Abstract
The agents responsible for transmissible spongiform encephalopathies (TSEs), or prion diseases, contain as a major component PrPSc, an abnormal conformer of the host glycoprotein PrPC. TSE agents are distinguished by differences in phenotypic properties in the host, which nevertheless can contain PrPSc with the same amino-acid sequence. If PrP alone carries information defining strain properties, these must be encoded by post-translational events. Here we investigated whether the glycosylation status of host PrP affects TSE strain characteristics. We inoculated wild-type mice with three TSE strains passaged through transgenic mice with PrP devoid of glycans at the first, second or both N-glycosylation sites. We compared the infectious properties of the emerging isolates with TSE strains passaged in wild-type mice by in vivo strain typing and by the standard scrapie cell assay in vitro. Strain-specific characteristics of the 79A TSE strain changed when PrPSc was devoid of one or both glycans. Thus infectious properties of a TSE strain can be altered by post-translational changes to PrP which we propose result in the selection of mutant TSE strains.
Keywords: glycosylation, phenotypic change, prion, transmissible spongiform encephalopathy
Introduction
The transmissible spongiform encephalopathies (TSEs) or prion diseases are a group of fatal, infectious, neurodegenerative diseases that include Creutzfeldt-Jakob disease (CJD) in humans, scrapie in sheep, bovine spongiform encephalopathy (BSE) in cattle and chronic wasting disease (CWD) in cervids. There are many distinct strains of TSEs that differ in incubation time, pathology, clinical presentation and biochemical properties in the infected host (Bruce, 2003; Morales et al, 2007). Remarkably, the infectious properties of many of these strains can be maintained and propagated indefinitely in the same host. On the other hand some strains have been shown to mutate, generating variants according to the characteristics of the environment in which they replicate (Kimberlin et al, 1987; Li et al, 2010). More than 15 strains have been experimentally derived from transmission of different scrapie, BSE or human CJD sources into a number of species such as rodents or sheep (Dickinson, 1976; Kimberlin and Walker, 1978; Bruce and Dickinson, 1987; Foster and Dickinson, 1988; Bruce et al, 1991; Manuelidis, 1998). Moreover, different natural strains have been identified in humans (Will, 2003; Ironside et al, 2005; Bishop et al, 2010), sheep (Bruce, 2003; Buschmann et al, 2004), cattle (Biacabe et al, 2004; Casalone et al, 2004) and most recently cervids (Angers et al, 2010).
In viral and microbial diseases different strains of the infectious agent are common. The emergence of new strains is associated with mutations in their nucleic acid genomes that give rise to infectious agents with an altered ability to invade the host, cause disease or resist drugs (Domingo and Gomez, 2007; Hawkey, 2008). The failure to detect any nucleic acid associated with TSE infectivity, however, renders the existence of TSE strains difficult to explain and has given rise to several hypotheses. The most popular of these, the prion hypothesis, proposes that the agent consists of misfolded isoforms (PrPSc) of the ubiquitous cellular glycoprotein, PrPC, which encode the phenotypic properties of TSE strains. PrPC and PrPSc differ in their conformation and biochemical properties, including resistance to proteolytic cleavage and solubility in detergents (Harris, 1999; Silveira et al, 2004). TSE replication is thought to depend on the capacity of PrPSc to infect a host, bind host PrPC and induce its misfolding in a self-perpetuating process (Prusiner, 1998).
If a protein is the only component of the infectious agent, it must be able to encode different information for each strain of the agent. If this is the case, the existence of TSE strains with well-defined, distinct infectious characteristics has to be considered a unique feature in microbiology, and understanding the nature of these strains and their ability to mutate is important in order to prevent new outbreaks of these lethal diseases in humans and animals (Manson et al, 2006; Telling, 2010). Given these challenges to the prion hypothesis, consideration is also given to the hypothesis that TSE agents have nucleic acid genomes (Rohwer, 1991; Chesebro, 1998; Somerville, 2002; Manuelidis, 2011).
To explain the existence of different TSE strains in the absence of any associated informational nucleic acid, it has been postulated that distinct PrPSc conformations may dictate strain characteristics (Bessen and Marsh, 1994; Peretz et al, 2002; Jones and Surewicz, 2005; Aguzzi et al, 2007; Angers et al, 2010). The quasi-species hypothesis (Eigen, 1996) predicts that PrPSc with different conformations may be present at low levels in an infectious inoculum and that the variant most suitable for replication in a particular host is selected to become the dominant component of the population (Collinge and Clarke, 2007; Li et al, 2010).
Murine PrP has two potential sites for N-linked glycosylation, at amino acids 180 and 196, which are variably occupied, giving rise to di-, mono- and unglycosylated PrPC and PrPSc. All three glycoforms are commonly observed within a host, however, the degree of glycosylation can vary within and between infected tissues (Somerville et al, 1997; Hill et al, 1999). The glycosylation status of host PrPC has been shown by us and others to be a key component in determining the infectious process in the brain (DeArmond et al, 1997; Neuendorf et al, 2004; Tuzi et al, 2008). Moreover, glycans on PrPC have a strong impact in dictating the propagation and/or transport of infectivity from the periphery to the CNS (Cancellotti et al, 2010). Glycosylation of PrPSc may also have an important role in defining strain characteristics (DeArmond et al, 1997; Aguzzi et al, 2007). However, amplification of two TSE strains (RML and 301C) using serial protein misfolding amplification (sPMCA) and enzymatically deglycosylated mouse PrPC in vitro has been demonstrated with limited phenotypic characterisation of the product, which failed to show any changes in TSE agent phenotype (Piro et al, 2009).
A possible role for N-linked glycans on PrPSc in determining TSE strain characteristics is indicated mainly by two observations: (i) in general, glycosylation is an important factor in determining and maintaining conformation, function and interactions of glycoproteins (O’Connor and Imperiali, 1996; Helenius and Aebi, 2001); (ii) different TSE strains are usually associated with different ratios of di-, mono- and unglycosylated PrP, and it has been suggested that these different glycotypes are responsible for determining infectious properties of the prion strain (Collinge et al, 1996; DeArmond et al, 1997). For this reason, PrPSc glycoform analysis in the infected host is one of the criteria used to distinguish between TSE strains (Collinge et al, 1996; Parchi et al, 1996; Casalone et al, 2004; Head et al, 2004).
We have previously developed gene-targeted mice in which endogenous PrPC was replaced with an altered PrPC sequence designed to prevent attachment of the sugars at the first (G1, N180T), second (G2, N196T) or both (G3, N180T and N196T) N-glycan sites (Cancellotti et al, 2005). As reported previously, G1, G2 and G3 transgenic mice inoculated with a number of classical murine TSE strains (79A, ME7 or 301C) accumulated novel forms of glycosylation-deficient PrPSc in their brains. We have demonstrated that host PrP plays a major role in TSE disease susceptibility and incubation period and that TSE strains differ dramatically in their requirements for host PrP glycosylation for TSE disease to occur. Moreover, glycosylation of PrPSc is not necessary for the transmission of TSE infectivity to a new host, and the glycotype of the host PrP has a major influence on the de novo generated PrPSc (Tuzi et al, 2008).
These novel sources of infectivity produced in this first study provide us with a valuable tool to investigate the effect of glycosylation of PrP associated with the infecting TSE strain to determine strain characteristics. To do so, we have injected these brain materials, which have strikingly different glycoform profiles, into wild-type mice and utilised our well-characterised in vivo strain-typing method to investigate TSE strain properties (Bruce and Dickinson, 1987; Bruce, 2003). In addition, the characteristics of the strains were assessed in vitro by the standard scrapie cell assay (SSCA) using strain-specific inhibitors (Mahal et al, 2008; Browning et al, 2011; Oelschlegel et al, 2012). Here we show that the infectious characteristics of the 79A strain changed dramatically after passage in mice with PrP lacking glycans, as determined by both strain typing and SSCA, and in some cases exhibiting phenotypic properties typical of the 139A strain (Dickinson, 1976; Dickinson et al, 1984). In contrast, the characteristics of the ME7 and 301C strains were not affected by the lack of one of the sugars on PrP. These results demonstrate that the presence or absence of oligosaccharides on host PrP sometimes influences the phenotypic variability of the infectious agent. They highlight a role of N-linked glycans in defining strain characteristics for some but not all TSE strains.
Results
Infectious isolates and mouse lines
Three TSE strains, 79A, ME7 and 301C, were previously passaged in glycosylation-deficient mice (the G1, G2 and G3 mice) and in 129Ola mice as wild-type mouse controls (Tuzi et al, 2008). Modifications to these strains, which occurred in the glycosylation-deficient hosts, are now analysed here by passage in a wild-type mouse panel and by examination of changes taking place at second pass in the transgenic mice.
To test the effect of N-linked glycan chains in determining TSE strain properties, brain homogenates from TSE-infected mice with altered PrP glycotype profiles (illustrated in Figure 1) were injected into the strain-typing panel of mice (C57, VM and CVF1) (Table I). The inocula were named after the mouse genotype from which the infectious brain was derived and the name of the strain used in the original inoculum. The inocula used were: G1-79A (79A passaged through a G1 host, with PrPSc lacking glycans at the first site); G2-79A (79A passaged through a G2 host with PrPSc lacking glycans at the second site); G3-79A (79A passaged through a G3 host with totally unglycosylated PrPSc); G2-ME7 (ME7 passaged through a G2 host); and G2-301C (301C passaged through a G2 host). The PrPSc glycotype profile of the inocula used was confirmed by western blotting analysis (Figure 1). All inocula were prepared from animals with confirmed clinical and pathological TSE disease or in one case with PrPSc deposition in the brain but no recorded clinical signs of TSE infection (Table I). No transmissions were performed from brains from G1 and G3 mice infected with ME7 or 301C as these mice had shown no clinical or pathological signs of TSE infection (Tuzi et al, 2008). Because the effect of removing sugars from PrPSc on the infectious characteristics of the different strains was unknown and unpredictable, two separate infectious inocula expressing glycosylation-deficient PrPSc were analysed in each experiment. A single inoculum from wild-type 79A, ME7 or 301C (termed 129-79A, 129-ME7 and 129-301C, respectively) was considered sufficient because the infectious properties in these cases are well characterised.
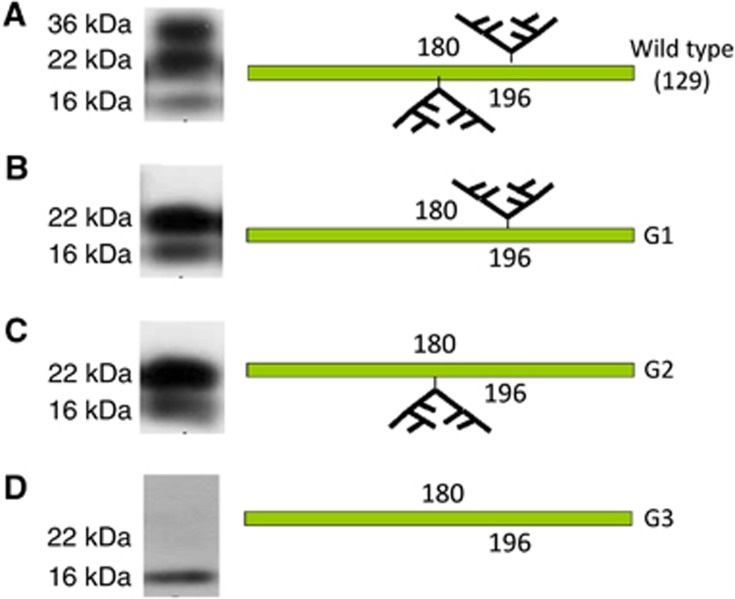
Figure 1.
Western blot analysis of differentially glycosylated PrPSc in the inocula. Example of PrPSc from 79A infected brains after passage through 129Ola wild-type, G1, G2 and G3 mice, and a diagram of the glycosylation pattern of PrP in the four mouse genotypes.(A) Wild-type brain after PK digestion showing PrPSc that comprises di, mono and unglycosylated isoforms because both glycosylation sites at codons 180 and 196 are preserved. (B) G1 inoculum expressing PrPSc lacking the diglycosylated isoform because the 180 glycosylation site has been abolished. (C) G2 inoculum with PrPSc lacking the diglycosylated isoform because the 196 glycosylation site has been abolished. (D) G3 inoculum expressing totally unglycosylated PrPSc because both 180 and 196 glycosylation sites have been modified.
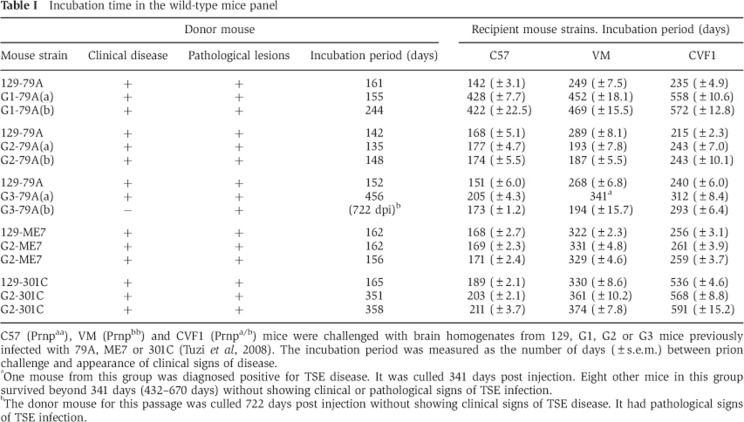
Table 1. Incubation time in the wild-type mice panel.
C57 (Prnpaa), VM (Prnpbb) and CVF1 (Prnpa/b) mice were challenged with brain homogenates from 129, G1, G2 or G3 mice previously infected with 79A, ME7 or 301C (Tuzi et al, 2008). The incubation period was measured as the number of days (±s.e.m.) between prion challenge and appearance of clinical signs of disease.
aOne mouse from this group was diagnosed positive for TSE disease. It was culled 341 days post injection. Eight other mice in this group survived beyond 341 days (432–670 days) without showing clinical or pathological signs of TSE infection.
bThe donor mouse for this passage was culled 722 days post injection without showing clinical signs of TSE disease. It had pathological signs of TSE infection.
Glycosylation determines the characteristics of TSE strain 79A
As 79A produced clinical disease in G1, G2 and G3 mice in the previously reported primary passage experiments (Tuzi et al, 2008), we were able to analyse the effect of sugars at both N-linked glycosylation sites of PrPSc by strain typing in the wild-type panel (C57, VM and CVF1 mice).
G1-79A. Wild-type mice (129Ola) were injected intracerebrally (i.c.) with 129-79A or with independent replicates of G1-79A (G1-79A (a) and G1-79A (b)) (n=10–12). Incubation periods in all three wild-type mouse strains (C57, VM, CVF1) were much longer after injection of G1-79A than of 129-79A (Table I, Figure 2). The ranking of incubation periods also differed for G1-79A inocula compared with the 129-79A inoculum, indicating a change in strain properties. Specifically, with G1-79A inocula the incubation periods in CVF1 mice were longer than in either C57 or VM mice, but with 129-79A the CVF1 incubation period was between the two homozygote strains (Table I). The degree of spongiform degeneration in the brain was also analysed. Lesion profiles from G1-79A- and 129-79A-infected brains differed in the location and degree of lesions (Figure 3A–E). Specifically, in C57 brains, differences were observed in several areas including the superior colliculus (position G3), hypothalamus (G4), hippocampus (G6), septum (G7) and mesencephalic tegmentum (W2) (Figure 3A); in VM mice in the hypothalamus (G4), hippocampus (G6) and forebrain cortex (G9) (Figure 3B); in CVF1 mice mostly in the superior colliculus (G3), mesencephalic tegmentum (W2) and pyramidal tract (W3) (Figure 3C); and in 129Ola mice only in the superior colliculus (G3) (Figure 3D). By contrast, no differences were noted between first and second passages of G1-79A in G1 mice (Figure 3E) indicating the strain had stabilised after the first passage in G1 mice.
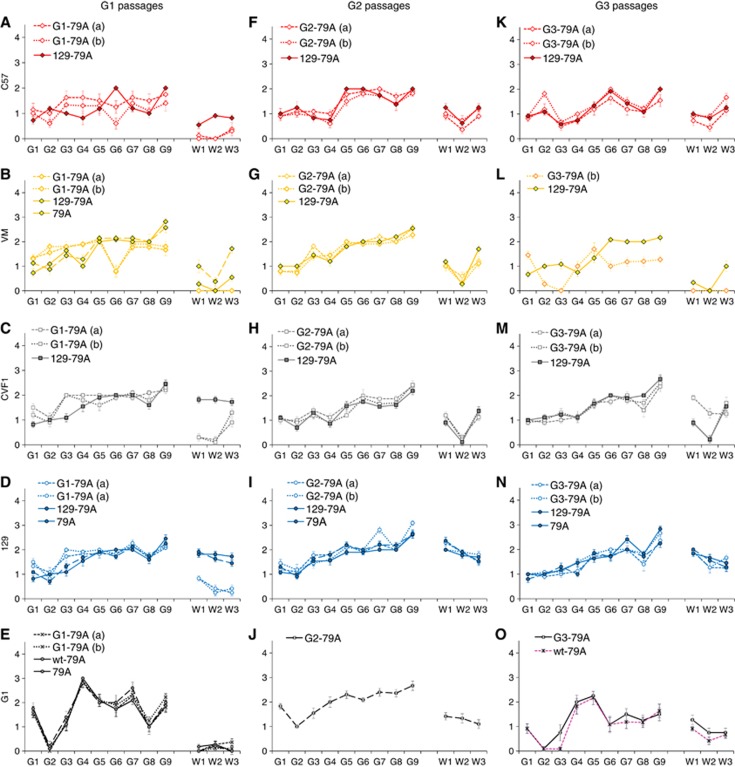
Figure 2.
Incubation period analyses of passages of 79A through G1, G2 and G3 mice. The 79A TSE strain was passaged in glycosylation-deficient mice (first pass). Passages were established from one 129 mouse (passage from 129) or two G1, G2 or G3 mice (pass from G1/G2/G3) in C57, VM or CVF1 mice or repassaged in glycosylation-deficient mice. The graphs show the incubation period for each mouse strain for each passage. (A) Passage through G1 mice, (B) G2 passage through G2 mice and (C) passage through G3 mice. The small yellow triangle and arrow indicate the single positive case in VM mice in the G3-79A(a) passage.
Figure 3.
Lesion profile analyses of passages of 79A through G1, G2 and G3 mice. Lesion profiles are shown for each passage of 79A in individual mouse strains (G1-79A, A–E; G2-79A, F–J, and G3-79A, K–O). Spongiform degeneration was scored in nine grey matter areas: G1, dorsal medulla; G2, cerebellar cortex; G3, superior colliculus; G4, hypothalamus; G5, medial thalamus; G6, hippocampus; G7, septum; G8, cerebral cortex; and G9, forebrain cortex, and in three white matter areas: W1, cerebellar white matter; W2, mesencephalic tegmentum; and W3, pyramidal tract (x axis) of mice scored clinically positive.
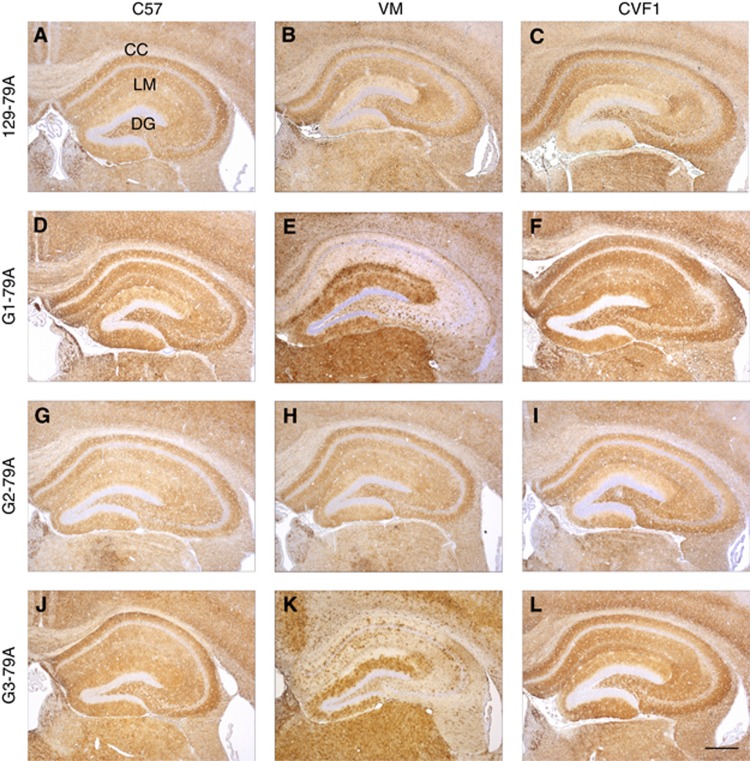
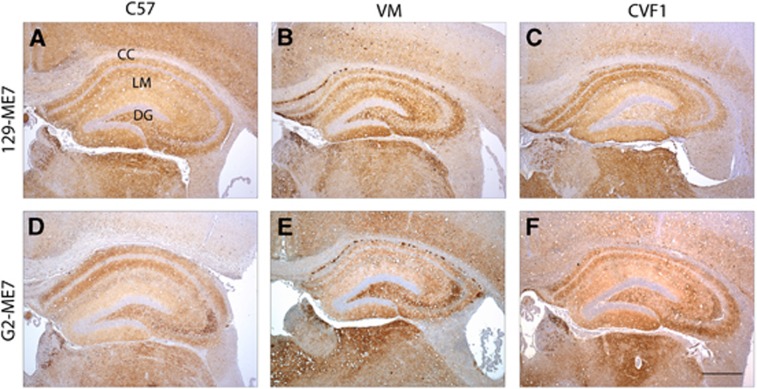
Immunohistochemical analysis of 129-79A injected into the wild-type panel showed diffuse, widespread accumulation of fine punctate deposits of PrP in the hippocampus (Figure 4A–C). Multiple areas were affected including the cortex, caudate, septum, hippocampus, hypothalamus, thalamus, midbrain, brain stem and cerebellum (not shown). G1-79A inoculated into C57 and CVF1 mice showed a similar pattern and distribution of PrP immunostaining (Figure 4D and F). Again the hippocampus and thalamus were consistently targeted. In addition, a peculiar patchy pattern of PrP deposition was observed in several areas of the brain, particularly the hippocampus and cortex in VM mice injected with both G1-79A (Figure 4E). A marked intense staining was observed in the dentate gyrus of the hippocampus often accompanied by a milder staining pattern in the stratum lacunosum-moleculare (Figure 4E). Overall, alterations in ranking of incubation period, lesion profile and PrP deposition strongly suggest that G1-79A exhibits novel strain properties, clearly distinct from 129-79A.
Figure 4.
PrP deposition in wild-type mouse brains after infection with 79A passaged in G1 G2, G3 or 129 mice. (A–C) 129-79A, (D–F) G1-79A, (G–I) G2-79A, (J–L) G3-79A passaged through C57, VM and CVF1 mice respectively. Widespread diffuse pattern of PrP deposition was observed in all brains irrespective of the inoculum with which they had been infected, with the hippocampus consistently targeted. A patchy pattern of PrP staining was observed in the hippocampus in VM mice infected with G1-79A (E) and G3-79A (K), intense band of staining observed in the dentate gyrus. CC indicates the corpus callosum, LM: lacunosum-moleculare, DG: Dentate gyrus. Sections stained with 6H4 antibody. Scale bar=400 μm.
G2-79A. Two independent brain homogenates from 79A-infected G2 mice (G2-79A(a) and G2-79A(b)) and one from wild-type mice (129-79A) were injected into the strain-typing panel (n=10–12). There was a distinct difference in ranking of incubation times in the different mouse lines between the 129-79A and G2-79A transmissions (Table I, Figure 2B). C57 and CVF1 mice inoculated with G2-79A(a) and (b) brain homogenates succumbed to disease with slightly longer incubation times than 129-79A. However, in the VM mice ∼100 days difference in incubation times were observed between the 129-79A and G2-79A inocula (a) and (b). No difference of lesion profiles in the brain was observed between the wild-type mouse strains, C57, VM, CVF1, and 129 and G2 mice infected with 129-79A or G2-79A (Figure 3F–J). In addition, there were no consistent differences in C57, VM or CVF1 mice infected with 129-79A or G2-79A observed by immunohistochemistry (Figure 4A–C and G–I). Thus, while an important component of strain properties namely the ranking of incubation periods between mouse strains is altered here, other criteria namely vacuolation profile and PrP deposition remain constant. The incubation period properties of G2-79A observed here resembled those of the 139A TSE strain, namely a shorter incubation period in VM mice than 79A, and specifically a shorter incubation period than in CVF1 mice for both strains (Bruce, 2003). No other phenotypic differences, for example, in lesion profiles, between 79A and 139A have been identified in these mouse strains and this was the case for the G2-79A passes.
G3-79A. The effect of removing both glycans from PrPSc on the properties of 79A was also determined by comparing two separate G3-79A brain homogenates with one sample of fully glycosylated 129-79A in the strain-typing panel. The ranking of incubation periods in the different mouse strains differed both between the two G3-79A isolates and the 129-79A sample (Table I, Figure 2C). Of note, VM mice developed clinical disease in a much shorter incubation period with the G3-79A(b) inoculum than the 129-79A inoculum (Table I). In the case of the second isolate G3-79A(a) only one VM mouse succumbed to infection after 341 days while eight mice showed no clinical or pathological signs of infection, five mice surviving more than 600 days.
The spongiform degeneration (Figure 3K–N) in C57, CVF1 and 129 mice inoculated with 129-79A and G3-79A was remarkably similar in C57 and CVF1 mice (Figure 3K and M). However, differences were observed in the lesion profile in VM mice inoculated with G3-79A(b) when compared to VM mice inoculated with 129-79A. Vacuolation was targeted to different areas including cerebellar cortex (G2), superior colliculus (G3), hippocampus (G6), septum (G7), cerebral (G8) and forebrain cortex (G9) and the pyramidal tract (W3) (Figure 3L). There was little difference in the lesion profiles between the first and second passages of 79A in G3 mice (Figure 3O). Results from immunohistochemical analysis were similar to those for G1-79A (see above) (Figure 4J–L), in particular, a widespread patchy pattern of PrP deposition was seen in VM mice inoculated with G3-79A similar to that observed in VM mice inoculated with G1-79A (Figure 4E). This pattern of PrP accumulation was particularly evident in the hippocampus with the most intense staining in the dentate gyrus. Overall these results suggest that G3-79A also exhibits phenotypic properties clearly distinct from 129-79A and resembling those of the 139A TSE strain, but that it was present at low titre in the inoculum since only one out of eight VM mice succumbed to infection with G3-79A(a) (Table I). Incubation periods in the other mouse strains were longer than in mice infected with 129-79A by similar amounts. However, it cannot be ruled out that another novel strain may also be present.
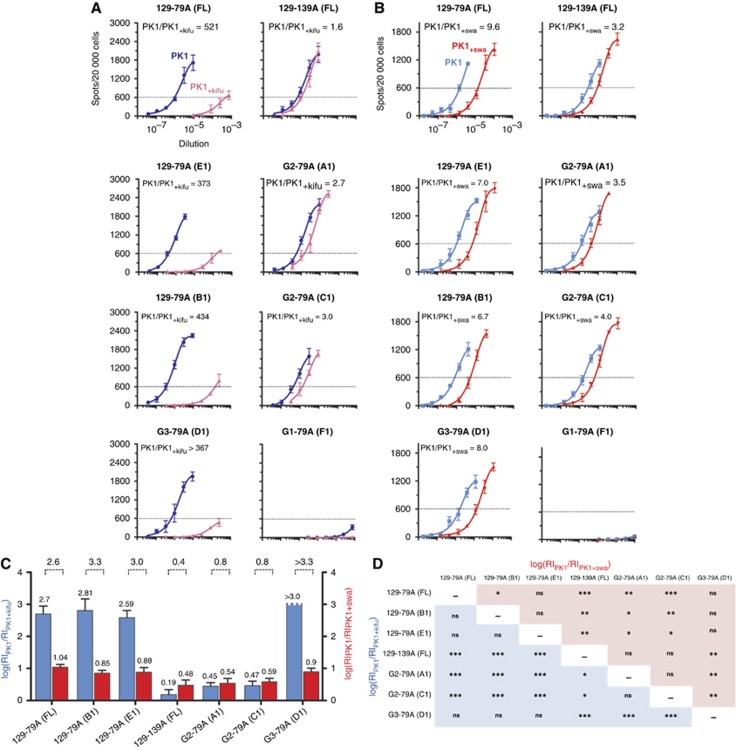
Strain characterisation by the SSCA. To determine whether 79A had altered its properties after passage through G1, G2 or G3 mice and resembled either 79A or 139A, we examined the different TSE strains by the SSCA. In this assay, N2a neuroblastoma-derived PK1 cells were exposed to a serial dilution of a TSE-infected sample in the presence or absence of specific inhibitors, passaged for three splits and the proportion of PrPSc-containing cells was plotted against the sample dilution (Klohn et al, 2003; Mahal et al, 2010; Browning et al, 2011; Oelschlegel et al, 2012). We arbitrarily define the response index (RI) as the reciprocal of the dilution at which 600 of 20 000, or 3% of the cells, became infected. We have reported that 79A and 139A can be distinguished by their distinct susceptibility to two inhibitors, kifunensine (kifu) and swainsonine (swa), when assayed on PK1 cells (Browning et al, 2011; Oelschlegel et al, 2012). Figure 5A shows that kifu strongly inhibited infection of PK1 cells by 129-79A but had little effect on 139A. G3-79A did not differ from the original 129-79A, however, G2-79A was largely kifu resistant and thus distinct from 129-79A, and, while resembling 139A, was not identical with it. As summarised in the bar graph (Figure 5C) log[RIPK1/ RIPK1+kifu] (blue bars) of G2-79A (0.45, 0.47) was clearly distinct from 129-79A and G3-79A (2.6 to >3.0), and, while closer to the value of 139A (0.19), still significantly differed from it (P=0.05; Figure 5D). By the same token, log[RIPK1/RIPK1+swa] (red bars) for 129-79A and G3-79A were similar (1.04, 0.85, 0.88 and 0.90) and significantly different from G2-79A (0.54, 0.59) and 129-139A (0.48) (Figure 5D). Due to the low levels of infectivity of G1-79A brains (Figure 5A and B), we were unable to determine whether the strain characteristics were different from those of 129-79A. In summary, the SSCA confirms the conclusion from strain typing, namely that G2-79A differs from the original 129-79A and resembles 139A, without being identical to it.
Figure 5.
Characterisation of 129-79A, G1-79A, G2-79A and G3-79A by the SSCA. The SSCA was performed on PK1 cells in the absence (blue) or presence of the glycosylation inhibitor kifunensine (kifu, pink). The RI is the reciprocal of the brain homogenate dilutions giving rise to 600 spots. (A) Samples were assayed in the absence (blue) or presence (red) of kifunensine (kifu). The ratios RI−kifu/RI+kifu for 129-79A and G3-79A are similar (>370), and more than 200 times higher than for 129-139A (1.6) and G2-79A (2.7, 3.0). The difference between 129-139A and G2-79A is statistically significant (see D). (B) Samples were assayed in the absence (blue) or presence (red) of swainsonine (swa). The ratios RI−swa/RI+swa for 129-79A (9.6, 7.0, 6.7) and G3-79A (8.0) are indistinguishable, but about twice as high as for 129-139A (3.2) and G2-79A (3.5, 4.0), which are also indistinguishable. (C) Bar diagram showing the log RI ratios of PK1/PK1kifu (blue bars) and PK1/PK1swa (red bars). The ratios of the log RIs are also shown above the bars indicating two distinct groups: ratios >2.6 for 79A-like strains and <1.0 for 139-like strains. (D) Matrix of log RI ratios of PK1/PK1kifu (blue squares) and PK1/PK1swa (red squares). Statistical analysis (unpaired t-test) was performed using raw data from each well of the assay after removing background signal. * To *** represent P-values from 0.03 to <0.0001, respectively; ns, not significant. The apparent non-identity (*) between 129-79A (FL) and 129-79A(B1) is unexplained. The figure shows that 129-79A and G3-79A are indistinguishable with both kifu and swa and differ from 129-139A and G2-79A, which are similar but not identical.
Lack of glycans at PrP position 196 does not change the infectious properties of TSE strain ME7
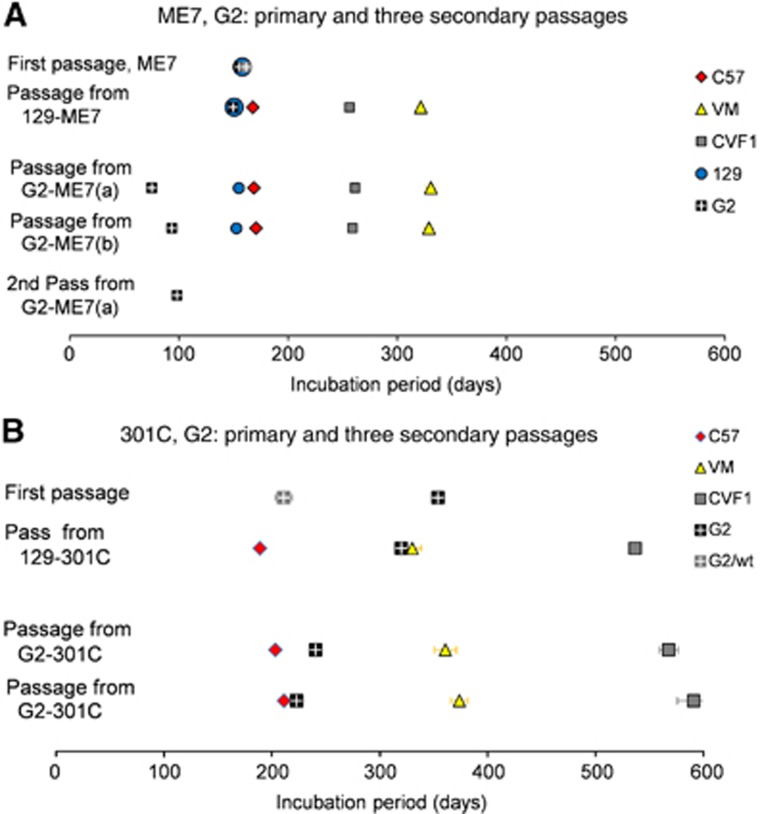
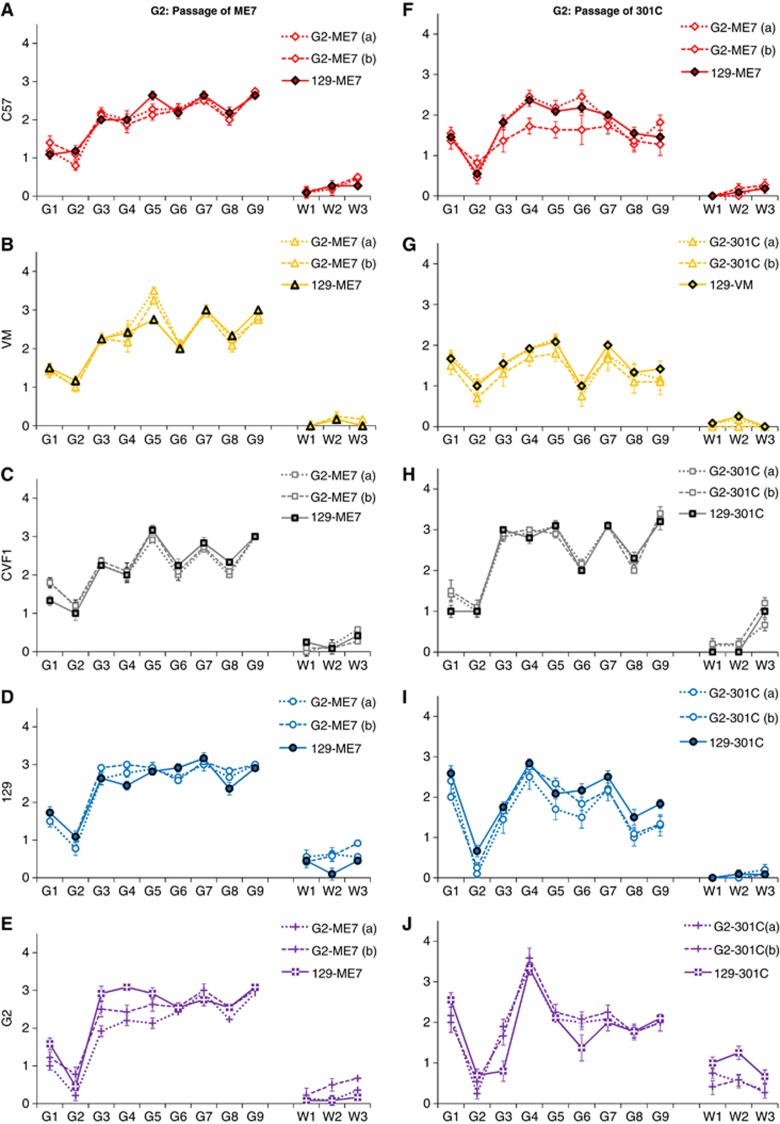
In order to analyse the effect of passaging ME7 in G2 mice, two distinct G2-ME7(a) and (b) and one 129-ME7 isolate were injected i.c. into the wild-type mouse panel (Table I, Figure 6A). The ranking of incubation periods was the same regardless of the glycosylation status of PrPSc in the ME7 inoculum, namely C57<CVF1<VM. No differences were observed between the spongiform profiles generated by 129-ME7 and G2-ME7 in the brains of the wild-type C57 and VM mice (Figure 7A–E). However, the second passage in G2 mice resulted in shorter incubation periods (Table I, Figure 6A) and altered lesion profiles (Figure 7E: G3, superior colliculus, G4, hypothalamus and G5, medial thalamus), suggesting a change in phenotype was detectable only after second passage in the G2 mice.
Figure 6.
Incubation period analyses of passages of ME7 and 301C through G2 mice. The ME7 TSE strain (A) and 301C, BSE-derived strain (B) were passaged in glycosylation-deficient mice (first pass). Passes were established from one 129 mouse (pass from 129) or two G2 mice (pass from G2) in C57, VM or CVF1 mice or repassaged in glycosylation-deficient mice. The graphs show the incubation period for each mouse strain for each passage.
Figure 7.
Lesion profile analyses of ME7 and 301C passaged through G2 mice. Lesion profiles are shown for each passage of ME7 (A–E) and 301C (F–J) in individual mouse strains. Spongiform degeneration was scored in nine grey matter areas: G1, dorsal medulla; G2, cerebellar cortex; G3, superior colliculus; G4, hypothalamus; G5, medial thalamus; G6, hippocampus; G7, septum; G8, cerebral cortex; and G9, forebrain cortex, and in three white matter areas: W1, cerebellar white matter; W2, mesencephalic tegmentum; and W3, pyramidal tract (x axis) of mice scored clinically positive.
Immunohistochemical analysis showed that the anatomical distribution of PrP in brains of mice infected with 129-ME7 or G2-ME7 was similar in every host analysed. All brains showed a typical ME7 deposition pattern characterised by a widespread, diffuse fine punctate PrP deposition in various areas of the cerebrum, cerebellum and brain stem; again, similarly to 129-79A, the hippocampus was consistently targeted (Figure 8A). In some animals small PrP-positive plaques were seen in the corpus callosum (Figure 8B, E and F). Therefore, the PrPSc targeting associated with the ME7 strain was not affected by the PrP glycosylation status of the ME7-derived inoculum.
Figure 8.
PrP deposition in wild-type mouse brains after infection with ME7 passed in G2 or 129 mice. Diffuse widespread accumulation of abnormal PrP was observed throughout the hippocampus, this distribution was observed in both wt (A–C) and G2-ME7 (D–F) inoculated animals. Small plaques where also observed in the corpus callosum (B, E, F) but this was variable. CC indicates the corpus callosum, LM: lacunosum-moleculare, DG: Dentate gyrus. Sections stained with 6H4 antibody. Scale bar=400 μm.
Lack of glycans at PrP position 196 in G2 mice does not change the infectious properties of TSE strain 301C
The BSE-derived strain 129-301C and two distinct G2-301C inocula, (a) and (b), were injected into wild-type mice (Table I, Figure 6B). As for ME7, the ranking of incubation periods for 301C in the different lines of wild-type mice was the same regardless of the glycosylation status of PrPSc in the infectious inoculum, namely C57<VM<CVF1 (Figure 6B). The distribution of vacuolar lesions in the brain of clinically sick animals infected with 129-301C or G2-301C inocula was similar in all the grey and white matter areas (Figure 7F–J), showing that they were similar despite different glycosylation status. Overall there was no change in the incubation period or lesion profile phenotype of G2-301C when passaged back into wild-type mice. However, the incubation period in G2 mice shortened at second passage, although there was little change in the lesion profiles. This may be due to a change in phenotype expressed in G2 mice only.
There was a similar pattern of PrP deposition in the wild-type panel of mice inoculated with 129-301C and G2-301C (Figure 9). In most animals analysed widespread accumulation of fine punctate PrP deposits were seen in the cerebrum cerebellum and brainstem. The brain areas most consistently affected are the thalamus, midbrain and brainstem.
Figure 9.
PrP deposition in wild-type mouse brains after infection with 301C passed in G2 or 129 mice. There was a similar pattern of PrP deposition in both groups of mice inoculated with 129-301C (A–C) and G2-301C (D–F), with a targeted pericellular form observed in the dentate gyrus and CA2 sector of the hippocampus. CC indicates the corpus callosum, LM: lacunosum-moleculare, DG: Dentate gyrus. Sections stained with BH1 antibody. Scale bar=400 μm.
Discussion
Here we have demonstrated both in vivo and in vitro that partial or complete removal of N-glycosyl moieties from PrP allowed change to occur to the phenotypic characteristics of one TSE agent strain, but did not affect the properties of two other strains in wild-type mice. These changes could be successfully retained on passage in mice, that is, they demonstrated heritable change of information. In describing these changes, we use the word ‘mutation’ to designate heritable changes regardless of the underlying molecular mechanism.
The 79A strain readily infected G1 mice but with an altered phenotype, suggesting that there was mutational change on passage of 79A in G1 mice that resulted in a short incubation period phenotype in G1 but a long incubation period phenotype in the C57, VM and CVF1 mouse panel. This conclusion is supported by the data from the SSCA, which showed a novel response to G1-79A brain homogenate. The barely detectable signal in SSCA could mean either there is very little infectivity in the sample, or that it is a novel strain which cannot infect PK1 cells as efficiently as 129-79A, G2-79A or G3-79A. However, high levels of infectivity are indicated by the transmissions to mice of the same samples assayed in SSCA, thus indicating a novel strain with lower specific infectivity for PK1 cells.
79A also readily infected G2 mice, with little apparent difference in incubation period and no change in lesion profiles compared to the wild-type mouse panel. After one passage in G2 mice, G2-79A showed the 139A incubation period phenotype when tested in C57, VM and CVF1 mice. These data are consistent with preferential selection in G2 mice of the strain that gives rise to the 139A phenotype (Bruce, 2003). This conclusion is supported by the data from the SSCA analysis, where G2-79A brain homogenate showed properties resembling those of 139A more than 79A.
The incubation period data from the G3-79A(b) isolate are consistent with a low titre of 139A giving longer incubation periods for this sample in all mouse strains than is observed with the higher titre samples that are normally assayed. The titre of the G3-79A(a) sample may be even lower, as indicated by low levels of infection of VM mice (one positive case in nine). It is not possible from the incubation period of the single VM mouse to say whether the 79A or the 139A phenotype was observed. It is likely that titre differences were primarily responsible for the difference between the two G3-79A passages. The donor for G3-79A(a) had a much shorter incubation period and much less vacuolation, indicated by a lower lesion profile than the donor for G3-79A(b). Both G3-79A(a) and G3-79(b) resembled those observed with low titres of a 79A/139A-like strain in C57 and CVF1 mice. The original strain may have been carried over or replicated very poorly in the G3 mice. This interpretation is supported by the very long incubation period at first passage in G3 mice, with longer incubation periods at second passage. This contrasts with the passages in G1 mice. Thus, either a low titre of 139A was detected or a novel strain had emerged. The detection of 79A properties by SSCA favours the survival and limited replication of the 79A and/or 139A at low titres in G3 mice, rather than the emergence of a new strain. In the SSCA, G3-79A was indistinguishable from 129-79A. There was no indication of a low titre of 139A since the 79A pattern was dominant.
G2-ME7 readily passaged into G2 mice, with similar incubation periods to wild-type mice. There was no change in lesion profile in wild-type mice, suggesting no change in phenotypic properties. There was a shortening of incubation periods at second passage in G2 mice, with concomitant change in lesion profile. These data suggest a mutational change of ME7 on passage of ME7 in G2 mice, which only affects phenotypic properties in G2 mice. Similarly no differences in the phenotypic properties of G2-301C were detected in the wild-type mouse panel after passage in G2 mice. However, incubation periods of first passage of 129-301C in G2 were longer than those in wild-type mice but were shorter at second passage, with little change in the lesion profiles. These data are consistent with a mutational change to G2-passaged 301C, which does not affect lesion profiles or interactions with wild-type mice.
Thus, changing the glycosylation status of the host PrP resulted in a significant phenotypic change to 79A, particularly when passaged in G1 mice. Changes to ME7 and 301C passaged in G2 mice were not detected on subsequent passages in wild-type mice. This study is the first to demonstrate, both in vivo and in vitro, that post-translational modifications can significantly change the phenotypic characteristics of a TSE agent.
Do glycan or amino acid changes cause changes in TSE agents?
Because glycosylation was prevented by substituting threonine (Thr) for asparagine (Asn) at positions 180 (G1), 196 (G2) or at both positions (G3), we must consider the possibility that the effects ascribed to lack of glycosylation were partly or entirely due to the changes in the amino-acid sequence per se (Tuzi et al, 2008). Previous studies performed by us in these transgenic mice, and by others using different amino-acid substitutions to generate glycosylation-deficient PrP (Neuendorf et al, 2004), produced similar results at primary passages in three of the six combinations of TSE strains and mice with glycosylation-deficient PrP. These comparisons show that the changes in glycosylation status of PrP rather than the changes in amino-acid sequence were probably responsible for the change in TSE agent interaction with the host (Neuendorf et al, 2004; Tuzi et al, 2008), but do not completely rule out the possibility that amino-acid changes per se were responsible for the changes in TSE agent properties, as is the case for the three other combinations of TSE agent and mice with glycosylation-deficient PrP.
The present study also showed a dramatic difference between G1-79A and G3-79A, and also a difference between G2-79A and G3-79A despite G3 mice having the same amino-acid changes to PrP as G1 and G2. Ikeda et al (2008) reported that preventing glycosylation by replacing the Asn residues at both glycosylation sites with a series substitutions at the Asn sites (N180 or N196) or Thr sites (T182 or T198) did not prevent TSE-infected ScN2a cells from producing PrPSc. Moreover, amplification of two TSE strains (RML and 301C) using sPMCA and enzymatically deglycosylated mouse PrPC in vitro failed to show any changes in TSE agent phenotype (Piro et al, 2009). Changes in the strain phenotype observed are therefore probably due to the changes in the glycosylation status and not amino-acid replacement.
Switching between 79A and 139A phenotypes
In the glycosylation-deficient mice we have observed switching between the two closely related strains 79A and 139A, a phenomenon previously observed several times in other contexts. These two strains and RML all came from the Drowsy Goat source of scrapie. RML has shown properties similar to 79A rather than 139A on two occasions (Carlson et al, 1986; Oelschlegel et al, 2012). Switching of phenotypic properties between 79A and 139A has been observed several times in the Edinburgh series of passages of these strains (Dickinson et al, 1984) and by Carp et al (1997). When 139A was passaged in PK1 cells and returned to mice, a strain indistinguishable from 79A was recovered (Oelschlegel et al, 2012). In the present experiments, G2 mice appear to favour the replication of the 139A phenotype. However, there is some evidence of both strains being present in G3 mice, albeit at very low effective titres since the incubation period data for G3-79(b) appear similar to the 139A phenotype allowing for a very low titre, whereas the 79A phenotype was observed by SSCA.
Switching between strains may be due to a single heritable change or mutational switch that occurs at a sufficiently high frequency for both strains to be present in samples with high titres. Subtle advantages for one or the other strain can allow either strain to emerge from the donor strain. Replication of the ‘genetic’ information of TSE agents therefore proceeds at rates dependent on the environment in which replication takes place. In the current experiments removing the carbohydrate moieties of PrP changes the environment in which replication takes place and in the case of 79A and 139A this change favours 139A.
Conclusion
Mutational change to TSE strains has long been recognised, probably occurring stochastically (Bruce and Dickinson, 1987; Kimberlin et al, 1987). Recently, it has been shown that TSE strains can mutate and be subject to selection in the presence of drugs (Ghaemmaghami et al, 2009; Li et al, 2010) or other alterations in the host environment, such as glycosylation status (Li et al, 2010; Mahal et al, 2010). We have shown that glycosylation can be important in allowing selection of a mutant strain from 79A. According to the prion hypothesis, a population of TSE agent infectious units comprises many different PrPSc conformations, of which only one or a few form a majority in a particular host, constituting a quasi-species (Peretz et al, 2002; Collinge, 2010; Li et al, 2010; Weissmann et al, 2011). However, evidence for large numbers of conformations is not established nor is it clear whether all the required conformations would be sufficiently thermodynamically stable. Alternatively, heritable change, that is, mutation, would require change, insertion or deletion of one or more nucleotides in a putative TSE nucleic acid genome (Somerville, 2002), as is the case for any virus or other living organism with a nucleic acid-based genome.
Our data for 79A can be explained if the 79A population is a quasi-species comprising multiple sub-species, including 79A and 139A, some of which replicate more efficiently than others in the presence of mono- or unglycosylated PrP. In populations of other microorganisms, such as RNA viruses, the concomitant presence of multiple genotypes, or quasi-species, has been detected, with the fittest genotype being the prevalent one (Domingo et al, 1978; Eigen, 1996). The two other strains, ME7 and 301C, appeared to be unaffected by the change in glycosylation status, suggesting these strains did not produce mutants that replicated more successfully in the glycosylation-deficient mice.
In summary, the experiments reported here show that TSE infectivity can replicate in mice with glycosylation-deficient PrP (G1, G2 and G3), but with TSE strain-specific responses. Changes in the glycosylation status of PrP in the inoculum affected strain properties in some cases but not others, demonstrating that the carbohydrate moieties are not essential to TSE replication or retention of strain-specific properties. Indeed despite the partial or complete absence of the carbohydrate moieties on PrP, TSE strain properties were maintained, or in the case of 79A altered to 139A in a similar fashion to changes taking place in wild-type mice. In other cases strain properties changed and a new TSE agent phenotype emerged. Overall these results demonstrate that the glycosylation status of host PrP can affect the replication and selection of the fittest TSE strain in that environment but that TSE strain properties are independent from host PrP.
Material and methods
Mouse strains
C57BL/Dk (referred to as C57 in this work), VM/Dk (referred to as VM) and C57BLxVM (referred to as CVF1) wild-type mouse strains were used. These mouse strains carry different alleles of murine PrP, defined by polymorphisms at codons 108 (leucine or phenylalanine) and 189 (threonine or valine) that distinguish the Prnpa allele (108L/189T) from the Prnpb allele (108F/189V) (Westaway et al, 1987). These amino acids have a role in determining the incubation period of the host to different TSE isolates (Moore et al, 1998; Barron et al, 2005). The C57 strain is classified as Prnpaa, VM as Prnpbb and CVF1 resulting from the cross between C57 and VM is a Prnpa/b heterozygous line. These particular lines of mice are used because the combinations of TSE strain and mouse genotype produce very characteristic disease phenotypes that define the different strains (Bruce et al, 1991; Bruce, 2003).
Preparation of the inoculum and injections
Inocula (0.1% weight/volume (w/v) in physiological saline) were prepared from brain tissues of 129/Ola (referred to as 129) wild-type mice or G1 (with the N180T mutation that abolishes the first glycosylation site on PrPC), G2 (with the N196T mutation that abolishes the second glycosylation site on PrPC) and G3 (with the mutations N180T and N196T that abolish both glycosylation consensus sites on PrPC) transgenic mice infected with ME7, 79A or 301C TSE strains (Tuzi et al, 2008). Inocula for the first passes of 79A, ME7 and 301C were prepared from standard pass lines of these strains maintained in C57BL/Dk mice. C57, VM and CVF1 wild-type mice (Bruce et al, 1991) were challenged by i.c. injection with 20 μl of inoculum. All groups were age and sex matched. Full local ethical approval for use of experimental animals was acquired. All experiments were carried out under licence and in accordance with the UK Home Office Regulations (Animals (Scientific Procedures Act) 1986).
Scoring of clinical TSE disease
The presence of clinical TSE disease was assessed as described previously (Fraser and Dickinson, 1967) without reference to the genotype of the mouse. Genotypes were confirmed for each animal by PCR analysis of tail DNA at the end of the experiment. Incubation times were calculated as the interval between inoculation and cull due to terminal TSE disease. Mice were killed by cervical dislocation at the terminal stage of disease, at termination of the experiment (between 500 and 700 days), or for welfare reasons due to intercurrent illness.
Western blotting
Mice were killed by cervical dislocation and brains were removed, flash frozen in liquid nitrogen and then stored at −70°C until required. Brain homogenates (10% w/v) were prepared in NP40 lysis buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, 150 mM NaCl, 50 mM Tris-HCl pH 7.5 and 1 mM PMSF). The homogenate was centrifuged at 16 000 r.p.m. for 10 min at 4°C and supernatant fluid isolated. Total protein was denatured in 1 × Novex Tris-Glycine SDS Sample Buffer (Invitrogen Life Technologies) and 1 × NuPage Sample Reducing Agent (Invitrogen Life Technologies) for 30 min at 95°C. Proteins were separated by gel electrophoresis at 125 V using 12% Novex Tris-Glycine gel (Invitrogen Life Technologies) and transferred to polyvinylidene fluoride membrane at 2 mA/cm2 gel using a semi-dry transfer blotter (Biorad) in 1 × Transfer Solution (48 mM Tris, 39 mM glycine, 0.375% sodium dodecyl sulphate, 20% methanol). Presence of PrP was assessed using the anti-PrP monoclonal antibodies 8H4 or 7A12 (1/10 000; kind gift of M.S Sy) (Zanusso et al, 1998).
Lesion profiles
Lesion profile analysis in nine areas of the grey matter and three areas of the white matter of the brain is extensively used to characterise TSE strains (Fraser and Dickinson, 1967, 1973). Sections were haematoxylin and eosin stained and scored for vacuolar degeneration on a scale of 0–5 in nine standard grey matter areas: G1, dorsal medulla; G2, cerebellar cortex; G3, superior colliculus; G4, hypothalamus; G5, medial thalamus; G6, hippocampus; G7, septum; G8, cerebral cortex; and G9, forebrain cortex, and in three white matter areas: W1, cerebellar white matter; W2, mesencephalic tegmentum; and W3, pyramidal tract of mice scored clinically positive (Fraser and Dickinson, 1967).
Immunohistochemical analysis in brain
Sections were stained for disease-associated PrP using monoclonal antibody 6H4 (Figures 4 and 8) (1/1000; Prionics) or BH1 (Figure 9) (1:1000) (McCutcheon et al, unpublished). Antigen retrieval by autoclaving at 121°C for 15 min and 5 min in formic acid (98%) was used to facilitate detection of the antigens. Sections were then blocked with normal serum prior to incubation with the primary antibody. Antibody binding was detected with the Catalysed Signal Amplification System (Dakocytomation) (6H4) or the Vectastain Elite ABC kit (Vector Labs) (BH1), and visualised with 3,3′-diaminobenzidine. In all experiments normal brain homogenate inoculum and Edinburgh PrP knock-out mice (Manson et al, 1994) were used as negative controls. Images were taken using a Nikon Eclipse E800 microscope. Sections were analysed by an observer blinded to the mouse genotype and type of inoculum.
Standard scrapie cell assay
Infected brains were sent to Scripps Florida from Edinburgh, UK, and assayed by the SSCA in a blind study. Half of each brain was homogenised in PBS to give a 10% homogenate using a ribolyser (Mahal et al, 2010), and stored at −80°C. After thawing, samples were rehomogenised using a 28-gauge needle immediately prior to infecting cells. Murine neuroblastoma N2a-derived PK1 cells were propagated in OptiMEM (Invitrogen), 4.5% BGS (HyClone, UT) with 1% penicillin/streptomycin (Invitrogen). The SSCA was performed as described on PK1 cells (Mahal et al, 2010), in the absence or presence of the inhibitors kifunensine (5 μg/ml; Toronto Research Chemicals Inc.) or swainsonine (2 μg/ml; Toronto Research Chemicals Inc.). The inhibitors were added at infection and at the first split. In summary, 5000 cells were infected with serial 1:3 dilutions of the infectious brain homogenate from the different TSE strains and split 1:7 every 3–4 days using a Freedom Evo robotic workstation (Tecan). After the third split, cells were propagated to confluence, 20 000 cells were aspirated onto the membranes of a 96-well Immobilon-P membrane plate (Millipore) and the plates were baked at 50°C for 1–2 h. The PK-ELISA assay was performed with primary anti-PrP antibody D18 (Williamson et al, 1998) and anti-human AP-conjugated secondary antibody as described (Mahal et al, 2010). PrPSc-positive cells were counted using the BioReader 5000-Eb (Biosys) and spot number was plotted against log10 of sample dilution, using GraphPad Prism. The RI, a relative measure of the level of infectivity on cells under the conditions of the assay, is the reciprocal of the dilution that yields an arbitrary percentage (in this paper, 3%) of PrPSc-positive cells (Mahal et al, 2007, 2010).
Supplementary Material
Acknowledgments
The work in Edinburgh was supported by a Roslin Institute Strategic Grant funding from the BBSRC. PP was supported by funding from the National Institutes of Health: NIH-NIAID Y1-A1-4893-02 and the Food and Drugs Administration: FDA 224-05-1307. The work in TSRI Florida was supported by grants from the National Institutes of Health 1RO1NSO59543, 1RO1NS067214 and from the Alafi Family Foundation to CW. We thank Dorothy Kisielewski for mouse genotyping, Aileen Boyle for the excellent work on lesion profile analysis, and Anne Coghill, Sandra Mack and Gillian McGregor for assistance with the pathology analysis. Irene McConnell, Val Thompson, Simon Cumming, Leanne Frame and Kris Hogan for the breeding, care, injections and the clinical scoring of the mice. We are very grateful to Dr Andreas Lengeling for critical reading of the manuscript. 8H4 and 7A12 antibodies were kindly provided by Dr Man-Sun Sy (Case Western Reserve University) and the BH1 antibody by Dr Sandra McCutcheon (The Roslin Institute). The findings and conclusions in this article have not been formally disseminated by the Food and Drug Administration and should not be construed to represent any Agency determination or policy.
Author contributions: EC and JCM conceived and planned the research. SPM and CW planned and performed the SSCA experiments. AD, DB and PP performed the pathological analyses. RS collated and analysed the data. All authors contributed to the writing of the paper.
Footnotes
The authors declare no conflict of interest.
References
- Aguzzi A, Heikenwalder M, Polymenidou M (2007) Insights into prion strains and neurotoxicity. Nat Rev Mol Cell Biol 8: 552–561 [DOI] [PubMed] [Google Scholar]
- Angers RC, Kang HE, Napier D, Browning S, Seward T, Mathiason C, Balachandran A, McKenzie D, Castilla J, Soto C, Jewell J, Graham C, Hoover EA, Telling GC (2010) Prion strain mutation determined by prion protein conformational compatibility and primary structure. Science 328: 1154–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron RM, Baybutt H, Tuzi NL, McCormack J, King D, Moore RC, Melton DW, Manson JC (2005) Polymorphisms at codons 108 and 189 in murine PrP play distinct roles in the control of scrapie incubation time. J Gen Virol 86: 859–868 [DOI] [PubMed] [Google Scholar]
- Bessen RA, Marsh RF (1994) Distinct PrP properties suggest the molecular-basis of strain variation in transmissible mink encephalopathy. J Virol 68: 7859–7868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biacabe A-G, Laplanche J-L, Ryder S, Baron T (2004) Distinct molecular phenotypes in bovine prion diseases. EMBO Rep 5: 110–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop MT, Will RG, Manson JC (2010) Defining sporadic Creutzfeldt-Jakob disease strains and their transmission properties. Proc Natl Acad Sci USA 107: 12005–12010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning S, Baker CA, Smith E, Mahal SP, Herva ME, Demczyk CA, Li J, Weissmann C (2011) Abrogation of complex glycosylation by swainsonine results in strain- and cell-specific inhibition of prion replication. J Biol Chem 286: 40962–40973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce ME (2003) TSE strain variation. Br Med Bull 66: 99–108 [DOI] [PubMed] [Google Scholar]
- Bruce ME, Dickinson AG (1987) Biological evidence that scrapie agent has an independent genome. J Gen Virol 68: 79–89 [DOI] [PubMed] [Google Scholar]
- Bruce ME, McConnell I, Fraser H, Dickinson AG (1991) The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J Gen Virol 72: 595–603 [DOI] [PubMed] [Google Scholar]
- Buschmann A, Biacabe AG, Ziegler U, Bencsik A, Madec JY, Erhardt G, Luhken G, Baron T, Groschup MH (2004) Atypical scrapie cases in Germany and France are identified by discrepant reaction patterns in BSE rapid tests. J Virol Methods 117: 27–36 [DOI] [PubMed] [Google Scholar]
- Cancellotti E, Bradford BM, Tuzi NL, Hickey RD, Brown DR, Brown KL, Barron RM, Kisielewski D, Piccardo P, Manson JC (2010) Glycosylation of PrPC determines timing of neuroinvasion and targeting in the brain following transmissible spongiform encephalopathy infection by a peripheral route. J Virol 84: 3464–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancellotti E, Wiseman F, Tuzi NL, Baybutt H, Monaghan P, Aitchison L, Simpson J, Manson JC (2005) Altered glycosylated PrP proteins can have different neuronal trafficking in brain but do not acquire scrapie-like properties. J Biol Chem 280: 42909–42918 [DOI] [PubMed] [Google Scholar]
- Carlson GA, Kingsbury DT, Goodman PA, Coleman S, Marshall ST, DeArmond S, Westaway D, Prusiner SB (1986) Linkage of prion protein and scrapie incubation time genes. Cell 46: 503–511 [DOI] [PubMed] [Google Scholar]
- Carp RI, Meeker H, Sersen E (1997) Scrapie strains retain their distinctive characteristics following passages of homogenates from different brain regions and spleen. J Gen Virol 78: 283–290 [DOI] [PubMed] [Google Scholar]
- Casalone C, Zanusso G, Acutis P, Ferrari S, Capucci L, Tagliavini F, Monaco S, Caramelli M (2004) Identification of a second bovine amyloidotic spongiform encephalopathy: molecular similarities with sporadic Creutzfeldt-Jakob disease. Proc Natl Acad Sci USA 101: 3065–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B (1998) BSE and prions: uncertainties about the agent. Science 279: 42–43 [DOI] [PubMed] [Google Scholar]
- Collinge J (2010) Prion strain mutation and selection. Science 328: 1111–1112 [DOI] [PubMed] [Google Scholar]
- Collinge J, Clarke AR (2007) A general model of prion strains and their pathogenicity. Science 318: 930–936 [DOI] [PubMed] [Google Scholar]
- Collinge J, Sidle KCL, Meads J, Ironside J, Hill AF (1996) Molecular analysis of prion strain variation and the aetiology of ‘new variant' CJD. Nature 383: 685–690 [DOI] [PubMed] [Google Scholar]
- DeArmond SJ, Sanchez H, Yehiely F, Qiu Y, Ninchak-Casey A, Daggett V, Camerino AP, Cayetano J, Rogers M, Groth D, Torchia M, Tremblay P, Scott MR, Cohen FE, Prusiner SB (1997) Selective neuronal targeting in prion disease. Neuron 19: 1337–1348 [DOI] [PubMed] [Google Scholar]
- Dickinson AG (1976) Scrapie in sheep and goats. Front Biol 44: 209–241 [PubMed] [Google Scholar]
- Dickinson AG, Bruce ME, Outram GW, Kimberlin RH (1984) Scrapie strain differences: the implications of stability and mutation. In Proceedings of Workshop on Slow Transmissible Diseases Tateishi J (eds) pp 105–118Japanese Ministry of Health and Welfare: Tokyo, [Google Scholar]
- Domingo E, Gomez J (2007) Quasispecies and its impact on viral hepatitis. Virus Res 127: 131–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E, Sabo D, Taniguchi T, Weissmann C (1978) Nucleotide sequence heterogeneity of an RNA phage population. Cell 13: 735–744 [DOI] [PubMed] [Google Scholar]
- Eigen M (1996) On the nature of virus quasispecies. Trends Microbiol 4: 216–218 [DOI] [PubMed] [Google Scholar]
- Foster JD, Dickinson AG (1988) The unusual properties of CH1641, a sheep-passaged isolate of scrapie. Vet Rec 123: 5–8 [DOI] [PubMed] [Google Scholar]
- Fraser H, Dickinson AG (1967) Distribution of experimentally induced scrapie lesions in the brain. Nature 216: 1310–1311 [DOI] [PubMed] [Google Scholar]
- Fraser H, Dickinson AG (1973) Scrapie in mice. Agent-strain differences in the distribution and intensity of grey matter vacuolation. J Comp Pathol 83: 29–40 [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Ahn M, Lessard P, Giles K, Legname G, DeArmond SJ, Prusiner SB (2009) Continuous quinacrine treatment results in the formation of drug-resistant prions. PLoS Pathog 5: e1000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris DA (1999) Cellular biology of prion diseases. Clin Microbiol Rev 12: 429–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkey PM (2008) Molecular epidemiology of clinically significant antibiotic resistance genes. Br J Pharmacol 153(Suppl 1): S406–S413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head MW, Bunn TJR, Bishop MT, McLoughlin V, Lowrie S, McKimmie CS, Williams MC, McCardle L, MacKenzie J, Knight R, Will RG, Ironside JW (2004) Prion protein heterogeneity in sporadic but not variant Creutzfeldt-Jakob disease: UK cases 1991-2002. Ann Neurol 55: 851–859 [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M (2001) Intracellular functions of N-linked glycans. Science 291: 2364–2369 [DOI] [PubMed] [Google Scholar]
- Hill AF, Butterworth RJ, Joiner S, Jackson G, Rossor MN, Thomas DJ, Frosh A, Tolley N, Bell JE, Spencer M, King A, Al-Sarraj S, Ironside JW, Lantos PL, Collinge J (1999) Investigation of variant Creutzfeldt-Jakob disease and other human prion diseases with tonsil biopsy samples. Lancet 353: 183–189 [DOI] [PubMed] [Google Scholar]
- Ikeda S, Kobayashi A, Kitamoto T (2008) Thr but Asn of the N-glycosylation sites of PrP is indispensable for its misfolding. Biochem Biophys Res Commun 369: 1195–1198 [DOI] [PubMed] [Google Scholar]
- Ironside JW, Ritchie DL, Head MW (2005) Phenotypic variability in human prion diseases. Neuropathol Appl Neurobiol 31: 565–579 [DOI] [PubMed] [Google Scholar]
- Jones EM, Surewicz WK (2005) Fibril conformation as the basis of species- and strain-dependent seeding specificity of mammalian prion amyloids. Cell 121: 63–72 [DOI] [PubMed] [Google Scholar]
- Kimberlin RH, Cole S, Walker CA (1987) Temporary and permanent modifications to a single strain of mouse scrapie on transmission to rats and hamsters. J Gen Virol 68: 1875–1881 [DOI] [PubMed] [Google Scholar]
- Kimberlin RH, Walker CA (1978) Evidence that the transmission of one source of scrapie agent to hamsters involves separation of agent strains from a mixture. J Gen Virol 39: 487–496 [DOI] [PubMed] [Google Scholar]
- Klohn PC, Stoltze L, Flechsig E, Enari M, Weissmann C (2003) A quantitative, highly sensitive cell-based infectivity assay for mouse scrapie prions. Proc Natl Acad Sci USA 100: 11666–11671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Browning S, Mahal SP, Oelschlegel AM, Weissmann C (2010) Darwinian evolution of prions in cell culture. Science 327: 869–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahal SP, Baker CA, Demczyk CA, Smith EW, Julius C, Weissmann C (2007) Prion strain discrimination in cell culture: the cell panel assay. Proc Natl Acad Sci U S A 104: 20908–20913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahal SP, Browning S, Li JL, Suponitsky-Kroyter I, Weissmann C (2010) Transfer of a prion strain to different hosts leads to emergence of strain variants. Proc Natl Acad Sci USA 107: 22653–22658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahal SP, Demczyk CA, Smith EW Jr., Klohn PC, Weissmann C (2008) Assaying prions in cell culture: the standard scrapie cell assay (SSCA) and the scrapie cell assay in end point format (SCEPA). Methods Mol Biol 459: 49–68 [DOI] [PubMed] [Google Scholar]
- Manson JC, Cancellotti E, Hart P, Bishop MT, Barron RM (2006) The transmissible spongiform encephalopathies: emerging and declining epidemics. Biochem Soc Trans 34: 1155–1158 [DOI] [PubMed] [Google Scholar]
- Manson JC, Clarke AR, McBride PA, McConnell I, Hope J (1994) PrP gene dosage determines the timing but not the final intensity or distribution of lesions in scrapie pathology. Neurodegen 3: 331–340 [PubMed] [Google Scholar]
- Manuelidis L (1998) Vaccination with an attenuated Creutzfeldt-Jakob disease strain prevents expression of a virulent agent. Proc Natl Acad Sci USA 95: 2520–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L (2011) Nuclease resistant circular DNAs copurify with infectivity in scrapie and CJD. J Neurovirol 17: 131–145 [DOI] [PubMed] [Google Scholar]
- Moore RC, Hope J, McBride PA, McConnell I, Selfridge J, Melton DW, Manson JC (1998) Mice with gene targetted prion protein alterations show that Prnp, Sinc and Prni are congruent. Nat Genet 18: 118–124 [DOI] [PubMed] [Google Scholar]
- Morales R, Abid K, Soto C (2007) The prion strain phenomenon: molecular basis and unprecedented features. Biochim Biophys Acta 1772: 681–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuendorf E, Weber A, Saalmueller A, Schatzl HM, Reifenberg K, Pfaff E, Groschup MH (2004) Glycosylation deficiency at either one of the two glycan attachment sites of cellular prion protein preserves susceptibility to bovine spongiform encephalopathy and scrapie infections. J Biol Chem 279: 53306–53316 [DOI] [PubMed] [Google Scholar]
- Oelschlegel AM, Fallahi M, Ortiz-Umpierre S, Weissmann C (2012) The Extended Cell Panel Assay (ECPA) characterizes the relationship of RML, 79A and 139A prion strains and reveals conversion of 139A to 79A-like prions in cell culture. J Virol 86: 5297–5303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor SE, Imperiali B (1996) Modulation of protein structure and function by asparagine-linked glycosylation. Chem Biol 3: 803–812 [DOI] [PubMed] [Google Scholar]
- Parchi P, Castellani R, Capellari S, Ghetti B, Young K, Chen SG, Farlow M, Dickson DW, Sima AA, Trojanowski JQ, Petersen RB, Gambetti P (1996) Molecular basis of phenotypic variability in sporadic Creutzfeldt-Jakob disease. Ann Neurol 39: 767–778 [DOI] [PubMed] [Google Scholar]
- Peretz D, Williamson RA, Legname G, Matsunaga Y, Vergara J, Burton DR, DeArmond SJ, Prusiner SB, Scott MR (2002) A change in the conformation of prions accompanies the emergence of a new prion strain. Neuron 34: 921–932 [DOI] [PubMed] [Google Scholar]
- Piro JR, Harris BT, Nishina K, Soto C, Morales R, Rees JR, Supattapone S (2009) Prion Protein Glycosylation Is Not Required for Strain-Specific Neurotropism. J Virol 83: 5321–5328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB (1998) Prions. Proc Natl Acad Sci USA 95: 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer RG (1991) The scrapie agent: ‘a virus by any other name’. Curr Top Microbiol Immunol 172: 195–232 [PubMed] [Google Scholar]
- Silveira JR, Caughey B, Baron GS (2004) Prion protein and the molecular features of transmissible spongiform encephalopathy agents. Curr Top Microbiol Immunol 284: 1–50 [DOI] [PubMed] [Google Scholar]
- Somerville RA (2002) TSE agent strains and PrP: reconciling structure and function. Trends Biochem Sci 27: 606–612 [DOI] [PubMed] [Google Scholar]
- Somerville RA, Chong A, Mulqueen OU, Birkett CR, Wood SCER, Hope J (1997) Biochemical typing of scrapie strains. Nature 386: 564. [DOI] [PubMed] [Google Scholar]
- Telling GC (2010) Nucleic acid-free mutation of prion strains. Prion 4: 252–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzi NL, Cancellotti E, Baybutt H, Blackford L, Bradford B, Plinston C, Coghill A, Hart P, Piccardo P, Barron RM, Manson JC (2008) Host PrP glycosylation: a major factor determining the outcome of prion infection. PLoS Biol 6: e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann C, Li J, Mahal SP, Browning S (2011) Prions on the move. EMBO Rep 12: 1109–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westaway D, Goodman PA, Mirenda CA, McKinley MP, Carlson GA, Prusiner SB (1987) Distinct prion proteins in short and long scrapie incubation period mice. Cell 51: 651–662 [DOI] [PubMed] [Google Scholar]
- Will RG (2003) Acquired prion disease: iatrogenic CJD, variant CJD, kuru. Brit Med Bull 66: 255–265 [DOI] [PubMed] [Google Scholar]
- Williamson RA, Peretz D, Pinilla C, Ball H, Bastidas RB, Rozenshteyn R, Houghten RA, Prusiner SB, Burton DR (1998) Mapping the prion protein using recombinant antibodies. J Virol 72: 9413–9418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanusso G, Liu D, Ferrari S, Hegyi I, Yin X, Aguzzi A, Hornemann S, Liemann S, Glockshuber R, Manson JC, Brown P, Petersen RB, Gambetti P, Sy MS (1998) Prion protein expression in different species: analysis with a panel of new mAbs. Proc Natl Acad Sci USA 95: 8812–8816 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.