Abstract
The guanine nucleotide exchange factor C3G (RAPGEF1) regulates proliferation, migration, and differentiation of cells and is essential for mammalian embryonic development. The molecular effectors of C3G dependent functions are poorly understood. Here we report that C3G functions as a negative regulator of β-catenin, a major player in pathways known to be deregulated in human cancers. In mammalian cells, C3G is present in a complex with cellular β-catenin. The proline rich Crk binding region of C3G and residues 90-525 of β-catenin are sufficient for the interaction. Knockdown of cellular C3G stimulated, and its overexpression repressed, β-catenin/TCF transcription activity. C3G acts by destabilizing β-catenin protein and inhibiting its nuclear accumulation. Nuclear extracts of C3G overexpressing cells showed reduced binding to TCF consensus oligos. C3G exerts its effects independent of its function as an exchange factor. It also inhibits stability and activity of an N-terminal deletion construct of β-catenin that is not subject to GSK3β dependent phosphorylation, suggesting that C3G exerts its effect independent of GSK3β. β-catenin repression by C3G was not significantly altered in the presence of proteasome inhibitors, MG132 or lactacystin, suggesting that alternate mechanisms are engaged by C3G to cause β-catenin turnover. C3G expression represses β-catenin target gene expression, and stable clones of MCF-7 breast cancer cells expressing C3G showed reduced migration. Activation of cellular β-catenin or expression of constitutively active β-catenin resulted in reduced C3G expression, indicating that C3G gene expression is negatively regulated by β-catenin. Our results identify a novel property of C3G in functioning as a negative regulator of β-catenin signaling by promoting its degradation. In addition, we show that β-catenin inhibits C3G expression, forming a feedback loop.
Keywords: C3G, β-catenin, guanine nucleotide exchange factor, signaling, GSK3β
Introduction
By virtue of its function as a component of cell-cell junctions and as a modulator of transcription, β-catenin is involved in regulating a variety of cellular properties. The pool of cytosolic β-catenin that is a component of the degradation complex consisting of Axin, APC, and GSK3β is under rapid turnover, being controlled by phosphorylation and the ubiquitin proteasome system.1 CK1 and GSK3β phosphorylation sites, as well as ubiquitin sites targeted by β-TrCP, are present in the N-terminal of β-catenin.2 In response to Wnt/TGF-β signaling or inactivation of the kinase GSK3β, β-catenin is stabilized and activated by nuclear translocation to regulate transcription of downstream targets by complexing with TCF/LEF. N-terminal deletion of β-catenin makes it refractory to phosphorylation and shows constitutive activity.3 Major targets are genes that regulate embryonic development, cell proliferation, survival, and migration like cyclin D1, matrilysin, cMyc, Id2, transferrin receptor (TfR), Akt, Cox2, and Survivin.4-6 Therefore, aberrant β-catenin activation is associated with tumorigenesis and metastasis. Mutations in β-catenin or components of its degradation complex result in aberrant overexpression of β-catenin seen in a large number of human solid tumors.7
Regulation of gene expression by β-catenin has primarily been associated with transcriptional activation, but a few studies have shown that β-catenin can also repress target genes.8,9 β-catenin signaling is evolutionarily conserved and regulates embryonic development. Knockout mice show early embryonic lethality with embryos not developing beyond 6.5 to 7.5 days post coitum.10 Transgenic mice with constitutively active β-catenin and tissue specific activation of β-catenin show embryonic lethality, indicating that maintaining optimal levels of β-catenin is essential for proper cell fate determination.11,12 In adult cells, β-catenin regulates morphology, proliferation, and differentiation. Proteins interacting with β-catenin can modulate its stability and function. Some molecules like 14-3-3ζ are activators, and others like ICAT, Dickkopf, Axin, and HIF-1α are inhibitors.13-15 β-catenin function can be repressed by several mechanisms, such as decrease in protein stability, inhibition of its nuclear translocation, inhibition of binding to TCF, enabling engagement with repressive factors, and exosome mediated release.16 Alternate mechanisms of β-catenin degradation, independent of GSK3β phosphorylation, also have been described. The p53 target Siah-1, which binds ubiquitin conjugating enzymes, has been shown to cause β-catenin degradation independent of GSK3β phosphorylation and β-TrCP mediated proteolysis.17 Understanding β-catenin regulation by cellular proteins belonging to various signaling pathways would help in delineating its functions related to development, cell fate determination, and tumor growth.
The guanine nucleotide exchange factor (GEF) C3G is ubiquitously expressed and relays a variety of extracellular signals to regulate functions like cytoskeletal remodeling, adhesion, migration, proliferation, apoptosis, and transformation.18 C3G is a multidomain protein and has functions dependent on its catalytic domain and protein interaction domains. It has been shown to function as an exchange factor for Ras family GTPases, Rap1, Rap2, R-Ras, and the Rho family GTPase, TC10.18 Proline rich sequences present in its central domain are responsible for interaction with SH3 domain containing proteins like Crk, Grb2, p130Cas, Hck, and c-Abl.19-23 More recently, this domain has been shown to interact with actin and TC-PTP, molecules lacking SH3 domains.24,25 Sequences in the N-terminus are involved in its interaction with E-cadherin,26 a molecule involved in maintenance of cell junctions. Endogenous as well as overexpressed C3G localizes primarily to the cytoplasm27 and shows membrane association upon receptor activation.28,29 Its association with endosomes has been shown in neuronal cells.30 C3G function is regulated through protein-interaction, phosphorylation, and actin cytoskeletal dynamics.18
C3G is essential for mammalian embryonic development as C3G knockout mice show early embryonic lethality.31 Lack of C3G appears to result in defects in multiple systems during development.32 C3G null fibroblasts are defective in cell adhesion and migration.31 C3G dependent signaling has been shown to promote or inhibit cell proliferation in a cell type dependent manner.33-36 Independent of its catalytic function, C3G shows transformation suppression properties that may be significant in the context of cancer development.37 Recent studies have shown an association of deregulated C3G levels with certain human cancers.38,39 To understand the molecular effectors and pathways engaged by C3G in signaling to inhibit cell proliferation and migration, we examined its ability to modulate β-catenin signaling functions. We show that C3G physically and functionally interacts with β-catenin and plays a repressive role in its activation by enhancing degradation, independent of GSK3β mediated phosphorylation. In addition, we show that β-catenin activation results in repression of C3G mRNA and protein levels, suggesting reciprocal regulation between C3G and β-catenin.
Results
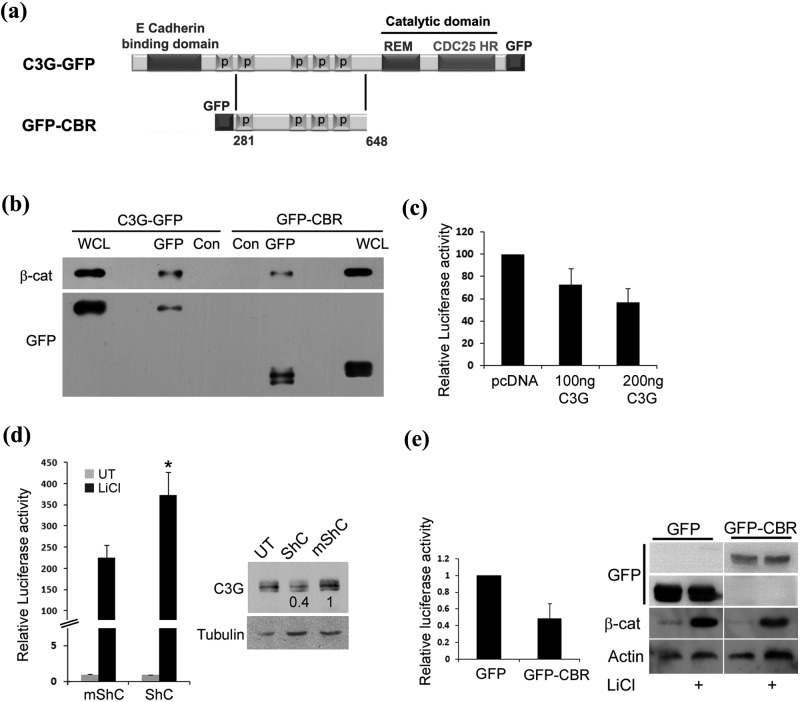
C3G complexes with and inhibits transcriptional activity of β-catenin
A large number of cellular proteins that regulate β-catenin activity form complexes with β-catenin. We therefore investigated whether C3G could interact with cellular β-catenin by examining its co-precipitation with C3G from cell lysates. GFP-tagged full-length C3G and a deletion variant having only its proline-rich central Crk binding region (CBR) (Fig. 1a) were expressed in HEK 293T cells and lysates used for immunoprecipitation, followed by Western blotting. Cellular β-catenin co-precipitated with both C3G and CBR, indicating that the central domain of C3G was sufficient for the interaction (Fig. 1b). A higher amount of β-catenin complexed with full-length C3G, suggesting that other domains of C3G may also aid in complex formation. Deletion constructs of β-catenin lacking the N-terminal regulatory domain or C-terminal transactivation domain could also form a complex with CBR, indicating that interaction was not dependent on N-terminal or C-terminal domains of β-catenin (Suppl. Fig. S1, b and d). The interaction between C3G and β-catenin was examined in an in vitro binding assay using purified recombinant GST-CBR fusion protein and cell lysates expressing ΔN β-catenin or ΔC 525 β-catenin. Both deletion proteins interacted with GST-CBR but not with GST (Suppl. Fig. S1, c and e).
Figure 1.
C3G interacts with and downregulates transcription activity of β-catenin/TCF. (a) Schematic shows domains of C3G in C3G-GFP and GFP-CBR constructs. (b) Cellular β-catenin co-precipitates with C3G. Cell lysates of HEK 293T cells transfected with C3G-GFP or GFP-CBR were subjected to immunoprecipitation with either GFP antibody or normal mouse IgG (Con), followed by Western blotting to detect the indicated proteins with the corresponding antibodies. (c) Effect of C3G on transcription activity of β-catenin induced by GSK3β inhibition. Lysates of HEK 293T cells transfected with reporter constructs along with pcDNA or C3G expression vector and treated with LiCl were used for luciferase assays. Relative luciferase activity after normalization with β-Gal is shown as mean ± SD. (d) Knockdown of endogenous C3G levels causes enhanced β-catenin dependent promoter activation. HEK 293T cells transfected with reporter constructs along with shRNA construct targeting C3G (ShC) or mutant shRNA (mShC) were either left untreated or treated with LiCl. Cell lysates, made after 48 hours of expression, were used for luciferase assays and Western blotting to detect C3G (right panel). Tubulin was used as a loading control. Relative luciferase activity after normalization with β-Gal is shown as mean ± SD in the bar diagram.*P < 0.01. (e) Crk binding region (CBR) of C3G is sufficient for repressing TCF activity. β-catenin dependent luciferase assay in HEK 293T cells expressing GFP or GFP-CBR and treated with LiCl. Western blot was carried out using the lysates to confirm an increase in β-catenin levels upon LiCl treatment. Expression of GFP and GFP-CBR was detected by probing the corresponding regions of the blot with GFP antibody.
To test the functional relevance of the interaction of C3G with β-catenin, we examined the possibility of C3G affecting β-catenin transcriptional activity. Assays using β-catenin/TCF-responsive luciferase reporter constructs were used, which is a well-established readout for β-catenin activity. Cells were transfected with either control or C3G plasmid along with reporter constructs and cellular β-catenin activation induced by LiCl, an inhibitor of GSK3β.40 C3G expression caused a dose-dependent decrease in TCF reporter activity (Fig. 1c). Knockdown of cellular C3G levels using small hairpin RNA (ShRNA) enhanced LiCl induced cellular β-catenin activation (Fig. 1d). These results indicated that cellular C3G negatively regulates β-catenin activity. Crk binding region of C3G, which was sufficient for interaction with β-catenin, also showed repression of LiCl induced TCF promoter activity (Fig. 1e). An increase in endogenous β-catenin in response to LiCl treatment was confirmed by Western blotting. These results indicated that the effect of C3G on β-catenin activity was independent of its exchange factor activity.
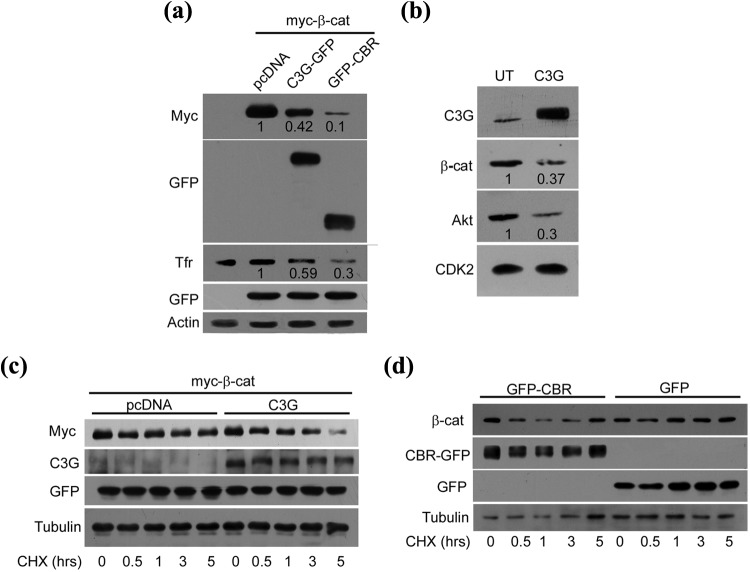
C3G reduces protein levels of β-catenin and its target genes
The mechanism involved in C3G mediated reduction in β-catenin activity was investigated by examining the effect of C3G expression on β-catenin protein levels and turnover. Myc-β-catenin was co-expressed with pcDNA, C3G-GFP, or GFP-CBR and 20 ng of GFP, and protein expression was examined. C3G as well as CBR caused a reduction in levels of ectopically expressed β-catenin (Fig. 2a). No effect was seen on GFP or on cellular actin. C3G expression also caused a reduction in cellular TfR levels, a target of β-catenin that has a role in malignancy.41 To rule out the possibility of variable β-catenin plasmid transfection, the effect of C3G overexpression on endogenous β-catenin was examined in MDA-MB-231 breast cancer cells. C3G overexpression resulted in a reduction in β-catenin and also a concomitant decrease in its target, Akt (Fig. 2b). C3G expression did not alter β-catenin transcript levels as examined by quantitative and semiquantitative RT-PCR (Suppl. Fig. S2, a and b).
Figure 2.
C3G reduces β-catenin protein and expression of β-catenin/TCF target genes. (a) Effect of C3G on exogenously expressed β-catenin protein levels. HEK 293T cells transfected with 200 ng of β-catenin expression vector and 20 ng of GFP along with 200 ng of pcDNA, C3G-GFP, or GFP-CBR expression vectors were lysed after 30 hours of expression and subjected to Western blotting with the indicated antibodies. TfR is a β-catenin target gene. GFP and actin were used as transfection and loading controls, respectively. Numbers indicate decrease in β-catenin and TfR levels in C3G expressing cells relative to levels in control plasmid transfected cells. (b) Effect of C3G expression on cellular β-catenin protein. MDA-MB-231 cells were transfected with 400 ng of C3G expression vector, and cell lysates made after 48 hours of expression were subjected to Western blotting, along with lysates from untransfected cells (UT), with the indicated antibodies. Numbers indicate quantitation as in (a). (c) HEK 293T cells were transfected with 200 ng of β-catenin and 200 ng of pcDNA or C3G and 20 ng of GFP vectors. Cells were treated with 40 µg/mL of cycloheximide after 10 hours of expression for the indicated time periods. Cell lysates made at each time point were subject to Western blotting with the indicated antibodies. Tubulin was used as loading control. (d) CBR domain of C3G enhances turnover of endogenous β-catenin. MDA-MB-231 cells transfected with 400 ng of GFP or GFP-CBR were treated with 40 µg/mL of cycloheximide after 30 hours of expression for the indicated time periods. Cell lysates made at each time point were subject to Western blotting with the indicated antibodies.
Cellular β-catenin protein is under constant turnover due to proteasome-mediated degradation. We examined whether C3G altered β-catenin protein stability by analyzing β-catenin protein levels in the presence of cycloheximide (CHX), an inhibitor of protein synthesis. Exogenously expressed β-catenin showed enhanced degradation in C3G expressing HEK 293T cells compared with control plasmid expressing cells (Fig. 2c). C3G levels were not significantly altered during the course of CHX treatment. C3G expression did not alter exogenously expressed GFP levels, indicating that the effect of C3G was specific to β-catenin. Half-life of overexpressed β-catenin calculated from multiple experiments in C3G expressing cells was 2.1 hours compared with 4.9 hours seen in control cells. Expression of CBR decreased the stability of cellular β-catenin in MDA-MB-231 cells, indicating that C3G mediated its effect on β-catenin turnover independent of its catalytic activity (Fig. 2d). These results also indicated that C3G exerts its effects on β-catenin through a post-translational mechanism.
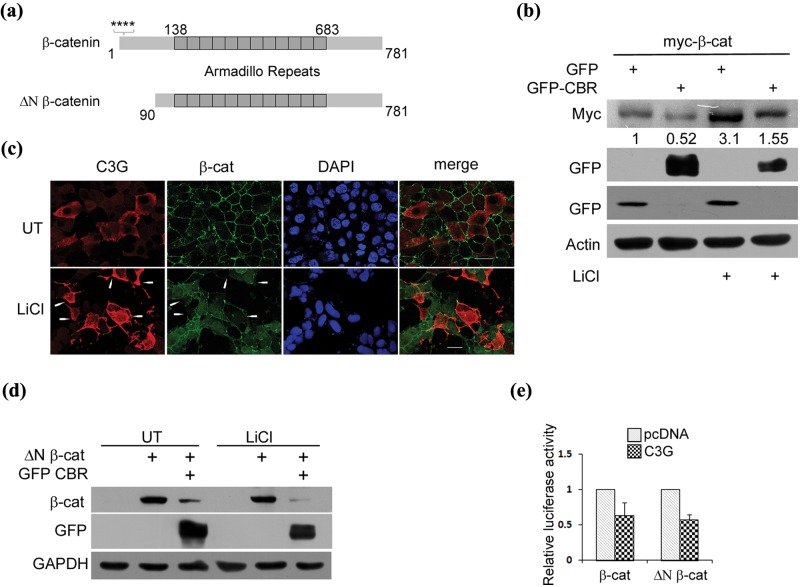
C3G represses β-catenin independent of GSK3β
C3G caused a repression in β-catenin activity induced by LiCl, suggesting that C3G induced effects on β-catenin were not mediated through altering GSK3β activity. LiCl treatment resulted in an increase in cellular β-catenin levels (Fig. 3b). Expression of C3G in HEK 293T cells reduced LiCl induced β-catenin protein to an extent similar to that seen in untreated cells (Fig. 3b). When examined by indirect immunofluorescence, C3G expressing cells showed reduction in LiCl induced β-catenin staining in HEK 293T (Fig. 3c) as well as MCF-7 breast cancer cells (Suppl. Fig. S2c). C3G induced reduction in endogenous β-catenin was seen in both nuclear and cytoplasmic compartments. The effect of C3G on an N-terminal deletion mutant of β-catenin (ΔN β-catenin) (Fig. 3a) that is refractory to GSK3β mediated regulation was examined. C3G repressed TCF promoter activity induced by β-catenin as well as ΔN β-catenin (Fig. 3e). A decrease in ΔN β-catenin protein observed upon coexpression of CBR in HEK 293T cells suggested that N-terminal regulatory domain of β-catenin is not necessary for C3G mediated destabilization of β-catenin protein (Fig. 3d). As expected, LiCl treatment had no effect on ΔN β-catenin levels, and CBR expression resulted in reduced ΔN β-catenin levels even in the presence of LiCl. These results indicated that C3G exerts its effects on β-catenin downstream of GSK3β.
Figure 3.
C3G represses β-catenin independent of GSK3β. (a) A schematic showing full-length β-catenin and constitutively active ΔN β-catenin. Stars indicate serine residues phosphorylated by CK1 and GSK3β. (b) CBR represses exogenously expressed β-catenin protein under conditions of GSK3β inhibition. Lysates of HEK 293T cells expressing myc tagged full-length β-catenin along with GFP or GFP-CBR and treated with LiCl were subject to Western blotting to detect the indicated proteins with antibodies. (c) C3G overexpressing cells showed reduced staining for cellular β-catenin induced by LiCl treatment. HEK 293T cells transfected with pcDNA C3G were treated with 50 mM LiCl after 6 hours of expression and fixed after 30 hours of expression followed by indirect immunofluorescence to detect C3G and β-catenin expression. Arrows indicate some C3G expressing cells. The C3G antibody used does not detect endogenous C3G under the conditions of our experiment. Bar, 20 µm. (d) CBR domain of C3G represses β-catenin protein lacking its N-terminal regulatory domain. ΔN β-catenin protein levels were examined in HEK 293T cells co-expressing pcDNA or GFP-CBR in the presence or absence of LiCl. (e) C3G inhibits activity of ΔN β-catenin. Lysates of HEK 293T cells transfected with reporter constructs and 100 ng of pcDNA or C3G along with 100 ng of β-catenin or ΔN β-catenin expression vectors were used for luciferase assays. Relative luciferase counts after normalization with β-Gal are shown as mean ± SD in the bar diagram.
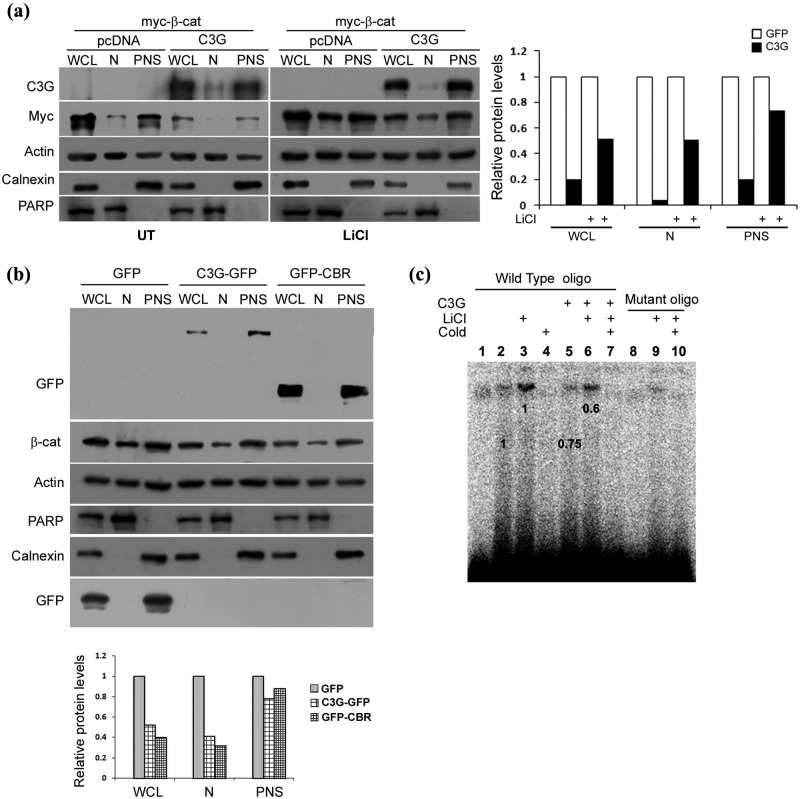
It is known that many proteins that inhibit β-catenin activity, like APC and Axin, do so by reducing its nuclear levels.42 We examined whether C3G expression, in addition to reducing β-catenin stability, has an effect on its nucleocytoplasmic exchange. Cytoplasmic and nuclear fractions of cells with or without C3G overexpression were examined for co-expressed β-catenin in untreated and LiCl treated cells. LiCl causes nuclear translocation of β-catenin in addition to stabilization of the protein. C3G expression reduced nuclear levels of β-catenin in both untreated and LiCl treated cells (Fig. 4a). PARP and calnexin were used as markers for nuclear and cytoplasmic fractions, respectively. Endogenous levels of β-catenin protein in the nucleus induced by LiCl treatment were also significantly reduced (Fig. 4b). Quantitation showed that C3G induced overall reduction in cellular β-catenin levels was primarily due to reduction in nuclear levels.
Figure 4.
C3G reduces nuclear levels of β-catenin. (a) Effect of C3G expression on exogenously expressed β-catenin in nuclear and postnuclear fractions (PNS). C3G was co-expressed with myc β-catenin in HEK 293T cells and β-catenin protein analyzed in whole cell lysate (WCL), nuclear (N), and postnuclear fractions (PNS) in untreated or LiCl treated cells. Actin was used as loading control. Calnexin and PARP are used as markers for establishing purity of postnuclear and nuclear fractions, respectively. Relative change in β-catenin in subcellular fractions upon C3G expression is shown in bar diagram. (b) Effect of C3G and CBR expression on endogenous nuclear β-catenin levels in cells treated with LiCl. HEK 293T cells expressing GFP, C3G-GFP, or GFP-CBR were treated with LiCl, and cellular β-catenin levels in nuclear and postnuclear fraction were examined by Western blotting. Calnexin and PARP are used to ascertain the purity of PNS and N fractions, respectively. Relative change in β-catenin in subcellular fractions upon C3G-GFP or GFP-CBR expression is shown in bar diagram. (c) Nuclear extracts from C3G expressing cells show reduced binding to TCF binding consensus oligo. EMSA was performed using nuclear extracts of untransfected or C3G transfected HEK 293T cells with or without LiCl treatment and TCF binding consensus (wild-type) or mutant oligo. Specificity of the band is shown by using 100-fold molar ratio excess of unlabeled oligo (cold). Lanes 1 and 10 are devoid of nuclear extracts. Numbers on the figure indicate relative change in band intensity upon C3G expression.
Because C3G reduced β-catenin in the nucleus, we examined whether it affected binding of β-catenin-TCF complexes to target sequences. Electrophoretic mobility shift assay (EMSA) was carried out using oligonucleotides corresponding to TCF binding consensus sequence and nuclear lysates from C3G expressing and nonexpressing cells, with and without LiCl treatment. C3G expressing nuclear lysates showed reduced binding to TCF target sequence (Fig. 4c). Specificity of the interaction was shown by competition with cold probe and using mutant oligos.
Proteasome inhibition does not significantly affect C3G mediated β-catenin repression
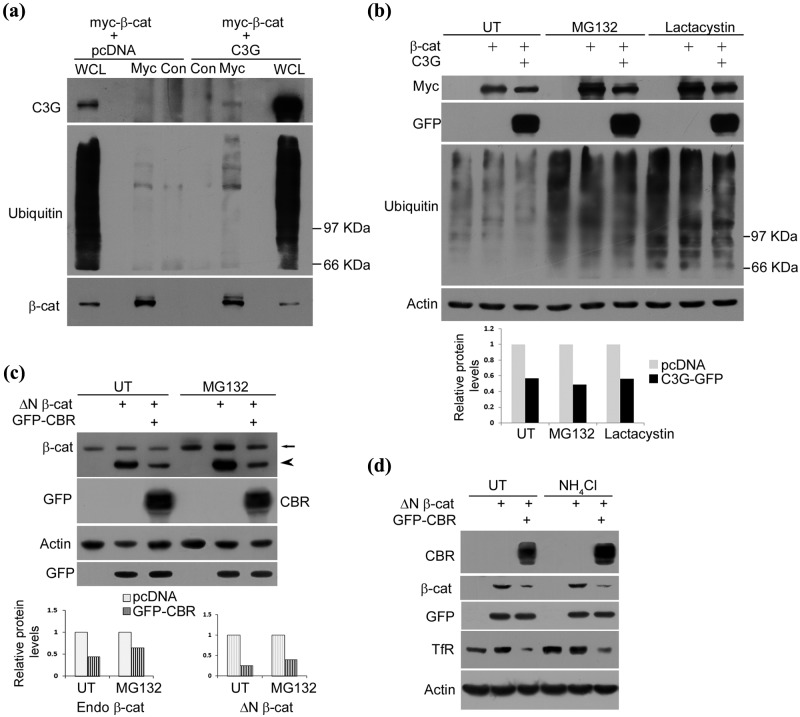
Ubiquitin dependent proteasomal degradation is the primary mechanism in regulating cellular β-catenin levels. One possible mechanism by which C3G enhances β-catenin turnover could be by increasing β-catenin ubiquitination. This was tested by immunoblotting β-catenin immunoprecipitates from MG132 treated cells expressing β-catenin with or without C3G. Increased levels of high molecular weight ubiquitin conjugates were seen in β-catenin immunoprecipitates from cells expressing C3G compared with those not expressing C3G (Fig. 5a).
Figure 5.
C3G causes repression of β-catenin levels under conditions of proteasomal inhibition. (a) C3G co-expression enhances ubiquitination of β-catenin. HEK 293T cells transfected with myc tagged β-catenin along with pcDNA or C3G expression constructs were treated with MG132 and subject to immunoprecipitation using myc antibody or normal mouse IgG (Con) followed by Western blotting to detect the indicated proteins with antibodies. (b) C3G represses co-expressed β-catenin protein independent of proteasomal degradation. HEK 293T cells were transfected with 200 ng of β-catenin and 200 ng of pcDNA or C3G expression constructs and were either left untreated (UT) or treated with MG132 or lactacystin for 8 hours after 22 hours of expression. Cell lysates were subject to Western blotting to detect the indicated proteins. Actin was used as loading control. Bar diagram shows relative change in β-catenin levels due to C3G expression. (c) HEK 293T cells were transfected with 200 ng of ΔN β-catenin and 200 ng of pcDNA or CBR-GFP along with 20 ng of GFP expression constructs and treated as in (b). Cell lysates were subject to Western blotting to detect the indicated proteins. Arrow and arrowhead indicate endogenous β-catenin and overexpressed ΔN β-catenin, respectively. GFP was used as transfection control and actin as loading control. Bar diagram shows densitometric analysis of relative change in endogenous β-catenin and ΔN β-catenin levels due to GFP-CBR expression. (d) CBR induced β-catenin repression is independent of lysosomal degradation. HEK 293T cells were transfected as in (c) and treated with 20 mM NH4Cl after 6 hours of expression. Cell lysates made after 30 hours of expression were subject to Western blotting to detect the indicated proteins with antibodies. Cellular TfR was examined to show effect of NH4Cl treatment.
We therefore examined whether C3G mediated β-catenin turnover was mediated through proteasomal degradation. Treatment of HEK 293T cells with 10 µM MG132 or 60 µM lactacystin for 8 hours resulted in enhanced β-catenin protein levels (Fig. 5b). We observed that C3G expression caused repression of exogenously expressed β-catenin protein to a similar extent in the presence or absence of both the proteasome inhibitors. Similar effect of CBR was seen on ΔN β-catenin (Fig. 5c). Endogenous β-catenin levels were also repressed by CBR in the presence of MG132 (Fig. 5c and Suppl. Fig. S3). A partial increase in endogenous β-catenin seen in MG132 treated C3G expressing cells compared with untreated cells is attributed to the enhanced β-catenin levels in cells that do not express C3G. Transfection efficiency under our experimental conditions is about 50% in HEK 293T and 20% in MDA-MB-231 cells. These results suggest that C3G engaged proteasome independent mechanisms to cause β-catenin degradation. It was also seen that the effect of CBR on ΔN β-catenin was not altered in the presence of lysosomal inhibitor NH4Cl (Fig. 5d). Lysosomal inhibition did not rescue ΔN β-catenin activity as seen by its effects on the expression of cellular TfR. As expected, constitutive levels of TfR show an increase upon lysosomal inhibition,43 but no effect was seen on ΔN β-catenin induced levels.
C3G expression reduces motility of breast cancer cells
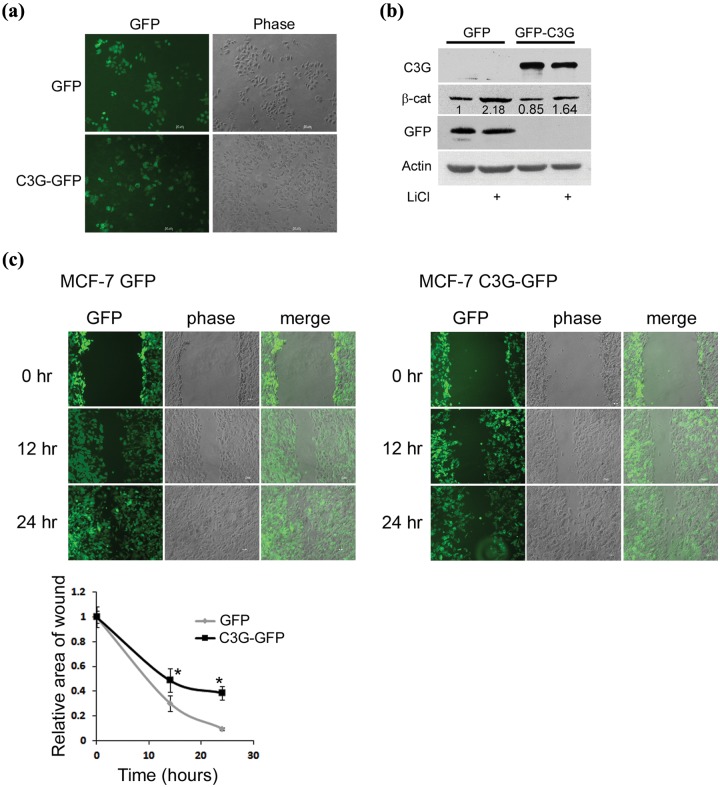
The consequence of C3G expression in regulating cellular behavior was examined by generating stable clones of MCF7 cells expressing GFP or C3G-GFP (Fig. 6a). β-catenin levels were examined and found to be 85% of those seen in GFP expressing clones (Fig. 6b). C3G expression resulted in reduced levels of β-catenin even under conditions of LiCl stimulation. The consequence of C3G expression was examined on cell migration, a property dependent on β-catenin activity, using wound healing assay. C3G expressing cells showed a significantly lower rate of wound healing compared with GFP expressing clones (Fig. 6c). In the C3G-GFP clones, cells with high levels of expression showed slower migration compared with weakly expressing cells.
Figure 6.
Stable expression of C3G inhibits motility of MCF-7 cells. (a) Live cell images of MCF-7 clones stably expressing GFP or C3G-GFP. Bar, 50 µm. (b) Endogenous β-catenin expression in MCF-7 clones. MCF-7 clones stably expressing GFP or C3G-GFP were either left untreated or treated with LiCl and lysed after 24 hours of treatment. Cell lysates were subject to Western blotting to detect the indicated proteins with antibodies. (c) Effect of C3G expression on cell motility. Wound healing assay was performed on MCF-7 clones stably expressing GFP or C3G-GFP. Live cell images were taken after indicated time periods of making the wound. Graph shows relative wound area with respect to time averaged from 3 wounds each from 2 independent experiments. Student t test was performed to test the significance of difference in wound area. Bar, 50 µm. *P < 0.05.
β-catenin activation causes reduction in cellular C3G levels
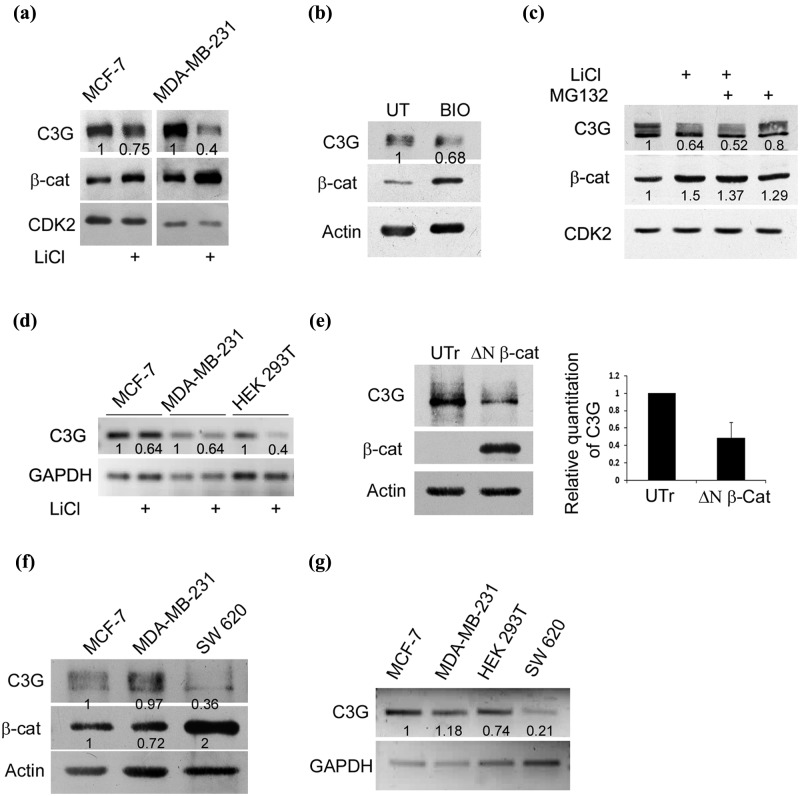
Since C3G showed properties of negatively regulating β-catenin function, we examined the consequence of activating β-catenin signaling on cellular C3G levels. β-catenin signaling can be initiated either by GSK3β inhibition or by expressing constitutively active β-catenin mutants. LiCl treatment resulted in an increase in cellular β-catenin levels and a decrease in C3G levels in both MCF-7 and MDA-MB-231 cells (Fig. 7a). Reduction in C3G levels was also observed upon treatment of MCF-7 cells with another GSK3β inhibitor, BIO (Fig. 7b). C3G levels in both nuclear and cytoplasmic compartments showed a decrease upon LiCl treatment in MDA-MB-231 cells (Suppl. Fig. S4). The decrease in C3G level was not due to proteasome dependent degradation, as treatment with MG132 did not reverse the effects of LiCl (Fig. 7c). We therefore tested whether LiCl effects on C3G were mediated at the transcription level. C3G gene expression was examined by semiquantitative RT-PCR in cells treated with or without LiCl. C3G transcript levels showed a decrease upon LiCl treatment in MCF7, MDA-MB-231, and HEK 293T cells (Fig. 7d), indicating that cellular C3G levels are repressed at the transcriptional level upon stimulation of β-catenin signaling. To confirm that the effect on C3G transcription was mediated through activation of β-catenin, we examined cellular C3G levels upon expression of a constitutively active variant of β-catenin, ΔN β-catenin. HEK 293T cells expressing ΔN β-catenin showed a significant reduction in C3G protein levels, indicating that β-catenin negatively regulates C3G (Fig. 7e). We therefore hypothesized that C3G expression would be repressed in cells that show constitutively high β-catenin levels. For this purpose we compared protein and mRNA expression in SW620 colon cancer cells that possess APC mutation resulting in high β-catenin activity.44 C3G protein levels (Fig. 7f) as well as transcript levels (Fig. 7g) were low in SW620 cells compared with those in other cell types with moderate β-catenin expression, indicating that β-catenin functions to transcriptionally repress C3G.
Figure 7.
β-catenin activation reduces cellular C3G levels. (a) Increase in cellular β-catenin levels by GSK3β inhibition results in reduced C3G levels. C3G and β-catenin levels determined by Western blotting in MCF-7 and MDA-MB-231 cells treated with LiCl. CDK2 was used as a loading control. (b) C3G and β-catenin levels in MCF-7 cells treated with BIO, a specific inhibitor of GSK3β. UT, untreated. (c) β-catenin activation induces reduction in C3G levels independent of proteasomal degradation. C3G and β-catenin levels were examined under conditions of LiCl and MG132 treatment in MCF-7 cells. CDK2 was used as internal control. (d) Activation of β-catenin by LiCl treatment reduces C3G transcript levels. MCF-7, MDA-MB-231, and HEK 293T cells were treated with 50 mM LiCl, and cells were lysed after 24 hours for RNA isolation. cDNA was prepared from RNA followed by semiquantitative PCR using primers designed for GAPDH and C3G. (e) Expression of a constitutively active variant of β-catenin reduces cellular β-catenin protein levels. Cell lysates of HEK 293T cells transfected with ΔN β-catenin for 48 hours were subject to Western blotting to detect C3G and β-catenin expression. Actin was used as a loading control. UTr, untransfected. Bar diagram shows relative C3G levels as mean ± SD from 3 experiments. (f) Comparative C3G expression in cell lines. Cell lysates of MCF-7, MDA-MB-231, and SW620 cells were subject to Western blotting to detect the indicated proteins with antibodies. SW620 cells show constitutively high β-catenin activity due to APC mutation. (g) Semiquantitative PCR to detect C3G and GAPDH transcripts in MCF-7, MDA-MB-231, HEK 293T, and SW620 cells.
Discussion
Disruption of β-catenin signaling is a primary strategy for prevention of carcinomas.45 Therefore, understanding how cellular β-catenin stability and function are regulated has been of key importance. Using ectopic expression and knockdown strategies, we reveal a role for the guanine nucleotide exchange factor C3G as a negative regulator of β-catenin. There were 6 principle findings indicating physical and functional interaction between C3G and β-catenin: (1) C3G and β-catenin co-precipitate from cell lysates; (2) C3G overexpression represses and its knockdown enhances β-catenin/TCF transcription activity; (3) C3G induced inhibition of β-catenin activity correlates with reduction in β-catenin/TCF target gene expression; (4) C3G expression decreases stability of overexpressed as well as endogenous β-catenin; (5) C3G expression decreases nuclear levels of β-catenin; and (6) nuclear extracts from C3G transfected cells show weaker interaction with TCF target sequences compared with untransfected extracts. As increase in cell migration is an important property of β-catenin, effects of C3G on β-catenin are reflected in the reduced motility of stable clones of MCF-7 cells expressing C3G.
Using immunoprecipitation assays, we show that cellular β-catenin forms a complex with C3G and that the central proline rich Crk binding region of C3G is sufficient for the interaction. The N-terminal regulatory sequences or C-terminal transactivation domain of β-catenin and the catalytic domain of C3G are not essential for the interaction. It therefore appears that the CBR domain of C3G can mediate interactions with molecules lacking an SH3 domain. The exact residues of β-catenin involved in the interaction are yet to be deciphered, but we could conclude that residues between 90 and 525 (which encompass ARM repeats 1 to 10) are sufficient. As a consequence of the interaction, C3G functionally regulates β-catenin dependent transcription independent of its catalytic activity as a GEF. ShRNA mediated knockdown of C3G resulted in enhanced LiCl induced promoter activity, suggesting that endogenous C3G, which shows predominant cytoplasmic localization similar to that of overexpressed protein, also functions to repress transcription activity of β-catenin.
Expression of C3G reduced cellular as well as overexpressed β-catenin protein without any effect on transcript levels, indicating that regulation was mediated at a post-translational level and not by affecting β-catenin transcription. C3G accelerates β-catenin protein degradation as seen by decreased half-life of exogenously expressed as well as endogenous β-catenin in C3G expressing cells. In accordance with its ability to interact with and affect β-catenin transcription activity, the CBR domain was sufficient for decreasing β-catenin stability. A role for C3G in limiting proliferation of neural precursor cells in response to FGF2 has been described.36 Reduced C3G levels resulted in inhibition of GSK3β, leading to enhanced β-catenin levels. These events were shown to be dependent on Rap1 activation. It therefore appears that C3G can regulate β-catenin activity by engaging mechanisms dependent on its catalytic function as well as independent of its catalytic activity.
C3G mediated its effect on β-catenin independent of GSK3β activity, as it was observed that C3G or CBR could repress β-catenin protein as well as activity under conditions of treatment with an inhibitor of GSK3β. This was also evident from the fact that N-terminus of β-catenin, which enables its regulation by GSK3β and the proteasome, was dispensable for the inhibitory effect of C3G on β-catenin expression. Similar regulation has been shown with respect to other inhibitor molecules like En-1, Psoriasin, and Nur77.46-48 Modification by ubiquitination of lysines in the N-terminus of β-catenin has been the major mechanism involved in degradation of cellular β-catenin through the proteasome.49 Coexpression of C3G enhanced β-catenin ubiquitination, but it was seen that C3G could cause β-catenin degradation even under conditions of proteasome inhibition. C3G could decrease levels of ΔN β-catenin, which lacks phosphorylation and ubiquitin sites for β-TrCP mediated degradation. This degradation was not rescued by proteasomal or lysosomal inhibitors, indicating that alternate mechanisms may be engaged by C3G. Although the proteases involved have not been defined, ubiquitinated targets degraded by proteasome independent pathways are known.50 As C3G primarily affected nuclear levels of β-catenin, it is possible that C3G enhances its nuclear export, or presently unknown mechanisms of protein degradation may be engaged by C3G in the nucleus. A decrease in nuclear but not cytoplasmic β-catenin levels upon curcumin treatment has been shown.51 Further work is required to understand the mechanism involved in reduction of cellular β-catenin levels by C3G.
Many proteins in signaling pathways are often subject to feedback regulation to either enhance or repress downstream effector functions. In our experiments, we observed reduction in cellular C3G levels under conditions of β-catenin activation and in cells that possess constitutively high β-catenin activity. Although the C3G protein has been studied extensively, its regulation at the transcriptional level is poorly understood. Our results suggest that C3G transcription is negatively regulated by β-catenin. β-catenin mediated signaling primarily involves activation of genes involved in cell proliferation and migration, but genes that inhibit these processes like IGFBP-6, E-cadherin, and 15PGDH are repressed.8,9,52 Several studies have shown that in mammalian cells, C3G primarily functions to inhibit cell proliferation and migration. An important role for C3G in differentiation of various cell types has also been shown.18 Activation of β-catenin signaling to achieve cell proliferation can therefore keep C3G levels repressed, thereby preventing differentiation.
C3G can exert its effects as a negative regulator of cell proliferation through MAPK as well as β-catenin signaling. It can therefore be hypothesized that loss or downregulation of C3G seen in certain human cancers may be associated with deregulated β-catenin function. Although more work is needed to elucidate the mechanism involved in C3G mediated β-catenin degradation, our results suggest that C3G’s role in embryonic development, migration, and differentiation may be mediated through β-catenin.
The present study therefore provides evidence for a novel mechanism by which C3G can exert its effects as an inhibitor of cell proliferation, migration, and oncogene induced transformation independent of its catalytic activity. C3G as well as β-catenin is essential for embryonic development. Reciprocal regulation between them may aid in suppression of unscheduled proliferation of cells required for tissue differentiation during development. The repression of cellular C3G levels upon GSK3β inhibition not only will enable efficient accumulation of functional β-catenin but also may lower the threshold of signals required for activating the pathway, thereby aiding in sustained and amplified signaling.
Materials and Methods
Cell culture, transfections, and treatments
HEK 293T, MDA-MB-231, MCF-7, and SW620 were obtained from American Type Culture Collection (ATCC) (Manassas, VA, USA) and cultured in DMEM containing 10% serum. Transient transfections were performed using Lipofectamine Plus reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. Stable clones of MCF-7 cells expressing GFP or C3G-GFP were generated by selection of transiently transfected cells in 500 µg/mL G418 for 14 days. Clones were maintained in 100 µg/mL G418. Cells were treated with LiCl (50 mM) or BIO (5 µM) (Calbiochem, San Diego, CA, USA) after 6 hours of transfections for 24 hours. MG132 was from Calbiochem. Cycloheximide (Sigma, St.Louis, Missouri, USA) was used at 40 µg/mL. Protease inhibitor cocktail was from (Roche, Basel, Switzerland). Lactacystin (Calbiochem, San Diego, CA, USA) was used at 60 µM for 8 hours.
Plasmids and antibodies
C3G, C3G-GFP, and myc tagged full-length β-catenin have been described previously.53 Reporter constructs PGL3-OT having 3 copies of consensus TCF binding sites upstream to luciferase and PGL3-OF (having mutant sites) were a kind gift from Dr. Bert Vogelstein. pCMV sport β Gal construct was from (Gibco, Carlsbad, CA, USA). ΔN β-catenin construct expressing constitutively active β-catenin, lacking the first 89 residues, was a kind gift from Dr. Lynn Matrisian. Myc tagged ΔC 525 β-catenin construct containing amino acids 1 to 525 was a kind gift from Dr. Marian Waterman. GFP-CBR, which expresses the central Crk binding region of C3G encompassing residues 281 to 648, was made by releasing HindIII and XhoI fragment from pcDNA CBR and cloning it into SalI and HindIII sites of EGFPC3. GST-CBR has been described earlier.21 ShC, expressing small hairpin RNA targeting C3G and mShC (expressing ShRNA mutant), has been described earlier.54 C3G (sc-869), β-catenin, myc, α-tubulin, GFP, calnexin, and CDK2 antibodies were from (Santa Cruz, California, USA). GAPDH and actin antibodies were from (Millipore, San Diego, CA, USA). Transferrin receptor, PARP, ubiquitin, and Akt antibodies were from Invitrogen, Roche, Calbiochem, and (Cell Signaling, Danvers, Massachusetts, USA), respectively. Lamin B1 antibody was from(Abcam, Cambridge, UK).
Western blotting, immunofluorescence, and imaging
Cell lysates were made after 30 hours of transfections unless mentioned otherwise. Immunofluorescence and Western blotting methods have been described earlier.22 Secondary antibodies used were rabbit or mouse IgG tagged with Alexa 488 (Invitrogen) and Cy3 (Amersham, Amersham, UK). Quantitation of protein and RNA levels was determined by densitometric analysis of bands normalized with the corresponding loading control. Images were taken using Leica TCS SP-5 AOBS confocal microscope (Leica Microsystems, Mannheim, Germany) or Carl Zeiss Apotome with Axioimager Z1 upright microscope and were analyzed by Leica Application Suite or by Axiovision 4.4 (Carl Zeiss Microscopy GmbH, Gottingen, Germany). For wound healing assay, confluent monolayer of cells was scratched with a micropipette tip, and images of the wound were taken at an interval of 12 hours using the 10× objective of a Carl Zeiss Axiovert Live microscope. The average wound area at various time points was determined from 2 independent experiments conducted with at least 3 wounded areas.
Luciferase reporter assays
For luciferase reporter assay, HEK 293T cells were transfected with 50 ng of pCMV-SPORT-β-galactosidase, 150 ng of reporter construct (PGL3-OT or PGL3-OF), and other expression constructs. Reporter assay was performed using (Promega, Fitchburg, Wisconsin, USA) luciferase assay kit following the manufacturer’s instructions. Luciferase reporter activity is shown as mean ± SD of results from at least 3 experiments after normalization for corresponding β-galactosidase activity.
Quantitative and semiquantitative RT PCR
RNA was made using Trizol reagent (Sigma). cDNA was made using Superscript first strand synthesis kit (Invitrogen). cDNA was subjected to PCR for C3G and GAPDH using the same master mix of template. Primers used for C3G were forward, 5′ TCCTCCTTCCGAGCCTAC 3′, and reverse, 5′ CCACCGCTTGGAGAAGTT 3′; those for GAPDH were forward, 5′ CACCAGGGCTGCTTTTAACTCT 3′, and reverse, 5′ TTCCCGTTCTCAGCCTTGAC 3′. For quantitative real-time PCR, the RNA was subjected to DNAse treatment. Quantitative PCR for β-catenin and GAPDH was performed. Primers used for β-catenin were forward, 5′ CGGCTTTCAGTTGAGCTGAC 3′, and reverse, 5′ CAAGGCATCCTGGCCATATC 3′.
Immunoprecipitation and cell fractionation
Nuclear and postnuclear fractions of cells were prepared as described.55 HEK 293T cells transfected with the required plasmids were lysed, and immunoprecipitation was carried out as described.21
GST pull-down assay
Escherichia coli BL-21 DE-3 cells expressing GST or GST-CBR were induced by 1 mM IPTG (Calbiochem) for 3 hours at 37°C. Preparation of cell lysates and coupling of recombinant protein to glutathione-agarose used for in vitro binding assay were performed as described.54
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts of HEK 293T cells were made as described.56 EMSA was performed as described57 with some modifications. In the binding reaction buffer, 50 ng of poly dI-dC and 2 ng of labeled oligo were used along with 3 µg of nuclear extract. 100-fold molar excess of unlabeled oligos were used for competition. The oligos, 5′-CCCTTTGATCTTACC (wild-type) having the consensus TCF binding site and 5′-CCCTTTGGCCTTACC (mutant) having mutated binding site, were end-labeled in a poly nucleotide kinase reaction using γp32ATP.
Supplementary Material
Acknowledgments
We thank Dr. Lynn Matrisian (Vanderbilt University, Nashville, Tennessee), Dr. Marian Waterman (University of California, Irvine), and Dr. Bert Vogelstein (Howard Hughes Medical Institute, Baltimore, Maryland) for the gift of various constructs.
Footnotes
Supplementary Material: Supplementary material for this article is available on the Genes & Cancer website at http://ganc.sagepub.com/supplemental.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: K.D. was supported by a fellowship from the Council for Scientific and Industrial Research and K.S. from the Indian Council of Medical Research. A research grant awarded to V.R. from the Department of Biotechnology, Government of India, is acknowledged.
References
- 1. Liu C, Li Y, Semenov M, et al. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108(6):837-47 [DOI] [PubMed] [Google Scholar]
- 2. Verheyen EM, Gottardi CJ. Regulation of Wnt/beta-catenin signaling by protein kinases. Dev Dyn. 2010;239(1):34-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Munemitsu S, Albert I, Rubinfeld B, Polakis P. Deletion of an amino-terminal sequence beta-catenin in vivo and promotes hyperphosporylation of the adenomatous polyposis coli tumor suppressor protein. Mol Cell Biol. 1996;16(8):4088-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dihlmann S, Kloor M, Fallsehr C, von Knebel Doeberitz M. Regulation of AKT1 expression by beta-catenin/Tcf/Lef signaling in colorectal cancer cells. Carcinogenesis. 2005;26(9):1503-12 [DOI] [PubMed] [Google Scholar]
- 5. Huang M, Wang Y, Sun D, et al. Identification of genes regulated by Wnt/beta-catenin pathway and involved in apoptosis via microarray analysis. BMC Cancer. 2006;6:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rohrs S, Kutzner N, Vlad A, Grunwald T, Ziegler S, Muller O. Chronological expression of Wnt target genes Ccnd1, Myc, Cdkn1a, Tfrc, Plf1 and Ramp3. Cell Biol Int. 2009;33(4):501-8 [DOI] [PubMed] [Google Scholar]
- 7. Polakis P. The many ways of Wnt in cancer. Curr Opin Genet Dev. 2007;17(1):45-51 [DOI] [PubMed] [Google Scholar]
- 8. Jamora C, DasGupta R, Kocieniewski P, Fuchs E. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature. 2003;422(6929):317-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smartt HJ, Greenhough A, Ordonez-Moran P, et al. Beta-catenin represses expression of the tumour suppressor 15-prostaglandin dehydrogenase in the normal intestinal epithelium and colorectal tumour cells. Gut. 2012;61(9):1306-14 [DOI] [PubMed] [Google Scholar]
- 10. Haegel H, Larue L, Ohsugi M, Fedorov L, Herrenknecht K, Kemler R. Lack of beta-catenin affects mouse development at gastrulation. Development. 1995;121(11):3529-37 [DOI] [PubMed] [Google Scholar]
- 11. Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8(5):739-50 [DOI] [PubMed] [Google Scholar]
- 12. Grigoryan T, Wend P, Klaus A, Birchmeier W. Deciphering the function of canonical Wnt signals in development and disease: conditional loss- and gain-of-function mutations of beta-catenin in mice. Genes Dev. 2008;22(17):2308-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee CW, Ito K, Ito Y. Role of RUNX3 in bone morphogenetic protein signaling in colorectal cancer. Cancer Res. 2010;70(10):4243-52 [DOI] [PubMed] [Google Scholar]
- 14. Filipovich A, Gehrke I, Poll-Wolbeck SJ, Kreuzer KA. Physiological inhibitors of Wnt signaling. Eur J Haematol. 2011;86(6):453-65 [DOI] [PubMed] [Google Scholar]
- 15. Tian Q, Feetham MC, Tao WA, et al. Proteomic analysis identifies that 14-3-3zeta interacts with beta-catenin and facilitates its activation by Akt. Proc Natl Acad Sci U S A. 2004;101(43):15370-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chairoungdua A, Smith DL, Pochard P, Hull M, Caplan MJ. Exosome release of beta-catenin: a novel mechanism that antagonizes Wnt signaling. J Cell Biol. 2010;190(6):1079-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu J, Stevens J, Rote CA, et al. Siah-1 mediates a novel beta-catenin degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol Cell. 2001;7(5):927-36 [DOI] [PubMed] [Google Scholar]
- 18. Radha V, Mitra A, Dayma K, Sasikumar K. Signalling to actin: role of C3G, a multitasking guanine-nucleotide-exchange factor. Biosci Rep. 2011;31(4):231-44 [DOI] [PubMed] [Google Scholar]
- 19. Kirsch KH, Georgescu MM, Hanafusa H. Direct binding of p130(Cas) to the guanine nucleotide exchange factor C3G. J Biol Chem. 1998;273(40):25673-9 [DOI] [PubMed] [Google Scholar]
- 20. Knudsen BS, Feller SM, Hanafusa H. Four proline-rich sequences of the guanine-nucleotide exchange factor C3G bind with unique specificity to the first Src homology 3 domain of Crk. J Biol Chem. 1994;269(52):32781-7 [PubMed] [Google Scholar]
- 21. Mitra A, Radha V. F-actin-binding domain of c-Abl regulates localized phosphorylation of C3G: role of C3G in c-Abl-mediated cell death. Oncogene. 2010;29(32):4528-42 [DOI] [PubMed] [Google Scholar]
- 22. Shivakrupa R, Radha V, Sudhakar C, Swarup G. Physical and functional interaction between Hck tyrosine kinase and guanine nucleotide exchange factor C3G results in apoptosis, which is independent of C3G catalytic domain. J Biol Chem. 2003;278(52):52188-94 [DOI] [PubMed] [Google Scholar]
- 23. Tanaka S, Morishita T, Hashimoto Y, et al. C3G, a guanine nucleotide-releasing protein expressed ubiquitously, binds to the Src homology 3 domains of CRK and GRB2/ASH proteins. Proc Natl Acad Sci U S A. 1994;91(8):3443-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martin-Encabo S, Santos E, Guerrero C. C3G mediated suppression of malignant transformation involves activation of PP2A phosphatases at the subcortical actin cytoskeleton. Exp Cell Res. 2007;313(18):3881-91 [DOI] [PubMed] [Google Scholar]
- 25. Mitra A, Kalayarasan S, Gupta V, Radha V. TC-PTP dephosphorylates the guanine nucleotide exchange factor C3G (RapGEF1) and negatively regulates differentiation of human neuroblastoma cells. PLoS One. 2011;6(8):e23681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Asuri S, Yan J, Paranavitana NC, Quilliam LA. E-cadherin dis- engagement activates the Rap1 GTPase. J Cell Biochem. 2008; 105(4):1027-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hirata T, Nagai H, Koizumi K, et al. Amplification, up-regulation and over-expression of C3G (CRK SH3 domain-binding guanine nucleotide-releasing factor) in non-small cell lung cancers. J Hum Genet. 2004;49(6):290-5 [DOI] [PubMed] [Google Scholar]
- 28. Mor A, Wynne JP, Ahearn IM, Dustin ML, Du G, Philips MR. Phospholipase D1 regulates lymphocyte adhesion via upregulation of Rap1 at the plasma membrane. Mol Cell Biol. 2009;29(12):3297-306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Deevi RK, Koney-Dash M, Kissenpfennig A, et al. Vasodilator-stimulated phosphoprotein regulates inside-out signaling of beta2 integrins in neutrophils. J Immunol. 2010;184(12):6575-84 [DOI] [PubMed] [Google Scholar]
- 30. Hisata S, Sakisaka T, Baba T, et al. Rap1-PDZ-GEF1 interacts with a neurotrophin receptor at late endosomes, leading to sustained activation of Rap1 and ERK and neurite outgrowth. J Cell Biol. 2007;178(5):843-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ohba Y, Ikuta K, Ogura A, et al. Requirement for C3G-dependent Rap1 activation for cell adhesion and embryogenesis. EMBO J. 2001;20(13):3333-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Voss AK, Britto JM, Dixon MP, et al. C3G regulates cortical neuron migration, preplate splitting and radial glial cell attachment. Development. 2008;135(12):2139-49 [DOI] [PubMed] [Google Scholar]
- 33. Ishimaru S, Williams R, Clark E, Hanafusa H, Gaul U. Activation of the Drosophila C3G leads to cell fate changes and overproliferation during development, mediated by the RAS-MAPK pathway and RAP1. EMBO J. 1999;18(1):145-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Radha V, Rajanna A, Gupta RK, Dayma K, Raman T. The guanine nucleotide exchange factor, C3G regulates differentiation and survival of human neuroblastoma cells. J Neurochem. 2008;107(5):1424-35 [DOI] [PubMed] [Google Scholar]
- 35. Stork PJ, Schmitt JM. Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol. 2002;12(6):258-66 [DOI] [PubMed] [Google Scholar]
- 36. Voss AK, Krebs DL, Thomas T. C3G regulates the size of the cerebral cortex neural precursor population. EMBO J. 2006;25(15):3652-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guerrero C, Fernandez-Medarde A, Rojas JM, Font de, Mora J, Esteban LM, Santos E. Transformation suppressor activity of C3G is independent of its CDC25-homology domain. Oncogene. 1998;16(5):613-24 [DOI] [PubMed] [Google Scholar]
- 38. Gutierrez-Berzal J, Castellano E, Martin-Encabo S, et al. Characterization of p87C3G, a novel, truncated C3G isoform that is overexpressed in chronic myeloid leukemia and interacts with Bcr-Abl. Exp Cell Res. 2006;312(6):938-48 [DOI] [PubMed] [Google Scholar]
- 39. Okino K, Nagai H, Nakayama H, et al. Inactivation of Crk SH3 domain-binding guanine nucleotide-releasing factor (C3G) in cervical squamous cell carcinoma. Int J Gynecol Cancer. 2006;16(2):763-71 [DOI] [PubMed] [Google Scholar]
- 40. Stambolic V, Ruel L, Woodgett JR. Lithium inhibits glycogen synthase kinase-3 activity and mimics wingless signalling in intact cells. Curr Biol. 1996;6(12):1664-8 [DOI] [PubMed] [Google Scholar]
- 41. Singh M, Mugler K, Hailoo DW, et al. Differential expression of transferrin receptor (TfR) in a spectrum of normal to malignant breast tissues: implications for in situ and invasive carcinoma. Appl Immunohistochem Mol Morphol. 2011;19(5):417-23 [DOI] [PubMed] [Google Scholar]
- 42. Krieghoff E, Behrens J, Mayr B. Nucleo-cytoplasmic distribution of beta-catenin is regulated by retention. J Cell Sci. 2006;119(Pt 7):1453-63 [DOI] [PubMed] [Google Scholar]
- 43. Matsui T, Itoh T, Fukuda M. Small GTPase Rab12 regulates constitutive degradation of transferrin receptor. Traffic. 2011;12(10):1432-43 [DOI] [PubMed] [Google Scholar]
- 44. Ilyas M, Tomlinson IP, Rowan A, Pignatelli M, Bodmer WF. Beta-catenin mutations in cell lines established from human colorectal cancers. Proc Natl Acad Sci U S A. 1997;94(19):10330-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Clapper ML, Coudry J, Chang WC. beta-catenin-mediated signaling: a molecular target for early chemopreventive intervention. Mutat Res. 2004;555(1-2):97-105 [DOI] [PubMed] [Google Scholar]
- 46. Bachar-Dahan L, Goltzmann J, Yaniv A, Gazit A. Engrailed-1 negatively regulates beta-catenin transcriptional activity by destabilizing beta-catenin via a glycogen synthase kinase-3beta-independent pathway. Mol Biol Cell. 2006;17(6):2572-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhou G, Xie TX, Zhao M, et al. Reciprocal negative regulation between S100A7/psoriasin and beta-catenin signaling plays an important role in tumor progression of squamous cell carcinoma of oral cavity. Oncogene. 2008;27(25):3527-38 [DOI] [PubMed] [Google Scholar]
- 48. Sun Z, Cao X, Jiang MM, et al. Inhibition of beta-catenin signaling by nongenomic action of orphan nuclear receptor Nur77. Oncogene. 2012;31(21):2653-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kikuchi A, Kishida S, Yamamoto H. Regulation of Wnt signaling by protein-protein interaction and post-translational modifications. Exp Mol Med. 2006;38(1):1-10 [DOI] [PubMed] [Google Scholar]
- 50. Zhang DD, Lo SC, Sun Z, Habib GM, Lieberman MW, Hannink M. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J Biol Chem. 2005;280(34):30091-9 [DOI] [PubMed] [Google Scholar]
- 51. Park CH, Hahm ER, Park S, Kim HK, Yang CH. The inhibitory mechanism of curcumin and its derivative against beta-catenin/Tcf signaling. FEBS Lett. 2005;579(13):2965-71 [DOI] [PubMed] [Google Scholar]
- 52. Denys H, Jadidizadeh A, Amini Nik S, et al. Identification of IGFBP-6 as a significantly downregulated gene by beta-catenin in desmoid tumors. Oncogene. 2004;23(3):654-64 [DOI] [PubMed] [Google Scholar]
- 53. Dayma K, Radha V. Cytoskeletal remodeling by C3G to induce neurite-like extensions and inhibit motility in highly invasive breast carcinoma cells. Biochim Biophys Acta. 2011;1813(3):456-65 [DOI] [PubMed] [Google Scholar]
- 54. Radha V, Rajanna A, Mitra A, Rangaraj N, Swarup G. C3G is required for c-Abl-induced filopodia and its overexpression promotes filopodia formation. Exp Cell Res. 2007;313(11):2476-92 [DOI] [PubMed] [Google Scholar]
- 55. Radha V, Nambirajan S, Swarup G. Subcellular localization of a protein-tyrosine phosphatase: evidence for association with chromatin. Biochem J. 1994;299(Pt 1):41-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19(9):2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Surendran K, Simon TC. CNP gene expression is activated by Wnt signaling and correlates with Wnt4 expression during renal injury. Am J Physiol Renal Physiol. 2003;284(4):F653-62 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.