Abstract
Breast cancer is a group of heterogeneous diseases that show substantial variation in their molecular and clinical characteristics. This heterogeneity poses significant challenges not only in breast cancer management, but also in studying the biology of the disease. Recently, rapid progress has been made in understanding the genomic diversity of breast cancer. These advances led to the characterisation of a new genome-driven integrated classification of breast cancer, which substantially refines the existing classification systems currently used. The novel classification integrates molecular information on the genomic and transcriptomic landscapes of breast cancer to define 10 integrative clusters, each associated with distinct clinical outcomes and providing new insights into the underlying biology and potential molecular drivers. These findings have profound implications both for the individualisation of treatment approaches, bringing us a step closer to the realisation of personalised cancer management in breast cancer, but also provide a new framework for studying the underlying biology of each novel subtype.
Keywords: breast cancer, classification, genomic
Introduction
Breast cancer remains one of the leading causes of cancer death in women, despite significant improvements in survival over the past 25 years. One of the greatest challenges faced by clinicians and researchers in this field is that breast cancer is not a single entity, but rather a heterogeneous group of several subtypes displaying distinct differences in biological and clinical behaviour. A primary aim in cancer management is to tailor clinical decisions to the individual, based on a detailed understanding of the molecular profile of the tumour and the likely clinical outcome of the individual’s disease. This progress will facilitate personalised treatment approaches that are more targeted, have superior efficacy and are associated with less toxicity. Our increased knowledge of the genomic aberrations underlying human breast cancers, and the molecular processes that are disrupted, are key to understanding the diversity of the disease and achieving the aims of personalised medicine. Over the past decade, the development of high-throughput technologies to study genetic, epigenetic and proteomic changes has allowed for rapid progress in our understanding of the complexity of breast cancer biology. Here, we review recent advances that have led to the integration of information on the genomic and transcriptomic landscapes of breast cancers to refine the molecular classification of the disease.
Current histopathological classification of breast cancer
The classification of invasive breast cancer currently involves the assessment of histological criteria encompassing both morphology-based and immunohistochemical (IHC) analyses. Traditional pathological parameters such as histological type, tumour size, histological grade and axillary lymph-node involvement have been shown to correlate with clinical outcome and provide the basis for prognostic evaluation (Elston et al, 1999). IHC markers such as the expression of hormone receptors (oestrogen (ER) and progesterone receptors (PR)) and the overexpression and/or amplification of the human epidermal growth factor receptor 2 (HER2) provide additional therapeutic predictive value and are of key importance in guiding treatment selection (Harris et al, 2007).
Histopathological subtypes and tumour grade
The vast majority of breast carcinomas (∼70–80%) are described as invasive ductal carcinomas not otherwise specified (IDC-NOS) based on architectural patterns and cytological features (Ellis, 2003). In contrast, around 25% of breast cancers are characterised according to ‘histological special types’ such as lobular, tubular, medullary and metaplastic carcinomas (Ellis, 2003). At the molecular level, each histological special type appears to be more homogenous than IDC-NOS and is likely to be driven by key underlying molecular mechanisms (Weigelt et al, 2008). However, the majority of the special types are rare and to date, this has limited their analysis in large-scale molecular studies. In addition to histological tumour type, tumour grade is the other important intrinsic tumour characteristic that can be assessed by histopathological analysis. Tumour grade is an assessment of differentiation (tubule formation and nuclear pleomorphism) and proliferative activity (mitotic index), allowing tumours to be further stratified and providing key prognostic information (Rakha et al, 2010).
ER, PR and HER2
In conjunction with histopathological assessment, the standard evaluation of breast cancer for clinical purposes involves IHC characterisation of ER, PR and HER2 status. Hormone receptor-positive breast cancers account for around 75–80% of all cases and standardised IHC assays for the routine testing of ER and PR are used to guide the selection of patients for hormonal-based therapies. HER2 represents the only additional predictive marker currently in routine use. Approximately 10–15% of breast cancers have HER2 overexpression and/or amplification with around half of these co-expressing hormone receptors (Konecny et al, 2003). These patients are selected for anti-HER2 based therapies, including the humanised monoclonal HER2 antibody, trastuzumab, which targets the extracellular domain of the HER2 receptor. The remaining 10–15% of breast cancers are defined by hormone receptor and HER2 negativity (i.e., triple negative cancers), which represent a key clinical entity given their lack of therapeutic options (Dawson et al, 2009).
While the current classification of human breast tumours has been fundamental for prognostic and predictive evaluation, there remain a number of important limitations. First, considerable variation in response to therapy and clinical outcome still exists, even for tumours with apparent similarities in clinical and pathological characteristics. Second, this classification continues to provide limited insight into the complex underlying biology and the molecular pathways driving the disease in different subtypes.
Molecular classification of breast cancer
Gene expression profiling and the identification of intrinsic subtypes
Expression analysis using microarray-based technology has provided researchers with an opportunity to begin moving towards comprehensive molecular profiling of breast cancer. These efforts have resulted in the identification of clinically relevant molecular subtypes, and have provided early insights into the molecular heterogeneity of the disease (Perou et al, 2000; Sorlie et al, 2001, 2003; Hu et al, 2006). Five distinct intrinsic subtypes have been identified based solely on gene expression: luminal A, luminal B, HER2 overexpressing, basal-like and normal breast tissue-like. Differences in gene expression patterns reflect basic alterations in the cell biology of the tumours and importantly are associated with significant variation in clinical outcome (Sorlie et al, 2003). The prognosis of patients with ER-positive disease is largely determined by the expression of genes related to proliferation (Hu et al, 2006). More recently, the intrinsic classification has been refined in a PAM50 assay based on the expression of 50 genes designed to classify single samples into each of the five intrinsic subtypes (Parker et al, 2009; Nielsen et al, 2010).
Following the initial identification of the intrinsic molecular subtypes, gene expression studies have evolved and further sub-classification of breast cancers into new molecular entities have been proposed. For example, a detailed analysis of genes differentially expressed in ER-negative tumours has demonstrated that basal breast cancers are a heterogeneous group with at least four main subtypes (Teschendorff et al, 2007). Furthermore, this analysis revealed an immune response gene expression module, which identifies a good prognosis subtype in ER-negative disease. Other recent studies have also identified a new breast cancer intrinsic subtype known as Claudin-low or mesenchymal-like (Prat et al, 2010). This subtype is characteristically negative for ER, PR and HER2 and carries an intermediate prognosis between basal and luminal subtypes. Importantly, Claudin-low/mesenchymal tumours appear to be enriched with cells showing distinct biological properties associated with mammary stem cells and tumour initiating potential (Hennessy et al, 2009; Lim et al, 2009; Lehmann et al, 2011; Bruna et al, 2012).
In parallel with the identification of the intrinsic subtypes, gene expression profiling has also been used by several groups to identify distinct prognostic signatures (van de Vijver et al, 2002; van’t Veer et al, 2002; Paik et al, 2004). Two of these signatures, Mammaprint (a microarray-based assay of the Amsterdam 70-gene breast cancer signature) and OncotypeDX (a PCR-based assay of a panel of 21 genes) have been approved for clinical use and are now being tested in randomised clinical trials (Cardoso et al, 2008; Sparano and Paik, 2008).
Subtypes defined through IHC markers
IHC can reproduce a similar molecular taxonomy of the disease (Callagy et al, 2003; Abd El-Rehim et al, 2005; Jacquemier et al, 2005; Blows et al, 2010). The largest IHC study involving close to 12 000 samples (Blows et al, 2010) showed that the luminal and non-luminal subtypes are recognised primarily by the presence or absence of ER and PR expression, respectively. These two groups can be further separated on the basis of HER2 expression with luminal HER2-positive tumours most closely resembling the luminal B subtype and non-luminal HER2 expressing tumours representing the HER2 molecular subtype. The triple negative subtype is characteristically negative for ER, PR and HER2 expression, but importantly, can also be divided into two further subgroups based on the expression of basal cytokeratins (such as CK5-6) and EGFR (epidermal growth factor receptor) (Nielsen et al, 2004; Blows et al, 2010). The six subtypes of breast cancer defined by this approach demonstrate distinct differences in terms of breast cancer survival (Blows et al, 2010). Furthermore, similarly to gene expression profiling, the IHC expression of proliferation markers such as Ki67 and Aurora A kinase is associated with prognosis in ER-positive disease (Cheang et al, 2009; Ali et al, 2012). In addition, BCL2 expression, as assessed by IHC, is a powerful predictor of favourable prognosis in breast cancer across different molecular subtypes (Dawson et al, 2010). Despite these findings, the routine assessment of IHC markers in addition to ER, PR and HER2 has not yet been implemented into standard clinical treatment guidelines.
Integrating changes at the genomic level into classification
The varied genomic landscape of breast carcinomas is not fully captured using histopathological or transcriptomic analysis. Distinct patterns of genomic rearrangements in breast cancer have been characterised (Stephens et al, 2009) and molecular portraits of breast cancer can be identified from studying the spectrum of copy number alterations using array comparative genomic hybridisation (aCGH) (Chin et al, 2007). For example, aCGH analysis has identified a novel genomic subtype of ER-negative breast cancer characterised by low genomic instability (Chin et al, 2007). Changes in gene expression patterns are influenced by the underlying genomic architecture and some features associated with the intrinsic subtypes have been defined by copy number profiling (Chin et al, 2006; Ding et al, 2010). Furthermore, measures of genomic complexity, such as the complex arm aberration index (CAAI), have been shown to provide important prognostic information in both ER-positive and ER-negative diseases (Russnes et al, 2010).
The emergence of next-generation sequencing technologies has now allowed the characterisation of the mutational landscape of the disease. These analyses have identified novel cancer genes found to be recurrently mutated in breast cancer (Shah et al, 2009, 2012; Banerji et al, 2012; Ellis et al, 2012; Stephens et al, 2012; TCGA, 2012). Although mutations in many of these genes are relatively infrequent, specific patterns of somatic mutations can be grouped according to their association with cellular pathways, underlying tumour biology and distinct clinical phenotypes. Furthermore, these studies have demonstrated the extent of heterogeneity across breast cancer genomes and have allowed further exploration into the role of intratumour heterogeneity (Shah et al, 2009, 2012; Nik-Zainal et al, 2012). The limitation of most of these first generation sequencing studies is the relatively modest number of samples analysed making it difficult to integrate this information with other IHC-based or expression-based classifiers.
Moving towards an integrated classification: METABRIC
All breast carcinomas show significant genetic diversity due to both inherited genetic variation and acquired genomic aberrations (Table I). Inherited variants consist of single-nucleotide polymorphisms (SNPs) and copy number variants (CNVs) and these changes form the background germline genetic landscape of the individual where a cancer might develop. Somatic genomic changes, which include single-nucleotide variants (mutations) and copy number aberrations (CNAs) are acquired and contribute to the initiation and progression of sporadic breast cancers. Genomic aberrations can contribute to carcinogenesis by inducing abnormal gene expression. Through the integrated analysis of both genomic and transcriptomic data across large numbers of breast cancers, the impact of genomic aberrations on the transcriptome can be appreciated. We have recently used this approach to characterise the genomic and transcriptomic architecture of 2000 breast tumours as part of METABRIC (Molecular Taxonomy of Breast Cancer International Consortium) (Curtis et al, 2012).
Table 1. Definition of genomic alterations.
| Genomic alterations | Definition | Description | |
|---|---|---|---|
| SNP | Single-nucelotide polymorphism | Germiline | Inherited genetic variation in the DNA sequence that occurs when a single nucleotide is altered |
| CNV | Copy number variant | Germline | Inherited alteration of DNA that results in an abnormal number of copies of one or more segments of DNA (1 kilobase or larger) |
| SNV | Single-nucleotide variant | Somatic | Acquired genetic variation in the DNA sequence that occurs when a single nucleotide is altered (i.e., point mutation) |
| CNA | Copy number aberration | Somatic | Acquired alteration of DNA that results in an abnormal number of copies of one or more segments of DNA (1 kilobase or larger) |
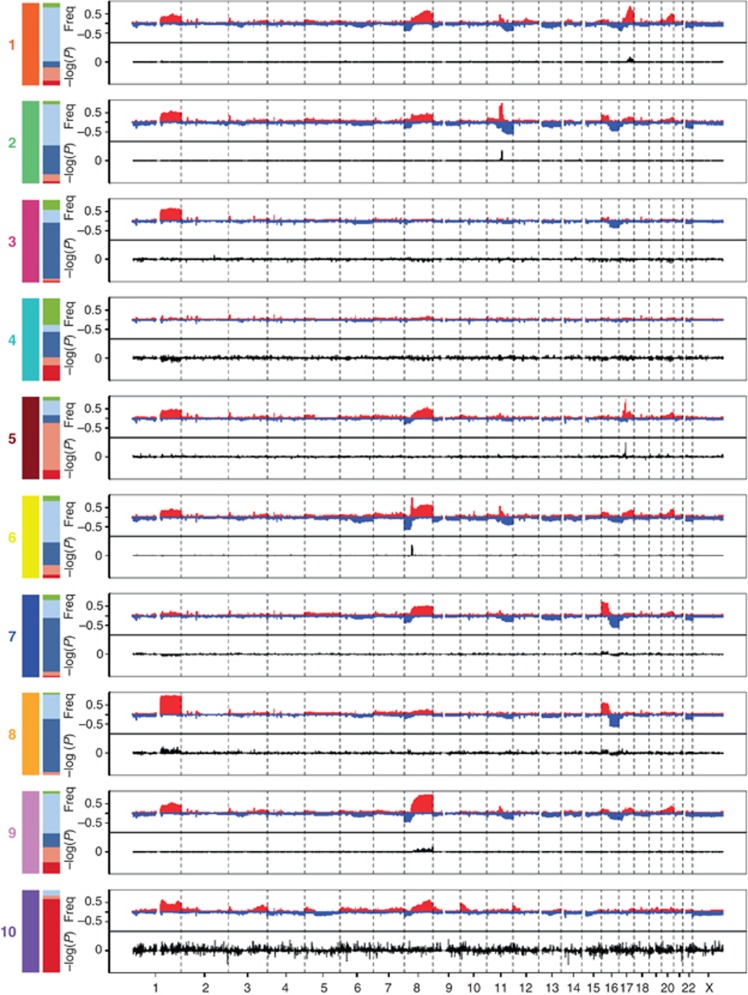
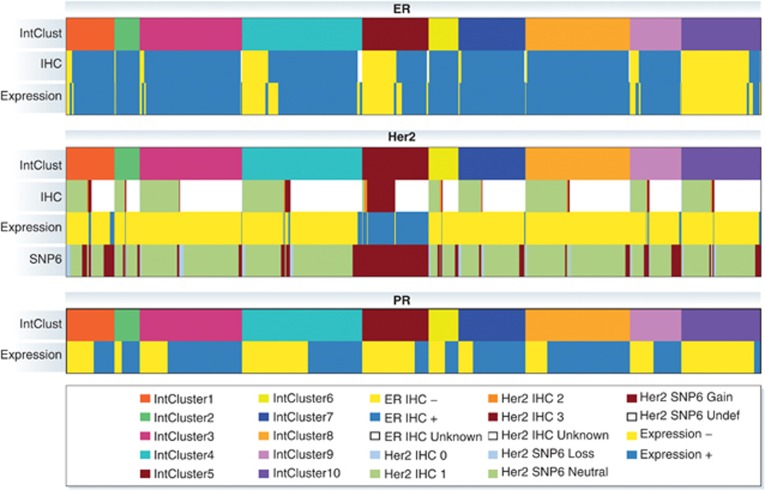
In this analysis, both germline variants (CNVs and SNPs) and somatic aberrations (CNAs) were found to be associated with alterations in gene expression. However, CNAs accounted for the greatest variability in gene expression. Somatic CNAs were shown to modify the expression of genes located both in cis (nearby to the genomic aberration) and trans (distant to the genomic aberration), but the effects of cis-acting CNAs dominated. Clustering analysis of joint copy number and gene expression data from the cis-associated genes revealed 10 novel molecular subgroups (Figure 1; Table II). The 10 integrative clusters (IntClust 1–10) were each associated with distinct CNAs and gene expression changes (Figure 1). These clusters clearly demonstrated the heterogeneity present within tumours classified according to ER, PR and HER2 expression, and they divided all of the previously identified intrinsic subtypes into separate groups (Figure 2). Furthermore, the 10 groups were associated with distinct clinical features and outcomes (Figure 3). Here, we will provide an overview of each of the novel subtypes, including an analysis of the distribution of mutations in the TCGA data set (TCGA, 2012) (Figure 4), and summarise the new insights gained from this classification relating to the underlying biology and potential molecular drivers in each group.
Figure 1.
The genomic and transcriptomic landscape of the 10 integrative clusters. Genome-wide frequencies (Freq) of somatic copy number alterations (Y axis, upper plot) and the subtype-specific association (−log10 P-value) of gene expression (Y axis, bottom plot) based on the differential expression in each of the 10 integrative clusters compared to the rest. Regions of copy number gain are indicated in red and regions of loss in blue in the frequency plot (upper plot). Overexpressed genes are represented as positive and underexpressed genes as negative (lower plot). The left bars show the composition of each cluster in terms of the intrinsic subtypes (red: basal, pink: HER2, dark blue: luminal A, light blue: luminal B and green: normal-like).
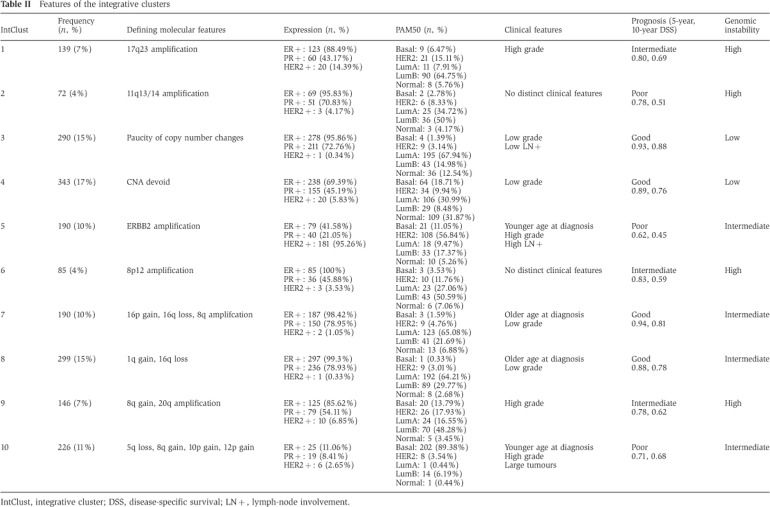
Table 2. Features of the integrative clusters.
IntClust, integrative cluster; DSS, disease-specific survival; LN+, lymph-node involvement.
Figure 2.
Relationship between the 10 integrative clusters and ER expression, HER2 expression and PR expression. IHC, immunohistochemistry; Expression, mRNA gene expression; SNP6=copy number alteration as assessed by Affymetrix SNP 6.0 array.
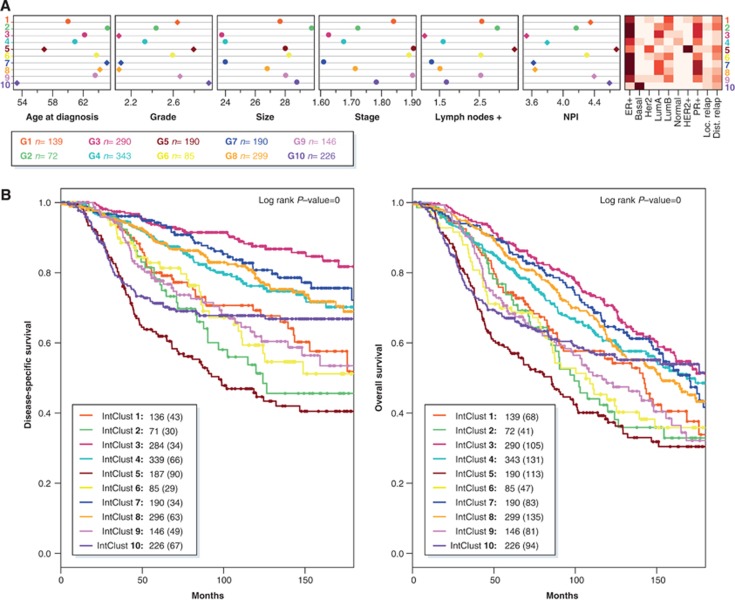
Figure 3.
The 10 integrative clusters represent distinct entities. (A) Distinct clinical features across the 10 integrated clusters. In the left plot, the coloured diamonds mark clinical features demonstrating significant subtype-specific associations. NPI, Nottingham prognostic index. The intensity of the colours in the right plot represents the frequency of each variable in every cluster. (B) Kaplan–Meier curves of disease-specific survival (left) and overall survival (right) across the 10 integrated clusters (truncated at 15 years). For each cluster, the number of cases at risk is indicated as well as the total number of deaths (in parentheses).
Figure 4.
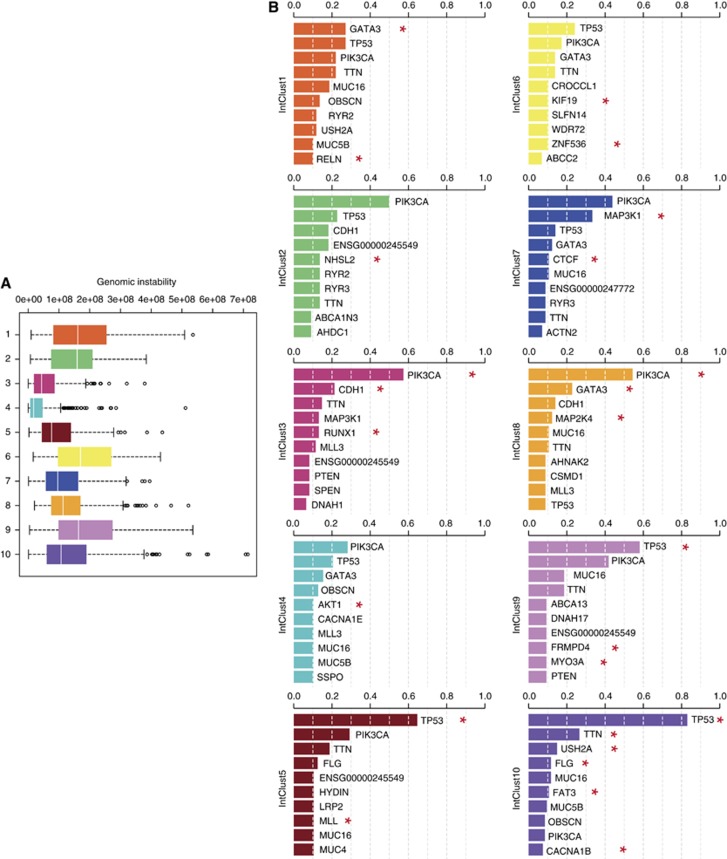
The genomic instability and mutational landscape of the 10 breast cancer subtypes. (A) Genomic instability across the 10 integrative clusters. Genomic instability was measured by the area under the segmented means of the log ratios. (B) Somatic mutation spectrum across the 10 integrative clusters. The Cancer Genome Atlas Network has characterised the mutational landscape of ∼500 breast cancers, also classified into the 10 integrative clusters, using exome sequencing (TCGA, 2012). The top 10 genes mutated in each integrative cluster are represented. Mutational frequency is displayed on the Y axis. Red asterisks indicate genes where the mutational frequency shows a significant subtype-specific association.
The ten integrative clusters
IntClust 1
Integrative cluster 1 is constituted by ER-positive tumours, predominantly classified into the luminal B intrinsic subtype. The subgroup typically has an intermediate prognosis, similar to that of IntClust 6 and 9 (Figure 3). All encompass a high proportion of higher proliferation ER+/luminal B tumours, and are characterised by relatively high levels of genomic instability (Figure 4). The defining molecular feature of IntClust 1 is amplification of the 17q23 locus (Figure 1), a region of amplification previously well described (Sinclair et al, 2003; Parssinen et al, 2007). IntClust 1 also has the highest prevalence of GATA3 mutations across all of the 10 clusters (Figure 4). These features separate IntClust 1 tumours from other ER-positive tumours previously grouped together within the luminal B intrinsic subtype.
Amplification of 17q23 in these tumours is associated with cis-driven overexpression of several adjacent genes including RPS6KB1, PPM1D, PTRH2 and APPBP2 (Figure 1). In particular, RPS6KB1 (ribosomal protein S6 kinase 1) and PPM1D (protein phosphatase 1D), show high cis outlying expression in this group and both have previously been implicated as potential oncogenic drivers (Sinclair et al, 2003). RPS6KB1 is a serine/threonine protein kinase that acts downstream of mammalian target of rapamycin (mTOR) signalling to regulate cell cycle, cell growth, proliferation and migration through translational control (Fingar et al, 2002, 2004; Hannan et al, 2003). PPM1D is a p53-inducible serine/threonine protein phosphatase known to dephosphorylate p38 mitogen-activated protein kinase (MAPK) and inhibit p38 MAPK-dependent phosphorylation of p53 leading to downregulation of p53-dependent transcription, inhibition of cell-cycle arrest and apoptosis (Bulavin et al, 2002). Both of these oncogene candidates represent novel potential therapeutic targets, highlighting the importance of understanding key genomic drivers within the subtypes and using this information to improve substratification within the disease.
IntClust 2
Integrative cluster 2 is comprised of ER-positive tumours and includes both luminal A and luminal B tumours. Remarkably, this subgroup is associated with the worst prognosis of all ER-positive tumours with a 10-year disease-specific survival rate of only around 50% (Figure 3). The defining molecular feature of this subtype is amplification of 11q13/14 (Figure 1) showing a characteristic ‘firestorm’ pattern identified by clustered narrow peaks of relatively high copy number gains (Hicks et al, 2006). This is reflected in the relatively high levels of genomic instability in this group (Figure 4). Amplification of 11q13/14 is well recognised in breast cancer, with several known and putative driver genes residing in this region including CCND1 (11q13.3), EMSY (11q13.5) and PAK1 (11q14.1) (Hughes-Davies et al, 2003; Santarius et al, 2010). Analysis of copy number data suggests two separate amplicons in this region; one amplicon centred around CCND1 at 11q13.3 and the other spanning UVRAG-GAB2 between 11q13.5 and 11q14.1 encompassing multiple genes that show strong cis outlying gene expression including PAK1, RSF1, EMSY, C11orf67 and INTS4 (Figure 1). Distinguishing a single driver in the region of 11q13/14 is challenging as the majority of individuals in this subgroup have amplifications involving multiple genes, suggesting that a combination of drivers are likely to be important rather than just a single oncogene. Pathway analysis in this subtype shows enrichment of genes involved in cell-cycle regulation, particularly the G1/S transition as exemplified by CCND1. These alterations are likely to explain the aggressive pathophysiology of this cluster and emphasise the importance of identifying this poor prognostic group within ER-positive subtypes.
IntClust 3
Integrative cluster 3 is composed primarily of luminal A cases and is enriched for histopathological subtypes that have a good prognosis such as invasive lobular and tubular carcinomas. Clinically, individuals within this subtype often present with small low-grade tumours and a low incidence of regional lymph-node involvement (Figure 3). At the molecular level, the subtype is characterised by low genomic instability, a very low prevalence of TP53 mutations, and a paucity of copy number and cis-acting alterations (Figures 1 and 4). However, of note, tumours within this subtype have the highest frequency of PIK3CA, CDH1 and RUNX1 mutations (Figure 4). Importantly, the subgroup is associated with the best prognosis of all the 10 integrative clusters with a 10-year disease-specific survival of around 90% (Figure 3). The excellent prognosis of this subtype emphasises the importance of identifying this cluster within the previously defined luminal A intrinsic subtype, as these individuals represent a distinct group that could potentially be spared treatment with systemic chemotherapy.
IntClust 4
Integrative cluster 4 is a unique cluster incorporating both ER-positive (n=238/343) and ER-negative (n=105/343) cases, including 26% of all triple negative tumours, and a mixture of intrinsic subtypes including basal-like cases (Table II). Importantly, the subtype is associated with favourable outcome and a 10-year disease-specific survival of around 80% (Figure 3). Similarly to IntClust 3, IntClust4, the largest subtype of breast cancer (up to 17% of cases), is characterised molecularly by low levels of genomic instability and a ‘CNA-devoid’ flat copy number landscape (Figures 1 and 4). Around 20% of cases within this subtype demonstrate deletions at the T-cell receptor (TCR) loci on chromosomes 7 (TRG) and 14 (TRA), in the background of an otherwise genomically quiescent subtype. Many of the tumours within this subgroup show evidence of extensive lymphocytic infiltration and the observed deletions are the consequence of the somatic TCR rearrangement present in the infiltrating T cells. This was previously reported using single-cell sequencing in one case of triple negative breast cancer with extensive lymphocytic infiltration (Navin et al, 2011), but is now demonstrated using array profiling and a fairly large number of tumours. The genomic copy number loss at the TCR loci is associated in trans with an immune response expression signature mirroring the lymphocytic infiltration and probably explaining the favourable prognosis seen, in particular, for the triple negative tumours classified into this subtype. These findings, in a subset of basal triple negative tumours, support earlier observations using gene expression analysis (Teschendorff et al, 2007; Teschendorff and Caldas, 2008) and are also corroborated by a recent combined analysis of imaging and expression data (Yuan et al, 2012). The observations suggest that the presence of mature T lymphocytes in the tumour represents a specific immunological response to the cancer, a finding that could potentially be exploited in the development of future therapeutics.
IntClust 5
Integrative cluster 5 encompasses the ERBB2 amplified cancers composed of both HER2-enriched ER-negative (58%) and luminal ER-positive cases (42%). Women in the METABRIC study were enrolled before the general availability of trastuzumab, and as expected, this group demonstrated the worst disease-specific survival at 10 years of around 45% (Figure 3). In keeping with common clinical features of HER2-positive breast cancers, individuals within this subtype often present at a younger age, with high-grade tumours and involvement of regional lymph nodes (Figure 3). In addition to specific ERBB2 amplification at 17q12 (Figure 1), these tumours demonstrate intermediate levels of genomic instability and a high proportion of TP53 mutations (in >60% cases) (Figure 4). In contrast to the HER2-enriched intrinsic subtype, the IntClust 5 identifies almost all cases with ERBB2 amplification, thus grouping all individuals that might benefit from HER2-related targeted therapy into a single subtype.
IntClust 6
Integrative cluster 6 represents a distinct subgroup of ER-positive tumours, comprising both luminal A and luminal B cases. Clinically, this cluster shows an intermediate prognosis and a 10-year disease-specific survival of around 60% (Figure 3). Molecularly, this subtype is characterised by specific amplification of the 8p12 locus (Figure 1) and high levels of genomic instability (Figure 4). Notably, tumours within this cluster demonstrate the lowest levels of PIK3CA mutations across all of the ER-positive cancers (Figure 4). The genomic landscape is dominated by cis-acting alterations associated with the 8p12 amplicon, a region previously known to be commonly amplified in ER-positive breast cancers, which encompasses the known oncogenic driver ZNF703 (Holland et al, 2011; Sircoulomb et al, 2011; Slorach et al, 2011; Figure 1). ZNF703 (zinc finger protein 703) is a transcriptional repressor that regulates genes involved in key cancer phenotypes such as increased proliferation, invasion and the balance of the progenitor/stem cell compartment (Holland et al, 2011). Similarly to IntClust 2, identification of this more aggressive group of ER-positive/HER2-negative tumours within the luminal intrinsic subtypes may assist in improving the stratification and prediction of outcome in women with ER-positive disease.
IntClust 7
Integrative cluster 7 is comprised predominately of ER-positive luminal A tumours and identifies a good prognostic subgroup with 10-year disease-specific survival rates of around 80% (Figure 3). As for cases in IntClust 3, the majority of individuals within this cluster present with low-grade well-differentiated tumours that display both ER and PR positivity (Figure 3). However, unlike the paucity of copy number changes seen in association with IntClust 3, IntClust 7 is characterised by intermediate levels of genomic instability, specific 16p gain and 16q loss, as well as a higher frequency of 8q amplification (Figures 1 and 4). Interestingly, tumours within IntClust 7 also demonstrate the highest frequency of MAP3K1 and CTCF mutations across all clusters (Figure 4).
IntClust 8
Integrative cluster 8 shares similarities with IntClust7 and encompasses ER-positive tumours predominately of the luminal A intrinsic subtype. As described for IntClust 7, individuals within IntClust 8 also present with low-grade well-differentiated tumours (Figure 3), and the subgroup is associated with a good prognosis and similar 10-year disease-specific survival rates of around 80% (Figure 3). This subgroup, however, is characterised molecularly by the classical 1q gain/16q loss event that corresponds to a common unbalanced translocation event (Kokalj-Vokac et al, 1993; Russnes et al, 2010), unlike IntClust 7 that lacks the 1q alteration but maintains the 16q changes (Figure 1). Although genomic instability is generally lower in well-differentiated breast carcinomas (Figure 4), the frequent identification of 1q gains and 16q losses that characterise the IntClust 8 subtype is well recognised in low-grade invasive ductal carcinomas (Roylance et al, 1999). Furthermore, tumours within IntClust 8 demonstrate high levels of PIK3CA, GATA3 and MAP2K4 mutations (Figure 4). Together with IntClust 3 and IntClust 7, IntClust 8 separates tumours previously grouped under the luminal A intrinsic subtype into three distinct biological subgroups driven by specific genomic aberrations (Figure 3).
IntClust 9
Integrative cluster 9 is comprised of a mixture of intrinsic subtypes, but includes a large number of ER-positive cases of the luminal B subgroup. Similarly to IntClust 6, IntClust 9 shows an intermediate prognosis with a 10-year disease-specific survival of around 60% (Figure 3). This cluster is characterised by high levels of genomic instability and the highest level of TP53 mutations among the ER-positive subtypes (Figure 4). Molecularly, it is defined by 8q cis-acting alterations and 20q amplification (Figure 1). In conjunction with IntClust 1 and IntClust 6, IntClust 9 shows a high proportion of cases with deletions of PPP2R2A, on chromosome 8p. PP2R2A (protein phosphatase 2 regulatory subunit B alpha) is a serine/threonine phosphatase integral to several signal transduction pathways. Loss of transcript expression of PPP2R2A appears to predominate in mitotic ER-positive breast cancers typically of the luminal B intrinsic subtype, such as those grouped into IntClust 1, 6 and 9. Mutations and methylation silencing of PPP2R2A have recently been reported in other solid malignancies (Tan et al, 2010; McConechy et al, 2011), suggesting a possible role for PPP2R2A as a putative tumour suppressor in tumours associated with the luminal B subtype, in particular those associated with this integrative cluster.
IntClust 10
Integrative cluster 10 incorporates mostly triple negative tumours (n=190/320 classify into this cluster) from the core basal-like intrinsic subtype. Although the subtype represents a high-risk group in the first 5 years after diagnosis, beyond 5 years the prognosis for this subgroup is relatively good (Blows et al, 2010; Figure 3). Clinically, these women usually present at a younger age with high-grade and poorly differentiated tumours (Figure 3). These breast cancers have the highest rates of TP53 mutations despite displaying only intermediate levels of genomic instability (Figure 4). Molecularly, the subtype is characterised by copy number alterations involving 5q loss and gains at 8q, 10p and 12p (Figure 1). In particular, 5q deletions are associated with a basal-specific trans gene expression module enriched for many checkpoint, DNA damage repair and apoptosis genes such as AURKB, BCL2, BUB1, CDCA3, CDCA4, CDC20, CDC45, CHEK1, FOXM1, HDAC2, IGF1R, KIF2C, KIFC1, MTHFD1L, RAD51AP1, TTK and UBE2C. These transcriptional changes reflect the high mitotic index typically associated with this subgroup. Of note, TTK (MPS1) a dual specificity kinase that assists AURKB in chromosome alignment during mitosis is upregulated in association with 5q loss and high levels of TTK have recently been reported to promote aneuploidy in breast cancer (Daniel et al, 2011). These findings suggest that 5q deletions modulate the landscape of genomic instability and cell-cycle regulation alterations observed within this subgroup.
Conclusions
The future of breast cancer classification will involve multiple levels of assessment incorporating clinical information about the patient, tumour-specific information determined by histopathology, and molecular information revealed by genomic, transcriptomic and proteomic profiling to provide subtype-specific diagnostic, prognostic and predictive tests. At the genomic level, next generation sequencing will allow the complete genomic landscape of somatic mutations, structural rearrangements, copy number alterations and epigenetic events to be assessed adding increasing complexity, yet helping to further elucidate the mechanisms driving each subtype. As well as focussing on understanding intertumour heterogeneity, the context of intratumour heterogeneity will also require consideration in the interpretation and implementation of molecular classification systems (Caldas, 2012). Integrating multiple layers of complex data into robust classifiers and the clinical implementation of these into the routine management of breast cancer patients will present many challenges. However, rapid progress is being made, allowing us to move closer to the realisation of individualising the diagnosis and treatment of breast cancer.
Through the integrated analysis of CNAs and their effect on gene expression, novel molecular subgroups have been identified that help refine our understanding of breast cancer heterogeneity. The integrative clusters provide important biological insights into the potential molecular drivers and pathways underlying certain groups, and these have distinct implications for the rationale development of targeted therapeutics. In addition the analysis highlights key subtypes, such as those devoid of somatic CNAs that will require more extensive molecular profiling. As we gain a better appreciation of the heterogeneity of breast cancer, it is clear that thousands of patients must be studied to fully appreciate the clinical implications of novel and rare subgroups of the disease. Furthermore, more sophisticated model systems, both in vitro and in vivo, including the use of xenograft tumours derived directly from primary clinical material, will be needed to dissect the biological complexities of this heterogeneity. These approaches will provide the opportunity to understand the molecular events and pathways underpinning each group, and potentially allow the identification of the cell of origin or tumour-initiating cell in each subtype. These findings will undoubtedly lead to fundamental advances in our approach to the classification, biological characterisation and management of breast cancer.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abd El-Rehim DM, Ball G, Pinder SE, Rakha E, Paish C, Robertson JF, Macmillan D, Blamey RW, Ellis IO (2005) High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int J Cancer 116: 340–350 [DOI] [PubMed] [Google Scholar]
- Ali HR, Dawson SJ, Blows FM, Provenzano E, Pharoah PD, Caldas C (2012) Aurora kinase A outperforms Ki67 as a prognostic marker in ER-positive breast cancer. Br J Cancer 106: 1798–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY, Sougnez C, Zou L, Cortes ML, Fernandez-Lopez JC, Peng S, Ardlie KG, Auclair D, Bautista-Pina V, Duke F, Francis J, Jung J, Maffuz-Aziz A et al. (2012) Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 486: 405–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blows FM, Driver KE, Schmidt MK, Broeks A, van Leeuwen FE, Wesseling J, Cheang MC, Gelmon K, Nielsen TO, Blomqvist C, Heikkila P, Heikkinen T, Nevanlinna H, Akslen LA, Begin LR, Foulkes WD, Couch FJ, Wang X, Cafourek V, Olson JE et al. (2010) Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med 7: e1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruna A, Greenwood W, Le Quesne J, Teschendorff A, Miranda-Saavedra D, Rueda OM, Sandoval JL, Vidakovic AT, Saadi A, Pharoah P, Stingl J, Caldas C (2012) TGFbeta induces the formation of tumour-initiating cells in claudinlow breast cancer. Nat Commun 3: 1055. [DOI] [PubMed] [Google Scholar]
- Bulavin DV, Demidov ON, Saito S, Kauraniemi P, Phillips C, Amundson SA, Ambrosino C, Sauter G, Nebreda AR, Anderson CW, Kallioniemi A, Fornace AJ Jr, Appella E (2002) Amplification of PPM1D in human tumors abrogates p53 tumor-suppressor activity. Nat Genet 31: 210–215 [DOI] [PubMed] [Google Scholar]
- Caldas C (2012) Cancer sequencing unravels clonal evolution. Nat Biotechnol 30: 408–410 [DOI] [PubMed] [Google Scholar]
- Callagy G, Cattaneo E, Daigo Y, Happerfield L, Bobrow LG, Pharoah PD, Caldas C (2003) Molecular classification of breast carcinomas using tissue microarrays. Diagn Mol Pathol 12: 27–34 [DOI] [PubMed] [Google Scholar]
- Cardoso F, Van’t Veer L, Rutgers E, Loi S, Mook S, Piccart-Gebhart MJ (2008) Clinical application of the 70-gene profile: the MINDACT trial. J Clin Oncol 26: 729–735 [DOI] [PubMed] [Google Scholar]
- Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, Watson M, Davies S, Bernard PS, Parker JS, Perou CM, Ellis MJ, Nielsen TO (2009) Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 101: 736–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, Chen F, Feiler H, Tokuyasu T, Kingsley C, Dairkee S, Meng Z, Chew K, Pinkel D, Jain A, Ljung BM et al. (2006) Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 10: 529–541 [DOI] [PubMed] [Google Scholar]
- Chin SF, Teschendorff AE, Marioni JC, Wang Y, Barbosa-Morais NL, Thorne NP, Costa JL, Pinder SE, van de Wiel MA, Green AR, Ellis IO, Porter PL, Tavare S, Brenton JD, Ylstra B, Caldas C (2007) High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol 8: R215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Langerod A, Green A, Provenzano E, Wishart G et al. (2012) The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 486: 346–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J, Coulter J, Woo JH, Wilsbach K, Gabrielson E (2011) High levels of the Mps1 checkpoint protein are protective of aneuploidy in breast cancer cells. Proc Natl Acad Sci USA 108: 5384–5389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson SJ, Makretsov N, Blows FM, Driver KE, Provenzano E, Le Quesne J, Baglietto L, Severi G, Giles GG, McLean CA, Callagy G, Green AR, Ellis I, Gelmon K, Turashvili G, Leung S, Aparicio S, Huntsman D, Caldas C, Pharoah P (2010) BCL2 in breast cancer: a favourable prognostic marker across molecular subtypes and independent of adjuvant therapy received. Br J Cancer 103: 668–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson SJ, Provenzano E, Caldas C (2009) Triple negative breast cancers: clinical and prognostic implications. Eur J Cancer 45(Suppl 1): 27–40 [DOI] [PubMed] [Google Scholar]
- Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, Abbott RM, Hoog J, Dooling DJ, Koboldt DC, Schmidt H, Kalicki J, Zhang Q, Chen L, Lin L, Wendl MC et al. (2010) Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464: 999–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, Ng S, Lin L, Crowder R, Snider J, Ballman K, Weber J, Chen K, Koboldt DC, Kandoth C, Schierding WS et al. (2012) Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486: 353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis P (2003) WHO Classification of tumours. Pathology and Genetics of Tumours of the Breast and Female Genital Organs Lyon Press: Lyon, 13–59 [Google Scholar]
- Elston CW, Ellis IO, Pinder SE (1999) Pathological prognostic factors in breast cancer. Crit Rev Oncol Hematol 31: 209–223 [DOI] [PubMed] [Google Scholar]
- Fingar DC, Richardson CJ, Tee AR, Cheatham L, Tsou C, Blenis J (2004) mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol Cell Biol 24: 200–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingar DC, Salama S, Tsou C, Harlow E, Blenis J (2002) Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev 16: 1472–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur GA, Pearson RB, Hannan RD (2003) mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol 23: 8862–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, Somerfield MR, Hayes DF, Bast RC Jr. (2007) American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol 25: 5287–5312 [DOI] [PubMed] [Google Scholar]
- Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, Fridlyand J, Sahin A, Agarwal R, Joy C, Liu W, Stivers D, Baggerly K, Carey M, Lluch A, Monteagudo C, He X, Weigman V, Fan C, Palazzo J et al. (2009) Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res 69: 4116–4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J, Krasnitz A, Lakshmi B, Navin NE, Riggs M, Leibu E, Esposito D, Alexander J, Troge J, Grubor V, Yoon S, Wigler M, Ye K, Borresen-Dale AL, Naume B, Schlicting E, Norton L, Hagerstrom T, Skoog L, Auer G et al. (2006) Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res 16: 1465–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland DG, Burleigh A, Git A, Goldgraben MA, Perez-Mancera PA, Chin SF, Hurtado A, Bruna A, Ali HR, Greenwood W, Dunning MJ, Samarajiwa S, Menon S, Rueda OM, Lynch AG, McKinney S, Ellis IO, Eaves CJ, Carroll JS, Curtis C et al. (2011) ZNF703 is a common Luminal B breast cancer oncogene that differentially regulates luminal and basal progenitors in human mammary epithelium. EMBO Mol Med 3: 167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, Nobel A, Parker J, Ewend MG, Sawyer LR, Wu J, Liu Y, Nanda R, Tretiakova M, Ruiz Orrico A, Dreher D et al. (2006) The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics 7: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes-Davies L, Huntsman D, Ruas M, Fuks F, Bye J, Chin SF, Milner J, Brown LA, Hsu F, Gilks B, Nielsen T, Schulzer M, Chia S, Ragaz J, Cahn A, Linger L, Ozdag H, Cattaneo E, Jordanova ES, Schuuring E et al. (2003) EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell 115: 523–535 [DOI] [PubMed] [Google Scholar]
- Jacquemier J, Ginestier C, Rougemont J, Bardou VJ, Charafe-Jauffret E, Geneix J, Adelaide J, Koki A, Houvenaeghel G, Hassoun J, Maraninchi D, Viens P, Birnbaum D, Bertucci F (2005) Protein expression profiling identifies subclasses of breast cancer and predicts prognosis. Cancer Res 65: 767–779 [PubMed] [Google Scholar]
- Kokalj-Vokac N, Alemeida A, Gerbault-Seureau M, Malfoy B, Dutrillaux B (1993) Two-color FISH characterization of i(1q) and der(1;16) in human breast cancer cells. Genes Chromosomes Cancer 7: 8–14 [DOI] [PubMed] [Google Scholar]
- Konecny G, Pauletti G, Pegram M, Untch M, Dandekar S, Aguilar Z, Wilson C, Rong HM, Bauerfeind I, Felber M, Wang HJ, Beryt M, Seshadri R, Hepp H, Slamon DJ (2003) Quantitative association between HER-2/neu and steroid hormone receptors in hormone receptor-positive primary breast cancer. J Natl Cancer Inst 95: 142–153 [DOI] [PubMed] [Google Scholar]
- Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA (2011) Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 121: 2750–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, Feleppa F, Huschtscha LI, Thorne HJ, Fox SB, Yan M, French JD, Brown MA, Smyth GK, Visvader JE, Lindeman GJ (2009) Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med 15: 907–913 [DOI] [PubMed] [Google Scholar]
- McConechy MK, Anglesio MS, Kalloger SE, Yang W, Senz J, Chow C, Heravi-Moussavi A, Morin GB, Mes-Masson AM, Carey MS, McAlpine JN, Kwon JS, Prentice LM, Boyd N, Shah SP, Gilks CB, Huntsman DG (2011) Subtype-specific mutation of PPP2R1A in endometrial and ovarian carcinomas. J Pathol 223: 567–573 [DOI] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, Muthuswamy L, Krasnitz A, McCombie WR, Hicks J, Wigler M (2011) Tumour evolution inferred by single-cell sequencing. Nature 472: 90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, Akslen LA, Ragaz J, Gown AM, Gilks CB, van de Rijn M, Perou CM (2004) Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res 10: 5367–5374 [DOI] [PubMed] [Google Scholar]
- Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, Davies SR, Snider J, Stijleman IJ, Reed J, Cheang MC, Mardis ER, Perou CM, Bernard PS, Ellis MJ (2010) A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res 16: 5222–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J, Ramakrishna M, Shlien A, Cooke SL, Hinton J, Menzies A, Stebbings LA, Leroy C, Jia M, Rance R, Mudie LJ, Gamble SJ et al. (2012) The life history of 21 breast cancers. Cell 149: 994–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N (2004) A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351: 2817–2826 [DOI] [PubMed] [Google Scholar]
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS (2009) Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 27: 1160–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parssinen J, Kuukasjarvi T, Karhu R, Kallioniemi A (2007) High-level amplification at 17q23 leads to coordinated overexpression of multiple adjacent genes in breast cancer. Br J Cancer 96: 1258–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D (2000) Molecular portraits of human breast tumours. Nature 406: 747–752 [DOI] [PubMed] [Google Scholar]
- Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM (2010) Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 12: R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakha EA, Reis-Filho JS, Baehner F, Dabbs DJ, Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR, Palacios J, Richardson AL, Schnitt SJ, Schmitt FC, Tan PH, Tse GM, Badve S, Ellis IO (2010) Breast cancer prognostic classification in the molecular era: the role of histological grade. Breast Cancer Res 12: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roylance R, Gorman P, Harris W, Liebmann R, Barnes D, Hanby A, Sheer D (1999) Comparative genomic hybridization of breast tumors stratified by histological grade reveals new insights into the biological progression of breast cancer. Cancer Res 59: 1433–1436 [PubMed] [Google Scholar]
- Russnes HG, Vollan HK, Lingjaerde OC, Krasnitz A, Lundin P, Naume B, Sorlie T, Borgen E, Rye IH, Langerod A, Chin SF, Teschendorff AE, Stephens PJ, Maner S, Schlichting E, Baumbusch LO, Karesen R, Stratton MP, Wigler M, Caldas C et al. (2010) Genomic architecture characterizes tumor progression paths and fate in breast cancer patients. Sci Transl Med 2: 38ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS (2010) A census of amplified and overexpressed human cancer genes. Nat Rev Cancer 10: 59–64 [DOI] [PubMed] [Google Scholar]
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, Steidl C, Holt RA, Jones S, Sun M, Leung G, Moore R, Severson T, Taylor GA, Teschendorff AE, Tse K et al. (2009) Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461: 809–813 [DOI] [PubMed] [Google Scholar]
- Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, Bashashati A, Prentice LM, Khattra J, Burleigh A, Yap D, Bernard V, McPherson A, Shumansky K, Crisan A, Giuliany R et al. (2012) The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486: 395–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair CS, Rowley M, Naderi A, Couch FJ (2003) The 17q23 amplicon and breast cancer. Breast Cancer Res Treat 78: 313–322 [DOI] [PubMed] [Google Scholar]
- Sircoulomb F, Nicolas N, Ferrari A, Finetti P, Bekhouche I, Rousselet E, Lonigro A, Adelaide J, Baudelet E, Esteyries S, Wicinski J, Audebert S, Charafe-Jauffret E, Jacquemier J, Lopez M, Borg JP, Sotiriou C, Popovici C, Bertucci F, Birnbaum D et al. (2011) ZNF703 gene amplification at 8p12 specifies luminal B breast cancer. EMBO Mol Med 3: 153–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slorach EM, Chou J, Werb Z (2011) Zeppo1 is a novel metastasis promoter that represses E-cadherin expression and regulates p120-catenin isoform expression and localization. Genes Dev 25: 471–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98: 10869–10874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 100: 8418–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparano JA, Paik S (2008) Development of the 21-gene assay and its application in clinical practice and clinical trials. J Clin Oncol 26: 721–728 [DOI] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, Greenman CD, Jia M, Latimer C, Teague JW, Lau KW, Burton J, Quail MA, Swerdlow H, Churcher C, Natrajan R et al. (2009) Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462: 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, Yates LR, Papaemmanuil E, Beare D, Butler A, Cheverton A, Gamble J, Hinton J, Jia M, Jayakumar A, Jones D et al. (2012) The landscape of cancer genes and mutational processes in breast cancer. Nature 486: 400–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Lee PL, Li Z, Jiang X, Lim YC, Hooi SC, Yu Q (2010) B55beta-associated PP2A complex controls PDK1-directed myc signaling and modulates rapamycin sensitivity in colorectal cancer. Cancer Cell 18: 459–471 [DOI] [PubMed] [Google Scholar]
- TCGA. (2012) Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Caldas C (2008) A robust classifier of high predictive value to identify good prognosis patients in ER-negative breast cancer. Breast Cancer Res 10: R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C (2007) An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol 8: R157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415: 530–536 [DOI] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH et al. (2002) A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347: 1999–2009 [DOI] [PubMed] [Google Scholar]
- Weigelt B, Horlings HM, Kreike B, Hayes MM, Hauptmann M, Wessels LF, de Jong D, Van de Vijver MJ, Van’t Veer LJ, Peterse JL (2008) Refinement of breast cancer classification by molecular characterization of histological special types. J Pathol 216: 141–150 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Failmezger H, Rueda OM, Ali HR, Graf S, Chin SF, Schwarz RF, Curtis C, Dunning MJ, Bardwell H, Johnson N, Doyle S, Turashvili G, Provenzano E, Aparicio S, Caldas C, Markowetz F (2012) Quantitative image analysis of cellular heterogeneity in breast tumours complements genomic profiling. Sci Transl Med 4: 157ra143. [DOI] [PubMed] [Google Scholar]