Abstract
In Caenorhabditis elegans the Toll-interleukin receptor domain adaptor protein TIR-1 via a conserved mitogen-activated protein kinase (MAPK) signaling cascade induces innate immunity and upregulates serotonin (5-HT) biosynthesis gene tph-1 in a pair of ADF chemosensory neurons in response to infection. Here, we identify transcription factors downstream of the TIR-1 signaling pathway. We show that common transcription factors control the innate immunity and 5-HT biosynthesis. We demonstrate that a cysteine to tyrosine substitution in an ARM motif of the HEAT/Arm repeat region of the TIR-1 protein confers TIR-1 hyperactivation, leading to constitutive tph-1 upregulation in the ADF neurons, increased expression of intestinal antimicrobial genes, and enhanced resistance to killing by the human opportunistic pathogen Pseudomonas aeruginosa PA14. A forward genetic screen for suppressors of the hyperactive TIR-1 led to the identification of DAF-19, an ortholog of regulatory factor X (RFX) transcription factors that are required for human adaptive immunity. We show that DAF-19 concerts with ATF-7, a member of the activating transcription factor (ATF)/cAMP response element-binding B (CREB) family of transcription factors, to regulate tph-1 and antimicrobial genes, reminiscent of RFX-CREB interaction in human immune cells. daf-19 mutants display heightened susceptibility to killing by PA14. Remarkably, whereas the TIR-1-MAPK-DAF-19/ATF-7 pathway in the intestinal immunity is regulated by DKF-2/protein kinase D, we found that the regulation of tph-1 expression is independent of DKF-2 but requires UNC-43/Ca2+/calmodulin-dependent protein kinase (CaMK) II. Our results suggest that pathogenic cues trigger a common core-signaling pathway via tissue-specific mechanisms and demonstrate a novel role for RFX factors in neuronal and innate immune responses to infection.
Author Summary
Toll-interleukin receptor (TIR)–domain adaptor proteins are keys to activate signaling cascades inducing transcriptional responses to internal and external pathogenic signals in evolutionary disparate organisms. Despite lacking a homolog of the mammalian innate immunity transcriptional regulator nuclear factor-kappaB (NF-κB), the nematode Caenorhabditis elegans responds to infections by activating TIR-1 signaling targets in the innate immune system and in neurons. Through a genetic screen for factors required for TIR-1 signaling to upregulate the serotonin biosynthesis gene tph-1, we identified DAF-19, an ortholog of regulatory factor X (RFX) transcription factors that were initially discovered in human immune cells. We show that DAF-19 concerts with ATF-7, a member of the activating transcription factor (ATF)/cAMP response element-binding B (CREB) family of transcription factors, to upregulate tph-1 in the ADF chemosensory neurons and antimicrobial genes in the intestine in response to bacterial infection, reminiscent of RFX-CREB interaction in human immune cells. daf-19 mutants display heightened susceptibility to killing by the human pathogen Pseudomonas aeruginosa PA14. Our studies suggest that RFX transcriptional regulation, which is essential for human adaptive immunity, has an ancient role in controlling serotonin biosynthesis and innate immunity.
Introduction
Innate immunity is an integral part of the stress response program in which the host activates a range of defense genes to enhance the chance of survival against internal and environmental threats. In mammals, signals associated with pathogenic microbes trigger Toll-like receptors to recruit Toll-interleukin receptor (TIR) domain adaptor proteins, thereby forming scaffolds with downstream signaling cascades leading to transcriptional upregulation of defense genes. A growing body of evidence indicates that classical immune proteins including Toll-like receptors and TIR domain adaptor proteins are expressed in the developing and mature brain in mammals [1], [2]. It has been proposed that certain common molecular mechanisms may function in neurons and non-neuronal tissues to induce physiologically distinct responses to aversive cues [1], [2], [3]. Except a few cases, the gene targets of immune factors in neurons are not known and it is unclear whether those immune signaling cascades are differentially regulated in neurons and non-neuronal tissues. Consequently, identification of upstream regulators and downstream effectors of conserved core immune signaling pathways may provide insights into the regulation of the immunity as well as the regulation of neural plasticity.
Our laboratory has focused on genetic dissection of environment-dependent transcriptional regulation of the tph-1 gene, encoding the rate-limiting serotonin (5-HT) biosynthesis enzyme tryptophan hydroxylase, in the nematode Caenorhabditis elegans. Previously, we showed that tph-1 expression in a pair of ADF chemosensory neurons in the head sensory organ Amphid is modulated by two layers of transcriptional regulation according to growth conditions: signaling through the OCR-2/OSM-9 TRPV channel turns on the basal tph-1 expression under optimal growth conditions, and aversive growth conditions further upregulate tph-1 expression independently of OCR-2/OSM-9 [4], [5]. Work from several laboratories suggests that tph-1 expression in the ADF neurons responds to pathogenic food. C. elegans feeds on bacteria and is killed by a large number of pathogenic microbes in its natural environment [6]. In an elegant study, it showed that feeding worms with the human opportunistic pathogen Pseudomonas aeruginosa PA14 triggers upregulation of tph-1 in the ADF neurons leading to aversive learning and avoidance behavior [7]. A subsequent study indicated that the TIR-domain adaptor protein TIR-1, which was initially identified as an upstream regulator of a conserved mitogen-activated protein kinase (MAPK) signaling pathway in the innate immunity [8], is required for PA14-induced tph-1 upregulation and PA14 avoidance behavior [9]. However, the C. elegans genome lacks a homolog of nuclear factor-kappaB (NF-κB), the major transcriptional activator of the mammalian innate immunity [10]. In addition, deletion of the sole C. elegans Toll receptor gene tol-1 did not affect the intestinal immunity [11] or tph-1 expression [7]. These observations suggest that the TIR-1 signaling cascade may involve evolutionarily more ancient upstream players and downstream transcription factors.
Activating transcription factor (RFX) transcription factors were first identified in human subjects of bare lymphocyte syndrome, a hereditary immunodeficiency disease, and are required for the expression of the major histocompatibility complex class II (MHC II) genes [12]. RFX proteins bind to the X-box motif on the MHC II promoters and interact with cAMP response element-binding (CREB) protein and other cofactors to form a higher order “enhanceosome”, which then recruits the non-DNA-binding transcriptional activator CIITA to turn on MHC II expression [13]. RFX factors have since been identified in diverse eukaryotic species [14], [15], [16] and are expressed broadly in neuronal and non-neuronal cells in animals, suggesting additional roles for RFX factors in biological processes of multiple tissues. Studies of the sole C. elegans RFX factor daf-19 have uncovered its role in the development of dendritic cilia of sensory neurons [17], [18]. Subsequent studies found RFX factors regulating ciliogenesis in Drosophila [19] and mouse [20], demonstrating one aspect of RFX function conserved across phyla.
In this paper, we identified DAF-19 as a key transcriptional regulator of tph-1 in the ADF neurons and intestinal antimicrobial genes in C. elegans. We found that, analogous to the RFX-CREB interaction for MHC II expression in human immune cells, DAF-19 concerts with ATF-7, a member of activating transcription factor (ATF)/CREB superfamily of transcription factors, acting downstream of the TIR-1 signaling cascade to control transcriptional responses to pathogenic bacterial food in C. elegans. We show that the TIR-1-DAF-19/ATF-7 pathway is differentially regulated to induce tph-1 upregulation and intestinal immunity in response to P. aeruginosa PA14. Thus, our data suggest that pathogenic signals may trigger a common core signaling pathway via cell-specific mechanisms and a RFX transcription factor acts in an ancient host to regulate 5-HT biosynthesis and the innate immunity.
Results
Isolation of tir-1(yz68) gain-of-function mutation
We carried out a forward genetic screen to identify components underling aversive environment-induced tph-1 upregulation in C. elegans. The levels of tph-1 expression in identified neurons in living C. elegans can be estimated by quantifying fluorescence intensity of a green fluorescence protein (GFP) driven by the tph-1 promoter (tph-1::gfp) [4]. A pair of ADF neurons is the only chemosensory neurons producing 5-HT in a hermaphrodite C. elegans [21]. Each ADF neuron projects a single dendrite to the tip of the nose where the ciliated sensory endings are exposed to the external environment and its axon extends to the nerve ring, the brain of C. elegans [22]. We started with a strain expressing a stably chromosomally integrated tph-1::gfp transgene in ocr-2(yz5) mutant background, in which tph-1::gfp expression in the ADF neurons is visible under aversive growth conditions but not under optimal growth conditions, providing a visual assay for environment-dependent changes in tph-1 expression [5]. We isolated mutagenized worms with enhanced ADF tph-1::gfp under optimal growth conditions, and analyzed the mutants in the ocr-2 background as well as after the ocr-2 mutation being outcrossed. yz68 is one of the mutants retrieved from the screen.
Through single nucleotide polymorphism-based (SNP) mapping, RNA-interference (RNAi)-mediated inactivation of candidate genes in the mapped contig, and sequencing the yz68 mutant genome, we identified a nucleic acid change predicting a substitution of cysteine426 by tyrosine (C426Y) in the fourth ARM motif of the HEAT/Arm repeat region of the TIR-1 protein (Figure 1A). Several experimental data suggest that the C426Y substitution causes TIR-1 constitutive activation. First, whole mount anti-5-HT-antibody staining showed that ADF 5-HT immunoreactivity in tir-1(yz68);ocr-2 double mutants was elevated compared to the ocr-2 single mutant (Figure 1D). As 5-HT is being secreted, 5-HT immunostaining does not fully reflect the rate of 5-HT biosynthesis. With this caveat in mind, the results suggest that tir-1(yz68) enhanced 5-HT in the ADF neurons. Second, transgenic expression of tir-1(yz68) cDNA under the gpa-13 promoter (Pgpa-13::tir-1(yz68c)), which is expressed in ADF, AWC and ASH sensory neurons in the head region, recapitulated tph-1::gfp upregulation (Figure 1B). Third, RNAi of tir-1 in tir-1(yz68);ocr-2 mutants blocked the tph-1 upregulation (Figure 1C). The tir-1(ok1052) mutation, which causes mixed gain- and loss-of-function tir-1 phenotypes in the AWC neuron development [23] but does not affect the innate immunity [24], caused only a modest increase in ADF tph-1::gfp (Figure 1C). Collectively, these data suggest that the C426Y substitution alters a site critical for TIR-1 activation in the ADF neurons.
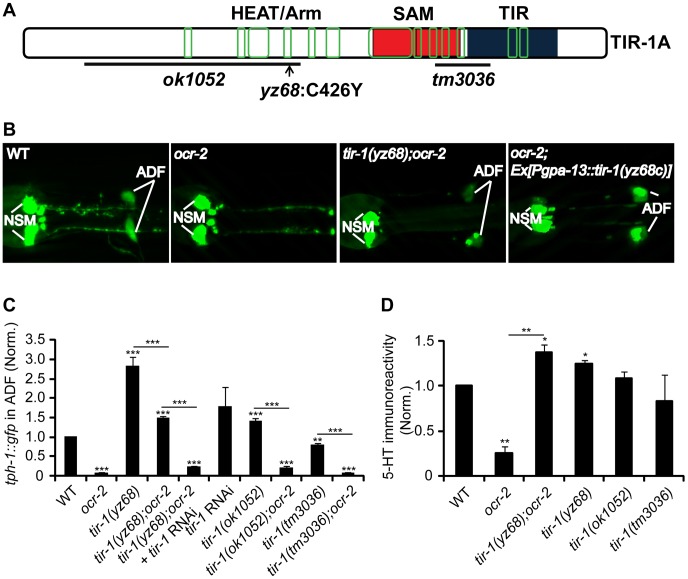
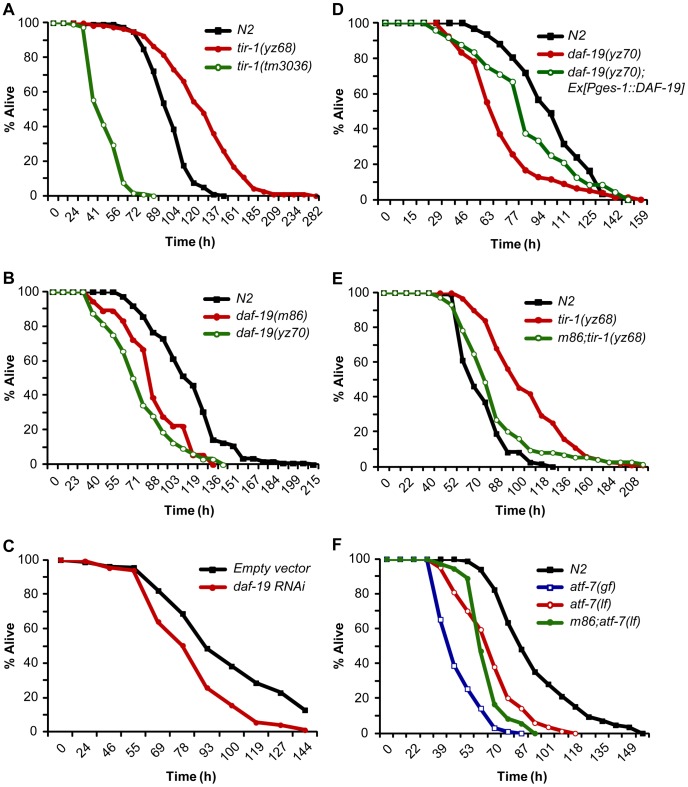
Figure 1. tir-1(yz68gf) mutants constitutively upregulate tph-1::gfp expression in ADF neurons.
A. Mutations in predicted tir-1 protein. The TIR-1 structure is adapted from a previous report [8]. Blue, red and green denote the TIR domain, SAM domains, and regions containing the HEAT/Arm motifs, respectively. B. Photomicrographs showing that tir-1(yz68gf) mutation and transgenic expression of tir-1(yz68gf) cDNA enhanced ADF tph-1::gfp expression. ocr-2 background was used to reduce basal ADF tph-1::gfp for a better detection of tph-1::gfp upregulation by tir-1(yz68gf). C. Quantification of ADF tph-1::gfp fluorescence in tir-1(lf) and tir-1(gf) mutants under optimal growth conditions. tir-1(yz68gf) mutants displayed enhanced ADF fluorescence on its own and in ocr-2 backgrounds relative to corresponding controls, and RNAi of tir-1 abolished tph-1::gfp upregulation by tir-1(yz68gf). The value of GFP fluorescence in the ADF neurons of mutants is normalized to that of WT animals. For RNAi experiments, the value of tir-1 RNAi is normalized to that in the same strain but treated with an empty vector. Each bar represents the average of at least three trials ± SEM. Throughout of this paper, statistics between WT and individual mutants is marked on the top of each bar, and that between two mutant strains or two treatments is indicated on the top of the line across compared strains, *** p<0.001. D. Quantification of 5-HT immunoreactivity in ADF neurons of tir-1 mutants. Data represent the average of two trials, each with at least 20 L4-stage animals per strain ± SEM, and the value of the mutants is normalized to that of WT animals stained in parallel, *p<0.05, ** p<0.01.
Unlike the ocr-2 mutation, tir-1(tm3036) loss-of-function (lf) mutants and RNAi of tir-1 do not lead to a dramatic reduction in ADF tph-1::gfp (Figure 1C). Likewise, inactivation of TIR-1 downstream MAPKKK nsy-1 or MAPKK sek-1 by RNAi and deletion, respectively, did not downregulate tph-1::gfp, although RNAi of nsy-1 did abolish tph-1::gfp upregulation by tir-1(yz68gf) (Figure S1). These observations are consistent with a published work [9]. These data suggest that TIR-1 signaling is not a major regulator of basal tph-1 expression in the ADF neurons under optimal growth conditions.
TIR-1 signaling pathway selectively regulates serotonergic response to pathogenic food
Previously, we identified that a number of aversive conditions may induce tph-1 upregulation in the ADF neurons [5]. We asked whether tir-1 signaling specifically mediates the response to pathogenic bacteria, or it is responsible for tph-1 upregulation under all aversive conditions. We first analyzed the intensity of tph-1::gfp in the ADF neurons in tir-1(lf) and tir-1(gf) mutants fed with the pathogenic P. aeruginosa strain PA14. Following feeding on PA14 for 6 hr, ADF fluorescence in WT animals was ∼1.4-fold higher than their sibling on OP50, but tir-1(lf) mutants failed to upregulate tph-1::gfp under the same conditions (Figure 2A), consistent with prior reports [7], [9]. The tir-1(yz68gf) mutant also did not exhibit a significant increase in tph-1::gfp following PA14 feeding, suggesting that the pathogen signals cannot further enhance tir-1(yz68gf) protein activity (Figure 2A).
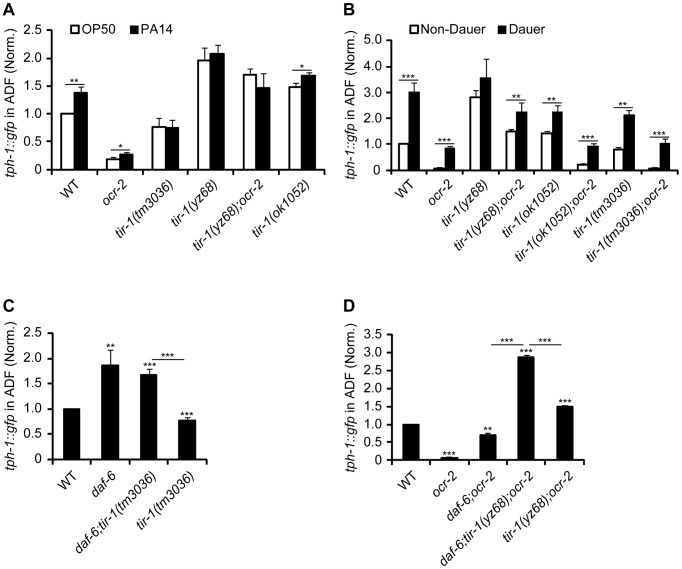
Figure 2. TIR-1 signaling selectively regulates tph-1::gfp response to pathogenic bacterial food.
A. PA14-induced ADF tph-1::gfp upregulation was impaired in tir-1(tm3036lf) and tir-1(yz68gf) mutants. B. tir-1(tm3036lf) and tir-1(yz68gf) mutations did not block tph-1::gfp upregulation during dauer formation. C. TIR-1 is not required for tph-1::gfp upregulation caused by changes in cilial morphology. Mutations in daf-6/Patched alter the morphology of dendritic cilia of all chemosensory neurons including ADF and cause ADF tph-1::gfp upregulation on its own as well as in tir-1(lf) background. D. tir-1(yz68gf) and daf-6 mutations confer additive upregulation of tph-1::gfp in the ADF neurons. Data represent the average of three trials each with at least 20 animals per strain per condition ± SEM. The value of ADF GFP fluorescence in WT animals under a stress paradigm and that of mutants is normalized to the value of WT animals under optimal conditions. *p<0.05, ** p<0.01, *** p<0.001.
We next tested if TIR-1 function is required for tph-1 upregulation during dauer formation. Under the conditions of starvation, high growth temperature and high levels of pheromones, C. elegans develops into a stress-resistant dauer larva through a series of cellular and physiological remodeling and turns on a battery of stress genes [25]. We previously showed that WT and ocr-2 mutant worms upregulated ADF tph-1::gfp when they entered the dauer stage [5]. We therefore induced tir-1(lf) and tir-1(gf) mutants to form dauers by treating the worms with dauer pheromones. We observed ADF tph-1::gfp upregulation in both tir-1(tm3036lf) and tir-1(tm3036lf);ocr-2 double mutant dauers as compared with corresponding L4-stage animals (Figure 2B). ADF tph-1::gfp was increased in tir-1(yz68gf) mutants during dauer formation and this increase was more evident in the tir-1(yz68gf);ocr-2 double mutant dauers (Figure 2B). Thus neither TIR-1 deficiency nor TIR-1 hyperactivation can block tph-1::gfp upregulation induced by dauer formation.
We previously showed that mutations that alter the morphology of dendritic cilia of ADF neurons cause tph-1 upregulation [5]. We therefore tested whether aberrant cilia trigger TIR-1 leading to tph-1::gfp upregulation. If this were the case, then we can expect that inactivation of tir-1 blocks tph-1::gfp upregulation in cilial mutants. Mutations of daf-6/Patched, which is expressed in the glia ensheathing the dendritic cilia of the chemosensory neurons [26], and the Intraflagellar Transport (IFT) gene dyf-1 that is essential for cilia formation [27] conferred ADF tph-1::gfp upregulation [5]; however, inactivation of tir-1 did not block tph-1::gfp upregulation in daf-6 (Figure 2C) or dyf-1 mutants (Figure S2). Furthermore, a triple mutant of daf-6, tir-1(yz68gf);ocr-2 showed higher ADF tph-1::gfp than daf-6;ocr-2 and tir-1(yz68gf);ocr-2 double mutants (Figure 2D). Together these data suggest that the ADF neurons can detect and discriminate multiple aversive cues, and indicate that TIR-1 is selectively involved in the pathogen signaling transduction pathway.
daf-19 is a suppressor of tir-1(yz68gf)
To identify the effectors of TIR-1 signaling, we carried out a suppressor screen for mutants that abrogate tph-1 expression in tir-1(yz68gf) mutants. Using a combination of SNP mapping, non-complementation tests and sequencing the mutant genomes, we identified that two mutations, yz69 and yz70, are allelic to the daf-19 gene, encoding the sole C. elegans ortholog of the RFX transcription factors (Figure 3A). Subsequent experiments with our alleles and the previously existing daf-19(m86)-null mutation revealed that DAF-19 function is required not only for tir-1(yz68gf) to upregulate tph-1::gfp, but also for ADF tph-1::gfp expression under optimal growth conditions (Figure 3B), during dauer formation (data not shown) and in aberrant cilia backgrounds (Figure 3E; Figure S2). The reduced tph-1::gfp in daf-19 mutants was fully rescued by transgenic expression of WT daf-19 genomic sequence (Figure 3B) or daf-19 cDNA driven by the gpa-13 promoter (Figure 3D).
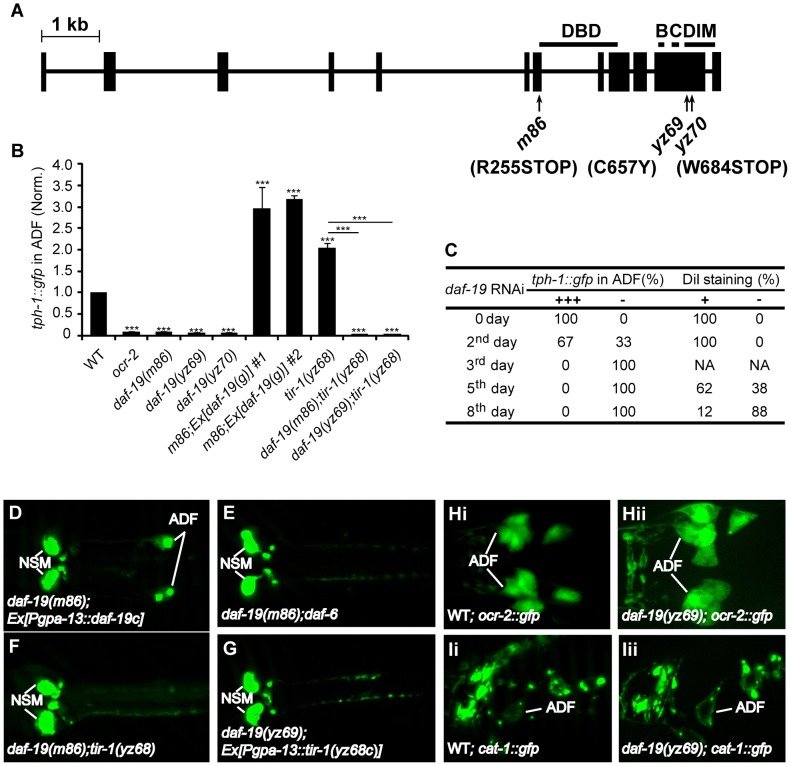
Figure 3. daf-19 is required for tph-1 expression in the ADF neurons.
A. Mutations in the daf-19 gene. The daf-19 gene structure is adapted from a previous report [17]. Boxes denote exons and the line denotes introns. The areas containing the DNA binding domain (DBD), dimerization domain (DIM) and conserved B and C regions are indicated. B. ADF tph-1::gfp is dramatically reduced in daf-19 mutants and daf-19;tir-1(yz68gf) double mutants. Two transgenic lines expressing WT daf-19 genomic sequence rescued tph-1::gfp expression in daf-19(m86)-null mutants. *** p<0.001. C. RNAi of daf-19 after embryogenesis. L1 larvae with normal ADF tph-1::gfp and cilial morphology were fed E. coli expressing RNAi clone against daf-19. tph-1::gfp was quantified on indicated days, and dye filling of chemosensory neurons with fluorescence dye DiI was performed to monitor gross cilia morphology of the chemosensory neurons. daf-19 RNAi eliminated ADF tph-1::gfp (-) and eliminated dye filling of the cilia, but tph-1::gfp was abolished in many animals prior to a detectable change in the cilia morphology. -, No discernible fluorescence of tph-1::gfp or DiI staining in the neurons. D–G. Photomicrographs showing tph-1::gfp in daf-19 mutants under various genetic backgrounds. daf-19 deficiency abolished tph-1::gfp expression in the ADF neurons of both daf-6 and tir-1(yz68gf) backgrounds. H–I. Expressions of ADF marker genes in daf-19 mutants. ocr-2::gfp and VMAT cat-1::gfp were comparable between WT (Hi and Ii) and daf-19 mutant (Hii and Iii) animals, suggesting that daf-19 deficiency does not grossly alter ADF cell fates.
An implicit concern was that daf-19 deficiency alters ADF cell fates. To directly analyze the effect of daf-19 deficiency on tph-1 expression, we inactivated daf-19 by RNAi after 5-HT phenotypes established. Figure 3C shows that 100% of larval stage 1 (L1) worms expressed tph-1::gfp prior to RNAi treatment, 33% of the animals lost ADF GFP after 24 hr RNAi treatment, and by 48 hr, 100% of the animals showed no GFP in the ADF neurons. DAF-19 is required for the expression of cilia IFT components [17]. Although we showed that aberrant cilia induced ADF tph-1::gfp upregulation [5] (Figure S2), complete lacking cilia could inhibit tph-1 expression. To rule out this possibility, we used lipophilic dye DiI staining to examine the dendritic cilia morphology of the chemosensory neurons in worms treated with daf-19 RNAi. We observed that RNAi of daf-19 eliminated ADF tph-1::gfp prior to a detectable change in the cilia (Figure 3C).
We investigated whether the reduced tph-1 expression is a secondary consequence of reduced ocr-2 and tir-1 expression in daf-19 mutants. daf-19(yz69) (Figure 3Hii) and daf-19(m86)-null (not shown) mutants expressed a GFP reporter for ocr-2 (ocr-2::gfp) in ADF and other chemosensory neurons indistinguishable from WT animals. Diminished tph-1::gfp expression also cannot be ascribed to reduced tir-1 expression, as transgenic expressing tir-1(yz68) cDNA by the gpa-13 promoter failed to increase ADF tph-1::gfp in daf-19 mutants (Figure 3G), although the Pgpa-13::daf-19c transgene did (Figure 3D), indicating that the gpa-13 promoter is expressed in daf-19 mutants but that the tir-1(yz68gf) protein cannot stimulate tph-1 expression in the absence of DAF-19 function.
To assess whether daf-19 deficiency abolishes all 5-HT phenotype genes, we analyzed the expression of cat-1, encoding the vesicular monoamine transporter required for 5-HT synaptic release [28]. GFP-tagged CAT-1 was expressed and localized properly in the ADF neurons of daf-19 mutants (Figure 3Iii). Thus, diminished tph-1::gfp in daf-19 mutants cannot be attributed to altered ADF cell fate.
TIR-1-induced tph-1 upregulation requires DAF-19 and ATF-7
Because daf-19 appeared to be required for tph-1 expression controlled by multiple mechanisms, we hypothesized that DAF-19 may concerts with other transcriptional regulators to confer specificity to particular signaling pathway activation. In light of the partnership of RFX and CREB proteins for MHC II expression, we further hypothesized that an analogous mechanism might underscore transcriptional responses to pathogenic signals in C. elegans. ATF-7, an ortholog of the mammalian ATF2/ATF7/CREB5 family of transcription factors [29], has been implicated as a transcriptional repressor of antimicrobial genes in the intestine; activation of PMK-1 p38 MAPK de-represses ATF-7, hence upregulating the antimicrobial genes [30]. We therefore analyzed tph-1::gfp expression in atf-7(gf) and atf-7(lf) mutants. atf-7(gf) mutation presumably constitutively represses TIR-1 signaling targets. atf-7(gf) did not downregulate ADF tph-1::gfp under optimal growth conditions (Figure 4A), as seen in mutants with reduced tir-1, TIR-1 downstream nsy-1/MAPKKK and sek-1/MAPKK (Figure 1C, Figure S1) [9]. Like tir-1(lf) mutants, atf-7(gf), as well as daf-19 mutants, failed to upregulate ADF tph-1::gfp following 6 hr feeding on PA14 (Figure 2A, Figure 4B). By contrast, both atf-7(gf) and atf-7(lf) mutants upregulated ADF tph-1::gfp during dauer formation (Figure 4C). These data suggest that ATF-7 confers the specificity to upregulate tph-1 in response to pathogenic bacteria.
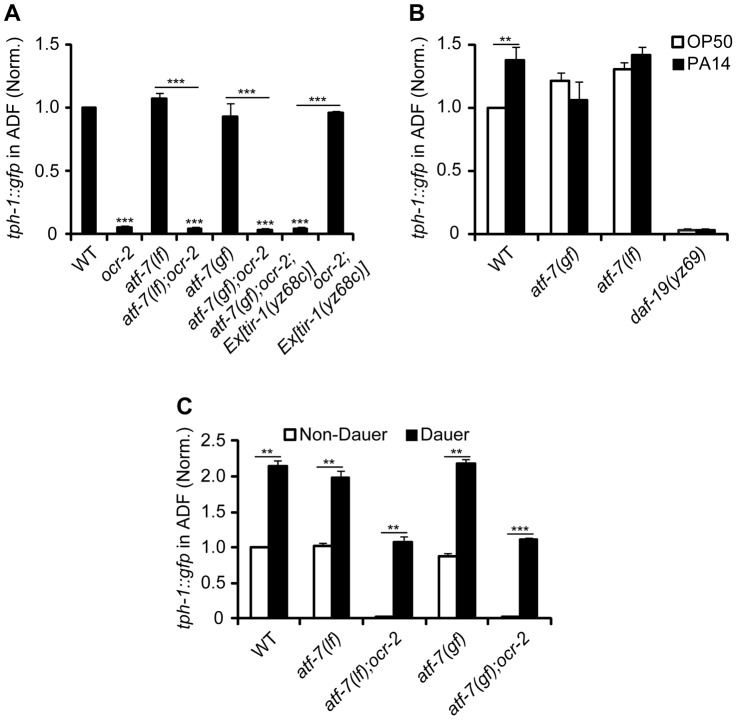
Figure 4. Constitutive repressor function of atf-7(gf) mutation blocks tph-1::gfp upregulation induced by tir-1(gf) and PA14.
A. Quantification of ADF tph-1::gfp in atf-7(lf), atf-7(gf) and atf-7(gf) carrying the Ex[tir-1(yz68gf)] transgene under optimal growth conditions, on their own or in the background of ocr-2 mutation. B. atf-7(lf), atf-7(gf) and daf-19 mutants failed to upregulate ADF tph-1::gfp following 6 hr incubation on a lawn of PA14 relative to their sibling incubated on OP50. One-day-old adults were tested. C. atf-7(lf) and atf-7(gf) mutations, on their own or in ocr-2 mutation background, did not prevent ADF tph-1::gfp upregulation during dauer formation, as compared to L4 worms. Data represent the average of three trials each with at least 20 animals per strain per condition ± SEM. The value of GFP fluorescence in the ADF neurons of WT animals under a stress paradigm and that of mutants is normalized to the value of WT animals under optimal conditions. ** p<0.01, *** p<0.001.
We tested further whether constitutive repression function of atf-7(gf) could suppress tph-1::gfp upregulation by tir-1(yz68gf). We crossed the Pgpa-13::tir-1(yz68c) transgene into atf-7(gf) mutants. The Pgpa-13::tir-1(yz68c) transgene conferred a significantly increase of ADF tph-1::gfp as tested in ocr-2 mutant background but not in ocr-2;atf-7(gf) double mutants (Figure 4A). Thus, tph-1 upregulation induced by PA14 and tir-1(yz68gf) requires both DAF-19 and ATF-7.
daf-19 deficiency suppresses the enhanced innate immunity of tir-1(yz68gf) mutants
daf-19 is expressed broadly in neurons but also in the intestine (Figure S3) [31], [32]. We used epistasis analysis to investigate the role of DAF-19 in TIR-1-mediated innate immunity. When incubated on a lawn of PA14, tir-1(yz68gf) mutants exhibited enhanced resistance to killing by PA14, in contrast to the heightened susceptibility of tir-1(lf) mutants (Figure 5A). daf-19(m86) and daf-19(yz70) mutants exhibited heightened susceptibility to PA14 compared to WT (Figure 5B). Similarly, RNAi of daf-19 enhanced the susceptibility to PA14 compared to mock RNAi (Figure 5C). Transgenic expression of daf-19 cDNA in the intestine partially rescued PA14 resistance in daf-19 mutants (Figure 5D). daf-19(m86)-null mutation suppressed enhanced immune resistance of the tir-1(yz68gf) mutants, showing that the enhanced pathogen resistance of tir-1(yz68gf) mutants also requires DAF-19 function (Figure 5E). However, the daf-19(m86);tir-1(yz68gf) double mutants did not display heightened pathogen susceptibility as seen in daf-19(m86) single mutants, suggesting additional transcriptional regulators involved in the immunity induced by TIR-1 activation.
Figure 5. DAF-19 regulation of TIR-1-mediated innate immunity.
A. Survival rates of WT and loss- and gain-of-function mutants of tir-1 fed PA14 under standard slow killing pathogenesis assay conditions. B. daf-19 mutants displayed elevated susceptibility to killing by PA14. C. Survival rates of vector control and daf-19 RNAi-treated worms fed PA14. D. Transgenic expression of daf-19 in the intestine by the ges-1 promoter partially rescued the elevated PA14 susceptibility of daf-19 mutants. E. daf-19 deficiency suppressed the enhanced PA14 resistance of tir-1(yz68gf) mutants. F. daf-19;atf-7(lf) mutants displayed elevated susceptibility to killing by PA14 similarly to atf-7(lf) mutants. The fraction of live animals was determined at each indicated time points. The experiments were performed in triplicates. Results are representative of at least two independent experiments.
We tested the functional relationship between DAF-19 and ATF-7 in the innate immunity. Both atf-7(gf) and atf-7(lf) mutants exhibited heightened susceptibility to killing by PA14, although the immunodeficiency phenotype of atf-7(lf) mutants is weaker [30]. If ATF-7 and DAF-19 function in parallel, we could expect a stronger immunodeficiency phenotype in a double mutant of atf-7(lf) and daf-19 relative to the single mutants. Contrary to the prediction, atf-7(lf);daf-19 double mutants displayed a survival rate on PA14 comparable to the atf-7(lf) single mutant (Figure 5F). This result is more consistent with the model in which DAF-19 regulates ATF-7 targets in the immune system.
TIR-1- and ATF-7-regulated immune reporter genes are also regulated by DAF-19
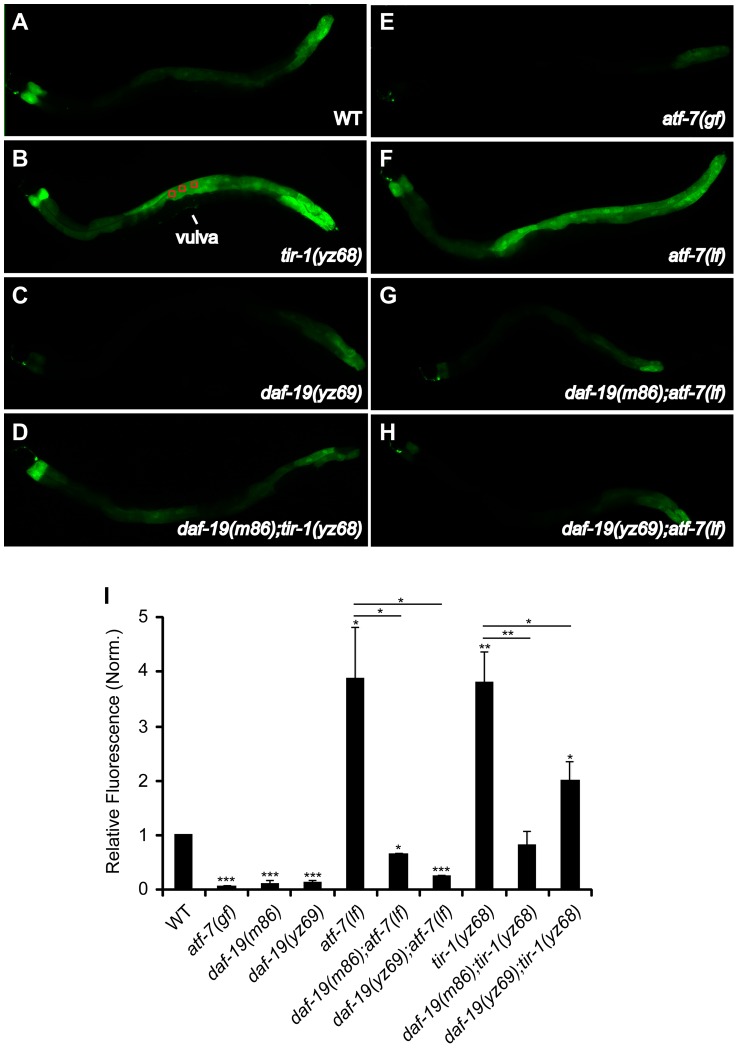
The exact detoxification mechanisms of the C. elegans immunity are not known. To validate the role of DAF-19 in the innate immunity, we made use of the fact that bacterial infections cause intestine to induce the transcription of a battery of secretory proteins that are thought to produce antimicrobial effects [33], [34]. Transcriptional regulation of those candidate antimicrobial genes has been used as an assay for genetic delineation of C. elegans immune pathways [35], [36]. For example, the atf-7(gf) allele, as well as a number of tir-1(lf) alleles, were identified based on the diminished intestinal expression levels of a GFP reporter for the ShK-like toxin peptide gene T24B8.5 (T24B8.5::gfp), and atf-7(lf) alleles were identified as suppressors of the diminished T24B8.5::gfp of atf-7(gf) [30]. We therefore analyzed the same integrated T24B8.5::gfp reporter in tir-1 and daf-19 mutants. On a lawn of standard bacterial food E. coli OP50, T24B8.5::gfp intensity in the intestine of tir-1(yz68) was substantially enhanced relative to WT animals (Figure 6B, 6I), further validating the constitutive activity of tir-1(yz68gf). By contrast, intestinal T24B8.5::gfp was markedly reduced in daf-19 mutants as in atf-7(gf) and tir-1(lf) mutants (Figure 6C, 6E, 6I). Thus, DAF-19 deficiency results in downregulation of an immune gene marker regulated by TIR-1 and ATF-7.
Figure 6. Reduced immune gene marker in the intestine of daf-19 mutants.
A–H. Representative photomicrographs showing two-day-old adult WT and indicated mutants expressing a GFP reporter for the ShK-like toxin peptide gene T24B8.5 (T24B8.5::gfp) in the intestine, all the animals shown with the anterior to the left. I. Quantification of T24B8.5::gfp intensity in the intestine of two-day-old adults. Fluorescence was quantified by measuring pixel intensity of three areas along the body (as depicted in B). T24B8.5::gfp intensity was reduced in atf-7(gf) and two daf-19 alleles, but increased in tir-1(yz68gf) and atf-7(lf) mutants. daf-19 deficiency diminished the increased T24B8.5::gfp in tir-1(yz68gf) as well as atf-7(lf) mutants. Data represent the average of three trials each with 20 animals per strain ± SEM, and the value of GFP fluorescence in mutants is normalized to that of WT animals analyzed on the same day. Statistics between WT and mutants is marked on the top of each bar, and that between two indicated groups is marked on the top of lines, * p<0.05, ** p<0.01, *** p<0.001.
Similar to the effect of the daf-19 mutation on tir-1(yz68gf) immunity (Figure 5E), the daf-19(m86) and daf-19(yz69) mutations diminished the increased T24B8.5::gfp expression in tir-1(yz68gf) mutants, although the GFP level in the daf-19; tir-1(yz68gf) double mutants was higher compared to the daf-19 single mutants (Figure 6D, 6I). We also detected an increase in T24B8.5::gfp in the atf-7(lf) intetine relative to WT animals; two tested daf-19 alleles both reversed the increased T24B8.5::gfp in atf-7(lf) mutants (Figure 6F, 6G, 6H, 6I).
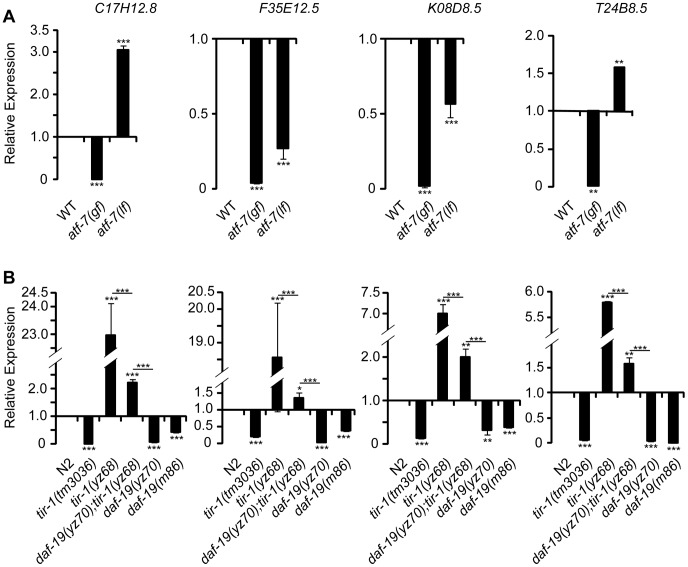
To confirm the role for DAF-19 in the regulation of candidate antimicrobial genes, we used quantitative real-time RT-PCR (qPCR) to measure the expression of endogenous T24B8.5 as well as three other ATF-7-regulated candidate antimicrobial genes. Every tested antimicrobial gene was reduced in two tested daf-19 alleles compared to WT animals (Figure 7B). In contrast, the message levels of these antimicrobial genes were increased in tir-1(yz68gf) mutants (Figure 7B). daf-19 mutation diminished the increases of those antimicrobial genes in tir-1(yz68gf) mutants (Figure 7B), similar to that seen with T24B8.5::gfp in the intestine. As the controls, we analyzed atf-7(lf) and atf-7(gf) mutants. We observed that the message levels of K08D8.5 and F35E12.5 were significantly reduced, and C17H12.8 elevated in the atf-7(lf) mutant (Figure 7A), as previously reported [30]. Consistent with our observation of enhanced T24B8.5::gfp in atf-7(lf) intestine, we found T24B8.5 message level increased in atf-7(lf) mutants (Figure 7A). We did observe dramatically reduced message levels of all tested genes in atf-7(gf) and tir-1(lf) mutants (Figure 7A, 7B), as previously reported [30], [36].
Figure 7. qPCR analysis of predicted antimicrobial genes in daf-19 mutants.
A. Expression levels of four predicted antimicrobial genes in adult atf-7(gf) and atf-7(lf) mutants. B. Expression levels of the predicted antimicrobial genes in adult tir-1(lf), tir-1(yz68gf), two daf-19 alleles and daf-19;tir-1(yz68gf) double mutants. The levels of all tested genes were reduced in both daf-19 alleles and tir-1(lf) alleles. daf-19(lf) diminished the enhanced expression of the antimicrobial genes in tir-1(yz68gf) mutants. For each gene, the expression level in WT is defined as 1, and that of mutants is relative to WT. Experiments were carried out in triplicates. Results are representative of two independent experiments. Analysis of WT and indicated mutants at the L4 stage showed comparable results. Statistics between WT and mutants is marked on the top of each bar, and that between two indicated groups is marked on the top of lines, ** p<0.01, *** p<0.001.
Our data thus far indicated that DAF-19 is required for TIR-1 signaling to upregulate tph-1 in the ADF neurons and candidate antimicrobial genes in the intestine. We wished to determine whether DAF-19 regulates every TIR-1 target, or it selectively mediates TIR-1 regulation of pathogen inducible genes. It has been well established that, TIR-1 specifies asymmetrical expression of the olfactory receptor STR-2 in one of two AWC olfactory neurons during the development [23]. We observed that neither AWC neuron expressed str-2::gfp in the tir-1(yz68gf) mutant, as seen in mutants with excessive TIR-1 activity [23]. However, daf-19 mutants did not exhibit the AWC phenotype seen in tir-1(lf) mutants (Figure S4). Thus, DAF-19 is critical for TIR-1 signaling to induce transcriptional responses to pathogenic bacteria in the 5-HT neurons and the intestine, but is not required for TIR-1 to regulate neural development.
PA14 triggers TIR-1 signaling via tissue-specific mechanisms
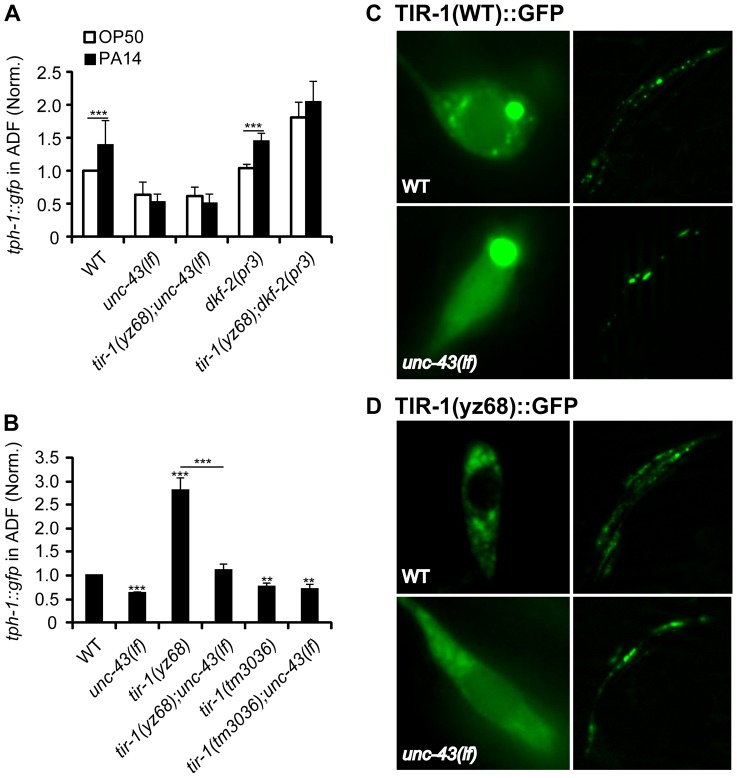
The finding of the shared transcriptional effectors of TIR-1 signaling in the ADF neurons and intestine raised an intriguing question as to whether pathogenic bacterial signals trigger the neurons and immune cells in the same manner in C. elegans. Although little is known about how the worm senses the presence of pathogens and relays the signals to TIR-1, the protein kinase D DKF-2 is thought to promote transcriptional responses to PA14 by activating the TIR-1 signaling pathway [37]. However, we found that the dkf-2(pr3)-null mutation did not suppress tph-1::gfp upregulation by tir-1(yz68gf) or PA14 (Figure 8A).
Figure 8. Regulation of TIR-1 signaling in ADF neurons.
A. tph-1::gfp in the ADF neurons of CaMKII unc-43(lf) and PKD dkf-2(lf) mutants fed PA14 for 6 hr compared with corresponding sibling fed OP50. dkf-2 deficiency cannot block tph-1::gfp upregulation by PA14 or tir-1(yz68gf). unc-43(lf) mutants failed to upregulate ADF tph-1::gfp following PA14 treatment. B. tph-1::gfp in the ADF neurons of unc-43(lf) mutants compared with that in tir-1(lf) and tir-1(gf) mutants under optimal growth conditions. unc-43(lf) suppressed increased tph-1::gfp expression by tir-1(yz68gf). Data represent the average of three trials each with at least 15 animals per strain ± SEM. The value of GFP fluorescence in WT animals fed PA14 and that of mutants fed OP50 or PA14 is normalized to the value of WT animals fed OP50. Statistics between WT and individual mutants is marked on the top of each bar, and that between two indicated groups is marked on the top of lines, ** p<0.01, *** p<0.001. C. Representative images of TIR-1(WT)::GFP driven by the gpa-13 promoter in ADF, ASH and AWC neurons in WT and unc-43(lf) backgrounds. The left panels show the cell bodies of ADF neurons, and the right panels show the axons of the neurons around the nerve ring. In unc-43(lf) background, the number of punctate structures in the ADF cell body was reduced. The number of puncta in the axons was also reduced although the changes appeared to be less pronounced. D. Images of TIR-1(yz68)::GFP driven by the gpa-13 promoter in WT and unc-43(lf) backgrounds. In WT background, TIR-1(yz68)::GFP showed increased punctate structures as compared to TIR-1(WT)::GFP. In unc-43(lf) background TIR-1(yz68)::GFP punctate structures were diminished and the fluorescence in the cell bodies became diffuse.
Previously, we showed that a gain-of-function mutation in the calcium/calmodulin-dependent protein kinase II (CaMKII) UNC-43 upregulates ADF tph-1::gfp [4]. We hypothesized that UNC-43 could be a component of the TIR-1 signaling pathway in the ADF neurons, similar to its involvement in TIR-1-mediated AWC development [23]. To test this hypothesis, we analyzed tph-1::gfp in unc-43(lf);tir-1(yz68) double mutant. We observed that unc-43(lf) abrogated ADF tph-1::gfp upregulation in tir-1(yz68) mutants (Figure 8B). Furthermore, PA14 failed to induce ADF tph-1::gfp upregulation in both the unc-43(lf) single and unc-43(lf);tir-1(yz68gf) double mutants (Figure 8A). We previously showed that unc-43(lf) does not block ADF tph-1::gfp upregulation during dauer formation [5]. These observations together suggest that UNC-43 selectively regulates TIR-1-mediated tph-1 upregulation and that tir-1(yz68gf) cannot bypass UNC-43 function.
In cultured mammalian neurons, CaMKII activation alters the subcellular localization of signaling components to initiate cellular responses [38]. UNC-43 has been shown enriched in postsynaptic sites of a number of neuronal types and was co-immunopecipitated with TIR-1 when co-expressed in cultured mammalian cells [23], [39]. If UNC-43 regulates TIR-1 subcellular distribution, then it is possible that the subcellular distribution of the TIR-1(yz68gf) protein also depends on UNC-43. We tested this idea by comparing the subcellular distribution of GFP-tagged TIR-1 and TIR-1(yz68gf) proteins expressed in chemosensory neurons by the gpa-13 promoter in WT and unc-43(lf) backgrounds. In WT animals, TIR-1(WT)::GFP was observed in punctate structures in the axons around nerve ring, with more diffused fluorescence seen in the cell bodies (Figure 8C). By contrast, TIR-1(yz68gf)::GFP displayed increased punctate structures in the axons as well as in the cell bodies (Figure 8D). Importantly, TIR-1(WT)::GFP and TIR-1(yz68gf)::GFP punctate structures were reduced in unc-43(lf) mutants (Figure 8C, 8D). Based on these observations, we speculate that the C426Y substitution of TIR-1(yz68gf) alters the protein conformation, thereby enhancing its interaction with UNC-43 in particular cellular compartments where TIR-1 interacts with MAPK signaling components, but that TIR-1(yz68gf) cannot efficiently interact with its downstream components in the absence of UNC-43. Collectively, these results suggest that PA14 triggers distinct mechanisms to activate TIR-1 signaling to induce DAF-19/ATF-7 targets in the ADF neurons and the intestine. Our data indicate that UNC-43 is required for TIR-1 signaling in ADF, and DKF-2 is not.
Discussion
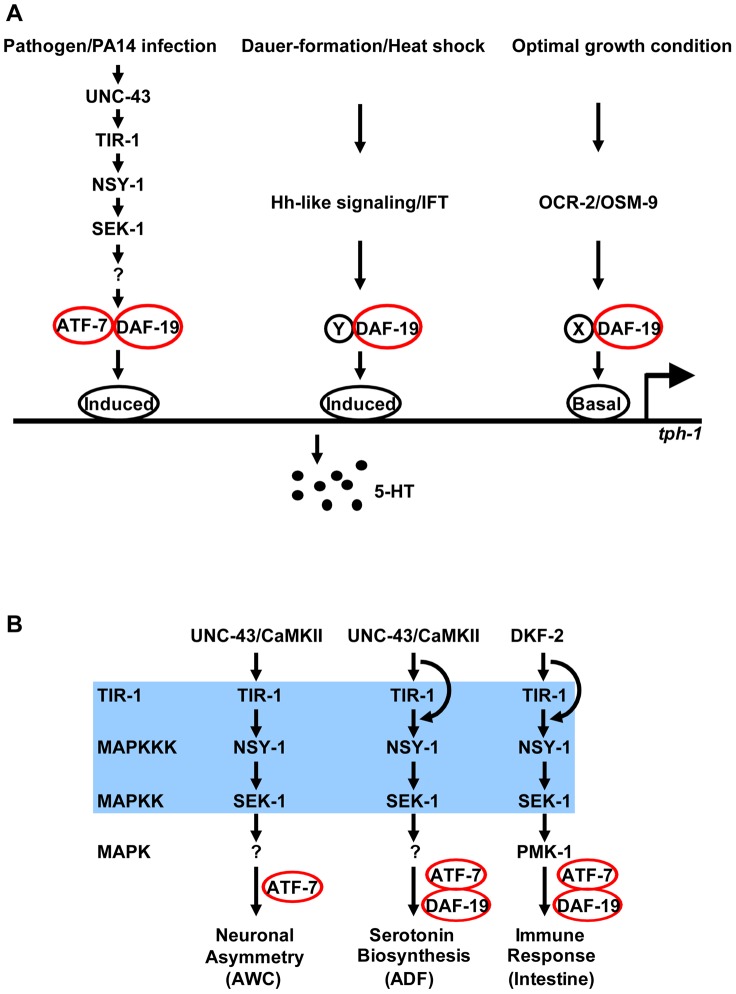
Here, we used an unbiased genetic approach to identify molecular mechanisms underlying transcriptional responses to pathogenic bacterial infection in C. elegans. We identified DAF-19, the ortholog of RFX transcription factors. We showed that DAF-19 acts downstream in the TIR-1 pathway to induce tph-1 in the ADF neurons and antimicrobial genes in the intestine. Our genetic analyses suggest that bacterial infection may trigger the TIR-1 signaling cascade via cell-specific mechanisms, but common TIR-1 downstream transcription factors regulate neuronal and immune responses to infection. Our data demonstrat that RFX gene function, which is required for human adaptive immunity, regulates 5-HT biosynthesis and innate immunity in C. elegans (Figure 9).
Figure 9. DAF-19 regulation of 5-HT biosynthesis and innate immunity.
A. DAF-19 is engaged in multiple independent signaling pathways that regulate tph-1 expression in ADF neurons. Based on the genetic interaction of DAF-19 and ATF-7 in the TIR-1 signaling pathway, we speculate that DAF-19 interacts with different cofactors at different cis-elements of the tph-1 promoter in response to different environmental cues. DAF-19 may simultaneously interact with multiple cofactors to confer additive upregulation of tph-1 in response to multiple aversive cues. B. DAF-19 regulates TIR-1 targets in response to pathogenic bacterial infection, but is not required for TIR-1 to regulate AWC neuron development. However, pathogenic bacterial infection may trigger the TIR-1-DAF-19/ATF-7 pathway via cell-specific mechanisms.
DAF-19 is an ancient transcriptional regulator of innate immunity
Parallel to the C. elegans NSY-1/MAPKKK-SEK-1/MAPKK-PMK-1/p38 MAPK pathway, the ASK-1/MAPKKK to p38 MAPK pathway regulates mouse innate immunity [40]. While members of the NF-κB family of transcription factors are the major effectors in mammalian innate immune responses, the ASK-1-p38 innate immune pathway is independent of NF-κB [40]. Since C. elegans lacks NF-κB, this MAPK pathway was proposed to act via effectors evolutionarily more ancient than NF-κB in the host defense systems [40]. In this study, we identify that DAF-19 RFX is a transcription factor downstream of this core innate immune pathway in C. elegans. Our genetic analyses suggest that DAF-19 concerts with ATF-7, a member of the ATF/CREB superfamily of transcription factors, to upregulate tph-1 in the ADF neurons and antimicrobial factors in the intestine in response to the pathogenic P. aeruginosa strain PA14, reminiscent of the RFX-CREB partnership for human MHC II gene expression.
Several lines of evidence point to RFX factors as an ancient mechanism for enhancing survival under aversive conditions, one predating the divergence of stress responses and immunity. First, RFX is a regulator of cell cycle in a nutrient sensing pathway in Schizosaccharomyces pombe [41] and an effector of the DNA damage and replication checkpoint pathway essential for Saccharomyces cerevisiae survival under replicative stress [42]. Second, biochemical experiments have identified a role for RFX factors in RAS signaling-regulated transcription in mammalian epithelial cells [43]. Third, in addition to mediate transcriptional responses to PA14, daf-19 controls the decision to enter the stress-resistant dauer stage that is specialized for enduring aversive environmental conditions [17]. Fourth, 5-HT is perhaps one of the most ancient mechanisms of stress responses conserved across phyla [44]. Pharmacological and biophysical experiments have long demonstrated that 5-HT biosynthesis in mammals is highly sensitive to environmental conditions and can be upregulated by a variety of metabolic, psychological and physical stressors in a region-specific manner [45], [46], although systematic dissection of 5-HT biosynthesis in the mammalian brain has not been feasible. Our results raise the possibility that the mechanisms that regulate 5-HT biosynthesis and certain aspects of innate immunity may be interrelated.
What could be the mechanism by which DAF-19 regulates tph-1 and those antimicrobial genes? Earlier studies of RFX regulation of human MHC II genes provided evidence that RFX factors regulate gene expression indirectly through recruiting additional transcriptional regulators to the promoters [13]. In a recent study, RFX factors were shown to protect promoters against epigenetic silencing by DNA methylation through recruiting chromatin-remodeling factors [47]. Thus one plausible mechanism could be that DAF-19 and ATF-7 bind to a promoter element shared in common among the pathogen-inducible genes, and infection induces the TIR-1 signaling cascade leading to phosphorylation of ATF-7 [30] and transcriptional activation of the targets. However, genomewide X-box motif search identified hundreds of candidate genes, only a few antimicrobial genes are among them [48] and no X-box can be recognized in the promoter of the tph-1, T24B8.5, C17H12.8, K08D8.5 or F35E12.5 genes. Thus, DAF-19 is likely to bind non-consensus X-boxes on the promoters via co-regulators. Alternatively, DAF-19 may modulate chromatin structure, facilitating the binding of other transcriptional regulators to the promoters. We favor the binding-via-cofactor model because our data thus far suggest that DAF-19 is involved in multiple environment-dependent regulations of tph-1 expression, whereas ATF-7 selectively regulates the response to PA14. We speculate that DAF-19 interacts with distinct cofactors that are regulated by distinct environmental cues (Figure 9A). Further experiments are required to determine whether DAF-19 directly interacts with ATF-7. In addition, the possibility that DAF-19 and ATF-7 regulate yet unidentified transcription factor(s), which in turn regulate pathogen-inducible genes, cannot be excluded. It is perhaps interesting to note that our two daf-19 alleles both are located in the predicted dimerization (DIM) domain, suggestive for the importance of protein-protein interaction in DAF-19 function.
While the exact mechanism of DAF-19 on the target gene promoters remains to be elucidated, our data showed that DAF-19 and ATF-7 regulate common immune gene markers. Remarkably, by comparing the list of TIR-1 signaling targets identified by microarray analysis [36] with the database of several independent expression profiling of daf-19 mutants [49], [50], [51], we found that 102 out of 215 TIR-1 gene targets are among those differentially expressed in daf-19 mutants; one of them is C17H12.8, which was confirmed by our qPCR. We showed that daf-19 mutations can suppress increased expression of immune gene markers in tir-1(yz68gf) mutants. However, daf-19-null;tir-1(yz68gf) double mutants did not display reduced expression of those immune genes as seen in the daf-19 single mutants, judging by T24B8.5::gfp in living worms and qPCR. Consistent with the gene expression analyses, the daf-19-null mutation blocked the enhanced resistance to killing by PA14 in tir-1(yz68gf) mutant but the daf-19;tir-1(yz68gf) double mutant did not exhibit heightened susceptibility as daf-19 single mutants did. These observations may be consistent with the model in which DAF-19 and ATF-7 interact with additional transcription factor(s) that also contribute to the regulation of those antimicrobial genes.
Multiple mechanisms relay pathogen signals to the TIR-1-DAF-19 signaling pathway
There is converging evidence from neurobiology and immunology suggesting that the brain immune privilege is not absolute [52]. Internal and external pathogenic products can infiltrate into the CNS and there is extensive bi-directional communications between the CNS and immune systems [53], [54]. The finding of a large number of classical immune proteins in the CNS has led to the idea that common molecular mechanisms may be involved in neuronal and immune responses to pathogenic signals [1], [2], [53]. Our identification of the TIR-1-DAF-19/ATF-7 pathway in regulating tph-1 and antimicrobial genes supports this idea. Moreover, our genetic analysis suggests that activation mechanisms of this core signaling pathway in the neurons and intestine during infection differ. A prior work showed that DKF-2 is required for TIR-1 signaling cascade to induce intestinal immunity [37]. We found that dkf-2-null did not prevent PA14-induced tph-1::gfp upregulation. Instead, we identified a requirement of UNC-43 CaMKII for tph-1::gfp upregulation induced by PA14 and tir-1(yz68gf). Analysis of TIR-1(WT)::GFP and TIR-1(yz68)::GFP suggests that UNC-43 regulates subcellular distribution of TIR-1. While several mammalian TIR domain adaptor proteins have been shown to translocate to the plasma membrane following Toll-like receptor activation, SARM, the ortholog of TIR-1, is activated in the brain by neural toxicity via a yet unidentified mechanism [55], [56]. Our results raise the possibility that the cue associated with pathogenic bacterial infection triggers Ca2+ signaling to activate TIR-1 in the ADF neurons. It may be sensible that neurons and immune cells detect distinct molecular cues associated with infection thereby coordinating neuronal and physiological responses.
Materials and Methods
Strains
C. elegans strains were maintained at 20°C on NGM agar plates seeded with a lawn of Escherichia coli OP50 as a food source. WT animals were Bristol strain N2. The Hawaiian isolate CB4856 was used in genetic mapping of the daf-19 and tir-1 mutations. The following existing mutant strains were used in this study: atf-7(qd22gf), atf-7(qd22 qd130lf), daf-6(e1377), daf-19(m86), dkf-2(pr3), dyf-1(yz66), eri-1(mg366);lin-15B(n744), ocr-2(yz5), sek-1(km4), tir-1(ok1052), tir-1(tm3036lf), unc-43(e408). Transgenic strains used in this study were: agIs219: Is[T24B8.5::gfp; ttx-3::gfp] [30], CX3695: kyIs140[str-2::gfp; lin-15(+)] [57], GR1333: yzIs71[tph-1::gfp; Rol-6(d)] [21], Is[cat-1::gfp] [58], JY222: ExZ042[ocr-2::gfp; Rol-6(d)] [4], JY449: ExX002[str-2::gfp; Rol-6(d)] [59].
Identification of tir-1(yz68), daf-19(yz69), and daf-19(yz70) mutations
yz68 is a dominant mutation isolated from a genetic screen for mutants with enhanced GFP expression in ADF chemosensory neurons after ethyl methane sulfonate (EMS) mutagenesis of ocr-2(yz5) mutant carrying an integrated tph-1::gfp transgene as described previously [5]. Genetic mapping using single-nucleotide polymorphisms (SNP) of CB4856 localized yz68 to a contig of 1.43 map on the chromosome III. To identify the mutant gene, 174 genes located in the contig were individually inactivated in yz68 mutants by RNAi, and the clone F13B10.1 expressing double stranded(ds)-RNA of tir-1 suppressed the tph-1::gfp upregulation of the yz68 mutant. Sequencing yz68 genomic DNA revealed a G to A transition resulting in a cysteine426 to tyrosine substitution in the fourth ARM motif of the HEAT/Arm repeat region of TIR-1. The amino acid altered in yz68 is in reference of the tir-1a isoform.
yz69 and yz70 mutants are recessive mutations isolated from an EMS mutagenesis screen for mutants with dramatically reduced/absence of ADF tph-1::gfp. Analysis of the amphid morphology with fluorescence dye DiI revealed that ciliated neurons in yz69 and yz70 mutants were dye filling defective. CB4856 SNP mapping localized yz69 to a contig of 1.28 map units between the polymorphisms in the clones C18D1 and F44F4 on the chromosome II. Non-complementation assays with dye-filling mutants within the region indicated that both yz69 and yz70 were allelic to daf-19. Sequencing the daf-19 gene of the mutants revealed in yz69 a G to A transition predicting a cysteine to tyrosine substitution at the conserved dimerization domain, and in yz70 a G to A transition predicting an opal mutation in the dimerization domain. The amino acid changes depicted in Figure 3A are in reference of daf-19a isoform.
Generation of transgenic animals
All constructs were generated by PCR. daf-19(g) was a ∼14.8 kb genomic fragment amplified from the WT genome encompassing 2.9 kb 5′-upstream promoter sequence, exons/introns, and 574 bp 3′-UTR of the daf-19 gene.
To express tir-1 and daf-19 in specific neurons, we fused tir-1 and daf-19 cDNA sequences individually to the gpa-13 promoter, which is expressed in three pairs of amphidal ciliated sensory neurons ADF, ASH and AWC. The gpa-13 promoter is expressed additionally in PHA and PHB phasmid neurons located in the tail [60]. tir-1(WT) and tir-1(yz68) cDNA were amplified from cDNA mixture prepared from total RNA of WT and tir-1(yz68) animals, respectively, using primers corresponding to the tir-1a isoform. The cDNA of the daf-19c isoform was amplified from the plasmid PS0243 (kindly provided by P. Swoboda). The 2.6 kb gpa-13 promoter sequence amplified from the plasmid PS0243 was fused to the sequences in the order of a cDNA, GFP and unc-54 3′UTR or a cDNA, mCherry and unc-54 3′UTR. To express DAF-19 in the intestine, the cDNA sequence of daf-19a isoform was fused to the 2.9 kb ges-1 promoter. For each construct, products from three independent PCR reactions were pooled to reduce potential PCR errors. The pooled PCR products were purified (Qiagen) and microinjected at the concentration of 50 ng/µl into worms. The plasmid containing either a dominant rol-6 gene (Rol-6(d)), elt-2::gfp or unc-122::RFP was co-injected as a transgenic marker.
RNAi
All RNA interference (RNAi) experiments were done in the background of eri-1;lin-15B, which enhances sensitivity to RNA interference in neurons [61]. RNAi assays were carried out by feeding worms E. coli HT1115 expressing dsRNA of a target gene or the control empty L4440 vector (Ahringer RNAi library, University of Cambridge, England). RNAi clones were individually cultured overnight in Luria broth containing 100 µg/ml ampicillin, 500 µl of the bacterial culture were seeded onto agar plates containing NGM supplemented with 1 mM IPTG and 25 µg/ml carbenicillin to induce dsRNA expression, and incubated overnight at room temperature. For RNAi of tir-1 and nsy-1, about 60 eggs were placed onto each plate and allowed to hatch, grow to adults and lay eggs. F1 progeny were transferred to a fresh plate, and tph-1::gfp in the ADF neurons of L4-stage worms of F2 generation quantified. For developmental RNAi of daf-19, synchronized L1 worms were transferred to the plates (day 0), the worms were transferred to freshly prepared plates every day, and the expression of tph-1::gfp in ADF neurons or DiI staining of cilia morphology were analyzed on indicated days. DiI staining was done as previously described [5]. For each RNAi experiment, three independent trials each with three replicates were performed, and data from one representative trial presented.
Indirect immunofluorescence microscopy
Whole-mount staining of worms with anti-5-HT antibody was performed as described previously [21]. The staining patterns were visualized via Alexa Fluor 594 or 488 conjugated secondary antibodies (Molecular Probe) under an AxioImager Z1 microscope equipped with proper filters, and images were captured using AxioCam MR digital camera (Zeiss, Northwood, NY). To quantify the intensity of 5-HT immunoreactivity, images of ADF neurons in individual worms were captured under a 40× objective lens at a fixed exposure time of 3 ms with 100% UV exposure level. For each image, fluorescence intensity of a circular 10 pixels area within the ADF cell body was quantified using the ImageJ software (National Institute of Health, Bethesda, Maryland). To exclude the background, fluorescence intensity over a circular 10 pixels area posterior to the ADF cell body in the same image was quantified, and the value of the background was subtracted from the value of the ADF area.
Assessment of GFP reporter levels and statistics
The expression of a chromosomally integrated tph-1::gfp reporter in ADF neurons in living WT or mutant worms was evaluated by measuring GFP fluorescence intensity. Images of ADF neurons in individual animals were captured at a fixed exposure time. The external contour of each ADF neuron was delineated, and fluorescence intensity within the entire neuron was quantified by using the ImageJ software. L4-stage animals were examined, unless noted otherwise. Ideally mutants that reduce and that upregulate tph-1::gfp were assayed in parallel, thus the exposure time was designed to detect reduction as well as upregulation of tph-1::gfp. This setting was most reliable at the range of 1–3 fold higher than WT; consequently, we used ocr-2 mutant background to lower the basal tph-1::gfp to evaluate the changes of mutants with constitutively increased ADF tph-1::gfp, and the double mutants were compared with ocr-2 single mutants assayed on the same day.
For quantifying tph-1::gfp intensity following PA14 treatment, one-day-old young adult worms were transferred to standard slow killing assay plates [62] seeded with either PA14 or control OP50, incubated at 25°C for 6 hr, images of the ADF neurons were captured and GFP intensity quantified.
For quantifying tph-1::gfp intensity in dauers, WT and mutants were induced to form dauers by dauer pheromones. Dauer pheromone and pheromone-containing plates were prepared as we have done previously [5], following an established protocol [63]. 20–25 gravid worms from each strain were transferred to NGM plates supplied with 1 unit of dauer pheromone, allowed to lay eggs for 2–3 hr in a 25°C incubator, the adults were then removed from the plates, and dauers developed from hatched eggs on the plates were analyzed 72 hr later. For each strain the value of dauers was compared to that of L4 grown on the plates without pheromone and assayed on the same day.
The expression of T24B8.5::gfp in the intestine was analyzed in two-day-old adult worms carrying the integrated agIs219[T24B8.5::gfp; ttx-3::gfp] transgene cultured on OP50 at 20°C. Images of the intestine were captured under a 10× objective lens at a fixed exposure time, and fluorescence intensity was quantified by measuring pixel intensity of three areas along the body of each animal as depicted in Figure 6B.
Data represent the average of at least three trials unless specified otherwise. For each trial, 15–20 animals per strain per condition/treatment were analyzed and compared to the controls assayed on the same day. WT animals under the same conditions and treatments were analyzed for every experiment. Unpaired Student's t-test was used for comparisons between a mutant and WT and between two mutants or two treatments.
P. aeruginosa PA14 pathogenesis assays
The standard PA14 slow killing assays were performed as previously described [62]. Briefly, PA14 was cultured in King's broth overnight and the culture was seeded at the center of 3.5 cm diameter assay plates and incubated at 37°C for 20 hr followed by 20 hr incubation at room temperature. 40–50 L4 worms per strain were transferred onto each assay plate, incubated at 25°C and scored for dead or live every 8 hr. Worms were scored as dead if no response was detected after prodding with a platinum wire. daf-19 mutants tend to claw off the plate. So for each assay, more than 300 L4 daf-19 mutants were transferred to each plate, and live and dead animals on the agar surface were scored at indicated time points; dead animals on the wall of the plate not counted. For each strain, three replicates were analyzed for each experiment. To test the effect of RNAi of daf-19, 4 to 6 gravid animals were grown on the RNAi plates as described above seed with either E-coli HT1115 harboring empty control plasmid L4440 or plasmids expressing RNAi against daf-19 gene. F1 progeny at the L4 stage were used to test the susceptibility to killing by PA14.
Quantitative real-time PCR (qPCR) analysis
Total RNA from 100 one-day-old adults of WT and indicated mutant strains was extracted using Trizol (Invitrogen), reserve transcribed to cDNA using the SuperScript III system (Invitrogen), and the cDNA was used for qPCR analyses using the StepOnePlus machine (Applied Biosystems) and SYBR Green detection system (Applied Biosystems) in triplicated reactions. The primers for qPCR were designed using Primer Premier 5.0 (Premier Biosoft) (Figure S5). Values were normalized against the reference gene act-1 [37]. gpd-2 was analyzed as a second control showing no change relative to act-1. Fold change was calculated using the delta Ct method [64]. qPCR analysis of L4 worms of those strains showed comparable results; data of the adults are presented.
Supporting Information
Regulation of tph-1::gfp expression in the ADF neurons by TIR-1 downstream nsy-1 MAPKKK and sek-1 MAPKK RNAi of nsy-1 suppressed tph-1::gfp upregulation by tir-1(yz68gf). RNAi of nsy-1 and loss-of-function mutation of sek-1 did not confer tph-1::gfp reduction, indicating that the TIR-1 signaling pathway is designated primarily to upregulate tph-1 expression in response to pathogen infection. Data represent the average of three trials each with at least 15 animals per strain ± SEM. The value of GFP fluorescence in mutants was normalized to that of WT animals, and the value of RNAi-treated animals is normalized to that of mock RNAi with an empty vector. Statistics between WT and individual mutants and RNAi-treated animals is marked on the top of each bar, and that between two indicated groups is marked on the top of lines, *** p<0.001, unpaired student's t test.
(TIF)
tph-1::gfp in the ADF neurons of the IFT mutant dyf-1. dyf-1 mutant animals were treated with a mock RNAi with an empty vector or a vector expressing RNAi against tir-1 or daf-19. RNAi of tir-1 did not reduce tph-1::gfp in the ADF neurons, compared to mock RNAi. By contrast, RNAi of daf-19 abolished ADF tph-1::gfp. Data represent the average of three trials each with at least 15 animals per strain ± SEM. The value of RNAi of tir-1 and daf-19 is normalized to that of dyf-1 mutants treated with the empty vector. *** p<0.001, unpaired student's t test.
(TIF)
daf-19 is expressed in the intestine. Top, an image of L4 worm expressing GFP driven by a genomic fragment encompassing 2.9 kb 5′-upstream sequence to exon 8 of daf-19. Bottom, a bright field image showing the position of the same worm. Anterior is to the left, and arrows point to the nuclei of the intestinal cells.
(TIF)
DAF-19 is not required for TIR-1 regulation of AWC cell fates, while atf-7 is. A–D. Photomicrographs showing L4 animals expressing an integrated GFP reporter for the olfactory receptor str-2 (str-2::gfp). daf-19 and WT animals expressed str-2::gfp stochastically in one of two AWC neurons. Neither AWC expressed str-2::gfp in tir-1(yz68gf) mutants. E. Quantification of str-2::gfp expression in AWC neurons in daf-19, atf-7 and tir-1 signaling mutants. atf-7(gf) mutants exhibited str-2::gfp expression pattern as seen in tir-1(lf) mutants, but atf-7(lf) showed mixed str-2::gfp patterns of tir-1(lf) and tir-1(yz68gf). daf-19 did not display the str-2::gfp phenotype of tir-1(lf), nor suppressed the str-2::gfp phenotype of tir-1(yz68gf). Data represent the percentage of the animals of a strain showing each AWC phenotype. n, number of animals analyzed.
(TIF)
Primer sequences used for qPCR analysis of candidate antimicrobial genes.
(TIF)
Acknowledgments
We thank D. Kim, C. Rubin, and the Caenorhabditis Genetics Center for C. elegans strains. Special thanks to P. Swoboda for sending us daf-19 expressing vectors and to Shohei Mitani and Tokyo Women's Medical College (Tokyo, Janpan) for tir-1 deletion alleles.
Funding Statement
This work is supported by the NIMH grant MH64747. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Boulanger LM (2009) Immune proteins in brain development and synaptic plasticity. Neuron 64: 93–109. [DOI] [PubMed] [Google Scholar]
- 2. Veerhuis R, Nielsen HM, Tenner AJ (2011) Complement in the brain. Molecular immunology 48: 1592–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shatz CJ (2009) MHC class I: an unexpected role in neuronal plasticity. Neuron 64: 40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang S, Sokolchik I, Blanco G, Sze JY (2004) Caenorhabditis elegans TRPV ion channel regulates 5HT biosynthesis in chemosensory neurons. Development 131: 1629–1638. [DOI] [PubMed] [Google Scholar]
- 5. Moussaif M, Sze JY (2009) Intraflagellar transport/Hedgehog-related signaling components couple sensory cilium morphology and serotonin biosynthesis in Caenorhabditis elegans. The Journal of neuroscience : the official journal of the Society for Neuroscience 29: 4065–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sifri CD, Begun J, Ausubel FM (2005) The worm has turned–microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol 13: 119–127. [DOI] [PubMed] [Google Scholar]
- 7. Zhang Y, Lu H, Bargmann CI (2005) Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438: 179–184. [DOI] [PubMed] [Google Scholar]
- 8. Liberati NT, Fitzgerald KA, Kim DH, Feinbaum R, Golenbock DT, et al. (2004) Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proceedings of the National Academy of Sciences of the United States of America 101: 6593–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shivers RP, Kooistra T, Chu SW, Pagano DJ, Kim DH (2009) Tissue-specific activities of an immune signaling module regulate physiological responses to pathogenic and nutritional bacteria in C. elegans. Cell Host Microbe 6: 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Newton K, Dixit VM (2012) Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol 5 doi: 10.1101/cshperspect.a011247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, et al. (2001) A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Current biology : CB 11: 809–821. [DOI] [PubMed] [Google Scholar]
- 12. Reith W, Mach B (2001) The bare lymphocyte syndrome and the regulation of MHC expression. Annu Rev Immunol 19: 331–373. [DOI] [PubMed] [Google Scholar]
- 13. Reith W, LeibundGut-Landmann S, Waldburger JM (2005) Regulation of MHC class II gene expression by the class II transactivator. Nat Rev Immunol 5: 793–806. [DOI] [PubMed] [Google Scholar]
- 14. Reith W, Ucla C, Barras E, Gaud A, Durand B, et al. (1994) RFX1, a transactivator of hepatitis B virus enhancer I, belongs to a novel family of homodimeric and heterodimeric DNA-binding proteins. Mol Cell Biol 14: 1230–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chu JS, Baillie DL, Chen N (2010) Convergent evolution of RFX transcription factors and ciliary genes predated the origin of metazoans. BMC evolutionary biology 10: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Piasecki BP, Burghoorn J, Swoboda P (2010) Regulatory Factor X (RFX)-mediated transcriptional rewiring of ciliary genes in animals. Proceedings of the National Academy of Sciences of the United States of America 107: 12969–12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Swoboda P, Adler HT, Thomas JH (2000) The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol Cell 5: 411–421. [DOI] [PubMed] [Google Scholar]
- 18. Wang J, Schwartz HT, Barr MM (2010) Functional specialization of sensory cilia by an RFX transcription factor isoform. Genetics 186: 1295–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dubruille R, Laurencon A, Vandaele C, Shishido E, Coulon-Bublex M, et al. (2002) Drosophila regulatory factor X is necessary for ciliated sensory neuron differentiation. Development 129: 5487–5498. [DOI] [PubMed] [Google Scholar]
- 20. El Zein L, Ait-Lounis A, Morle L, Thomas J, Chhin B, et al. (2009) RFX3 governs growth and beating efficiency of motile cilia in mouse and controls the expression of genes involved in human ciliopathies. Journal of cell science 122: 3180–3189. [DOI] [PubMed] [Google Scholar]
- 21. Sze JY, Victor M, Loer C, Shi Y, Ruvkun G (2000) Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature 403: 560–564. [DOI] [PubMed] [Google Scholar]
- 22. White JG, Southgate E, Thomson JN, Brenner S (1986) The Structure of the Nervous-System of the Nematode Caenorhabditis-Elegans. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences 314: 1–340. [DOI] [PubMed] [Google Scholar]
- 23. Chuang CF, Bargmann CI (2005) A Toll-interleukin 1 repeat protein at the synapse specifies asymmetric odorant receptor expression via ASK1 MAPKKK signaling. Genes Dev 19: 270–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kurz CL, Shapira M, Chen K, Baillie DL, Tan MW (2007) Caenorhabditis elegans pgp-5 is involved in resistance to bacterial infection and heavy metal and its regulation requires TIR-1 and a p38 map kinase cascade. Biochemical and biophysical research communications 363: 438–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu PJ (2007) Dauer. WormBook 1–19 Available: http:///www.wormbase.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Perens EA, Shaham S (2005) C. elegans daf-6 encodes a patched-related protein required for lumen formation. Dev Cell 8: 893–906. [DOI] [PubMed] [Google Scholar]
- 27. Starich TA, Herman RK, Kari CK, Yeh WH, Schackwitz WS, et al. (1995) Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics 139: 171–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Duerr JS, Frisby DL, Gaskin J, Duke A, Asermely K, et al. (1999) The cat-1 gene of Caenorhabditis elegans encodes a vesicular monoamine transporter required for specific monoamine-dependent behaviors. The Journal of neuroscience : the official journal of the Society for Neuroscience 19: 72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Amoutzias GD, Veron AS, Weiner J 3rd, Robinson-Rechavi M, Bornberg-Bauer E, et al. (2007) One billion years of bZIP transcription factor evolution: conservation and change in dimerization and DNA-binding site specificity. Molecular biology and evolution 24: 827–835. [DOI] [PubMed] [Google Scholar]
- 30. Shivers RP, Pagano DJ, Kooistra T, Richardson CE, Reddy KC, et al. (2010) Phosphorylation of the conserved transcription factor ATF-7 by PMK-1 p38 MAPK regulates innate immunity in Caenorhabditis elegans. PLoS Genet 6: e1000892 doi:10.1371/journal.pgen.1000892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Senti G, Swoboda P (2008) Distinct isoforms of the RFX transcription factor DAF-19 regulate ciliogenesis and maintenance of synaptic activity. Mol Biol Cell 19: 5517–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reece-Hoyes JS, Shingles J, Dupuy D, Grove CA, Walhout AJ, et al. (2007) Insight into transcription factor gene duplication from Caenorhabditis elegans Promoterome-driven expression patterns. BMC genomics 8: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kurz CL, Tan MW (2004) Regulation of aging and innate immunity in C. elegans. Aging Cell 3: 185–193. [DOI] [PubMed] [Google Scholar]
- 34. Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, et al. (2002) Inducible antibacterial defense system in C. elegans. Current biology : CB 12: 1209–1214. [DOI] [PubMed] [Google Scholar]
- 35. Styer KL, Singh V, Macosko E, Steele SE, Bargmann CI, et al. (2008) Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science 322: 460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, et al. (2006) p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet 2: e183 doi:10.1371/journal.pgen.0020183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ren M, Feng H, Fu Y, Land M, Rubin CS (2009) Protein kinase D is an essential regulator of C. elegans innate immunity. Immunity 30: 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wayman GA, Tokumitsu H, Davare MA, Soderling TR (2011) Analysis of CaM-kinase signaling in cells. Cell calcium 50: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rongo C, Whitfield CW, Rodal A, Kim SK, Kaplan JM (1998) LIN-10 is a shared component of the polarized protein localization pathways in neurons and epithelia. Cell 94: 751–759. [DOI] [PubMed] [Google Scholar]
- 40. Matsuzawa A, Saegusa K, Noguchi T, Sadamitsu C, Nishitoh H, et al. (2005) ROS-dependent activation of the TRAF6-ASK1-p38 pathway is selectively required for TLR4-mediated innate immunity. Nat Immunol 6: 587–592. [DOI] [PubMed] [Google Scholar]
- 41. Wu SY, McLeod M (1995) The sak1+ gene of Schizosaccharomyces pombe encodes an RFX family DNA-binding protein that positively regulates cyclic AMP-dependent protein kinase-mediated exit from the mitotic cell cycle. Mol Cell Biol 15: 1479–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang M, Zhou Z, Elledge SJ (1998) The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94: 595–605. [DOI] [PubMed] [Google Scholar]
- 43. Maijgren S, Sur I, Nilsson M, Toftgard R (2004) Involvement of RFX proteins in transcriptional activation from a Ras-responsive enhancer element. Archives of dermatological research 295: 482–489. [DOI] [PubMed] [Google Scholar]
- 44. Azmitia EC (2007) Serotonin and brain: evolution, neuroplasticity, and homeostasis. International review of neurobiology 77: 31–56. [DOI] [PubMed] [Google Scholar]
- 45. Chaouloff F, Berton O, Mormede P (1999) Serotonin and stress. Neuropsychopharmacology 21: 28S–32S. [DOI] [PubMed] [Google Scholar]
- 46. Chen GL, Miller GM (2012) Advances in tryptophan hydroxylase-2 gene expression regulation: new insights into serotonin-stress interaction and clinical implications. Am J Med Genet B Neuropsychiatr Genet 159B: 152–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Seguin-Estevez Q, De Palma R, Krawczyk M, Leimgruber E, Villard J, et al. (2009) The transcription factor RFX protects MHC class II genes against epigenetic silencing by DNA methylation. Journal of immunology 183: 2545–2553. [DOI] [PubMed] [Google Scholar]
- 48. Efimenko E, Bubb K, Mak HY, Holzman T, Leroux MR, et al. (2005) Analysis of xbx genes in C. elegans. Development 132: 1923–1934. [DOI] [PubMed] [Google Scholar]
- 49. Chen N, Mah A, Blacque OE, Chu J, Phgora K, et al. (2006) Identification of ciliary and ciliopathy genes in Caenorhabditis elegans through comparative genomics. Genome biology 7: R126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Phirke P, Efimenko E, Mohan S, Burghoorn J, Crona F, et al. (2011) Transcriptional profiling of C. elegans DAF-19 uncovers a ciliary base-associated protein and a CDK/CCRK/LF2p-related kinase required for intraflagellar transport. Developmental biology 357: 235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Blacque OE, Perens EA, Boroevich KA, Inglis PN, Li C, et al. (2005) Functional genomics of the cilium, a sensory organelle. Current biology : CB 15: 935–941. [DOI] [PubMed] [Google Scholar]
- 52. McAllister AK, van de Water J (2009) Breaking boundaries in neural-immune interactions. Neuron 64: 9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sternberg EM (2006) Neural regulation of innate immunity: a coordinated nonspecific host response to pathogens. Nat Rev Immunol 6: 318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Irwin MR, Cole SW (2011) Reciprocal regulation of the neural and innate immune systems. Nat Rev Immunol 11: 625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim Y, Zhou P, Qian L, Chuang JZ, Lee J, et al. (2007) MyD88-5 links mitochondria, microtubules, and JNK3 in neurons and regulates neuronal survival. The Journal of experimental medicine 204: 2063–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. O'Neill LA, Bowie AG (2007) The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nature reviews Immunology 7: 353–364. [DOI] [PubMed] [Google Scholar]
- 57. Troemel ER, Sagasti A, Bargmann CI (1999) Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell 99: 387–398. [DOI] [PubMed] [Google Scholar]
- 58. Sze JY, Zhang S, Li J, Ruvkun G (2002) The C. elegans POU-domain transcription factor UNC-86 regulates the tph-1 tryptophan hydroxylase gene and neurite outgrowth in specific serotonergic neurons. Development 129: 3901–3911. [DOI] [PubMed] [Google Scholar]
- 59. Zheng X, Chung S, Tanabe T, Sze JY (2005) Cell-type specific regulation of serotonergic identity by the C. elegans LIM-homeodomain factor LIM-4. Dev Biol 286: 618–628. [DOI] [PubMed] [Google Scholar]
- 60. Jansen G, Thijssen KL, Werner P, van der Horst M, Hazendonk E, et al. (1999) The complete family of genes encoding G proteins of Caenorhabditis elegans. Nature genetics 21: 414–419. [DOI] [PubMed] [Google Scholar]
- 61. Sieburth D, Ch'ng Q, Dybbs M, Tavazoie M, Kennedy S, et al. (2005) Systematic analysis of genes required for synapse structure and function. Nature 436: 510–517. [DOI] [PubMed] [Google Scholar]
- 62. Tan MW, Mahajan-Miklos S, Ausubel FM (1999) Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proceedings of the National Academy of Sciences of the United States of America 96: 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vowels JJ, Thomas JH (1994) Multiple chemosensory defects in daf-11 and daf-21 mutants of Caenorhabditis elegans. Genetics 138: 303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Regulation of tph-1::gfp expression in the ADF neurons by TIR-1 downstream nsy-1 MAPKKK and sek-1 MAPKK RNAi of nsy-1 suppressed tph-1::gfp upregulation by tir-1(yz68gf). RNAi of nsy-1 and loss-of-function mutation of sek-1 did not confer tph-1::gfp reduction, indicating that the TIR-1 signaling pathway is designated primarily to upregulate tph-1 expression in response to pathogen infection. Data represent the average of three trials each with at least 15 animals per strain ± SEM. The value of GFP fluorescence in mutants was normalized to that of WT animals, and the value of RNAi-treated animals is normalized to that of mock RNAi with an empty vector. Statistics between WT and individual mutants and RNAi-treated animals is marked on the top of each bar, and that between two indicated groups is marked on the top of lines, *** p<0.001, unpaired student's t test.
(TIF)
tph-1::gfp in the ADF neurons of the IFT mutant dyf-1. dyf-1 mutant animals were treated with a mock RNAi with an empty vector or a vector expressing RNAi against tir-1 or daf-19. RNAi of tir-1 did not reduce tph-1::gfp in the ADF neurons, compared to mock RNAi. By contrast, RNAi of daf-19 abolished ADF tph-1::gfp. Data represent the average of three trials each with at least 15 animals per strain ± SEM. The value of RNAi of tir-1 and daf-19 is normalized to that of dyf-1 mutants treated with the empty vector. *** p<0.001, unpaired student's t test.
(TIF)
daf-19 is expressed in the intestine. Top, an image of L4 worm expressing GFP driven by a genomic fragment encompassing 2.9 kb 5′-upstream sequence to exon 8 of daf-19. Bottom, a bright field image showing the position of the same worm. Anterior is to the left, and arrows point to the nuclei of the intestinal cells.
(TIF)
DAF-19 is not required for TIR-1 regulation of AWC cell fates, while atf-7 is. A–D. Photomicrographs showing L4 animals expressing an integrated GFP reporter for the olfactory receptor str-2 (str-2::gfp). daf-19 and WT animals expressed str-2::gfp stochastically in one of two AWC neurons. Neither AWC expressed str-2::gfp in tir-1(yz68gf) mutants. E. Quantification of str-2::gfp expression in AWC neurons in daf-19, atf-7 and tir-1 signaling mutants. atf-7(gf) mutants exhibited str-2::gfp expression pattern as seen in tir-1(lf) mutants, but atf-7(lf) showed mixed str-2::gfp patterns of tir-1(lf) and tir-1(yz68gf). daf-19 did not display the str-2::gfp phenotype of tir-1(lf), nor suppressed the str-2::gfp phenotype of tir-1(yz68gf). Data represent the percentage of the animals of a strain showing each AWC phenotype. n, number of animals analyzed.
(TIF)
Primer sequences used for qPCR analysis of candidate antimicrobial genes.
(TIF)