Abstract
Seed development in Arabidopsis and in many dicots involves an early proliferation of the endosperm to form a large embryo sac or seed cavity close to the size of the mature seed, followed by a second phase during which the embryo grows and replaces the endosperm. SHORT HYPOCOTYL UNDER BLUE1 (SHB1) is a member of the SYG1 protein family in fungi, Caenorhabditis elegans, flies, and mammals. SHB1 gain-of-function enhances endosperm proliferation, increases seed size, and up-regulates the expression of the WRKY transcription factor gene MINISEED3 (MINI3) and the LRR receptor kinase gene HAIKU2 (IKU2). Mutations in either IKU2 or MINI3 retard endosperm proliferation and reduce seed size. However, the molecular mechanisms underlying the establishment of the seed cavity and hence the seed size remain largely unknown. Here, we show that the expression of MINI3 and IKU2 is repressed before fertilization and after 4 days after pollination (DAP), but is activated by SHB1 from 2 to 4 DAP prior to the formation of the seed cavity. SHB1 associates with their promoters but without a recognizable DNA binding motif, and this association is abolished in mini3 mutant. MINI3 binds to W-boxes in, and recruits SHB1 to, its own and IKU2 promoters. Interestingly, SHB1, but not MINI3, activates transcription of pMINI3::GUS or pIKU2::GUS. We reveal a critical developmental switch through the activation of MINI3 expression by SHB1. The recruitment of SHB1 by MINI3 to its own and IKU2 promoters represents a novel two-step amplification to counter the low expression level of IKU2, which is a trigger for endosperm proliferation and seed cavity enlargement.
Author Summary
Seed development in many dicots is characterized by a rapid proliferation of the endosperm and growth of integument to form a large embryo sac or seed cavity. In Arabidopsis, the seed cavity is generated at the globular stage or 4 days after pollination. The subsequent growth of the embryo replaces the endosperm during the second phase. Therefore, the volume of the initial seed cavity correlates closely with the final seed size. In shb1-D, an even larger seed cavity is created at 4 DAP due to an up-regulated expression of MINI3 and IKU2 by SHB1. We report that the expression of MINI3 and IKU2 coincides with the formation of the seed cavity. SHB1 is anchored to these promoters by MINI3 to activate their expression in a W-box-dependent manner. Spatiotemporal regulation of gene expression is a crucial mechanism that controls embryo development in many organisms. This interaction of SHB1 with MINI3 should impact studies of their homologs in many other organisms, including humans. Seed development in major seed crops, such as soybean and canola, follows a very similar path to that of Arabidopsis. Our results should lead to an increase in agricultural yields and concomitant increases in the proteins and oil content per seed.
Introduction
In angiosperms, double fertilization leads to the formation of a diploid embryo and a triploid endosperm, and the endosperm arises from the central cell that contains two identical haploid genomes. In many dicots, such as Arabidopsis, seed development follows two distinct phases and the embryo grows to full size and replaces most of the endosperm at maturity [1], [2]. During the first phase, the syncytial phase, a rapid growth and proliferation of the endosperm occurs, which generates a large multinucleate cell and results in a larger embryo sac or seed cavity by 4 DAP [3]–[5]. This syncytium is then partitioned into individual cells by a specific type of cytokinesis called cellularization [6]. During the second phase, embryo growth takes place at the expense of the endosperm. Upon maturity, the seed contains only a single layer of endosperm cells in Arabidopsis, and the maternal integument ultimately becomes the seed coat [6], [7]. The seed coat and endosperm growth in Arabidopsis precedes embryo growth, and the seed reaches almost its final size before the embryo enlarges. Both maternal and non-maternal factors are involved in seed size regulation [8].
In Arabidopsis, increased dosage of the paternal genome in the endosperm increases seed size whereas increased dosage of the maternal genome causes the opposite effect [9]. Defects in endosperm development include delayed cellularization of the peripheral endosperm and hypertrophy of the chalazal endosperm and associated nodules [7]. Mutations in DNA METHYLTRANSFERASE1 (MET1) and DECREASE IN DNA METHYLATION1 (DDM1) reduce DNA methylation dramatically and cause parent-of-origin effects on F1 seed size [9]. Pollination of met1-6 or ddm1-2 pistils with wild-type pollens produced large F1 seeds, and reciprocal crosses generated small F1 seeds. Mutations in the transcription factor APETALA2 (AP2) increased seed size due to an increase in both embryo cell number and cell size [10], [11]. The seed trait is passed via the maternal sporophyte and endosperm genomes.
In contrast, mutations in either IKU (HAIKU) or MINI3 (MINISEED3) reduce seed size, and the mutant seed phenotypes depend on the genotype of the embryo and the endosperm but not on the genotype of the maternal ovule [12]. In Arabidopsis, a larger embryo sac or seed cavity, coordinated by the growth of the maternal integument and the endosperm, is created at 4 DAP [1], [2]. TRANSPARENT TESTA GLABRA2 (TTG2), METHYLTRANSFERASE1 (MET1), and MEGAINTEGUMENTA/AUXIN RESPONSE FACTOR 2 (MNT/ARF2) regulate integument growth [8], [13], [14] whereas MINI3, IKU2, IKU1, and SHB1 control endosperm proliferation [12], [15], [16]. Mutations in either MINISEED3 (MINI3) or HAIKU2 (IKU2) cause premature cellularization of the endosperm, reduced growth of the endosperm including the chalazal endosperm, and reduced proliferation of the embryo after the early torpedo stage [6], [12]. SHB1 was initially isolated from a gain-of-function overexpression mutant, short hypocotyl under blue 1 Dominant (shb1-D), based on its long hypocotyl phenotype under red, far-red, and blue light [17]. shb1-D significantly increased seed mass and the total seed yield compared with Ws wild type [15]. In shb1-D, an even larger seed cavity is created at 4 DAP along with an enlarged chalazal endosperm and a delay in endosperm cellularization. AGL62 also regulates endosperm cellularization and the endosperm cellularizes prematurely in agl62 seeds [18].
MINI3 or the WRKY10 gene encodes a WRKY family transcription factor [12]. MINI3 is expressed in the endosperm and the embryo from 12 to 96 hr after fertilization but not in the late-heart embryo at 110 hr after fertilization or the unfertilized ovule [12]. IKU2 encodes a leucine-rich repeat (LRR) receptor kinase. IKU2 expression was visible in the endosperm at 12 and 48 hr post-fertilization but not in the embryo or elsewhere in the plant [12]. SHB1 contains an N-terminal SPX domain and a C-terminal EXS domain and is homologous to the SYG1 protein family members of fungi, C. elegans, flies, and mammals [17], [19]. Unlike the location of many SYG1-like proteins to the membrane, SHB1 is localized to the nucleus in Arabidopsis. IKU1 is a nuclear protein with an N-terminal VQ motif [17]. IKU1 shows a similar expression pattern to MINI3 and IKU2 in the early developing endosperm before cellularization as MINI3 and IKU2 but GFP signals in pIKU1:GFP:IKU1 plants were also detected in the integument or seed coats [16].
The expression of IKU2 is reduced in mini3, iku1, and shb1, and the expression of MINI3 is reduced in iku1 and shb1 [12], [15]. In this study, we observed that the expression of MINI3 and IKU2 is regulated stringently before fertilization and after 4 DAP, and it is activated by SHB1 at 2 and 3 DAP during endosperm proliferation. SHB1 associates with the promoters of MINI3 and IKU2, and this association requires MINI3. MINI3 binds to its target W-boxes in these two promoters and interacts with the N-terminus of SHB1. Furthermore, SHB1, but not MINI3, trans-activates the expression from either a MINI3 or an IKU2 promoter-GUS construct.
Results
SHB1 regulates the spatio-temporal expression of MINI3 and IKU2
The expression of MINI3 and IKU2 peaks during a narrow window of up to 4 DAP, coincides with the formation of a large seed cavity, and is regulated by SHB1 [12], [15]. GUS is expressed in MINI3::GUS transgenic plants at 12 to 96 hr after fertilization in the globular and early-heart embryo and endosperm but not in the late-heart embryo at 110 hr post-fertilization [12]. GUS activity in IKU2::GUS plants was visible in endosperm at 12 and 48 hr post-fertilization and before cellularization but not in the embryo [12]. The expression of SHB1 has been shown to overlap with MINI3 and IKU2 in the endosperm [15]. To refine the dynamics of MINI3 and IKU2 expression after pollination, we generated four independent pMINI3::GFP or pIKU2::GFP transgenic plants with a single t-DNA insertion in Ws and Col, respectively. To learn when and where SHB1 activates the expression of MINI3 and IKU2, we crossed these transgenes into shb1-D, shb1, or mini3 background (Figure 1 and Figure 2).
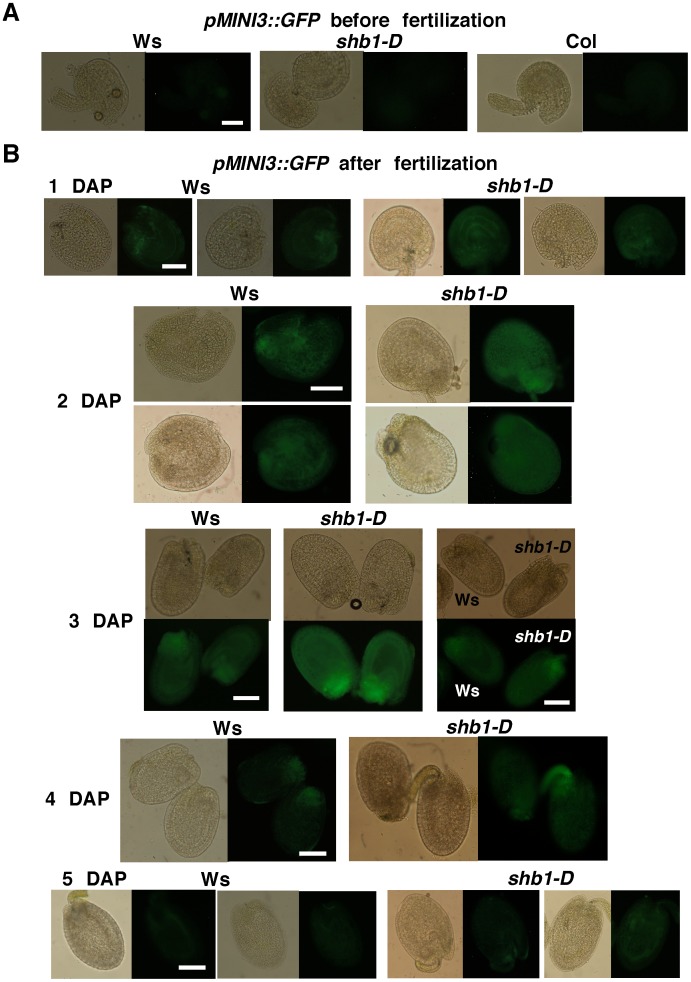
Figure 1. SHB1 regulates the expression of pMINI3::GFP.
Expression of pMINI3::GFP in Ws wild type and shb1-D seeds before fertilization (A) and from 1 to 5 DAP (B). Images on the left (1 to 2 and 4 to 5 DAP) or at the top (3 DAP) are of bright field, and images on the right (1 to 2 and 4 to 5 DAP) or at the bottom (3 DAP) are of GFP fluorescence. Scale bars, 100 µm.
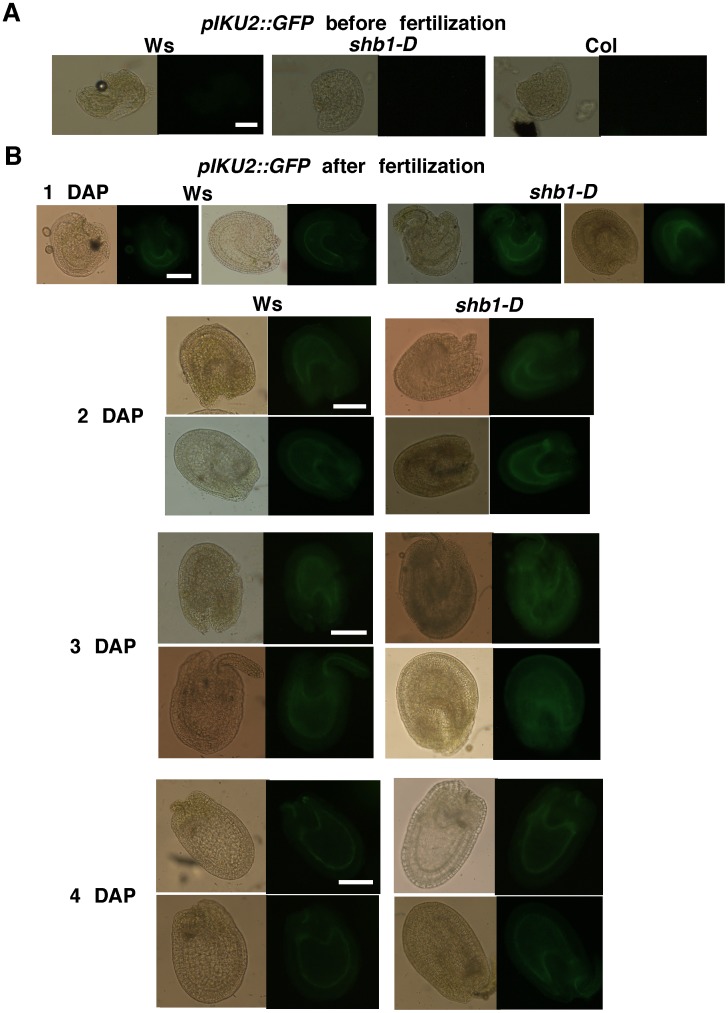
Figure 2. SHB1 regulates the expression of pIKU2::GFP.
The expression of pIKU2::GFP in Ws wild type and shb1-D seeds before fertilization (A) and from 1 to 5 DAP (B). Images on the left are of bright field, and images on the right are of GFP fluorescence. Scale bars, 100 µm.
We examined the expression of GFP in several seeds from the same silique for each genotype and examined the expression levels in several flowers of individual plants. When recording and processing fluorescent images, the illumination was adjusted and the gain used was similar for all. GFP expression was similar within individual plants, and two representative seeds from the same silique are shown, to indicate the consensus pattern of GFP expression, in Figures 1 and 2. In either Ws or Col, pMINI3::GFP signals were not detectable in the ovules before fertilization and was weak at 1 DAP (Figures 1 and Figure S1). Activity of the MINI3 promoter increased at 2 DAP, reached a maximum at 3 DAP, declined at 4 DAP, and was barely detectable or at background level by 5 DAP (Figure 1B and Figure S1). Activity of the MINI3 promoter was observed in the embryo, the peripheral endosperm, and the chalazal endosperm (Figure 1B and Figure S1). In shb1-D, GFP signals were enhanced slightly at 1 DAP, enhanced robustly at 2 and 3 DAP, and remained enhanced at 4 and 5 DAP (Figure 1B). In contrast, activity of the MINI3 promoter was reduced in shb1 seeds compared with Col seeds, particularly at 2 and 3 DAP (Figure S1). Activity of pMINI3::GFP was not affected significantly in mini3 (Figure S1), which is consistent with a previous report showing that MINI3 may possess an auto-regulatory function to repress its own expression [12].
Activity of pIKU2::GFP was not detectable in ovules before fertilization, and was weak at 1 DAP, stronger at 2 DAP, strong at 3 DAP, weak again at 4 DAP, and barely detectable at 5 DAP in Ws plants (Figure 2). GFP signals were primarily observed in the peripheral endosperm and the chalazal endosperm (Figure 2). Based on 4 independent transgenic lines for each construct and each ecotype (Figure 1 and Figure 2), the signals generated by pIKU2::GFP were in general much weaker than those of pMINI3::GFP plants. Stronger GFP activity was observed in shb1-D seeds compared with Ws seeds, particularly at 2 and 3 DAP (Figure 2A). Due to the weaker activity of the IKU2 promoter, we were unable to detect a consistent difference in GFP signals in shb1 or mini3 seeds compared with Col seeds. Importantly, SHB1 activates the expression of MINI3 and IKU2 within a very narrow window at approximately 2 or 3 DAP and prior to the enlargement of the seed cavity.
MINI3 ChIPs to MINI3 and IKU2 promoters
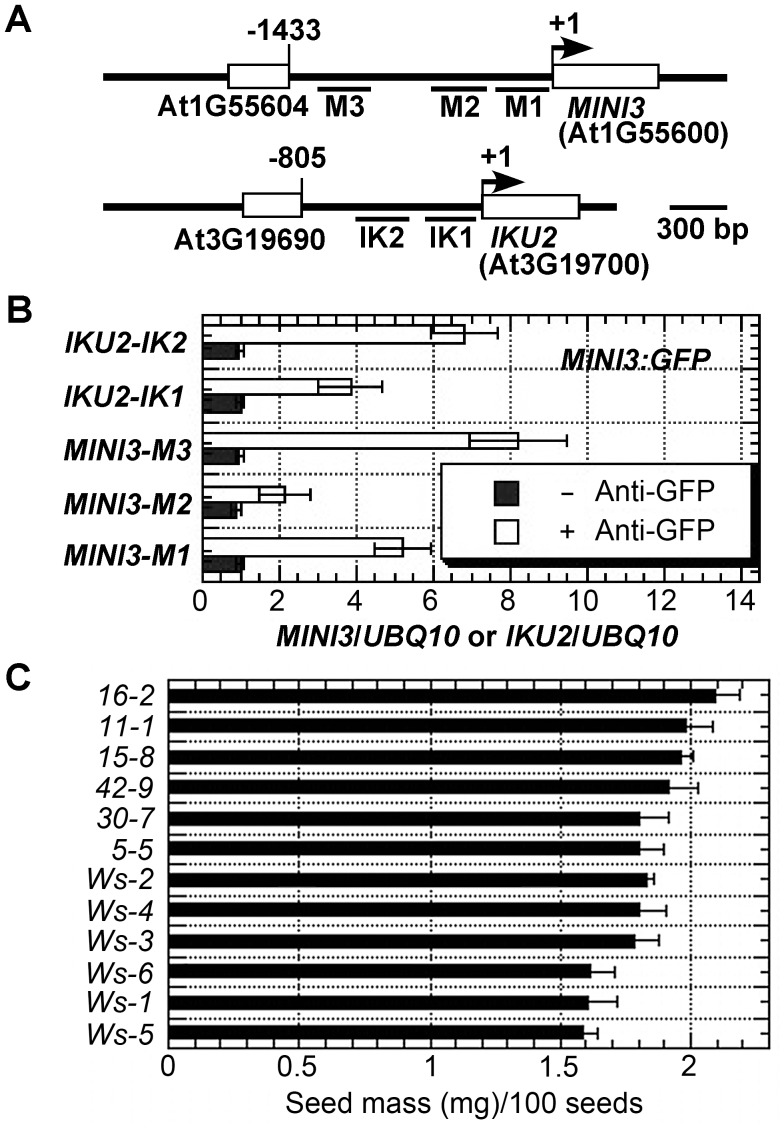
SHB1 regulates the expression of MINI3 and IKU2 and associates with their promoters in vivo [15]. To examine whether MINI3 is the transcription factor that anchors SHB1 to these promoters, we performed a chromatin immunoprecipitation quantitative PCR (ChIP-qPCR) assay in transgenic plants expressing MIN3:GFP under the control of a 35S promoter. Figure 3A shows the various amplicons in the MINI3 and IKU2 promoters that were used for the ChIP analysis: M1 from +6 to −348, M2 from −398 to −619, M3 from −985 to −1214, IK1 from −4 to −297, and IK2 from −278 to −566. Interestingly, MINI3 also associated with the MINI3 promoter because both amplicon M1 and M3 were highly enriched whereas amplicon M2 in the MINI3 promoter was not enriched significantly (Figure 3B). Amplicon 1 in the IKU2 promoter was moderately enriched and amplicon 2 was highly enriched (Figure 3B). To determine whether the level of GFP-tagged MINI3 produced in the endosperm of 35S::MINI3:GFP transgenic plants is sufficient for ChIP analysis, we examined the seed mass of six independent 35S::MINI3:GFP transgenic lines (Figure 3C). Compared with six independently propogated Ws lines, the six 35S::MINI3:GFP transgenic lines produced relatively larger seeds.
Figure 3. MINI3 ChIPs to the promoters of MINI3 and IKU2.
(A) Schematic diagram of the MINI3 and IKU2 loci and five amplicons (M1, M2, M3, IK1, and IK2) used for the ChIP-qPCR analysis. Rectangles represent genes and numbers indicate genomic nucleotide sequence coordination. Arrowheads indicate transcription start sites. (B) ChIP-qPCR analysis of the amplicons in the MINI3 and IKU2 promoters using anti-GFP monoclonal antibody in samples from 35S:MINI3:GFP transgenic plants. BSA was used as a mock control, and the fold enrichment of the specific chromatin fragments was normalized to the UBQ10 amplicon. Means were calculated from three biological samples, each of which was calculated from three technical replicates. The calculated standard errors comprise the technical error and biological error. (C) Seed masses were examined for six independently propagated Ws lines and six independent 35::MINI3:GFP transgenic lines (5-5 to 42-9) and expressed as mg/100 seeds.
The association of SHB1 with MINI3 and IKU2 promoters requires MINI3
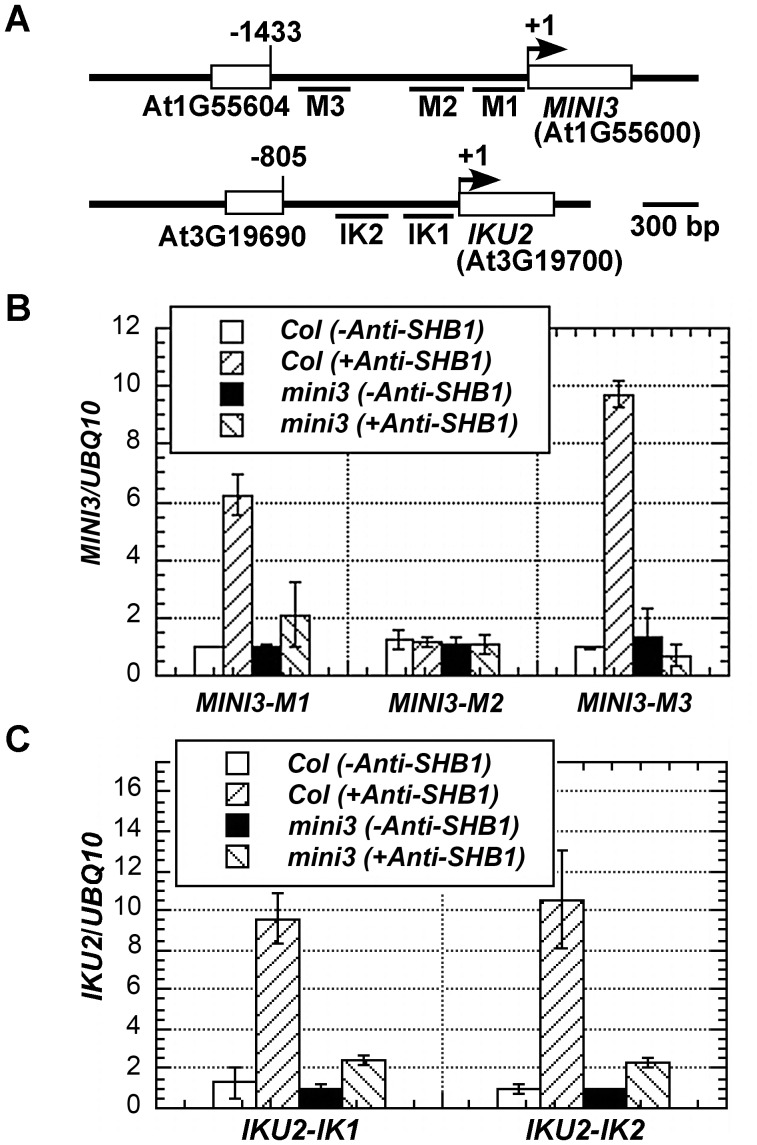
A similar pattern of fragment enrichment in the MINI3 and IKU2 promoters was also observed in the SHB1 ChIP assay [15]. SHB1 does not contain a recognizable DNA binding motif, and MINI3 is a WRKY transcription factor [12], [15]. MINI3 may mediate the in vivo association of SHB1 to the MINI3 and IKU2 promoters. We performed a ChIP analysis of SHB1 over the promoters of MINI3 and IKU2 in wild type and mini3 mutant samples (Figure 4B and 4C). SHB1 associated similarly with the promoters of both MINI3 and IKU2 in wild-type MINI3 siliques but failed to associate with either the MINI3 or the IKU2 promoter when MINI3 is mutated (Figure 4B and 4C) [15]. SHB1 is most likely recruited to either the MINI3 or the IKU2 promoter by MINI3.
Figure 4. Association of SHB1 with MINI3 and IKU2 promoters requires MINI3.
(A) A schematic diagram of the MINI3 and IKU2 loci and the five amplicons (M1, M2, M3, IK1, and IK2) used for the ChIP-qPCR analysis. Rectangles represent genes and numbers indicate genomic nucleotide sequence coordination. Arrowheads indicate transcription start sites. ChIP-qPCR analysis of the amplicons from the promoters of MINI3 (B) and IKU2 (C) using an anti-SHB1 antibody in wild type or mini3 mutant samples. Preimmune serum was used as a mock control, and the fold enrichment of the specific chromatin fragments was normalized to the UBQ10 amplicon. Means were calculated from three biological samples, each of was calculated from three technical replicates. The calculated standard errors comprise the technical error and biological error.
MINI3 binds W-boxes in the MINI3 or IKU2 promoters
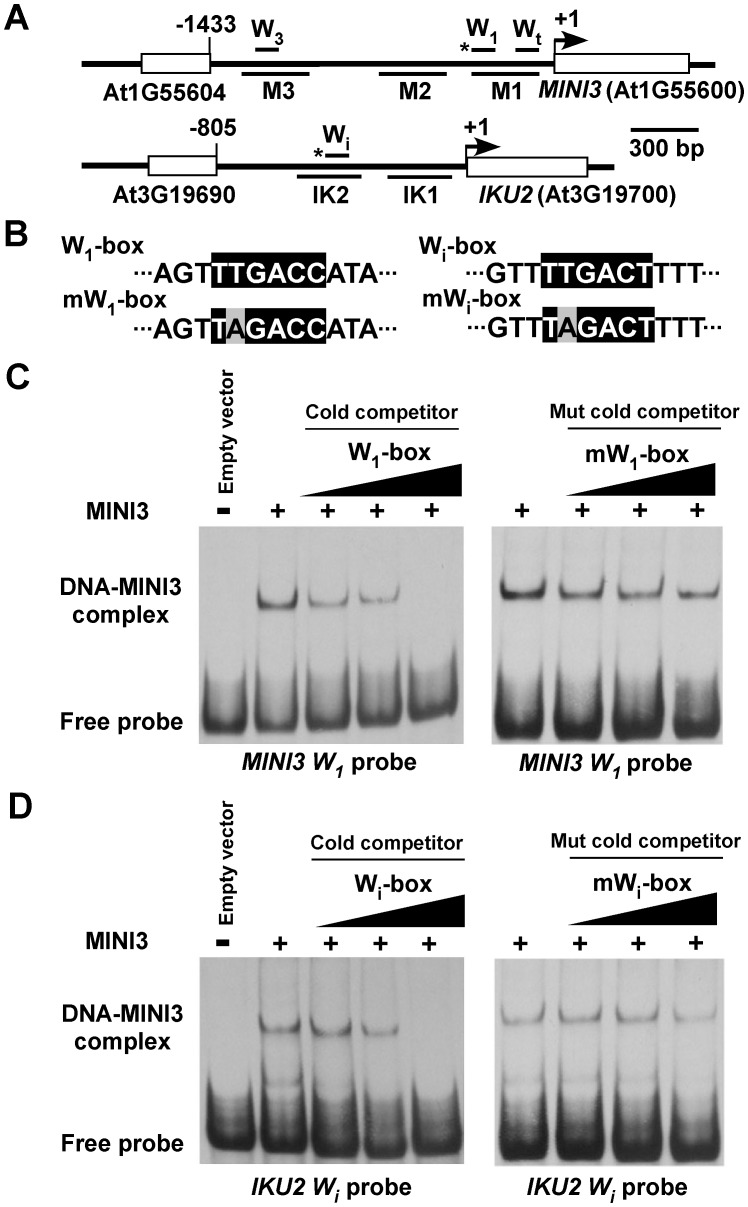
MINI3 is encoded by the WRKY10 gene, and the preferential binding site of WRKY factors is TTGACC (A/T) or a W-box [12], [20]. There are three putative W-boxes in the MINI3 promoter: W1-Box TTGACCA from nucleotides −257 to −262, W3-Box TTGACAA from nucleotides −1332 to −1337, and Wt-Box TTGACAT from nucleotides −19 to −24 (Figure 5A). The IKU2 promoter contains only one W-box, Wi-Box TTGACTT from nucleotides −386 to −391 (Figure 5A). To examine whether MINI3 binds to the W-boxes, we examined the affinity of a recombinant MINI3 protein to four genomic DNA fragments that contained the corresponding W-boxes using an electrophoretic mobility shift assay (EMSA).
Figure 5. MINI3 binds to W-boxes in the MINI3 and IKU2 promoters.
(A) A schematic diagram of the MINI3 and IKU2 loci, the five amplicons (M1, M2, M3, IK1, and IK2) used for the ChIP-qPCR analysis, and the position of the W-boxes in the MINI3 and IKU2 promoters. Rectangles represent genes and numbers indicate genomic nucleotide sequence coordination. Arrowheads indicate transcription start sites, and the asterisk indicates W-boxes recognized by MINI3. (B) The nucleotide sequences of the W1-box, mutated W1-box (mW1-box), Wi-box, and the mutated Wi-box (mWi-box). Core sequences are shaded in black, and mutated nucleotides are shaded in gray. EMSA analysis of the binding of MINI3 to W1-box in the MINI3 promoter (C) or Wi-box in the IKU2 promoter (D). Cold wild type or mutated W1-box and Wi-box competitors were used at a molar excess of 5X, 10X, or 50X.
MINI3 bound directly to the W1-box in the MINI3 promoter and the Wi-box in the IKU2 promoter (Figure 5C and 5D) but had a much weaker affinity with the W3-box in the MINI3 promoter (Figure S2C). To confirm that MINI3 indeed bound W1-box or Wi-box, we mutated the second T in the W-boxes to A and performed EMSA competition analyses (Figure 5B). The EMSA fragments that contained the mutated W1-box or Wi-box failed to compete effectively with radio-labeled cognate wild type fragments (Figure 5C and 5D). Intriguingly, cold EMSA fragments that contained either wild type or mutated W3-box competed well with the radio-labeled cognate wild type fragment (Figure S2C). Either the binding of MINI3 to the W3-box was not specific or the second T in the W3-box is not essential for binding with MINI3. The binding of MINI3 to Wt-box was clearly nonspecific because excess wild type cold Wt-box failed to compete with radio-labeled Wt-box (Figure S2D).
SHB1 interacts with MINI3
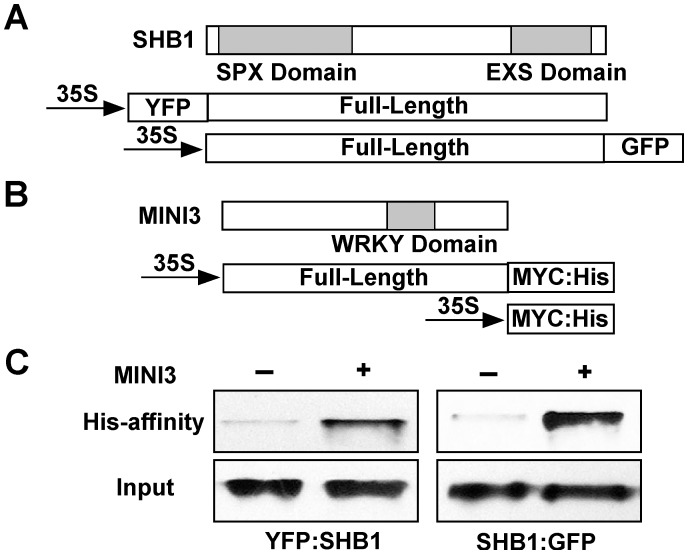
Both SHB1 and MINI3 associated with the same fragments in both MINI3 and IKU2 promoters, and the association of SHB1 with both promoters required MINI3 (Figure 3 and Figure 4). SHB1 and MINI3 may interact physically, and we performed an in vivo affinity-precipitation of MINI3:MYC:His for YFP:SHB1 in protein extracts prepared from Arabidopsis siliques (Figure 6A and 6B). MINI3:MYC:His did precipitate YFP:SHB1 but not MYC:His (Figure 6C, left). We then performed a parallel affinity-precipitation in a Nicotiana transient expression system. MINI3:MYC:His was also capable of precipitating SHB1:GFP in this system (Figure 6C, right).
Figure 6. SHB1 interacts with MINI3.
(A) A schematic drawing of full-length SHB1 fused to YFP at its N-terminus or GFP at its C-terminus under the control of the CaMV 35S promoter. The SPX and EXS domains of SHB1 are shaded in gray. (B) A schematic drawing of full-length MINI3 fused to MYC:His at its C-terminus under the control of the CaMV 35S promoter. The WRKY domain of MINI3 is shaded in gray. (C) His affinity-precipitation of YFP:SHB1 using MYC:His or MINI3:MYC:His from protein extracts prepared from Arabidopsis siliques (left), and of SHB1:GFP using MYC:His or MINI3:MYC:His from protein extracts prepared from Nicotiana leaves (right).
SHB1 N-terminus interacts with MINI3 N-terminus
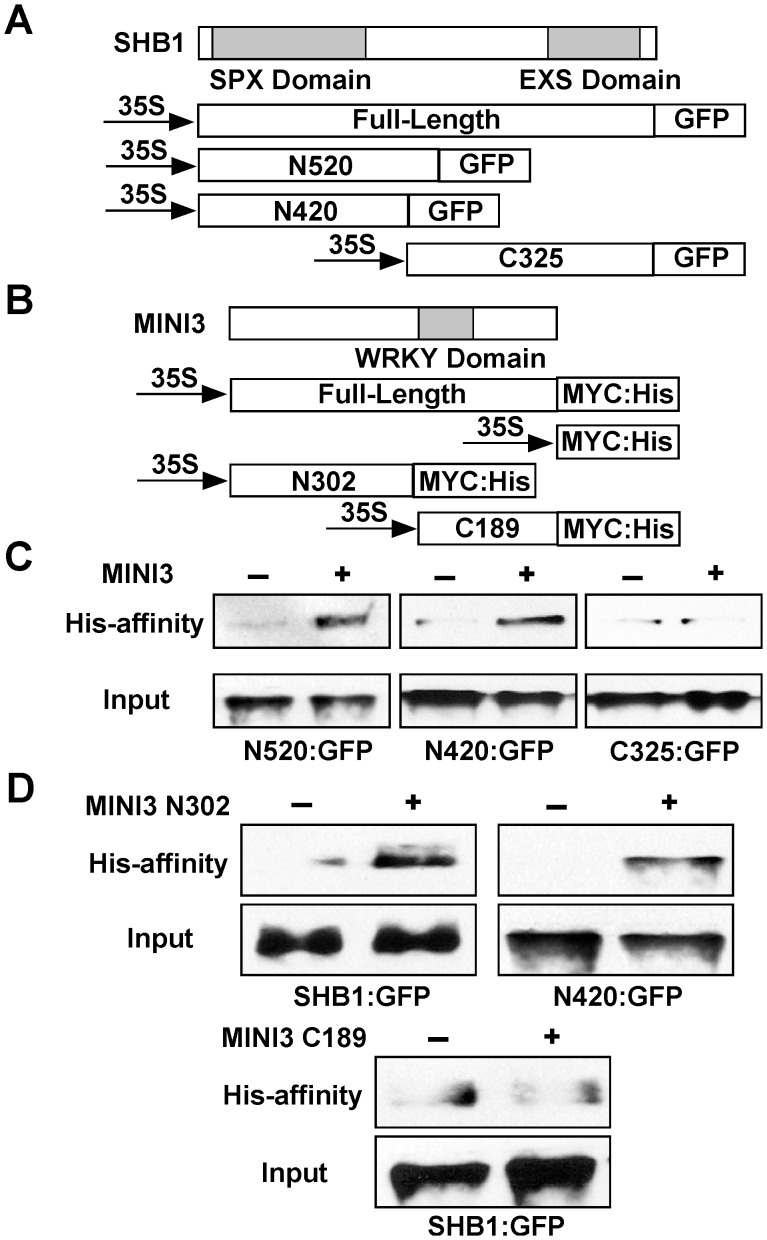
The most likely function of the SPX domain in SHB1 is to mediate protein-protein interactions [19]. The EXS domain contains several predicted trans-membrane helices, suggesting a membrane localization of the proteins [19], [21], [22]. However, SHB1 localizes to the nucleus, and the EXS domain in SHB1 may have a function distinct from those in other SYG1–like proteins. To map the interaction domain of SHB1 with MINI3, we performed an affinity precipitation of three truncated SHB1:GFP fusions, N520:GFP, N420:GFP, and C325:GFP, with MINI3:MYC:His in the Nicotiana transient expression system (Figure 7A and 7B). The N520:GFP and N420:GFP fusion proteins contain the N-terminal 520 or 420 amino acids, respectively, and include the putative SPX domain. The C325:GFP fusion protein contains the C-terminal 325 amino acids including the EXS domain (Figure 7A). MINI3:MYC:His precipitated both N520:GFP and N420:GFP (Figure 7C, left and middle) but not C325:GFP (Figure 7C, right). We conclude that SHB1 N-terminus mediates its interaction with MINI3, and its SPX domain may play an important role in this interaction.
Figure 7. The SHB1 N-terminus interacts with the MINI3 N-terminus.
(A) A schematic drawing of full-length and truncated SHB1 fused to GFP at its C-terminus under the control of the CaMV 35S promoter. The SPX and EXS domains of SHB1 are shaded in gray. (B) A schematic drawing of full-length and truncated MINI3 fused to MYC:His at its C-terminus under the control of the CaMV 35S promoter. The WRKY domain of MINI3 is shaded in gray. (C) His affinity-precipitation of SHB1 N520:GFP (left), N420:GFP (middle), or C325:GFP (right) using MYC:His or MINI3:MYC:His from protein extracts prepared from Nicotiana leaves. (D) His affinity-precipitation of SHB1:GFP (top left) or N420:GFP (top right) using MYC:His or MINI3 N302:MYC:His from protein extracts prepared from Nicotiana leaves. His affinity-precipitation of SHB1:GFP (bottom) using MYC:His or MINI3 C189:MYC:His from protein extracts prepared from Nicotiana leaves.
The MINI3 protein contains 485 amino acids and belongs to the WRKY family of plant transcription factors [23]. MINI3 contains a conserved WRKY DNA binding domain between amino acids 306 and 365 and the remaining amino acid sequence does not reveal any significant sequence homologies to others in the database. We made an N-terminal MINI3 construct that contains the first 302 amino acids (N302:MYC:His) and a C-terminal MINI3 construct that contains the C-terminal 189 amino acids including the WRKY domain (C189:MYC:His; Figure 7B). The N302:MYC:His precipitated SHB1:GFP and the N420:GFP fusion protein (Figure 7D, top left and right). In contrast, the C189:MYC:His protein failed to precipitate SHB1:GFP (Figure 7D, bottom). The N-terminus of MINI3 but not its WRKY DNA binding domain is important for its interaction with SHB1.
SHB1 activates expression from MINI3 or IKU2 promoters in a W-box-dependent manner
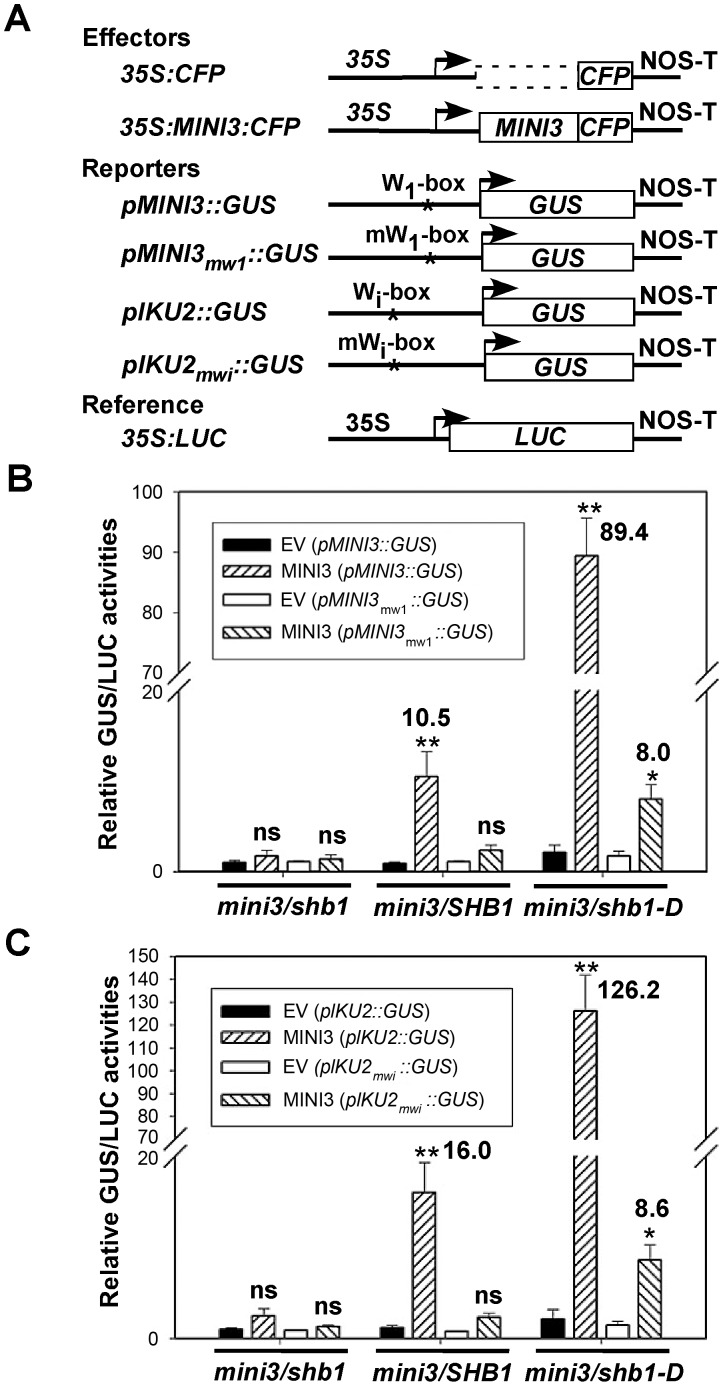
MINI3 does not contain a recognizable trans-activation motif, and MINI3 may not activate the expression of MINI3 or IKU2 alone. We conducted in vivo trans-activation experiments in Arabidopsis siliques by bombarding 35S CaMV::MINI3 effectors together with MINI3- or IKU2 promoter-GUS reporters into mini3/shb1, mini3/SHB1, or mini3/shb1-D developing siliques (Figure 8A). A CaMV 35S::LUC vector was also co-transformed to control for differences in transformation efficiency. When delivered back to mini3/shb1 double mutant, MINI3 alone activated very little transcription from either the MINI3 or the IKU2 promoter over that of the empty vector (EV) control (Figure 8B and 8C). In contrast, SHB1 activated expression from both the MINI3 and the IKU2 promoters by 11- and 16-fold, respectively, when MINI3 was delivered to the mini3/SHB1 background. Transcription from either the MINI3 or the IKU2 promoter was further activated up to 89- and 126-fold, respectively, in mini3/shb1-D.
Figure 8. SHB1 activates transcription from MINI3 and IKU2 promoters.
(A) A diagram showing the effector, reporter, and reference constructs used in the transient trans-activation assays. Full-length MINI3 fused to CFP was used as effector and was driven by the CaMV 35S promoter, and an empty vector (EV) was used as a control. For reporters, the GUS gene was driven by either the wild-type MINI3 or IKU2 promoter or mutated W1-box in the MINI3 promoter or mutated Wi-box in the IKU2 promoter and used as a reporter. Arrowheads indicate transcription start sites, and NOS-T represents polyadenylation signal from the nopaline synthase gene. The asterisk indicates the location of W-boxes in the MINI3 or IKU2 promoters. LUC gene driven by the CaMV 35S promoter was used as an internal reference. (B) GUS expression from the MINI3 promoter with a wild-type (pMINI3::GUS) or a mutated (pMINI3m1::GUS) W1-box in the presence of an empty vector (EV) or MINI3 in mini3/shb1, mini3/SHB1 or mini3/shb1-D backgrounds. (C) GUS expression from the IKU2 promoter with a wild-type (pIKU2::GUS) or a mutated (pIKU2mwi::GUS) Wi-box in the presence of an EV or MINI3 in mini3/shb1, mini3/SHB1 or mini3/shb1-D bckgrounds. GUS activities were expressed in pico-moles per min per mg protein relative to LUC activities, which were expressed in light units per mg protein. The relative GUS/LUC activities for the EV and pMINI3::GUS pair (24 pico-moles/min) in B or the EV and pIKU2::GUS pair (52 pico-moles/min) in C were set to 1, and the remaining values were expressed as fold trans-activation. Data are calculated from at least three independent experiments as the mean plus or minus standard error (n≥3). The levels of significance of differences were determined using Student's t-test. ** P<0.01, * P<0.05, and ns, not significantly different.
When the high-affinity W1-box binding site in the MINI3 promoter was mutated, the activation of expression from the MINI3 promoter was almost undetectable in mini3/SHB1 plants and decreased to 8.0-fold in mini3/shb1-D plants (Figure 8B). When the MINI3 Wi-box binding site in the IKU2 promoter was mutated, the activation of expression from the IKU2 promoter by SHB1 was similarly undetectable in mini3/SHB1 plants and was reduced 8.6-fold in mini3/shb1-D plants (Figure 8C). A mutation in the low-affinity W3-box binding site slightly reduced the activation of MINI3 expression by SHB1 to 6.3-fold in mini3/SHB1 plants and 47.2-fold in mini3/shb1-D plants (Figure S3A and S3B). A mutation in the non-specific Wt-box binding site had little affect on the activation by SHB1 (Figure S3A and S3C). Thus, the W1-box is the major binding site for MINI3, and the W3-box may play a subsidiary role to stabilize the DNA-protein interaction. In addition, the flanking sequences upstream or downstream of the core W-box may also exert secondary effects on the affinity of MINI3 to each particular W-box [23].
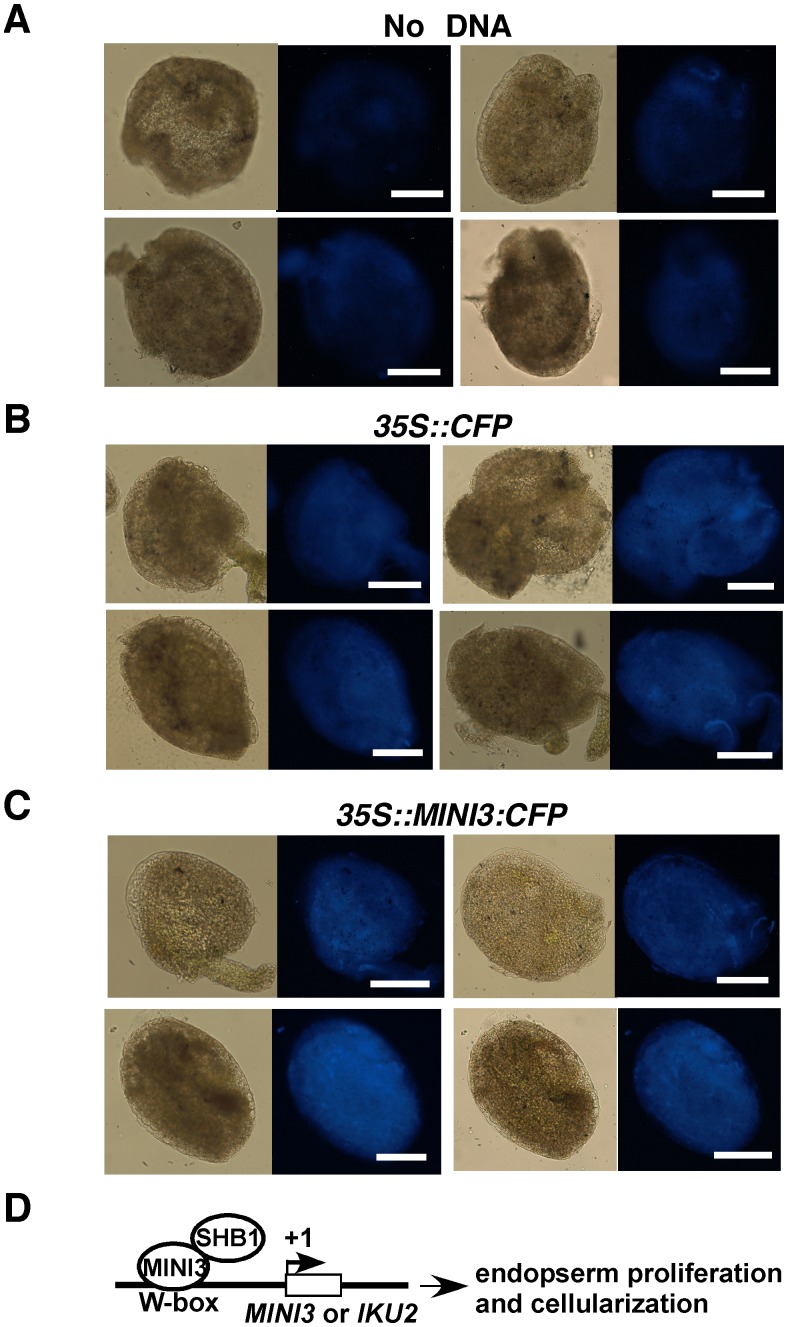
In the absence of MINI3, SHB1 induced little activation of expression from the promoters of MINI3 and IKU2 because SHB1 failed to target to these promoters (Figure S4). There was no notable activation of pMINI3::GUS or pIKU2::GUS expression in MINI3/shb1 plants, and MINI3 alone was unable to activate transcription from either promoter (Figure S5). We did not observe an inhibitory effect of MINI3 on its own expression in this transient system as has been shown by others in stable transgenic plants [12]. To determine whether the 35S promoter drives the expression of CFP or MINI3:CFP in this transient system, we delivered no DNA, 35S::CFP vector, or 35S::MINI3:CFP vector to Ws seeds at various DAP but primarily at 3 to 4 DAP (Figure 9A to 9C). Figure 9A shows background signals in various seed tissues in the control (no DNA vector delivered). CFP signals were robust in tissues bombarded with either 35S::CFP or 35S::MINI3:CFP vector (Figure 9B and 9C).
Figure 9. The CaMV 35S promoter drives expression of CFP or MINI3:CFP in transient bombardment experiments.
CFP signals in seeds bombarded without DNA (A) and with 35::CFP (B) or 35S::MINI3:CFP (C) construct. (D) A model depicting the function of SHB1 in MINI3 and IKU2 expression. SHB1 is likely recruited to the MINI3 and IKU2 promoters by MINI3, which specifically recognizes particular W-box sequences in these promoters. SHB1 or SHB1 together with other proteins activates expression from the MINI3, IKU2, and/or other target gene promoters required for endosperm proliferation and seed cavity enlargement.
Discussion
SHB1 activates the expression of MINI3 and IKU2 to trigger endosperm proliferation
In most species, endosperm in the syncytial phase undergoes cell expansion and proliferation to generate a large multinucleate cell close to the size of the mature seed [4], [5]. At 4 DAP, cellularization then partitions this syncytium into individual cells and marks the end of the maximum rate of endosperm proliferation [6]. Mutations in MINI3 and IKU2 cause premature cellularization of the endosperm and attenuate further proliferation of the cellularized endosperm [6], [12]. In contrast, the timing of endosperm cellularization is delayed in shb1-D [15]. The expression of MIN3 and IKU2 falls within a relatively narrow window, and the expression pattern of SHB1 overlaps with MINI3 and IKU2 in the endosperm [12], [15]. The expression of MIN3 and IKU2 is undetectable before fertilization (Figures 1 and 2), and one of the most compelling reasons is that MINI3 inhibits its own expression [12]. Consequently, the expression of IKU2 is also repressed because MINI3 is required for the proper expression of IKU2 [12], [15]. The expression of MINI3 and IKU2 peaks at 3 DAP, before the formation of a large seed cavity at 4 DAP, and declines or becomes repressed again after 4 DAP (Figure 1 and Figure 2). It is not known how MINI3 represses its own expression before fertilization and 4 DAP. The activation of MINI3 and IKU2 expression between 2 and 3 DAP may involve both the de-repression of MINI3 expression and an increase in SHB1 activity. The trigger for these two events remains unknown.
MINI3 recruits SHB1 as a transcription activator
In the absence of SHB1, MINI3 induces little activation of the expression of MINI3 or IKU2 and the expression of MINI3 and IKU2 is enhanced in mini3 SHB1 or mini3 shb1-D plants (Figure 8). MINI3 may function as a scaffold protein, and its N-terminus interacts with the N-terminus of SHB1 (Figure 7). Together with other proteins, the C-terminus of SHB1 may be capable of trans-activation. We propose a model where SHB1 is recruited by MINI3, which specifically binds W-box sequences in the promoters of MINI3 and IKU2, to activate the transcription from the MINI3 and IKU2 promoters and/or other target genes required for endosperm development (Figure 9D). IKU1 is a nuclear VQ motif protein that also interacts with the N-terminus of MINI3 via its N-terminal VQ motif [16]. IKU1 is expressed in the early developing endosperm prior to cellularization but GFP signals in PIKU1:GFP:IKU1 plants are also detected in the integument or seed coats [16].
Amount of stable and transient expression driven by the CaMV 35S promoter in the endosperm
It has been reported previously that cauliflower mosaic virus 35S promoter was not active during embryogenesis prior to the torpedo stage and that it did not show any activity in the syncytial endosperm [3]. We performed a MINI3 ChIP analysis in 35S::MINI3:GFP transgenic plants. Although we observed slightly higher MINI3:GFP expression compared with the background fluorescence of non-transgenic Ws plants, we are not convinced and do not present these images. Instead, we present data from six independent transgenic lines that each shows a consistently increase in seed mass compared with non-transgenic Ws plants (Figure 3C). MINI3 is normally expressed in the endosperm and the embryo, and its expression in the endosperm is required to maintain IKU2 expression for endosperm proliferation and seed cavity enlargement. The observed seed phenotype may be due to the low expression of MINI3:GFP driven by the CaMV 35S promoter in the endosperm. In addition, in our experiments, we harvested siliques at approximately 4 DAP or at the early heart stage, and the embryo constitutes only a small volume of the seed at this stage. The weak expression of MINI3:GFP in embryo only does not account for our positive ChIP results. Therefore, the 35S::MINI:GFP transgenic plants may produce sufficient levels of tagged MINI3 for the ChIP analysis.
We also used a MINI3:CFP construct driven by the 35S promoter as an effector in our transient trans-activation analysis that was transfected via particle bombardment (Figure 8). We delivered CFP and MINI3:CFP vectors driven by the 35S promoter to seeds at 3 to 4 DAP, and crobust CFP signals were detected in tissues bombarded with the 35S::CFP or 35S::MINI3:CFP vector (Figure 9B and 9C). The strength of the CaMV 35S promoter in the endosperm may be low in stable transgenic plants [3]. The low expression level in the endosperm conferred by the CaMV 35S promoter may be due to specific chromatin structures surrounding the transcription factor genes that act on this promoter. The mechanism for transient expression may be different. In the case of agrobacterium-mediated transient expression, only a small percentage of the DNA molecules integrate into the host chromosomes but the pieces of DNA that do not integrate remain transcriptionally competent [24]. In a bombardment experiment, DNA vectors are delivered into the integument cells, the endosperm nucleus, and the large cytosolic fraction of the endosperm at 4 DAP, which is when the endosperm begins to cellularize and that embryo is tiny and at the globular or early heart stage. The DNA vectors bombarded into the endosperm nucleus may not integrate into the host chromosomes but are transcriptionally competent. RNA polymerase and transcription factors that are translated in the cytosol could act on the DNA vectors delivered to the cytosolic fraction of the endosperm. A strong indication of this possibility is the transcription from the IKU2::GUS construct in our transient system because the IKU2 promoter confers expression that is endosperm-specific (Figure 2 and Figure 8C).
Redundant pathways regulate endosperm proliferation
After cellularization, further proliferation and expansion of the endosperm ceases, and the timing of endosperm cellularization may shape the final seed size. Many genes that influence seed size and the timing of endosperm cellularization fall into three major categories. These three redundant pathways ensure a robust seed set for success in reproduction. AP2 and some MADS-box transcription factors comprise the first category. AP2 encodes a plant-specific transcription factor with an AP2 DNA binding motif of 68 amino acids [25]. In an ap2 mutant, the cellularization process is delayed and prolonged resulting in a larger embryo sac [26]. Several MADS-box proteins regulate endosperm development maternally. FEM111/AGAMOUS-LIKE80 (AGL80) regulates central cell development and the subsequent endosperm cellularization [27]. An agl61 mutant shows a similar central cell and endosperm phenotype to fem111/agl80 [28]. AGL80 interacts with AGL61 and appears to recruit AGL61 to the nucleus [29]. AGL80 also interacts with AGL62, and the endosperm undergoes early cellularization in an agl62 mutant [18].
The second category includes five polycomb (PcG) proteins to form repressive FERTILIZATION INDEPENDENT SEED (FIS) complexes via histone methylation, and the imprinting of the polycomb genes by METHYL TRANSFERASE1 (MET1) and DECREASE IN DNA METHYLATION 1 (DDM1) [30]. In fis mutants, the female gametophytes initiate endosperm hyperproliferation without fertilization but the endosperm fails to cellularize. The paternal imprinting of MEDEA (MEA or FIS1) and FIS2 is mediated by MET1, and loss of MET1 activity in Arabidopsis causes genome wide DNA hypomethylation at CpG dinucleotides [31], [32]. The met1 mutation in the maternal genome causes larger seeds and results in a delay in endosperm cellularization and a larger endosperm volume [9]. In contrast, the met1 mutation in the paternal genome produces smaller seeds due to early cellularization of the endosperm. A similar result was also observed in a reciprocal cross between wild type and ddm1, which carries a mutation in a gene required for genomic cytosine methylation [9].
SHB1 co-activator, a WRKY transcriptional factor, and a receptor protein kinase belong to the third category. In this report, we reveal the molecular actions and interactions of the key players but their connections to other pathways remain elusive. In addition, how the three pathways interact and integrate remains largely unknown. Previously, an interesting story had emerged that AGL62 could mediate FIS protein function [18]. AGL62 is strongly expressed in the endosperm during the syncytial phase, and its expression declines sharply before cellularization. In some fis mutants, the endosperm fails to cellularize and AGL62 expression fails to decline [18]. The misexpression of AGL62 in fis mutants may account for the cellularization phenotype of the fis mutants. Furthermore, whether the three pathways target the same set or different sets of genes required for endosperm development awaits further investigation.
Materials and Methods
Plant materials and mutant genotyping
Arabidopsis thaliana ecotype Wassilewskija (Ws) and Columbia-0 (Col) were used as wild type plants. The mutant lines shb1-D, shb1 (SALK_128406), mini3-2 (SALK_050364), and the mini3-2 shb1-D double mutant were described previously [15], [17]. The shb1 mini3-2 double mutant was generated by crossing shb1 to mini3-2 and PCR-genotyped using the primers described previously [15]. Plants were grown under continuous light in a growth chamber at 21°C.
Expression of MINI3 and IKU2
The entire promoter regions of MINI3 from −1398 to −4 and of IKU2 from −806 to −1 were PCR-amplified from Arabidopsis genomic DNA. The stop codons of the adjacent genes are a few bp further upstream, and the regions used likely contain the majority of the regulatory information. The primer pairs used are TCAGGAGGTGACATCGAA and GACAAATCCTTAGGATGTC for MINI3, and TACGTACGTGTTGGTGGT and TGTTCTCTACGTCGGAAGGA for IKU2. The PCR fragments were cloned into the pCR8/GW/TOPO entry vector, and then recombined into the pMDC107 vector [33]. The constructs were transformed into Arabidopsis Ws or Col backgrounds by vacuum infiltration [34]. Four independent transgenic plants for each construct were further selected from multiple lines for a single T-DNA insertion based on their hygromycin resistance. The transgene in each of the four independent lines of either Ws or Col background was crossed into corresponding shb1-D, shb1, or mini3 mutant backgrounds. Plants homozygous for the transgene were used for all analyses. Images were acquired using a Nikon C1si Laser Scanning Confocal Microscope (Nikon, Melville, NY) equipped with a three-channel PMT detector. The tissue sample was excited at 488 nm for GFP fluorescence.
ChIP analysis
MINI3 genomic DNA was PCR-amplified and cloned into the pCR8/GW/TOPO vector, and then recombined into the pMDC83 vector [33]. The primers used were CGCGGATCCGATGAGTGATTTTGATC and CGCGGATCCCATGTCGACACCAAACT. This 35S:MINI3:GFP construct was transformed into Arabidopsis by vacuum infiltration [34]. The ChIP-PCR experiment was described previously, and chromatin DNA was sonicated and sheared to approximately 0.3 to 2 kb [15], [35]. The seed mass of several Ws and independent 35S::MINI3:GFP transgenic lines was measured as described previously [15].
Electrophoretic Mobility Shift Assay (EMSA)
Full-length MINI3 cDNA was cloned into the pDEST17 vector with a His-tag at its C-terminus. Total soluble protein was prepared from E. coli in binding buffer that contained 25 mM Hepes-KOH pH 7.2, 40 mM KCl, 1 mM DTT, 1 mM EDTA, 10% glycerol, and a complete set of protease inhibitors. The recombinant proteins were purified as pdescribed previously, and were dialyzed against the binding buffer [36]. An empty pDEST17 vector was used as a negative control.
EMSA was performed as described previously [36]. Three DNA fragments from the MINI3 promoter and one fragment from the IKU2 promoter were approximately 150 bp in length and were PCR-labeled with alpha-35S dATP. The Primer pairs used were GAGGATATAGTGGGTCT and GACAAATCCTTAGGATGTC for fragment containing the W1-box, TCAGGAGGTGACATCGAA and GACATTACTGTCATCGT for fragment containing the W3-box, ATTTGCTGCCACTCTTTGA and GCTGCTAGCGTTTTCTGA for fragment containing the Wt-box, and TACGTACGTGTTGGTGGT and CCAACGGATATGATTCA for fragment containing the Wi-box. The binding reaction was initiated by mixing 2 µg poly-dIdC, 3 ng radiolabeled probe (300 cpm/ng), 0.01 µM ZnCl2, various amounts of cold competitor, and 1 µg purified protein in 20 µl binding buffer. The reaction was incubated at room temperature for 30 min and separated on a 6% polyacrylamide gel in 0.25× TBE buffer. All W-boxes were also mutated by changing the second T to A using the QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA).
In vivo affinity-precipitation
SHB1 genomic DNA was PCR-amplified and cloned into the pCR8/GW/TOPO vector, and then recombined into the pEARLEYGATE104 (35S:YFP:SHB1) [37]. The primers used were CATGCCATGGTGAGGTTTGGGAAAGA and ATTGTTATGATGATCTCCA. MINI3 genomic DNA in the pCR8/GW/TOPO vector, as described in the Methods for the ChIP assay, was recombined into a modified pCAMBIA1390 vector, C-terminal to a MYC:His tag [38]. All constructs were introduced to Arabidopsis using the Agrobacterium-mediated vacuum infiltration method [34]. Either the 35S:MYC:His or the 35S:MINI3:MYC:His transgene was crossed into the 35S:YFP:SHB1 background. Approximately 4 g of developing siliques at 3 to 4 DAP were harvested, frozen in liquid nitrogen, and ground in 8 ml of cold co-precipitation buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 10% glycerol, 0.5% NP-40, 10 mM imidazole, and a full set of proteinase inhibitors). After centrifuging at 30,000 g and 4°C for 30 minutes, the supernatant was incubated with 150 µl pre-equilibrated nickel-agarose beads (Qiagen, Valencia, CA) at 4°C for 2 hr [38]. The incubation mixture was washed 5 times using cold co-precipitation buffer, and the proteins attached to the column were eluted using 100 µl cold co-precipitation buffer containing 200 mM imidazole. Approximately 48 µl of the elution was mixed with 12 µl 5x SDS loading buffer and loaded into a 10% SDS-PAGE gel. After blotting onto a membrane, the blotted proteins were probed using an anti-GFP antibody or anti-MYC primary antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
In vivo affinity-precipitation in a transient Nicotiana expression system was performed as described [39]. MINI3 truncated derivatives, N302 and C189, were PCR-amplified and cloned into a pCR8/GW/TOPO vector and recombined into the modified pCAMBIA1390 vector as described above. Primer pairs used were CGCGGATCCGATGAGTGATTTTGATG and GTCGCTTTCCATCTGAAG for N302, and CTTCAGATGGAAAGCGAC and CATGTCGACACCAAACTT for C189. The SHB1 full-length and SHB1 truncated derivatives, N520:GFP, N420:GFP and C325:GFP, in the pEZT-NL vector have been described previously [40]. Approximately 10 g of Nicotiana leaves were harvested 5 days after the initial infiltration, frozen in liquid nitrogen, and ground in 10 ml of cold co-precipitation buffer [41].
Transient trans-activation assay
The promoter regions of MINI3 and IKU2 in pCR8/GW/TOPO entry vectors were recombined into the pMDC163 vector as reporter constructs [33]. The mutations in each W-box were introduced as described for the MSA assay. MINI3 genomic DNA in the pCR8/GW/TOPO vector, as described for the ChIP assay, was recombined into the pEARLAYGATE102 vector as an effector together with the empty vector as a control [37]. A 35S::LUC vector harboring firefly luciferase under the control of the cauliflower mosaic virus 35S promoter was co-bombarded as an internal standard.
Transient expression analysis in Arabidopsis siliques was performed as described previously [42]. Tungsten particles (Bio-Rad, Hercules, CA) were coated with 1 µg of the reporter plasmid, 1 µg of the effector plasmid, and 30 ng of internal control plasmid suspended in ethanol. Arabidopsis siliques at 3 to 4 DAP were harvested, placed in a 3-cm-diameter circle in the center of a 10-cm Petri plate with ½ MS and 0.8% (w/v) agar, and then incubated for 2 h in a growth chamber prior to bombardment. The tungsten particles were delivered using the PDS-1000/He Biolistic Particle Delivery System (Bio-Rad, Hercules, CA). After bombardment, the siliques were incubated for 20 h (12 h light/8 h dark) at 21°C in a growth chamber, harvested, and then frozen in liquid nitrogen. Plant extracts were prepared for GUS activity assay [41]. The LUC activity in the extracts was measured using a Luciferase Assay System (Promega, Madison, WI). GUS activity was normalized to LUC activity.
Accession numbers
The sequences of the genes described in this report can be found in the Arabidopsis Genome Initiative database with the following accession numbers: SHB1 (At4G25350), MINI3 (At1G55600), IKU2 (At3G19700), and UBQ10 (At4G05320).
Supporting Information
The SHB1 mutation shows a partial affects on the expression of pMINI3::GFP. Expression of pMINI3::GFP in Col wild type, shb1, and mini3 seeds from 1 to 5 days after pollination (DAP). Images on the left (1 to 2 DAP) or at the top (3 to 5 DAP) are of bright field and images on the right (1 to 2 DAP) or at the bottom (3 to 5 DAP) are of GFP fluorescence. Scale bars, 100 µm.
(TIF)
MINI3 binds the W3-box weakly and the Wt-box nonspecifically. (A) A schematic diagram of the MINI3 loci, the three amplicons (M1, M2, and M3) used for the ChIP-qPCR analysis, and the position of the W-boxes in the MINI3 promoter. Rectangles represent genes and numbers indicate genomic nucleotide sequence coordination. The arrowhead indicates the transcription start site, and the asterisk indicates the W-boxes likely recognized by MINI3. (B) The nucleotide sequences of W3-box, mutated W3-box (mW3-box), Wt-box, and mutated Wt-box (mWt-box). Core sequences are shaded in black, and mutated nucleotides are shaded in gray. (C and D) EMSA analysis of the binding of MINI3 to W3-box (C) or Wt-box (D) in the MINI3 promoter. Cold wild type or mutated W3-box and Wt-box competitors were used at a molar excess of 5X, 10X, or 50X.
(TIF)
A mutation in the W3-box, but not the Wt-box, shows a partial affect on transient expression from the MINI3 or IKU2 promoters. (A) A diagram showing the effector, reporter, and reference constructs used in the transient trans-activation assays. Full-length MINI3 fused to CFP was driven by the CaMV 35S promoter as an effector, and an empty vector (EV) was used as a control. GUS gene was driven by either wild type or a mutated W3-box or Wt-box in the MINI3 promoter and used as a reporter. Arrowheads indicate transcription start sites, and NOS-T represents polyadenylation signal from the nopaline synthase gene. The asterisk indicates the location of the W-boxes in the MINI3 promoter. The LUC gene driven by the CaMV 35S promoter was used as an internal reference. (B) GUS expression from the MINI3 promoter with a wild-type (pMINI3::GUS) or a mutated (pMINI3mw3::GUS) W3-box in the presence of an empty vector (EV) or MINI3 in mini3/shb1, mini3/SHB1 or mini3/shb1-D backgrounds. (C) GUS expression from the MINI3 promoter with a wild-type (pMINI3::GUS) or a mutated (pMINI3mwt::GUS) Wt-box in the presence of an EV or MINI3 in mini3/shb1, mini3/SHB1 and mini3/shb1-D backgrounds. GUS activities were expressed in pico-moles per min per mg protein relative to LUC activities, which were expressed in light units per mg protein. The relative GUS/LUC activity for the EV and pMINI3::GUS pair (24 pico-moles/min; shown in Figure 7B) was set to 1 and the remaining values were expressed as fold trans-activation. Data were calculated from at least three independent experiments as the mean plus or minus the standard error (n≥3). The levels of significance of differences were determined using the Student's t-test. ** P<0.01, * P<0.05, and ns, not significantly different.
(TIF)
SHB1 is not targeted to either the MINI3 or IKU2 promoter in the absence of MINI3. (A) GUS is expressed from a wild type MINI3 promoter (pMINI3::GUS) in the absence of MINI3:CFP in mini3/shb1, mini3/SHB1 and mini3/shb1-D. (B) GUS is expressed from a wild type IKU2 promoter (pIKU2::GUS) in the absence of MINI3:CFP in mini3/shb1, mini3/SHB1 and mini3/shb1-D. GUS activities were expressed in pico-moles per min per mg protein relative to LUC activities, which were expressed in light units per mg protein. The relative GUS/LUC activity in mini3/shb1 was set to 1 and the remaining values are expressed as fold trans-activation. Data were calculated from at least three independent experiments as the mean plus or minus the standard error (n≥3). The levels of significance of differences were determined using the Student's t-test.
(TIF)
MINI3 does not activate the expression of MINI3 or IKU2 in the absence of SHB1. (A) GUS is expressed from a wild type MINI3 promoter (pMINI3::GUS) in mini3/shb1 and MINI3/shb1. (B) GUS is expressed from a wild type IKU2 promoter (pIKU2::GUS) in mini3/shb1 and MINI3/shb1. GUS activities were expressed in pico-moles per min per mg protein relative to LUC activities, which were expressed in light units per mg protein. The relative GUS/LUC activity in mini3/shb1 was set to 1 and the remaining values were expressed as fold trans-activation. Data were calculated from at least three independent experiments as the mean plus or minus the standard error (n≥3). The levels of significance of differences were determined using the Student's t-test.
(TIF)
Acknowledgments
We thank Dr. Albrecht Von Arnim for kindly providing the 35S:LUC plasmid, Dr. Salomé Prat for kindly providing the pBin61-P19 plasmid, Dr. R. Scott Mclvor for help with the LUC assay, and Dr. Xiaodong Sun for valuable discussions.
Funding Statement
This research was supported by grants from the National Science Foundation (IOS0919886) and the USDA NIFA (2011-67013-30150) and by Plant Biological Sciences Summer Graduate Fellowships from the University of Minnesota. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sundaresan V (2005) Control of seed size in plants. Proc Natl Acad Sci USA 102: 17887–17888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sun X, Shantharaj D, Kang X, Ni M (2010) Transcriptional and hormonal signaling control of Arabidopsis seed development. Curr Opin Plant Biol 13: 611–620. [DOI] [PubMed] [Google Scholar]
- 3. Boisnard-Lorig C, Colon-Carmona A, Bauch M, Hodge S, Doerner P, et al. (2001) YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13: 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Olsen OA (2001) Endosperm development: cellularization and cell fate specification. Annu Rev Plant Phys Plant Mol Biol 52: 233–267. [DOI] [PubMed] [Google Scholar]
- 5. Berger F (2003) Endosperm, the crossroad of seed development. Curr Opin Plant Biol 6: 42–50. [DOI] [PubMed] [Google Scholar]
- 6. Garcia D, Saingery V, Chambrier P, Mayer U, Jurgens G, et al. (2003) Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol 131: 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Scott RJ, Spielman M, Bailey J, Dickinson HG (1998) Parent-of-origin effects on seed development in Arabidopsis thaliana . Development 125: 3329–3341. [DOI] [PubMed] [Google Scholar]
- 8. Garcia D, Fitz Gerald JN, Berger F (2005) Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xiao W, Brown RC, Lemmon BE, Harada JJ, Goldberg RB, et al. (2006) Regulation of seed size by hypomethylation of maternal and paternal genomes. Plant Physiol 142: 1160–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jofuku KD, Omidyar PK, Gee Z, Okamuro JK (2005) Control of seed mass and seed yield by the floral homeotic gene APETALA2 . Proc. Natl. Acd. Sci. USA 102: 3117–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ohto M, Fischer RL, Goldberg RB, Nakamura K, Harada JJ (2005) Control of seed mass by APETALA2 . Proc. Natl. Acd. Sci. USA 102: 3123–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Luo M, Elizabeth S, Dennis ES, Berger F, Peacock WJ, et al. (2005) MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis . Proc Natl Acd Sci USA 102: 17531–17536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. FitzGerald J, Luo M, Chaudhury A, Berger F (2008) DNA methylation causes predominant maternal controls of plant embryo growth. PLoS ONE 3: e2298 doi:10.1371/journal.pone.0002298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, et al. (2005) The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133: 251–261. [DOI] [PubMed] [Google Scholar]
- 15. Zhou Y, Zhang X, Kang X, Zhao X, Zhang X, et al. (2009) SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to control Arabidopsis seed development. Plant Cell 21: 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang A, Garcia D, Zhang H, Feng K, Chaudhury A, et al. (2010) The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J 63: 670–679. [DOI] [PubMed] [Google Scholar]
- 17. Kang X, Ni M (2006) Arabidopsis SHORT HYPOCOTYL UNDER BLUE 1 contains SPX and EXS domains and acts in cryptochrome signaling. Plant Cell 18: 921–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kang IH, Steffen JG, Portereiko MF, Lloyd A, Drews GN (2008) The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis . Plant Cell 20: 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Spain BH, Koo D, Ramakrishnan M, Dzudzor B, Colicelli J (1995) Truncated forms of a novel yeast protein suppress the lethality of a G protein α subunit deficiency by interacting with the β subunit. J Biol Chem 270: 25435–25444. [DOI] [PubMed] [Google Scholar]
- 20. Ciolkowski I, Wanke D, Birkenbihl RP, Somssich IE (2008) Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol Biol 68: 81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tailor CS, Nouri A, Lee CG, Kozak C, Kabat D (1999) Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc Natl Acad Sci USA 96: 927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang YL, Guo L, Xu S, Holland CA, Kitamura T, et al. (1999) Receptors for polytropic and xenotropic mouse leukemia viruses encoded by a single gene at Rmc1. Nat Genet 21: 216–219. [DOI] [PubMed] [Google Scholar]
- 23. Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206. [DOI] [PubMed] [Google Scholar]
- 24. Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, et al. (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13 doi:10.1186/1746-4811-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Riechmann JL, Meyerowitz EM (1998) The AP2/EREBP family of plant transcription factors. Biol Chem 379: 633–646. [DOI] [PubMed] [Google Scholar]
- 26. Ohto MA, Floyd SK, Fischer RL, Goldberg RB, Harada JJ (2009) Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis. Sex Plant Reprod 22: 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Portereiko MF, Lloyd A, Steffen JG, Punwani JA, Otsuga D, et al. (2006) AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell 18: 1862–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Steffen JG, Kang IH, Portereiko MF, Lloyd A, Drews GN (2008) AGL61 interacts with AGL80 and is required for central cell development in Arabidopsis. Plant Physiol 148: 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Folter S, Immink RG, Kieffer M, Parenicová L, Henz SR, et al. (2005) Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell 17: 1424–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hennig L, Derkacheva M (2009) Diversity of Polycomb group complexes in plants: same rules, different players? Trends Genet 25: 414–423. [DOI] [PubMed] [Google Scholar]
- 31. Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A (2000) Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci USA 97: 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xiao W, Gehring M, Choi Y, Margossian L, Pu H, et al. (2003) Imprinting of the MEA Polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev Cell 5: 891–901. [DOI] [PubMed] [Google Scholar]
- 33. Curtis MD, Grossniklaus U (2003) A Gateway Cloning Vector Set for High-Throughput Functional Analysis of Genes in Planta. Plant Physiol 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bent A, Kunkel B, Dahlbeck D, Brown K, Schmidt R, et al. (1994) RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265: 1856–1860. [DOI] [PubMed] [Google Scholar]
- 35. Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, et al. (2004) Chromatin techniques for plant cells. Plant J 39: 776–789. [DOI] [PubMed] [Google Scholar]
- 36. Sun X, Ni M (2011) HYPOSENSITIVE TO LIGHT, an Alpha/Beta Fold Protein, Acts Downstream of ELONGATED HYPOCOTYL 5 to Regulate Seedling De-Etiolation. Mol Plant 4: 116–126. [DOI] [PubMed] [Google Scholar]
- 37. Earley KW, Haag JR, Pontes O, Opper K, Juehne T, et al. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629. [DOI] [PubMed] [Google Scholar]
- 38. Tang W, Yuan M, Wang R, Yang Y, Wang C, et al. (2011) PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol 13: 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun X, Kang X, Ni M (2012) Hypersensitive to Red and Blue 1 and its modification by Protein Phosphatase 7 are implicated in the control of Arabidopsis stomatal aperture. PLoS Genet 8: e1002674 doi:10.1371/journal.pgen.1002674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou Y, Ni M (2010) SHB1 truncations and mutations alter its association with a signaling protein complex. Plant Cell 22: 703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wydro M, Kozubek E, Lehmann P (2006) Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotiana benthamiana . Acta Biochim Pol 53: 289–298. [PubMed] [Google Scholar]
- 42. Sessa G, Borello U, Morelli G, Ruberti I (1998) A Transient Assay for Rapid Functional Analysis of Transcription Factors in Arabidopsis. Plant Mol Biol Rep 16: 191–197. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The SHB1 mutation shows a partial affects on the expression of pMINI3::GFP. Expression of pMINI3::GFP in Col wild type, shb1, and mini3 seeds from 1 to 5 days after pollination (DAP). Images on the left (1 to 2 DAP) or at the top (3 to 5 DAP) are of bright field and images on the right (1 to 2 DAP) or at the bottom (3 to 5 DAP) are of GFP fluorescence. Scale bars, 100 µm.
(TIF)
MINI3 binds the W3-box weakly and the Wt-box nonspecifically. (A) A schematic diagram of the MINI3 loci, the three amplicons (M1, M2, and M3) used for the ChIP-qPCR analysis, and the position of the W-boxes in the MINI3 promoter. Rectangles represent genes and numbers indicate genomic nucleotide sequence coordination. The arrowhead indicates the transcription start site, and the asterisk indicates the W-boxes likely recognized by MINI3. (B) The nucleotide sequences of W3-box, mutated W3-box (mW3-box), Wt-box, and mutated Wt-box (mWt-box). Core sequences are shaded in black, and mutated nucleotides are shaded in gray. (C and D) EMSA analysis of the binding of MINI3 to W3-box (C) or Wt-box (D) in the MINI3 promoter. Cold wild type or mutated W3-box and Wt-box competitors were used at a molar excess of 5X, 10X, or 50X.
(TIF)
A mutation in the W3-box, but not the Wt-box, shows a partial affect on transient expression from the MINI3 or IKU2 promoters. (A) A diagram showing the effector, reporter, and reference constructs used in the transient trans-activation assays. Full-length MINI3 fused to CFP was driven by the CaMV 35S promoter as an effector, and an empty vector (EV) was used as a control. GUS gene was driven by either wild type or a mutated W3-box or Wt-box in the MINI3 promoter and used as a reporter. Arrowheads indicate transcription start sites, and NOS-T represents polyadenylation signal from the nopaline synthase gene. The asterisk indicates the location of the W-boxes in the MINI3 promoter. The LUC gene driven by the CaMV 35S promoter was used as an internal reference. (B) GUS expression from the MINI3 promoter with a wild-type (pMINI3::GUS) or a mutated (pMINI3mw3::GUS) W3-box in the presence of an empty vector (EV) or MINI3 in mini3/shb1, mini3/SHB1 or mini3/shb1-D backgrounds. (C) GUS expression from the MINI3 promoter with a wild-type (pMINI3::GUS) or a mutated (pMINI3mwt::GUS) Wt-box in the presence of an EV or MINI3 in mini3/shb1, mini3/SHB1 and mini3/shb1-D backgrounds. GUS activities were expressed in pico-moles per min per mg protein relative to LUC activities, which were expressed in light units per mg protein. The relative GUS/LUC activity for the EV and pMINI3::GUS pair (24 pico-moles/min; shown in Figure 7B) was set to 1 and the remaining values were expressed as fold trans-activation. Data were calculated from at least three independent experiments as the mean plus or minus the standard error (n≥3). The levels of significance of differences were determined using the Student's t-test. ** P<0.01, * P<0.05, and ns, not significantly different.
(TIF)
SHB1 is not targeted to either the MINI3 or IKU2 promoter in the absence of MINI3. (A) GUS is expressed from a wild type MINI3 promoter (pMINI3::GUS) in the absence of MINI3:CFP in mini3/shb1, mini3/SHB1 and mini3/shb1-D. (B) GUS is expressed from a wild type IKU2 promoter (pIKU2::GUS) in the absence of MINI3:CFP in mini3/shb1, mini3/SHB1 and mini3/shb1-D. GUS activities were expressed in pico-moles per min per mg protein relative to LUC activities, which were expressed in light units per mg protein. The relative GUS/LUC activity in mini3/shb1 was set to 1 and the remaining values are expressed as fold trans-activation. Data were calculated from at least three independent experiments as the mean plus or minus the standard error (n≥3). The levels of significance of differences were determined using the Student's t-test.
(TIF)
MINI3 does not activate the expression of MINI3 or IKU2 in the absence of SHB1. (A) GUS is expressed from a wild type MINI3 promoter (pMINI3::GUS) in mini3/shb1 and MINI3/shb1. (B) GUS is expressed from a wild type IKU2 promoter (pIKU2::GUS) in mini3/shb1 and MINI3/shb1. GUS activities were expressed in pico-moles per min per mg protein relative to LUC activities, which were expressed in light units per mg protein. The relative GUS/LUC activity in mini3/shb1 was set to 1 and the remaining values were expressed as fold trans-activation. Data were calculated from at least three independent experiments as the mean plus or minus the standard error (n≥3). The levels of significance of differences were determined using the Student's t-test.
(TIF)