Abstract
Multiple biotic and abiotic environmental stress factors affect negatively various aspects of plant growth, development, and crop productivity. Plants, as sessile organisms, have developed, in the course of their evolution, efficient strategies of response to avoid, tolerate, or adapt to different types of stress situations. The diverse stress factors that plants have to face often activate similar cell signaling pathways and cellular responses, such as the production of stress proteins, upregulation of the antioxidant machinery, and accumulation of compatible solutes. Over the last few decades advances in plant physiology, genetics, and molecular biology have greatly improved our understanding of plant responses to abiotic stress conditions. In this paper, recent progresses on systematic analyses of plant responses to stress including genomics, proteomics, metabolomics, and transgenic-based approaches are summarized.
1. Introduction
Plants and animals share some response mechanisms to unfavorable environmental conditions; however, plants, being sessile organisms, have developed, in the course of their evolution, highly sophisticated and efficient strategies of response to cope with and adapt to different types of abiotic and biotic stress imposed by the frequently adverse environment.
Stress can be understood as a stimulus or influence that is outside the normal range of homeostatic control in a given organism: if a stress tolerance is exceeded, mechanisms are activated at molecular, biochemical, physiological, and morphological levels; once stress is controlled, a new physiological state is established, and homeostasis is reestablished. When the stress is retired, the plant may return to the original state or to a new physiological situation [1].
In the last years, and because of the great interest for both basic and applied research, there has been an important progress in the understanding of the mechanisms and processes underlying abiotic stress adaptation and defense in different plant species [1, 2]. The sensing of biotic or abiotic stress conditions induces signaling cascades that activate ion channels, kinase cascades, production of reactive oxygen species (ROS), accumulation of hormones such as salicylic acid, ethylene, jasmonic acid, and abscisic acid. These signals ultimately induce expression of specific subsets of defense genes that lead to the assembly of the overall defense reaction [3].
The emergence of the novel “omics” technologies, such as genomics, proteomics, and metabolomics, is now allowing researchers to identify the genetic behind plant stress responses (Figure 1). These omics technologies enable a direct and unbiased monitoring of the factors affecting plant growth and development and provide the data that can be directly used to investigate the complex interplay between the plant, its metabolism, and also the stress caused by the environment or the biological threats (insects, fungi, or other pathogens). Plant responses to stress are mediated via profound changes in gene expression which result in changes in composition of plant transcriptome, proteome, and metabolome [4].
Figure 1.
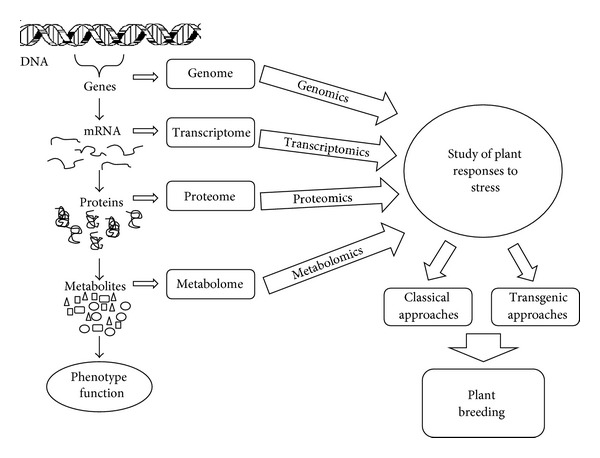
Plant response to abiotic stress factors. Genomics, transcriptomics, proteomics, and metabolomics have enabled active analyses of regulatory networks that control abiotic stress responses. Such analyses increase our knowledge on plant responses and adaptation to stress conditions and allow improving plant breeding.
In this work, the main biotechnological approaches to study plant responses to stress are reviewed.
2. Genomics
A gene by gene approach has been typically used to understand its function. In Table 1, some of the genes involved in plant responses to stress are listed. Functional genomics allows large-scale gene function analysis with high throughput technology and incorporates interaction of gene products at cellular and organism level. The information coming from sequencing programs is providing enormous input about genes to be analyzed. The availability of many plant genomes nowadays (reviewed in [5, 6]) facilitates studying the function of genes on a genomewide scale. The lack of information from other plant genomes will also be compensated in part by the availability of large collection of expressed sequence tags (ESTs) and cDNA sequences [7]. The basic interest behind these EST projects is to identify genes responsible for critical functions. ESTs, cDNA libraries, microarray, and serial analysis of gene expression (SAGE) are used to analyze global gene expression profiles in a functional genomics program. Large mutant collections are tools that complement large-scale expression studies. Gene identification through physical and chemical mutagens has become amenable for large-scale analysis with the availability of markers [8], but gene tagging is more promising for functional analysis on a wider scale. Moreover, the understanding of the complexity of stress signaling and plant adaptive processes would require the analysis of the function of numerous genes involved in stress response.
Table 1.
Genes involved in plant responses to stress.
| Stress | Reference | |
|---|---|---|
| 14.3.3 gene family (GF14b, GF14c) | Salinity, drought, fungal | [9] |
| MAPK | Abiotic and biotic stresses | [10] |
| MEKK1 and ANP1 | Environmental stress | [11, 12] |
| MPK3, MPK4 and MPK6 | Abiotic stress (pathogens) and oxidative stress | [10, 13] |
| CBF/DREB families (CBF1, CBF2, DREB2A) | Drought, cold, salinity | [47, 82] |
| HVA1 | Salinity and drought | [83] |
| Glycerol-3-phosphate acyltransferase gene | Cold | [84] |
| ICS | Pathogens, UV light | [85] |
| LOX | Wounding, drought, and pathogens | [86, 87] |
| bZIPs family (e.g., ABF1, ABF2) | Drought, temperature, salinity | [24–33] |
| WRKY family (AtWRKY2, AtWRKY6, AtWRKY18) | Pathogens, wounding, salinity, temperature, drought, oxidative stress | [37] |
| ATAF | Wounding, drought, salinity, cold, pathogens |
[88] |
Numerous investigations show that plant defense response genes are transcriptionally activated by pathogens and also by different types of abiotic stress. It has been described that the induction of specific defense genes, in the response against certain pathogens, is dependent on specific environmental conditions, suggesting the existence of a complex signaling network that allows the plant to recognize and protect itself against pathogens and environmental stress [3]. Similar induction patterns of members of the 14.3.3 gene family (GF14b and GF14c) by abiotic and biotic stresses such as salinity, drought, ABA, and fungal inoculation have been documented in rice [9]. The rice GF14 genes contain cis-elements in their promoter regions that are responsive to abiotic stress and pathogen attack. The 14-3-3s family genes are also subject to the regulation by certain transcript factors [9].
On the other hand, kinase cascades of the mitogen-activated protein kinase (MAPK) class play a remarkably important role in plant signaling of a variety of abiotic and biotic stresses, and it is an essential step in the establishment of resistance to pathogens [10]. It has been described that in Arabidopsis MEKK1 and ANP1 act in the environmental stress response [11, 12], and MPK3, MPK4, and MPK6, are activated by a diversity of stimuli including abiotic stresses, pathogens, and oxidative stress [13].
Elucidating the molecular mechanism that mediates the complex stress responses in plants system is an important step to develop improved variety of stress tolerant crops. Many crop traits are quantitative, complex, and controlled by multiple interacting genes. Recent progress in molecular biology provides the tools to study the genetical make-up of plants, which allows us to unravel the inheritance of all traits whether they are controlled by single genes or many genes acting together, known as the quantitative trait loci (QTL). The molecular marker technologies available since the 1980s allows dissecting the variation in traits. With the progress of QTL mapping, new breeding approaches such as marker-assisted selection and breeding by design have emerged [14].
Advances in plant genomics research have opened up new perspectives and opportunities for improving crop plants and their productivity. The genomics technologies have been found useful in deciphering the multigenicity of biotic and abiotic plant stress responses through genome sequences, stress-specific cell and tissue transcript collections, protein and metabolite profiles and their dynamic changes, protein interactions, and mutant screens.
3. Proteomics
The adaptation of plants to biotic or abiotic stress conditions is mediated through deep changes in gene expression which result in changes in composition of plant transcriptome, proteome, and metabolome. Since proteins are directly involved in plant stress response, proteomics studies can significantly contribute to elucidate the possible relationships between protein abundance and plant stress acclimation. Several studies [15] have already proven that the changes in gene expression at transcript level do not often correspond with the changes at protein level. The investigation of changes in plant proteome is highly important since proteins, unlike transcripts, are direct effectors of plant stress response. Proteins not only include enzymes catalyzing changes in metabolite levels, but also include components of transcription and translation machinery (Table 2).
Table 2.
Proteins and enzymes involved in plant responses to stress.
| Stress | Reference | |
|---|---|---|
| ERF family | Cold, drought, pathogen infection, wounding, ET, SA, and JA | [19] |
| bZIPs family (e.g., ABF1, ABF2) | Drought, temperature, salt | [24–33] |
| WRKY | Pathogens, wounding, salinity, temperature, drought, oxidative stress | [36–39] |
| MYB family (AtMYB15, AtMYB30, AtMYB33 AtMYB60, AtMYB96, AtMYB101 AtMYB15, and AtMYB108) | Biotic and abiotic stress (pathogens, drought, cold) | [40–46] |
| ABF | Drought | [79] |
| NAC | Drought, salinity, cold | [22, 23] |
| MYC | Environmental stresses | [47] |
| LEA family (PMA 80, PMA 1959) | Salinity and drought | [71] |
| Heat shock proteins | Temperatures | [74, 75] |
| LOX family (e.g., LOX1) | Wounding, drought, and pathogens | [86, 87] |
| Glutathione peroxidase, superoxide dismutase, ascorbate peroxidases, and glutathione reductases | Oxidative stress | [68, 70] |
In the last years, there has been an important progress in the knowledge of several families of plant transcription factors linked to plant stress responses, such as responses to ultraviolet light, wounding, anaerobic stress, and pathogens [16]. The most important ones are as follows.
The ethylene-responsive-element-binding factors (ERFs). This protein family has been linked to a wide range of stresses; the RNA levels of specific ERF genes are regulated by cold, drought, pathogen infection, wounding or treatment with ethylene, SA or JA [17]. ERF proteins are shown to function as either activators or repressors of transcription, which is of great relevance in all processes related to plant development and its responses to adverse growing conditions due to both biotic and abiotic factors [18]. It has been reported that ERF proteins from one plant species function in other plant species, enhancing their potential utility in increasing the stress tolerance of plants [19, 20]. However, constitutive overexpression of ERF genes generally causes deleterious effects. To overcome this problem, the use of stress-inducible promoters to control the expression of the ERF genes has been successfully used (reviewed in [21]).
NAC proteins are plant-specific transcription factors having a variety of important functions not only in plant development but also in abiotic stress tolerance [22]. NAC domain-containing proteins represent one of the largest TF families, firstly identified in model plants as Arabidopsis and rice but also recently characterized in woody fruit species [23].
Another important family of transcription factors is the called “basic-domain leucine-zipper (bZIP)” which are regulators of important plant processes such as organ and tissue differentiation [24], cell elongation [25], nitrogen/carbon balance control [26], pathogen defense [27], energy metabolism [28], unfolded protein response [29], hormone and sugar signaling [30], light response [31], osmotic control [32], and seed storage protein gene regulation [33]. One class of bZIP proteins that is linked to stress responses comprises the TGA/octopine synthase (ocs)-element-binding factor (OBF) proteins. These bind to the activation sequence-1 (as-1)/ocs element, which regulates the expression of some stress-responsive genes [34]. A major advance was the discovery that TGA/OBF family members interact with nonexpressor of PR1 (NPR1), a key component in the SA defense signaling pathway [35].
WRKY proteins are a family of transcription factors that are unique to plants specific WRKY family members show enhanced expression and/or DNA-binding activity following induction by a range of pathogens, defense signals, and wounding (reviewed in [36]). Significant progress has been made in the past years in identifying target genes for WRKY factors. WRKY proteins bind to the W box, which is found in the promoters of many plant defense genes [37]. WRKY proteins also regulate the expression of regulatory genes such as receptor protein kinases [38]. Positive and negative regulation of WRKY promoters by specific WRKY proteins has been observed, and the promoters of many of the pathogen- and/or SA-regulated AtWRK genes are rich in W boxes [39].
MYB proteins are key factors in regulatory networks controlling development [40], metabolism [41], and responses to biotic and abiotic stresses [42]. Since the Arabidopsis genome sequence was published, some years ago, an important amount of data has accumulated on the roles of MYB transcription factors in plants and some members of this family are involved in these responses. Therefore, AtMYB30 encodes an activator of the hypersensitive cell death program in response to pathogen attack [43]; AtMYB96 acts through the ABA signaling cascade to regulate water stress and disease resistance [44]. AtMYB33 and AtMYB101 are involved in ABA-mediated responses to environmental signals. AtMYB15 is also involved in cold stress tolerance [45]. AtMYB108 in both biotic and abiotic stress responses [46]. The elucidation of MYB protein function and regulation that is possible in Arabidopsis will allow predicting the contributions of MYB proteins to the responses to biotic and abiotic stress conditions in other plant species.
MYC proteins are involved in the response of plants to unfavorable environmental conditions. This transcription factor family plays a role in the induction of apoptosis, important in the hypersensitive cell death program in response to pathogen attack. Another putative MYC target is the ornithine decarboxylase gene, involved in polyamines synthesis. On the other hand, MYC proteins activate the major ABA-dependent stress response (reviewed in [47]).
Therefore, studies of plant reaction upon stress conditions at protein level can significantly contribute to our understanding of physiological mechanisms underlying plant stress tolerance. Proteomics studies could thus lead to identification of potential protein markers whose changes in abundance can be associated with quantitative changes in some physiological parameters related to stress tolerance (reviewed in [16]).
4. Metabolomics
The possibility of monitoring a complete set of metabolites could largely improve the understanding of many physiological plant processes. This systematic study, defined as “metabolomics,” is intended to provide an integrated view of the functional status of an organism. Besides its use as a breeding or selection tool, metabolomics techniques have also been used to evaluate stress responses in barley [48], Citrus [49], Medicago truncatula [50], and Arabidopsis thaliana [51]. Some of the metabolites that have been involved in the plant responses to stress are listed in Table 3.
Table 3.
Metabolites and hormones involved in plant responses to stress.
| Stress | Reference | |
|---|---|---|
| Abscisic acid, jasmonic acid, salicylic acid, polyamines, and others | Drought, salinity, cold | [3, 58, 65–67] |
| Proline, glycine-betaine, and other compatible osmolytes | Environmental stresses: drought, salinity, osmotic | [62–64] |
| Phytoalexins | Microbial pathogens | [89] |
| Terpenes | Toxins and pathogens | [89] |
| Phenolic compounds (coumarin, lignin, flavonoids, tannins, isoflavonoids) | Pathogens, oxidative stress, UV light | [89, 90] |
| Alkaloids | Pathogens (predators) | [87] |
| Unsaturated fatty acids | Environmental stresses | [73] |
| ROS, malondialdehyde | Biotic and abiotic stresses | [3, 58, 68] |
| Phytochelatins and metallothioneins | Heavy metal intoxication | [76, 77] |
This technological tool, recently developed, includes different approaches, namely, targeted analysis, metabolic fingerprinting, and metabolite profiling.
Targeted analysis is the most developed analytical approach in metabolomics [49]. It is used to measure the concentration of a limited number of known metabolites precisely, by using either gas chromatography (GC) or liquid chromatography (LC) coupled to mass spectrometry (MS) or nuclear magnetic resonance spectroscopy (NMR).
Other approaches using high throughput metabolite analysis focus on a subset of useful information while avoiding the difficulties of comprehensive metabolite characterization; metabolic fingerprinting uses signals from hundreds to thousands of metabolites for rapid sample classification via statistical analysis [52]. In the last years, metabolite profiling attempts to identify and quantify a specific class or classes of chemically related metabolites that often share chemical properties that facilitate simultaneous analysis (reviewed in [53]).
The metabolome represents the downstream result of gene expression and is closer to phenotype than transcript expression or proteins. Extensive knowledge on metabolic flows could allow assessment of genotypic or phenotypic differences between plant species or among genotypes exhibiting different tolerance to some biotic or abiotic stresses. In addition, target metabolites have been analyzed as nutritional and/or agronomical biomarkers to classify different crop cultivars or to optimize growth conditions [54].
In contrast to high throughput methodology for the analysis of DNA, RNA, and proteins, current strategies for metabolite characterization still face significant obstacles. These challenges are largely caused by the high degree of chemical diversity among metabolite pools as well as the complexity of spatial and temporal distribution within living tissues. Plant metabolomics methodology and instrumentation are being developed at a rapid pace to address these analytical challenges [55].
Like other functional genomics research, metabolomics generates large amounts of data. Handling, processing, and analyzing this data is a clear challenge for researchers and requires specialized mathematical, statistical, and bioinformatic tools [56]. Further developments in this area require improvements in both analytical science and bioinformatics. Development of new analytical techniques is largely focused on increasing resolution and comprehensiveness, increasing speed and throughput of analytical assays and equipment miniaturization.
5. Transgenic Approaches: From the Study of Stress Tolerance Mechanisms in Plants to Crop Genetic Improvement
Use of modern molecular biology tools for elucidating the control mechanisms of stress tolerance and for engineering stress tolerant plants is based on the expression of specific stress-related genes. To date, successes in genetic improvement of environmental stress resistance have involved manipulation of a single or a few genes involved in signaling/regulatory pathways or that encode enzymes involved in these pathways [57].
The plant hormone abscisic acid (ABA) regulates the adaptive response of plants to environmental stresses such as drought, salinity, and chilling via diverse physiological and developmental processes [58]. The ABA biosynthetic pathway has been deeply studied, and many of the key enzymes involved in ABA synthesis have been used in transgenic plants to improve abiotic stress tolerance [59]. Transgenic plants overexpressing the genes involved in ABA synthesis showed increased tolerance to drought and salinity stress [59]. Similarly, many studies have illustrated the potential of manipulating CBF/DREB genes to confer improved drought tolerance [60].
Another mechanism involved in plant protection to osmotic stress associated to many abiotic stresses such as drought and salinity implies the accumulation of compatible solutes involved in avoiding oxidative damage and chaperoning through direct stabilization of membranes and/or proteins [61]. Many genes involved in the synthesis of these osmoprotectants have been explored for their potential in engineering plant abiotic stress tolerance [61]. The amino acid proline is known to occur widely in higher plants and normally accumulates in large quantities in response to environmental stresses [62]. The osmoprotectant role of proline has been verified in some crops by overexpressing genes involved in proline synthesis [63]. The results of transgenic modifications of biosynthetic and metabolic pathways in most of the previously mentioned cases indicate that higher stress tolerance and the accumulation of compatible solutes may also protect plants against damage by scavenging of reactive oxygen species (ROS) and by their chaperone-like activities in maintaining protein structures and functions [64].
Polyamines, being polycationic compounds of low molecular weight, are involved in many cellular processes, such as replication, transcription, translation, membrane stabilization, enzyme activity modulation, plant growth, and development [65]. It has been reported that stress results in an accumulation of free or conjugated polyamines, indicating that polyamine biosynthesis might serve as an integral component of plant response to stress [66, 67].
Polyamines metabolic pathways are regulated by a limited number of key enzymes, among them ornithine decarboxylase (ODC) and arginine decarboxylase (ADC). Transgenic plants overexpressing ADC gene showed increase in biomass and better performance under salt stress conditions. It has also been described that genetic transformation with genes encoding ADC improved environmental stress tolerance in various plant species [66].
A common factor among most stresses is the active production of reactive oxygen species [2]. ROS are not only toxic to cells but also play an important role as signaling molecules. Under normal growth conditions, there is equilibrium between the production and the scavenging of ROS, but abiotic stress factors may disturb this equilibrium, leading to a sudden increase in intracellular levels of ROS.
In order to control the level of ROS and protect the cells from oxidative injury, plants have developed a complex antioxidant defense system to scavenge them [68]. These antioxidant systems include various enzymes and nonenzymatic metabolites that may also play a significant role in ROS signaling in plants. A number of transgenic improvements for abiotic stress tolerance have been achieved through detoxification strategy [69]. These include transgenic plants over expressing enzymes involved in oxidative protection, such as glutathione peroxidase, superoxide dismutase, ascorbate peroxidases, and glutathione reductases [70].
LEA proteins, including several groups of high molecular weight, accumulate in response to different environmental stresses. It has been reported that constitutive overexpression of the HVA1, a group 3 LEA protein from barley, conferred tolerance to soil water deficit and salt stress in transgenic rice plants [71]. It has also been reported that plants expressing a wheat LEA group 2 protein (PMA80) gene or the wheat LEA group 1protein (PMA1959) gene resulted in increased tolerance to dehydration and salt stresses [69].
An important strategy for achieving greater tolerance to abiotic stress is to help plants to reestablish homeostasis under stressful environments, restoring both ionic and osmotic homeostasis. This is a major approach to improve salt tolerance in plants through genetic engineering, where the target is to achieve Na+ excretion out of the root, or their storage in the vacuole [72].
Transgenic approaches also aim to improve photosynthesis under abiotic stress conditions through changes in the lipid biochemistry of the membranes. Genetically engineered plants overexpressing chloroplast glycerol-3-phosphate acyltransferase gene (involved in phosphatidyl glycerol fatty acid desaturation) showed an increase in the number of unsaturated fatty acids and a corresponding decrease in the chilling sensitivity [73].
The heat shock response is a highly conserved biological response, occurring in all organisms in response to heat or other toxic agent exposures [74]. Genetic engineering for increased thermotolerance by enhancing heat shock protein synthesis in plants has been achieved in a number of plant species. Some authors have reported the positive correlation between the levels of heat shock proteins and stress tolerance (reviewed in [75]).
A special case of study is the heavy metal contamination. In spite of the natural occurrence of heavy metals as rare elements, diverse anthropogenic practices have contributed to spread them in the environment. Plants have developed mechanisms that can protect cells from heavy metal cytotoxicity, as the cytosolic detoxification by binding to the metal-binding molecules as phytochelatins, and metallothioneins which play an important role in heavy metal detoxification and homeostasis of intracellular metal ions in plant tissues. Overexpression of phytochelatin synthase in Arabidopsis leads to enhanced arsenic tolerance but surprisingly to cadmium hypersensitivity [76]. Therefore, new approaches could contribute to uncovering the complexity of plant tolerance to heavy metal stress [77].
The transcription factors activate cascades of genes that act together in enhancing tolerance towards multiple stresses as indicated before. On the other hand, some stress responsive genes may share the same transcription factors, as indicated by the significant overlap of the gene expression profiles that are induced in response to different stresses [37]. Transcriptional activation of stress-induced genes has been possible in transgenic plants over expressing one or more transcription factors that recognize promoter regulatory elements of these genes [75, 78]. Two families, bZIP and MYB, are involved in ABA signaling and its gene activation. Introduction of transcription factors in the ABA signaling pathway can also be a mechanism of genetic improvement of plant stress tolerance. Constitutive expression of ABF3 or ABF4 demonstrated enhanced drought tolerance in Arabidopsis, with altered expression of ABA/stress-responsive genes, for example, rd29B, rab18, ABI1, and ABI2 [79].
It is important to point that genetic modification of higher plants by introducing DNA into their cells is a highly complex process. Practically any plant transformation experiment relies at some point on cell and tissue culture. Although the development transformation methods that avoid plant tissue culture have been described for Arabidopsis and have been extended to a few crops, the ability to regenerate plants from isolated cells or tissues in vitro is needed for most plant transformation systems. Not all plant tissue is suited to every plant transformation method, and not all plant species can be regenerated by every method [80]. There is, therefore, a need to find both a suitable plant tissue culture/regeneration regime and a compatible plant transformation methodology [81].
6. Conclusions
To understand how plants respond to stress, it must be considered that they are subjected to a combination of adverse conditions. This preliminary consideration is essential to understand the performance of plants under stress and also to identify strategies to improve stress tolerance.
The integration of the omics approaches is likely to enable researchers to reconstruct the whole cascade of cellular events leading to rapid responses and adaptation to the various abiotic stimuli. A well-focused approach combining molecular, physiological, and metabolic aspects of plant stress tolerance is required to increase knowledge on the effects of gene expression and to understand whole plant phenotype under stress. A better understanding of the underlying physiological processes in response to different abiotic stresses can drive the selection of the appropriate promoter or transcription factor to be used for transformation.
In addition, the use of genetic and genomic analysis to identify DNA molecular markers associated to stress resistance can facilitate breeding strategies for crop improvement. This approach is particularly useful when target characters are controlled by several genes, as in the case of abiotic stress tolerance. These omics approaches could be combined with the potential to map different QTLs contributing to a given agronomical trait and to identify linked molecular markers. This will open the possibility to transfer simultaneously several QTLs and to pyramid QTLs for several agronomical traits in one improved cultivar.
Acknowledgments
This work was supported by the Spanish Ministerio de Economía y Competitividad (MINECO) and Universitat Jaume I/Fundació Bancaixa through Grants no. AGL2010-22195-C03-01/AGR and P11B2009-01, respectively.
References
- 1.Fraire-Velázquez S, Rodríguez-Guerra R, Sánchez-Calderón L. Abiotic and biotic stress response crosstalk in plants. In: Shanker A, editor. Abiotic Stress Response in Plants—Physiological, Biochemical and Genetic Perspectives. Rijeka, Croatia: InTech; 2011. pp. 3–26. [Google Scholar]
- 2.Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant Journal. 2010;61(6):1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x. [DOI] [PubMed] [Google Scholar]
- 3.Jaspers P, Kangasjärvi J. Reactive oxygen species in abiotic stress signaling. Physiologia Plantarum. 2010;138(4):405–413. doi: 10.1111/j.1399-3054.2009.01321.x. [DOI] [PubMed] [Google Scholar]
- 4.Pérez-Alfocea F, Ghanem ME, Gómez-Cadenas A, Dodd I. Omics of root-to-shoot signaling under salt stress and water deficit. OMICs A Journal of Integrative Biology. 2011;15(12):893–901. doi: 10.1089/omi.2011.0092. [DOI] [PubMed] [Google Scholar]
- 5.Feuillet C, Leach JE, Rogers J, Schnable PS, Eversole K. Crop genome sequencing: lessons and rationales. Trends in Plant Science. 2010;16(2):77–88. doi: 10.1016/j.tplants.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Chain PSG, Grafham DV, Fulton RS, et al. Genome project standards in a new era of sequencing. Science. 2009;326(5950):236–237. doi: 10.1126/science.1180614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marques MC, Alonso-Cantabrana H, Forment J, et al. A new set of ESTs and cDNA clones from full-length and normalized libraries for gene discovery and functional characterization in citrus. BMC Genomics. 2009;10(article 428) doi: 10.1186/1471-2164-10-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lukowitz W, Gillmor CS, Scheible WR. Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiology. 2000;123(3):795–805. doi: 10.1104/pp.123.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen F, Li Q, Sun L, He Z. The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Research. 2006;13(2):53–63. doi: 10.1093/dnares/dsl001. [DOI] [PubMed] [Google Scholar]
- 10.Pitzschke A, Schikora A, Hirt H. MAPK cascade signalling networks in plant defence. Current Opinion in Plant Biology. 2009;12(4):421–426. doi: 10.1016/j.pbi.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Nakagami H, Soukupová H, Schikora A, Žárský V, Hirt H. A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis . Journal of Biological Chemistry. 2006;281(50):38697–38704. doi: 10.1074/jbc.M605293200. [DOI] [PubMed] [Google Scholar]
- 12.Suarez-Rodriguez MC, Adams-Phillips L, Liu Y, et al. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiology. 2007;143(2):661–669. doi: 10.1104/pp.106.091389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu JL, Zhou L, Yun BW, et al. Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiology. 2008;148(1):212–222. doi: 10.1104/pp.108.120006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peleman JD, van der Voort JR. Breeding by design. Trends in Plant Science. 2003;8(7):330–334. doi: 10.1016/S1360-1385(03)00134-1. [DOI] [PubMed] [Google Scholar]
- 15.Bogeat-Triboulot MB, Brosché M, Renaut J, et al. Gradual soil water depletion results in reversible changes of gene expression, protein profiles, ecophysiology, and growth performance in Populus euphratica, a poplar growing in arid regions. Plant Physiology. 2007;143(2):876–892. doi: 10.1104/pp.106.088708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosová K, Vítámvás P, Prášil IT, Renaut J. Plant proteome changes under abiotic stress—contribution of proteomics studies to understanding plant stress response. Journal of Proteomics. 2011;74(8):1301–1322. doi: 10.1016/j.jprot.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Oñate-Sánchez L, Singh KB. Identification of Arabidopsis ethylene-responsive element binding factors with distinct induction kinetics after pathogen infection. Plant Physiology. 2002;128(4):1313–1322. doi: 10.1104/pp.010862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell. 2000;12(3):393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiology. 2006;140(2):411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu L, Chen X, Ren H, et al. ERF protein JERF1 that transcriptionally modulates the expression of abscisic acid biosynthesis-related gene enhances the tolerance under salinity and cold in tobacco. Planta. 2007;226(4):815–825. doi: 10.1007/s00425-007-0528-9. [DOI] [PubMed] [Google Scholar]
- 21.Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta. 2012;1819(2):86–96. doi: 10.1016/j.bbagrm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 22.Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. NAC transcription factors in plant biotic and abiotic stress responses. Biochimica et Biophysica Acta. 2011;1819(2):97–103. doi: 10.1016/j.bbagrm.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 23.de Oliveira TM, Cidade LC, Gesteira AS, Coelho Filho MA, Soares Filho WS, Costa MGC. Analysis of the NAC transcription factor gene family in citrus reveals a novel member involved in multiple abiotic stress responses. Tree Genetics and Genomes. 2011;7(6):1123–1134. [Google Scholar]
- 24.Abe M, Kobayashi Y, Yamamoto S, et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309(5737):1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- 25.Corrêa LG, Riaño-Pachón DM, Schrago CG, dos Santos RV, Mueller-Roeber B, Vincentz M. The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS ONE. 2008;3(8, article e2944) doi: 10.1371/journal.pone.0002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weltmeier F, Ehlert A, Mayer CS, et al. Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO Journal. 2006;25(13):3133–3143. doi: 10.1038/sj.emboj.7601206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaminaka H, Näke C, Epple P, et al. bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO Journal. 2006;25(18):4400–4411. doi: 10.1038/sj.emboj.7601312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baena-González E, Rolland F, Thevelein JM, Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448(7156):938–942. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- 29.Liu JX, Srivastava R, Che P, Howell SH. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant Journal. 2007;51(5):897–909. doi: 10.1111/j.1365-313X.2007.03195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nieva C, Busk PK, Domínguez-Puigjaner E, et al. Isolation and functional characterisation of two new bZIP maize regulators of the ABA responsive gene rab28. Plant Molecular Biology. 2005;58(6):899–914. doi: 10.1007/s11103-005-8407-x. [DOI] [PubMed] [Google Scholar]
- 31.Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Molecular Cell. 2006;23(3):439–446. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 32.Satoh R, Fujita Y, Nakashima K, Shinozaki K, Yamaguchi-Shinozaki K. A novel subgroup of bZIP proteins functions as transcriptional activators in hypoosmolarity-responsive expression of the ProDH gene in Arabidopsis . Plant and Cell Physiology. 2004;45(3):309–317. doi: 10.1093/pcp/pch036. [DOI] [PubMed] [Google Scholar]
- 33.Alonso R, Oñate-Sánehez L, Weltmeier F, et al. A Pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell. 2009;21(6):1747–1761. doi: 10.1105/tpc.108.062968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fode B, Siemsen T, Thurow C, Weigel R, Gatz C. The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters. Plant Cell. 2008;20(11):3122–3135. doi: 10.1105/tpc.108.058974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Henanff G, Heitz T, Mestre P, Mutterer J, Walter B, Chong J. Characterization of Vitis vinifera NPR1 homologs involved in the regulation of pathogenesis-related gene expression. BMC Plant Biology. 2009;9(article 54) doi: 10.1186/1471-2229-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends in Plant Science. 2010;15(5):247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Qiu D, Xiao J, Xie W, Cheng H, Li X, Wang S. Exploring transcriptional signalling mediated by OsWRKY13, a potential regulator of multiple physiological processes in rice. BMC Plant Biology. 2009;9(article 74) doi: 10.1186/1471-2229-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan J, Zhang XC, Stacey G. Chitin signaling and plant disease resistance. Plant Signaling and Behavior. 2008;3(10):831–833. doi: 10.4161/psb.3.10.5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang SG, Park E, Do KS. Identification of a pathogen-Induced glycine max transcription factor GmWRKY1. Plant Pathology Journal. 2009;25(4):381–388. [Google Scholar]
- 40.Mandaokar A, Browse J. MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis . Plant Physiology. 2009;149(2):851–862. doi: 10.1104/pp.108.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lepiniec L, Debeaujon I, Routaboul JM, et al. Genetics and biochemistry of seed flavonoids. Annual Review of Plant Biology. 2006;57:405–430. doi: 10.1146/annurev.arplant.57.032905.105252. [DOI] [PubMed] [Google Scholar]
- 42.Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis . Trends in Plant Science. 2010;15(10):573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 43.Raffaele S, Vailleau F, Léger A, et al. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis . Plant Cell. 2008;20(3):752–767. doi: 10.1105/tpc.107.054858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seo PJ, Park CM. MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis . New Phytologist. 2010;186(2):471–483. doi: 10.1111/j.1469-8137.2010.03183.x. [DOI] [PubMed] [Google Scholar]
- 45.Agarwal M, Hao Y, Kapoor A, et al. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. Journal of Biological Chemistry. 2006;281(49):37636–37645. doi: 10.1074/jbc.M605895200. [DOI] [PubMed] [Google Scholar]
- 46.Zhang L, Zhao G, Jia J, Liu X. Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. Journal of Experimental Botany. 2011;63(1):203–214. doi: 10.1093/jxb/err264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agarwal PK, Jha B. Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biologia Plantarum. 2010;54(2):201–212. [Google Scholar]
- 48.Widodo, Patterson JH, Newbigin E, Tester M, Bacic A, Roessner U. Metabolic responses to salt stress of barley (Hordeum vulgare L.) cultivars, Sahara and Clipper, which differ in salinity tolerance. Journal of Experimental Botany. 2009;60(14):4089–4103. doi: 10.1093/jxb/erp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Djoukeng JD, Arbona V, Argamasilla R, Gomez-Cadenas A. Flavonoid profiling in leaves of citrus genotypes under different environmental situations. Journal of Agricultural and Food Chemistry. 2008;56(23):11087–11097. doi: 10.1021/jf802382y. [DOI] [PubMed] [Google Scholar]
- 50.Broeckling CD, Huhman DV, Farag MA, et al. Metabolic profiling of Medicago truncatula cell cultures reveals the effects of biotic and abiotic elicitors on metabolism. Journal of Experimental Botany. 2005;56(410):323–336. doi: 10.1093/jxb/eri058. [DOI] [PubMed] [Google Scholar]
- 51.Fukushima A, Kusano M, Redestig H, Arita M, Saito K. Metabolomic correlation-network modules in Arabidopsis based on a graph-clustering approach. BMC Systems Biology. 2011;5(article no. 1) doi: 10.1186/1752-0509-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chatterjee S, Srivastava S, Khalid A, et al. Comprehensive metabolic fingerprinting of Withania somnifera leaf and root extracts. Phytochemistry. 2010;71(10):1085–1094. doi: 10.1016/j.phytochem.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 53.Seger C, Sturm S. Analytical aspects of plant metabolite profiling platforms: current standings and future aims. Journal of Proteome Research. 2007;6(2):480–497. doi: 10.1021/pr0604716. [DOI] [PubMed] [Google Scholar]
- 54.Arbona V, Argamasilla R, Gómez-Cadenas A. Common and divergent physiological, hormonal and metabolic responses of Arabidopsis thaliana and Thellungiella halophila to water and salt stress. Journal of Plant Physiology. 2010;167(16):1342–1350. doi: 10.1016/j.jplph.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 55.Hegeman AD. Plant metabolomics-meeting the analytical challenges of comprehensive metabolite analysis. Briefings in Functional Genomics and Proteomics. 2010;9(2):139–148. doi: 10.1093/bfgp/elp053. [DOI] [PubMed] [Google Scholar]
- 56.Shulaev V. Metabolomics technology and bioinformatics. Briefings in Bioinformatics. 2006;7(2):128–139. doi: 10.1093/bib/bbl012. [DOI] [PubMed] [Google Scholar]
- 57.Jewell MC, Campbell BC, Godwin ID. Transgenic plants for abiotic stress resistance. In: Kole C, et al., editors. Transgenic Crop Plants. Heidelberg, Germany: Springer; 2010. [Google Scholar]
- 58.Arbona V, Gómez-Cadenas A. Hormonal modulation of citrus responses to flooding. Journal of Plant Growth Regulation. 2008;27(3):241–250. [Google Scholar]
- 59.Ji X, Dong B, Shiran B, et al. Control of abscisic acid catabolism and abscisic acid homeostasis is important for reproductive stage stress tolerance in cereals. Plant Physiology. 2011;156(2):550–563. doi: 10.1104/pp.111.176164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiong Y, Fei SZ. Functional and phylogenetic analysis of a DREB/CBF-like gene in perennial ryegrass (Lolium perenne L.) Planta. 2006;224(4):878–888. doi: 10.1007/s00425-006-0273-5. [DOI] [PubMed] [Google Scholar]
- 61.Zhang YY, Li Y, Gao T, et al. Arabidopsis SDIR1 enhances drought tolerance in crop plants. Bioscience, Biotechnology and Biochemistry. 2008;72(8):2251–2254. doi: 10.1271/bbb.80286. [DOI] [PubMed] [Google Scholar]
- 62.Ashraf M, Foolad MR. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environmental and Experimental Botany. 2007;59(2):206–216. [Google Scholar]
- 63.Hmida-Sayari A, Gargouri-Bouzid R, Bidani A, Jaoua L, Savouré A, Jaoua S. Overexpression of D1-pyrroline-5-carboxylate synthetase increases proline production and confers salt tolerance in transgenic potato plants. Plant Science. 2005;169(4):746–752. [Google Scholar]
- 64.Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K. Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Current Opinion in Biotechnology. 2006;17(2):113–122. doi: 10.1016/j.copbio.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 65.Liu JH, Moriguchi T. Changes in free polyamines and gene expression during peach flower development. Biologia Plantarum. 2007;51(3):530–532. [Google Scholar]
- 66.Wang J, Sun PP, Chen CL, Wang Y, Fu XZ, Liu JH. An arginine decarboxylase gene PtADC from Poncirus trifoliata confers abiotic stress tolerance and promotes primary root growth in Arabidopsis . Journal of Experimental Botany. 2011;62(8):2899–2914. doi: 10.1093/jxb/erq463. [DOI] [PubMed] [Google Scholar]
- 67.Liu JH, Nakajima I, Moriguchi T. Effects of salt and osmotic stresses on free polyamine content and expression of polyamine biosynthetic genes in Vitis vinifera . Biologia Plantarum. 2011;55(2):340–344. [Google Scholar]
- 68.Hossain Z, López-Climent MF, Arbona V, Pérez-Clemente RM, Gómez-Cadenas A. Modulation of the antioxidant system in citrus under waterlogging and subsequent drainage. Journal of Plant Physiology. 2009;166(13):1391–1404. doi: 10.1016/j.jplph.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 69.Amudha J, Balasubramani G. Recent molecular advances to combat abiotic stress tolerance in crop plants. Biotechnology and Molecular Biology Review. 2011;6(2):31–58. [Google Scholar]
- 70.Tang L, Kwon SY, Kim SH, et al. Enhanced tolerance of transgenic potato plants expressing both superoxide dismutase and ascorbate peroxidase in chloroplasts against oxidative stress and high temperature. Plant Cell Reports. 2006;25(12):1380–1386. doi: 10.1007/s00299-006-0199-1. [DOI] [PubMed] [Google Scholar]
- 71.Rohila JS, Jain RK, Wu R. Genetic improvement of Basmati rice for salt and drought tolerance by regulated expression of a barley Hva1 cDNA. Plant Science. 2002;163(3):525–532. [Google Scholar]
- 72.Wu YY, Chen QJ, Chen M, Chen J, Wang XC. Salt-tolerant transgenic perennial ryegrass (Lolium perenne L.) obtained by Agrobacterium tumefaciens-mediated transformation of the vacuolar Na+/H+ antiporter gene. Plant Science. 2005;169(1):65–73. [Google Scholar]
- 73.Sui N, Li M, Zhao SJ, Li F, Liang H, Meng QW. Overexpression of glycerol-3-phosphate acyltransferase gene improves chilling tolerance in tomato. Planta. 2007;226(5):1097–1108. doi: 10.1007/s00425-007-0554-7. [DOI] [PubMed] [Google Scholar]
- 74.Miller G, Mittler R. Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Annals of Botany. 2006;98(2):279–288. doi: 10.1093/aob/mcl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bhatnagar-Mathur P, Vadez V, Sharma KK. Transgenic approaches for abiotic stress tolerance in plants: retrospect and prospects. Plant Cell Reports. 2008;27(3):411–424. doi: 10.1007/s00299-007-0474-9. [DOI] [PubMed] [Google Scholar]
- 76.Li Y, Dhankher OP, Carreira L, et al. Overexpression of phytochelatin synthase in Arabidopsis leads to enhanced arsenic tolerance and cadmium hypersensitivity. Plant and Cell Physiology. 2004;45(12):1787–1797. doi: 10.1093/pcp/pch202. [DOI] [PubMed] [Google Scholar]
- 77.Chaffai R, Koyama H. Heavy metal tolerance in Arabidopsis thaliana . Advances in Botanical Research. 2011;6:1–49. [Google Scholar]
- 78.Munns R. Genes and salt tolerance: bringing them together. New Phytologist. 2005;167(3):645–663. doi: 10.1111/j.1469-8137.2005.01487.x. [DOI] [PubMed] [Google Scholar]
- 79.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 80.Tzfira T, Citovsky V. Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Current Opinion in Biotechnology. 2006;17(2):147–154. doi: 10.1016/j.copbio.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 81.Pérez-Clemente RM, Gómez-Cadenas A. In vitro tissue culture, a tool for the study and breeding of plants subjected to abiòtic stress conditions. Recent Advances in Plant In Vitro Culture. 2012:91–108. [Google Scholar]
- 82.Trujillo LE, Sotolongo M, Menéndez C, et al. SodERF3, a novel sugarcane ethylene responsive factor (ERF), enhances salt and drought tolerance when overexpressed in tobacco plants. Plant and Cell Physiology. 2008;49(4):512–525. doi: 10.1093/pcp/pcn025. [DOI] [PubMed] [Google Scholar]
- 83.Fu D, Huang B, Xiao Y, Muthukrishnan S, Liang GH. Overexpression of barley hva1 gene in creeping bentgrass for improving drought tolerance. Plant Cell Reports. 2007;26(4):467–477. doi: 10.1007/s00299-006-0258-7. [DOI] [PubMed] [Google Scholar]
- 84.Yan QC, Kuo MS, Li S, et al. AGPAT6 is a novel microsomal glycerol-3-phosphate acyltransferase. Journal of Biological Chemistry. 2008;283(15):10048–10057. doi: 10.1074/jbc.M708151200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Catinot J, Buchala A, Abou-Mansour E, Métraux JP. Salicylic acid production in response to biotic and abiotic stress depends on isochorismate in Nicotiana benthamiana . FEBS Letters. 2008;582(4):473–478. doi: 10.1016/j.febslet.2007.12.039. [DOI] [PubMed] [Google Scholar]
- 86.Yang XY, Jiang WJ, Yu HJ. The expression profiling of the lipoxygenase (LOX) family genes guring fruit development, abiotic stress and hormonal treatments in cucumber (Cucumis sativus L.) International Journal of Molecular Sciences. 2012;13:2481–2500. doi: 10.3390/ijms13022481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Andreou A, Feussner I. Lipoxygenases—structure and reaction mechanism. Phytochemistry. 2009;70(13-14):1504–1510. doi: 10.1016/j.phytochem.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 88.Christianson JA, Dennis ES, Llewellyn DJ, Wilson IW. ATAF NAC transcription factors: regulators of plant stress signaling. Plant Signaling and Behavior. 2010;5(4):428–432. doi: 10.4161/psb.5.4.10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mazid M, Khan TA, Mohammad F. Role of secondary metabolites in defense mechanisms of plants. Biology and Medicine. 2011;3(2):232–249. [Google Scholar]
- 90.Edreva A, Velikova V, Tsonev T, et al. Stress-protective role of secondary metabolites: diversity of functions and mechanisms. Genetics and Plant Physiology. 2008;34(1):67–78. [Google Scholar]


