Abstract
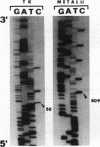
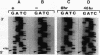
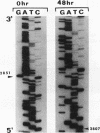
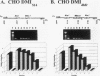
Densely methylated DNA sequence islands, designated DMIs, have been observed in two Chinese hamster cell chromosomal replication origins by using a PCR-based chemical method of detection. One of the origins, oriS14, is located within or adjacent to the coding sequence for ribosomal protein S14 on chromosome 2q, and the other, ori-beta, is approximately 17 kbp downstream of the dhfr (dihydrofolic acid reductase) locus on chromosome 2p. The DMI in oriS14 is 127 bp long, and the DMI in ori-beta is 516 bp long. Both DMIs are bilaterally methylated (i.e., all dCs are modified to 5-methyl dC) only in cells that are replicating their DNA. When cell growth and DNA replication are arrested, methylation of CpA, CpT, and CpC dinucleotides is lost and the sequence islands display only a subset of their originally methylated CpG dinucleotides. Several possible roles for DMI-mediated regulation of mammalian chromosomal origins are considered.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anachkova B., Hamlin J. L. Replication in the amplified dihydrofolate reductase domain in CHO cells may initiate at two distinct sites, one of which is a repetitive sequence element. Mol Cell Biol. 1989 Feb;9(2):532–540. doi: 10.1128/mcb.9.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker A., Smith D. W. Methylation of GATC sites is required for precise timing between rounds of DNA replication in Escherichia coli. J Bacteriol. 1989 Oct;171(10):5738–5742. doi: 10.1128/jb.171.10.5738-5742.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992 Jul 10;70(1):5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Blow J. J., Laskey R. A. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature. 1988 Apr 7;332(6164):546–548. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- Burhans W. C., Selegue J. E., Heintz N. H. Isolation of the origin of replication associated with the amplified Chinese hamster dihydrofolate reductase domain. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7790–7794. doi: 10.1073/pnas.83.20.7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burhans W. C., Vassilev L. T., Caddle M. S., Heintz N. H., DePamphilis M. L. Identification of an origin of bidirectional DNA replication in mammalian chromosomes. Cell. 1990 Sep 7;62(5):955–965. doi: 10.1016/0092-8674(90)90270-o. [DOI] [PubMed] [Google Scholar]
- Burhans W. C., Vassilev L. T., Wu J., Sogo J. M., Nallaseth F. S., DePamphilis M. L. Emetine allows identification of origins of mammalian DNA replication by imbalanced DNA synthesis, not through conservative nucleosome segregation. EMBO J. 1991 Dec;10(13):4351–4360. doi: 10.1002/j.1460-2075.1991.tb05013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caddle M. S., Lussier R. H., Heintz N. H. Intramolecular DNA triplexes, bent DNA and DNA unwinding elements in the initiation region of an amplified dihydrofolate reductase replicon. J Mol Biol. 1990 Jan 5;211(1):19–33. doi: 10.1016/0022-2836(90)90008-A. [DOI] [PubMed] [Google Scholar]
- Chen I. T., Roufa D. J. The transcriptionally active human ribosomal protein S17 gene. Gene. 1988 Oct 15;70(1):107–116. doi: 10.1016/0378-1119(88)90109-6. [DOI] [PubMed] [Google Scholar]
- Diaz J. J., Rhoads D. D., Roufa D. J. Genetic analysis of a vital mammalian housekeeping locus using CHO cells that express a transfected mutant allele. Somat Cell Mol Genet. 1990 Nov;16(6):517–528. doi: 10.1007/BF01233092. [DOI] [PubMed] [Google Scholar]
- Dijkwel P. A., Hamlin J. L. Matrix attachment regions are positioned near replication initiation sites, genes, and an interamplicon junction in the amplified dihydrofolate reductase domain of Chinese hamster ovary cells. Mol Cell Biol. 1988 Dec;8(12):5398–5409. doi: 10.1128/mcb.8.12.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss H. M., Roberts C. J., Claeys K. M., Selker E. U. Abnormal chromosome behavior in Neurospora mutants defective in DNA methylation. Science. 1993 Dec 10;262(5140):1737–1741. doi: 10.1126/science.7505062. [DOI] [PubMed] [Google Scholar]
- Frommer M., McDonald L. E., Millar D. S., Collis C. M., Watt F., Grigg G. W., Molloy P. L., Paul C. L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillin F. D., Roufa D. J., Beaudet A. L., Caskey C. T. 8-Azaguanine resistance in mammalian cells. I. Hypoxanthine-guanine phosphoribosyltransferase. Genetics. 1972 Oct;72(2):239–252. doi: 10.1093/genetics/72.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handeli S., Klar A., Meuth M., Cedar H. Mapping replication units in animal cells. Cell. 1989 Jun 16;57(6):909–920. doi: 10.1016/0092-8674(89)90329-2. [DOI] [PubMed] [Google Scholar]
- Huberman J. A., Riggs A. D. On the mechanism of DNA replication in mammalian chromosomes. J Mol Biol. 1968 Mar 14;32(2):327–341. doi: 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- Kao F. T., Johnson R. T., Puck T. T. Complementation analysis on virus-fused Chinese hamster cells with nutritional markers. Science. 1969 Apr 18;164(3877):312–314. doi: 10.1126/science.164.3877.312. [DOI] [PubMed] [Google Scholar]
- Leu T. H., Hamlin J. L. High-resolution mapping of replication fork movement through the amplified dihydrofolate reductase domain in CHO cells by in-gel renaturation analysis. Mol Cell Biol. 1989 Feb;9(2):523–531. doi: 10.1128/mcb.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. D., Meehan R. R., Henzel W. J., Maurer-Fogy I., Jeppesen P., Klein F., Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992 Jun 12;69(6):905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Ma C., Leu T. H., Hamlin J. L. Multiple origins of replication in the dihydrofolate reductase amplicons of a methotrexate-resistant chinese hamster cell line. Mol Cell Biol. 1990 Apr;10(4):1338–1346. doi: 10.1128/mcb.10.4.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki C., Rhoads D. D., Stewart M. J., Van Slyke B., Roufa D. J. The Drosophila melanogaster RPS17 gene encoding ribosomal protein S17. Gene. 1989 Jul 15;79(2):289–298. doi: 10.1016/0378-1119(89)90211-4. [DOI] [PubMed] [Google Scholar]
- Malki A., Kern R., Kohiyama M., Hughes P. Inhibition of DNA synthesis at the hemimethylated pBR322 origin of replication by a cell membrane fraction. Nucleic Acids Res. 1992 Jan 11;20(1):105–109. doi: 10.1093/nar/20.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan R. R., Lewis J. D., Bird A. P. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992 Oct 11;20(19):5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan R. R., Lewis J. D., McKay S., Kleiner E. L., Bird A. P. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989 Aug 11;58(3):499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- Meinkoth J., Killary A. M., Fournier R. E., Wahl G. M. Unstable and stable CAD gene amplification: importance of flanking sequences and nuclear environment in gene amplification. Mol Cell Biol. 1987 Apr;7(4):1415–1424. doi: 10.1128/mcb.7.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer W., Noyer-Weidner M. Timing and targeting: the biological functions of Dam methylation in E. coli. Cell. 1988 Sep 9;54(6):735–737. doi: 10.1016/s0092-8674(88)90911-7. [DOI] [PubMed] [Google Scholar]
- Nakamichi N. N., Kao F. T., Wasmuth J., Roufa D. J. Ribosomal protein gene sequences map to human chromosomes 5, 8, and 17. Somat Cell Mol Genet. 1986 May;12(3):225–236. doi: 10.1007/BF01570781. [DOI] [PubMed] [Google Scholar]
- Nakamichi N., Rhoads D. D., Roufa D. J. The Chinese hamster cell emetine resistance gene. Analysis of cDNA and genomic sequences encoding ribosomal protein S14. J Biol Chem. 1983 Nov 10;258(21):13236–13242. [PubMed] [Google Scholar]
- Ogden G. B., Pratt M. J., Schaechter M. The replicative origin of the E. coli chromosome binds to cell membranes only when hemimethylated. Cell. 1988 Jul 1;54(1):127–135. doi: 10.1016/0092-8674(88)90186-9. [DOI] [PubMed] [Google Scholar]
- Rhoads D. D., Dixit A., Roufa D. J. Primary structure of human ribosomal protein S14 and the gene that encodes it. Mol Cell Biol. 1986 Aug;6(8):2774–2783. doi: 10.1128/mcb.6.8.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads D. D., Roufa D. J. A cloned human ribosomal protein gene functions in rodent cells. Mol Cell Biol. 1987 Oct;7(10):3767–3774. doi: 10.1128/mcb.7.10.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads D. D., Roufa D. J. Emetine resistance of Chinese hamster cells: structures of wild-type and mutant ribosomal protein S14 mRNAs. Mol Cell Biol. 1985 Jul;5(7):1655–1659. doi: 10.1128/mcb.5.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads D. D., Roufa D. J. Molecular evolution of the mammalian ribosomal protein gene, RPS14. Mol Biol Evol. 1991 Jul;8(4):503–514. doi: 10.1093/oxfordjournals.molbev.a040665. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975 May 25;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Selker E. U., Fritz D. Y., Singer M. J. Dense nonsymmetrical DNA methylation resulting from repeat-induced point mutation in Neurospora. Science. 1993 Dec 10;262(5140):1724–1728. doi: 10.1126/science.8259516. [DOI] [PubMed] [Google Scholar]
- Shapiro R., Braverman B., Louis J. B., Servis R. E. Nucleic acid reactivity and conformation. II. Reaction of cytosine and uracil with sodium bisulfite. J Biol Chem. 1973 Jun 10;248(11):4060–4064. [PubMed] [Google Scholar]
- Stark G. R., Debatisse M., Giulotto E., Wahl G. M. Recent progress in understanding mechanisms of mammalian DNA amplification. Cell. 1989 Jun 16;57(6):901–908. doi: 10.1016/0092-8674(89)90328-0. [DOI] [PubMed] [Google Scholar]
- Tasheva E. S., Roufa D. J. A mammalian origin of bidirectional DNA replication within the Chinese hamster RPS14 locus. Mol Cell Biol. 1994 Sep;14(9):5628–5635. doi: 10.1128/mcb.14.9.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasheva E. S., Roufa D. J. Deoxycytidine methylation and the origin of spontaneous transition mutations in mammalian cells. Somat Cell Mol Genet. 1993 May;19(3):275–283. doi: 10.1007/BF01233075. [DOI] [PubMed] [Google Scholar]
- Toth M., Müller U., Doerfler W. Establishment of de novo DNA methylation patterns. Transcription factor binding and deoxycytidine methylation at CpG and non-CpG sequences in an integrated adenovirus promoter. J Mol Biol. 1990 Aug 5;214(3):673–683. doi: 10.1016/0022-2836(90)90285-T. [DOI] [PubMed] [Google Scholar]
- Vassilev L. T., Burhans W. C., DePamphilis M. L. Mapping an origin of DNA replication at a single-copy locus in exponentially proliferating mammalian cells. Mol Cell Biol. 1990 Sep;10(9):4685–4689. doi: 10.1128/mcb.10.9.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn J. P., Dijkwel P. A., Hamlin J. L. Replication initiates in a broad zone in the amplified CHO dihydrofolate reductase domain. Cell. 1990 Jun 15;61(6):1075–1087. doi: 10.1016/0092-8674(90)90071-l. [DOI] [PubMed] [Google Scholar]
- Wang R. Y., Gehrke C. W., Ehrlich M. Comparison of bisulfite modification of 5-methyldeoxycytidine and deoxycytidine residues. Nucleic Acids Res. 1980 Oct 24;8(20):4777–4790. doi: 10.1093/nar/8.20.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock D. M., Crowther P. J., Diver W. P. The majority of methylated deoxycytidines in human DNA are not in the CpG dinucleotide. Biochem Biophys Res Commun. 1987 Jun 15;145(2):888–894. doi: 10.1016/0006-291x(87)91048-5. [DOI] [PubMed] [Google Scholar]
- Worton R. G., Duff C., Campbell C. E. Marker segregation without chromosome loss at the emt locus in Chinese hamster cell hybrids. Somatic Cell Genet. 1980 Mar;6(2):199–213. doi: 10.1007/BF01538796. [DOI] [PubMed] [Google Scholar]