Abstract
Mouse telomeres have been suggested to resemble common fragile sites (CFS), showing disrupted TTAGGG fluorescent in situ hybridization signals after aphidicolin treatment. This “fragile” telomere phenotype is induced by deletion of TRF1, a shelterin protein that binds telomeric DNA and promotes efficient replication of the telomeric ds[TTAGGG]n tracts. Here we show that the chromosome-internal TTAGGG repeats present at human chromosome 2q14 form an aphidicolin-induced CFS. TRF1 binds to and stabilizes CFS 2q14 but does not affect other CFS, establishing 2q14 as the first CFS controlled by a sequence-specific DNA binding protein. The data show that telomeric DNA is inherently fragile regardless of its genomic position and imply that CFS can be caused by a specific DNA sequence.
Introduction
Common fragile sites (CFS) are non-random chromosomal loci known to be hotspots for DNA breakage under conditions that induce replication stress (Sutherland et al. 1998). In response to replication inhibitors, fragile sites become unstable, exhibit frequent sister chromatid exchanges (Glover and Stein 1987; Feichtinger and Schmid 1989), are preferential sites for DNA integration (Rassool et al. 1991; Wilke et al. 1996; Thorland et al. 2003; Matzner et al. 2003; Bester et al. 2006), and induce rearrangements, including deletions, translocations (Glover and Stein 1988), and regional amplifications (Coquelle et al. 1997; Ciullo et al. 2002; Hellman et al. 2002; Zimonjic et al. 2003; Miller et al. 2006; Reshmi et al. 2007; Pelliccia et al. 2010). In precancerous lesions, aberrant stimulation of cell proliferation can induce DNA replication stress that preferentially targets common fragile sites (Bartkova et al. 2006; Gorgoulis et al. 2005; Di Micco et al. 2006; Tsantoulis et al. 2008; Bignell et al. 2010; Dereli-Oz et al. 2011; Bester et al. 2011). Therefore, CFS are thought to make an important contribution to the complex genomic rearrangements in cancer.
Rare fragile sites are caused by an expansion of arrays of CGG or AT repeats beyond a critical size, leading to impairment of DNA replication by non-B DNA structures or intra-strand hairpins formed by the repeats (Schwartz et al. 2006). CFS, on the other hand, generally lack such repeats but are often characterized by a high AT content and regions with high flexibility that might similarly interfere with DNA replication by forming stable secondary structures (Mishmar et al. 1998; Zlotorynski et al. 2003). The idea that the DNA sequence itself is a critical factor in CFS is consistent with the fragile nature of FRA3B sequences placed at ectopic sites (Ragland et al. 2008). Moreover, within FRA16C, replication fork stalling occurs at/near AT-rich sequences (Ozeri-Galai et al. 2011).
For both the common and rare fragile sites, impaired replication could result in incompletely replicated and/or partially condensed regions, explaining the apparent breaks or gaps in metaphase chromosomes. Consistent with replication playing a role in CFS stability, CFS expression is exacerbated by deficiency in the ATR kinase and its effector kinase CHK1, which function to respond to replication stress, and by diminished function of recombination factors such as BRCA1, RAD51, and the Bloom’s syndrome RecQ helicase BLM, which might facilitate the recovery of stalled replication forks or enable restart events (Durkin and Glover 2007; Chu and Hickson 2009; Fundia et al. 1995).
However, the idea that most CFS are primarily caused by sequences whose secondary structure hampers replication was recently challenged. Several CFS associated with very long transcription units were shown to be due to the collision between transcription and replication (Helmrich et al. 2011). Furthermore, FRA3B was found to be situated in a region with a very low density of replication origins in lymphocytes, resulting in the late replication of its core sequences and incomplete replication of FRA3B upon aphidicolin-induced fork slowing (Letessier et al. 2011). The effects of large transcription units on DNA replication and/or the local paucity of origins could explain why the expression levels of CFS can vary in different cell types.
Recent studies have uncovered a striking similarity between fragile sites and telomeres, the elements that protect chromosome ends from the DNA damage response (de Lange 2009). Mammalian telomeric DNA consists of a long array of duplex TTAGGG repeats that challenge the DNA replication machinery. In cells treated with aphidicolin, telomeres display aberrant structures in metaphase chromosomes, referred to as fragile telomeres, which resemble those of aphidicolin-induced common fragile sites (Sfeir et al. 2009; Martinez et al. 2009). The fragile telomere phenotype may be due to secondary structures formed by the G-rich repeats, including the G-quartet structures (G4 DNA), that are reminiscent of the secondary structures invoked as the culprit at some common fragile sites (Sfeir et al. 2009; Vannier et al. 2012; Salvati et al. 2010). Efficient replication of telomeric DNA is promoted by TRF1, a double-stranded TTAGGG repeat binding protein that is part of the telomeric shelterin complex (Palm and de Lange 2008).When TRF1 is deleted from mouse cells, the fragile telomere phenotype is strongly enhanced both in the presence and absence of aphidicolin. TRF1 was shown to act in conjunction with two helicases, BLM and RTEL1, which are implicated in the removal of G4 DNA (Sfeir et al. 2009; Vannier et al. 2012). While these data indicated that telomeres resemble CFS, it was not clarified whether this attribute is due to the terminal position of the telomeric DNA or due to the nature of the TTAGGG repeats per se. Indeed, while it was suggested that internal telomeric sequence might cause chromosomal breakage (Hastie and Allshire 1989; Ashley and Ward 1993; Day et al. 1998; Bertoni et al. 1994; Slijepcevic et al. 1996), no direct evidence has linked interstitial telomeric repeats to fragile site behavior.
In order to determine whether telomeric DNAs can cause a CFS at a chromosome-internal site, we focused on human chromosome 2q14, which contains two stretches of TTAGGG repeats in inverted orientation. These internal telomeric repeat sequences are the remnant of a telomere–telomere fusion of two ancestral ape chromosomes (IJdo et al. 1991). We document that this locus behaves as a common fragile site that is sensitive to aphidicolin and requires TRF1 for its stability. These findings reveal that telomeric DNA can induce a CFS and argue for a sequence-dependent component in the expression of this CFS.
Results
Identification of a novel common fragile site at 2q14
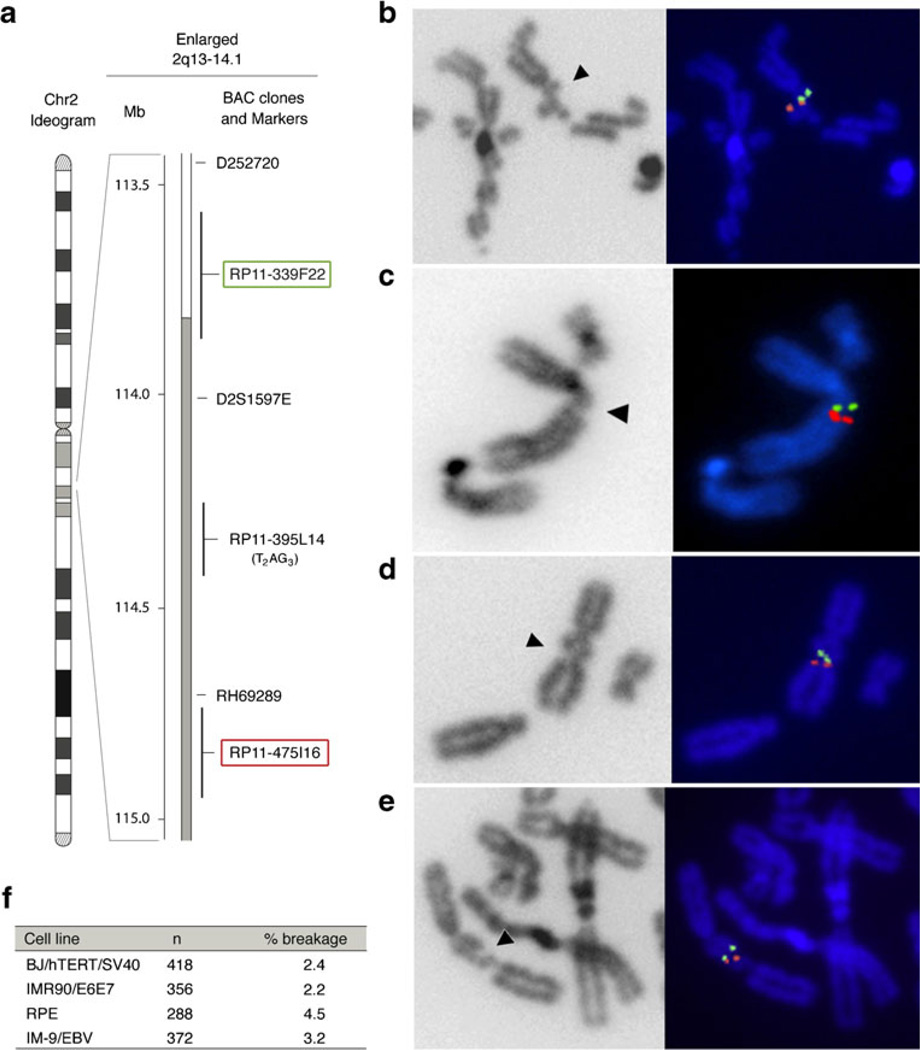
The relevant region in 2q14 is a 1-kb segment which contains exact copies of the telomeric TTAGGG repeats and TTAGGG repeat permutations interspersed with TTAGGG-related repeats (Fig. S1). To test whether this sequence is associated with a fragile site, we used two BAC-derived fluorescent in situ hybridization (FISH) probes (RP11-339F22 and RP11-475I16) residing approximately 0.5 Mb on either side of the 2q14 telomeric repeats locus (Fig. 1a–f). When SV40-transformed, telomerase-immortalized human BJ fibroblasts (BJ/hTERT/SV40) were treated with aphidicolin to induce replication stress, the fluorescent signals of the two BAC probes flanked a chromosome break in 2.4 % of the meta-phases (Fig. 1b, f; Fig. S2). This fragile region was also observed in HPV-E6/E7 transformed human IMR90 fibroblast (IMR90/E6E7) treated with aphidicolin (Fig. 1c, f). IMR90/E6E7 and BJ/hTERT/SV40 showed approximately the same frequency of breakage (Fig. 1f), whereas the primary human retinal pigment epithelial (RPE) cell line and the Epstein–Barr virus transformed lymphoblastoid IM-9 cell line (IM-9/EBV) showed aphidicolin-induced breakage at 2q14 at a higher frequency (4.5 and 3.2 %, respectively) (Fig. 1d–f). These data identify a new aphidicolin-inducible CFS near the chromosome-internal telomeric repeats in 2q14.
Fig. 1.
A newly identified common fragile site at human chromosome 2q14. a Ideogram of the human chromosome 2 with the physical map of the 2q13-q14.1 region enlarged. The BAC clones used for the FISH analysis (RP11-339F22 and the RP11-475I16 highlighted in green and red, respectively), the RP11-395L14 harboring the telomeric DNA, and same markers are shown. b–d FISH of the RP11-339F22 (green) and RP11-475I16 (red) probes on human chromosome 2 expressing the fragile site at 2q14 (arrow head) in aphidicolin-induced BJ/hTERT/SV40 (b), IMR90/E6E7 (c), RPE cells (d), and IM-9/EBV (e). f Frequency of the expression of the breakage at 2q14 in the analyzed cell lines after aphidicolin treatment. n is the number of chromatids analyzed by FISH, % breakage refers to the percentage of chromatids that showed a gap/break between the two probes
TRF1 and other shelterin proteins reside at 2q14
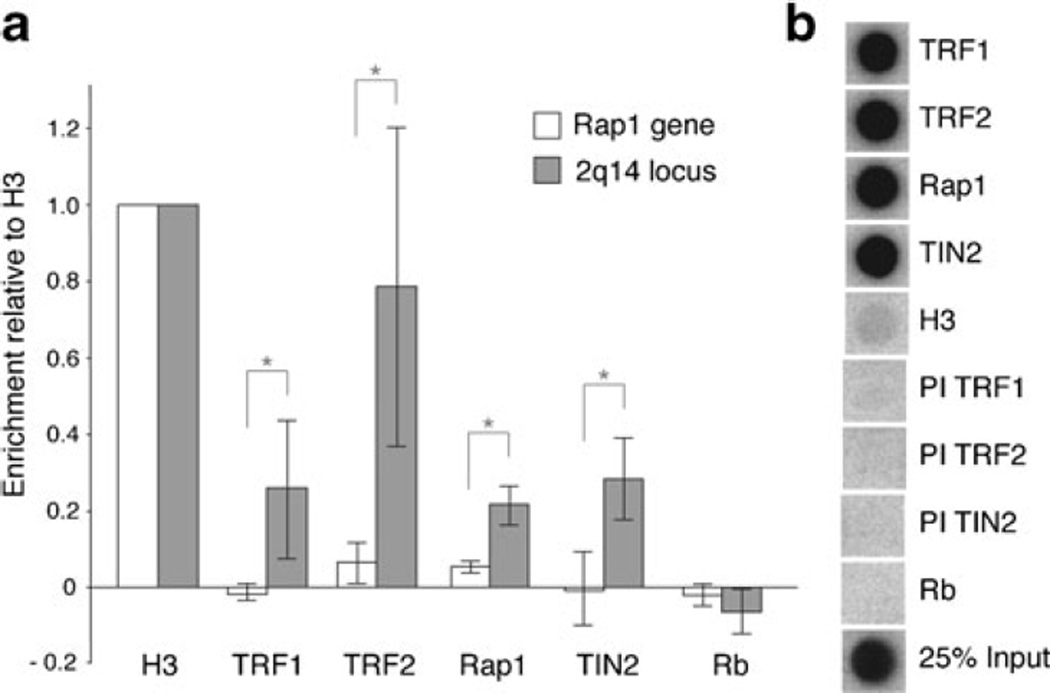
The 2q14 region is highly enriched for the telomeric sequences that are recognized by the shelterin protein TRF1. It contains 27 copies of the sequence 5′-TAGGGTT-3′ binding site for the TRF1 Myb/SANT DNA binding domain in either the G-strand or C-strand orientation within 1 kb (Fig. S1a). Although 2q14 lacks the long arrays of tandem TTAGGG repeats found at telomeres, the presence of multiple 5′-TAGGGTT-3′ sequences is predicted to allow binding of TRF1 which binds DNA as a flexible dimer, engaging two 5′-TAGGGTT-3′ sites at variable distance and orientation (Bianchi et al. 1997, 1999). To test whether TRF1 and other shelterin components are indeed bound to this region, we performed ChIP for TRF1 and its interacting partner TIN2 as well as for a second telomeric DNA binding protein in shelterin, TRF2, and its binding partner Rap1. Quantitative PCR (qPCR) was used to evaluate the recovery of the 2q14 fusion site in the ChIPs, while a chromosome-internal region lacking TTAGGG repeats (part of the Rap1 gene; Fig. S1b) was used as a negative control. For both loci, ChIPs for histone H3 were used as a positive control and for normalization. The results showed an enrichment of the 2q14 region in the ChIPs for the shelterin proteins TRF1, TRF2, Rap1, and TIN2 (Fig. 2a). In contrast, ChIPs for these shelterin proteins did not show an enrichment at the Rap1 gene control locus (Fig. 2a). The specificity of the ChIPs with shelterin proteins was confirmed by hybridization to a TTAGGG repeat probe (Fig. 2b), whereas pre-immune sera and an antibody for Rb failed to bring down the 2q14 region or telomeric DNA (Fig. 2a, b). These results indicate that the telomeric repeats at 2q14 associate with TRF1, TRF2, and their interacting factors, in agreement with previous reports of the association of TRF1 and TRF2 with chromosome-internal telomeric sequences (Simonet et al. 2011; Yang et al. 2011).
Fig. 2.
Internal telomeric repeats associate with TRF1 and other shelterin proteins. a Quantification of the recovery of the 2q14 fusions site (2q14 locus) and Rap1 control region (Rap1 gene) in the ChIPs using the indicated antibodies or pre-immune serum (PI) by qPCR in BJ/hTERT/SV40 cells. Ct values collected from each qPCR analysis performed in triplicate were normalized to H3. Ct values were than corrected for the averaged Ct values obtained with the pre-immune antibodies that yielded background signals (PI TRF1, PI TRF2, PI TIN2). Bars represent mean values of three independent experiments with SD. Brackets with asterisks indicate p values below 0.05 (Student’s t test). b Dot blots monitoring the specificity of ChIP with antibodies to shelterin proteins and control sera by hybridization with a TTAGGG repeats probe
TRF1, BLM, and ATR affect the stability of the CFS at 2q14
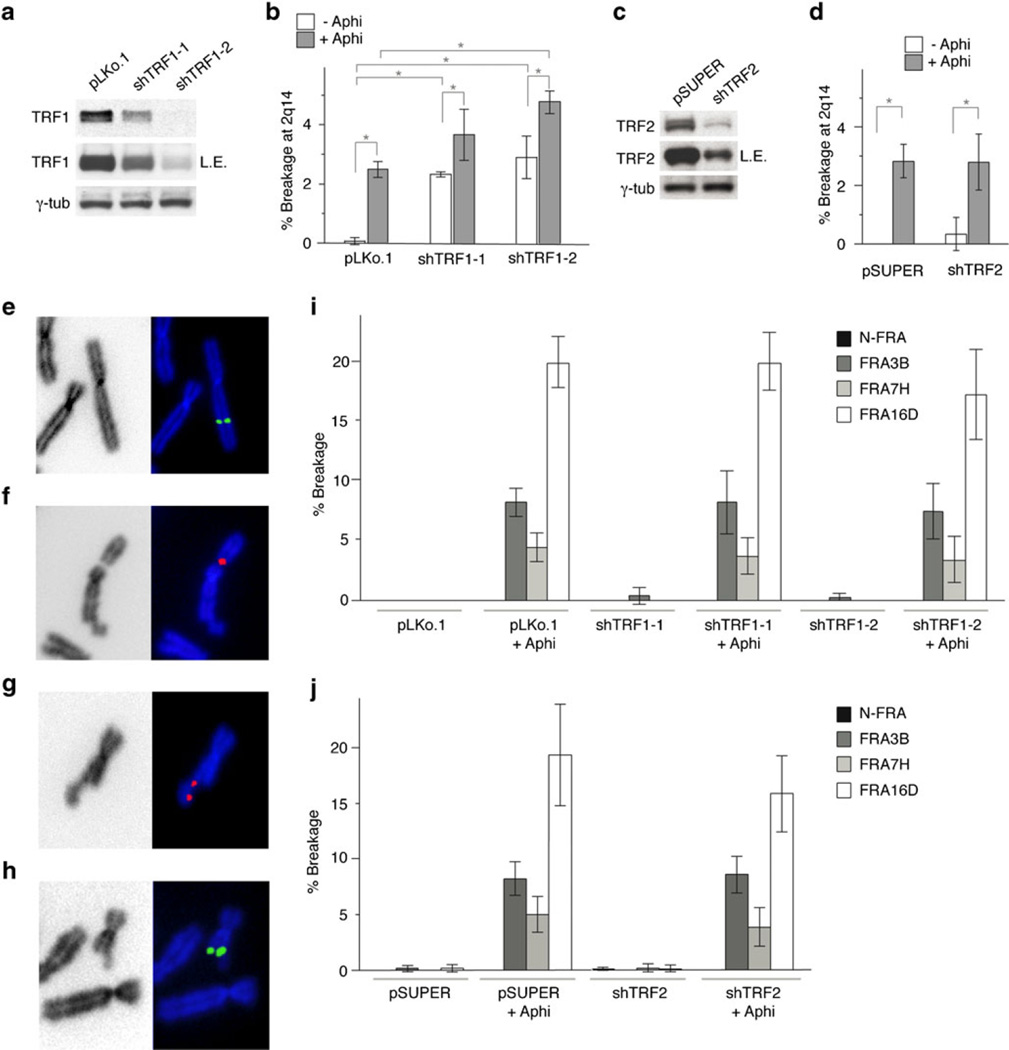
To determine whether TRF1 affects the breakage at 2q14, we measured the expression of this fragile site in metaphase spreads from BJ/hTERT/SV40 cells treated with two TRF1 shRNAs. Suppression of TRF1 was confirmed by immunoblotting (Fig. 3a), IF analysis for TRF1 IF signals, induction of telomere dysfunction-induced foci, and induction of fragile telomeres (Fig. S3a–c). Importantly, the reduction in TRF1 levels resulted in a 1.5–2-fold increase in the frequency of aphidicolin-induced breaks at 2q14 (Fig. 3b, Suppl. Table 1), whereas repression of TRF2 with an shRNA had no effect (Fig. 3c, d, Suppl. Table 2). Even in the absence of aphidicolin-induced replication stress, the break at 2q14 was observed at significant frequencies when TRF1 was suppressed (Fig. 3b, Suppl. Table 1). We assume that the breakage at CFS 2q14 is less frequent than the fragile telomere phenotype because of the much lower density and copy number of the telomeric sequences at the internal site.
Fig. 3.
TRF1 modulation of the common fragile site at 2q14. a Immunoblot monitoring the reduction of TRF1 upon shRNA treatment (2 days after infection). γ-Tubulin was used as loading control. LE long exposure. b Quantification of the breakage at 2q14 observed after inhibition of TRF1, with or without aphidicolin treatment. Bars represent mean values of three independent experiments with SD (see Suppl. Table 1). c Immunoblot showing shRNA-mediated repression of TRF2. γ-Tubulin is used as loading control. LE long exposure. d Frequency of breakage at CFS 2q14 after inhibition of TRF2, with or without aphidicolin treatment. Bars represent mean values of three independent experiments with SD (see Suppl. Table 2). e–h FISH signals showing, in order, the non-fragile NFRA control region, FRA3B, FRA7H, and FRA16D in BJ/hTERT/SV40 after aphidicolin treatment. i, j Quantification of the breakage observed at NFRA, FRA3B, FRA7H, and FRA16D, after inhibition of TRF1 (i) and TRF2 (j), with or without aphidicolin treatment. Bars represent mean values of three independent experiments with SD (see Suppl. Tables 1 and 2). Brackets with asterisks indicate p values below 0.05 (Student’s t test)
TRF1 suppression did not affect several non-telomeric common fragile sites (FRA3B, FRA7H, and FRA16D) or a region on human chromosome 2 known to be not fragile (NFRA; Pelliccia et al. 2008) (Fig. 3e–i, Suppl. Table 1). As expected, the shRNA to TRF2 also did not affect telomere fragility or the breakage at FRA3B, FRA7H, and FRA16D (Fig. S3d, Fig. 3j, Suppl. Table 2).
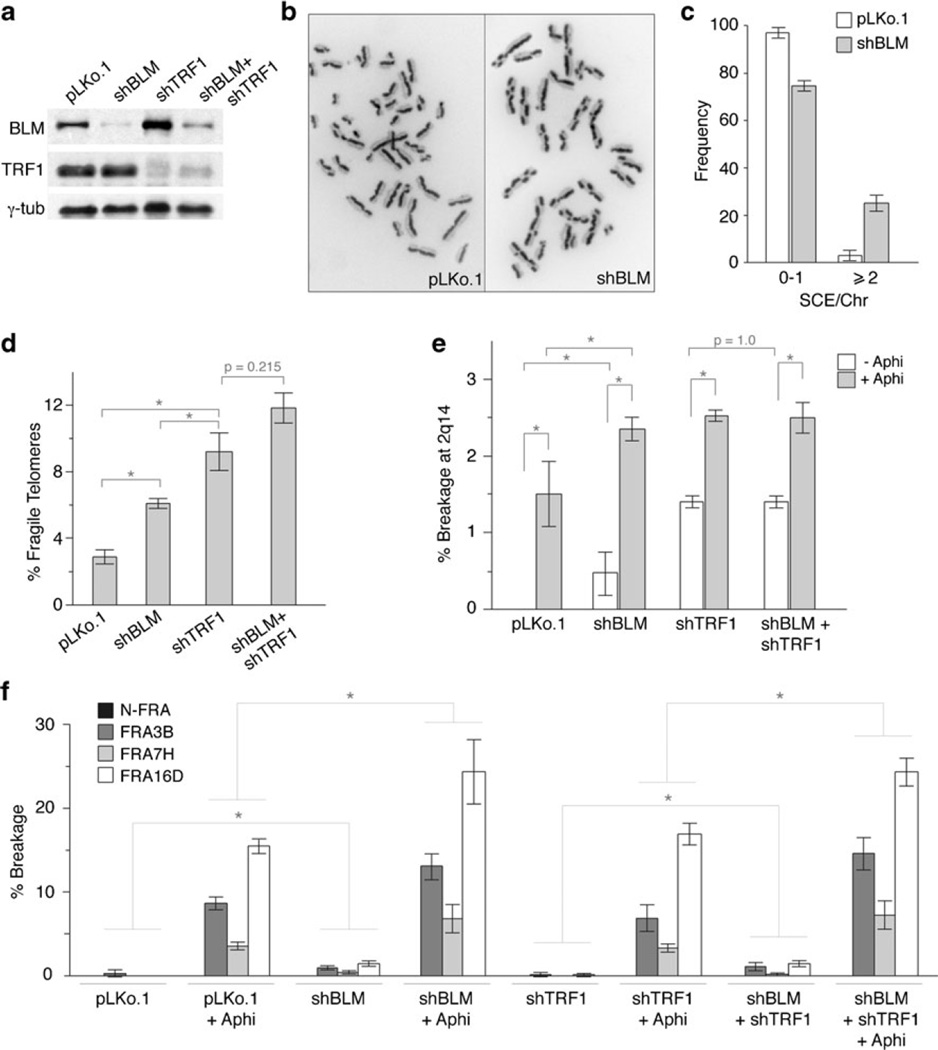
The Bloom’s syndrome helicase (BLM) was shown to act downstream of TRF1 to prevent telomere fragility (Sfeir et al. 2009). We therefore analyzed the effect of BLM on CFS 2q14 using a BLM shRNA that reduced BLM protein levels (Fig. 4a), induced sister chromatid exchanges (SCEs, Fig. 4b, c), and increased the frequency of telomere fragility (Fig. 4d). BLM suppression increased the frequency of common fragile sites as well as the breakage at 2q14 either in the presence or absence of aphidicolin (Fig. 4e, f, Suppl. Tables 3 and 4). Moreover, when cells were treated with shRNAs to both TRF1 and BLM, the breakage at 2q14 was similar to that observed with either shRNA alone (Fig. 4e, Suppl. Table 3). As expected from the lack of effect of TRF1 on the non-telomeric common fragile sites, dual suppression of TRF1 and BLM had no further effect on FRA3B, FRA7H, and FRA16D compared to suppression of BLM alone (Fig. 4f, Suppl. Table 4). Thus, the effect of TRF1 and BLM is epistatic at CFS 2q14 as it is at telomeres. This result is consistent with TRF1 promoting the function of BLM at sites containing telomeric repeats.
Fig. 4.
TRF1 and BLM affect stability of the common fragile site at 2q14. a Immunoblot monitoring shRNA-mediated repression of BLM and TRF1 (day 2 post-infection). γ-Tubulin was used as loading control. b Example of SCEs in control (left panel) and shBLM-treated cells (right panel) and c quantification of SCEs after BLM loss. d Frequency of fragile telomeres after repression of BLM, TRF1, or both. e Quantification of the breakage at 2q14 and f frequency of breakage at NFRA, FRA3B, FRA7H, and FRA16D after repression of BLM, TRF1, or both with or without aphidicolin treatment. In all panels, bars represent mean values of three independent experiments with SD (see Suppl. Tables 3 and 4). Brackets with asterisks indicate p values below 0.05 (Student’s t test)
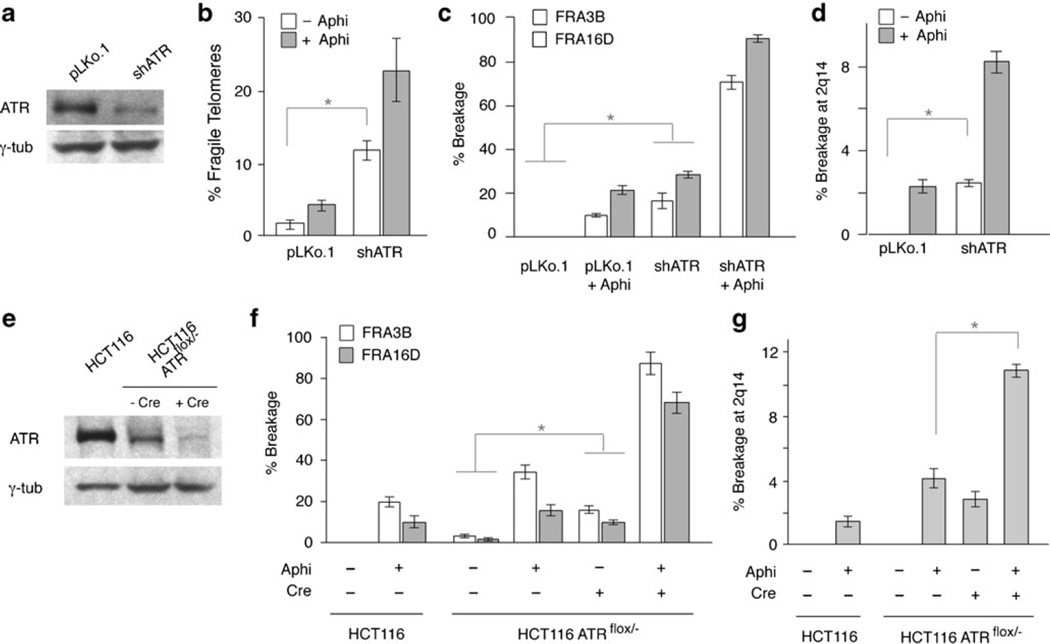
In addition to BLM, ATR affects the expression of the fragile telomere phenotype (Sfeir et al. 2009). To determine the effect of ATR on the CFS 2q14, we reduced the expression of the ATR kinase with an shRNA in BJ/hTERT/SV40 fibroblasts or deleted of ATR from ATRflox/− HCT116 cells with Cre recombinase (Cortez et al. 2001). In both settings, reduction of ATR proteins level (Fig. 5a, e) resulted in the expected increase in the frequency of fragile telomeres (Fig. 5b) and the expression of FRA3B and FRA16D (Fig. 5c, f, Suppl. Tables 5 and 6). Importantly, inhibition of ATR also affected the expression of CFS 2q14 (Fig. 5d, g, Suppl. Tables 5 and 6), indicating that CFS 2q14 behaves like the other CFS in this regard. Due to lack of appropriate reagents to diminish the level of human RTEL1, we were unable to test the effect of this helicase on CFS 2q14.
Fig. 5.
ATR regulates CFS 2q14 stability. a Immunoblot monitoring repression of ATR at day 2 after shRNA infection of BJ/hTERT/SV40 cells. γ-Tubulin was used as loading control. b Frequency of fragile telomeres after repression of ATR in BJ/hTERT/SV40 cells. c Quantification of the breakage at FRA3B and FRA16D and d at 2q14 after repression of ATR in BJ/hTERT/SV40 cells. Bars represent mean values of three independent experiments with SD (see Suppl. Table 5). Brackets with asterisks indicate p values below 0.05 (Student’s t test). e Immunoblot monitoring repression of ATR in HCT116 and HCT116 ATRflox/− cells with Cre recombinase (day 3). γ-Tubulin was used as loading control. f Quantification of the breakage at FRA3B and FRA16D and g at 2q14 in HCT116 and HCT116 ATRflox/− cells. Bars represent mean values of two independent experiments with SEM (see Suppl. Table 6). Brackets with asterisks indicate p values below 0.05 (Wilcoxon rank sum test)
Discussion
These data identify a previously unrecognized aphidicolin-inducible common fragile site at chromosome 2q14 (CFS 2q14). Conventional cytogenetic techniques have not detected CFS 2q14 probably due to its low expression (Le Tallec et al. 2011) although one report noted this site (Mrasek et al. 2010). A detailed analysis of the CFS in the long arm of chromosome 2 reported a novel CFS at 2q12-14 (Brueckner et al. 2012) as the third most frequently expressed fragile site on chromosome arm 2q which could be due to two different CFS residing at this site: the CFS at 2q13 (Sutherland and Mattei 1987) and the less frequently expressed CFS 2q14.
CFS 2q14 is expressed at very low levels because under normal conditions this fragile site is repressed by TRF1. However, when TRF1 levels are diminished, the aphidicolin-induced expression of CFS 2q14 is significantly increased and on par with FRA7H. The dependence of CFS 2q14 on TRF1 strongly argues that the telomeric sequences in the 2q14 locus are the cause of breakage in this region. Removal of TRF1 from mouse telomeres leads to replication fork pausing, activation of the ATR kinase, and fragile telomeres, suggesting that replication problems originating from the nature of the TTAGGG repeats, including their ability to form G4 DNA, are causing the fragile phenotype of the telomeres (Sfeir et al. 2009; Martinez et al. 2009; Salvati et al. 2010; Vannier et al. 2012). TRF1 was proposed to facilitate the replication of telomeric DNA through the recruitment of BLM and thereby prevent the occurrence of fragile telomeres (Sfeir et al. 2009). Similarly, we propose that at CFS 2q14, the telomeric and other G-rich repeats hinder replication fork progression, most likely by forming G4 DNA, and that TRF1 facilitates the replication of 2q14, in part by promoting the action of BLM and possibly RTEL1 although we were unable to test the involvement of RTEL1 in this study.
CFS 2q14 is the first common fragile site known to depend on a sequence-specific DNA binding protein. This finding indicates that this particular common fragile site and possibly others is indeed due to the nature of the local sequence impeding replication fork progression. At telomeres, it was shown that G4 stabilizing ligands induce the fragile telomere phenotype (Salvati et al. 2010; Vannier et al. 2012), providing direct evidence for a secondary structure-based impairment of replication. Furthermore, in the telomeric setting, TRF1 acts together with BLM and RTEL1 (Sfeir et al. 2009; Vannier et al. 2012), most likely preventing the occurrence of replication fork stalling events due to these structures. Indeed, when TRF1 is removed from telomeres, fork stalling is strongly induced (Sfeir et al. 2009). It is reasonable to assume that these molecular principles, worked out at telomeres, also hold for CFS 2q14. Other models, such as effects on the local density of replication origins or the size of transcription units, which explain the behavior of other CFS, are unlikely to come into play at CFS 2q14, since TRF1 is not known to affect either.
Many vertebrates, including hamster, chicken, and reptiles, have large blocks of TTAGGG-like sequences often associated with the pericentric heterochromatin. As in the case of 2q14, the interstitial telomeric DNA in hamster cells is associated with TRF1 (Smogorzewska et al. 2000; Krutilina et al. 2001). Our data predict that these clusters would have the tendency to break, especially when TRF1 becomes limiting. Indeed, cytogenetic analysis has implicated the chromosome-internal telomeric repeat regions in hamster cells in spontaneous and induced breakage (Alvarez et al. 1993; Fernandez et al. 1995; Slijepcevic et al. 1996; Kilburn et al. 2001). Why are these fragile sequences maintained although they can easily be removed by unequal sister chromatid exchange? We have previously argued that the fragile nature of telomeres might lure transposable elements and other invasive DNA to telomeres where their insertion is relatively harmless (Sfeir et al. 2009). Similarly, the chromosome-internal blocks of fragile TTAGGG repeats might form a safe sink for mobile DNA, placing them in a setting where they are not disruptive and are likely to become repressed by their heterochromatic environment. In this manner, TTAGGG repeats may not only protect the ends of the chromosomes but also safeguard chromosome-internal sequences.
Materials and methods
Cell culture
SV40-transformed, hTERT immortalized BJ fibroblasts (BJ/hTERT/SV40) cells were cultured in complete DMEM-containing 199 medium (4:1) with 10 % bovine calf serum. HPV E6/E7 transformed IMR90 fibroblasts (IMR90/E6E7) and primary RPE cells were cultured in DMEM with 10 % bovine calf serum. EBV immortalized IM-9 lymphoblastoid cells were grown in RPMI1640 with 10 % FBS. HCT116 and HCT116flox/− cells (Cortez et al. 2001) were grown in McCoy's 5a Medium with 10 % bovine calf serum. Cre recombinase was introduced in HCT116flox/− cells with Hit&Run-Cre to allow Cre-lox removal of the remaining ATR allele and cells were harvested 3 days post-infection. Aphidicolin (Sigma) was added (0.3 µmol/l) for 20 h before fixing. Colchicine (10−4 mM, Roche) was added 2 h before metaphase was prepared using standard procedures.
ChIP-qPCR
ChIP and dot blot analysis were performed as previously described (Loayza and de Lange 2003) with the following sera: TRF1 (#371, crude rabbit polyclonal); TRF2 (#647, crude rabbit polyclonal); TIN2 (#865, crude rabbit polyclonal); Rap1 (#765, crude rabbit polyclonal); Rb (#554136, BD Pharmingen); and H3 (ab1791, Abcam). For qPCR, samples were amplified by quantitative real-time PCR (ABI PRISM 7700 Sequence Detection System) using specific primers for Chr2 (Chr2-F 5′-GCATTCCCCTAAGCACAGAG-3′ and Chr2-R 5′-TCACCCTCACCCTGCAAT-3′) and Rap1 (Rap1-F 5′-AGCTGC CATTAAGATCATTCGGCAG-3′ and Rap1-R 5′-CGAAATTCAATCCTCCGAGC-3′). PCR products were run on agarose gel to check for correct amplification fragment size.
shRNAs
TRF1 (shTRF1-1 5′-CCCAGCAACAAGACCTTAATA-3′ and shTRF1-2 5′-CCCTTGATGCACAGTTTGAAA-3′), BLM (5′-GCCTTTATTCAATACCCATTT-3′), and ATR (5′-CTGTGGTTGTATCTGTTCAAT-3′) shRNAs in pLKo.1 were introduced using three infections at 12 h intervals with supernatant from transfected 293 T cells. For TRF2 analysis (shTRF2 5′-TCACAGGAGCATGGTTCCTAATA-3′), the shRNA in pSUPERIOR was introduced with four retroviral infections at 12 h intervals with supernatant from transfected PhoenixA cells.
Immunoblotting
Immunoblots were performed on whole cell lysates as described previously (Takai et al. 2010) using the following antibodies: TRF1 (#371, purified rabbit polyclonal); TRF2 (#647, purified rabbit polyclonal); BLM (#ab2179, Abcam); γ-tubulin (clone GTU88, Sigma); and ATR (N-19, goat polyclonal, Santa-Cruz).
BAC probes and FISH
BAC clones (Children’s Hospital Oakland Research Institute, Oakland, CA, USA) used were: 2q14, RPCI-11 339F22 (AC016724), and RPCI-11 475I16 (AC010982); FRA3B, RPCI-11 94D19 (AC AC096917); FRA7H, RPCI-11 36B6 (AC016831); FRA16D, RPCI-11 264L1 (AC046158); and non-fragile control, BAC RPCI-11 284E18, lying approximately 1 Mb from the fragile site FRA2G (AC008065). Probes were labeled by nick-translation with biotin-16-dUTP or digoxigenin-11-dUTP (Roche). RNase-treated slides were dehydrated with 5 min washes in 70, 95, and 100 % ethanol and allowed to air dry. Slides were incubated with hybridization solution containing 200 ng of each labeled BAC probe, mixed with 8 µg of Cot-1 DNA (Invitrogen), 10 % dextran sulfate, 50%formamide, and 2× SSC (pH 7.0). Slides were denatured at 80 °C for 2 min, hybridized at 37 °C for 18 h, and washed in 2× SSC at 65 °C, and signals were detected with Cy3-conjugated avidin and FITC-conjugated antidigoxigenin antibodies. Telomeric FISH was performed using a FITC-OO-(CCCTAA)3 PNA probe (Applied Biosystems) according to the manufacturer’s instructions. Digital images were captured with a Zeiss Axioplan II microscope with a Hamamatsu C4742-95 camera using Improvision OpenLab software for merging the images.
IF-FISH
Telomeric DNA FISH was combined with IF using a rabbit polyclonal 53BP1 antibody (100-304, Novus Biological) or TRF1 (Ab 371) as previously described (Takai et al. 2010).
SCE assay
Subconfluent cells were incubated with BrdU (20 µM) for 48 h before preparation of metaphase spreads. Slides were rehydrated in PBS and incubated in 2× SSC, Hoechst 33258 (25 µg/ml) in the dark for 30 min and exposed to UV light (10 min at 5.4×103 J/m2). Slides were washed in PBS; dehydrated in 70, 95, and 100 % ethanol; and counterstained with DAPI in anti-fade embedding medium (ProLong Gold, Invitrogen). Digital images were captured as above.
Supplementary Material
Acknowledgments
We thank Agnel Sfeir for the help in the early stages of this work and members of the de Lange lab for comments on this manuscript. We are grateful to Eugene Rudensky for the help with qPCR. This work was supported by a grant from the NIH (AG16642) to TdL. TdL is an American Cancer Society Research Professor. NB is supported by “The Alessandro & Catherine di Montezemolo” American-Italian Cancer Foundation Post-Doctoral Research Fellowship.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00412-012-0377-6) contains supplementary material, which is available to authorized users.
Conflict of interest None declared.
References
- Alvarez L, Evans JW, Wilks R, Lucas JN, Brown JM, Giaccia AJ. Chromosomal radiosensitivity at intrachromosomal telomeric sites. Genes Chromosomes Cancer. 1993;8:8–14. doi: 10.1002/gcc.2870080103. [DOI] [PubMed] [Google Scholar]
- Ashley T, Ward DC. A "hot spot" of recombination coincides with an interstitial telomeric sequence in the Armenian hamster. Cytogenet Cell Genet. 1993;62:169–171. doi: 10.1159/000133464. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, Takaoka M, Nakagawa H, Tort F, Fugger K, Johansson F, Sehested M, Andersen CL, Dyrskjot L, Orntoft T, Lukas J, Kittas C, Helleday T, Halazonetis TD, Bartek J, Gorgoulis VG. Oncogeneinduced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Bertoni L, Attolini C, Tessera L, Mucciolo E, Giulotto E. Telomeric and nontelomeric (TTAGGG)n sequences in gene amplification and chromosome stability. Genomics. 1994;24:53–62. doi: 10.1006/geno.1994.1581. [DOI] [PubMed] [Google Scholar]
- Bester AC, Schwartz M, Schmidt M, Garrigue A, Hacein-Bey-Abina S, Cavazzana-Calvo M, Ben-Porat N, Von Kalle C, Fischer A, Kerem B. Fragile sites are preferential targets for integrations of MLV vectors in gene therapy. Gene Ther. 2006;13:1057–1059. doi: 10.1038/sj.gt.3302752. [DOI] [PubMed] [Google Scholar]
- Bester AC, Roniger M, Oren YS, ImMM, Sarni D, ChaoatM, Bensimon A, Zamir G, Shewach DS, Kerem B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Smith S, Chong L, Elias P, de Lange T. TRF1 is a dimer and bends telomeric DNA. Embo J. 1997;16:1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Stansel RM, Fairall L, Griffith JD, Rhodes D, de Lange T. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. Embo J. 1999;18:5735–5744. doi: 10.1093/emboj/18.20.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S,Andrews JM, Buck G, Chen L, Beare D, Latimer C, Widaa S, Hinton J, Fahey C, Fu B, Swamy S, Dalgliesh GL, Teh BT, Deloukas P, Yang F, Campbell PJ, Futreal PA, Stratton MR. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueckner LM, Sagulenko E, Hess EM, Zheglo D, Blumrich A, Schwab M, Savelyeva L. Genomic rearrangements at the FRA2H common fragile site frequently involve non-homologous recombination events across LTR and L1(LINE) repeats. Hum Genet. 2012 doi: 10.1007/s00439-012-1165-3. [DOI] [PubMed] [Google Scholar]
- Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- Ciullo M, Debily MA, Rozier L, Autiero M, Billault A, Mayau V, El Marhomy S, Guardiola J, Bernheim A, Coullin P, Piatier-Tonneau D, Debatisse M. Initiation of the breakage-fusion-bridge mechanism through common fragile site activation in human breast cancer cells: the model of PIP gene duplication from a break at FRA7I. Hum Mol Genet. 2002;11:2887–2894. doi: 10.1093/hmg/11.23.2887. [DOI] [PubMed] [Google Scholar]
- Coquelle A, Pipiras E, Toledo F, Buttin G, Debatisse M. Expression of fragile sites triggers intrachromosomal mammalian gene amplification and sets boundaries to early amplicons. Cell. 1997;89:215–225. doi: 10.1016/s0092-8674(00)80201-9. [DOI] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- Day JP, Limoli CL, Morgan WF. Recombination involving interstitial telomere repeat-like sequences promotes chromosomal instability in Chinese hamster cells. Carcinogenesis. 1998;19:259–65. doi: 10.1093/carcin/19.2.259. [DOI] [PubMed] [Google Scholar]
- de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereli-Oz A, Versini G, Halazonetis TD. Studies of genomic copy number changes in human cancers reveal signatures of DNA replication stress. Mol Oncol. 2011;5:308–314. doi: 10.1016/j.molonc.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre' M, Nuciforo PG, Bensimon A, Maestro R, Pelicci PG, d'Adda di Fagagna F. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- Feichtinger W, Schmid M. Increased frequencies of sister chromatid exchanges at common fragile sites (1)(q42) and (19)(q13) Hum Genet. 1989;83:145–147. doi: 10.1007/BF00286707. [DOI] [PubMed] [Google Scholar]
- Fernandez JL, Gosalvez J, Goyanes V. High frequency of mutagen-induced chromatid exchanges at interstitial telomere-like DNA sequence blocks of Chinese hamster cells. Chromosome Res. 1995;3:281–284. doi: 10.1007/BF00713065. [DOI] [PubMed] [Google Scholar]
- Fundia A, Gorla N, Larripa I. Non-random distribution of spontaneous chromosome aberrations in two Bloom syndrome patients. Hereditas. 1995;122:239–243. doi: 10.1111/j.1601-5223.1995.00239.x. [DOI] [PubMed] [Google Scholar]
- Glover TW, Stein CK. Induction of sister chromatid exchanges at common fragile sites. Am J Hum Genet. 1987;41:882–890. [PMC free article] [PubMed] [Google Scholar]
- Glover TW, Stein CK. Chromosome breakage and recombination at fragile sites. Am J Hum Genet. 1988;43:265–273. [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RAJ, Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, Kittas C, Halazonetis TD. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Hastie ND, Allshire RC. Human telomeres: fusion and interstitial sites. Trends Genet. 1989;5:326–331. doi: 10.1016/0168-9525(89)90137-6. [DOI] [PubMed] [Google Scholar]
- Hellman A, Zlotorynski E, Scherer SW, Cheung J, Vincent JB, Smith DI, Trakhtenbrot L, Kerem B. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell. 2002;1:89–97. doi: 10.1016/s1535-6108(02)00017-x. [DOI] [PubMed] [Google Scholar]
- Helmrich A, Ballarino M, Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell. 2011;44:966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- IJdo JW, Baldini A, Ward DC, Reeders ST, Wells RA. Origin of human chromosome 2: an ancestral telomere–telomere fusion. Proc Natl Acad Sci U S A. 1991;88:9051–9055. doi: 10.1073/pnas.88.20.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilburn AE, Shea MJ, Sargent RG, Wilson JH. Insertion of a telomere repeat sequence into a mammalian gene causes chromosome instability. Mol Cell Biol. 2001;21:126–35. doi: 10.1128/MCB.21.1.126-135.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutilina RI, Oei S, Buchlow G, Yau PM, Zalensky AO, Zalenskaya IA, Bradbury EM, Tomilin NV. A negative regulator of telomere-length protein trf1 is associated with interstitial (TTAGGG)n blocks in immortal Chinese hamster ovary cells. Biochem Biophys Res Commun. 2001;280:471–45. doi: 10.1006/bbrc.2000.4143. [DOI] [PubMed] [Google Scholar]
- Le Tallec B, Dutrillaux B, Lachages AM, Millot GA, Brison O, Debatisse M. Molecular profiling of common fragile sites in human fibroblasts. Nat Struct Mol Biol. 2011;18:1421–1423. doi: 10.1038/nsmb.2155. [DOI] [PubMed] [Google Scholar]
- Letessier A, Millot GA, Koundrioukoff S, Lachages AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470:120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- Loayza D, de Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;424:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- Martinez P, Thanasoula M, Munoz P, Liao C, Tejera A, McNees C, Flores JM, Fernandez-Capetillo O, Tarsounas M, Blasco MA. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009;23:2060–2075. doi: 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzner I, Savelyeva L, Schwab M. Preferential integration of a transfected marker gene into spontaneously expressed fragile sites of a breast cancer cell line. Cancer Lett. 2003;189:207–219. doi: 10.1016/s0304-3835(02)00504-9. [DOI] [PubMed] [Google Scholar]
- Miller CT, Lin L, Casper AM, Lim J, Thomas DG, Orringer MB, Chang AC, Chambers AF, Giordano TJ, Glover TW, Beer DG. Genomic amplification of MET with boundaries within fragile site FRA7G and upregulation of MET pathways in esophageal adenocarcinoma. Oncogene. 2006;25:409–418. doi: 10.1038/sj.onc.1209057. [DOI] [PubMed] [Google Scholar]
- Mishmar D, Rahat A, Scherer SW, Nyakatura G, Hinzmann B, Kohwi Y, Mandel-Gutfroind Y, Lee JR, Drescher B, Sas DE, Margalit H, Platzer M, Weiss A, Tsui LC, Rosenthal A, Kerem B. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc Natl Acad Sci U S A. 1998;95:8141–8146. doi: 10.1073/pnas.95.14.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrasek K, Schoder C, Teichmann AC, Behr K, Franze B, Wilhelm K, Blaurock N, Claussen U, Liehr T, Weise A. Global screening and extended nomenclature for 230 aphidicolin-inducible fragile sites, including 61 yet unreported ones. Int J Oncol. 2010;36:929–940. doi: 10.3892/ijo_00000572. [DOI] [PubMed] [Google Scholar]
- Ozeri-Galai E, Lebofsky R, Rahat A, Bester AC, Bensimon A, Kerem B. Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. Mol Cell. 2011;43:122–131. doi: 10.1016/j.molcel.2011.05.019. [DOI] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Pelliccia F, Bosco N, Curatolo A, Rocchi A. Replication timing of two human common fragile sites: FRA1H and FRA2G. Cytogenet Genome Res. 2008;121:196–200. doi: 10.1159/000138885. [DOI] [PubMed] [Google Scholar]
- Pelliccia F, Bosco N, Rocchi A. Breakages at common fragile sites set boundaries of amplified regions in two leukemia cell lines K562—molecular characterization of FRA2H and localization of a new CFS FRA2S. Cancer Lett. 2010;299:37–44. doi: 10.1016/j.canlet.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Ragland RL, Glynn MW, Arlt MF, Glover TW. Stably transfected common fragile site sequences exhibit instability at ectopic sites. Genes Chromosomes Cancer. 2008;47:860–872. doi: 10.1002/gcc.20591. [DOI] [PubMed] [Google Scholar]
- Rassool FV, McKeithan TW, Neilly ME, van Melle E, Rr E, Le Beau MM. Preferential integration of marker DNA into the chromosomal fragile site at 3p14: an approach to cloning fragile sites. Proc Natl Acad Sci U S A. 1991;88:6657–6661. doi: 10.1073/pnas.88.15.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshmi SC, Huang X, Schoppy DW, Black RC, Saunders WS, Smith DI, Gollin SM. Relationship between FRA11F and 11q13 gene amplification in oral cancer. Genes Chromosomes Cancer. 2007;46:143–154. doi: 10.1002/gcc.20394. [DOI] [PubMed] [Google Scholar]
- Salvati E, Scarsella M, PorruM, Rizzo A, Iachettini S, Tentori L, Graziani G, D'Incalci M, Stevens MF, Orlandi A, Passeri D, Gilson E, Zupi G, Leonetti C, Biroccio A. PARP1 is activated at telomeres upon G4 stabilization: possible target for telomere-based therapy. Oncogene. 2010;29:6280–6293. doi: 10.1038/onc.2010.344. [DOI] [PubMed] [Google Scholar]
- Schwartz M, Zlotorynski E, Kerem B. The molecular basis of common and rare fragile sites. Cancer Lett. 2006;232:13–26. doi: 10.1016/j.canlet.2005.07.039. [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonet T, Zaragosi LE, Philippe C, Lebrigand K, Schouteden C, Augereau A, Bauwens S, Ye J, Santagostino M, Giulotto E, Magdinier F, Horard B, Barbry P, Waldmann R, Gilson E. The human TTAGGG repeat factors 1 and 2 bind to a subset of interstitial telomeric sequences and satellite repeats. Cell Res. 2011;21:1028–1038. doi: 10.1038/cr.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slijepcevic P, Xiao Y, Dominguez I, Natarajan AT. Spontaneous and radiation-induced chromosomal breakage at interstitial telomeric sites. Chromosoma. 1996;104:596–604. doi: 10.1007/BF00352299. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T. Control of human telomere length by TRF1 and TRF2. Mol Cell Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland GR, Mattei JF. Report of the committee on cytogenetic markers. Cytogenet Cell Genet. 1987;46:316–324. doi: 10.1159/000132482. [DOI] [PubMed] [Google Scholar]
- Sutherland GR, Baker E, Richards RI. Fragile sites still breaking. Trends Genet. 1998;14:501–506. doi: 10.1016/s0168-9525(98)01628-x. [DOI] [PubMed] [Google Scholar]
- Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T. In vivo stoichiometry of shelterin components. J Biol Chem. 2010;285:1457–1467. doi: 10.1074/jbc.M109.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorland EC, Myers SL, Gostout BS, Smith DI. Common fragile sites are preferential targets for HPV16 integrations in cervical tumors. Oncogene. 2003;22:1225–1237. doi: 10.1038/sj.onc.1206170. [DOI] [PubMed] [Google Scholar]
- Tsantoulis PK, Kotsinas A, Sfikakis PP, Evangelou K, Sideridou M, Levy B, Mo L, Kittas C, Wu XR, Papavassiliou AG, Gorgoulis VG. Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Oncogene. 2008;27:3256–3264. doi: 10.1038/sj.onc.1210989. [DOI] [PubMed] [Google Scholar]
- Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- Wilke CM, Hall BK, Hoge A, ParadeeW, Smith DI, Glover TW. FRA3B extends over a broad region and contains a spontaneous HPV16 integration site: direct evidence for the coincidence of viral integration sites and fragile sites. Hum Mol Genet. 1996;5:187–195. doi: 10.1093/hmg/5.2.187. [DOI] [PubMed] [Google Scholar]
- Yang D, Xiong Y, Kim H, He Q, Li Y, Chen R, Songyang Z. Human telomeric proteins occupy selective interstitial sites. Cell Res. 2011;21:1013–1027. doi: 10.1038/cr.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimonjic DB, Durkin ME, Keck-Waggoner CL, Park SW, Thorgeirsson SS, Popescu NC. SMAD5 gene expression, rearrangements, copy number, and amplification at fragile site FRA5C in human hepatocellular carcinoma. Neoplasia. 2003;5:390–396. doi: 10.1016/s1476-5586(03)80041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotorynski E, Rahat A, Skaug J, Ben-Porat N, Ozeri E, Hershberg R, Levi A, Scherer SW, Margalit H, Kerem B. Molecular basis for expression of common and rare fragile sites. Mol Cell Biol. 2003;23:7143–7151. doi: 10.1128/MCB.23.20.7143-7151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.