Abstract
Faithful transmission of chromosomes during eukaryotic cell division requires sister chromatids to be paired from their generation in S phase until their separation in M phase. Cohesion is mediated by the cohesin complex, whose Smc1, Smc3 and Scc1 subunits form a tripartite ring that entraps both DNA double strands. Whereas centromeric cohesin is removed in late metaphase by Scc1 cleavage, metazoan cohesin at chromosome arms is displaced already in prophase by proteolysis-independent signalling. Which of the three gates is triggered by the prophase pathway to open has remained enigmatic. Here, we show that displacement of human cohesin from early mitotic chromosomes requires dissociation of Smc3 from Scc1 but no opening of the other two gates. In contrast, loading of human cohesin onto chromatin in telophase occurs through the Smc1–Smc3 hinge. We propose that the use of differently regulated gates for loading and release facilitates unidirectionality of DNA’s entry into and exit from the cohesin ring.
Keywords: cell cycle, cohesin, mitosis, prophase pathway, separation of chromosome arms
Introduction
A dividing eukaryotic cell has to faithfully segregate one copy of each chromosome into the two newly forming daughter cells. The challenge to identify matching sister chromatids is mastered by keeping them paired from the time of their generation in S phase until their separation in M phase. Cohesion is mediated by cohesin. Within this heterotetrameric complex, Smc1 and -3 form long intramolecular, anti-parallel coiled coils that heterodimerise at one end via a hinge domain. At the other end, the N- and C-terminal domain of each Smc subunit forms a nucleotide binding domain (NBD). Smc1 and -3 interact with (or near) their NBDs with C- and N-terminal domains of the α-kleisin Scc1 (also known as Rad21), respectively. Together, these interactions result in a heterotrimeric ring with a diameter of 50 nm (Anderson et al, 2002; Haering et al, 2002, 2004). A forth, peripheral subunit, SA1 or -2 is tightly associated with Scc1. Studies on budding yeast cohesin showed that the ring topologically entraps sister chromatids rather than holding them together by physical protein–DNA interactions (Haering et al, 2002, 2008; Gruber et al, 2003; Ivanov and Nasmyth, 2005, 2007).
Cohesin is loaded onto chromatin by Scc2–Scc4, the kollerin complex, which in Xenopus is recruited to DNA in telophase by prereplicative complexes (Ciosk et al, 2000; Takahashi et al, 2004). Loading of budding yeast cohesin requires the Smc NBDs to interact and compose an active ATPase and it requires the opening of the Smc1–Smc3 hinge (Weitzer et al, 2003; Gruber et al, 2006). Whether loading of vertebrate cohesin also involves dissociation of Smc1 from Smc3 has not yet been studied. Cohesion establishment occurs during S phase in a co-replicative manner and requires Esco1/2-dependent acetylation of two adjacent lysine residues within the head domain of Smc3. This counteracts an anti-establishment activity of the Scc1-bound releasin complex consisting of Pds5 and Wapl and results in replacement of Wapl with cohesion conserving sororin (Gandhi et al, 2006; Kueng et al, 2006; Zhang et al, 2008; Rowland et al, 2009; Shintomi and Hirano, 2009; Lafont et al, 2010; Nishiyama et al, 2010).
Premise for sister chromatid segregation is removal of cohesin from chromatin. In metazoans, this occurs in two waves (Waizenegger et al, 2000). While most cohesin from chromosome arms is removed in prophase, full separation of sister chromatids occurs only at the metaphase-to-anaphase transition when Scc1 of persisting centromeric cohesin is endoproteolytically cleaved by separase (Uhlmann et al, 2000).
Prophase pathway signalling involves the releasin complex, kinase activity of Plk1 as well as Aurora B and phosphorylation of SA1 and -2 (Sumara et al, 2002; Giménez-Abián et al, 2004; Hauf et al, 2005; Kueng et al, 2006; Shintomi and Hirano, 2009). It culminates in the proteolysis-independent opening of the cohesin ring, which raises a number of interesting, as yet unresolved questions: Which two subunits of the tripartite Smc1–Smc3–Scc1 ring detach from each other in early mitosis? Are there alternatively used gates or might arm cohesin even fall apart entirely in prophase? Does DNA enter and leave cohesin through the same or a different gate? Very recently, the Nasmyth group demonstrated a surprisingly dynamic interaction of S. cerevisiae cohesin with interphase chromatin, which depends on releasin complex mediated dissociation of Scc1 from Smc3 (Chan et al, 2012). Although S. cerevisiae lacks a prophase pathway, the conservation of cohesin and many associated proteins might suggest a functional homology between cohesion dynamics in yeast interphase and the prophase pathway in vertebrate mitosis.
To address these issues, we have exploited the rapamycin-induced heterodimerisation of FKBP (FK506 binding protein 12) and FRB (FKBP-rapamycin binding domain of mTOR) to block DNA exit from individual gates of human cohesin. Subsequent cohesin chromatin retention assays revealed that the prophase pathway failed if—and only if—Smc3-FKBP and FRB-Scc1 replaced their endogenous counterparts and rapamycin was present. Furthermore and similar to the situation in budding yeast (Gruber et al, 2006), rapamycin interfered with cohesin loading when the hinge domains of Smc1 and -3 were tagged with FKBP and FRB. Thus, joining and disjoining of sister chromatids involves DNA to enter and exit the cohesin ring through different gates.
Results
Adopting the FRB-FKBP system to human cohesin
The bulk of human cohesin is removed from chromosome arms by action of the prophase pathway but re-associates with chromatin already in telophase (Figure 1A). In contrast, the kinetochore protein Hec1 associates with chromatin only in mitosis, that is, at times when most of cohesin does not. RNAi-dependent depletion of Wapl, a releasin and crucial player of the prophase pathway (Gandhi et al, 2006; Kueng et al, 2006), extends cohesin’s residence time on chromatin such that it can now be detected at Hec1-positive pro- and prometaphase chromosomes until cleaved by separase in metaphase (Figure 1B). Moreover, this prolonged association mediates close cohesion of sister chromatids along their entire length resulting in a ‘zipped’ rather than ‘butterfly-like’ appearance of chromosomes in spreads (Figure 1C).
Figure 1.
Localisation and function of cohesin during mitosis. (A, B) RNAi of the prophase pathway component Wapl results in prolonged chromatin association of cohesin during mitosis. HeLa L cells were transfected with siRNA directed against GL2 (Luciferase, control) (A) or Wapl (B). Two days later, their DNA (Hoechst 33342), Hec1 (mitosis-specific kinetochore marker) and Smc1 were visualised by (immuno-) fluorescence microscopy. Scale bars=10 μm. (C) Wapl depletion causes tight cohesion of sister chromatids along their entire length. Two days after transfection of GL2 or Wapl siRNA, HeLa L cells were enriched in prometaphase by a 15-h nocodazole treatment and then subjected to chromosome spreading. Graph depicts the relative number of spreads displaying a primarily ‘Butterfly-like’ or ‘Zipped’ chromosome morphology as exemplified by the images on the right. n=100.
If the non-proteolytic removal of human cohesin in prophase involves ring opening, then it should fail when the relevant gate is artificially kept closed. To test this prediction, we sought to fuse cohesin subunits with FRB and FKBP since these 11–12 kDa tags are induced by the small molecule rapamycin to form highly stable heterodimers (Banaszynski et al, 2005; Gruber et al, 2006). Demonstrating the applicability of the FRB-FKBP system to living human cells, FKBP-mCherry co-localised with chromatin-bound histone 2B-FRB only when rapamycin was added to doubly transfected Hek293T cells (Figure 2A). Guided by existing structural informations and a previous study on yeast cohesin (Haering et al, 2002, 2004; Gruber et al, 2006), we then fused FKBP to the 5′ end of SCC1 and SMC1 while FRB was attached to the 3′ end of SCC1 and SMC3 (Figure 2B). Two additional variants of Smc1 and -3 were created by inserting FKBP and FRB, respectively, within their hinge domains but at sites predicted to oppose the interaction surface. Pairwise expression of corresponding tailored subunits should each give rise to a cohesin complex with one of its three gates being FKBP-FRB tagged and, hence, rapamycin lockable (illustrated in Figure 2C using the example of Smc3 and Scc1). We then created three corresponding stable cell lines, each of which inducibly expresses one matched pair of FKBP-FRB-fused cohesin subunits from corresponding, siRNA-resistant transgenes (Figure 2D). Preventing the expression of the endogenous genes by RNAi and simultaneously inducing the siRNA-resistant transgenes with doxycycline resulted in partial or even extensive replacement of the wild-typic cohesin subunits with their engineered counterparts (Figure 2E).
Figure 2.
In vivo expression of tailored cohesins with individually lockable gates. (A) Rapamycin can induce heterodimerisation of FRB/FKBP-tagged proteins at human chromatin. Hek293T cells transiently transfected to co-express histone 2B-FRB (H2B-FRB) and FKBP-mCherry fusions were arrested in prometaphase in the presence or absence of rapamycin (±rapa) before mitotic chromatin was analysed by fluorescence microscopy for co-localisation of mCherry (right panels) with Hoechst 33342-stained DNA (left panels). Scale bar=10 μm. (B) Drawn to scale cartoons of the matched pairs of FRB/FKBP-tagged cohesin subunits used in this study. The lines connecting two boxes represent linker peptides of 9 or 29 amino acids, respectively, in Smc1-int. (internal) FKBP and Smc3-int. FRB or in FKBP-Scc1 and FKBP-Smc1. (C) Rationale of one of the three gates of recombinant cohesin being lockable by rapamycin. As illustrated here for the Smc3–Scc1 contact site, each of the three pairs of tailored cohesin subunits will present the FRB and FKBP tags closely juxtaposed at a given gate of the tripartite ring. The rapamycin-induced tight FRB-FKBP interaction should prevent passage of DNA (not shown) through the gate even if the corresponding cohesin subunits detach from each other in response to prophase pathway signalling. (D) Three double-transgenic cell lines inducibly co-express FKBP-Scc1 and Smc3-FRB, Scc1-FRB and FKBP-Smc1 or Smc1-int. FKBP and Smc3-int. FRB. The corresponding Hek293 lines were cultured in the presence or absence of doxycycline (dox) for 24 h before being analysed by FKBP and FRB immunoblots for transgene expression. An anti-α-tubulin western served as loading control. (E) RNAi and transgene induction result in efficient replacement of endogenous cohesin subunits by their FRB/FKBP-tagged counterparts. The double-transgenic Hek293 lines were transfected with siRNAs targeting GL2 (luciferase, control) or the indicated endogenous cohesin subunits and incubated for 3 days in the presence (+) or absence (−) of transgene-inducing doxycycline. Note that Smc3-FRB, Scc1-FRB and Smc3-int. FRB migrate only slightly above the untagged proteins and, thus, are difficult to discern from the endogenous subunits in the mock-depleted samples. Note also that the western signals for Scc1-FRB and Smc3-FRB do not accurately reflect their expression levels because the corresponding antibodies display a greatly reduced sensitivity when their antigens are C-terminally tagged.
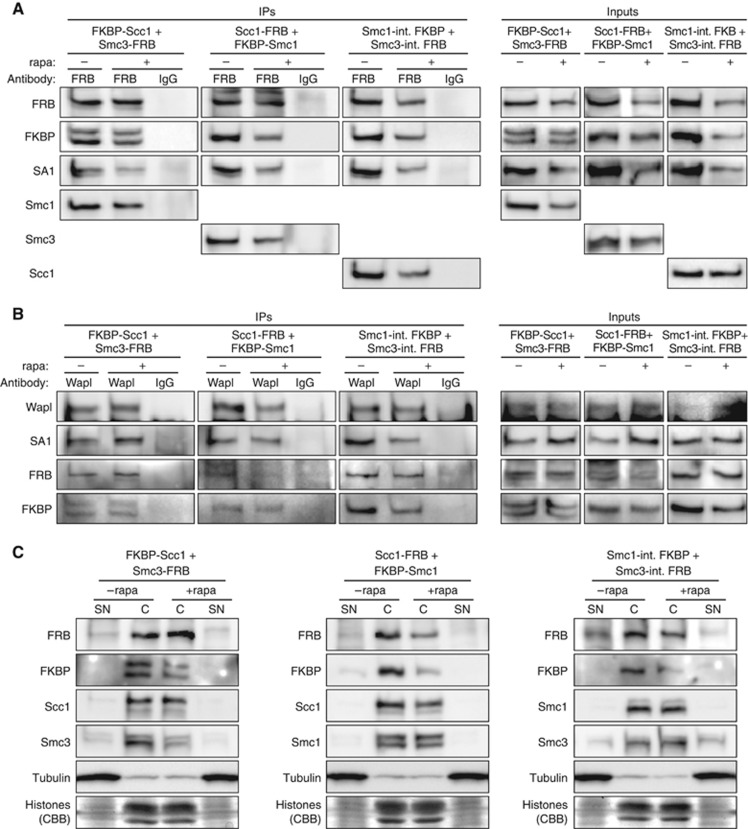
To assess the functionality of the modified cohesin subunits, we first conducted immunoprecipitation experiments (IPs) using FRB and Wapl antibodies. Subsequent western analyses revealed that the tagging did not interfere with the interaction of cohesin subunits with each other and with the essential prophase pathway component Wapl (Figure 3A and B). Importantly, addition of rapamycin prior to cell lysis had no impact on the outcome of the IPs, thereby excluding the remote possibility that the tags might impair proper cohesin interactions only upon their induced heterodimerisation. We then prepared chromatin-enriched as well as cytoplasmic fractions from unsynchronised cells and analysed them by immunoblotting to detect FRB/FKBP-tagged as well as endogenous cohesin subunits. These experiments demonstrated that the tagged cohesin subunits behave just like the endogenous counterparts with respect to their pronounced association with interphase chromatin (Figure 3C). The results were independently confirmed by immunofluorescence (IF) microscopy, which demonstrated the association of FRB-tagged cohesin with pre-extracted interphase nuclei (Supplementary Figure S1). Thus, the engineered cohesin complexes seem to retain normal protein–protein and protein–DNA interaction abilities.
Figure 3.
The FRB/FKBP-tagged cohesin subunits incorporate into bona fide cohesin complexes capable of interacting with Wapl and chromatin. (A) FRB-tagged cohesin subunits are proficient in rapamycin-independent association with FKBP-tagged and endogenous cohesin subunits. The double-transgenic Hek293 lines were transfected with siRNAs targeting GL2 (luciferase, control) or the respective pair of cohesin subunits that was being replaced by the engineered variants and incubated for 3 days in the presence of doxycycline. Forty-one hours after siRNA transfection, cells were treated with rapamycin (rapa) or DMSO as control. The cells were arrested in prometaphase by a 16-h nocodazole treatment before they were harvested. Cell lysates were cleared by high-speed centrifugation to remove chromatin (Inputs) and then subjected to immunoprecipitation (IPs) with anti-FRB or unspecific IgG. Inputs and immunoprecipitates were finally analysed by western using the indicated antibodies. (B) The essential prophase pathway factor Wapl interacts with FRB/FKBP-tagged cohesin subunits irrespective of the presence or absence of rapamycin. The experiment was conducted as described in (A) with the exception that anti-Wapl was used for the IP instead of anti-FRB. (C) The FRB/FKBP-tagged cohesin subunits retain their ability to bind chromatin. The double-transgenic Hek293 lines were largely treated as described in (A) but without the nocodazole-mediated synchronisation. Cell lysates were prepared in the presence of nuclei permeabilising detergent and centrifuged to separate soluble supernatant (SN) from pelleting chromatin (C), both of which were subsequently analysed by western blot using the specified antibodies and by staining with Coomassie Brilliant Blue (CBB) to visualise histones.
DNA exits the cohesin ring through the Smc3–Scc1 gate
To test, which of the intersubunit contacts (Smc1–Smc3, Smc3–Scc1 or Scc1–Smc1) has to dissolve during dissociation of cohesin from chromatin in prophase, our double transgenic cell lines were treated as following (Figure 4A): the corresponding endogenous cohesin subunits were depleted by RNAi. At the same time, the (siRNA-resistant) transgenes were induced to express the two FKBP-FRB-tagged cohesin variants. Two days later, thymidine was added to synchronise the cells in early S phase. After additional 16 h, cells were released for 7 h into early G2 phase. Then, nocodazole was added for another 7 h to arrest cells in prometaphase of the following mitosis. Finally, the cells were fixed and processed for either in situ IF microscopy or chromosome spreading. Rapamycin (or DMSO as a control) was added at two alternative time points during the experiment, i.e., together with thymidine (‘Early’) or together with nocodazole (‘Late’). Notably, using a Smc1 antibody for IF, cohesin could be detected on Hec1-positive, condensed chromatin if—and only if—FKBP-Scc1 and Smc3-FRB were expressed and rapamycin was present (Figure 4C). The same observation was made when transgenic cohesin was specifically visualised with an FRB antibody (Figure 4D). Quantifications of three independent IF experiments each led to very similar results for both antibodies (Figure 4E and F). Our data strongly suggest that displacement of cohesin from early mitotic chromatin requires the ability to open the Smc3–Scc1 gate.
Figure 4.
DNA exits the cohesin ring through the Smc3–Scc1 gate. (A) Assay timeline for results shown in (B–F). (B) Indicated double-transgenic cell lines were treated as illustrated in (A) and then subjected to chromosome spreading and quantitative assessment of arm cohesion. The relative numbers of cells displaying zipped prometaphase chromosomes are plotted. Each column represents the mean of three independent data points (represented by circles, triangles and squares) totalling 600 analysed cells. Corresponding data sets across all cell lines and conditions are identified by identical shapes. (C, D) Indicated double-transgenic cell lines were treated as illustrated in (A) and then Hoechst 33342- and immunostained to in situ-visualise DNA, Hec1 and Smc1 (C) or FRB (D). Exemplary images are shown. Scale bars=10 μm. (E, F) Quantification of the data shown in (C) and (D), respectively. The relative numbers of cells with prometaphase chromatin positive for Smc1 (E) or FRB (F) are plotted. Circles, triangles and squares correspond to individual data points of three independent reiterations while columns represent means. Data sets of each experiment across all cell lines and conditions are represented by identical shapes. Between 293 and 600 cells were analysed per column.
When rapamycin was added late, that is, merely 7 h before analysis, around 55% of all FKBP-Scc1 and Smc3-FRB expressing prometaphase cells exhibited persistence of chromosomal cohesin (Figure 4E and F). If loading/establishment of cohesion did require opening of the Smc3–Scc1 gate, then this fraction would be expected to decrease when rapamycin was already present as most cells traversed telophase and G1 phase. Instead, the fraction of prometaphase cells with cohesin-positive chromosomes increased to around 70% when rapamycin was added 30 h prior to fixation (Figure 4E and F) suggesting that DNA does not enter the cohesin ring through the Smc3–Scc1 gate (see below).
Extending the IF-based localisation studies we also conducted morphological analyses of spread prometaphase chromosomes. Due to action of the prophase pathway these chromosomes usually exhibit fully separated arms and appear butterfly-like. In contrast, close pairing of sister chromatids along their entire length indicates that DNA entrapment by cohesin persisted beyond prophase. These zipped chromosomes were detected in appreciable amounts (40–50%) only when FKBP-Scc1 and Smc3-FRB were expressed and rapamycin was present (Figure 4B). Under all other conditions the typical butterfly-like chromosome morphology predominated (85–90%). Taken together, our results demonstrate that entrapped DNA leaves the cohesin ring through the Smc3–Scc1 gate during mitotic prophase in human cells.
The prophase pathway does not require opening of the Scc1–Smc1 gate
The data presented above did not indicate any involvement of the Smc1–Smc3 or Scc1–Smc1 gate in the displacement of cohesin from prophase chromosomes. While we can largely rule out functional inactivation of the tagged cohesin subunits (see Figure 3), these negative results could still be accredited to improper relative orientation of FRB and FKBP that prevented their rapamycin-induced heterodimerisation. To address these issues, we first re-investigated the putative role of the Scc1–Smc1 gate for the prophase pathway by creating a stable cell line that inducibly expressed a direct in-frame fusion of both open reading frames (ORFs) (Figure 5A). Like endogenous Smc1, the transgenic Scc1–Smc1 fusion protein also co-purified with chromatin isolated from unsynchronised cells indicating that it can readily be loaded onto DNA (Figure 5B). Similarly to endogenous SA1, it also co-immunoprecipitated with endogenous Smc3 and Wapl, thereby signifying its incorporation into a bona fide cohesin ring (Figure 5C and D). Depletion of endogenous Scc1 and Smc1 by RNAi and simultaneous induction of the Scc1–Smc1 chimera did not result in prolonged association of cohesin with chromatin in prometaphase-arrested cells (Figure 5E). Furthermore, in spreads we could not detect any increase in tightly cohesed, zipped chromosomes upon expression of the fusion construct (Figure 5F). Thus, based on these observations we propose that Scc1 does not need to dissociate from Smc1 for cohesin to leave prophase chromatin.
Figure 5.
DNA does not leave cohesin through the Scc1–Smc1 gate. (A) A stable transgenic Hek293 cell line that inducibly expresses a covalent Scc1–Smc1 fusion. The siRNA-resistant expression of the Scc1–Smc1 fusion protein with respect to the endogenous Smc1 that persists after 2 days of RNAi is shown. (B) Covalent fusion of Scc1 and Smc1 is compatible with chromatin association. Insoluble chromatin (C) and soluble supernatant (SN) fractions of the Scc1–Smc1 expressing cell line were analysed by Smc1- and α-tubulin-westerns and by Coomassie Brilliant Blue (CBB) staining of chromatin bound histones. (C, D) The Scc1–Smc1 fusion protein interacts with endogenous cohesin subunits and Wapl. The transgenic Hek293 line was largely treated as described in Figure 3A (only without addition of rapamycin). The respective mitotic cell lysate (Input) was cleared by high-speed centrifugation to remove chromatin and subjected to immunoprecipitations (IPs) with anti-Smc3 (C) or anti-Wapl (D). Co-purifying proteins were detected by western analysis using the indicated antibodies. Mock-IPs using unspecific rabbit IgG served as controls. (E, F) A covalent Scc1–Smc1 fusion does not impair release of cohesin from prophase chromosomes. Scc1–Smc1 expressing cells were treated largely as described in Figure 4A (only without addition of rapamycin) and then analysed by IF (E) and chromosome spreading (F). (E) Averaged over two independent experiments, 99.5% of a total of 200 Hoechst 33342, anti-Hec1, and anti-Smc1 stained cells did not exhibit any signs of Smc1–chromatin association beyond prophase. (F) Averaged over three independent experiments, 94.7% of a total of 300 spread nuclei displayed butterfly-like chromosomes. An increase in tightly cohesed (‘zipped’) chromosomes could not be detected.
DNA enters the cohesin ring through the Smc1–Smc3 hinge
Finally, we wanted to address whether the internally FKBP/FRB-tagged Smc1 and -3 remain proficient in mediating cohesion and whether the specific sterical orientation of the tags allows their near-irreversible heterodimerisation by rapamycin. It is known that opening of the hinge region is important for loading of S. cerevisiae cohesin (Gruber et al, 2006). If this process is conserved in humans, then addition of rapamycin prior to telophase should impair cohesin loading. To test this prediction, endogenous Smc1 and Smc3 were knocked down by RNAi while Smc1-int. FKBP and Smc3-int. FRB were simultaneously expressed. Two days thereafter, the cells were synchronised in prometaphase by a 16-h nocodazole treatment, during which rapamycin (or DMSO) was added after 14 h. Then, the cells were released for 2.5 h into nocodazole-free medium (containing rapamycin or DMSO) before they were fixed in early G1 phase and analysed by IF for their ability to reload FRB-tagged cohesin (Figure 6A). Importantly, rapamycin reduced the fraction of cells that exhibited FRB-positive postmitotic chromatin by 30% (Figure 6B). While Smc1-int. FKBP and Smc3-int. FRB containing cohesin complexes should be prevented from reloading in the presence of rapamycin, those that contain at least one residual endogenous SMC subunit should be unaffected by rapamycin in their re-association with DNA. Consistent with these notions, the signals for Scc1 remained unaffected by rapamycin with respect to the number of cells that exhibited reloading (around 80%) but exhibited a drop in intensity at the level of the individual cells (Figure 6C and D). Together, these data argue that the Smc1-int. FKBP and Smc3-int. FRB are functional and rapamycin lockable, thereby corroborating the conclusion that opening of the hinge domain is dispensable for the release of arm cohesin in prophase (see Figure 4 above). Moreover, they also demonstrate for the first time that opening of the Smc1–Smc3 gate is necessary for proper loading of cohesin onto chromatin in humans.
Figure 6.
Cohesin loading requires opening of the Smc1–Smc3 hinge. (A, B) A double-transgenic cell line induced to co-express Smc1-int. FKBP and Smc3-int. FRB was simultaneously depleted of endogenous Smc1 and -3 by transfection of corresponding siRNAs, nocodazole arrested in prometaphase and finally released into telophase/early G1 phase in the presence or absence of rapamycin (±rapa). (A) Following pre-extraction, fixation and staining of DNA and FRB, only those cells were analysed that were still in telophase or formed closely coupled pairs with similar FRB levels. Exemplary images are shown. Scale bar=10 μm. (B) Quantitative assessment of recombinant cohesin loading. The relative numbers of cell pairs exhibiting FRB staining in telo-/early G1 phase are plotted. Columns correspond to means of three independent experiments with each triplicate totalling 600 cells. Circles, triangles and squares represent data points and identify respective sets from individual experiments. (C, D) Cells were treated as described in (A, B) and stained for FRB and endogenous Scc1. Most of the early G1-phase nuclei that lacked an FRB signal could still reload endogenous Scc1 in the presence of Smc1-int. FKBP, Smc3-int. FRB and rapamycin (due to incomplete knockdown of endogenous Smc1/3 and the rapamycin-independent loading of cohesin complexes that retain at least one untagged SMC subunit). However, the intensities of the Scc1 signals were reduced consistent with the transgenic proteins exerting a dominant-negative effect on cohesin reloading (see text for details). Scale bar=10 μm. For each quantification, 100 cells were counted.
Discussion
Here, we explore rapamycin-mediated in vivo bonding of FRB/FKBP-tagged recombinant proteins to demonstrate that preventing ring opening at specific sites disturbs proper chromatin–cohesin interactions also in human cells. Our findings therefore endorse the ring model, which has so far been exclusively based on budding yeast studies, and argue for its general applicability. We demonstrate that blocking the passage through the Smc1–Smc3 interface compromises cohesin loading onto human chromatin. Studying S. cerevisiae Gruber et al (2006) made a similar observation leading us to conclude that the Smc1–Smc3 hinge constitutes the universal DNA entry gate into the cohesin ring. Moreover, we provide evidence for prophase pathway failure upon sealing of Smc3–Scc1 but not the other two gates in cycling human cells. Supporting this notion, an accompanying Drosophila study shows that the dissociation of cohesin from prophase chromosome arms is prevented in neuroblasts if Smc3 and Scc1 are covalently fused and restored if the linker peptide is proteolytically cleaved (Eichinger et al, co-submitted manuscript). The above findings are summarised in an updated model of the dynamic cohesin–chromosome interactions within the metazoan cell cycle (Figure 7): DNA enters the ring through the Smc1–Smc3 gate, when cohesin is first loaded in telophase, and leaves it through the Smc3–Scc1 gate, when the bulk of cohesin is displaced from chromosome arms in prophase. Interestingly, transient opening of the Smc3–Smc1 interface is no speciality of metazoan cells cycling through early mitosis. In S. cerevisiae, which lacks a prophase pathway, and in Drosophila salivary gland cells, which are no longer dividing, a large fraction—if not most—of chromosomal cohesin exhibits dynamics that also depends on the ability of Smc3 and Scc1 to disengage (Chan et al, 2012; Eichinger et al, co-submitted manuscript). Taken together, this means that the exit gate of DNA from cohesin is conserved from yeast to man and represented by the Smc3–Scc1 interface.
Figure 7.
Model of the dynamic cohesin–chromosome interactions throughout the metazoan cell cycle. DNA enters the tripartite cohesin ring through the Smc1–Smc3 gate in telophase. Cohesion is then established during DNA replication in S phase. The bulk of cohesin is released from chromosome arms in prophase of mitosis when DNA exits the ring through the Smc3–Scc1 gate. (Note that sister chromatid separation occurs only when prevailing centromeric cohesin is proteolytically cleaved by separase at the metaphase-to-anaphase transition. This step was omitted for sake of clarity.)
Why does DNA enter and exit the cohesin ring through different gates? One obvious advantage is that the use of two functionally distinct gates offers a simpler, more explicit regulation of the individual gate in response to input signals, which can now be integrated more readily. Another consideration relates to the thrilling question of whether (and—if yes—how) the cell knows in which direction to pass DNA through a given opening in the cohesin ring. With two separate gates this problem could elegantly be solved if both gates would work like turnstile doors allowing DNA to pass only in one direction. Such a model has essentially been proposed for the Smc1–Smc3 hinge on the basis of available structural informations (Gruber et al, 2006). It is currently unknown whether the efficient dissociation of arm cohesin in prophase requires the Smc3–Scc1 interface to (also) operate in a unidirectional manner or whether in this case random diffusion of DNA through the exit gate might be sufficient. Determining the as yet unknown structure of the Smc3–Scc1 interface will be an important first step towards clarifying this issue.
Materials and methods
Generation of double-transgenic cell lines inducibly expressing two tailored cohesin subunits
An SV40 promoter-loxP-ATG cassette was inserted between the ampicillin resistance gene and the CMV promoter of pcDNA5/FRT/TO (Life Technologies). The resulting pcDNAL/FRT/TO was used to accommodate in its multiple cloning site (MCS) the ORF of the first FRB- or FKBP-tagged cohesin subunit. Flp-recombinase mediated integration of this construct into the unique FRT site of a host cell genome (here: Flp-In T-Rex Hek293 line from Life Technologies) allowed for a second round of site-specific integration by Cre recombinase. The corresponding vector, pcDNA5/loxP/TO, had the FRT-hygromycin cassette replaced by loxP-neomycin and carried within its MCS the ORF of the second FRB- or FKBP-tagged cohesin subunit. Details of this method will be described elsewhere.
Antibodies
For staining/binding of FRB in western blots (1.41 μg/ml), IFM (2.82 μg/ml) and IP (10 μg/2 × 107 cells), a polyclonal rabbit antibody was used, which has been raised against the bacterially expressed full-length protein (Coring System Diagnostix). For staining of FKBP12 in western blots (1:1000 or 1:500), we used a monoclonal mouse antibody from Abcam (ab58072). For staining of Smc1 in western blots (1:4000) and IFM (1:500), we used a rabbit antibody from Bethyl Laboratories (A300-055A). Scc1 was stained in western blots either with a rabbit peptide antibody (1:1000 of serum) (Stemmann et al, 2001) or with a commercial mouse monoclonal antibody from Millipore (1:1000) (05-908), which was also used for IFM (1:500). Smc3 was detected either with a rabbit peptide antibody (Schöckel et al, 2011) or with a rabbit antibody raised against the C-terminal 180 amino acids of Xenopus Smc3 (kindly provided by Susannah Rankin) (1 μg/ml). The latter was used for IP at 10 μg/2 × 107 cells. Susannah Rankin further supported us with rabbit antibodies raised against the C-terminus of Wapl (IP: 5 μg/2 × 107 cells; western blot: 1.3 μg/ml) and the C-terminal 199 amino acids of SA1 (3.5 μg/ml). Both were raised against the Xenopus orthologues but are cross-reactive with the human proteins. For α-tubulin staining in western blots (1:200 of hybridoma supernatant), we used a mouse monoclonal antibody from the Developmental Studies Hybridoma Bank (12G10). Hec1 was stained in IFM experiments (1:500) using a mouse monoclonal antibody from Genetex (GTX70268). Unspecific rabbit IgG was from Bethyl. Secondary antibodies for western blot (all 1:20 000): HRP conjugated goat anti-rabbit and anti-mouse IgG (Sigma Aldrich). Secondary antibodies for IFM (all 1:500): goat anti-rabbit- and anti-mouse-Alexa Fluor 488, as well as goat anti-rabbit and anti-mouse-Cy3 (Life Technologies).
siRNAs
In the GL2/Wapl RNAi experiment, 70 nM siRNA was used. For all other RNAi knockdowns, 57 nM siRNA was used (when two siRNAs were used for one target 28.5 nM of each were used). siRNAs targeting Scc1, Smc3, Wapl and Luciferase GL2 were described previously (Schöckel et al, 2011). The sequence of the siRNA targeting the ORF of Smc1 was 5′-GGAAGAAAGUAGAGACAGA-3′.
Cell culture
Cells (Hek293T and Hek293 FlpIn cells stably expressing one or two transgenes) were cultured at 37°C and 5% CO2. Normally, cells were grown in Dulbecco’s Modified Eagle Medium (DMEM)/10% Fetal Bovine Serum (FBS)/5% Penicillin/Streptomycin. During transfections only FBS was added to the medium. In the GL2/Wapl RNAi experiment, siRNAs were transfected using Invitrogen Lipofectamine 2000 (Life Technologies) according to manufacturer’s instructions. All other transfections were conducted using the calcium phosphate-based method. To synchronise cells, 2 mM Thymidine was added to the medium for 16 h, arresting the cells in early S phase. Cells were then released into fresh medium. To arrest cells in prometaphase, 200 ng/ml nocodazole was added for up to 16 h. Rapamycin was added at 100 nM. Experiments in Figure 4 were performed as described in the text and displayed in Figure 4A. The experiments in Figure 5E and F were performed largely as described in the text. Only, nocodazole was added 49 h after siRNA transfection.
Chromatin isolation
When chromatin was to be isolated for fluorescence microscopy, cells were harvested and swollen in 1 × PME (5 mM PIPES/NaOH pH 7.2, 5 mM NaCl, 5 mM MgCl2, 1 mM EGTA) for 5 min at RT. After pelleting (300 g, 2 min, RT), this step was repeated. Cells were again pelleted (300 g, 2 min, RT), resuspended in PME lysis buffer (1 × PME, 1% (v/v) thiodiethylene glycol, 10 μg/ml Cytochalasin B, 0.2% (v/v) digitonin, 1 tablet/50 ml Complete protease inhibitors (Roche)) and incubated for 5 min on ice. Chromatin was eventually spun onto poly-L-lysine coated coverslips through an LSS cushion (1 × PME, 1% (v/v) thiodiethylene glycol, 0.9 M sucrose, 0.2% (v/v) digitonin) in a swing-out rotor at 2900 g and 4°C for 30 min. Coverslips were retrieved, washed with PBS and then fixed with PBS/4% (w/v) paraformaldehyde for 15 min. After another PBS wash, the fixing reaction was quenched with PBS/50 mM NH4Cl for 5 min followed by an additional PBS wash. DNA was stained with 2 μg/ml Hoechst 33342 in PBS for 20 min. The coverslips were washed twice with PBS and then mounted (see above).
When chromatin was isolated for western blot analysis, we used a method described by Méndez and Stillman (2000). Histones as a DNA marker were stained in the same gel used for the western blot using colloidal Coomassie Brilliant Blue (CBB).
Immunoprecipitation
Cells were harvested and lysed in lysis buffer 2 (20 mM Tris/HCl pH 7.7, 100 mM NaCl, 5 mM MgCl2, 0.1% (w/v) Triton X-100, 5% (v/v) glycerol, 10 mM NaF, 20 mM β-glycerophosphate, 1 tablet/50 ml Complete protease inhibitors (Roche)). The corresponding lysates were then incubated with the respective antibodies coupled to protein G sepharose 4 Fast Flow beads (GE Life Science) overnight at 4°C. The beads were washed six times with lysis buffer 2 (200 g, 1.5 min, 4°C), then supplemented with 2 × SDS sample buffer and brought to 95°C to elute the desired protein(s). This suspension was transferred to Mobicol columns (MoBiTec) and centrifuged (200 g, 2 min, RT) to separate the eluate from the beads.
IF microscopy
Cells grown on poly-L-lysine coated coverslips were pre-extracted with PBS-Tx (PBS/0.1% (w/v) Triton X-100) for 3 min, fixed with freshly prepared PBS/4% (w/v) paraformaldehyde for 15 min, treated with PBS/50 mM NH4Cl for 5 min to quench residual fixative and then incubated with PBS-Tx for 5 min. Each of these steps was followed by a brief PBS wash. The samples were blocked with PBS/3% (w/v) BSA/0.02% (v/v) NaN3 for ≥7 h at 4°C, moved to room temperature and then incubated for 1 h with the corresponding primary antibody in the same buffer. This was followed by three PBS washes, incubation with secondary antibody in blocking buffer for 1 h, two PBS washes, staining with 2 μg/ml Hoechst 33342 in PBS for 15 min and three final PBS washes. The coverslips were mounted onto glass slides in 2,33% (w/v) 1,4-diazabicyclo-[2,2,2]-octane/20 mM Tris–HCl, pH 8,0/78% (v/v) glycerol and imaged on an Axio Imager A1 microscope (Zeiss) using a 23.0 1.4 MP Spot Persuit camera system and Spot 4.5.9.1 software (Diagnostic Instruments).
Chromosome spreads
Chromosome spreads were performed as described in Boos et al (2008) but with the use of Hoechst 33342 (2 μg/ml) instead of Giemsa for DNA staining. Microscopy and image acquisition was performed as described above.
Supplementary Material
Acknowledgments
We would like to thank Valentina Ahl, Markus Hermann, Lisa Mohr and Annika Pfeiffer for technical assistance. Furthermore, we thank Susannah Rankin and Jan-Michael Peters for generously sharing reagents. This work was supported by the Bayreuth International Graduate School of Science (BIGSS) and a grant of the Deutsche Forschungsgemeinschaft (STE997/3-2 within the priority program SPP1384).
Author contributions: JB conducted the experiments and wrote the paper. OS designed the experiments and contributed to writing of the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anderson DE, Losada A, Erickson HP, Hirano T (2002) Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol 156: 419–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski LA, Liu CW, Wandless TJ (2005) Characterization of the FKBP.rapamycin.FRB ternary complex. J Am Chem Soc 127: 4715–4721 [DOI] [PubMed] [Google Scholar]
- Boos D, Kuffer C, Lenobel R, Körner R, Stemmann O (2008) Phosphorylation-dependent binding of cyclin B1 to a Cdc6-like domain of human separase. J Biol Chem 283: 816–823 [DOI] [PubMed] [Google Scholar]
- Chan K-L, Roig MB, Hu B, Beckouët F, Metson J, Nasmyth K (2012) Cohesin's DNA exit gate is distinct from its entrance gate and is regulated by acetylation. Cell 150: 961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Tóth A, Shevchenko A, Nasmyth K (2000) Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell 5: 243–254 [DOI] [PubMed] [Google Scholar]
- Gandhi R, Gillespie PJ, Hirano T (2006) Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol 16: 2406–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giménez-Abián JF, Sumara I, Hirota T, Hauf S, Gerlich D, la Torre de C, Ellenberg J, Peters J-M (2004) Regulation of sister chromatid cohesion between chromosome arms. Curr Biol 14: 1187–1193 [DOI] [PubMed] [Google Scholar]
- Gruber S, Arumugam P, Katou Y, Kuglitsch D, Helmhart W, Shirahige K, Nasmyth K (2006) Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell 127: 523–537 [DOI] [PubMed] [Google Scholar]
- Gruber S, Haering CH, Nasmyth K (2003) Chromosomal cohesin forms a ring. Cell 112: 765–777 [DOI] [PubMed] [Google Scholar]
- Haering CH, Farcas A-M, Arumugam P, Metson J, Nasmyth K (2008) The cohesin ring concatenates sister DNA molecules. Nature 454: 297–301 [DOI] [PubMed] [Google Scholar]
- Haering CH, Löwe J, Hochwagen A, Nasmyth K (2002) Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell 9: 773–788 [DOI] [PubMed] [Google Scholar]
- Haering CH, Schoffnegger D, Nishino T, Helmhart W, Nasmyth K, Löwe J (2004) Structure and stability of cohesin's Smc1-kleisin interaction. Mol Cell 15: 951–964 [DOI] [PubMed] [Google Scholar]
- Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters J-M (2005) Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol 3: e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D, Nasmyth K (2005) A topological interaction between cohesin rings and a circular minichromosome. Cell 122: 849–860 [DOI] [PubMed] [Google Scholar]
- Ivanov D, Nasmyth K (2007) A physical assay for sister chromatid cohesion in vitro. Mol Cell 27: 300–310 [DOI] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters J-M (2006) Wapl controls the dynamic association of cohesin with chromatin. Cell 127: 955–967 [DOI] [PubMed] [Google Scholar]
- Lafont AL, Song J, Rankin S (2010) Sororin cooperates with the acetyltransferase Eco2 to ensure DNA replication-dependent sister chromatid cohesion. Proc Natl Acad Sci 107: 20364–20369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez J, Stillman B (2000) Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol 20: 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, Bando M, Shirahige K, Hyman AA, Mechtler K, Peters J-M (2010) Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell 143: 737–749 [DOI] [PubMed] [Google Scholar]
- Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, Beckouët F, Underwood P, Metson J, Imre R, Mechtler K, Katis VL, Nasmyth K (2009) Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol Cell 33: 763–774 [DOI] [PubMed] [Google Scholar]
- Schöckel L, Möckel M, Mayer B, Boos D, Stemmann O (2011) Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat Cell Biol 13: 966–972 [DOI] [PubMed] [Google Scholar]
- Shintomi K, Hirano T (2009) Releasing cohesin from chromosome arms in early mitosis: opposing actions of Wapl-Pds5 and Sgo1. Genes Dev 23: 2224–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmann O, Zou H, Gerber SA, Gygi SP, Kirschner MW (2001) Dual inhibition of sister chromatid separation at metaphase. Cell 107: 715–726 [DOI] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Redemann N, Nigg EA, Peters J-M (2002) The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol Cell 9: 515–525 [DOI] [PubMed] [Google Scholar]
- Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC (2004) Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol 6: 991–996 [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K (2000) Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103: 375–386 [DOI] [PubMed] [Google Scholar]
- Waizenegger IC, Hauf S, Meinke A, Peters JM (2000) Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 103: 399–410 [DOI] [PubMed] [Google Scholar]
- Weitzer S, Lehane C, Uhlmann F (2003) A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr Biol 13: 1930–1940 [DOI] [PubMed] [Google Scholar]
- Zhang J, Shi X, Li Y, Kim B-J, Jia J, Huang Z, Yang T, Fu X, Jung SY, Wang Y, Zhang P, Kim S-T, Pan X, Qin J (2008) Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell 31: 143–151 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.