Abstract
Human breast cancer is a heterogeneous disease composed of different histologies and molecular subtypes, many of which are not replicated in animal models. Here, we report a mouse model of breast cancer that generates unique tumor histologies including tubular, adenosquamous, and lipid-rich carcinomas. Utilizing a nononcogenic variant of polyoma middle T oncogene (PyMT) that requires a spontaneous base-pair deletion to transform cells, in conjunction with lentiviral transduction and orthotopic transplantation of primary mammary epithelial cells, this model sporadically induces oncogene expression in both the luminal and myoepithelial cell lineages of the normal mouse mammary epithelium. Microarray and hierarchical analyses using an intrinsic subtype gene set revealed that lentiviral PyMT generates both luminal and basal-like tumors. Cumulatively, these results show that low-level expression of PyMT in a broad range of cell types significantly increases tumor heterogeneity and establishes a mouse model of several rare human breast cancer subtypes.
Keywords: PyMT, lipid rich, breast cancer, cell of origin, mammary gland
Introduction
Molecular and histological classifications of breast cancer provide essential diagnostic information that is often used to determine the initial course of therapy for patients.1,2 Since these classifications differ in both etiology and therapeutic options, it is critical to develop mouse models that represent the full spectrum of the disease. Through the study of diverse mouse models, our understanding of cancer progression and the development of subtype-specific therapeutics can improve. However, currently available mouse models of breast cancer do not replicate the full spectrum of subtypes observed in human disease.3 This is in large part due to uncertainty in the cellular origins of human breast cancer, which prevents appropriately targeting oncogenic drivers to physiologically relevant cell populations. In addition, the necessity to limit oncogene expression to the mammary gland requires the use of hormonally controlled promoters, such as the mouse mammary tumor virus long terminal repeat enhancer (MMTV-LTR) and whey acidic protein (WAP) promoter.4,5 Therefore, oncogene expression is limited to a subset of more differentiated cells of the mammary gland. These constraints preclude investigation into how different mammary cell populations respond to oncogenic transformation.
The MMTV-LTR is one of the most common promoters used in directing transgene expression to the mammary gland. This promoter exhibits high expression in a subset of hormone-responsive cell types in mammary epithelium, with low or undetectable expression in other tissues.6 Because of its tissue specificity, the MMTV-LTR promoter has been widely used to establish oncogenic mouse models of breast cancer. One such model drives the polyoma middle T (PyMT) oncogene (MMTV-PyMT) to activate a signaling pathway composed of known effectors of breast carcinogenesis, such as Src, Ras, and PI3K.7-11 Tumors established by the MMTV-PyMT model mature rapidly and metastasize to the lung, a common site of breast cancer dissemination.12 The majority of tumors generated in MMTV-PyMT mice are subtyped as luminal, solid adenocarcinomas based on molecular and histological analyses.4,13 It is unknown whether the luminal-restricted tumor phenotype observed in MMTV-PyMT mice is inherent to PyMT oncogenic effects or occurs from the constrained expression of the oncogene in a subset of hormone-responsive luminal cells.14 Thus, we sought to elucidate whether the limited heterogeneity in the MMTV-PyMT model was intrinsic to the oncogene or a result of the cell type–restricted nature of the MMTV promoter.
For this study, we developed a novel lentiviral infection and transplantation method that targets PyMT expression to a broad range of cell types. Using this model, we demonstrate that under the control of nonhormone-specific promoters, the PyMT oncogene generates a wide diversity of tumor histologies and both basal-like and luminal molecular subtypes. A particularly notable cancer established by this model replicates a rare and highly aggressive human breast cancer subtype called lipid-rich carcinoma,15 which to our knowledge has not been previously modeled.
Results
Development of a sporadic mouse model of PyMT oncogenesis
We sought to establish a method that does not limit oncogene activity to a specific mammary cell population. Accordingly, we used lentiviral-mediated gene delivery and mammary gland reconstitution16,17 to induce gene expression in diverse mammary lineages. Prior work has demonstrated that lentiviral transgenes driven by the ubiquitous elongation factor 1 α (EF1α) promoter results in gene expression in all mammary lineages, including luminal, myoepithelial, progenitor, and stem cell populations within mammary outgrowths.17 We employed this methodology to develop a model that enabled unrestricted oncogene expression within these various mammary cell populations.
We first confirmed the EF1α promoter activity in luminal and myoepithelial lineages by utilizing a fluorescent reporter lentivirus, EF1α-ZsGreen17 (Suppl. Fig. S1). To evaluate the mammary cell types that expressed this promoter in vitro, mouse mammary epithelial cells (MECs) were infected with high-titer EF1α-ZsGreen lentivirus and subsequently embedded as single cells in Matrigel (BD Biosciences, Franklin Lakes, NJ).18 After 2 weeks in 3-dimensional culture, the cells developed into clonally derived organoids that were fixed and stained for myoepithelial marker cytokeratin 14 (K14). As expected and consistent with previously published data,17 expression of the ZsGreen reporter was observed in both K14-positive myoepithelial cells and K14-negative luminal lineages (Fig. 1A, white arrows).
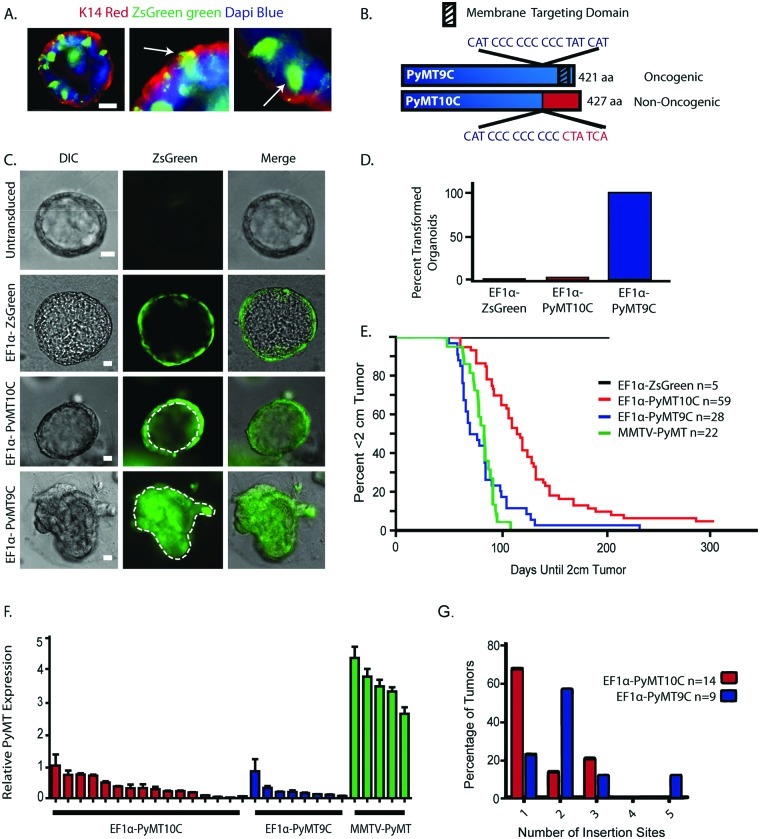
Figure 1.
Characterization of a sporadic breast cancer model. (A) EF1α-ZsGreen is expressed in both myoepithelial and luminal lineages. MECs were infected in suspension with EF1α-ZsGreen and grown in Matrigel for 2 weeks. Immunofluorescence was performed using a marker for myoepithelial cells (K14, red). White arrows highlight one co-stained ZsGreen-expressing K14-positive cell (myoepithelial cell) and one K14-negative ZsGreen-expressing cell (luminal cell). Scale bar is 20 µm. (B) Comparison of the EF1α-PyMT9C and EF1α-PyMT10C variants, highlighting changes in gene length, targeting domain changes, and oncogenic potential. (C) To assess the oncogenic potential of the PyMT variants, primary MECs were infected in monolayer with EF1α-ZsGreen, EF1α-PyMT10C, or EF1α-PyMT9C and grown in Matrigel for 2 weeks. The EF1α-ZsGreen–infected cyst has dotted lines to indicate a hollow lumen. The EF1α-PyMT10C–infected cyst also has a hollow lumen. The EF1α-PyMT9C–infected cyst has transformed characteristics of a solid lumen. Scale bar is 25 µm. (D) Proportion of infected organoids with a transformed phenotype. (E) Survival curves comparing time until a 2-cm tumor. Latency between EF1α-PyMT10C and EF1α-PyMT9C tumor models was significant (P ≤ 0.05). (F) RT-PCR quantification of relative PyMT expression levels in EF1α-PyMT10C, EF1α-PyMT9C, and MMTV-PyMT transgenic tumors, normalized to the RPLP0 housekeeping gene. (G) Number of insertion sites in EF1α-PyMT10C– and EF1α-PyMT9C–generated tumors.
In humans, carcinogenesis occurs through sporadic genetic events. To replicate the spontaneous nature of breast cancer, we generated a nononcogenic variant of PyMT (called PyMT10C) that sporadically converts during cell proliferation to the oncogenic variant, PyMT9C. The PyMT10C isoform is nononcogenic due to a single nucleotide insertion in a string of cytosines, which causes a frameshift in the coding sequence near the C-terminus of the protein.19 The result is both a loss of the membrane-targeting domain and gain of a nuclear localization sequence19 (Fig. 1B). This prevents transformation by PyMT10C, as the protein can no longer effectively act as a membrane scaffold for the upstream effectors of Ras and PI3K pathways.19 However, it has been proposed that DNA polymerase slippage at the 10-cytosine string during genomic replication sporadically results in the deletion of 1 cytosine, restoring the membrane localization of PyMT (PyMT9C) and its oncogenic activity.20 Reversion does not commonly occur during the production of lentiviral particles but at a high frequency in transduced cells undergoing proliferation.19 Consequently, in this system, reversion back to oncogenic PyMT9C is both sporadic and restricted to proliferating cells, creating a model that resembles spontaneous transformation in humans.21,22
To verify the immediate transforming activity of EF1α-PyMT9C and the delayed transformation of EF1α-PyMT10C, primary MECs were infected in monolayer with EF1α-ZsGreen, EF1α-PyMT9C, or EF1α-PyMT10C lentivirus (Suppl. Fig. S1) and subsequently embedded as single cells in Matrigel (BD Biosciences). Uninfected MECs, or MECs infected with either EF1α-ZsGreen (control virus) or EF1α-PyMT10C, developed into cyst-like organoids that contained a hollow lumen, similar to the normal morphology observed in untransduced cells (Fig. 1C). Conversely, all organoids generated from EF1α-PyMT9C–transduced cells developed solid, multilobular organoids with an infiltration of cells within their lumen (Fig. 1C and 1D), which is a phenotype consistent with transformation (P ≤ 0.0001 for PyMT9C and PyMT10C).18 In addition, infected MECs were transplanted in vivo and analyzed 4 weeks later at a time when ducts begin to arise from the site of injection. While both EF1α-PyMT9C and EF1α-PyMT10C injection sites still had unorganized morphology, duct-like structures emerging from the transplantation site were only observed in the EF1α-PyMT10C outgrowth, further demonstrating that EF1α-PyMT10C does not immediately transform cells (Suppl. Fig. S2A1-3).
To directly assay for conversion in vitro, we generated a PyMT10C hemagglutinin (HA)–tagged lentivirus (EF1α-PyMT10C-9CHA) where the HA epitope is in frame following conversion to PyMT9C (Suppl. Fig. S1). We infected MECs in vitro with the EF1α-PyMT10C-9CHA lentivirus and then assayed for conversion by immunofluorescence against HA at 72-hour and 1-week time points. At the 72-hour time point, only 1 cell in 15 fields expressed the PyMT9C HA-tagged protein. However, after 1 week, the number of converted cells had increased, with 44 detectable clones in 15 fields (Suppl. Fig. S2B1-3 and S2E). In addition, after analyzing K14 expression at the 1-week time point, we found that both luminal and basal cells expressed the ZsGreen reporter (Suppl. Fig. S2C1-D3). Taken together, these data demonstrate the sporadic nature of oncogenic conversion in both the luminal and myoepithelial cell lineages.
We reasoned that MECs expressing PyMT10C would undergo delayed tumor development since a conversion to the PyMT9C variant is necessary to elicit oncogenesis. To test this, primary MECs were infected with the EF1α-PyMT9C and EF1α-PyMT10C lentiviruses and subsequently transplanted into cleared mammary fat pads. As expected, tumor latency was significantly delayed (P ≤ 0.0001) in MECs transduced with EF1a-PyMT10C virus (median time to 2-cm tumor: 114.5 days) as compared to EF1α-PyMT9C–expressing cells (median time to 2-cm tumor: 71.5 days) (Fig. 1E). To determine whether a cytosine deletion occurred, we sequenced the polycytosine tract of PyMT in several tumors generated from EF1α-PyMT10C–transduced MECs. All tumors derived from EF1α-PyMT10C cells contained the PyMT9C oncogenic isoform (Suppl. Fig. S2F). Thus, PyMT10C undergoes a reversion mutation after transplantation, creating a tumor model of sporadic breast cancer.21,22
Integration of lentiviral transgenes into genomes results in single insertions at one, and sometimes several, chromosomal location within a transduced cell.23 Consequently, PyMT expression may vary between different tumors in the lentiviral-PyMT model as compared to the MMTV-PyMT model. Therefore, we characterized PyMT mRNA levels across EF1α-PyMT9C, EF1α-PyMT10C, and MMTV-PyMT tumors to assess differences in expression between these models. We found that PyMT expression varied significantly among tumors from the lentiviral-PyMT model (Fig. 1F); most likely because of different insertion sites, copy number, and proportion of uninfected host cells in tumors. In addition, PyMT expression was decreased 4- to 150-fold in the lentiviral-PyMT models when compared to MMTV-PyMT tumors (Fig. 1F).
During the transition from a normal cell to breast cancer, a mammary cell acquires sporadic transforming mutations, leading to its clonal expansion and subsequent development into a tumor.24 Thus, transformation initiates and progresses within a normal tissue environment. In contrast to this process, the MMTV-PyMT model generates multifocal hyperplasia throughout the mammary gland,25 which most likely leads to tumors derived from more than one clone. However, since the EF1α-PyMT10C–transduced cells must acquire a transforming mutation in the oncogene prior to initiating tumorigenesis, we predicted that tumors in this model would develop from a dominant clone. To test clonal dominance, we used ligation-mediated PCR to sequence lentiviral insertion sites from a subset of EF1α-PyMT9C and EF1α-PyMT10C tumors. Overall, only 22% of EF1α-PyMT9C tumors exhibited a single integration site, while 67% of EF1α-PyMT10C tumors had a single insertion site (Fig. 1G). Thus, the sporadic nature of EF1α-PyMT10C oncogenesis may contribute to the increased clonality in tumors generated from this model. Accordingly, we performed subsequent studies using single integration site tumors derived from the sporadic PyMT10C oncogenesis model. Studies performed using tumors with either uncharacterized integration sites or with the EF1α-PyMT9C lentivirus are noted accordingly.
Clonal EF1α-PyMT10C tumors display a range of unique histologies that resemble diverse human breast cancer subtypes
Our studies demonstrated that the EF1α-PyMT10C lentiviral infection and mammary reconstitution model could induce sporadic transformation and target diverse mammary cell populations. We next assessed whether the sporadic and diverse nature of transformation in the lentiviral-PyMT model would increase tumor heterogeneity as compared to the MMTV-PyMT transgenic mouse model. MECs were infected in suspension with the EF1α-PyMT10C lentivirus and subsequently injected into cleared mammary fat pads of recipient mice to generate tumors. As described below, several analyses were performed on these tumors to assess their cellular, molecular, and pathological heterogeneity in comparison to the MMTV-PyMT model.
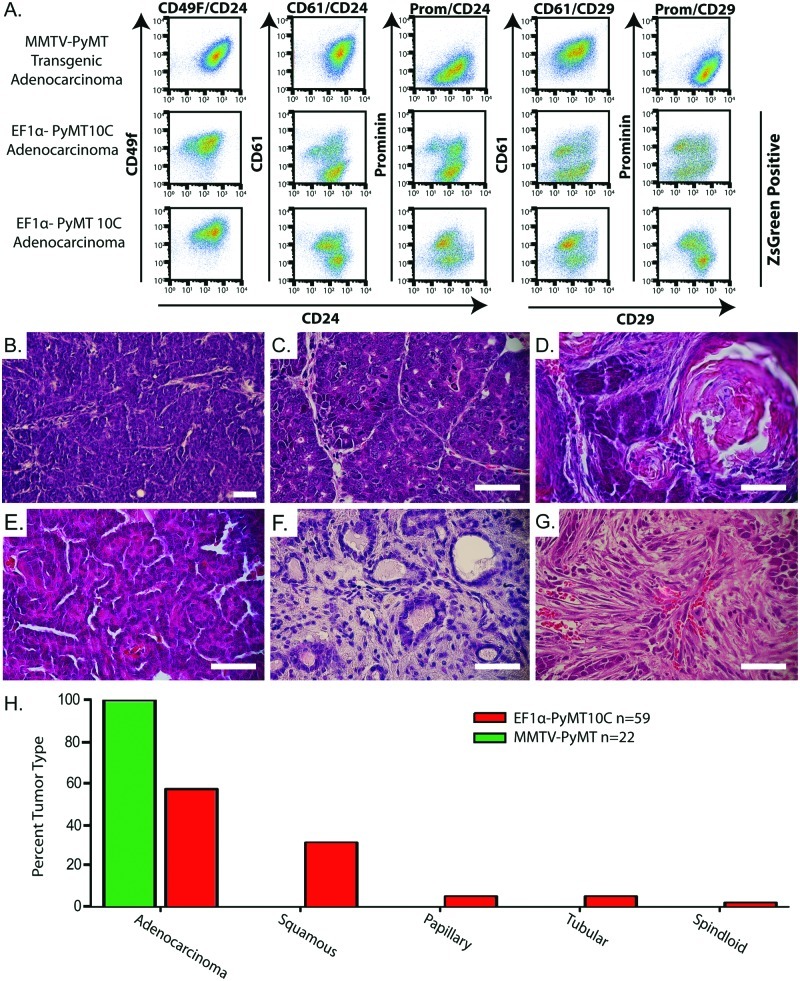
Breast cancer is known to establish heterogeneous cell populations within a single tumor.26,27 To examine cell population diversity, we used FACS analysis on secondary tumors derived from single integration site primary tumors. Specific cell populations were distinguished using the well-established mammary epithelial cell markers CD49f/CD24, CD61/CD24, prominin/CD24, CD61/CD29, and prominin/CD29.28-30 These markers can identify stem, luminal, myoepithelial, and progenitor cell populations within the normal mammary epithelium. While some of these populations have been implicated as representing cancer stem cells and noncancer stem cells,31,32 we used these markers simply as indicators of cellular diversity within and between tumors. We found that while MMTV-PyMT–derived tumors had only a single population for each set of markers, ZsGreen-gated tumor cells generated from the lentiviral-PyMT model were more diverse, showing several distinct cell populations (Fig. 2A and Suppl. Fig. S3). These data demonstrate that the EF1α-PyMT10C model is capable of generating significant intratumor cellular heterogeneity in contrast to the limited cellular diversity of the MMTV-PyMT model.
Figure 2.
Cellular and histological analyses of tumor types generated from the EF1α-PyMT10C–driven lentiviral tumor model. (A) FACS analysis of an MMTV-PyMT primary tumor and 2 clonal EF1α-PyMT10C secondary tumors. Populations were gated to exclude lineage-positive cells (CD45, CD31, and CD140) and apoptotic cells (7AAD) and then gated for ZsGreen expression (lentiviral tumors only) to ensure only tumor cells were represented in the final plot. (B) H&E stain of an adenocarcinoma from MMTV-PyMT mice. (C-G) H&E stain of histologies from single clone–dominant EF1α-PyMT10C–induced tumors. (C) Adenocarcinoma, (D) adenosquamous, (E) papillary, (F) tubular, and (G) spindloid. Scale bars are 25 µm. (H) Quantification showing the percentage of each histological subtype from all tumors (including tumors with unanalyzed integration sites) generated from the EF1α-PyMT10C and MMTV-PyMT tumor models. Tumor histology was scored if >50% of the tumor exhibited the specific histological feature by H&E staining.
Transgenic MMTV-PyMT–induced tumors usually produce solid, poorly differentiated adenocarcinomas (Fig. 2B).4,33 While some lentivirus-generated tumors shared this histology (Fig. 2C), about half the tumors displayed other unique but clinically relevant cancer pathologies. These included adenosquamous carcinomas that contained both keratin pearls and large sheets of keratinized cells (Fig. 2D), papillary tumors with thin epithelial cell layers surrounding the stroma (Fig. 2E), tubular tumors with dense stroma and well-differentiated duct-like structures (Fig. 2F), and poorly differentiated spindloid tumors (Fig. 2G). About 30% of EF1α-PyMT10C tumors were adenosquamous carcinomas, while papillary, tubular, and spindloid carcinomas combined made up 12% of the tumors (Fig. 2H). In contrast, all of the MMTV-PyMT tumors (n = 22) analyzed in this study exhibited solid adenocarcinoma histology. These data demonstrate that lentiviral-based tumorigenesis created tumors with a marked histological diversity when compared to the MMTV-PyMT transgenic model.
We next performed molecular analysis of the lentiviral-PyMT tumors to determine whether they expressed biomarkers associated with breast cancer. The lentiviral-derived tubular tumors resembled human tubular carcinomas with their well-differentiated morphology, extensive infiltration of stromal cells, and complete loss of adipocytes.34 These tumors were unique in their expression of the luminal differentiation markers cytokeratin 19 (K19) and progesterone receptor (Suppl. Fig. S4 and S5).
In contrast to the well-differentiated tubular tumors, adenosquamous carcinomas were poorly differentiated and exhibited dense clusters of keratinized tissue throughout the tumor. We examined cytokeratin 6 (K6) and cytokeratin 10 (K10), two common epidermal keratins that are expressed in adenosquamous breast cancer35 but only rarely observed in normal mammary tissue. EF1α-PyMT10C adenosquamous carcinomas were positive for both K6 and K10 (Suppl. Fig. S4), but both markers were absent in MMTV-PyMT tumors36 (Suppl. Fig. S4). Adenosquamous carcinomas have been described in the MMTV-Wnt1 transgenic mouse model but are rarely observed in MMTV-PyMT mice.33
It has been previously shown that late-stage MMTV-PyMT tumors exhibit an absence of myoepithelial cells.33 In contrast, we observed that solid adenocarcinomas generated by EF1α-PyMT10C often exhibited an abundant myoepithelial population (K14 expressing) (Suppl. Fig. S4), which was more similar to tumors derived from the MMTV-Wnt1 transgenic model.33
PyMT expression from the mucin-1 promoter generated a rare lipid-rich phenotype
We also evaluated whether the expression of PyMT10C under the control of a nonubiquitous promoter could establish a similar diversity observed with the EF1α promoter–driven model. We replaced the EF1α promoter with the well-characterized human mucin-1 (Muc1) promoter, which specifically targets cells in the luminal mammary epithelial lineage.37,38 We attempted to assess gene expression levels from this lentivirus using our 3-dimensional mammary organoid assay, but expression was extremely low, and we could not confirm that promoter activity was restricted to luminal cells.
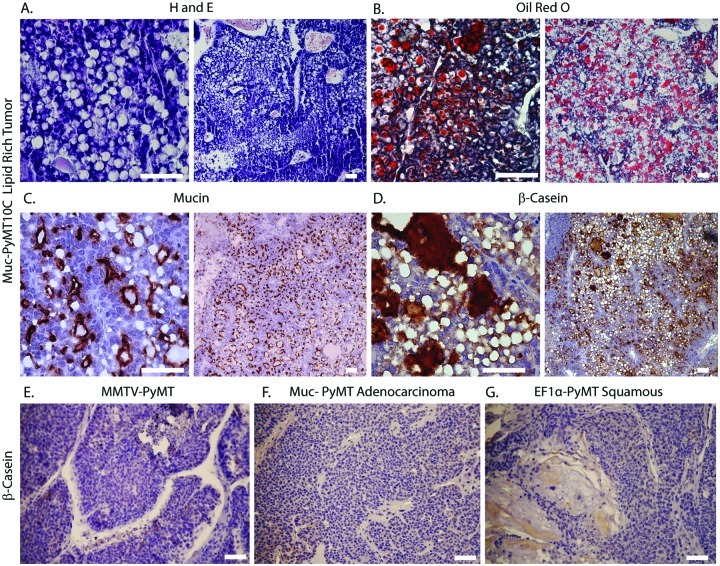
Interestingly, upon transplantation of Muc-PyMT10C–transduced MECs, we observed comparable tumor histologies with the EF1α-driven model, including adenocarcinoma, adenosquamous, tubular, and papillary. The average expression level of PyMT in tumors was not statistically different between the Muc and EF1α models, and Muc-PyMT10C–transduced MECs had a median tumor latency of 123.5 days, similar to EF1α-PyMT10C (Suppl. Table S1). In addition, several tumors were positive for estrogen receptor, especially well-differentiated tubular tumors, and tumors showed variation in luminal and basal keratin expression (Suppl. Fig. S6A-I). The most novel finding occurred when about 16% of Muc-PyMT10C tumors presented with a phenotype similar to lipid-rich carcinomas of the breast, which is a clinically relevant but rare cancer subtype.39 Histologically, the mouse tumors displayed characteristic large cytoplasmic vacuoles observed by H&E staining (Fig. 3A). Oil Red O staining confirmed that the vacuoles were filled with triglycerides and fats (Fig. 3B), and immunofluorescence against Muc1 verified that the tumors were of luminal origin (Fig. 3C). Moreover, some mouse lipid-rich tumors had lymph node or lung metastasis, which is consistent with the aggressive nature of lipid-rich carcinoma in humans.40 However, metastasis derived from lipid tumors did not exhibit a lipid-rich histology, suggesting the phenotype is lost in the process of dissemination (Suppl. Fig. S6J).
Figure 3.
Abundant lipid vacuoles and β-casein expression in lipid-rich carcinomas. (A-D) Histology of clone-dominant lipid-rich tumors. (A) H&E- and (B) Oil Red O–stained sections showing numerous lipid-filled vacuoles. (C) Immunohistochemistry for Muc1. (D) Immunohistochemistry for β-casein. (E-G) β-casein immunohistochemistry of non–lipid-rich tumors derived from different models. (E) MMTV-PyMT, (F) Muc-PyMT10C adenocarcinoma, and (G) EF1α-PyMT10C adenosquamous carcinoma. All scale bars are 25 µm.
During lactation, specialized luminal epithelial cells, called alveolar cells, produce copious amounts of lipid and milk.41 Since abundant lipid production in the mammary gland is observed in cells of alveolar origin,42 we asked whether lipid-rich tumors also exhibited milk-producing characteristics associated with alveolar cells. We assessed milk protein expression by immunohistochemistry against β-casein,43 which demonstrated that lipid-rich tumors expressed an abundance of milk protein as compared to other PyMT-induced tumors without lipid-rich pathology (Fig. 3D-G). The extensive lipid and milk protein production observed in these tumors is consistent with a possible cellular origin within the alveolar lineage.
Microarray analysis reveals unique gene expression pathways, and both luminal and basal-like subtypes are generated by the EF1α-PyMT lentiviral model
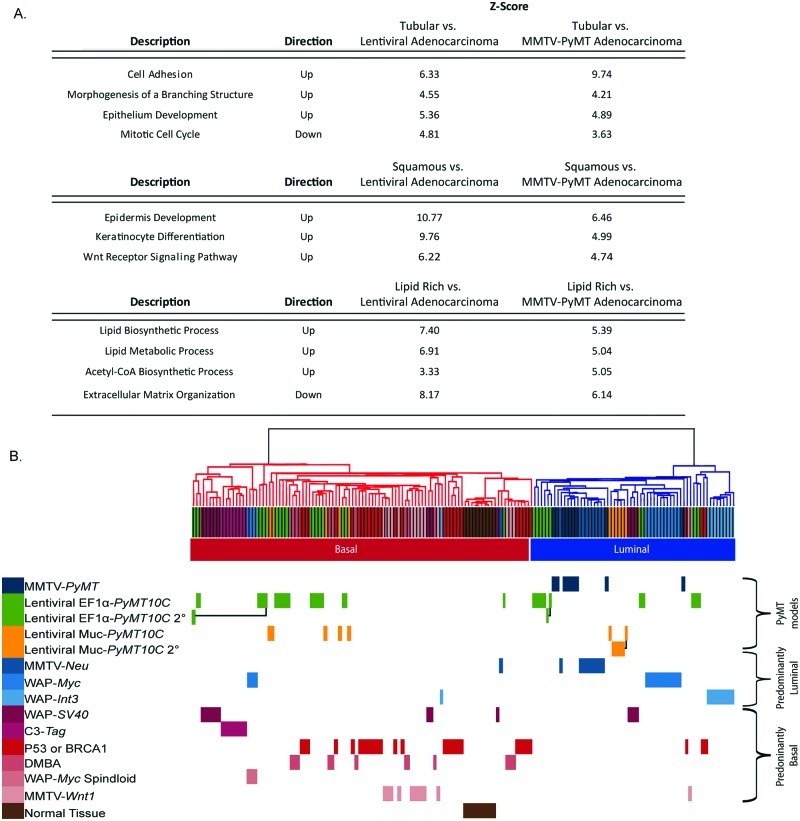
To molecularly identify the unique pathways associated with different tumor histologies, we performed a paired analysis of microarray profiles from adenocarcinomas, with tubular, adenosquamous, or lipid-rich tumors. In each comparison, we analyzed gene ontology (GO) pathways,44 examining expression profiles that differentiated between the tumor histologies (Fig. 4A). Tubular tumors had a mammary differentiation phenotype, with up-regulated pathways involved in cell adhesion, along with branching and epithelial cell differentiation, consistent with their well-differentiated pathology. In addition, tubular tumors exhibited down-regulation of the mitotic cell cycle pathway, indicating a reduced proliferation in this tumor type as compared to adenocarcinomas. In contrast, adenosquamous tumors revealed up-regulated pathways for epidermal development and keratinization. In this set of tumors, the Wnt pathway was also up-regulated, which is consistent with published data showing that MMTV-Wnt transgenic mice can generate adenosquamous tumors.33 The lipid-rich tumors exhibited up-regulation of metabolism and lipid biosynthesis pathways. These pathways included acetyl-CoA synthesis and genes involved in fatty acid biosynthesis and metabolism. Down-regulated pathways included extracellular matrix constituents, such as collagen, elastin, and laminin. Importantly, solid adenocarcinomas generated from either MMTV-PyMT or lentiviral-PyMT models were interchangeable in this analysis, with only minimal changes in the pathway expression between MMTV-PyMT adenocarcinomas and lentiviral adenocarcinomas. Taken together, these data demonstrate that tumors with tubular, adenosquamous, and lipid-rich pathology expressed unique GO pathways as compared to lentiviral-PyMT and MMTV-PyMT adenocarcinomas.
Figure 4.
Comparison of molecular pathways regulated in different subtypes using microarray analysis. (A) Gene pattern comparative analysis demonstrates pathways that are differentially regulated in tubular, adenosquamous, and lipid-rich tumor histologies when compared to adenocarcinomas generated by either lentiviral-PyMT or MMTV-PyMT models. (B) Hierarchical clustering of lentiviral-PyMT models demonstrates a greater subtype diversity than MMTV-PyMT transgenic mice. Secondary tumors cluster within the same molecular subtype as the primary tumor, as shown by black connectors. Microarray data for transgenic mouse models were obtained from Herschkowitz et al. 13
Using the microarray data, we directly compared molecular subtypes between 9 published mouse models of breast cancer13 with 24 lentivirally generated tumors (Fig. 4B and Suppl. Fig. S7). Of the lentiviral-PyMT–generated tumors, 14 clustered in the basal-like subtype, 9 were luminal, and 1 was an unclassified spindloid tumor. While genetically engineered mouse models produced tumors that classified predominantly into either a basal-like or luminal molecular subtype,13 tumors from the lentiviral-PyMT model were classified nearly equally into both subtypes. In addition, we evaluated whether secondary tumors generated from single insertion site luminal, basal-like, and lipid-rich subtypes exhibited the same molecular classification as their primary tumor. Hierarchical clustering demonstrated that primary and secondary tumors remained with a common molecular subtype (Fig. 4B, black connections between primary and secondary tumors). Thus, tumor phenotype was stable in secondary tumors derived from the lentiviral-PyMT model as observed by microarray gene expression.
Discussion
Here, we describe a lentiviral-PyMT oncogenesis model that sporadically transforms normal mammary epithelial cells, generating tumors with diverse histological and molecular subtypes. These data contrast the well-characterized pathology of MMTV-PyMT tumors, which are predominantly solid adenocarcinomas, exhibiting a histology and gene expression consistent with a luminal subtype.4 Two hypotheses have been proposed to explain the luminal tumor phenotype in the MMTV-PyMT model.14 First, the MMTV promoter could restrict oncogenesis to luminal cell types that have limited differentiation potential. Alternatively, the PyMT signaling pathway could force tumor cells to transform down a luminal tumor fate, regardless of the transformed cell lineage. Using our lentiviral-based method, we address these hypotheses by driving oncogenesis in both luminal and myoepithelial cell lineages. The EF1α-PyMT10C lentivirus generated several rare and clinically relevant breast cancer pathologies, and importantly, both luminal and basal-like tumors were observed in nearly equal ratios. These data suggest that the second hypothesis cannot entirely explain the limited tumor diversity in the MMTV-PyMT model since PyMT does not inherently restrict transformation toward a luminal subtype.
Instead, our data suggest a dual model where both the targeted cell type and expression level of PyMT contribute to influence tumor heterogeneity. While the MMTV promoter preferentially targets hormonally sensitive mammary epithelial cells in the mammary gland, the lentiviral-PyMT model broadly targets an array of adult mammary epithelial lineages, including luminal, myoepithelial, stem, and progenitor cell populations.17 In addition, tumors generated by the lentiviral-PyMT model showed significantly lower expression of the oncogene than in MMTV-PyMT tumors. Low PyMT expression may facilitate the cooperation between the oncogene and cell-specific signaling pathways within the tumor’s cell of origin. Such a scenario is analogous to estrogen-dependent breast cancer, where oncogenic driver mutations establish a transformed state with dependency on pre-existing signaling networks, such as those mediated by an estrogen receptor (ER). With such a dependence, the oncogenic drivers are not sufficient to maintain tumors in the absence of ER signaling, which is demonstrated by the regression of ER+ tumors in patients treated with tamoxifen.45 Thus, ER functions not as an oncogene but in concert with the oncogenic drivers, demonstrating the coordination between carcinogenic and normal signaling pathways. Similarly, low expression of PyMT may provide a growth advantage in the transformed cell but could require additional growth signals from pre-existing cellular networks, resulting in synergy between the signaling pathways inherent to both the oncogene and cell. Thus, the observed variation in tumor phenotype between the lentiviral-PyMT and MMTV-PyMT models may result from differences in both oncogene expression and cell types targeted by the model.
We questioned the ability of the luminal cell–driven Muc-PyMT10C lentivirus to generate tumors in the basal-intrinsic subtype. While we cannot identify the specific cellular origin of these tumors, it is possible that luminal cells acquired basal tumor traits during transformation. This hypothesis is supported by several recent reports that suggest luminal cells are the origins of some basal-like breast tumors, particularly those associated with the loss of BRCA1.46-48 Future studies are necessary to more specifically target oncogenes to distinct cell lineages to more accurately ascribe cellular origins with molecular and histological tumor subtypes.
Of particular interest was that the Muc-PyMT10C lentivirus generated a tumor representative of the clinically aggressive lipid-rich carcinoma subtype observed in human breast cancer. To our knowledge, this is the only mouse model that establishes this unique tumor pathology. We demonstrated that the lipid-rich tumors were distinct from other PyMT tumor subtypes, as evident by gene expression and histology showing abundant lipid and milk production. The characteristics of this tumor suggest that carcinogenesis occurred in a cell type with the capacity to produce copious lipid and milk, a feature of differentiated alveolar cells of the mammary gland.49 It is noteworthy that in human lipid-rich carcinomas, milk constituents including α-lactalbumin, human milk fat globule membrane antigen-2, and lactoferrin were shown to be highly expressed.50 Based on these data, it is intriguing to speculate the origin of these tumors as mature or immature alveolar cells of the breast. Normally, differentiated alveolar cells appear only transiently during pregnancy and lactation and require specific survival mechanisms, mediated through pathways such as JAK2, STAT5, and IGF, to prevent cell death.42,51 Thus, these tumors may be uniquely sensitive to therapeutic inhibitors of these pathways. A future goal of this study will be to further characterize the cellular etiology and transforming events that establish distinct tumor subtypes and evaluate the interaction between the cell of origin and oncogenic signaling networks.
We demonstrate that nonselectively targeting PyMT oncogenesis sporadically in the mouse mammary gland has a significant impact on both tumor subtype and pathology. Importantly, the expanded tumor heterogeneity generated by the lentiviral-based PyMT model resulted in several clinically significant and rare pathologies, including the first mouse model representation of lipid-rich carcinoma of the breast. Future studies will be necessary to more specifically assess how signaling networks within the transformed cell of origin contribute to breast cancer pathology.
Materials and Methods
Mice
FVB/N mice were obtained from Charles River Laboratories (Wilmington, MA) and maintained in a pathogen-free facility. Mice were handled according to University of Utah–approved Institutional Animal Care and Use Committee procedures.
Lentiviral vectors and virus production
The EF1α-ZsGreen (HIV-ZsGreen, plasmid 18121, Addgene, Cambridge, MA) plasmid was described previously.17 Both EF1α-PyMT9C and EF1α-PyMT10C plasmids were cloned by PCR amplification of PyMT and restriction site digest into the multi-cloning site of pEF1α-ZsGreen. To make EF1α-PyMT10C- 9CHA, PyMT was PCR amplified using an HA tagged reverse primer. Muc-PyMT10C was topo cloned after amplification of the human Muc1 promoter and generation of a topo entry vector. Removal of EF1α from EF1α-PyMT10C produced the destination vector for topo cloning of Muc-PyMT10C. Lentiviral production was performed as in Welm et al. 17 using BSL2-enhanced methods. Briefly, confluent plates of 293T cells covered with 9 mL of 293T media (DMEM + 10% FBS) were transfected with a polyethylenimine (PEI)52 and Opti-Mem mixture (30 µg PEI in 1 mL Opti-Mem [Invitrogen, Carlsbad, CA]) containing 1.7 µg of each packaging plasmid, pMDLg/pRRE, pVSVG, and pRSV-Rev, and 5 µg of transfer plasmid. Media were changed the next day, adding only 8 mL, and virus collection took place 24 and 48 hours later. To collect the virus, media were pooled, put into 50-mL conical vials, and kept on ice. Conical vials were spun at 2,000g for 10 minutes to remove large cell debris. Supernatant was then filtered through a 0.45-um celulose acetate filter to ensure the complete removal of debris. To collect a high-titer virus, virus-containing supernatant was spun in an ultracentrifuge at 35,000g for 1 hour 45 minutes at 4°C. Supernatant was removed, and the pellet was resuspended in 80 µL of DMEM media. Tubes were covered and allowed to sit overnight at 4°C. After concentrating the virus, infectious units were titered using either FACS or a commercial p24 ELISA kit (Cell Biolabs, San Diego, CA).
Mouse mammary epithelial cell preparations
Mouse mammary epithelial cells were collected from 8-week-old female FVB/n mice. Dissected mammary glands were minced with a razor blade for 5 minutes, forming a tissue slurry. Each gram of tissue was incubated with 5 mL of collagenase buffer (RPMI-1640, 25 mM HEPES, 5% FBS, 100 µg/mL streptomycin, 100 U/mL penicillin, and 2.5 mg/mL collagenase IV [Sigma, St. Louis, MO]) for 1.5 hours at 37°C with gentle rocking. Mammary organoids were separated from fat cells by two 10-minute centrifugations at 600g. Organoids were separated from stromal cells by 4 differential centrifugations at 500g for 45, 35, 30, and 30 seconds, respectively. Pelleted organoids were trypsinized (0.05% trypsin/EDTA, (Gibco, Burlington Ontario, Canada) for 15 minutes at 37°C or until organoids were dissociated into single cells and then filtered through a 70-µm filter to remove any residual cell clusters. Cells were frozen in freeze media (20% FBS, in DMEM/F12 + 10% DMSO) at a concentration of 4 × 106 cells/mL.
In vitro Matrigel transformation assay
Frozen mammary epithelial cells were thawed at 37°C and plated in suspension at 2 × 106 cells per well of a low-adhesion 24-well plate in MEC media (5 µg/mL ITS, 1 µg/mL hydrocortisone, 10 ng/mL mouse EGF, 10% FBS, 100 U/mL penicillin and streptomycin, in DMEM/F12), allowed to recover overnight, and then infected with lentivirus at a multiplicity of infection of 30. After overnight infection, cells were washed and then allowed to recover for 48 hours. After recovery, transduced cells were plated at a density of 3,000 cells per 20 µL of Matrigel on chamber slides (Millipore, Billerica, MA). Single cells were grown for 2 weeks, with a media change every 3 days, as they developed into cysts. Cell clusters were first assessed for ZsGreen expression and then counted as either a normal hollow cyst or as a solid, transformed organoid.
For immunofluorescence, Matrigel-embedded cells were fixed for 30 minutes with 4% paraformaldehyde at 26°C, followed by permeabilization with 0.5% Triton X-100 in PBS for 2 hours at 26°C. Cells were incubated in a blocking solution (5% BSA and 1% goat serum in PBS) for 1 hour at room temperature. Primary antibodies K8 (1:200, Troma-1, Developmental Studies Hybridoma Bank from The University of Iowa, Iowa City, IA) and K14 (1:500, PRB-155P-100, Covance, Princeton, NJ) were diluted in the blocking solution and left at 4°C overnight. The next day, primary antibody was washed 3 times for 10 seconds with PBS and then 2 times for 10 minutes with PBS. Secondary antibody Alexa Fluor 594 (Invitrogen) was diluted at 1:400 in blocking buffer and left at room temperature for 1 hour. Secondary antibody was removed and washed 3 times for 10 seconds with PBS and then 2 times for 10 minutes with PBS. Nuclei were stained with DAPI (1 µg/mL) in PBS for 20 minutes at 26°C. After rinsing with PBS, cells were mounted with ProLong Gold (Invitrogen).
In vitro PyMT10C to PyMT9C conversion assay
MECs were infected with EF1α-PyMT10C-9CHA in suspension overnight. Cells were then washed and plated on glass coverslips and allowed to grow for 72 hours or 1 week. At each time point, cells were fixed with 4% PFA in PBS for 15 minutes. After fixation, cells were washed with 50 mM NH4Cl to limit background fluorescence and permeabilized with 0.2% Triton for 8 minutes. Cells were blocked with 1% BSA in PBS for 10 minutes and then rocked for 1 hour with anti-HA and anti-K14 antibodies (MMS-101R and PRB-155P, Covance, Princeton, NJ). After primary antibody incubation, cells were washed 3 times with blocking solution, and cells were rocked with secondary antibody, Alexa Fluor 594 and 633 (Invitrogen), for 1 hour. After secondary antibody incubation, cells were washed and incubated with DAPI. Slides were imaged on an Olympus 1X81 microscope (Tokyo, Japan) using a Hamamatsu Photonics ORCA-ER camera (Hamamatsu, Japan). Images were taken and processed using Slidebook 64 version 5.0.0.24 (Denver, CO). Conversion was quantified in 15 random images at 10× magnification; a total of 47 ZsGreen-positive cells at the 72-hour time point and 396 ZsGreen-positive cells at the 1-week time point were analyzed.
Generation of mouse mammary tumors
For tumor generation, 2 × 106 primary MECs were suspended in a low-adhesion 24-well plate in 800 µL of MEC media (5 µg/mL ITS, 1 µg/mL hydrocortisone [in ethanol], 10 ng/mL mouse EGF, 10% FBS, 100 U/mL penicillin and streptomycin, in DMEM/F12) and infected with 1 × 107 high-titer lentiviral particles (MOI of 5) using BSL2-enhanced conditions. In suspension, cells were left to incubate at 37°C overnight, washed twice in 5 mL HBSS, and resuspended in Matrigel at a concentration of 1 × 105 cells per 10 µL of Matrigel (356234) and kept on ice until injection. There was 10 µL of the cell/Matrigel mix injected into cleared fat pads of 3-week-old FVB/N mice. Tumors were removed when they reached 2 cm in diameter. Portions were used for paraffin embedding and sectioning, RNA and DNA analysis, and processing into viable single cells. The viable cells were collected using the same protocol for MEC collection. These cells were used for secondary tumor generation. They were thawed, washed, and cultured overnight in MEC media in low-adhesion plates. The next day, tumor cells were washed and resuspended at a concentration of 6 × 104 cells per 10 µL of Matrigel. There was 10 µL of the cell/Matrigel mixture injected into cleared fat pads of 3-week-old FVB/N recipient mice. Tumor growth was monitored, and tumors were harvested at 2 cm.
Ligation-mediated PCR
Ligation-mediated PCR was performed as previously described.53,54 In brief, 100 ng of DNA, harvested from tumors, was completely fragmented by overnight digestion with the Mse1 restriction enzyme. Annealed adaptors (top: 5′-GTAATACGACTCACTATA GGGCTCCGCTTAAGGGAC-3′, bottom: 5′-PO4-TAGT CCCTTAAGCGGAG-NH2-3′) were then ligated to the sticky-ended fragments overnight using a high concentration ligase (New England Biolabs, Ipswich, MA). Fragments were then digested with a low-frequency cutter, Sac1, to eliminate any contaminating EF1α-PyMT plasmid. Next, we ran PCR on the fragments using an adaptor primer (5′-GTAATACGACTCACGATAGGGC-3′) and LTR primer 4 (5′-CAAGTAGTGTGTGCCCGTCT-3′) to amplify the insertion sites. The PCR mix was then diluted 1:10, and nested PCR was run using a nested adaptor primer (5′-AGGGCTCCGCTTAAGGGAC-3′) and LTR primer 3 (5′-CTGTTGTGTGACTCTGGTAAC-3′). Finally, samples were run on a 2.5% agarose gel at 100 V for 1.5 hours to separate fragments for direct cutting and sequencing using LTR primer 3. If direct sequencing was inconclusive, fragments were cloned into a pcr2.1 TOPO cloning vector (Invitrogen) and sequenced using the M13R primer. Sequences were analyzed using NCBI nucleotide BLAST and compiled to find the number of unique insertion sites for each tumor.
RT-PCR
RT-PCR was performed using Roche LightCycler (Basel, Switzerland) and Kapa SYBR Fast 2X Master Mix (Kapa Biosystems, Woburn, MA). PyMT primers were PyMT fwd (5′-ctgctactgcacccagacaa-3′) and PyMT rev (5′-gcaggtaagaggcattctgc-3′). Keratin 8 primers were keratin 8 fwd (5′-gagatcaccacctaccgcaa-3′) and keratin 8 rev (5′-cagtc ctcctgagtagccgc-3′). Keratin 14 primers were keratin 14 fwd (5′-aagctgctacatgctgctgctca-3′) and keratin 14 rev (5′-agggacaatacaggggctct-3′). RPLP0 was used as a reference gene, using primers RPLP0 fwd (5′-gatgcccagggaagacag-3′) and RPLP0 rev (5′-acaatgaagcattttggataatca-3′). Reactions were run according to the Kapa Biosystems protocol with a 60°C annealing temperature.
FACS analysis
Secondary tumor cells were dissociated from stroma and red blood cells using the protocol for mammary epithelial cell preparations. Once single cells were isolated, samples were kept on ice and aliquoted with 1 × 106 cells per 15-mL conical vial in 200 uL of wash solution (HBSS + 2% FBS). Staining consisted of 10 tubes: 1) a no-antibody control, 2) CD24-PE control, 3) CD24-biotin-streptavidin-APC control, 4) 7AAD control, 5) CD45-FITC/CD31-FITC/CD140a-FITC lineage control, 6) CD61-PE/CD29-biotin-streptavidin-APC/7AAD experimental sample, 7) CD144-PE/CD29-biotin-streptavidin-APC/7AAD experimental sample, 8) CD61-PE/CD24- biotin-streptavidin-APC/7AAD experimental sample, 9) CD49f-PE/CD24-biotin-streptavidin-APC/7AAD experimental sample, and 10) CD144-PE/CD24-biotin-streptavidin-APC/7AAD experimental sample. Antibodies were obtained from (BD Pharmingen, San Jose, CA).
Primary antibodies were added and incubated for 15 minutes on ice. To wash the cells, 1 mL of wash solution was added to each tube and centrifuged at 1,000g for 2 minutes. Cells were resuspended in 200 uL of wash solution, and if necessary, secondary antibody was added, and tubes were incubated for another 15 minutes on ice. The wash step was repeated, and cells were resuspended in 400 uL of wash solution, 7AAD was added, and cells were filtered through a 40-um filter.
Antibodies used included CD24-PE (1:1,000 dilution), CD24-biotin (1:100 dilution), streptavidin-APC (1:100 dilution), CD45-FITC (1:100 dilution), CD31-FITC (1:100 dilution), CD140a-FITC (1:100 dilution), CD61-PE (1:100 dilution), CD29-biotin (1:100 dilution), CD144-PE (1:100 dilution), CD49f-PE (1:50 dilution), and 7AAD (1:40 dilution).
Samples were run on a Becton Dickinson FACScan using BD CellQuest Pro version 5.2.1 and Cytek Rainbow version 1.2 software (Fremont, CA). Data were processed using Flowjo version 7.6.5 (Ashland, OR).
Histology
H&E staining was done on paraffin-embedded and sectioned tumor tissue according to standard hydrating, staining, and dehydrating protocols. Slides were imaged on an Olympus Bx50 microscope with a Canon EOS Rebel XSI camera (Tokyo, Japan) using EOS imaging software, and changes in contrast and brightness were performed in Photoshop CS4 (Adobe Systems, San Jose, CA). Immunofluorescence was also done on paraffin-embedded tumor sections. For antigen retrieval, slides were boiled in 10 mM sodium citrate for 20 minutes on high power in a microwave. Sections were blocked for 1 hour at 26°C using 5% BSA and 0.1% Tween in PBS. Primary antibodies keratin 14 (1:500, PRB-155P-100, Covance) and keratin 19 (1:200, Troma-3, Developmental Studies Hybridoma Bank from The University of Iowa) were incubated overnight at 4°C, followed by washing with blocking buffer and incubation for 1 hour with Alexa Fluor 594 Chicken Anti-Rat IgG and Alexa Fluor 633 Goat Anti-Rabbit IgG (Invitrogen) secondary antibody. Images were taken on an Olympus 1X81 microscope using a Hamamatsu Photonics ORCA-ER camera. Images were taken and processed using Slidebook 64 version 5.0.0.24.
Immunohistochemistry was performed on paraffin-embedded sections using the Vectastain ABC staining kit (Vector Labs, Burlingame, CA). Slides were deparaffinized, and antigen retrieval was done with 10 mM sodium citrate, as in immunofluorescence. Slides were blocked with peroxidase-blocking solution (90% MeOH + 3% hydrogen peroxide) for 5 minutes and then blocked in 5% serum for 1 hour at 26°C. Sections were incubated with primary antibody progesterone receptor (1:50, A0098, Dako, Glostrup, Denmark), ER (1:300, Santa Cruz Biotechnology, Santa Cruz, CA), K6 (1:300, PRB-169P, Covance), K10 (1:50, MMS-159S, Covance), β-casein (1:200, C 2206, Sigma), or Muc1 (1:200, ab15481, Abcam, Cambridge, UK) overnight at 4°C and washed in PBS-T (PBS + 0.1% Tween-20). Sections were then incubated for 30 minutes with biotin- sp–conjugated secondary antibody (1:1,000, Jackson ImmunoResearch, West Grove, PA). Slides were again washed in PBS-T. Next, slides were incubated with Vectastain ABC reagent (supplied in kit) for 30 minutes, washed in PBS, and incubated with DAB reagent (Vector Labs) for 5 minutes. Slides were then rinsed, dehydrated, and mounted. Slides were imaged on an Olympus Bx50 microscope with a Canon EOS Rebel XSI camera using EOS imaging software, and changes in contrast and brightness were done in Photoshop CS4 (Adobe Systems).
Oil Red O staining was carried out on OCT-embedded sections. Sections were fixed in 4% paraformaldehyde for 15 minutes and then rinsed in running water for 10 minutes. Sections were then rinsed with 60% isopropanol and incubated with Oil Red O working solution (30 mL stock solution of 0.5 g Oil Red O in 100 mL isopropanol diluted with 20 mL distilled water and filtered) for 15 minutes. After incubation, sections were washed with isopropanol, counterstained with hematoxylin, rinsed in distilled water, and mounted in permount. Slides were imaged on an Olympus Bx50 microscope with a Canon EOS Rebel XSI camera using EOS imaging software.
Microarray processing
Tumors were selected from both single insertion site and multiple insertion site tumors. Total RNA was isolated using the Qiagen RNeasy kit (Hilden, Germany) and was assayed for quality on the Bioanalyzer RNA 6000 nano chip (Agilent, Santa Clara, CA). Samples were labeled with the Agilent One-Color Quick Amp Labeling Kit. Labeled samples were hybridized to the Agilent Whole Mouse Gene Expression Array (microarray design ID 014868, GEO platform ID GPL7202) using the standard Agilent one-color gene expression hybridization and wash protocols. Microarray slides were scanned on an Agilent G2505B scanner at 5-µm resolution. The slide images were processed using Agilent Feature Extraction software version 9.5.1.1.
Microarray data filtering and normalization
The mean signal intensity from the arrays was filtered to remove control features and features flagged as nonuniform or feature population outliers. Remaining features for any probe were averaged to yield a single value for each unique probe sequence. These data were merged with the mean Cy5 signal from arrays described by Herschkowitz et al.13 The 14,509 microarray probes shared by the Welm laboratory arrays and Perou laboratory arrays were identified, and data from these probes were selected from both sets of arrays. As the Perou laboratory arrays were 2-color hybridizations, only the Cy5 mean signal was used. The combined data set was quantile normalized and then adjusted for batch effects using ComBat analysis from the Broad Institute’s Gene Pattern web site (Cambridge, MA).55 During batch adjustment, there were 6 total batches: the Perou laboratory Agilent 22K published array data (Gene Expression Omnibus series GSE3165) were treated as a single batch, the lentiviral-PyMT samples run at the University of North Carolina (UNC) were treated as a second batch, and each run at the Huntsman Cancer Institute (HCI) was treated as a single batch, for a total of 4 HCI batches (Gene Expression Omnibus series GSE40001).
Microarray analysis
The normalized data set was loaded into GeneSifter software (Geospiza, Seattle, WA) for analysis. Differentially expressed genes were selected using a t test, and multiple test correction was performed with the Benjamini-Hochberg method. Genes found significant at an adjusted P value of ≤0.05 and showing at least 2-fold differential expression were selected.
Hierarchical clustering on normalized data was conducted using a gene pattern hierarchical clustering program. The settings were as follows: column distance measure and row distance measure of Pearson correlation, pairwise-centroid clustering method, and median centering for rows. To ensure normalization properly removed batch effects between the UNC and HCI microarray facilities, microarrays on technical replicates of 4 tumor RNA samples were run independently at UNC and HCI. Hierarchical clustering of microarray data from these technical replicates demonstrated that they clustered together following normalization and analysis.
Supplementary Material
Acknowledgments
The authors acknowledge Alana Welm for helpful suggestions.
Footnotes
Supplementary material for this article is available on the Genes & Cancer website at http://ganc.sagepub.com/supplemental.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received the following financial support for the research, authorship, and/or publication of this article: Funding from the National Institutes of Health (R01CA143815-01 and R01CA140296) supported this study.
References
- 1. Parker JS, Mullins M, Cheang MCU, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Elston C, Ellis I. Pathological prognostic factors in breast cancer: the value of histological grade in breast cancer. Experience from a large study with long-term follow-up. Histopathology. 1991;19(5):403-10 [DOI] [PubMed] [Google Scholar]
- 3. Vargo-Gogola T, Rosen JM. Modelling breast cancer: one size does not fit all. Nat Rev. 2007;7:659-72 [DOI] [PubMed] [Google Scholar]
- 4. Guy C, Cardiff R, Muller W. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12(3):954-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pittius CW, Hennighausen L, Lee E, et al. A milk protein gene promoter directs the expression of human tissue plasminogen activator cDNA to the mammary gland in transgenic mice. Proc Natl Acad Sci U S A. 1988;85(16):5874-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wagner KU, Ward T, Davis B, Wiseman R, Hennighausen L. Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Res. 2001;10(6):545-53 [DOI] [PubMed] [Google Scholar]
- 7. Raptis L, Marcellus R, Corbley MJ, et al. Cellular ras gene activity is required for full neoplastic transformation by polyomavirus. J Virol. 1991;65(10):5203-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Webster MA, Hutchinson JN, Rauh MJ, et al. Requirement for both Shc and phosphatidylinositol 3′ kinase signaling pathways in polyomavirus middle T-mediated mammary tumorigenesis. Mol Cell Biol. 1998;18(4):2344-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wood L, Parsons D, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108-13 [DOI] [PubMed] [Google Scholar]
- 10. Fromowitz FB, Viola MV, Chao S, et al. Ras p21 expression in the progression of breast cancer. Hum Pathol. 1987;18(12):1268-75 [DOI] [PubMed] [Google Scholar]
- 11. Verbeek BS, Vroom TM, Adriaansen-Slot SS, et al. c-Src protein expression is increased in human breast cancer: an immunohistochemical and biochemical analysis. J Pathol. 1999;180(4):383-8 [DOI] [PubMed] [Google Scholar]
- 12. Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Herschkowitz JI, Simin K, Weigman VJ, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8(5):R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Visvader JE. Cells of origin in cancer. Nature. 2011;469(7330): 314-22 [DOI] [PubMed] [Google Scholar]
- 15. Ramos CV, Taylor HB. Lipid-rich carcinoma of the breast: a clinicopathologic analysis of 13 examples. Cancer. 1974;33(3):812-9 [DOI] [PubMed] [Google Scholar]
- 16. DeOme K, Faulkin L, Bern HA, Blair PB. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland- free mammary fat pads of female C3H mice. Cancer Res. 1959;19(5): 515-20 [PubMed] [Google Scholar]
- 17. Welm BE, Dijkgraaf GJP, Bledau AS, Welm AL, Werb Z. Lentiviral transduction of mammary stem cells for analysis of gene function during development and cancer. Cell Stem Cell. 2008;2(1):90-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30(3):256-68 [DOI] [PubMed] [Google Scholar]
- 19. Rodriguez-Viciana P, Collins C, Moule M, Fried M. Chromosomal instability at a mutational hotspot in polyoma middle T-antigen affects its ability to activate the ARF-p53 tumor suppressor pathway. Oncogene. 2005;25(10):1454-62 [DOI] [PubMed] [Google Scholar]
- 20. Streisinger G, Owen JE. Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics. 1985;109(4):633-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bouras T, Pal B, Vaillant F, et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3(4):429-41 [DOI] [PubMed] [Google Scholar]
- 22. Nik-Zainal S, Van Loo P, Wedge DC, et al. The life history of 21 breast cancers. Cell. 2012;149(5):994-1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Woods NB, Muessig A, Schmidt M, et al. Lentiviral vector transduction of NOD/SCID repopulating cells results in multiple vector integrations per transduced cell: risk of insertional mutagenesis. Blood. 2003;101(4):1284-9 [DOI] [PubMed] [Google Scholar]
- 24. Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481(7381):306-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maglione JE, Moghanaki D, Young LJT, et al. Transgenic polyoma middle-T mice model premalignant mammary disease. Cancer Res. 2001;61(22):8298-305 [PubMed] [Google Scholar]
- 26. Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park SY, Lee HE, Li H, Shipitsin M, Gelman R, Polyak K. Heterogeneity for stem cell–related markers according to tumor subtype and histologic stage in breast cancer. Clin Cancer Res. 2010;16(3):876-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stingl J, Eirew P, Ricketson I, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439(7079):993-7 [DOI] [PubMed] [Google Scholar]
- 29. Asselin-Labat ML, Sutherland KD, Barker H, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2006;9(2):201-9 [DOI] [PubMed] [Google Scholar]
- 30. Sleeman KE, Kendrick H, Robertson D, Isacke CM, Ashworth A, Smalley MJ. Dissociation of estrogen receptor expression and in vivo stem cell activity in the mammary gland. J Cell Biol. 2007;176(1):19-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vaillant F, Asselin-Labat ML, Shackleton M, Forrest NC, Lindeman GJ, Visvader JE. The mammary progenitor marker CD61/β3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res. 2008;68(19):7711-7 [DOI] [PubMed] [Google Scholar]
- 32. Abraham BK, Fritz P, McClellan M, Hauptvogel P, Athelogou M, Brauch H. Prevalence of CD44+/CD24-/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin Cancer Res. 2005;11(3):1154-9 [PubMed] [Google Scholar]
- 33. Rosner A, Miyoshi K, Landesman-Bollag E, et al. Pathway pathology: histological differences between ErbB/Ras and Wnt pathway transgenic mammary tumors. Am J Pathol. 2002;161(3):1087-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Holland DW, Boucher LD, Mortimer JE. Tubular breast cancer experience at Washington University: a review of the literature. Clin Breast Cancer. 2001;2(3):210-4 [DOI] [PubMed] [Google Scholar]
- 35. Geyer FC, Lambros MBK, Natrajan R, et al. Genomic and immunohistochemical analysis of adenosquamous carcinoma of the breast. Mod Pathol. 2010;23(7):951-60 [DOI] [PubMed] [Google Scholar]
- 36. Li Y, Welm B, Podsypanina K, et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci U S A. 2003;100(26):15853-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gendler S, Spicer A. Epithelial mucin genes. Annu Rev Physiol. 1995;57(1):607-34 [DOI] [PubMed] [Google Scholar]
- 38. Graham RA, Morris JR, Cohen EP, Taylor-Papadimitriou J. Up-regulation of MUC1 in mammary tumors generated in a double-transgenic mouse expressing human MUC1 cDNA, under the control of 1.4-kb 5′ MUC1 promoter sequence and the middle T oncogene, expressed from the MMTV promoter. Int J Cancer. 2001;92(3):382-7 [DOI] [PubMed] [Google Scholar]
- 39. Yerushalmi R, Hayes M, Gelmon K. Breast carcinoma—rare types: review of the literature. Ann Oncol. 2009;20(11):1763-70 [DOI] [PubMed] [Google Scholar]
- 40. Shi P, Wang M, Zhang Q, Sun J. Lipid-rich carcinoma of the breast: a clinicopathological study of 49 cases. Tumori. 2008;94(3):342-6 [DOI] [PubMed] [Google Scholar]
- 41. Sonnenberg A, Daams H, Van der Valk M, Hilkens J, Hilgers J. Development of mouse mammary gland: identification of stages in differentiation of luminal and myoepithelial cells using monoclonal antibodies and polyvalent antiserum against keratin. J Histochem Cytochem. 1986;34(8):1037-46 [DOI] [PubMed] [Google Scholar]
- 42. Hennighausen L, Robinson GW. Information networks in the mammary gland. Nat Rev Mol Cell Biol. 2005;6(9):715-25 [DOI] [PubMed] [Google Scholar]
- 43. Tei M, Uchida K, Chambers J, et al. Mammary lipid-rich carcinoma with extensive amyloid deposition in a dog. J Vet Med Sci. 2012;74(6):809-11 [DOI] [PubMed] [Google Scholar]
- 44. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stephens PJ, Tarpey PS, Davies H, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Molyneux G, Geyer FC, Magnay F-A, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7(3):403-17 [DOI] [PubMed] [Google Scholar]
- 47. Lim E, Vaillant F, Wu D, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15(8):907-13 [DOI] [PubMed] [Google Scholar]
- 48. Liu S, Ginestier C, Charafe-Jauffret E, et al. BRCA1 regulates human mammary stem/progenitor cell fate. Proc Natl Acad Sci U S A. 2008;105(5):1680-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23(22):2563-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wrba F, Ellinger A, Reiner G, Spona J, Holzner JH. Ultrastructural and immunohistochemical characteristics of lipid-rich carcinoma of the breast. Virchows Arch A Pathol Anat Histopathol. 1988;413(5):381-5 [DOI] [PubMed] [Google Scholar]
- 51. Watson CJ, Kreuzaler PA. Remodeling mechanisms of the mammary gland during involution. Int J Dev Biol. 2011;55(7-9):757-62 [DOI] [PubMed] [Google Scholar]
- 52. Godbey W, Wu KK, Mikos AG. Poly (ethylenimine) and its role in gene delivery. J Control Release. 1999;60(2):149-60 [DOI] [PubMed] [Google Scholar]
- 53. Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300(5626):1749-51 [DOI] [PubMed] [Google Scholar]
- 54. Winslow MM, Dayton TL, Verhaak RGW, et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473(7345):101-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118-27 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.