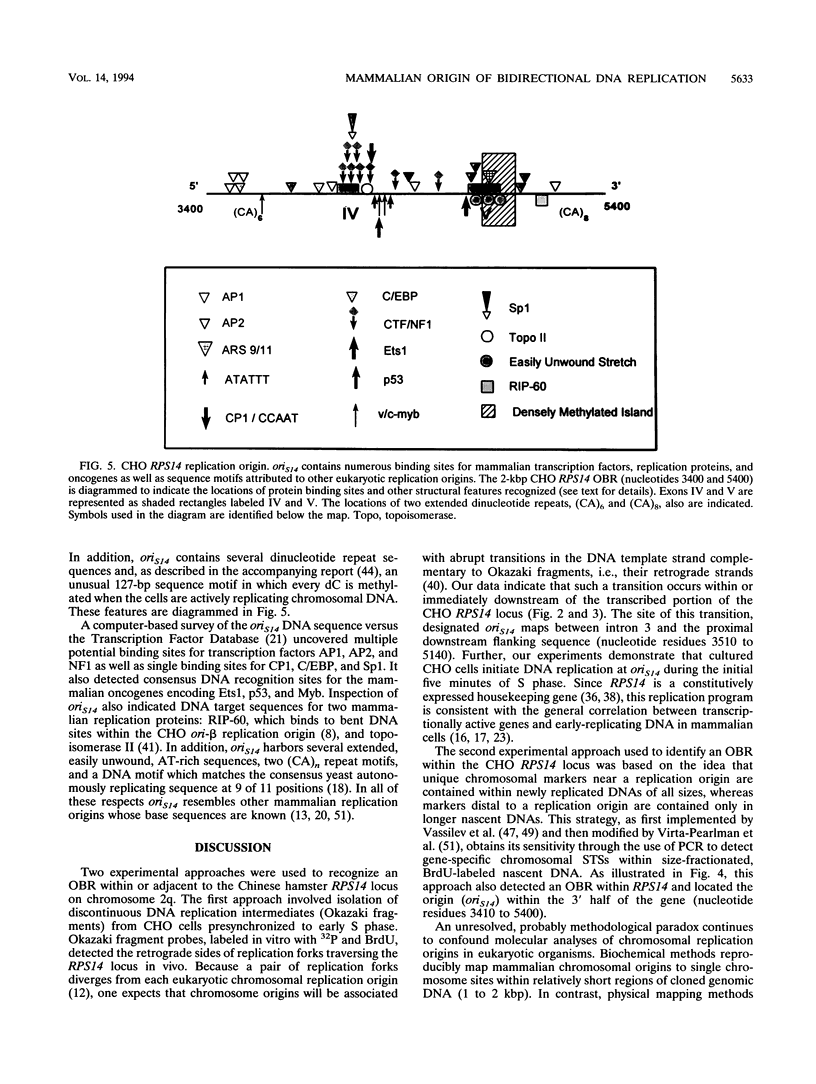
Abstract
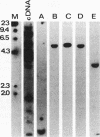
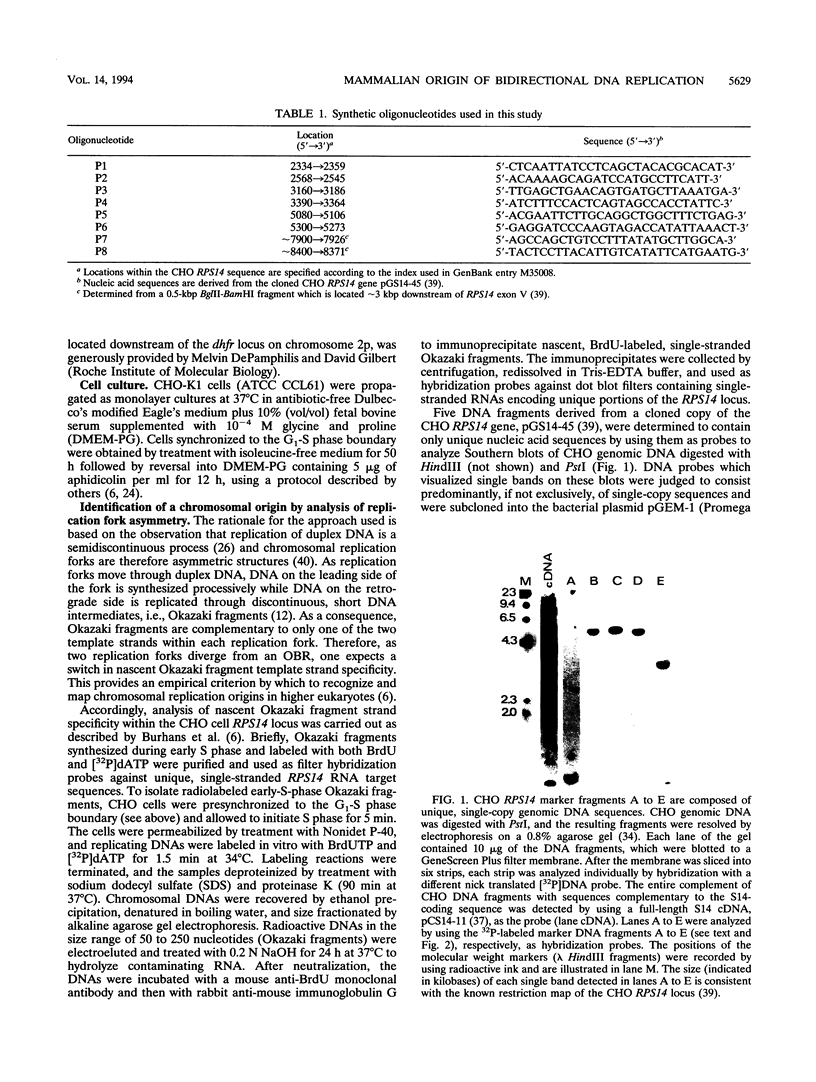
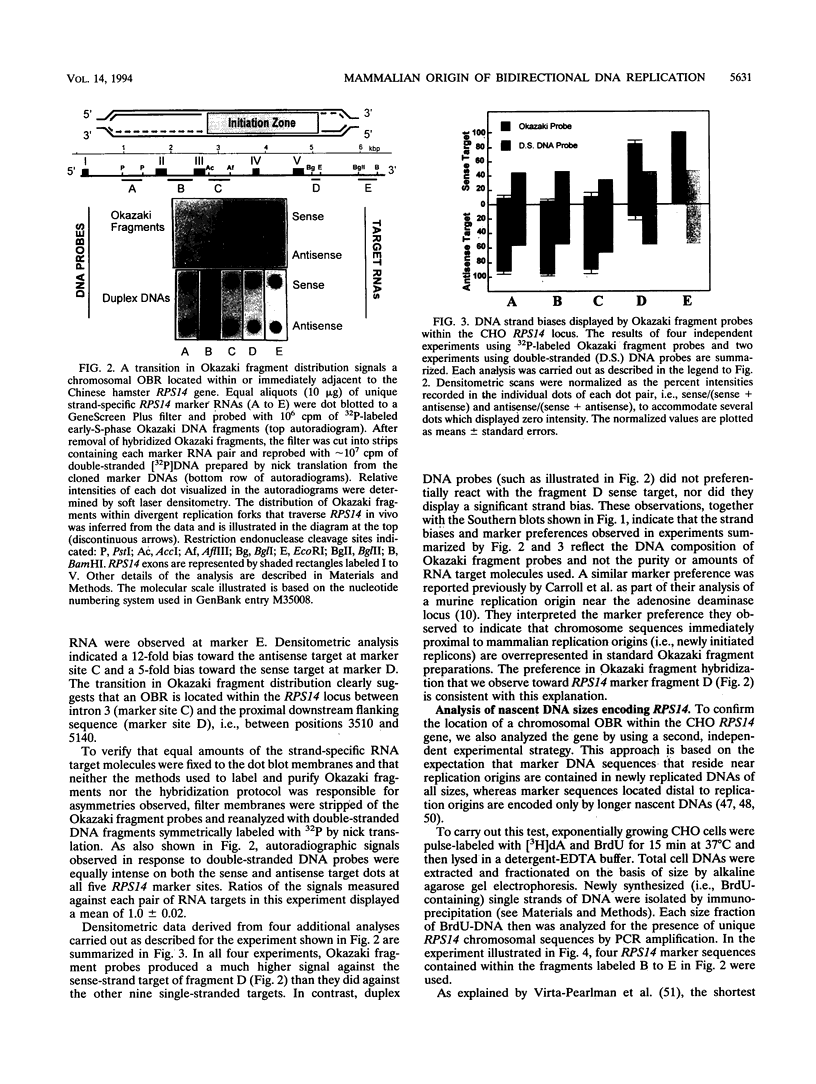
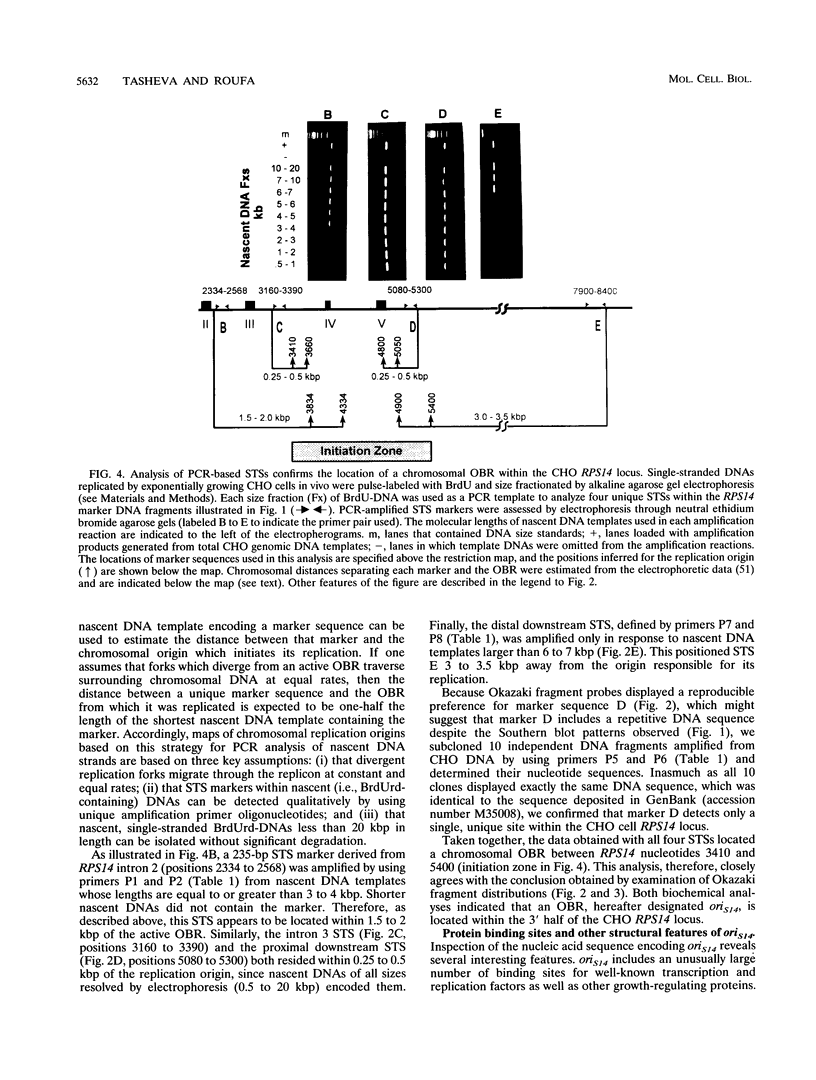
Two complementary experimental approaches have been used to identify a chromosomal origin of bidirectional DNA replication within or immediately downstream of the Chinese hamster ribosomal protein S14 gene (RPS14). The replication origin, designated oriS14, maps within a 1.6- to 2.0-kbp region of RPS14 that includes the gene's third and fourth introns, exons IV plus V, and approximately 500 bp of proximal downstream flanking DNA. The nucleic acid sequence encoding oriS14 closely resembles the other mammalian chromosomal replication origins whose primary structures are known. It contains DNA binding sites for a large number of transcription factors, replication proteins, and mammalian oncogenes as well as several dinucleotide repeat motifs, an AT-rich region, and a sequence that is likely to bend the DNA. In contrast to the other well-characterized mammalian replication origins, which are autosomal and therefore carried as two copies per somatic cell, oriS14 is encoded by single-copy DNA within a hemizygous segment of chromosome 2q in CHO-K1 cells. Also, other known mammalian replication origins are situated in nontranscribed, intergenic DNA, whereas the DNA sequence encoding oriS14 substantially overlaps the transcribed portion of a constitutively expressed housekeeping gene.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anachkova B., Hamlin J. L. Replication in the amplified dihydrofolate reductase domain in CHO cells may initiate at two distinct sites, one of which is a repetitive sequence element. Mol Cell Biol. 1989 Feb;9(2):532–540. doi: 10.1128/mcb.9.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi K., Wang Z., Tucker P. W. Immunoglobulin heavy chain enhancer is located near or in an initiation zone of chromosomal DNA replication. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3695–3699. doi: 10.1073/pnas.90.8.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E. H., Iqbal M. A., Stuart S., Hatton K. S., Valinsky J., Schildkraut C. L. Rate of replication of the murine immunoglobulin heavy-chain locus: evidence that the region is part of a single replicon. Mol Cell Biol. 1987 Jan;7(1):450–457. doi: 10.1128/mcb.7.1.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. J., Rhoads D. D., Stewart M. J., Van Slyke B., Chen I. T., Johnson T. K., Denell R. E., Roufa D. J. Ribosomal protein S14 is encoded by a pair of highly conserved, adjacent genes on the X chromosome of Drosophila melanogaster. Mol Cell Biol. 1988 Oct;8(10):4314–4321. doi: 10.1128/mcb.8.10.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burhans W. C., Selegue J. E., Heintz N. H. Isolation of the origin of replication associated with the amplified Chinese hamster dihydrofolate reductase domain. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7790–7794. doi: 10.1073/pnas.83.20.7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burhans W. C., Vassilev L. T., Caddle M. S., Heintz N. H., DePamphilis M. L. Identification of an origin of bidirectional DNA replication in mammalian chromosomes. Cell. 1990 Sep 7;62(5):955–965. doi: 10.1016/0092-8674(90)90270-o. [DOI] [PubMed] [Google Scholar]
- Burhans W. C., Vassilev L. T., Wu J., Sogo J. M., Nallaseth F. S., DePamphilis M. L. Emetine allows identification of origins of mammalian DNA replication by imbalanced DNA synthesis, not through conservative nucleosome segregation. EMBO J. 1991 Dec;10(13):4351–4360. doi: 10.1002/j.1460-2075.1991.tb05013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caddle M. S., Dailey L., Heintz N. H. RIP60, a mammalian origin-binding protein, enhances DNA bending near the dihydrofolate reductase origin of replication. Mol Cell Biol. 1990 Dec;10(12):6236–6243. doi: 10.1128/mcb.10.12.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calza R. E., Eckhardt L. A., DelGiudice T., Schildkraut C. L. Changes in gene position are accompanied by a change in time of replication. Cell. 1984 Mar;36(3):689–696. doi: 10.1016/0092-8674(84)90349-0. [DOI] [PubMed] [Google Scholar]
- Carroll S. M., DeRose M. L., Kolman J. L., Nonet G. H., Kelly R. E., Wahl G. M. Localization of a bidirectional DNA replication origin in the native locus and in episomally amplified murine adenosine deaminase loci. Mol Cell Biol. 1993 May;13(5):2971–2981. doi: 10.1128/mcb.13.5.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphili M. L. How transcription factors regulate origins of DNA replication in eukaryotic cells. Trends Cell Biol. 1993 May;3(5):161–167. doi: 10.1016/0962-8924(93)90137-p. [DOI] [PubMed] [Google Scholar]
- DePamphilis M. L. Eukaryotic DNA replication: anatomy of an origin. Annu Rev Biochem. 1993;62:29–63. doi: 10.1146/annurev.bi.62.070193.000333. [DOI] [PubMed] [Google Scholar]
- DePamphilis M. L. Origins of DNA replication in metazoan chromosomes. J Biol Chem. 1993 Jan 5;268(1):1–4. [PubMed] [Google Scholar]
- DePamphilis M. L. Origins of DNA replication that function in eukaryotic cells. Curr Opin Cell Biol. 1993 Jun;5(3):434–441. doi: 10.1016/0955-0674(93)90008-e. [DOI] [PubMed] [Google Scholar]
- DePamphilis M. L. Transcriptional elements as components of eukaryotic origins of DNA replication. Cell. 1988 Mar 11;52(5):635–638. doi: 10.1016/0092-8674(88)90398-4. [DOI] [PubMed] [Google Scholar]
- Dhar V., Mager D., Iqbal A., Schildkraut C. L. The coordinate replication of the human beta-globin gene domain reflects its transcriptional activity and nuclease hypersensitivity. Mol Cell Biol. 1988 Nov;8(11):4958–4965. doi: 10.1128/mcb.8.11.4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar V., Skoultchi A. I., Schildkraut C. L. Activation and repression of a beta-globin gene in cell hybrids is accompanied by a shift in its temporal replication. Mol Cell Biol. 1989 Aug;9(8):3524–3532. doi: 10.1128/mcb.9.8.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckdahl T. T., Anderson J. N. Conserved DNA structures in origins of replication. Nucleic Acids Res. 1990 Mar 25;18(6):1609–1612. doi: 10.1093/nar/18.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B. M., Fangman W. L. A position effect on the time of replication origin activation in yeast. Cell. 1992 Jan 24;68(2):333–339. doi: 10.1016/0092-8674(92)90474-q. [DOI] [PubMed] [Google Scholar]
- Gale J. M., Tobey R. A., D'Anna J. A. Localization and DNA sequence of a replication origin in the rhodopsin gene locus of Chinese hamster cells. J Mol Biol. 1992 Mar 20;224(2):343–358. doi: 10.1016/0022-2836(92)90999-z. [DOI] [PubMed] [Google Scholar]
- Ghosh D. A relational database of transcription factors. Nucleic Acids Res. 1990 Apr 11;18(7):1749–1756. doi: 10.1093/nar/18.7.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handeli S., Klar A., Meuth M., Cedar H. Mapping replication units in animal cells. Cell. 1989 Jun 16;57(6):909–920. doi: 10.1016/0092-8674(89)90329-2. [DOI] [PubMed] [Google Scholar]
- Hatton K. S., Dhar V., Brown E. H., Iqbal M. A., Stuart S., Didamo V. T., Schildkraut C. L. Replication program of active and inactive multigene families in mammalian cells. Mol Cell Biol. 1988 May;8(5):2149–2158. doi: 10.1128/mcb.8.5.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N. H., Hamlin J. L. An amplified chromosomal sequence that includes the gene for dihydrofolate reductase initiates replication within specific restriction fragments. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4083–4087. doi: 10.1073/pnas.79.13.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman J. A., Riggs A. D. On the mechanism of DNA replication in mammalian chromosomes. J Mol Biol. 1968 Mar 14;32(2):327–341. doi: 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- Kaufmann G., Bar-Shavit R., DePamphilis M. L. Okazaki pieces grow opposite to the replication fork direction during simian virus 40 DNA replication. Nucleic Acids Res. 1978 Jul;5(7):2535–2545. doi: 10.1093/nar/5.7.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachman H. M., Hatton K. S., Skoultchi A. I., Schildkraut C. L. c-myc mRNA levels in the cell cycle change in mouse erythroleukemia cells following inducer treatment. Proc Natl Acad Sci U S A. 1985 Aug;82(16):5323–5327. doi: 10.1073/pnas.82.16.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffak M., James C. D. Opposite replication polarity of the germ line c-myc gene in HeLa cells compared with that of two Burkitt lymphoma cell lines. Mol Cell Biol. 1989 Feb;9(2):586–593. doi: 10.1128/mcb.9.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens M. H., Huberman J. A. The two faces of higher eukaryotic DNA replication origins. Cell. 1990 Sep 7;62(5):845–847. doi: 10.1016/0092-8674(90)90258-g. [DOI] [PubMed] [Google Scholar]
- Ma C., Leu T. H., Hamlin J. L. Multiple origins of replication in the dihydrofolate reductase amplicons of a methotrexate-resistant chinese hamster cell line. Mol Cell Biol. 1990 Apr;10(4):1338–1346. doi: 10.1128/mcb.10.4.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki C. G., Rhoads D. D., Diaz J. J., Roufa D. J. A Drosophila ribosomal protein functions in mammalian cells. Mol Cell Biol. 1990 Sep;10(9):4524–4528. doi: 10.1128/mcb.10.9.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchionni M. A., Roufa D. J. Replication of viral DNA sequences integrated within the chromatin of SV40-transformed Chinese hamster lung cells. Cell. 1981 Oct;26(2 Pt 2):245–258. doi: 10.1016/0092-8674(81)90307-x. [DOI] [PubMed] [Google Scholar]
- Mullis K. B., Faloona F. A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- Nakamichi N., Rhoads D. D., Roufa D. J. The Chinese hamster cell emetine resistance gene. Analysis of cDNA and genomic sequences encoding ribosomal protein S14. J Biol Chem. 1983 Nov 10;258(21):13236–13242. [PubMed] [Google Scholar]
- Overman P. F., Rhoads D. D., Tasheva E. S., Pyle M. M., Roufa D. J. Multiple regulatory elements ensure accurate transcription of a human ribosomal protein gene. Somat Cell Mol Genet. 1993 Jul;19(4):347–362. doi: 10.1007/BF01232747. [DOI] [PubMed] [Google Scholar]
- Rhoads D. D., Dixit A., Roufa D. J. Primary structure of human ribosomal protein S14 and the gene that encodes it. Mol Cell Biol. 1986 Aug;6(8):2774–2783. doi: 10.1128/mcb.6.8.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads D. D., Roufa D. J. A cloned human ribosomal protein gene functions in rodent cells. Mol Cell Biol. 1987 Oct;7(10):3767–3774. doi: 10.1128/mcb.7.10.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads D. D., Roufa D. J. Emetine resistance of Chinese hamster cells: structures of wild-type and mutant ribosomal protein S14 mRNAs. Mol Cell Biol. 1985 Jul;5(7):1655–1659. doi: 10.1128/mcb.5.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads D. D., Roufa D. J. Molecular evolution of the mammalian ribosomal protein gene, RPS14. Mol Biol Evol. 1991 Jul;8(4):503–514. doi: 10.1093/oxfordjournals.molbev.a040665. [DOI] [PubMed] [Google Scholar]
- Roufa D. J., Marchionni M. A. Nucleosome segregation at a defined mammalian chromosomal site. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1810–1814. doi: 10.1073/pnas.79.6.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M., Hsieh T. S. Drosophila topoisomerase II double-strand DNA cleavage: analysis of DNA sequence homology at the cleavage site. Nucleic Acids Res. 1985 Feb 25;13(4):1057–1072. doi: 10.1093/nar/13.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975 May 25;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Tasheva E. S., Roufa D. J. Densely methylated DNA islands in mammalian chromosomal replication origins. Mol Cell Biol. 1994 Sep;14(9):5636–5644. doi: 10.1128/mcb.14.9.5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasheva E. S., Roufa D. J. Deoxycytidine methylation and the origin of spontaneous transition mutations in mammalian cells. Somat Cell Mol Genet. 1993 May;19(3):275–283. doi: 10.1007/BF01233075. [DOI] [PubMed] [Google Scholar]
- Ten Hagen K. G., Gilbert D. M., Willard H. F., Cohen S. N. Replication timing of DNA sequences associated with human centromeres and telomeres. Mol Cell Biol. 1990 Dec;10(12):6348–6355. doi: 10.1128/mcb.10.12.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe J. P., Lindstrom Y. I., Leffak M. Opposite replication polarities of transcribed and nontranscribed histone H5 genes. Mol Cell Biol. 1988 Apr;8(4):1657–1663. doi: 10.1128/mcb.8.4.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L. T., Burhans W. C., DePamphilis M. L. Mapping an origin of DNA replication at a single-copy locus in exponentially proliferating mammalian cells. Mol Cell Biol. 1990 Sep;10(9):4685–4689. doi: 10.1128/mcb.10.9.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L. T., DePamphilis M. L. Guide to identification of origins of DNA replication in eukaryotic cell chromosomes. Crit Rev Biochem Mol Biol. 1992;27(6):445–472. doi: 10.3109/10409239209082569. [DOI] [PubMed] [Google Scholar]
- Vassilev L., Johnson E. M. An initiation zone of chromosomal DNA replication located upstream of the c-myc gene in proliferating HeLa cells. Mol Cell Biol. 1990 Sep;10(9):4899–4904. doi: 10.1128/mcb.10.9.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L., Johnson E. M. Mapping initiation sites of DNA replication in vivo using polymerase chain reaction amplification of nascent strand segments. Nucleic Acids Res. 1989 Oct 11;17(19):7693–7705. doi: 10.1093/nar/17.19.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virta-Pearlman V. J., Gunaratne P. H., Chinault A. C. Analysis of a replication initiation sequence from the adenosine deaminase region of the mouse genome. Mol Cell Biol. 1993 Oct;13(10):5931–5942. doi: 10.1128/mcb.13.10.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worton R. G., Duff C., Campbell C. E. Marker segregation without chromosome loss at the emt locus in Chinese hamster cell hybrids. Somatic Cell Genet. 1980 Mar;6(2):199–213. doi: 10.1007/BF01538796. [DOI] [PubMed] [Google Scholar]
- Wu C., Zannis-Hadjopoulos M., Price G. B. In vivo activity for initiation of DNA replication resides in a transcribed region of the human genome. Biochim Biophys Acta. 1993 Sep 23;1174(3):258–266. doi: 10.1016/0167-4781(93)90194-i. [DOI] [PubMed] [Google Scholar]