Abstract
TET proteins convert 5-methylcytosine to 5-hydroxymethylcytosine, an emerging dynamic epigenetic state of DNA that can influence transcription. Evidence has linked TET1 function to epigenetic repression complexes, yet mechanistic information, especially for the TET2 and TET3 proteins, remains limited. Here, we show a direct interaction of TET2 and TET3 with O-GlcNAc transferase (OGT). OGT does not appear to influence hmC activity, rather TET2 and TET3 promote OGT activity. TET2/3–OGT co-localize on chromatin at active promoters enriched for H3K4me3 and reduction of either TET2/3 or OGT activity results in a direct decrease in H3K4me3 and concomitant decreased transcription. Further, we show that Host Cell Factor 1 (HCF1), a component of the H3K4 methyltransferase SET1/COMPASS complex, is a specific GlcNAcylation target of TET2/3–OGT, and modification of HCF1 is important for the integrity of SET1/COMPASS. Additionally, we find both TET proteins and OGT activity promote binding of the SET1/COMPASS H3K4 methyltransferase, SETD1A, to chromatin. Finally, studies in Tet2 knockout mouse bone marrow tissue extend and support the data as decreases are observed of global GlcNAcylation and also of H3K4me3, notably at several key regulators of haematopoiesis. Together, our results unveil a step-wise model, involving TET–OGT interactions, promotion of GlcNAcylation, and influence on H3K4me3 via SET1/COMPASS, highlighting a novel means by which TETs may induce transcriptional activation.
Keywords: chromatin, epigenetics, TET proteins
Introduction
Epigenetic marking of the genome and regulation of chromatin are central to establishing tissue-specific gene expression programs, and hence to several biological processes (Bonasio et al, 2010). Until recently, the only known epigenetic mark on DNA was 5-methylcytosine (mC), established and propagated by DNA methyltransferases, and generally associated with gene repression (Suzuki and Bird, 2008; Cedar and Bergman, 2009; Ndlovu et al, 2011). The discovery of cystosine 5-hydroxymethylation (hmC), and of the Ten-Eleven Translocation family of enzymes (TET1, TET2, and TET3) that catalyse the conversion of mC to hmC has sparked great interest in uncovering the roles played by this mark and these proteins (Kriaucionis and Heintz, 2009; Tahiliani et al, 2009; Ito et al, 2010). Genome-wide profiling of the distribution of Tet1 and hmC in mouse ES cells has shown that both are important in regulation of pluripotency and cellular differentiation (Ficz et al, 2011; Pastor et al, 2011; Williams et al, 2011; Wu et al, 2011). Other data have linked TET1 to epigenetic repression complexes, notably SIN3A and PRC2 (Williams et al, 2011; Wu et al, 2011). Regarding TET2 and TET3, mouse Tet2 has been implicated in haematopoiesis and human TET2 mutations have been found in various leukaemias (Langemeijer et al, 2009), while studies in mouse germ cells have shown the importance of Tet3 in epigenetic reprogramming (Gu et al, 2011). However the mode of action, particularly of the TET2 and TET3 family members, is still poorly understood.
Results
TET2 and TET3 associate with the O-GlcNAc transferase OGT
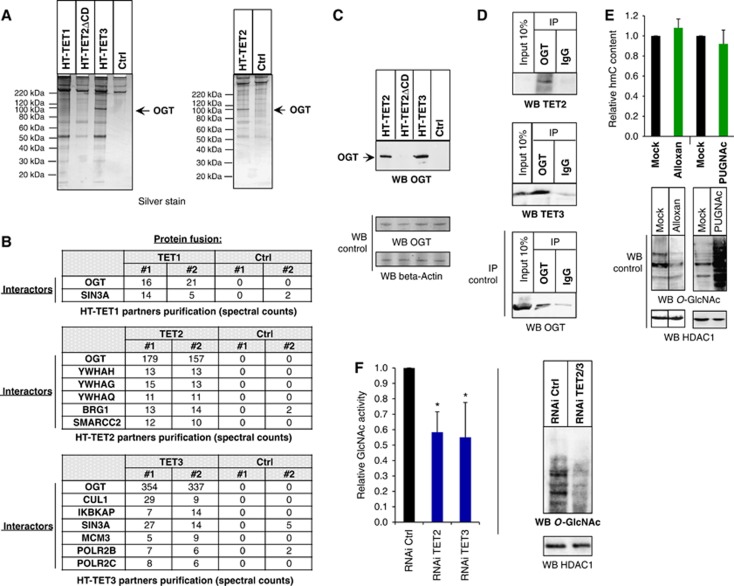
To aid in this understanding and further explore binding partners of these proteins, we performed an unbiased proteomic approach using HaloTag technology as previously described (Los et al, 2008; Daniels et al, 2012). To this end, we expressed the full-length TET1, TET2, and TET3 proteins as HaloTag (HT) fusions in HEK293T cells, covalently captured them on an HT affinity resin, eluted the interacting proteins, and purified these for mass spectrometry (LC/MS/MS) and spectral counting analysis (Materials and methods; Supplementary Figure 1). Figure 1A shows the silver stain gel for each complex isolation and the enrichment of numerous bands for each as compared to the HT alone control. Mass spectrometry of each revealed notably several transcriptional and epigenetic interacting proteins identified in biological replicates and enriched over the HT alone control, including the known TET1–SIN3A interaction and the absence of this with TET2 (Williams et al, 2011; Figure 1B). All TET proteins showed interaction with O-GlcNAc transferase, OGT, (Figure 1B), though TET2 and TET3 showed demonstrably higher enrichment. Given this, we focused all following experiments with the TET2 and TET3 family members. Two approaches were used to confirm the interaction between TET2 or TET3 and OGT: (i) western blotting with anti-OGT antibody, applied to samples of eluate from the above-mentioned HT-pulldown experiments (Figure 1C), and (ii) co-immunoprecipitations (Co-IPs) showing capture of both TET2 and TET3 from extracts of untransfected cells upon immunoprecipitation with an anti-OGT antibody, confirming the interaction of endogenous TET2 and TET3 with OGT (Figure 1D). Input loading controls of OGT are shown for each (Figures 1C and D). A variant of TET2 lacking the catalytic domain (TET2ΔCD) did not show interaction with OGT (Figures 1A and C; Supplementary Figure 2), indicating that the catalytic domain is important for the OGT interaction and demonstrating the specificity of the HT pulldown and mass spectrometry analysis. Supporting this, FLAG Co-IP experiments performed with FLAG-TET2 and -TET3 CD alone showed interaction with transfected or endogenous OGT (Supplementary Figure 3). Furthermore, to evaluate whether the interaction is direct, an in vitro protein isolation was performed using E. coli expressed and purified full-length OGT incubated with E. coli expressed HT-TET2CD or HT alone control covalently bound to resin. As shown in Supplementary Figure 3, OGT was specifically enriched on resin containing the TET2CD as compared to the control. Together, these data demonstrate that TET2 and TET3 interact directly with the OGT glycosyltransferase.
Figure 1.
TET2 and TET3 associate with the O-GlcNAc transferase OGT and promote GlcNAcylation. (A) Silver stain gel of HaloTag-TET protein complex isolations and HaloTag alone control (Ctrl). Protein pulldowns were performed from HEK293T cells overexpressing the indicated HT constructs (see Materials and methods and Supplementary Figure 1 for details). As not all of the indicated complex isolations were performed at the same time, two separate silver stain gels were run, as shown in this panel. (B) Table of transcriptional or chromatin protein interactors found in the various HaloTag-TET isolations. Spectral counts for each interacting protein are shown for biological replicates. TET1, but not TET2, as previously reported (Williams et al, 2011; Wu et al, 2011), shows interaction with SIN3A. OGT interacts with all TET proteins, though it is most highly abundant with TET2 and TET3. (C) Detection of OGT by western blotting from HT-TET2 and HT-TET3 pulldowns from (A). The indicated pulldowns were probed with an anti-OGT antibody to detect the presence of OGT. OGT and beta-Actin shown as input loading controls. (D) TET2 and TET3 co-immunoprecipitate (CoIP) with endogenous OGT from untransfected HEK293T cells. Cell extracts were immunoprecipitated with anti-OGT or rabbit IgG and probed with antibodies against the indicated proteins. An IP control of OGT alone is shown to demonstrate specific capture and enrichment of OGT. Inputs loading controls are shown for all. Note that in this experiment very weak expression of TET2 relative to TET3 is observed. (E) The global level of hmC does not change after cell treatment with Alloxan or PUGNAc. Dot blot quantification of global hmC after the indicated treatments. The hmC content is normalized with respect to the input DNA and to mock-treated cells, where the ratio is set at 1.00. Error bars indicate s.d. of three independent experiments. As controls, western blots using anti-O-GlcNAc antibody show the expected decrease in GlcNAcylation with Alloxan and increase with PUGNAc. HDAC1 input loading controls are also shown. Vertical line indicates juxtaposition of lanes non-adjacent within the same blot, exposed for the same time. (F) Global decrease in GlcNAcylation is observed in TET2/3 knockdowns. Left: TET2 kd or TET3 kd show decreased GlcNAc activity. Nuclear extracts were prepared from HEK293T cells expressing RNAi Ctrl, RNAi TET2, or RNAi TET3, and UDP-[3H]GlcNAc incorporation was measured. The amount incorporated into the control cells was set at 1. Error bars indicate s.d. of three independent experiments (*P<0.05). Right: Nuclear extracts were prepared from HEK293T cells expressing RNAi Ctrl or RNAi TET2/3 and global GlcNAcylation was visualized with an antibody against O-GlcNAc. HDAC1 input loading control is also shown.
TET2 and TET3 promote OGT-mediated GlcNAcylation
OGT places O-GlcNAc modifications on numerous proteins, including transcription factors, epigenetic regulators, and also histones (Hart et al, 2007). GlcNAcylation can activate or inhibit protein activity and serve as a mark to recruit other proteins (Slawson and Hart, 2011). As several proteins that interact with OGT are also regulated by GlcNAcylation (Hart et al, 2007), we analysed TET2 and TET3 by mass spectrometry for the presence of O-GlcNAc modifications. Despite high levels of spectral counts (∼400) for both proteins, GlcNAcylation could not be detected for either protein (data not shown). Based upon these results, we predict if TET proteins are GlcNAcylated, they would be at low levels. As TET and OGT are both enzymes, we next sought to determine if their respective activities are altered by each other. To evaluate if the enzymatic activity of OGT might influence hydroxymethylation by the TET enzymes, we measured the overall abundance of the hmC mark in dot blot assay using a specific hmC antibody (Supplementary Figure 4) in the absence or presence of either Alloxan, an OGT inhibitor, or PUGNAc, which prevents removal of the O-GlcNAc modification by inhibiting the enzyme O-linked N-acetylglucosaminidase (OGA) (Capotosti et al, 2011; Fujiki et al, 2009). As shown in Figure 1E, no changes in overall hmC level were observed with these respective treatments (upper panel), while a western blot of lysates revealed the expected impact on global levels of O-GlcNAc for each (lower panel). Similarly, overexpression of HT-OGT did not increase levels of hmC, while overexpression of a TET2CD alone control did (Supplementary Figure 4). Hence, cytosine hydroxylation does not appear to require the GlcNAcylating activity of OGT. We then investigated if GlcNAcylation might depend on TET2 and/or TET3. To answer this question, we measured overall OGT activity in lysates of cells where RNAi was used to knock down TET2 or TET3 (cf. Supplementary Figure 5 for RNAi efficiency and controls). As shown in Figure 1F (left panel), we observed lower GlcNAcylation activity in TET2 or TET3 RNAi than in the control cells. Following with this, in a TET2 and TET3 double RNAi kd (referred to as TET2/3 kd) a discernable decrease in O-GlcNAc modified proteins is seen as revealed by western blot analysis using an anti-O-GlcNAc antibody (Figure 1F, right panel; Supplementary Figure 5). To demonstrate that decreased OGT activity was not due to change in levels of OGT, analysis of lysates from mock RNAi and TET2/3 kd cells revealed that protein levels of OGT were unchanged after depletion of TET2 and TET3 (Supplementary Figures 5 and 6). These combined data suggest that TET2/3 positively impacts the activity of OGT.
TET2, TET3, and OGT show genome-wide co-localization, notably at CpG islands and at transcription start sites
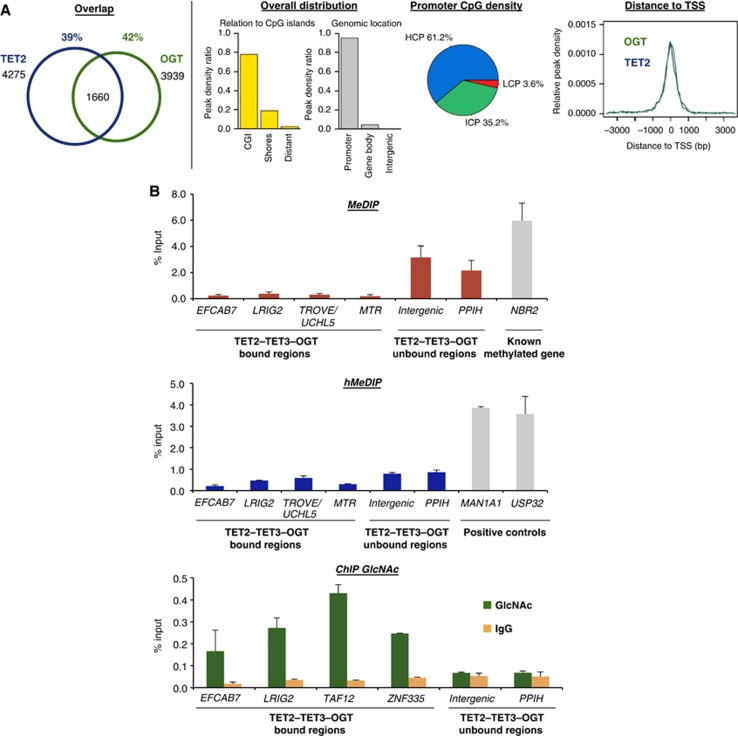
We next undertook to map genome-wide binding of TET2, TET3, and OGT, looking for their possible co-occurrence at genomic targets. We performed crosslinking and chromatin isolation from HEK293T cells, expressing HT fusion proteins (Los et al, 2008; Hartzell et al, 2009) coupled with high-throughput sequencing (Supplementary Figure 7). Our results show that TET2, TET3, and OGT all localize primarily to CpG islands (CGI) and promoter regions (Figure 2A; Supplementary Figure 8), similar to previously observed localization for Tet1 in mouse ES cells (Ficz et al, 2011; Williams et al, 2011; Wu et al, 2011). Co-occurrence analysis of OGT binding sites revealed statistically significant overlaps with TET2 (42%) (Figure 2A) and TET3 (55%) (Supplementary Figure 8; Supplementary Table 2). The TET2–OGT and TET3–OGT targets are primarily contained within promoter regions, tightly clustered around transcription start sites (TSSs), with intermediate to high CpG content (ICP and HCP) (Figure 2A; Supplementary Figures 8 and 9; Supplementary Table 2).
Figure 2.
TET2/3–OGT show genomic co-localization around TSSs and impact on H3K4me3 and transcriptional activation. (A) Left: Venn diagrams indicating significant overlap of TET2 and OGT bound regions (left part; P-value<10−10) identified after HaloCHIP-Seq in HEK293T cells expressing HT-TET2, or HT-OGT. Right: TET2–OGT targets are primarily found at TSSs and CpG-rich sequences. Similar profiles were also observed for TET3–OGT (Supplementary Figure 8). (B) An analysed subset of TET2–TET3–OGT targets show a lack of DNA methylation and hydroxymethylation, yet display GlcNAcylation. qPCR analysis of TET2–TET3–OGT binding and non-binding regions after MeDIP (top), hMeDIP (middle), or ChIP with an anti-O-GlcNAc antibody (bottom). ‘% Input’ represents real-time qPCR values normalized with respect to the input chromatin. Known methylated and hydroxymethylated regions are shown as positive controls in MeDIP and hMeDIP panels.
TET2, TET3, and OGT genomic targets are enriched for GlcNAcylation, but not for 5hmC or 5mC
We then examined the DNA methylation, DNA hydroxymethylation, and protein GlcNAcylation status of selected TET2/3–OGT target sequences. As shown in Figure 2B (upper and middle panels), methylated DNA and hydroxymethylated DNA immunoprecipitations (MeDIP and hMeDIP) followed by quantitative PCR (qPCR) analysis revealed essentially no enrichment of 5mC or 5hmC, respectively, at these targets in either immunoprecipitate. Significant enrichment of O-GlcNAc was found for TET2/3–OGT targets using chromatin immunoprecipitation (ChIP)-qPCR against O-GlcNAc (Figure 2B, lower panel), indicating OGT activity at these chromatin sites. Together, these data show co-occurrence of TET2, TET3, and OGT, which in a subset analysed, lack DNA methylation and hydroxymethylation, but display protein GlcNAcylation.
TET2, TET3, and OGT co-localize with and influence H3K4 trimethylation at active promoters in human cells
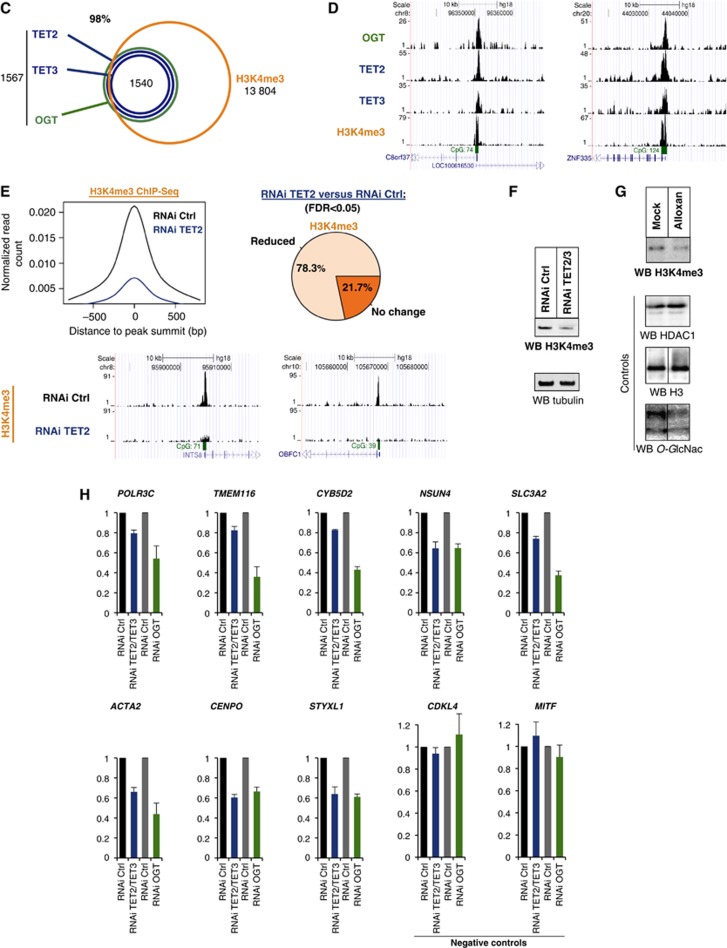
The clustering of TET2/3–OGT primarily at TSSs and HCP promoters (cf. Figure 2A) led us next to examine the genomic overlap at promoters with H3K4 trimethylation (H3K4me3), an expected mark at these sites which is also correlated with transcriptional activity. As depicted in Figures 2C and D, ChIP-Seq with an antibody against H3K4me3 confirmed an expected high overlap (98%) of TET2/3–OGT targets with the H3K4me3 mark. We then wondered whether TET proteins might be involved in regulating H3K4me3, and therefore repeated ChIP-Seq experiment in cells containing a TET2 RNAi kd. We observed a significant reduction of H3K4me3 at over 78% of the TET2/3–OGT genomic targets (Figure 2E). A similar impact on global reduction of H3K4me3 is observed in western blot analysis of TET2/3 knockdown cellular lysates (Figure 2F). As reduction in H3K4me3 could occur through a variety of mechanisms, we then sought to determine if there was a connection to OGT activity. Supporting this hypothesis and similar to the above data with TET2/3 kd (Figure 2F), cells treated with the OGT inhibitor, Alloxan, show also a decrease in global H3K4me3 as compared to controls (Figure 2G). Thus, TET2/3–OGT targets are enriched for H3K4me3, which is regulated by both the TET proteins and OGT activity.
Figure 2.
(C) TET2/3–OGT targets in HEK293T cells are enriched for H3K4me3 as depicted in a Venn diagram; P-value<10−10. (D) Examples of HaloCHIP-Seq OGT, TET2, TET3, and ChIP-Seq H3K4me3 profiles (UCSC tracks). (E) Decreased levels of H3K4me3 in TET2 kd cells. Upper-left: decrease in the normalized number of H3K4me3 reads in TET2/3–OGT-binding regions in TET2 kd cells versus control RNAi-treated cells. Upper-right: pie chart showing the percentage of TET2–TET3–OGT binding regions with a statistically significant reduction of the normalized number of H3K4me3 reads for TET2 kd versus control RNAi-treated cells. Lower-part: examples of H3K4me3 ChIP-Seq profiles (UCSC tracks) in TET2–TET3–OGT-binding regions for the RNAi control versus TET2 kd sample. (F) Western blot showing global decrease in H3K4me3 in a TET2/3 double kd cells. Lysates from mock HEK293T RNAi kd or TET2/3 kd cells were probed for H3K4me3 using an anti-H3K4 antibody in western blot. Tubulin is shown as a loading control. (G) OGT activity is important for H3K4me3. Cell extracts were prepared from HEK293T cells treated with or without the OGT inhibitor Alloxan, and then western blots for H3K4me3 were performed. HDAC1 and H3 are shown as loading controls and a western blot against O-GlcNAc was used to monitor specific GlcNAcylation inhibition by Alloxan. Vertical lines indicate juxtaposition of lanes non-adjacent within the same blot, exposed for the same time. (H) Decreases in transcription are observed in both TET2/3 knockdowns and an OGT knockdown. The indicated target genes (which showed decrease in H3K4me3 after TET2 kd; cf. E) and negative controls (unbound TET2/3–OGT–H3K4me3 targets), were analysed by RT–qPCR in HEK293T cells subjected to the various listed RNAi treatments. Independent experiments were performed in duplicates.
To correlate these data to gene activity, we found by ChIP-Seq a marked co-occupancy of TET2/3–OGT–H3K4me3 targets with RNA polymerase II (RNA Pol II) and by RNA Sequencing (RNA-Seq) analyses a significant and positive correlation with high expression levels (Supplementary Figure 10). To determine if there are effects on transcriptional levels after loss of TET and/or OGT, we then analysed by RT–qPCR a subset of TET2/3–OGT targets specifically decreased in H3K4me3 after TET kd, as identified in Figure 2E. As shown in Figure 2H, moderate, but reproducible and specific decreases in expression of targets after TET2/3 kd match patterns to those observed in OGT RNAi kd. Together, these data suggest that TET2/3–OGT–H3K4me3 targets are highly transcribed regions and for the subset studied, show concomitant transcriptional activation by both TET proteins and OGT.
TET2 and TET3 associate with HCF1, an OGT-modified target and component of the H3K4 methyltransferase complex SET1/COMPASS
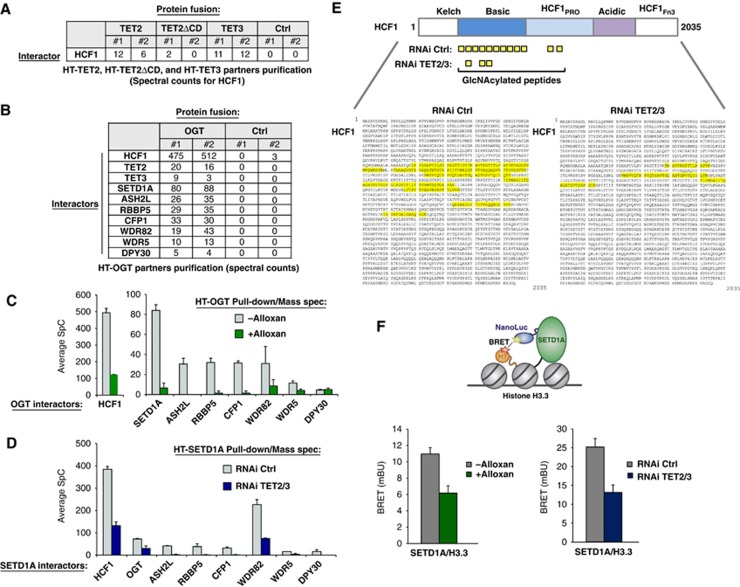
We next wished to extend our observed TET2/3–OGT interaction and identify potential specific downstream protein targets that may connect to H3K4me3 regulation. With this in mind, we noticed in our initial proteomic analysis for TET2 and TET3 that besides OGT (cf. Figure 1B), Host Cell Factor 1 (HCF1) was also identified and similarly to OGT, was decreased in the TET2ΔCD isolation (Figure 3A). HCF1 was of interest as not only is it a known GlcNAcylation target of OGT (Capotosti et al, 2011), but also a component of the histone H3K4 methyltransferase complex, SET1/COMPASS (Wysocka et al, 2003; Lee et al, 2010). ChIP-Seq experiments for endogenous HCF1 showed overlap of HCF1 target genes with those of Halo-TET2, TET3, OGT, and H3K4me3 (Supplementary Figure 11A). We then desired to further the mechanistic understanding of the TET2- or TET3-OGT–HCF1 interactions with relationship to H3K4me3, and therefore examined the interactions of OGT using a similar HT proteomics approach including mass spectrometry analysis. Interacting partners of HT-OGT included the previously identified HCF1 protein (Capotosti et al, 2011), the TET2 and TET3 proteins (confirming the TET–OGT interaction), and interestingly, all components of SET1/COMPASS (Figure 3B). To determine if the interactions were dependent on OGT activity, we repeated the above experiments in the presence of the OGT inhibitor Alloxan. Such treatment resulted not only in the expected decrease in OGT–HCF1 interaction (Capotosti et al, 2011), but also in significant loss of capture of the SET1/COMPASS components, including the H3K4 methyltransferase SETD1A (Figure 3C). Normalized spectral abundance factors (NSAFs) showed the same trend (Supplementary Figure 11B), therefore validating the direct comparison of respective loss of spectral counts for each protein within the samples. Mass spectrometry analysis of HCF1 peptides from HT-OGT protein isolations with or without Alloxan treatment showed high levels of GlcNAcylation in untreated cells and reduction after Alloxan treatment (Supplementary Figure 12A), matching previously published results (Capotosti et al, 2011).
Figure 3.
TET2/3 promotes GlcNAcylation of HCF1, and both TET and OGT activity favour the integrity of SET1/COMPASS and SETD1A binding to chromatin. (A) Mass spectrometry reveals HCF1, a known target of OGT (Wysocka et al, 2003) and component of SET1/COMPASS (Lee et al, 2010), as an interacting partner of HT-TET2 and HT-TET3. Biological duplicates and respective spectral counts (SpC) for HCF1 are shown. (B) Protein pulldowns of HT-OGT coupled with mass spectrometry identify HCF1, TET2, TET3, and all components of SET1/COMPASS as partners of OGT. Biological duplicates and SpC for each protein identified are shown for HT-OGT and Ctrl isolations. (C) The interaction of HCF1 and SET1/COMPASS components with HT-OGT depends on O-GlcNAc activity. Plot showing average SpCs for HCF1 and SET1/COMPASS components isolated from HT-OGT pulldowns of untreated (grey bars) and Alloxan-treated (green bars) HEK293T cells. Error bars represent s.d. of biological duplicates. Representative NSAF plots are shown in Supplementary Figure 11. (D) The interaction of HT-SETD1A with SET1/COMPASS components and OGT is reduced by a TET2/3 double kd. Plot showing average SpCs for SET1/COMPASS components and OGT isolated from HT-SETD1A pulldowns of control RNAi-treated (grey bars) and TET2/3 kd (blue bars) HEK293T cells. Error bars represent s.d. of biological duplicates. Representative NSAF plots are shown in Supplementary Figure 11. (E) A significant reduction in HCF1 GlcNAcylation is observed after TET2/3 double kd. Upper diagram shows a schematic representation of full-length HCF1 and its domains (Capotosti et al, 2011). The GlcNAcylated peptides identified by mass spectrometry from HT-SETD1A isolations from control RNAi-treated and TET2/3 kd cells are indicated below. The full-length HCF1 amino-acid sequence (NP_005325.2) shows the corresponding GlcNAcylated peptides highlighted in yellow with RNAi Ctrl on the left and RNAi TET2/3 kd on the right. (F) Bioluminescence resonance energy transfer (BRET) assays show reduction of SETD1A binding to histone H3.3 in the presence of an OGT inhibitor and in TET2/3 kd cells. Upper diagram showing the schematic of BRET energy transfer upon binding of a NanoLuc-SETD1A fusion donor and fluorescently labelled Histone H3.3-HaloTag fusion acceptor in live HEK293T cells (see Materials and methods for experimental details and calculation of BRET). Left: BRET measurements were calculated without treatment (grey) or with Alloxan treatment (green). Right: BRET measurement for RNAi control (grey) or RNAi TET2/3 (blue). Biological triplicates ±s.d. are shown.
TET2/3 promotes GlcNAcylation of HCF1, and both TET and OGT activity favour the integrity of SET1/COMPASS and SETD1A binding to chromatin
As these data suggested that GlcNAcylation of HCF1 might be important for its interaction with SET1/COMPASS, we then performed an HT-SETD1A pulldown to study the SET1/COMPASS complex. Mass spectrometry analysis of the interacting proteins revealed all the known members of the complex, as well as OGT (Figure 3D). Additional analysis of the HCF1 peptides from this experiment revealed extensive O-GlcNAc modifications, indicating that HCF1 is heavily modified when associated with SETD1A (Figure 3E, left panel ‘RNAi Ctrl’). We next examined whether the TET proteins might influence SET1/COMPASS complex formation and/or specific GlcNAcylation of HCF1. In order to do this, we performed HT-SETD1A isolations in cells harbouring the TET2/3 double kd, which were shown to decrease global H3K4me3 (cf. Figure 2F). As shown in Figure 3D (and Supplementary Figure 11B as NSAF values with the same trends), we observed a significant loss of SET1/COMPASS components and OGT in the TET2/3 RNAi kd cells, coinciding with a significant decrease in GlcNAcylation of HCF1 as determined by mass spectrometry (Figure 3E; Supplementary Methods). To show further the direct effect of TET proteins specifically on SET1/COMPASS complex formation, proteomics experiments were additionally performed with HT-WDR82, the only other specific component of SET1/COMPASS besides SETD1A. These data revealed the same disruption of capture of SET1/COMPASS components in cells harbouring a TET2/3 knockdown as compared to mock (Supplementary Figure 12B). These results indicate that TET2/3 proteins, and OGT GlcNAcylation of HCF1 are important for the integrity of the SET1/COMPASS complex, suggesting a means by which H3K4me3 may be regulated. To further this understanding, bioluminescence resonance energy transfer (BRET) experiments were carried out in living cells to monitor the interaction of SETD1A with chromatin. In short, SETD1A was designed as an energy donor, expressed as a fusion to NanoLuc luciferase, and Histone H3.3 as an energy acceptor, expressed and fluorescently labelled as a fusion to HT (Figure 3F). As shown in Figure 3F, treatment of cells with an OGT inhibitor, Alloxan, (left panel) or analysis in TET2/3 RNAi knockdown cells (right panel) result in specific reduction of BRET signal, indicating decrease in interaction between SETD1A and Histone H3.3 after these respective treatments. Together, these data support that TET2/3 proteins and OGT activity are important not only for the integrity of the H3K4 methyltransferase SET1/COMPASS complex, but also binding of the methyltransferase component, SETD1A, to chromatin.
Tet2 knockout mouse tissue shows that Tet2 is needed for global GlcNAcylation and H3K4me3 at target promoters
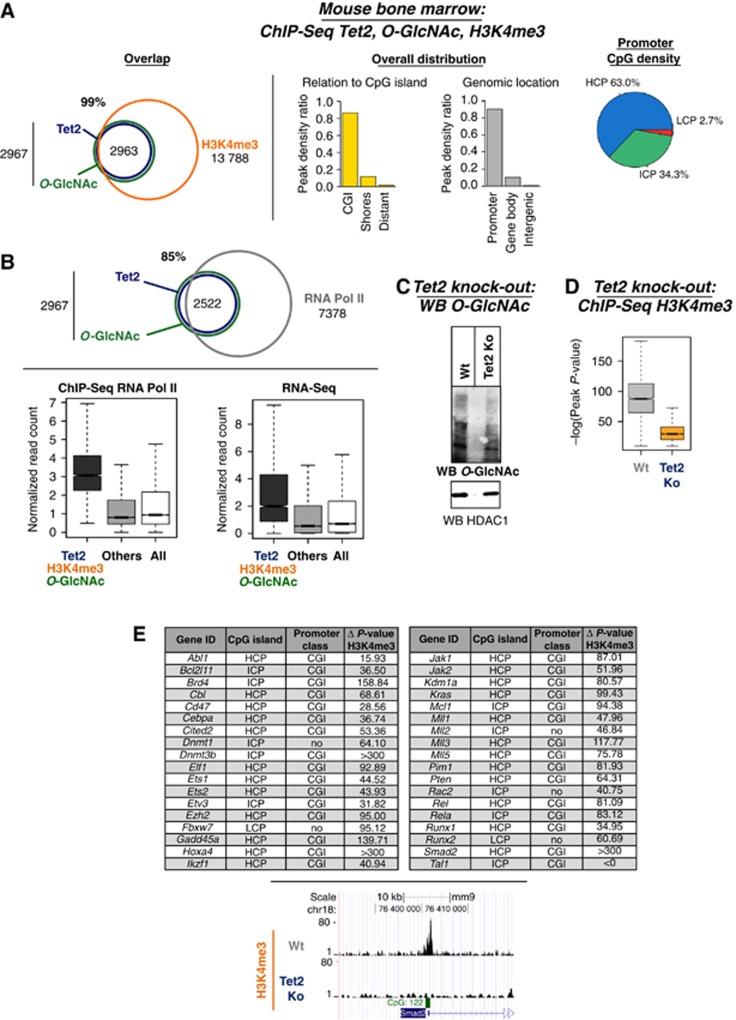
We then endeavoured to extend our findings to mouse bone marrow tissue, a model where Tet2 has been implicated in haematopoiesis and Tet2 mutations have been identified and linked to myeloid neoplasms (Langemeijer et al, 2009; Moran-Crusio et al, 2011; Quivoron et al, 2011). For this, we started by performing ChIP-Seq for Tet2, H3K4me3, and O-GlcNAc. First, ChIP-Seq analysis with an anti-Tet2 antibody in bone marrow tissue revealed genome-wide binding profile of endogenous Tet2 mostly at CpG-rich regions and TSSs (Supplementary Figure 13). This binding profile is similar to the genomic distribution observed for HT-TET2 in HEK293T cell lines (see Supplementary Figures 9B and 14). Next, we examined the genome-wide overlap between O-GlcNAc and H3K4me3 with Tet2. As shown in Figure 4A and Supplementary Table 3, a significant degree of overlap was observed between Tet2 and both O-GlcNac and H3K4me3 modifications, again predominately at CpG-rich promoters. Further, ChIP-Seq for RNA Pol II as well as RNA-Seq in mouse bone marrow indicated that the Tet2/O-GlcNAc/H3K4me3 co-bound targets were enriched at many active genes (Figure 4B). To determine the specific dependency of GlcNAcylation and H3K4me3 upon the Tet proteins in mouse bone marrow, experiments were then performed in a Tet2 knockout mouse (Moran-Crusio et al, 2011). Supporting the data that loss of human TET2/3 results in decreased OGT activity and global GlcNAcylation (Figure 1F), we observe in the Tet2 knockout mouse, a correlating decrease in global levels of O-GlcNAc (Figure 4C) as determined by a western blot using an anti-O-GlcNAc antibody. ChIP-Seq for H3K4me3 in the Tet2 knockout mouse bone marrow tissue, reveals a significant reduction in H3K4me3 (Figure 4D) (mirroring also the decrease observed in TET2 knockdown HEK293T cells; Figure 2E). Strikingly, it is noteworthy that among targets which showed decrease in H3K4me3 in a Tet2 knockout mouse, we found several key regulators of haematopoiesis, including the Cebpa, Tal1, Runx1, and several Mll genes (Figure 4E). Taken together, in this biologically relevant Tet system, we find genomic co-localization of Tet2, O-GlcNAc, and H3K4me3 on transcriptional active genes and most importantly, show an essential implication of Tet2 for both OGT activity and H3K4me3, notably on genomic targets regulating haematopoiesis.
Figure 4.
Tet2 knockout mouse tissue shows that Tet2 is needed for global GlcNAcylation and H3K4me3 at target promoters. (A) Genome-wide co-localization of endogenous Tet2 with O-GlcNAc and H3K4me3 at promoters and CpG-rich regions. Venn diagrams are shown (P-value overlap<10−10) as well as the indicated genome-wide distribution. (B) Tet2, O-GlcNAc, and H3K4me3 are enriched at many active genes, mirroring the presence of RNA Pol II. Upper panel: Venn diagram showing the overlap of Tet2 and O-GlcNAc with RNA Pol II (P-value overlap <10−10); lower panel: Box plots showing the reads density at targets and non-targets (others) for ChIP-Seq RNA Pol II or RNA-Seq in mouse bone marrow. (C) Global decrease in GlcNAcylation is observed in Tet2 knockout mouse bone marrow. Mouse bone marrow tissues with or without a Tet2 knockout were analysed by western blot for O-GlcNAc levels using an anti-O-GlcNAc antibody. HDAC1 is shown as loading control. (D) ChIP-Seq for H3K4me3 in Tet2 knockout mouse tissues shows reduced global H3K4me3 at target promoters. Overall impact on H3K4me3 peak significance (−log(P-value) of the peaks) between wild-type and Tet2 knockout bone marrow is shown. (E) Table showing key haematopoietic genes with specifically reduced H3K4me3 in Tet2 knockout as compared to wild type. The location at CpG islands and the promoter class for each is listed. Lower part: example of H3K4me3 ChIP-Seq profiles (UCSC tracks) in wild type versus Tet2 knockout.
Discussion
Here, we present data showing an unrecognized link between DNA modifying enzymes, TETs, a master cellular sensor protein, OGT, and a histone modifying complex, SET1/COMPASS. These new findings combined with our initial goal to further understanding of TET2 and TET3 proteins, have allowed us to propose a cohesive and hierarchical model of interactions and to determine the cascade of their respective activities leading to H3K4me3 and transcriptional activation (Figure 5). The first sequence of events is the formation of TET2 or TET3–OGT interaction, which promotes GlcNAcylation by OGT on numerous proteins, including HCF1. HCF1 has been shown to be required for recruitment of SET1/COMPASS and H3K4me3 (Narayanan et al, 2007; Tyagi and Herr, 2009; Liu et al, 2010) and our data further elucidate its role as we show a GlcNAcylated HCF1 is important for the integrity of the SET1/COMPASS complex. Lastly, both TET proteins and OGT activity favour binding of SETD1A to chromatin, an event necessary for histone H3K4me3 and subsequent transcriptional activation.
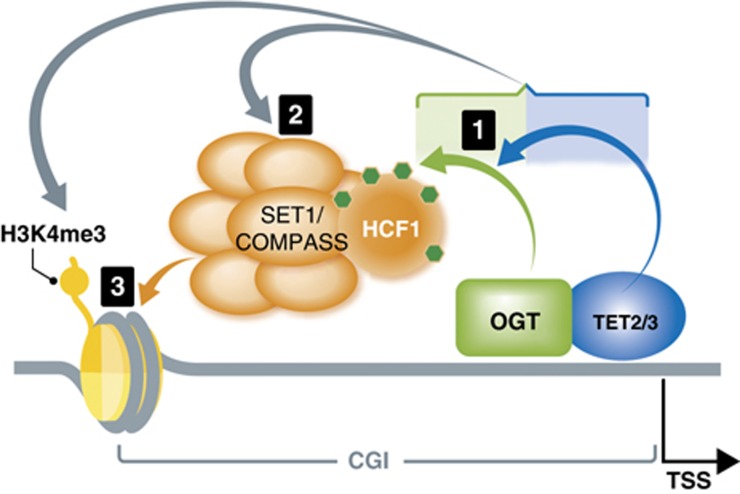
Figure 5.
Model connecting DNA modifying enzymes, TETs, a master cellular sensor protein, OGT, and a histone modifying complex, SET1/COMPASS. Based on our findings, a hierarchical model of the involved proteins, with the cascade of their respective activities, can be envisaged as followed: (1) The first sequence of events in the cascade is the formation of TET2/3–OGT interaction, which promotes OGT GlcNAcylation on numerous proteins, including HCF1; (2) In a TET-dependent manner, a GlcNAcylated HCF1 is important for the formation of the SET1/COMPASS; (3) In the last step, both TET proteins and OGT activity favour binding of SETD1A to chromatin, an event necessary for histone H3K4me3 and subsequent transcriptional activation.
As TET proteins have the potential to control DNA methylation fidelity by removing spurious or undesirable DNA methylation (Liu et al, 2010; Williams et al, 2011), the above model is likely not the only means these proteins utilize to influence H3K4me3. We hypothesize that TET proteins, in general, function as active guardians in promotion of transcriptional activation, which is supported by our data and previously published data of TET1 genomic localization primarily at regions high in H3K4me3 (Williams et al, 2011; Wu et al, 2011). In addition to their ability to reduce DNA methylation, the TET proteins might serve as scaffolding proteins beyond interaction with OGT as evidenced by the finding of several other interactors, and then recruit proteins required to establish a high-H3K4me3 chromatin environment. The TET–OGT interaction for example may be more broadly utilized to direct several epigenetic complexes to target sites or promote active transcription by GlcNAcylating not only HCF1, but also local histones or transcription factors at specific genomic localizations. Furthermore, similar to the dual function of TET1 in transcriptional regulation (Williams et al, 2011; Wu et al, 2011) our data suggest that TET3 might also provide a platform for repressive epigenetic complexes as it was shown also to interact with SIN3A. The TET proteins are thus more versatile and multifaceted than initially anticipated, performing important non-catalytic functions in addition to their known role of hydroxymethylation.
In addition to suggesting a broader role for the TET proteins, our study revealing a direct physical link between OGT and TET2/3 proteins provides new insight into the regulation and function of OGT in the cell. Although its connection with epigenetic regulatory events is by no means clear, new data are quickly emerging. In Drosophila, for example, OGT is reported to play a role in Polycomb function (Gambetta et al, 2009), and in human cells, O-GlcNAc modification on histone H2B is required for subsequent K120 monoubiquitination (Fujiki et al, 2011). The interaction evidenced here between TET2/3 and OGT may shed light on the link between the OGT glycosyltransferase and epigenetics and may add to the role of OGT observed in the regulation of MLL5 (Fujiki et al, 2009). Our study reveals an unforeseen genome-wide crosstalk between OGT and TET2/3 and a novel mode of regulation of OGT-directed O-GlcNAcylation. The precise mechanism of how TET impacts OGT activity is unclear and several possibilities exist (e.g., by other proteins associated with TET–OGT or by post-translational modifications). Our data allow however for a model wherein TET2 and TET3 might function as fine-tuning GlcNAcylation effectors, not only on epigenetic complexes such as SET1/COMPASS, but also other transcriptional regulation targets whose function is modulated by this modification.
In conclusion, our results reveal a relationship between TET2/3 and OGT with their direct interaction and influence on the H3K4 SET1/COMPASS complex shedding new light on the modes of action of TET proteins, impact upon chromatin biology, regulation of OGT activity, and the epigenetic mechanisms in which they participate.
Materials and methods
HT mammalian pulldown assay
HEK293T cells (12 × 106 cells) were plated in a 150-mm dish and grown to 70–80% confluency (∼18 h). The cells were then transfected with 30 μg of plasmid DNA using FuGENE HD Transfection Reagent (Promega) for 24 h, according to manufacturer’s protocol. Cells expressing HT-fusion proteins or HT-Ctrl were incubated in mammalian lysis buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, and 0.1% sodium deoxycholate) supplemented with Protease Inhibitor cocktail (Promega) and RQ1 RNase-Free DNase (Promega) for 10 min on ice. Lysate was then homogenized with a syringe and centrifuged at 14 000 g for 5 min to pellet cellular debris. Clarified lysate was incubated with HaloLink Resin (Promega) that had been pre-equilibrated in resin wash buffer (TBS and 0.05% IGEPAL CA-640; Sigma) for 15 min at 22°C with rotation. Resin was then washed five times with wash buffer after the initial binding of complexes to the resin and this, to remove non-specific interactions, and protein interactors were eluted with SDS elution buffer (50 mM Tris–HCl, pH 7.5, and 1% SDS). This number of washes reveals minimal non-specific interactions retained in control samples as determined by silver staining (Figure 1A). Such approach show quantitatively significantly lower levels of background as compared to standard Flag purifications using this affinity purification and washing procedure (Daniels et al, 2012). Affinity purified complexes were then analysed by nano LC/MS/MS (MSBioworks) and western blotting.
Chromatin immunoprecipitation
HEK293T or bone marrow cells were crosslinked 10 min at room temperature with 1% formaldehyde, then the reaction was quenched by addition of 0.125 M glycine and washed twice with 1 × cold PBS. ChIP experiments were performed according to the Transcription ChIP kit (Diagenode) protocol. Sonication was performed with bioruptor (Diagenode) in cold water+ice with the following settings: 2 cycles of 10 min, sonication strength set on high, with intervals of 30 s ON/OFF. 2 μg of mouse monoclonal antibody for H3K4me3 (ab1012; Abcam), 6 μg of mouse monoclonal antibody for O-linked N-acetylglucosamine (ab2739; Abcam), 3 μg of rabbit polyclonal antibody for Tet2 (sc-136926; Santa Cruz), 5 μg of rabbit polyclonal for HCF1 (A301-399A-1; Bethyl Lab), or the respective amount of control antibody was incubated with chromatin overnight at 4°C. After extensive washing steps, ChIPed DNA was eluted and de-crosslinked overnight at 65°C, then purified using QIAquick PCR purification kit (Qiagen). 3 μl of enriched fragmented DNA or 3 μl of input, supplemented with 0.5 μM of primers and SYBR Green master mix was subjected to 40 cycles of PCR using LightCycler 480 II (Roche). Percentage of input recovered after immunoprecipitation was calculated using the ΔCt formula: (2−(Ct IP—Ct Input)) × 100. Primer sequences are described in Supplementary Table 1.
Supplementary Material
Acknowledgments
RD, MD and MV were supported by the Belgian FNRS. FF is an FNRS ‘Senior Research Associate’. BD is an ‘FNRS Aspirant’. FF’s laboratory was funded by grants from the FNRS and Télévie, the ‘Interuniversity Attraction Poles’ (IAP Phase VII no P7/03) and by the ‘Action de Recherche Concertée’ (AUWB-2010-2015 ULB-No 7). MKS, JM, NM, MU, and DLD are supported by Promega Corporation. We wish to thank Kazusa DNA Research Institute for providing vectors, MSBioworks for all mass spectrometry analysis, Dr Martin Rosenberg for thoughtful advice and continued support, and Prof. David Jeruzalmi for critical reading of the manuscript.
Author contributions: RD, BD, and MKS designed experiments, performed research, and interpreted data. Protein partner purifications were done and analysed by MKS, JM, NM, MU, and DLD. Confirmations of interaction were done by RD and MKS. BD and MV performed the Dot Blot experiments. Cloning, RNAi experiments, and expression studies were done by RD, PP and EC. RD performed OGT enzymatic assays. ChIP was performed by BD and MAD. MeDIP and hMeDIP were performed by BD. ChIP-Seq experiments were performed by BD and bioinformatic analyses were conducted by MD. AHS, RLL, ES, TM, and OB provided mouse bone marrow tissues. BRET experiments were done by MKS. DLD and FF designed experiments, interpreted data, and directed the study. RD, BD, MD, and MKS prepared the figures. DLD and FF wrote the manuscript.
Footnotes
Promega Corporation is the commercial owner by assignment of patents of the HaloTag technology and its applications.
References
- Bonasio R, Tu S, Reinberg D (2010) Molecular signals of epigenetic states. Science 330: 612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capotosti F, Guernier S, Lammers F, Waridel P, Cai Y, Jin J, Conaway JW, Conaway RC, Herr W (2011) O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell 144: 376–388 [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y (2009) Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 10: 295–304 [DOI] [PubMed] [Google Scholar]
- Daniels DL, Mendez J, Mosley AL, Ramisetty SR, Murphy N, Benink H, Wood KV, Urh M, Washburn MP (2012) Examining the complexity of human RNA polymerase complexes using HaloTag technology coupled to label free quantitative proteomics. J Proteome Res 11: 564–575 [DOI] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W (2011) Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473: 398–402 [DOI] [PubMed] [Google Scholar]
- Fujiki R, Chikanishi T, Hashiba W, Ito H, Takada I, Roeder RG, Kitagawa H, Kato S (2009) GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature 459: 455–459 [DOI] [PubMed] [Google Scholar]
- Fujiki R, Hashiba W, Sekine H, Yokoyama A, Chikanishi T, Ito S, Imai Y, Kim J, He HH, Igarashi K, Kanno J, Ohtake F, Kitagawa H, Roeder RG, Brown M, Kato S (2011) GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature 480: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetta MC, Oktaba K, Muller J (2009) Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science 325: 93–96 [DOI] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, Iqbal K, Shi YG, Deng Z, Szabo PE, Pfeifer GP, Li J, Xu GL (2011) The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477: 606–610 [DOI] [PubMed] [Google Scholar]
- Hart GW, Housley MP, Slawson C (2007) Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446: 1017–1022 [DOI] [PubMed] [Google Scholar]
- Hartzell DD, Trinklein ND, Mendez J, Murphy N, Aldred SF, Wood K, Urh M (2009) A functional analysis of the CREB signaling pathway using HaloCHIP-chip and high throughput reporter assays. BMC Genomics 10: 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y (2010) Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324: 929–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, Kamping EJ, Verhoef GE, Verburgh E, Hagemeijer A, Vandenberghe P, de Witte T, van der Reijden BA, Jansen JH (2009) Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet 41: 838–842 [DOI] [PubMed] [Google Scholar]
- Lee JS, Smith E, Shilatifard A (2010) The language of histone crosstalk. Cell 142: 682–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, Desai A, Dorrestein PC, Glass CK, Rosenfeld MG (2010) PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 466: 508–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV (2008) HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol 3: 373–382 [DOI] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, Perna F, Pandey S, Madzo J, Song C, Dai Q, He C, Ibrahim S, Beran M, Zavadil J, Nimer SD et al. (2011) Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A, Ruyechan WT, Kristie TM (2007) The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc Natl Acad Sci USA 104: 10835–10840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndlovu MN, Denis H, Fuks F (2011) Exposing the DNA methylome iceberg. Trends Biochem Sci 36: 381–387 [DOI] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, Tahiliani M, Daley GQ, Liu XS, Ecker JR, Milos PM, Agarwal S, Rao A (2011) Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473: 394–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivoron C, Couronne L, Della VV, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, Godley L, Opolon P, Tilly H, Solary E, Duffourd Y, Dessen P, Merle-Beral H, Nguyen-Khac F, Fontenay M, Vainchenker W et al. (2011) TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 20: 25–38 [DOI] [PubMed] [Google Scholar]
- Slawson C, Hart GW (2011) O-GlcNAc signalling: implications for cancer cell biology. Nat Rev Cancer 11: 678–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A (2008) DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 9: 465–476 [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S, Herr W (2009) E2F1 mediates DNA damage and apoptosis through HCF-1 and the MLL family of histone methyltransferases. EMBO J 28: 3185–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K (2011) TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473: 343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y (2011) Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473: 389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W (2003) Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev 17: 896–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.