Abstract
To understand the molecular mechanism of ion permeation in pentameric ligand-gated ion channels (pLGIC), we solved the structure of an open form of GLIC, a prokaryotic pLGIC, at 2.4 Å. Anomalous diffraction data were used to place bound anions and cations. This reveals ordered water molecules at the level of two rings of hydroxylated residues (named Ser6′ and Thr2′) that contribute to the ion selectivity filter. Two water pentagons are observed, a self-stabilized ice-like water pentagon and a second wider water pentagon, with one sodium ion between them. Single-channel electrophysiology shows that the side-chain hydroxyl of Ser6′ is crucial for ion translocation. Simulations and electrostatics calculations complemented the description of hydration in the pore and suggest that the water pentagons observed in the crystal are important for the ion to cross hydrophobic constriction barriers. Simulations that pull a cation through the pore reveal that residue Ser6′ actively contributes to ion translocation by reorienting its side chain when the ion is going through the pore. Generalization of these findings to the pLGIC family is proposed.
Keywords: crystallography, electrophysiology, ion channels, molecular dynamics, permeation
Introduction
The molecular understanding of ion permeation is a central issue in the study of ion channels. Structural details at atomic resolution are often difficult to obtain for such transmembrane proteins. A large number of structural studies has been undertaken for the tetrameric potassium channel KcsA, complemented by computational approaches to simulate the dynamic aspects of permeation following the pioneering work of MacKinnon, Roux and others (Doyle et al, 1998; Zhou et al, 2001; Noskov et al, 2004; Kim and Allen, 2011). The pentameric ligand-gated ion channels (pLGICs), on the other hand, constitute a large family of ionotropic receptors for which little is known about the molecular mechanisms underpinning permeation and charge selectivity. pLGICs are ubiquitously represented in the animal kingdom and they all share a conserved molecular architecture based upon a five-fold symmetry (Corringer et al, 2012) with ions permeating through a central pore lying along the C5 symmetry axis. The pore of these ligand-gated ion channels is closed in their resting state and opens upon the binding of an agonist. In vertebrates, the family encompasses cation-selective 5HT3-type serotonin receptors and nicotinic acetylcholine receptors (nAChRs) on one hand, and anion-selective GABA and glycine receptors on the other hand (Corringer et al, 2012).
Recently, prokaryotic homologues to pLGICs have been found in the genomes of certain bacterial species (Tasneem et al, 2005; Bocquet et al, 2007). The structures of two distinct prokaryotic homologues have been solved by X-ray crystallography and revealed important insights on the structures of pLGICs: one from the plant pathogen Erwinia chrysanthemi (ELIC) at 3.3 Å resolution (Hilf and Dutzler, 2008) and another one from the cyanobacterium Gloeobacter violaceus (GLIC) at 2.9 Å (3EAM) and 3.1 Å (3EHZ) resolution (Bocquet et al, 2009; Hilf and Dutzler, 2009). Both structures display identical homo-pentameric organization with a highly conserved extracellular domain (ECD) folded as a β-sandwich and a transmembrane domain (TMD) consisting of four α-helices named M1 to M4. Interestingly, these structures differ in their conformations: while the ELIC structure shows a closed non-conductive conformation, the GLIC structure shows an open conductive conformation.
After determination of the first structure of an integral pLGIC at 4 Å using electron microscopy (Unwin, 2005), several groups have used computational methods to describe the permeation and charge selectivity of this family of ion channels. Early studies on the Torpedo nAChR revealed an energy barrier in the middle of the pore (9′ and 13′ level in prime notation counting from the N-terminus of M2) with a barrier height reaching ∼6 kcal/mol suggesting a hydrophobic gating model (Amiri et al, 2005; Beckstein and Sansom, 2006). Computational studies using a homology model of the human α7-nAChR (built from the Torpedo one), confirmed the presence of the 9′ and 13′ barrier, but with a lower energy cost (∼3 kcal/mol), compatible with a conductive state (Cheng et al, 2007; Ivanov et al, 2007). Clearly, higher quality structural data are required to investigate permeation in detail, the lack of which having long been an obstacle for deciphering the underlying mechanism of this particular family of ion channels. The crystal structures of the GLIC proton-gated ion channel were solved at about 3 Å resolution (Bocquet et al, 2009; Hilf and Dutzler, 2009) in an open form of the channel and provided an improved view of the pore structure and led to numerous computational permeation studies of GLIC (Ivanov et al, 2007; Cheng et al, 2010; Zhu and Hummer, 2010; Fritsch et al, 2011). The pore of GLIC is bordered by the M2 helices, which successively expose seven rings of residues to the lumen, from the top to the bottom, one ring of Glu19′ (Glu243), three rings of hydrophobic residues Ile16′ (Ile240), Ala13′ (Ala237), Ile9′ (Ile233), two rings of polar residues Ser6′ (Ser230), Thr2′ (Thr226) and finally Glu-2′ (Glu222). The pore is funnel-shaped, with a large upper diameter of 11 Å and a constriction of 5 Å diameter at the 2′ and −2′ level. Extensive site-directed mutagenesis in nAChR points to this constriction as constituting the charge selectivity filter (Corringer et al, 1999; Keramidas et al, 2004). Computational studies on GLIC revealed that the hydrophobic residues at the 9′ and 13′ levels act as the main permeation barrier in the TMD of GLIC (Fritsch et al, 2011). Importantly, three independent potential of mean-force studies on GLIC found a permeation barrier (comprised between 2 and 4 kcal/mol) for the transport of Na+ ions that is consistent with a physiologically open state of the channel (Cheng et al, 2010; Song and Corry, 2010; Zhu and Hummer, 2010). This finding is reinforced by Brownian Dynamics and Smoluchowsky model-diffusion studies that reproduced the experimental conductance of GLIC (Zhu and Hummer, 2012). The fact that the GLIC channel structure depicts an open conformation has subsequently been confirmed by the structure of an eukaryotic glutamate chloride channel (GluCl) solved at 3.3 Å in the presence of glutamate (the agonist) and ivermectin (an activator), which displays exactly the same arrangement of the pore helices (Hibbs and Gouaux, 2011). The GLIC structure thus provides a significant advance in the possibility to study ion permeation in pLGICs.
Understanding the molecular mechanisms of ion permeation in pLGICs requires documenting the interactions between ions, waters and the side chains of the M2 pore-lining residues that are thought to be crucial for ion transport. This kind of description is difficult to derive from existing pLGIC structures given the limited structural resolution achieved previously by X-ray crystallography. The 2.4 Å resolution GLIC structure presented here (hereafter referred to as GLIC_2.4) yields a more precise assignment of ions, water molecules and detergents within GLIC’s pore, when combined with the systematic use of anomalous diffraction data collected on crystals soaked with Br−, Cs+, Rb+ and Se-DDM (Dhakshnamoorthy et al, 2010) (Table I). In particular, we identify a self-stabilized tight water pentagon at the level of residue Ser6'. This position was mutated into Gly, Val and Thr: all mutants were expressed but produce no currents in whole-cell electrophysiology, while heteropentamers combining wild-type (wt) and Ser6’Gly subunits had a reduced but measurable single-channel conductance. Based on comparison to extensive molecular dynamics simulations (a total of 2 μs simulated) combined with electrostatic calculations, this study reveals novel insights on the permeation mechanism in pLGIC receptors.
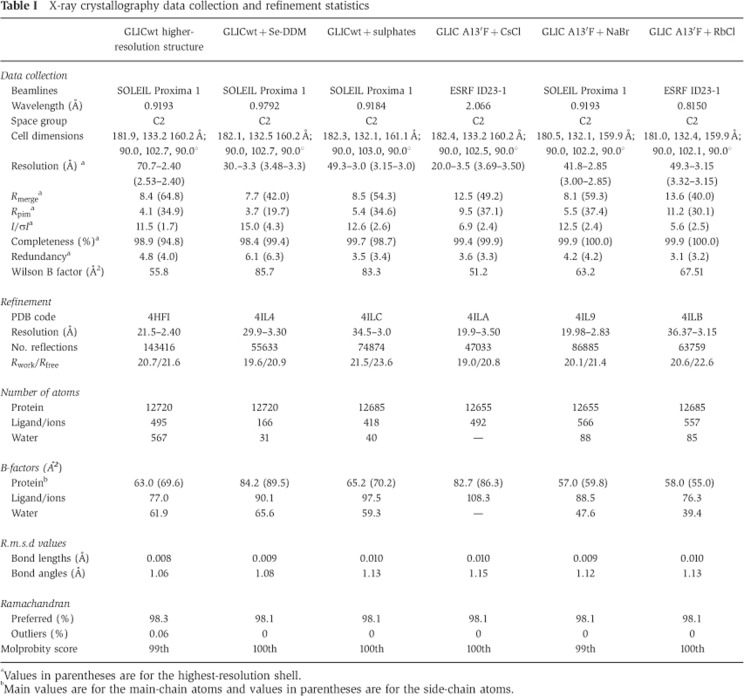
Table 1. X-ray crystallography data collection and refinement statistics.
aValues in parentheses are for the highest-resolution shell.
bMain values are for the main-chain atoms and values in parentheses are for the side-chain atoms.
Results
The 2.4 Å resolution GLIC structure
The GLIC_2.4 structure was obtained from crystals grown in conditions similar to the ones that were used previously, but the crystals were optimized by micro-seeding from a suspension of crushed crystals (Supplementary Information). The refined model shows a more complete description of protein-bound water molecules and presents significantly better refinement statistics (Table I). GLIC_2.4 is very similar to the previously published 3EAM structure and confirms some differences with the alternative 3EHZ structure that can be resolved by a re-refinement of 3EHZ (Supplementary Information). It also suggests that the cis conformation of the conserved proline located at the apex of the cys-loop is a universal feature of pLGICs (Corringer et al, 2012) (Supplementary Figure S1).
When the initial structure of GLIC was solved at 2.9 Å resolution, residual electron density was observed in the pore and interpreted as a bundle of six dodecyl-maltoside molecules (DDM) with the maltoside moiety pointing inward for the central detergent and outward for the five other detergents (Bocquet et al, 2009). However, the GLIC_2.4 structure reveals that the locus where the polar head of the central detergent was previously assigned is in fact occupied by a tight self-stabilized water pentagon (Figures 1 and 2). In order to confirm the presence of detergents in the upper half of the pore, GLIC was co-crystallized in selenium-derived DDM and single-wavelength anomalous data were collected at the peak value of selenium. This experiment unequivocally confirmed the presence of a detergent bundle in the pore of GLIC with the maltoside moieties pointing outward (Figure 3). The density for the aliphatic part of the central DDM is less well-defined compared with the five peripheral ones, possibly explaining the lack of anomalous density for the corresponding selenium atom by a higher flexibility and exposure to the solvent for this part of the central detergent (Figure 3).
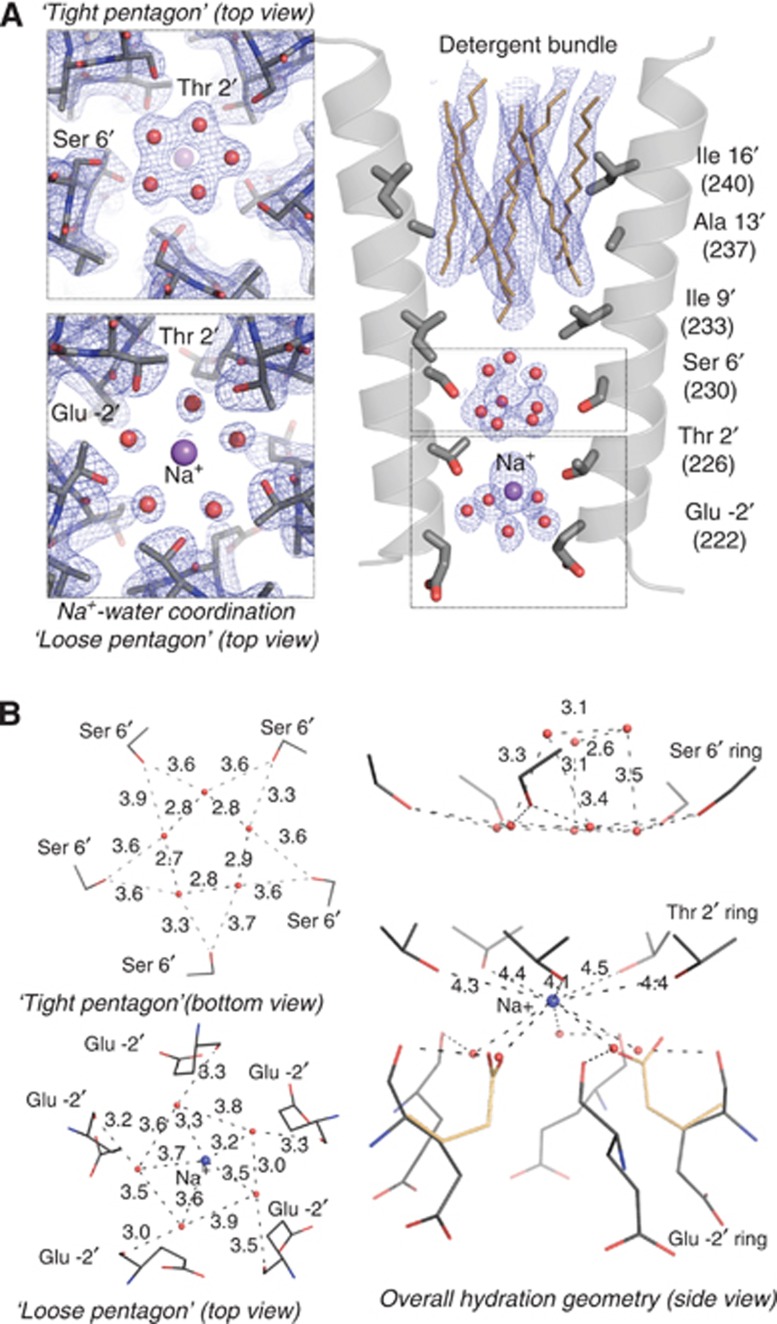
Figure 1.
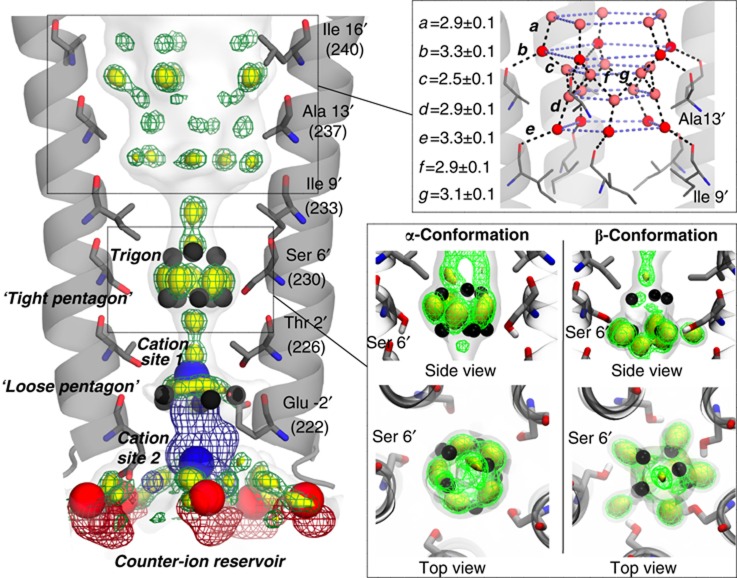
Structurally ordered water molecules in the pore of GLIC. (A) Enlarged representation of the pore with the M2 helices shown as a cartoon and the side chains of the pore-lining residues shown as sticks. A well resolved self-stabilized ‘tight pentagon’ of water molecules is present at the level of Ser6′ (left top). A Na+ atom and five bound water molecules are observed at the intracellular end of the pore (left bottom). Detergents are shown as sticks, waters and Na+ atoms as spheres. The blue mesh is the maximum likelihood 2mFo–DFc map contoured at a level of 1.5σ. For clarity, the electron density surrounding the Na+ has been removed from the left bottom panel. (B) Interaction network between the water molecules, the Na+ and the pore-lining M2 residues. The alternative conformation that is adopted by Glu-2′ is also represented (yellow).
Figure 2.
Residual electron density in the pore for a set of deposited GLIC structures at different resolutions (A, C, D, E and F) and for GluCl (B). The difference Fourrier (Fo–Fc) omit map (contoured at 3σ) as calculated from a model where detergents, Na+ and water molecules were not included during refinement. As the resolution decreases, the density around the water molecules no longer appears like individual spheres but rather like a fused blob. While the tight pentagon is systematically observed in the electron density, the second layer of water molecules located on top of it is unequally distributed among these structures, especially in the GLIC 3UUB entry, where a complete water pentagon can be built on top of the tight pentagon (blue). The Na+ ion is systematically observed in the residual electron density but with a corresponding peak of variable intensity and slightly different position. These differences might be explained by slight variations of the conditions used during crystal freezing. Superimposing the M2-helices in the GLIC (grey) and GluCl (pink) structures reveal a very strong structure conservation in this region of the pore (B). When the GluCl structure was solved, the authors observed residual electron density in the Fo–Fc map that was built tentatively as a chloride ion. We hypothesize that this residual density might as well be explained by several water molecules, presumably ordered in a similar pentagonal fashion as we observed in GLIC. The blue mesh is the Fo–Fc map (contoured at 2.5σ) obtained when the chloride ion built in the structure of GluCl was omitted during refinement. Iodide ions observed in GluCl (3RIA) are shown as purple circles.
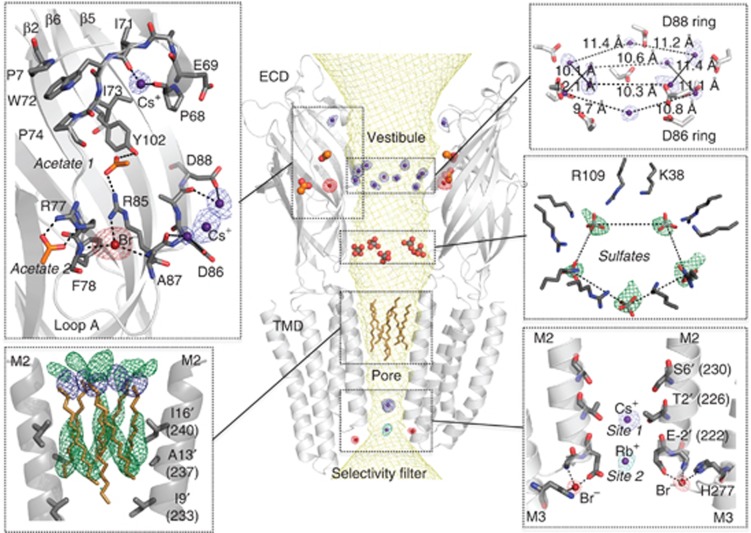
Figure 3.
Cation, anion and detergent binding sites in the pore of GLIC. Central view: Cartoon representation of the GLIC structure viewed from the side with the solvent-accessible surface of the channel shown as a mesh (yellow). The top and bottom panels represent enlarged views of the channel vestibule and of the transmembrane pore showing the interactions between the protein and the solvent molecules. Acetate (orange) and sulphate (grey) molecules are shown as ball and sticks. (Middle-right panel) The green mesh is the Fo–Fc difference map (contoured at 3σ) calculated when the sulphates were omitted during refinement. (Bottom-left panel) Orientation of the dodecyl-β-D-seleno-maltoside (Se-DDM). The green mesh is the Fo–Fc difference map (contoured at 3σ) calculated when detergents were omitted during refinement and the blue mesh is the anomalous map for selenium contoured at 4σ. (Bottom right) Ion binding sites at the selectivity filter. The anomalous maps calculated for Br− (red), Cs+ (blue) or Rb+ (cyan) are superimposed and shown as a mesh (contoured at a level of 4σ) with the corresponding ions shown as spheres.
Pentagonal geometry of hydration in the pore
The tightly ordered water molecules of the self-stabilized pentagon were already present in the 3EAM lower-resolution structure of GLIC but difficult to identify as they did not appear as individual spheres in the density but rather as a continuous crown (Figure 2). These five water molecules are coplanar with the γ-O atoms of Ser6′ (Figure 1). The mean distance between the two neighbouring water molecules in the tight pentagon is 2.76±0.06Å and is very similar to the 2.79±0.15 Å average distance that is observed for water pentagons in the hydration shell of very high-resolution protein structures (Teeter, 1984; Teeter et al, 2001; Lee and Kim, 2009) or ice (Jeffrey, 1997). The mean distance between a water molecule in the pentagon and the γ-O atom of the closest Ser6′ residue is of 3.57±0.12 Å, suggesting that the water pentagon interacts more strongly internally than with the ring of Ser6′ residues; in the rest of the discussion it will be referred to as the ‘self-stabilized’ or ‘tight’ water pentagon. In all structures of GLIC that we have solved (about 25), this pentagon is present (Figure 2).
In addition, three water molecules are observed ∼3 Å above the ‘tight’ water pentagon (Figure 1). Comparison of the different GLIC structures deposited in the PDB reveals that these ordered water molecules are systematically present but with some variability in their distribution along the z-axis and their exact number. In particular, it is possible to build a full pentagon of water molecules at this altitude in GLIC 3UUB. These small differences might be due to inherent variability in the kinetics of the crystal freezing process (Halle, 2004) (Figure 2).
On the cytoplasmic side of the pore, the strongest peak in the electron density (7σ) can be accounted for by a Na+ ion that is located on the five-fold symmetry axis at the level of the Thr2′ ring (Figures 1 and 2). The presence of a Na+ ion at this location has been confirmed by substituting Na+ with cations that display anomalous dispersion properties (see below). The ‘tight’ pentagon is too far apart from the Na+ ion (≈6.5 Å) to be considered part of its (upper) hydration shell. However, another water pentagon contributes directly to the hydration, underneath the Na+ ion (Figure 1A). These water molecules interact with both the Na+ ion (d=3.45±0.21 Å) and the main-chain carbonyl oxygen atom of Glu-2′ (d=3.25±0.18 Å) rather than with each other (d=3.67±0.23 Å) (Figure 1B). In the GLIC_3EHZ structure, the side chains of Glu-2′ assume an up-conformation that folds back into the pore at the level of this pentagon whereas in the GLIC_3EAM structure these side chains all point down toward the cytoplasm. In the GLIC_2.4 structure, the Glu-2′ carboxylate groups display significantly higher B-factors than the other residues of M2, suggesting that these residues are flexible and may adopt alternative conformations. The ε-O carboxylate atoms of the Glu-2′ residues probably continuously exchange their positions with individual water molecules of the sodium ion-bound water pentagon; therefore, we refer to this water pentagon as the ‘loose pentagon’.
Characterization of ion binding sites in GLIC by anomalous X-ray diffraction
In order to fully characterize the functional implications of the organized water molecules observed in the pore and their role in ion permeation, it is necessary to identify the monovalent ion binding sites in the GLIC structure. To this aim, the protein was co-crystallized with Br−, Cs+, or Rb+ and X-ray data were collected at optimized wavelengths for each of these derivatives so as to maximize the anomalous signal (Figure 3). Peaks with an anomalous signal higher than 4σ were used to identify ion binding sites. Ions located in between crystallographic symmetry-related molecules were included in the refined model but are not discussed here. The identified ion binding sites are essentially clustered in two distinct regions: in the ECD around the vestibule edge of the channel and in the TMD near the cytoplasmic end of the pore. Hereafter, we will focus on the ion binding sites that are located close to the channel lumen and especially in the transmembrane pore (Figure 3). Nevertheless, the other ion binding sites are important in at least two respects: (i) they validate computational studies that actually predict them quite accurately (see below) and (ii) ultimately they may prove to be important for understanding the gating process, once the structure of the resting state is known. These binding sites are described and discussed in Supplementary Information.
In the pore, the anomalous signal of Br− indicates a strong monovalent anion binding site located at the cytoplasmic side, stabilized by the main-chain amino group of Tyr-3′, the Nε imidazole atom of His277 from M3 and a carboxyl oxygen of Glu-2′ that is likely partly protonated due to the acidic pH of the crystallization mother liquor. For monovalent cations, we confirm the existence of a strong anomalous signal for Cs+ inside the pore at the level of Thr2′ ring of residues (cation site 1), already seen by other authors (Hilf et al, 2010), exactly where a Na+ ion was observed in the GLIC_2.4 structure (Figure 1). In the GLIC structure co-crystallized with Rb+, two strong peaks of 5 and 19σ are, respectively, observed in the Fo–Fc electron density map at cation site 1 and at the level of the Glu-2′ rings of residues (cation site 2). As the anomalous signal for Rb+ is only observed at the level of cation site 2, a Rb+ was modelled at that site, while cation site 1 was assigned to a Na+ ion that is also present in the crystallization liquor and that was already observed in the GLIC_2.4 structure (Figure 3). Overall, these results suggest that permeating ions occupy at least two preferential positions in this region of the pore that is thought to be the selectivity filter of these channels (Figure 3).
As discussed above, the GLIC_2.4 structure brings more details about the hydration architecture in this specific region of the pore. While the ‘loose pentagon’ appears to be directly involved in the hydration shell of the Na+ ion when occupying site 1, the ‘tight pentagon’ and the trigon are too far apart to be considered part of its hydration shell. In order to further explore the roles of these water molecules and that of the Ser6′ ring of residues, which side chains are coplanar with the ‘tight water pentagon’, we constructed and tested three mutants: Ser6′Gly (S6′G), Ser6′Val (S6′V) and Ser6′Thr (S6′T).
Ser6′ substitution has a major effect on GLIC single-channel conductance
DNA coding for each mutant was either injected in the nucleus of Xenopus oocytes or transfected into Baby Hamster Kidney cells (BHK cell line). Immunofluorescence microscopy revealed that all proteins express at the oocyte (Figure 4B) and BHK cell surfaces (Supplementary Figure S2B). In addition, all mutants are shown to be expressed in Escherichia coli (Figure 4C) and to form stable pentamers as revealed by analysis on size-exclusion chromatography (data not shown).
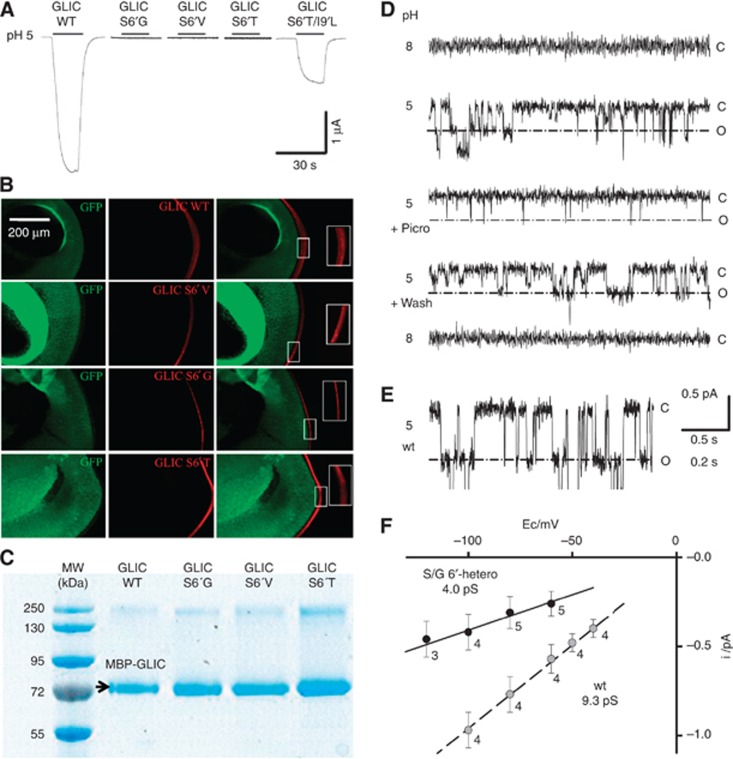
Figure 4.
Electrophysiology of Ser6′ mutants. (A) Voltage-clamp current traces recorded from GLIC wt and the S6′G, S6′T, S6′V and S6′T/I9′L variants expressed in Xenopus oocytes, showing the effect of switching extracellular pH from 8 to 5. (B) Immunofluorescence microscopy data showing that all GLIC variants express at the oocyte surface. (C) SDS denaturating gel experiment in reducing conditions showing that all GLIC variants express in E. coli. (D) Traces of currents recorded from an outside-out patch from BHK cells transfected with a mixture of cDNAs coding for GLIC S6′G and GLIC wt, in a 4 to 1 S6′G to wt DNA ratio (a=0.8). Currents recorded at extracellular pH 8 (upper trace), then pH 5, then pH 5 with 0.1 mM picrotoxinin, pH 5 after toxin wash-out, and pH 8, on the same patch. Holding potential −100 mV. The patch input resistance was 0.6 tΩ (closed level at pH 5). The current levels indicated right to the traces correspond to all channels closed (c), and to one channel open (o) for the prominent, large conductance events measured in F (4 pS), arising from S/G6′-heteropentamers. (E) Trace of current recorded from an outside-out patch from a cell expressing wt GLIC channels. Holding potential of −80 mV. The lower part of the recording has been skipped out from 1.4 pA under the closed channel current level, in order to emphasize the one open channel level for wt GLIC. (F) Plots of single-channel current versus holding potential values, for wt GLIC (grey circles) and for GLIC S/G6′-heteropentamers (black circles) corresponding to the open level emphasized in (D). Number of patches and error bars indicating standard deviations are shown for each voltage. Data were accumulated from four patches (wt), and from eight patches from cells with a=0.8 or a=0.94.
The three Ser6′ mutants were tested in oocytes under voltage-clamp. None of the oocytes tested produced GLIC current following changes in extracellular pH from 8 to 5 and 4 (n=9, 9 and 6 for S6′G, S6′V, and S6′T, respectively (Figure 4A)). The Ser 6′ mutants produced also no GLIC currents when tested in BHK cells (Supplementary Figure S2A).
To test whether the serine side chain determines the single-channel conductance, we generated heteropentamers of GLIC wt and S6′G mutant (S/G6′-heteropentamers) by co-injecting oocytes or co-transfecting BHK cells with mixtures of DNAs coding for wt and S6′G subunits, defined by the proportion (a) of mutated DNA versus total DNA. In oocytes, when the proportion of S6'G DNA increased, the amplitude of the GLIC currents decreased, the whole-cell current activation kinetics were slower and a shift of the proton's concentration—response curves was observed (Supplementary Figure S3). These data in oocytes strongly suggest that S/G6′-heteropentamers of GLIC are assembled, inserted in the membrane and functional with modified properties.
The properties of S/G6′-heteropentamers were further explored using analysis of single-channel currents recorded from BHK cells transfected with DNA containing a high proportion of S6'G versus total DNA (a=0.80 and a=0.94). In this type of experiment, a total of six types of S/G6′-heteropentamers can be a priori predicted, in four different stoichiometries. If the membrane protein synthesis, insertion and degradation systems were identical for wt and mutated subunits, a proportion a=0.8 or higher of mutant versus total DNA would result in a large majority of heteropentamers with 3, 4 or 5 mutated subunits. Consistently, no current events corresponding to those seen with wt GLIC channels could be detected, but single-channel currents with amplitudes smaller than those expected for wt GLIC were observed (Figure 4D–F). Actually, several types of current events were detected. The large current events were well resolved (Figure 4D) and corresponded to a single-channel conductance of 4pS for currents recorded with transmembrane voltages between −60 and −100 mV (Figure 4F). These 4pS events were reversibly activated by switching external pH from 8 to 5 (a=0.8) or 4.5 (a=0.94) and reversibly inhibited by picrotoxinin (0.1 mM) with the characteristic inhibition pattern (remaining events with very short open times) (Figure 4D, middle trace). These events are well accounted for by GLIC S/G6′-heteropentamers with a reduced, 4pS single-channel conductance. Smaller current events, with conductances near 2pS, were reversibly activated by lowering the external pH and reversibly inhibited by 0.1 mM picrotoxinin (Figure 4D) but were not fully characterized here due to their heterogeneity and low signal-to-noise ratio.
In conclusion, data obtained with proportions 0.80 and 0.94 of GLIC S6′G versus total DNA demonstrate that at least one type of GLIC S/G6′-heteropentamer is functional with a reduced single-channel conductance of 4pS, compared with 9.3pS for wt GLIC.
All-atom molecular dynamics simulations in a membrane environment
A 200-ns all-atom MD simulation was run on GLIC embedded in a DOPC membrane at 310 K in order to study the behaviour of ions and water molecules surrounding the protein and go beyond the averaged picture provided by the crystal structures. GLIC assumed a stable open conformation in the simulation, suggesting that the detergent bundle is not required to stabilize the open form of the channel, at least at this time scale. The stability is confirmed by the low structural drift of the protein over the second half of the simulation with root mean square deviations for Cα atoms of 1.64±0.10 Å, 1.88±0.14 Å, 1.05±0.06 Å, for the whole protein, the ECD and the TMD, respectively. Averaged-density maps were calculated for Na+, Cl− and water and compared with the experimentally observed locations (Figure 5A). Overall, the ion binding sites predicted by the MD simulation are in very good agreement with the various ions observed experimentally (Supplementary Figure S4B), giving us confidence on the use of the same simulation to study pore hydration.
Figure 5.
Ion atmosphere and water binding sites predicted by full-atom MD. Enlarged representation of the pore showing the averaged densities calculated over 200 ns for water molecules (green mesh and yellow surface contoured at increasing density starting from the bulk density shown as white transparent surface), Na+ (blue mesh) and Cl− (red mesh). For comparison, experimentally observed water molecules, anions and cations are shown as cpk spheres (coloured in black, red and blue, respectively). (Right bottom panel) Comparing the hydration profile observed when the side chain of Ser6′ adopts an α or a β conformation (see Supplementary Information for details). (Right top panel) Detailed view of the network of water molecules that covers the edges of the hydrophobic half of the channel (distances are measured in Å). Primary and secondary layers of water molecules are shown as red and pink spheres, respectively.
The simulation-derived mean water density calculated in the transmembrane pore showed peaks in the hydrophobic upper half region, where hydration cannot be studied by crystallography due to the presence of detergent molecules. In this region, water molecules are organized in five layers in a plane perpendicular to the five-fold symmetry axis (Figure 5). The first two layers of water molecules (hereafter referred to as primary layers), corresponding to the highest observed density of water, interact respectively with the main-chain carbonyl oxygen of the Ile9′ and Ala13′ rings of residues. These carbonyl oxygen atoms probably interact with water molecules through bifurcated hydrogen bonds as they already interact with the main-chain amide of residues at position n+4. Three layers of water molecules (hereafter referred to as secondary layers) were also observed that do not interact directly with the protein but do interact instead with the two primary layers of water molecules (Figure 5). This network of water molecules covers the edge of the hydrophobic half of the channel, thus rendering it polar.
In the lower half of the pore, the strongest averaged-density peaks are located at the level of the Ser6′ ring as expected. They are however slightly shifted by ∼ 1.4 Å upward on the z-axis and rotated in the xy plane by ∼30° compared with the ‘tight’ crystallographic pentagon, which could be reproduced by using MD conditions that resembled the crystallization conditions (presence of DDM, low temperature of 100 K and an appropriate model of ice-like water TIP4Pice (Abascal et al, 2005)). Moreover, it could be shown that Ser6′ side-chain orientation has a direct influence on the local water organization (Figure 5, for more details see Supplementary Information: Supplementary Methods and Supplementary Figures S5 and S6).
MD simulations of Ser6' mutants reveal dramatic changes in hydration of the pore
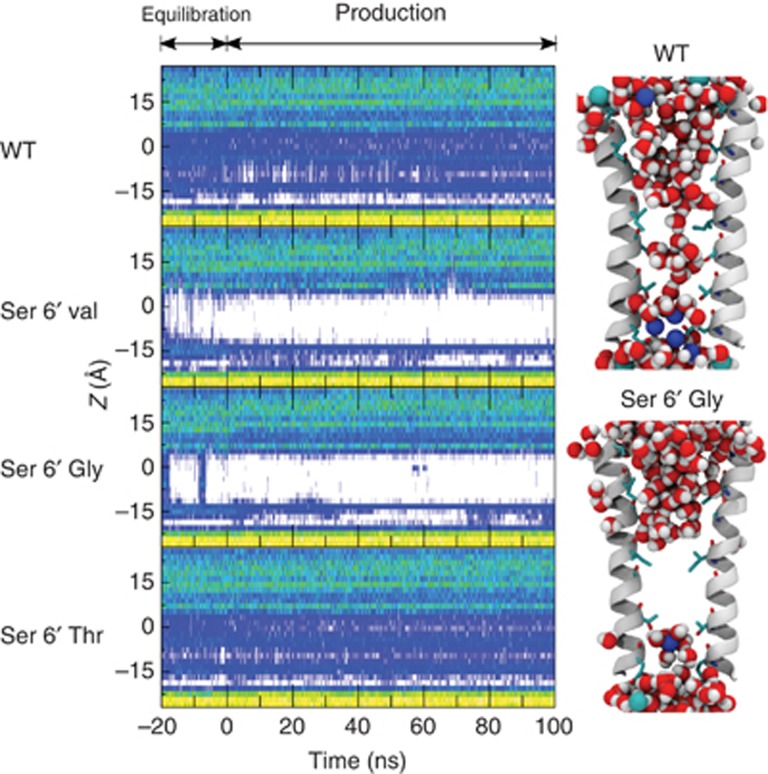
To further explore the role of the Ser6′ hydroxyl group, MD simulations of 100 ns duration were run on all three Ser6′ GLIC mutants studied by electrophysiology: S6′G, S6′V and S6′T. The pore remained stably open in all simulations and the hydration profile of each mutant is compared with that of the wt (Figure 6).
Figure 6.
Hydration profiles of GLIC′s pore in WT and Ser6′ mutant simulations. Hydration traces of the pore of GLIC during the 20-ns equilibration (with restraints on the Cα positions) and the following 100 ns production. The origin of the z-axis (0 Å) corresponds to the position of Ile9′. The hydration level is represented using a colour scale from dark blue (low hydration) to cyan, green and yellow (strong hydration). The absence of water is depicted by a white colour. On the right panel, snapshots corresponding to the last frame of the production are shown for the pore of GLIC wt and S6′T mutant. Water molecules are shown as spheres.
The S6′G and S6′V mutations lead to a local dehydration of the pore in a region encompassing residues 2′–9′ that could account for the loss-of-function phenotype observed for these mutants (Figure 6). The S6′T mutant revealed no such dramatic dehydration of the pore (Figure 6A), whereas voltage-clamp experiments suggested that this mutation leads to a non-functional channel (Figure 4A). To accurately investigate the possible more subtle differences between wt and S6′T mutant, we carried out three additional independent simulations for each system. The averaged hydration profiles for the four 100 ns simulations on GLIC wt and the S6′T mutant are very similar but indicate a slightly reduced hydration for the S6′T mutant at the levels of the 6′ and 9′ rings of residues (Supplementary Figure S7A). Interestingly, the pore radius at the level of 9′ was strongly affected by the mutation, being reduced from 2.28±0.06 Å to 1.91±0.03 Å, for GLIC wt and S6′T, respectively (Supplementary Figure S7A). This reduction of the radius of the pore might be due to the methyl group of the threonine side chains inducing a different rotamer of Ile9′ (Supplementary Figure S7B). The loss-of-function phenotype observed for the S6′T mutant might be accounted for by a significant reduction in pore radius at the 9′ level.
Interestingly, a multiple alignment of pLGICs indicates that while Thr6′ is very common, the combination of Thr6′–Ile9′ never occurs: Ser6′ appears to be paired with Ile9′ and Thr6′ is always paired with Leu9′. The finding that the methyl group of Thr6′ influences the rotamer conformation of the Ile9′ side chain during the MD simulations suggests that the apparent pairing of these important pore-lining residues might have conformational reasons. In order to test this hypothesis, the double-mutant I9′L/S6′T was expressed and tested under voltage-clamp in oocytes (Figure 4) and BHK cells (Supplementary Figure S2A). In both cases, the I9′L substitution restored the activity of the S6′T mutant but with a slightly reduced proton-sensitivity compared with GLIC wt (shifting EC50 from pH 5.4 to pH 4.7 (Supplementary Figure S8)). This experiment strongly suggests that distinct pairs of residues at 6′ and 9′ positions are crucial to the channel function. The combination of MD simulations performed on the S6′G and S6′V mutants and single-channel data strongly suggest that the crucial role played by residue Ser6′ in ion transport is coupled to its action on pore hydration. In order to get more insights at the molecular level about the role of Ser6′ and the ordered water molecules observed in the crystal structures, additional MD simulations where the ion is pulled linearly down the central pore axis were undertaken.
Pulling a Na + ion through the pore
Four independent 50 ns MD simulations were carried out pulling the ion down the central axis of the channel across its 40 Å-spanning TMD, leaving it free to move away from the symmetry axis. Na+ coordination and hydration were measured by counting water molecules, ions and protein oxygen atoms present in the first coordination shell (Figure 7). In the hydrophobic upper-half of the pore, the Na+ ion is highly hydrated. Only sporadic events of approaching counter ions were observed. The distribution of water around Na+ shows that ion hydration is almost uniform along the channel axis, similar to what could be observed in the bulk (Figure 7). This is partly due to the primary and secondary layers of water molecules observed in the MD simulations, which cover the pore and participate to hydrating the permeant Na+ ion as it crosses the upper half region of the pore.
Figure 7.
Hydration profile of Na+ crossing the TMD. (A) First hydration shell composition (calculated with a distance cutoff of 3.2 Å for all Na+ ligands and averaged over a time window of 0.25 ns). The colour code is the following: water (cyan), Cl− (red), Na+ (blue) and oxygen atoms from the side chains of Ser6′ (green), Thr2′ (yellow) and Glu-2′ (orange). (B) Z-dependent hydration profile of the pulled Na+. In each slice of 1 Å, the water density of the ion’s first water shell was computed along the z-axis. Selected pore structures (from a to f) are represented on the bottom panel (C) for six different snapshots; the pulled ion is displayed as a yellow sphere, water molecules are shown as red and white sticks, and water molecules present in the first hydration shell are represented by larger sticks. Pore-lining residues are shown in cyan. Transparent spheres represent waters and ions present in the crystal structure: small red spheres for water and large spheres for ions (red for Br− and blue for Cs+ and Rb+).
When the Na+ ion crosses the bottom half region of the pore that is more constricted, especially at the level of the Ile9′ and Thr2′ rings of residues, the distribution of water molecules around the permeant ion becomes strongly anisotropic (Figure 7). As the ion reaches the level of Ile9′, two distinct high-density peaks appear above and below the ion position in the z-axis water distribution. The water molecules constituting the upper peak are stabilized by the primary layer of water molecules at the Ile9′ level. The bottom peak involves water molecules just above the tight pentagon, where a triangle of ordered water molecules was observed in the GLIC_2.4 structure. When the ion permeated further down the channel, its hydration displayed the same anisotropic distribution, with peaks of high water-density corresponding to the ‘tight pentagon’ and the ‘loose pentagon’. Indeed, all four simulations show that the positions occupied by ordered water molecules observed in the GLIC_2.4 crystal structure are also occupied as the ion flows down the channel. Hence, these positions might be important in order to maintain the hydration shell of the permeant ion especially while it crosses the constriction points at the Ile9′ and Thr2′ levels.
In addition, the pulling simulation reveals a transient interaction of the permeant Na+ ion with the side chains of Ser6′, Thr2′ and Glu-2′ residues. Behaviour of Ser6′ during the pulling simulation displayed a particular correlation, as the side-chain orientation was sensitive to the ion position (Supplementary Figure S6). This suggests that Ser6′ might contribute directly and dynamically to ion transport by rearranging its side chain as the ion flows down the pore.
Electrostatic calculations illustrate the roles of residues 6′ and 2′ to accommodate ions during permeation in both cation- and anion-selective channels
MD simulation data presented above reveals that the relative orientation of the Hγ atom of Ser6′ influences locally the distribution of water molecules in the pore. This observation is also made when computing the pore electrostatic potential within the Poisson–Boltzmann–Langevin framework, where water molecules are simply modelled as freely orienting dipoles (Azuara et al, 2008). As the dihedral angle reaches a value where Hγ is located above (resp. below) Oγ, the water density at the level of Ser6′ increases (resp. decreases) while the cation density peak at the level of Thr2′ is strengthened (resp. spread upward) (Supplementary Figure S4C and D). These results illustrate that the orientation of the hydroxyl group of Ser6' greatly influences the electrostatic properties within the pore.
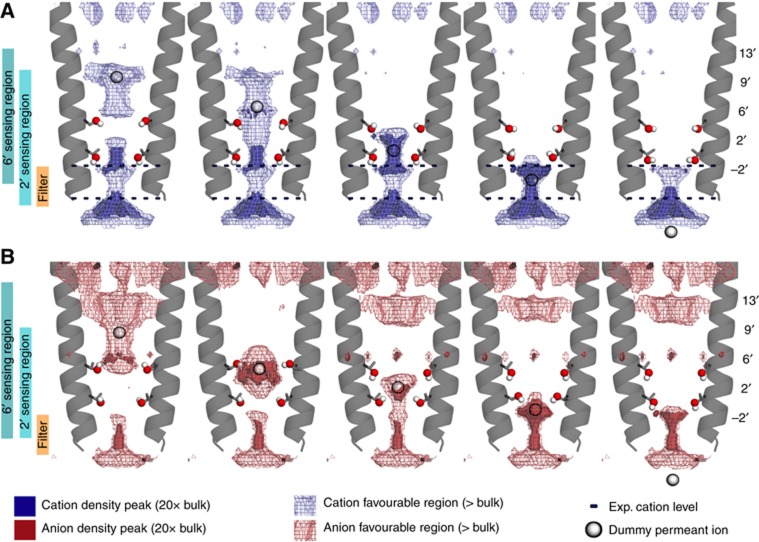
In order to further probe the effect of both Ser6′ and Thr2′ residues on the channel permeation, the consequence of changing their side-chain orientation was investigated through systematic generation of possible configurations (see Supplementary Figure S6). It proved possible to select a subset of these configurations in order to sequentially maximize the cation density along the permeation pathway. This crude sampling (see Figure 8A and Supplementary Movie SI1) reveals that it is at the same time possible to shift the maximum predicted density for cations in the pore and to reduce energy barriers between energy wells by choosing an appropriate sequence of orientations for the Oγ–Hγ dipole of Ser6′ (Supplementary Figure S9A) as the ion flows down the pore. Overall, Ser6′ (resp. Thr2') influences the ion density in a window much wider than expected: the region that exhibits the largest effect covers a 22-Å (resp. 24 Å) wide window along the pore axis, between Glu-2′ (resp. the M1–M2 tip) and Leu17′ (resp. Ala13′). The same protocol was applied on the structure of the anion-selective channel GluCl (Hibbs and Gouaux, 2011), in which residues 2′ and 6′ are both threonine, and the same observation was made, only this time the hydroxyl groups expose the positive partial charge of Hγ to the anion instead of the negative partial charge of Oγ to the cation in GLIC (Figure 8B; Supplementary Figure S9B; Supplementary Movie SI2).
Figure 8.
Optimized set of configurations of the Ser6′ and Thr2′ side chains. Five representative configurations out of the 72 generated ones of GLIC (A) and GluCl (B) are shown. They maximize the permeant ion (cation for GLIC, anion for GluCl) density at five altitudes in the pore (indicated by a white sphere on the pore axis), from its extracellular to its intracellular end (left to right). The hydroxyl groups of residues 2′ and 6′ are shown as spheres. The favourable region for the permeant ion is shown as a mesh isocontoured at the bulk density value (200 mM) (blue for cation, red for anion). The regions where the permeant ion density is higher than 20 times the bulk value are shown as a blue surface for cation, red surface for anion. The altitudes of experimentally observed ions in GLIC are indicated by dashed lines. On the left of the figure, vertical bars indicate the regions where the permeant ion density is affected by either residue 2′ or 6′.
Discussion
Combining X-ray crystallography, electrophysiology, MD simulations and electrostatic calculations provides novel insights on the molecular mechanisms of ion permeation in pLGICs, which are complex mechanisms as they involve protein residues, ions and water molecules that interact together dynamically and transiently as the ions flow down the channel.
Hydration profile in the pore of a pLGIC and its role in ion permeation
Diffraction experiments yield a time-averaged and space-averaged atomic description of the hydration geometry in the lower part of the pore. At 2.4 Å, we observe three layers of hydration: the first one can be a pentagon, as in 3UUB (Figure 3), but not always; it is present in all our crystal structures but its exact geometry varies from one crystal to the other. The second one, at the level of Ser6′ is a self-stabilized pentagon (‘tight pentagon’) whose dimensions and geometry are very close to water pentagons encountered in three types of ice: ice III with d(O–O)=2.70–2.85 Å, ice V with d(O–O)=2.76–2.86 Å and ice IX with d(O–O)=2.71–2.81 Å (Jeffrey, 1997). The third layer is also a water pentagon, but that differs from the ‘tight pentagon’ as it is not self-stabilized but interacts directly with a Na+ ion. This ‘loose pentagon’ might exchange with Glu-2′, a residue that is known to be important for the ion charge specificity (Imoto et al, 1991; Corringer et al, 1999; Fritsch et al, 2011). The observed mean distance between the Na+ ion and the water molecules from the ‘loose pentagon’ (3.45±0.21 Å) are higher than that of the bulk (2.43 Å) (Mahler and Persson, 2012). This suggests that ion hydration in the selectivity filter region of the pore differs significantly from that of the bulk and is reminiscent of the MD Na+-pulling experiment showing that the distribution of water molecules around the permeant ion becomes strongly anisotropic in this most constricted region of the pore.
MD studies provide a complementary (dynamic) interpretation of the crystallographic data. MD studies are also important as they confirm that the water molecules observed in the crystal structure are not an artefact that could have occurred during flash-cooling (Halle, 2004). In particular, the pulling simulation reveals a specific organization of water molecules as the Na+ ion crosses hydrophobic barriers and it shows that all three layers of hydration seen in the crystal structure are important at one point or another in ion permeation. MD simulations furthermore complement our view of the hydration in the upper part of the pore, namely in the hydrophobic upper half region where main-chain carbonyl oxygens of the Ile9′ and Ala13′ rings of residues contribute to organize water molecules in several layers that clearly differ from the bulk. Previous work has suggested that this part of the pore of pLGICs may collapse to close the gate and constitute a solid pore-obstructing plug between helices in the closed conformation (Cheng et al, 2007; Hilf and Dutzler, 2008; Nury et al, 2010b; Zheng and Auerbach, 2011; Prevost et al, 2012). In the conductive form of the channel, it is intuitively not straightforward to imagine how ions behave in this region, and if they remain hydrated; but MD simulations illustrate that both cations and anions can diffuse in this region, while maintaining their full hydration shell. This is due to the peculiar pattern of exposed carbonyl groups that maintains, at least in the open form, a good hydration in this region.
Description of important cations and anions binding sites in the pore of GLIC
Important anion and cations binding sites were identified in the cytoplasmic part of the pore by using anomalous diffraction data collected from crystals soaked with Br−, Cs+ and Rb+. The identified cation binding sites (cation site 1 and 2) display the highest electrostatic-attractive potential for cations, suggesting that both positions are preferentially occupied by the permeant cation during its translocation through the pore. Interestingly, the cation site 2 at the level of Glu-2′ ring was previously shown to bind the divalent transition metal Zn++ (Hilf et al, 2010). This region of the pore is particularly important as it comprises the charge selectivity filter of pLGICs, which has been shown long ago to be located in the most intracellular part of the pore (Imoto et al, 1991; Corringer et al, 1999), involving residues −1′ and 0′. In GLIC, a charged ring of glutamate residues is located at position −2′. As illustrated by the pulling experiment, Glu-2′ is able to pick-up a stabilized cation at the level of Thr2′ (cation site 1) and accommodate its trafficking trajectory toward cation site 2. Several glutamate residues can cooperate in this translocation, replacing the water molecules from the cation hydration shell, in line with the plausible exchange of this ‘loose’ water pentagon with the ε-O carboxylate atoms of Glu-2′ that was observed in the GLIC_2.4 structure. Consistently, a role for torsional flexibility in cation conduction was recently proposed for a ring of glutamates at position −1′ in the nAChR (Cymes and Grosman, 2012). Finally, the presence of a reservoir of counter ions (observed both experimentally (Figure 3) and in the MD simulation (Figure 5)) at the intracellular mouth of the transmembrane pore might help in either retaining the permeant ion out of the pore or creating a favourable electro-attractive corridor toward it.
This study has been mainly focused on the cation and anion binding sites observed in the pore even though several ion binding sites have been observed in the ECD of the receptor. However, the sites might appear to be functionally relevant, as it has been shown recently that the ECD of these receptors is also involved in ion permeation and ion selectivity (Hansen et al, 2008; Brams et al, 2011; Moroni et al, 2011). In particular, the two rings of Cs+ bound to Asp86 and Asp88 and the ring of sulphates bound to Lys38 and Arg109 are all exposed to the lumen of the channel and might have functional implications in ion permeation or selectivity. Important residues for ion permeation have already been identified in a similar region of the ECD from AchBP structures bound to ions (Hansen et al, 2008; Brams et al, 2011), and by site-directed mutagenesis studies carried out on the nACh (Hansen et al, 2008) or 5-HT3A (Livesey et al, 2011) receptors.
The key Ser6′ ring of residues displays multiple roles in ion transport
Site-directed mutagenesis experiments on Ser6′ residue revealed that this position is crucial for ion transport. This observation is consistent with previous mutational analysis of the polar rings of Ser6' and Thr2′ on nAChR (Imoto et al, 1991) showing that replacing them with hydrophobic residues lowers the conductance to an extent that increases with the size of the mutant side chain. Consistently, single-channel experiments performed on S/G6′-heteropentamers revealed that the S6′G substitution impacts the channel’s conductance.
The GLIC_2.4 structure reveals that Ser6′ is located next to the ordered water molecules from the ‘tight pentagon’, suggesting that its side chain might influence locally the pore hydration. This was confirmed by both electrostatics (Aquasol) calculations and MD simulations predicting that the local hydration is strongly influenced by the positions of the Oγ and Hγ atoms of Ser6′ (Figure 5; Supplementary Figure S2C and D). In addition, MD simulations performed on S6′G and S6′V revealed that removing the hydroxyl moiety of Ser6′ dramatically impacts the hydration in a region encompassing the 2′ and 9′ levels of the pore (Figure 6).
On the other hand, the S6′T mutation that preserves the hydroxyl moiety of serine does not produce such dramatic effect on hydration during the MD simulations, even though it was shown experimentally to express as an intact but inactive pentamer (Figure 4). The experiments on the S6′T/I9′L GLIC double-mutant are particularly interesting as the I9′L mutation restores the activity of the channel, suggesting that the residues at positions 6′ and 9′ are paired: S6′/I9′ in GLIC and S6′-T6′/L9′ in all other pLGICs. The coupling between the 6′ position (critical for pore hydration) and the 9′ position (critical for gating) might have important functional implications. One might ask if a change of conformation at 6′ might alter locally the water organization in a way that would facilitate the hydrophobic gating at the level of 9′. Consistently, it was previously proposed that pore dehydration is a required preamble to gating (Jha et al, 2009). However, other experiments, including a structure of the receptor in its resting state, will be necessary to formally demonstrate that point.
In addition to its influence on hydration, Ser6′ might facilitate ion transport by reducing the electrostatic free energy barriers encountered by the ion during its translocation. Depending on the orientations of the Ser6′ Hγ–Oγ bonds, a diffusing cation coming from the extracellular vestibule would either encounter a barrier at the level of the 9′ ring, or be attracted inward. In addition to their long-distance influence, the polar rings of Ser6′ might also be involved locally and directly in the control of ion permeation, as suggested by early experiments (Giraudat et al, 1986). The pulling simulation revealed that as soon as the cation reaches this level of the pore, it binds transiently to Ser6′ side chains, which accommodates its translocation, while retaining its full hydration shell.
In total, this study reveals that the Ser6′ ring of residues plays multiple roles in ion transport: (1) it influences locally the organization of important water molecules required for the ion to cross the hydrophobic 9′ constriction point, (2) it interacts transiently with the permeant ion thus facilitating its translocation, (3) it has a long-distance influence on the pore electrostatics. The critical nature of this portion of the pore, defined by the Ser6′ ring of residues is illustrated by recent works showing that it is the binding site of quaternary ammonium channel blockers (Hilf et al, 2010) and inhibitory general anaesthetics (Lebard et al, 2012).
A possibly conserved molecular mechanism of ion permeation in pLGIC
Due to the strong sequence conservation of M2 pore-lining residues within the pLGIC family, the conclusions presented in this study might be transferable to all the pLGIC channels. First, the water pentagon at the level of residue 6′ might be a general feature for all these channels. Indeed, a blob of electron density present at the level of Thr6′ in the structure of GluCl might be accounted for by the presence of a similar pentagon of water molecules (Figure 2). Second, the different steps involved do not depend on the charge of the ion, hence the scenario described above is readily transferable to any member of the pLGIC family, either cation or anion selective. The key element is a hydrophobic region exposing carbonyl atoms that can be solvated and therefore enable the permeant ion to remain hydrated, in conjunction with a water-rich self-stabilized region at the level of the highly conserved residue 6′, whose side chain is always of dipolar nature and able to adapt to any ion charge.
Materials and methods
Protein expression and purification
GLIC wt and GLIC A13′F were produced as previously described (Bocquet et al, 2009; Nury et al, 2010a, 2011). For the purpose of co-crystallization with selenium-derived DDM (Se-DDM), DDM was replaced by Se-DDM (Affymetrix) during purification. For the purpose of co-crystallization with Cs+ or Br−, NaCl was replaced by either 300 mM CsCl or 300 mM NaBr during the purification.
Crystallography
GLIC wt in its apo form or bound to selenium-derived DDM was crystallized as previously described (Bocquet et al, 2009; Nury et al, 2011) using a reservoir solution containing 12–15% PEG 4k, 400 mM NaSCN and 0.1 M NaAcetate pH 4.0. Crystals were cryoprotected using 20% glycerol. GLIC crystals grown in complex with sulphate were prepared in similar conditions but NaSCN was replaced by (NH4+)2SO42−. Crystals grown in complex with Cs+, Rb+ or Br−: to maximize chances of characterizing all possible cation binding sites in the pore, we used the A13′F mutant, for which it has been shown that the presence of a bulkier side chain at position 13′ destabilized the detergent bundle in the pore (Nury et al, 2010b). The protein was crystallized as described above but with the crystallization solution supplemented with 200–500 mM CsCl, RbCl or NaBr. Data sets were collected at optimized wavelengths for each derivative. Reflections were integrated and analysed using XDS (Kabsch, 2010) and CCP4 programs (Winn et al, 2011). The structure of GLIC (3EAM) served as a starting model that was improved and refined using BUSTER (Blanc et al, 2004) with NCS restraints. Structure validation was checked using Molprobity (Davis et al, 2007). Data collection and refinement statistics are summarized in Table I.
Electrophysiology
Protein expression. Separate plasmids coding for GLIC and GFP were co-injected in the nuclei of defolliculated stage 6 Xenopus oocytes or used for CaPO42− transfection of baby hamster kidney cells (BHK). Only GFP-positive cells were used.
Immunolabelling. Cells were fixed with 4% paraformaldehyde and immunolabelled using an anti-HA primary antibody and an anti-rabbit Cy5-coupled secondary antibody (Invitrogen). BHK cells and 40 μm sliced-oocytes were analysed using fluorescence microscopy.
Electrophysiology. The oocyte membrane was voltage-clamped at −50 mV using a Geneclamp 500 amplifier with two 3 M-KCl filled electrodes. For BHK cells, whole-cell and outside–out patch voltage-clamp were performed using an RK-400 patch-clamp amplifier (Bio-Logic). The programs pClamp (Axon Instruments) and SigmaPlot 11 (Systat) were used for acquisition and analysis. Recordings were performed using gravity-driven systems for oocyte chamber perfusion (4–10 ml/min) or local BHK cell superfusion (exchange time 1–2 s). The control extracellular solution (in mM: NaCl 100 (oocyte) or 160 (BHK), CaCl2 1, MgCl2 1, MES 10, pH adjusted to 8 using NaOH) was lowered to various pH values using HCl 2 M. Intracellular patch solution was composed of (mM): CsCl 145, MgCl2 1, MES 10, BAPTA 10, pH 7.2 (CsOH). Patch pipettes were pulled from thick wall borosilicate glass to resistances of 2–3 MΩ, used directly for whole-cell or fire-polished to 7–10 MΩ for single-channel recording (patch input resistance >40 GΩ). More details are in Supplementary Information.
Molecular Dynamics simulations and Electrostatics calculations are detailed in SI.
Supplementary Material
Acknowledgments
We thank the French National Agency for Research for funding (Grant ANR-2010-BLAN-1534). Molecular simulations were performed using HPC computing resources from GENCI-/TGCC/CINES/IDRIS (Grant 2011-072292 to MD). We thank Damien Laage and Fabio Sterpone for helpful discussions on water structure and Hugues Nury for help in early crystallographic work. We acknowledge the help of the Proteopole platform at Institut Pasteur. We thank the staffs of Proxima1 at Soleil and of ID23-1 at ESRF.
Author contributions: LS and LM performed the crystallography part of the work. CVL and GM performed the electrophysiology study. SM and FP performed, respectively, the molecular dynamic simulations and the electrostatic calculations. MD, MB and PJC supervised the study. All authors contributed to data analysis.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abascal JL, Sanz E, Garcia Fernandez R, Vega C (2005) A potential model for the study of ices and amorphous water: TIP4P/Ice. J Chem Phys 122: 234511. [DOI] [PubMed] [Google Scholar]
- Amiri S, Tai K, Beckstein O, Biggin PC, Sansom MS (2005) The alpha7 nicotinic acetylcholine receptor: molecular modelling, electrostatics, and energetics. Mol Membr Biol 22: 151–162 [DOI] [PubMed] [Google Scholar]
- Azuara C, Orland H, Bon M, Koehl P, Delarue M (2008) Incorporating dipolar solvents with variable density in Poisson-Boltzmann electrostatics. Biophys J 95: 5587–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckstein O, Sansom MS (2006) A hydrophobic gate in an ion channel: the closed state of the nicotinic acetylcholine receptor. Phys Biol 3: 147–159 [DOI] [PubMed] [Google Scholar]
- Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G (2004) Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr 60: 2210–2221 [DOI] [PubMed] [Google Scholar]
- Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux JP, Delarue M, Corringer PJ (2009) X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature 457: 111–114 [DOI] [PubMed] [Google Scholar]
- Bocquet N, Prado de Carvalho L, Cartaud J, Neyton J, Le Poupon C, Taly A, Grutter T, Changeux JP, Corringer PJ (2007) A prokaryotic proton-gated ion channel from the nicotinic acetylcholine receptor family. Nature 445: 116–119 [DOI] [PubMed] [Google Scholar]
- Brams M, Gay EA, Saez JC, Guskov A, van Elk R, van der Schors RC, Peigneur S, Tytgat J, Strelkov SV, Smit AB, Yakel JL, Ulens C (2011) Crystal structures of a cysteine-modified mutant in loop D of acetylcholine-binding protein. J Biol Chem 286: 4420–4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MH, Coalson RD, Tang P (2010) Molecular dynamics and brownian dynamics investigation of ion permeation and anesthetic halothane effects on a proton-gated ion channel. J Am Chem Soc 132: 16442–16449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Ivanov I, Wang H, Sine SM, McCammon JA (2007) Nanosecond-timescale conformational dynamics of the human alpha7 nicotinic acetylcholine receptor. Biophys J 93: 2622–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corringer PJ, Bertrand S, Galzi JL, Devillers-Thiery A, Changeux JP, Bertrand D (1999) Mutational analysis of the charge selectivity filter of the alpha7 nicotinic acetylcholine receptor. Neuron 22: 831–843 [DOI] [PubMed] [Google Scholar]
- Corringer PJ, Poitevin F, Prevost MS, Sauguet L, Delarue M, Changeux JP (2012) Structure and pharmacology of pentameric receptor channels: from bacteria to brain. Structure 20: 941–956 [DOI] [PubMed] [Google Scholar]
- Cymes GD, Grosman C (2012) The unanticipated complexity of the selectivity-filter glutamates of nicotinic receptors. Nat Chem Biol 8: 975–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB 3rd, Snoeyink J, Richardson JS, Richardson DC (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35: W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhakshnamoorthy B, Raychaudhury S, Blachowicz L, Roux B (2010) Cation-selective pathway of OmpF porin revealed by anomalous X-ray diffraction. J Mol Biol 396: 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R (1998) The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280: 69–77 [DOI] [PubMed] [Google Scholar]
- Fritsch S, Ivanov I, Wang H, Cheng X (2011) Ion selectivity mechanism in a bacterial pentameric ligand-gated ion channel. Biophys J 100: 390–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Dennis M, Heidmann T, Chang JY, Changeux JP (1986) Structure of the high-affinity binding site for noncompetitive blockers of the acetylcholine receptor: serine-262 of the delta subunit is labeled by [3H]chlorpromazine. Proc Natl Acad Sci USA 83: 2719–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halle B (2004) Biomolecular cryocrystallography: structural changes during flash-cooling. Proc Natl Acad Sci USA 101: 4793–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SB, Wang HL, Taylor P, Sine SM (2008) An ion selectivity filter in the extracellular domain of Cys-loop receptors reveals determinants for ion conductance. J Biol Chem 283: 36066–36070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbs RE, Gouaux E (2011) Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 474: 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilf RJ, Bertozzi C, Zimmermann I, Reiter A, Trauner D, Dutzler R (2010) Structural basis of open channel block in a prokaryotic pentameric ligand-gated ion channel. Nat Struct Mol Biol 17: 1330–1336 [DOI] [PubMed] [Google Scholar]
- Hilf RJ, Dutzler R (2008) X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature 452: 375–379 [DOI] [PubMed] [Google Scholar]
- Hilf RJ, Dutzler R (2009) Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature 457: 115–118 [DOI] [PubMed] [Google Scholar]
- Imoto K, Konno T, Nakai J, Wang F, Mishina M, Numa S (1991) A ring of uncharged polar amino acids as a component of channel constriction in the nicotinic acetylcholine receptor. FEBS Lett 289: 193–200 [DOI] [PubMed] [Google Scholar]
- Ivanov I, Cheng X, Sine SM, McCammon JA (2007) Barriers to ion translocation in cationic and anionic receptors from the Cys-loop family. J Am Chem Soc 129: 8217–8224 [DOI] [PubMed] [Google Scholar]
- Jeffrey GA (1997) An Introduction to Hydrogen Bonding Oxford University Press [Google Scholar]
- Jha A, Purohit P, Auerbach A (2009) Energy and structure of the M2 helix in acetylcholine receptor-channel gating. Biophys J 96: 4075–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010) Integration, scaling, space-group assignment and post-refinement. Acta Cryst D66: 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keramidas A, Moorhouse AJ, Schofield PR, Barry PH (2004) Ligand-gated ion channels: mechanisms underlying ion selectivity. Prog Biophys Mol Biol 86: 161–204 [DOI] [PubMed] [Google Scholar]
- Kim I, Allen TW (2011) On the selective ion binding hypothesis for potassium channels. Proc Natl Acad Sci USA 108: 17963–17968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebard DN, Henin J, Eckenhoff RG, Klein ML, Brannigan G (2012) General anesthetics predicted to block the GLIC pore with micromolar affinity. PLoS Comput Biol 8: e1002532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kim SH (2009) Water polygons in high-resolution protein crystal structures. Protein Sci 18: 1370–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey MR, Cooper MA, Lambert JJ, Peters JA (2011) Rings of charge within the extracellular vestibule influence ion permeation of the 5-HT3A receptor. J Biol Chem 286: 16008–16017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahler J, Persson I (2012) A study of the hydration of the alkali metal ions in aqueous solution. Inorg Chem 51: 425–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroni M, Meyer JO, Lahmann C, Sivilotti LG (2011) In glycine and GABA(A) channels, different subunits contribute asymmetrically to channel conductance via residues in the extracellular domain. J Biol Chem 286: 13414–13422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov SY, Berneche S, Roux B (2004) Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands. Nature 431: 830–834 [DOI] [PubMed] [Google Scholar]
- Nury H, Bocquet N, Le Poupon C, Raynal B, Haouz A, Corringer PJ, Delarue M (2010a) Crystal structure of the extracellular domain of a bacterial ligand-gated ion channel. J Mol Biol 395: 1114–1127 [DOI] [PubMed] [Google Scholar]
- Nury H, Poitevin F, Van Renterghem C, Changeux JP, Corringer PJ, Delarue M, Baaden M (2010b) One-microsecond molecular dynamics simulation of channel gating in a nicotinic receptor homologue. Proc Natl Acad Sci USA 107: 6275–6280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nury H, Van Renterghem C, Weng Y, Tran A, Baaden M, Dufresne V, Changeux JP, Sonner JM, Delarue M, Corringer PJ (2011) X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel. Nature 469: 428–431 [DOI] [PubMed] [Google Scholar]
- Prevost MS, Sauguet L, Nury H, Van Renterghem C, Huon C, Poitevin F, Baaden M, Delarue M, Corringer PJ (2012) A locally closed conformation of a bacterial pentameric proton-gated ion channel. Nat Struct Mol Biol 19: 642–649 [DOI] [PubMed] [Google Scholar]
- Song C, Corry B (2010) Ion conduction in ligand-gated ion channels: Brownian dynamics studies of four recent crystal structures. Biophys J 98: 404–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasneem A, Iyer LM, Jakobsson E, Aravind L (2005) Identification of the prokaryotic ligand-gated ion channels and their implications for the mechanisms and origins of animal Cys-loop ion channels. Genome Biol 6: R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter MM (1984) Water structure of a hydrophobic protein at atomic resolution: Pentagon rings of water molecules in crystals of crambin. Proc Natl Acad Sci USA 81: 6014–6018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter MM, Yamano A, Stec B, Mohanty U (2001) On the nature of a glassy state of matter in a hydrated protein: Relation to protein function. Proc Natl Acad Sci USA 98: 11242–11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin N (2005) Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol 346: 967–989 [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Auerbach A (2011) Decrypting the sequence of structural events during the gating transition of pentameric ligand-gated ion channels based on an interpolated elastic network model. PLoS Comput Biol 7: e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R (2001) Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature 414: 43–48 [DOI] [PubMed] [Google Scholar]
- Zhu F, Hummer G (2010) Pore opening and closing of a pentameric ligand-gated ion channel. Proc Natl Acad Sci USA 107: 19814–19819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Hummer G (2012) Theory and Simulation of Ion conduction in the Pentameric GLIC channel. JCTC 8: 3759–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.