Abstract
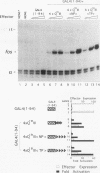
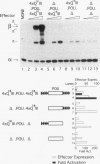
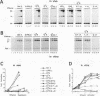
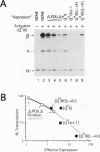
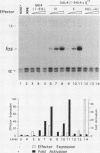
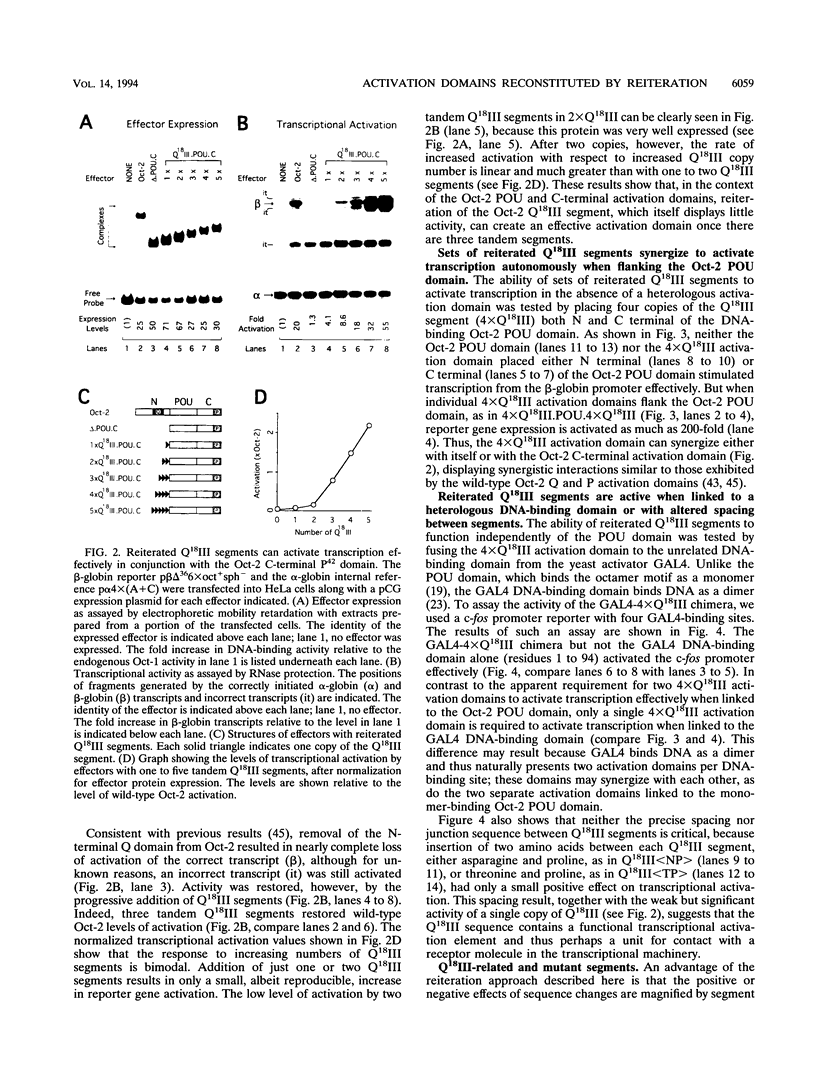
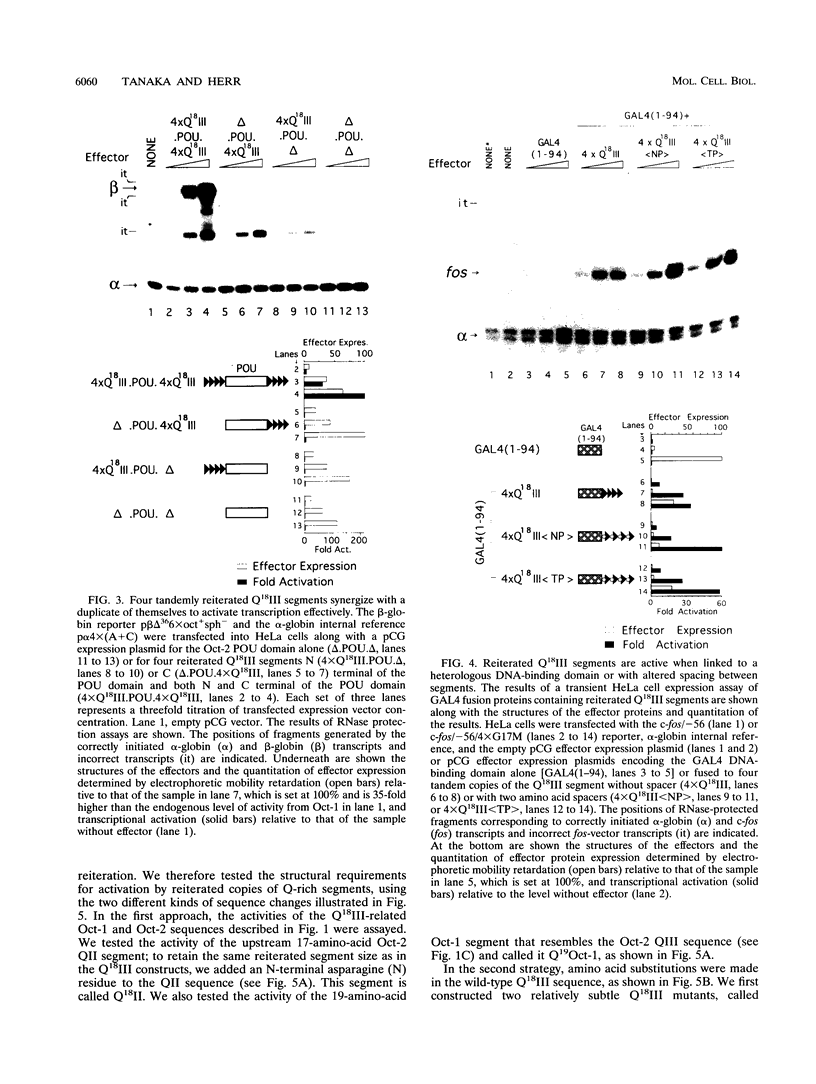
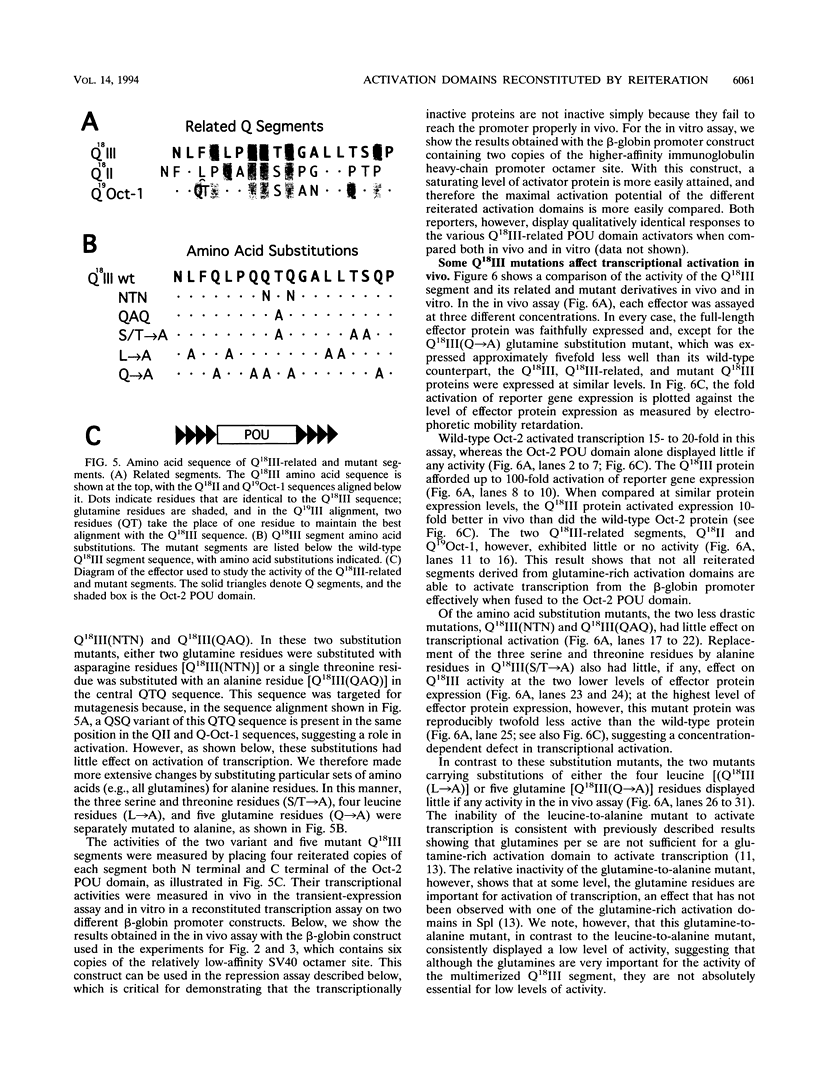
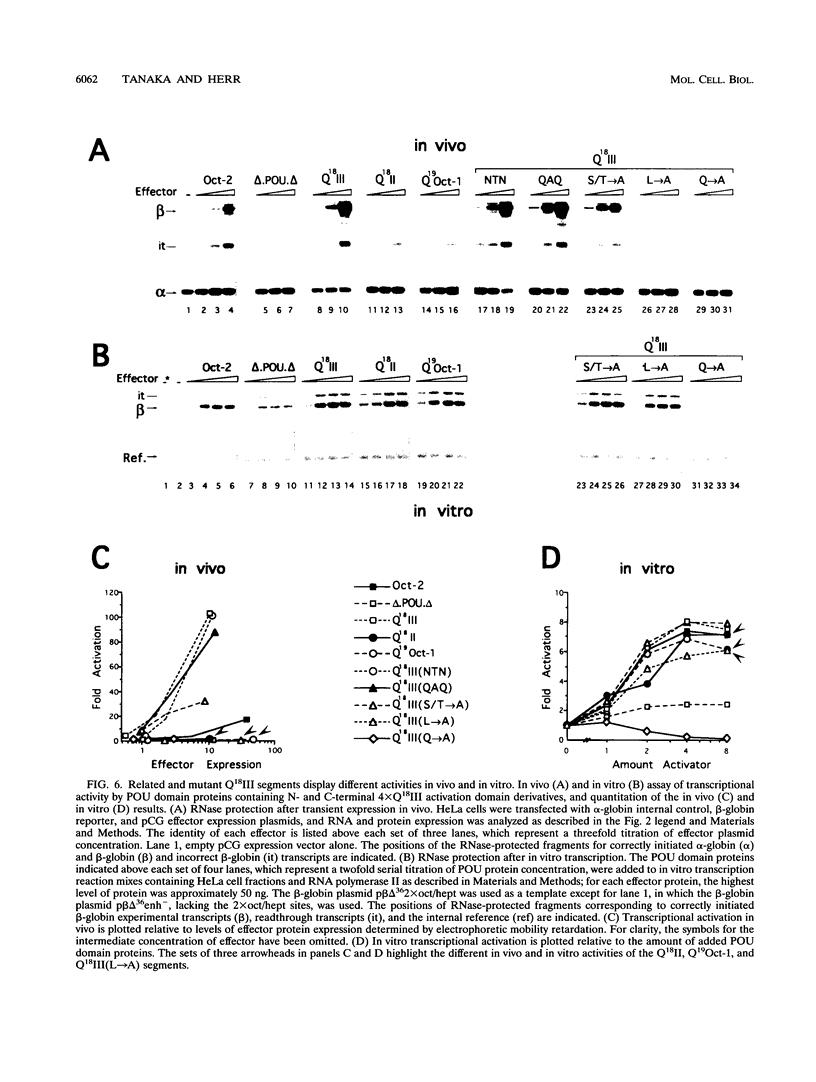
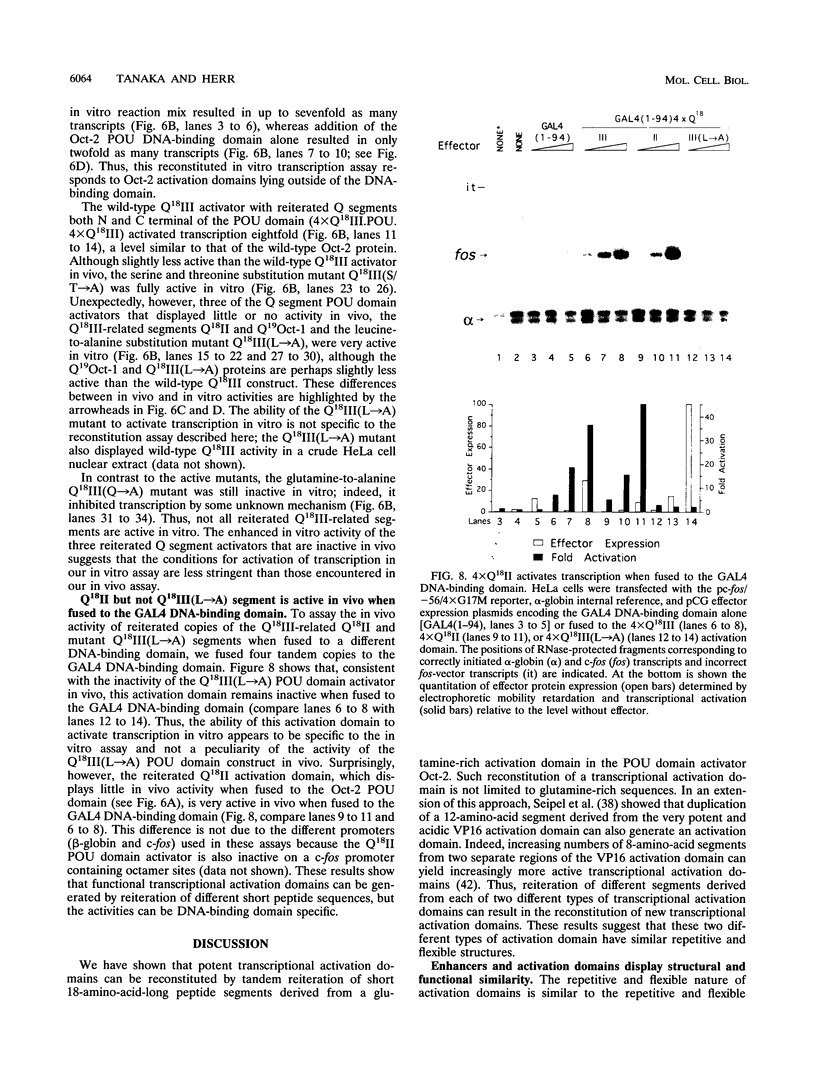
The POU domain activator Oct-2 contains an N-terminal glutamine-rich transcriptional activation domain. An 18-amino-acid segment (Q18III) from this region reconstituted a fully functional activation domain when tandemly reiterated and fused to either the Oct-2 or GAL4 DNA-binding domain. A minimal transcriptional activation domain likely requires three tandem Q18III segments, because one or two tandem Q18III segments displayed little activity, whereas three to five tandem segments were active and displayed increasing activity with increasing copy number. As with natural Oct-2 activation domains, in our assay a reiterated activation domain required a second homologous or heterologous activation domain to stimulate transcription effectively when fused to the Oct-2 POU domain. These results suggest that there are different levels of synergy within and among activation domains. Analysis of reiterated activation domains containing mutated Q18III segments revealed that leucines and glutamines, but not serines or threonines, are critical for activity in vivo. Curiously, several reiterated activation domains that were inactive in vivo were active in vitro, suggesting that there are significant functional differences in our in vivo and in vitro assays. Reiteration of a second 18-amino-acid segment from the Oct-2 glutamine-rich activation domain (Q18II) was also active, but its activity was DNA-binding domain specific, because it was active when fused to the GAL4 than to the Oct-2 DNA-binding domain. The ability of separate short peptide segments derived from a single transcriptional activation domain to activate transcription after tandem reiteration emphasizes the flexible and modular nature of a transcriptional activation domain.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brent R., Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985 Dec;43(3 Pt 2):729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- Brown J. H., Jardetzky T. S., Gorga J. C., Stern L. J., Urban R. G., Strominger J. L., Wiley D. C. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993 Jul 1;364(6432):33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- Clerc R. G., Corcoran L. M., LeBowitz J. H., Baltimore D., Sharp P. A. The B-cell-specific Oct-2 protein contains POU box- and homeo box-type domains. Genes Dev. 1988 Dec;2(12A):1570–1581. doi: 10.1101/gad.2.12a.1570. [DOI] [PubMed] [Google Scholar]
- Cress W. D., Triezenberg S. J. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991 Jan 4;251(4989):87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- Davidson I., Xiao J. H., Rosales R., Staub A., Chambon P. The HeLa cell protein TEF-1 binds specifically and cooperatively to two SV40 enhancer motifs of unrelated sequence. Cell. 1988 Sep 23;54(7):931–942. doi: 10.1016/0092-8674(88)90108-0. [DOI] [PubMed] [Google Scholar]
- Donaldson L., Capone J. P. Purification and characterization of the carboxyl-terminal transactivation domain of Vmw65 from herpes simplex virus type 1. J Biol Chem. 1992 Jan 25;267(3):1411–1414. [PubMed] [Google Scholar]
- Dynlacht B. D., Hoey T., Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991 Aug 9;66(3):563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- Fremont D. H., Matsumura M., Stura E. A., Peterson P. A., Wilson I. A. Crystal structures of two viral peptides in complex with murine MHC class I H-2Kb. Science. 1992 Aug 14;257(5072):919–927. doi: 10.1126/science.1323877. [DOI] [PubMed] [Google Scholar]
- Fromental C., Kanno M., Nomiyama H., Chambon P. Cooperativity and hierarchical levels of functional organization in the SV40 enhancer. Cell. 1988 Sep 23;54(7):943–953. doi: 10.1016/0092-8674(88)90109-2. [DOI] [PubMed] [Google Scholar]
- Gerber H. P., Seipel K., Georgiev O., Höfferer M., Hug M., Rusconi S., Schaffner W. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science. 1994 Feb 11;263(5148):808–811. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- Gerster T., Balmaceda C. G., Roeder R. G. The cell type-specific octamer transcription factor OTF-2 has two domains required for the activation of transcription. EMBO J. 1990 May;9(5):1635–1643. doi: 10.1002/j.1460-2075.1990.tb08283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G., Pascal E., Tseng Z. H., Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci U S A. 1994 Jan 4;91(1):192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G., Ptashne M. Mutants of GAL4 protein altered in an activation function. Cell. 1987 Oct 9;51(1):121–126. doi: 10.1016/0092-8674(87)90016-x. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Mahadevan S., Struhl K. Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature. 1988 Jun 16;333(6174):635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T., Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm J. D., Rould M. A., Aurora R., Herr W., Pabo C. O. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell. 1994 Apr 8;77(1):21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- LeBowitz J. H., Clerc R. G., Brenowitz M., Sharp P. A. The Oct-2 protein binds cooperatively to adjacent octamer sites. Genes Dev. 1989 Oct;3(10):1625–1638. doi: 10.1101/gad.3.10.1625. [DOI] [PubMed] [Google Scholar]
- Lobo S. M., Tanaka M., Sullivan M. L., Hernandez N. A TBP complex essential for transcription from TATA-less but not TATA-containing RNA polymerase III promoters is part of the TFIIIB fraction. Cell. 1992 Dec 11;71(6):1029–1040. doi: 10.1016/0092-8674(92)90397-u. [DOI] [PubMed] [Google Scholar]
- Madden D. R., Gorga J. C., Strominger J. L., Wiley D. C. The three-dimensional structure of HLA-B27 at 2.1 A resolution suggests a general mechanism for tight peptide binding to MHC. Cell. 1992 Sep 18;70(6):1035–1048. doi: 10.1016/0092-8674(92)90252-8. [DOI] [PubMed] [Google Scholar]
- Marmorstein R., Carey M., Ptashne M., Harrison S. C. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992 Apr 2;356(6368):408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Müller-Immerglück M. M., Schaffner W., Matthias P. Transcription factor Oct-2A contains functionally redundant activating domains and works selectively from a promoter but not from a remote enhancer position in non-lymphoid (HeLa) cells. EMBO J. 1990 May;9(5):1625–1634. doi: 10.1002/j.1460-2075.1990.tb08282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M. M., Ruppert S., Schaffner W., Matthias P. A cloned octamer transcription factor stimulates transcription from lymphoid-specific promoters in non-B cells. Nature. 1988 Dec 8;336(6199):544–551. doi: 10.1038/336544a0. [DOI] [PubMed] [Google Scholar]
- O'Hare P., Williams G. Structural studies of the acidic transactivation domain of the Vmw65 protein of herpes simplex virus using 1H NMR. Biochemistry. 1992 Apr 28;31(16):4150–4156. doi: 10.1021/bi00131a035. [DOI] [PubMed] [Google Scholar]
- Ondek B., Gloss L., Herr W. The SV40 enhancer contains two distinct levels of organization. Nature. 1988 May 5;333(6168):40–45. doi: 10.1038/333040a0. [DOI] [PubMed] [Google Scholar]
- Ondek B., Shepard A., Herr W. Discrete elements within the SV40 enhancer region display different cell-specific enhancer activities. EMBO J. 1987 Apr;6(4):1017–1025. doi: 10.1002/j.1460-2075.1987.tb04854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991 May 10;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Pugh B. F., Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990 Jun 29;61(7):1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- Regier J. L., Shen F., Triezenberg S. J. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):883–887. doi: 10.1073/pnas.90.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinberg D., Roeder R. G. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and functional analysis of initiation factors IIB and IIE. J Biol Chem. 1987 Mar 5;262(7):3310–3321. [PubMed] [Google Scholar]
- Scheidereit C., Cromlish J. A., Gerster T., Kawakami K., Balmaceda C. G., Currie R. A., Roeder R. G. A human lymphoid-specific transcription factor that activates immunoglobulin genes is a homoeobox protein. Nature. 1988 Dec 8;336(6199):551–557. doi: 10.1038/336551a0. [DOI] [PubMed] [Google Scholar]
- Schirm S., Jiricny J., Schaffner W. The SV40 enhancer can be dissected into multiple segments, each with a different cell type specificity. Genes Dev. 1987 Mar;1(1):65–74. doi: 10.1101/gad.1.1.65. [DOI] [PubMed] [Google Scholar]
- Seipel K., Georgiev O., Schaffner W. Different activation domains stimulate transcription from remote ('enhancer') and proximal ('promoter') positions. EMBO J. 1992 Dec;11(13):4961–4968. doi: 10.1002/j.1460-2075.1992.tb05603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Sturm R. A., Das G., Herr W. The ubiquitous octamer-binding protein Oct-1 contains a POU domain with a homeo box subdomain. Genes Dev. 1988 Dec;2(12A):1582–1599. doi: 10.1101/gad.2.12a.1582. [DOI] [PubMed] [Google Scholar]
- Sumimoto H., Ohkuma Y., Yamamoto T., Horikoshi M., Roeder R. G. Factors involved in specific transcription by mammalian RNA polymerase II: identification of general transcription factor TFIIG. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9158–9162. doi: 10.1073/pnas.87.23.9158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Clouston W. M., Herr W. The Oct-2 glutamine-rich and proline-rich activation domains can synergize with each other or duplicates of themselves to activate transcription. Mol Cell Biol. 1994 Sep;14(9):6046–6055. doi: 10.1128/mcb.14.9.6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Grossniklaus U., Herr W., Hernandez N. Activation of the U2 snRNA promoter by the octamer motif defines a new class of RNA polymerase II enhancer elements. Genes Dev. 1988 Dec;2(12B):1764–1778. doi: 10.1101/gad.2.12b.1764. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990 Feb 9;60(3):375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Lai J. S., Herr W. Promoter-selective activation domains in Oct-1 and Oct-2 direct differential activation of an snRNA and mRNA promoter. Cell. 1992 Feb 21;68(4):755–767. doi: 10.1016/0092-8674(92)90150-b. [DOI] [PubMed] [Google Scholar]
- Tanese N., Pugh B. F., Tjian R. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev. 1991 Dec;5(12A):2212–2224. doi: 10.1101/gad.5.12a.2212. [DOI] [PubMed] [Google Scholar]
- Van Hoy M., Leuther K. K., Kodadek T., Johnston S. A. The acidic activation domains of the GCN4 and GAL4 proteins are not alpha helical but form beta sheets. Cell. 1993 Feb 26;72(4):587–594. doi: 10.1016/0092-8674(93)90077-4. [DOI] [PubMed] [Google Scholar]
- Veldman G. M., Lupton S., Kamen R. Polyomavirus enhancer contains multiple redundant sequence elements that activate both DNA replication and gene expression. Mol Cell Biol. 1985 Apr;5(4):649–658. doi: 10.1128/mcb.5.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A. C., Zhang W., Sacchettini J. C., Nathenson S. G. The three-dimensional structure of H-2Db at 2.4 A resolution: implications for antigen-determinant selection. Cell. 1994 Jan 14;76(1):39–50. doi: 10.1016/0092-8674(94)90171-6. [DOI] [PubMed] [Google Scholar]