A ). The ErbB2‐overexpressing cell line SKBR3 showed an identical trend ( Figure 3B ). Heregulin enhanced colony formation by 40%, and the addition of GSI abolished the Heregulin‐induced colony growth. The GSI reduced the basal level of SKBR3 colony formation by 15%, consistent with the role of endogenous ErbB2‐dependent activation of Notch1 in SKBR3 cell colony formation.
Figure 3.
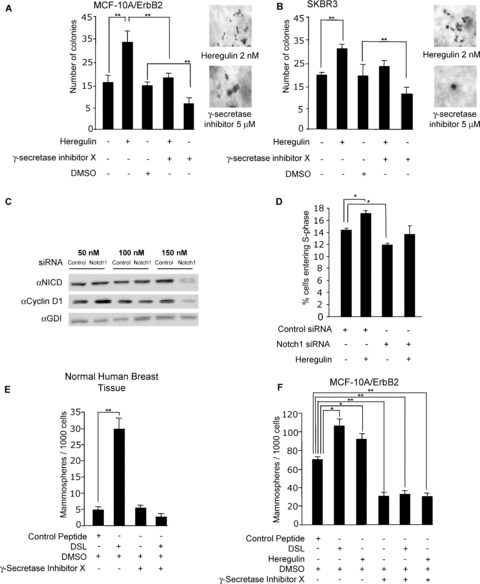
Heregulin induction of colony formation and DNA synthesis involves Notch signaling. (A) MCF‐10A/ErbB2 and (B), SKBR3 cells were grown in soft agar medium and treated with γ‐secretase X inhibitor (5 mM) and Heregulin (2 nM). Colonies were counted at 10 days (N= 3, **p < 0.001). (C) SKBR3 cells were treated with Notch1 siRNA or control siRNA for 72 hours, and Western blots were probed for the NICD, cyclin D1, and GDI (N= 6). (D) Serum‐starved SKBR3 cells were treated with either Notch1 siRNA (150 nM) or control siRNA. Following 72 hours of treatment with siRNA, the cells were stimulated with 2 nM Heregulin for 24 hours, and a cell‐cycle analysis was conducted. Data are represented as percentage of cells in S‐phase (mean ± SEM for N= 3, *p < 0.05). (E, F) Mammosphere induction by Notch1 ligand (DSL). Mammospheres were generated from normal breast tissue mastectomies and MCF‐10A/ErbB2 cells and were cultured in stem cell media. Mammospheres were treated with 2 nM DSL, 5 nM Heregulin, or γ‐secretase inhibitor X at 5 mM for 5 days. The mammosphere population/1,000 cells was determined (data are mean ± SEM for N= 3, *p < 0.05, **p < 0.001).
Notch1 siRNA reduced the NICD abundance with a corresponding reduction in cyclin D1 abundance ( Figure 3C ). Heregulin increased the proportion of SKBR3 cells in the DNA synthetic S‐phase of the cell cycle compared to control ( Figure 3D ). Notch1 siRNA reduced the proportion of cells in S‐phase by 15% ( Figure 3D ). Thus, Notch1 siRNA abrogated the Heregulin‐mediated induction of S‐phase, strongly suggesting the requirement for Notch1 in the Heregulin‐mediated induction of DNA synthesis.
Notch signaling plays an essential role in regulating cell fate and progenitor cell expansion, apoptosis, proliferation, and cellular migration. We determined the contribution of ErbB2 to Notch1 ligand (DSL)‐mediated induction of mammospheres in MCF‐10A and MCF‐10A/ErbB2 cells. Previous data have shown that human mammospheres are increased in number by DSL and reduced by GSI. 40 Our studies have demonstrated a requirement for Notch1 in the Heregulin‐mediated induction of DNA synthesis. To determine whether Heregulin mediates other Notch1 functions, we examined the role of Notch1 in regulating mammary progenitor cell expansion. The human mammospheres were used as a positive control ( Figure 3E ). The addition of DSL increased the number of mammospheres derived from primary human MECs, consistent with prior publications ( Figure 3E ). The addition of GSI abrogated the DSL‐induced mammosphere function. DSL increased the mammosphere population by 30% in MCF‐10A/ErbB2 cells ( Figure 3F ). GSI abrogated the DSL‐induced mammosphere formation ( Figure 3F ).
ErbB2 induction of Notch1 signaling requires cyclin D1
ErbB2 activation correlated with the induction of Notch1 activity. Therefore, we investigated the possibilities that ErbB2 may regulate downstream mediators of Notch1 signaling to induce Musashi1 (which enhances Notch1 signaling), inhibit Numb (an endogenous inhibitor of Notch1), or regulate the abundance of an additional factor. Our prior studies had suggested a relationship between Notch1 activity and cyclin D1. The activating mutants of Notch1 induce cyclin D1 expression in fibroblasts. 20
To examine the relationship between cyclin D1 and Notch1 in breast cancer cells, Western blot analysis was conducted on multiple cell lines. The cell lines with higher levels of active Notch1 showed a trend of higher levels of cyclin D1 ( Figure 4A ). ErbB2‐overexpressing cell lines had the highest expression of cyclin D1 and the NICD ( Figure 4B ). Our previous studies had shown a positive correlation between Notch1 and cyclin D1 during murine embryological development. 20 The possibility that cyclin D1 may also function upstream to regulate Notch1 activity had not previously been assessed. In order to determine whether cyclin D1 could regulate Notch1 activity, siRNA to cyclin D1 was used. NICD is released by proteolytic cleavage and translocates to the nucleus where it binds via the ankyrin repeats to transcription factor Suppressor of Hairless (Su(H), or CBF1 in invertebrates). Musashi functions to enhance Notch signaling in fibroblasts. In order to examine the effect of cyclin D1 on Notch intracellular activity, MCF7 cells were transduced with vectors expressing Musashi 1 and caN1 and treated with cyclin D1 siRNA. A luciferase assay was conducted using CBF(8)‐luc. The transfection of MCF‐7 cells with caN1 increased CBF(8)‐luc reporter activity 250‐fold. The additional transfection with an expression vector encoding Musashi 1 enhanced the CBF(8)‐luc activity to 500‐fold ( Figure 4C ). Thus, caN1 and Musashi 1 enhance Notch1 transcriptional activity in MCF‐7 cells. Cyclin D1 siRNA reduced CBF(8)‐luc activity 5‐fold ( Figure 4C ).
Figure 4.
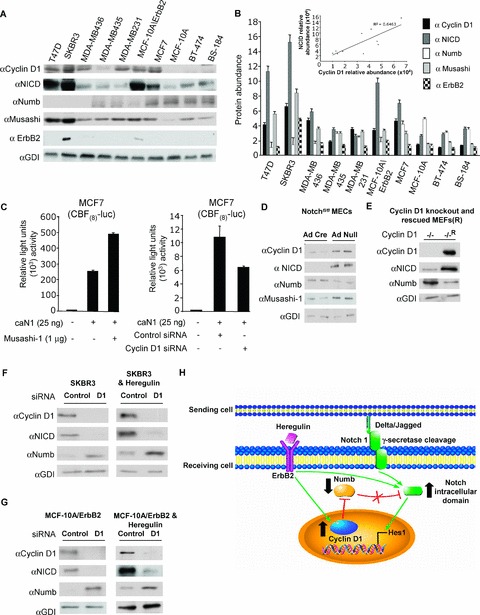
Cyclin D1 expression induces Notch activity. (A) Western blots of breast cancer cell lines for the NICD, cyclin D1, Musashi, Numb, and ErbB2, and GDI is a protein loading control. Densitometry of Western blots (N= 3) shown in (B) with relative abundance of cyclin D1 and the NICD shown as an inset. (C) The CBF(8)‐luc reporter was transfected into MCF7 cells with expression vectors for constitutively active Notch1 (caN1) or Musashi‐1. MCF‐7 cells were also transfected with 150 nM cyclin D1 siRNA to determine the role of endogenous cyclin D1 in regulating Notch1 activity (data are mean ± SEM for N > 5 separate transfections). (D) Notchfl/fl‐derived mammary epithelial cells were transduced with adenovirus expressing Cre recombinase or control adenovirus vector. Cell lysates were prepared and analyzed by Western blot using antibodies for indicated proteins. (E) Cyclin D1 KO (−/−) and Cyclin D1 KO rescued (−/−R) MEFs were analyzed by Western blot using antibodies for indicated proteins. (F, G) SKBR3 cells or MCF‐10A/ErbB2 cells were transfected with 150 nM cyclin D1 siRNA. Western blots were probed with antibodies directed to the NICD, Numb, or cyclin D1, and the protein loading control GDI (data representative for N= 3 experiments). (H) Schematic representation showing ErbB2 induction of cyclin D1 and Notch signaling and proposed mechanism by which cyclin D1 functions to enhance Notch1 activity.
To determine whether endogenous cyclin D1 is a physiological target of Notch1 in MECs, we examined Notchfl/fl transgenic mice. MECs were prepared from Notchfl/fl mice and treated with Ad‐Cre and Ad‐control. Excision of Notch1 with Ad‐Cre reduced the NICD abundance ( Figure 4D ). Compared to Ad‐Null, cyclin D1 was reduced by 90%. Upon deletion of Notch1, Numb abundance was increased by 60%, and Musashi 1 levels were reduced by 70% ( Figure 4D ).
To further examine the role of cyclin D1 in regulating the NICD, MEFs were prepared from cyclin D1 −/− mice. Western blot analysis showed that the NICD abundance was reduced in cyclin D1 −/− mice by 80%, compared to MEFS from cyclin D1‐rescued mice ( Figure 4E ). Cyclin D1 siRNA was used to transduce SKBR3 and MCF‐10A/ErbB2 cells. The Western blotting was conducted using an antibody specific to cyclin D1 and the NICD. siRNA to cyclin D1 reduced cyclin D1 abundance by >90% in SKBR3 cells ( Figure 4F ). The NICD abundance was reduced by >60%. This finding suggests that endogenous cyclin D1 increases Notch1 activity. In contrast, Numb abundance was increased by 60% by cyclin D1 siRNA ( Figure 4F ). MCF‐10A/ErbB2 cells were transfected with siRNA. Cyclin D1 abundance was reduced by approximately 95%. These results provide further support to the idea that cyclin D1 induces the NICD to enhance Notch signaling ( Figure 4G ).
Discussion
The current studies demonstrate that the activation of ErbB2 signaling, either through point mutation of the receptor or through addition of the ligand Heregulin, or the Notch ligand DSL, induces activity of the Notch signaling pathway. The ErbB2 kinase inhibitor CP 654577 inhibited Notch1 activity, assessed using the CBF(8)‐luc reporter assay. The activation of Notch signaling by ErbB2 was demonstrated through Western blot analysis, immunohistochemistry analysis for the NICD, and induction of the transcriptional activity using a CBF(8)‐luc reporter. The induction of Notch1 activity by ErbB2 was functionally significant as the ErbB2‐mediated induction of contact‐independent growth was reduced upon inhibition of Notch1 signaling using the GSI.
The Heregulin induction of a contact‐independent growth was blocked by the GSI. Currently, patients with ErbB2‐expressing breast cancer are treated with the immunoneutralizing antibody Herceptin. The resistance to Herceptin arises in 80% of patients, suggesting that alternative treatments are essential. The mechanism of resistance to Herceptin is poorly understood.
The finding that γ‐secretase inhibition blocked the growth of ErbB2‐expressing human breast cancer suggests that Notch1 inhibition may be a complementary approach for the Herceptin‐resistant breast cancer.
During analysis of the mechanism by which ErbB2 induced Notch1 activity, we observed that the expression of cyclin D1 was induced by ErbB2 and correlated with the activation of Notch1. The current studies demonstrate for the first time that Notch1 activity is induced by cyclin D1. The expression of cyclin D1 siRNA reduced Notch1 activity, assessed either using Western blot analysis of the NICD or using CBF(8)‐luc reporter activity. The reintroduction of cyclin D1 into cyclin D1‐defficient cells enhanced Notch1 activity. These studies have broad implications for breast tumorigenesis as cyclin D1 is induced by a variety of oncogenic signals and may, therefore, provide a mechanism by which multiple signaling pathways enhance Notch1 activity.
We hypothesized that cyclin D1 may induce Notch1 activity either by repressing Numb or by inducing Musashi 1 expression. In MCF7 and MCF‐10A/ErbB2 cells, cyclin D1 siRNA induced Numb and decreased the NICD; similar findings were made in MEFs. In MCF7 and MCF‐10A/ErbB2 cells, siRNA for cyclin D1 decreased Musashi 1 abundance, consistent with a model in which cyclin D1 induction of Notch1 activity may induce Musashi 1 in MECs. Musashi 1 levels were, however, unchanged upon reintroduction of cyclin D1 into cyclin D1 −/− fibroblasts. The mechanism by which cyclin D1 enhances Notch1 activity in different cell types remains to be determined. Collectively, these studies are consistent with the mechanism by which cyclin D1 enhances Notch1 activity through the inhibition of Numb expression ( Figure 4H ).
Several recent studies have provided evidence that Notch signaling is activated in breast cancer. In a recent immunohistochemical study using a tissue array of 98 cases of invasive breast cancer, ErbB2 status correlated with the Notch abundance. 41 The negative regulator of Notch signaling, Numb, is reduced in >50% of human breast cancers. Numb expression is lost in approximately 50% of breast tumors due to ubiquitination and proteosomal degradation. 22 The activation of Notch signaling was evidenced by an increase in the abundance of the NICD.
The current studies are consistent with the recent observations of 20 breast cancer samples showing the activation of Notch signaling. 21 Our studies confirm and extend these observations by demonstrating that Notch1 activity is induced by cyclin D1.
Conclusion
Notch activity is a key determinant of cellular development and differentiation. Recent studies have strongly implicated an increased Notch activity in promoting tumorigenesis, including breast cancer. The molecular mechanisms regulating Notch1 activity are, therefore, of fundamental importance. Herein, endogenous cyclin D1 enhanced Notch1 activity. Cyclin D1 is a labile protein that is induced by growth factors and oncogenes and repressed by several tumor suppressors. Cyclin D1 encodes a rate‐limiting component of the cell cycle and cellular proliferation pathway, encoding the regulatory subunit of the holoenzyme that phosphorylates and inactivates the retinoblastoma (pRb) protein. The current studies demonstrating the induction of Notch1 activity by cyclin D1 provide an important new mechanism for a cross‐talk between the cell cycle through cyclin D1 and the diverse roles of Notch1.
Acknowledgments
We thank Dr. A. Capobianco (Wistar Institute, Philadelphia, PA, USA) for the CBF(8)‐luc reporter construct, and Dr. T. Imai for the Musashi expression vector. This work was supported in part by grants R01CA70896, R01CA75503, R01CA86072, R01CA86071 (Richard G. Pestell), R01CA101841 (Hallgeir Rui), and R01CA111482, N01CN43309, and a grant from the Charlotte Geyer Foundation (Robert I. Glazer); the Kimmel Cancer Center is supported by the NIH Cancer Center Core grant P30CA56036 (Richard G. Pestell). Michael P. Lisanti and his laboratory were supported by grants from the NIH/NCI (R01‐CA‐80250; R01‐CA‐098779; R01‐CA‐120876), the American Association for Cancer Research (AACR), and the Department of Defense‐Breast Cancer Research Program (Synergistic Idea Award). This project is a generous grant from the Dr. Ralph and Marian C. Falk Medical Research Trust funded and supported, in part, by a grant from the Pennsylvania Department of Health. The Department disclaims responsibility for any analysis, interpretations, or conclusions.
References
- 1. Hunter T. Oncoprotein networks. Cell. 1997; 88: 333–346. [DOI] [PubMed] [Google Scholar]
- 2. Hanahan D, Weinberg R. The hallmarks of cancer. Cell, 2000. 100: p. 57–70. [DOI] [PubMed] [Google Scholar]
- 3. Kimble J, Simpson P. The LIN‐12/Notch signaling pathway and its regulation. Annu Rev Cell Dev Biol. 1997; 13: 333–361. [DOI] [PubMed] [Google Scholar]
- 4. Nickoloff BJ, Osborne BA, Miele L. Notch signaling as a therapeutic target in cancer: a new approach to the development of cell fate modifying agents. Oncogene. 2003; 22(42): 6598–6608. [DOI] [PubMed] [Google Scholar]
- 5. Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah NG, Israël A. The Notch1 receptor is cleaved constitutively by a furin‐like convertase. Proc Natl Acad Sci USA. 1998; 95(14): 8108–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schlondorff J, Blobel CP. Metalloprotease‐disintegrins: modular proteins capable of promoting cell‐cell interactions and triggering signals by protein‐ectodomain shedding. J Cell Sci. 1999; 112(Pt 21): 3603–3617. [DOI] [PubMed] [Google Scholar]
- 7. Nye JS. Developmental signaling: Notch signals Kuz it's cleaved. Curr Biol. 1997; 7(11): R716–R720. [DOI] [PubMed] [Google Scholar]
- 8. Shimizu K, Chiba S, Hosoya N, Kumano K, Saito T, Kurokawa M, Kanda Y, Hamada Y, Hirai H. Binding of Delta1, Jagged1, and Jagged2 to Notch2 rapidly induces cleavage, nuclear translocation, and hyperphosphorylation of Notch2 . Mol Cell Biol. 2000; 20(18): 6913–6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kurooka H, Honjo T. Functional interaction between the mouse Notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J Biol Chem. 2000; 275(22): 17211–1720. [DOI] [PubMed] [Google Scholar]
- 10. Ross DA, Kadesch T. The Notch intracellular domain can function as a coactivator for LEF‐1. Mol Cell Biol. 2001; 21(22): 7537–7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Allenspach EJ, Maillard I, Aster JC, Pear WS. Notch signaling in cancer. Cancer Biol Ther. 2002; 1(5): 466–476. [DOI] [PubMed] [Google Scholar]
- 12. Zagouras P, Stifani S, Blaumueller CM, Carcangiu ML, Artavanis‐Tsakonas S. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc Natl Acad Sci USA. 1995; 92(14): 6414–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aster JC, Pear WS. Notch signaling in leukemia. Curr Opin Hematol. 2001; 8(4): 237–244. [DOI] [PubMed] [Google Scholar]
- 14. Bocchetta M, Miele L, Pass HI, Carbone M. Notch1 induction, a novel activity of SV40 required for growth of SV40‐transformed human mesothelial cells. Oncogene. 2003; 22(1): 81–89. [DOI] [PubMed] [Google Scholar]
- 15. Tonon, G , Modi S, Wu L, Kubo A, Coxon AB, Komiya T, O‘Neil K, Stover K, El‐Naggar A, Griffin JD, Kirsch IR, Kaye FJ. t(11;19)(q21; p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat Genet. 2003; 33(2): 208–213. [DOI] [PubMed] [Google Scholar]
- 16. Brennan K, Brown AM. Is there a role for Notch signalling in human breast cancer? Breast Cancer Res. 2003; 5(2): 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Callahan R, Raafat A. Notch signaling in mammary gland tumorigenesis. J Mammary Gland Biol Neoplasia. 2001; 6(1): 23–36. [DOI] [PubMed] [Google Scholar]
- 18. Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, Osborne BA, Gottipati S, Aster JC, Hahn WC, Rudolf M, Siziopikou K, Kast WM, Miele L. Activation of Notch1 signaling maintains the neoplastic phenotype in human Ras‐transformed cells. Nat Med. 2002; 8(9): 979–986. [DOI] [PubMed] [Google Scholar]
- 19. Dievart A, Beaulieu N, Jolicoeur P. Involvement of Notch1 in the development of mouse mammary tumors. Oncogene. 1999; 18(44): 5973–5981. [DOI] [PubMed] [Google Scholar]
- 20. Stahl M, Ge C, Shi S, Pestell RG, Stanley P. Notch1‐induced transformation of RKE‐1 cells requires up‐regulation of cyclin D1. Cancer Res. 2006; 66(15): 7562–7570. [DOI] [PubMed] [Google Scholar]
- 21. Stylianou S, Clarke RB, Brennan K. Aberrant activation of Notch signaling in human breast cancer. Cancer Res. 2006; 66(3): 1517–1525. [DOI] [PubMed] [Google Scholar]
- 22. Pece S, Serresi M, Santolini E, Capra M, Hulleman E, Galimberti V, Zurrida S, Maisonneuve P, Viale G, Di Fiore PP. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol. 2004; 167(2): 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Soule HD, Maloney TM, Wolman SR, Peterson WD Jr, Brenz R, McGrath CM, Russo J, Pauley RJ, Jones RF, Brooks SC. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF‐10. Cancer Res. 1990; 50(18): 6075–6086. [PubMed] [Google Scholar]
- 24. Hung MC, Schechter AL, Chevray PY, Stern DF, Weinberg RA. Molecular cloning of the neu gene: absence of gross structural alteration in oncogenic alleles. Proc Natl Acad Sci USA. 1986; 83(2): 261–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zeng X, Xu H, Glazer RI. Transformation of mammary epithelial cells by 3‐phosphoinositide‐dependent protein kinase‐1 (PDK1) is associated with the induction of protein kinase C‐alpha. Cancer Res. 2002; 62: 3538–3543. [PubMed] [Google Scholar]
- 26. Small D, Kovalenko D, Soldi R, Mandinova A, Kolev V, Trifonova R, Bagala C, Kacer D, Battelli C, Liaw L, Prudovsky I, Maciag T. Notch activation suppresses fibroblast growth factor‐dependent cellular transformation. J Biol Chem. 2003; 278(18): 16405–16413. [DOI] [PubMed] [Google Scholar]
- 27. Sakamaki T, Casimiro MC, Ju X, Quong AA, Katiyar S, Liu M, Jiao X, Li A, Zhang X, Lu Y, Wang C, Byers S, Nicholson R, Link T, Shemluck M, Yang J, Fricke ST, Novikoff PM, Papanikolaou A, Arnold A, Albanese C, Pestell R. Cyclin D1 determines mitochondrial function in vivo . Mol Cell Biol. 2006; 26(14): 5449–5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Imai T, Tokunaga A, Yoshida T, Hashimoto M, Mikoshiba K, Weinmaster G, Nakafuku M, Okano H. The neural RNA‐binding protein Musashi1 translationally regulates mammalian Numb gene expression by interacting with its mRNA. Mol Cell Biol. 2001; 21(12): 3888–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Albanese C, Wu K, D'Amico M, Jarrett C, Joyce D, Hughes J, Hulit J, Sakamaki T, Fu M, Ben‐Ze‘ev A, Bromberg JF, Lamberti C, Verma U, Gaynor RB, Byers SW, Pestell RG. IKKalpha regulates mitogenic signaling through transcriptional induction of cyclin D1 via TCF. Mol Biol Cell. 2003; 14(2): 585–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ronchini C, Capobianco AJ. Notch(ic)‐ER chimeras display hormone‐dependent transformation, nuclear accumulation, phosphorylation and CBF1 activation. Oncogene. 2000; 19(34): 3914–3924. [DOI] [PubMed] [Google Scholar]
- 31. Wang P, Anton M, Graham FL, Bacchetti S. High frequency recombination between loxP sites in human chromosomes mediated by an adenovirus vector expressing Cre recombinase. Somat Cell Mol Genet. 1995; 21(6): 429–441. [DOI] [PubMed] [Google Scholar]
- 32. Wulf G, Garg P, Liou YC, Iglehart D, Lu KP. Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis. Embo J. 2004; 23(16): 3397–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li Z, Wang C, Jiao X, Lu Y, Fu M, Quong AA, Dye C, Yang J, Dai M, Ju X, Zhang X, Li A, Burbelo P, Stanley ER, Pestell RG. Cyclin D1 regulates cellular migration through the inhibition of thrombospondin 1 and ROCK signaling. Mol Cell Biol. 2006; 26(11): 4240–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Watanabe G, Albanese C, Lee RJ, Reutens A, Vairo G, Henglein B, Pestell RG. Inhibition of cyclin D1 kinase activity is associated with E2F‐mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol Cell Biol. 1998; 18(6): 3212–3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rui H, LeBaron MJ. Creating tissue microarrays by cutting‐edge matrix assembly. Expert Rev Med Devices. 2005; 2(6): 673–680. [DOI] [PubMed] [Google Scholar]
- 36. Wang C, Pattabiraman N, Zhou JN, Fu M, Sakamaki T, Albanese C, Li Z, Wu K, Hulit J, Neumeister P, Novikoff PM, Brownlee M, Scherer PE, Jones JG, Whitney KD, Donehower LA, Harris EL, Rohan T, Johns DC, Pestell RG. Cyclin D1 repression of peroxisome proliferator‐activated receptor gamma expression and transactivation. Mol Cell Biol. 2003; 23(17): 6159–6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barbacci EG, Pustilnik LR, Rossi AM, Emerson E, Miller PE, Boscoe BP, Cox ED, Iwata KK, Jani JP, Provoncha K, Kath JC, Liu Z, Moyer JD. The biological and biochemical effects of CP‐654577, a selective ErbB2 kinase inhibitor, on human breast cancer cells. Cancer Res. 2003; 63(15): 4450–4459. [PubMed] [Google Scholar]
- 38. MacAuley A, Pawson T. Cooperative transforming activities of ras, myc, and src viral oncogenes in nonestablished rat adrenocortical cells. J Virol. 1988; 62(12): 4712–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stingl J, Eaves CJ, Kuusk U, Emerman JT. Phenotypic and functional characterization in vitro of a multipotent epithelial cell present in the normal adult human breast. Differentiation. 1998; 63(4): 201–213. [DOI] [PubMed] [Google Scholar]
- 40. Dontu G, Jackson KW, McNicholas E, Kawamura MJ, Abdallah WM, Wicha MS. Role of Notch signaling in cell‐fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 2004; 6(6): R605–R615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Florena AM, Tripodo C, Guarnotta C, Ingrao S, Porcasi R, Martorana A, Lo Bosco G, Cabibi D, Franco V. Associations between Notch‐2, Akt‐1 and HER2/neu expression in invasive human breast cancer: a tissue microarray immunophenotypic analysis on 98 patients. Pathobiology. 2007; 74(6): 317–322. [DOI] [PubMed] [Google Scholar]


