Abstract
Pseudomonas aeruginosa strain PA14 is a multi-host pathogen that infects plants, nematodes, insects, and vertebrates. Many PA14 factors are required for virulence in more than one of these hosts. Noting that plants have a fundamentally different cellular architecture from animals, we sought to identify PA14 factors that are specifically required for plant pathogenesis. We show that synthesis by PA14 of the disaccharide trehalose is required for pathogenesis in Arabidopsis, but not in nematodes, insects, or mice. In-frame deletion of two closely-linked predicted trehalose biosynthetic operons, treYZ and treS, decreased growth in Arabidopsis leaves about 50 fold. Exogenously co-inoculated trehalose, ammonium, or nitrate, but not glucose, sulfate, or phosphate suppressed the phenotype of the double ΔtreYZΔtreS mutant. Exogenous trehalose or ammonium nitrate does not suppress the growth defect of the double ΔtreYZΔtreS mutant by suppressing the plant defense response. Trehalose also does not function intracellularly in P. aeruginosa to ameliorate a variety of stresses, but most likely functions extracellularly, because wild-type PA14 rescued the in vivo growth defect of the ΔtreYZΔtreS in trans. Surprisingly, the growth defect of the double ΔtreYZΔtreS double mutant was suppressed by various Arabidopsis cell wall mutants that affect xyloglucan synthesis, including an xxt1xxt2 double mutant that completely lacks xyloglucan, even though xyloglucan mutants are not more susceptible to pathogens and respond like wild-type plants to immune elicitors. An explanation of our data is that trehalose functions to promote the acquisition of nitrogen-containing nutrients in a process that involves the xyloglucan component of the plant cell wall, thereby allowing P. aeruginosa to replicate in the intercellular spaces in a leaf. This work shows how P. aeruginosa, a multi-host opportunistic pathogen, has repurposed a highly conserved “house-keeping” anabolic pathway (trehalose biosynthesis) as a potent virulence factor that allows it to replicate in the intercellular environment of a leaf.
Author Summary
Pseudomonas aeruginosa is an opportunistic human bacterial pathogen that infects a wide range of plants and animals, including the model laboratory plant Arabidopsis thaliana. P. aeruginosa utilizes many of the same virulence-related factors to infect both plants and animals. However, because plants have fundamentally different cellular architecture than animals, we hypothesized that P. aeruginosa synthesizes specific factors required for infecting plants but not animals. We found that synthesis of the sugar molecule trehalose, an unusual dimer of glucose, is required for plant but not animal pathogenesis. Although P. aeruginosa mutants defective in trehalose synthesis are non-pathogenic in Arabidopsis, Arabidopsis mutants that lack the polysaccharide xyloglucan in their cell walls can be infected by P. aeruginosa trehalose mutants. Moreover, application of ammonium nitrate overcomes the requirement for trehalose for infecting an Arabidopsis leaf. Our data suggest that trehalose promotes the acquisition of nitrogen-containing nutrients, thereby allowing P. aeruginosa to replicate in the nutrient-poor intercellular spaces in a leaf. This work shows how an opportunistic pathogen has repurposed a highly conserved “house-keeping” function (trehalose biosynthesis) as a potent virulence factor.
Introduction
The ubiquitous bacterium Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen that infects a wide diversity of hosts. For example, P. aeruginosa strain PA14 is infectious in several model genetic hosts including the plant Arabidopsis thaliana [1], the insect Drosophila melanogaster [2], and the nematode Caenorhabditis elegans [3]. Using these model hosts, we and others have sought to identify PA14 virulence-related factors that play key roles in pathogenesis with the goal of elucidating conserved mechanisms underlying the pathogenic process and to determine whether the spectrum of virulence-related genes in a multi-host opportunistic pathogen are distinct from the virulence genes in more specialized pathogens [1]–[7]. Our work to date suggests that PA14 virulence depends primarily on genes that are part of a conserved P. aeruginosa genome [8] rather than on an arsenal of host-specific virulence-related factors [8], [9].
One of the most unusual and unexpected features of P. aeruginosa is its ability to infect both plants and animals. Because plant cells are distinguished from metazoan cells primarily by their rigid and tough cellulosic walls, we reasoned that P. aeruginosa pathogenesis in plants may rely on plant-specific virulence factors related to the plant cell walls. Presumably as a consequence of these tough plant cell walls, most bacterial foliar pathogens replicate extracellularly in intercellular spaces and subvert plant cellular processes such as sugar transporters to obtain nutrients from mesophyll cells rather than attempting to directly breech plant cell walls. For example, Xanthomonas oryzae pv oryzae utilizes the Type III secretion system to inject transcriptional activators into plant mesophyll cells that upregulate the expression of sugar transporters that are not normally expressed in these cells [10].
In this paper, we report that the non-reducing disaccharide trehalose, made of two glucose residues joined by an atypical α,α-1,1-glucoside linkage, is a key virulence factor for P. aeruginosa PA14 pathogenesis in Arabidopsis leaves, but is not required for virulence in nematodes, flies, or mice. Trehalose is a common metabolite that has been shown to be involved in conferring tolerance to a variety of environmental stresses in diverse prokaryotic and eukaryotic species [17], [25], [26]. In PA14, trehalose is synthesized by enzymes encoded in two adjacent predicted operons, treYZ and treS, that utilize distinct mechanisms of synthesis. Deletion of these trehalose biosynthetic genes results in a highly attenuated non-pathogenic phenotype that can be rescued by trehalose and by various ammonium and nitrate sources, but not by sucrose or glucose. In addition, Arabidopsis mutants defective in the synthesis of the cell wall polymer xyloglucan also suppress the non-pathogenic phenotype of P. aeruginosa trehalose mutants. These data suggest that trehalose promotes the acquisition of nitrogen-containing nutrients and that the xyloglucan component of the plant cell wall is involved in this process, thereby allowing P. aeruginosa to replicate in the nutrient-poor intercellular spaces in a leaf. Our data show how pathogens can utilize what are normally considered to be “house-keeping” functions, such as the wide-spread ability to biosynthesize trehalose, as a potent virulence factor that allows them to replicate in the particular environment of a host.
Results
Trehalose production by PA14 is required for virulence in Arabidopsis
Reasoning that the tough cellulosic walls of plant cells may pose a unique challenge to plant pathogens, we surveyed the fully sequenced and annotated P. aeruginosa PA14 genome [4] to determine whether canonical cell wall degrading enzymes including cellulases, xylanases, and pectinases are encoded in the genome. In susceptible ecotypes (wild accessions) of Arabidopsis, P. aeruginosa PA14 causes soft-rot symptoms [1], typically caused by pathogens that secrete pectinases and other hydrolytic cell wall degrading enzymes. Moreover, PA14 infection causes extensive degradation of Arabidopsis mesophyll cell walls including the generation of “holes” approximately the diameter of P. aeruginosa through which the bacteria enter host cells [11]. We thus expected that the PA14 genome would encode a variety of cell wall degrading enzymes (CWDEs). However, our survey of the PA14 genome identified only a single, candidate cellulase, identified ambiguously as “cellulase/peptidase” (PA14_36500). Although PA14_36500 was upregulated two and three days post-inoculation in planta, correlating with the development of disease symptoms (Figure S1A), a transposon insertion in PA14_36500 (PA14_36500::MAR2xT7), in-frame deletion of the cellulose/peptidase gene (ΔPA14_36500), or in-frame deletion of a putative cellulase/peptidase operon (ΔPA14_36480-36520) did not cause a significant attenuation in virulence in Arabidopsis leaves (Table S1).
Because PA14_36500, which encodes the putative cellulose/peptidase, was induced during plant infection and because genes are often functionally clustered on bacterial genomes, we sought to identify genes adjacent to PA14_36500 that are co-regulated with PA14_36500. This led to the identification of a set of 38 genes (42.23 kb region; PA14_36375 to PA14_36830) spanning the cellulase/peptidase gene that is coordinately down-regulated in an mvfR (multiple virulence factor regulator) mutant grown under various culture conditions [12], [13]. Importantly, the quorum sensing-associated transcriptional regulator MvfR is required for maximum PA14 virulence in Arabidopsis [7]. Consistent with the in vitro transcriptional profiling data, cellulase/peptidase PA14_36500 expression was significantly reduced in planta in an mvfR mutant (Figure S1B).
Besides the putative cellulase/peptidase, the PA14_36375–36830 42.23 kb region encodes putative glucanolytic enzymes (PA14_36590, PA14_36630, PA14_36740) as well as two closely linked predicted operons (http://www.pseudomonas.com), PA14_36570-36630 consisting of six genes, and PA14_36710-37640 consisting of three genes, referred to hereafter as the “treYZ” and “treS” operons, respectively, that encode enzymes involved in two different trehalose biosynthetic pathways (Figure 1; Table S2). TreY and TreZ convert maltodextrins into trehalose in a two-step enzymatic reaction [14], whereas TreS catalyzes conversion of maltose into trehalose in a single reaction [15] (Figure S2). In addition to treY (PA14_36605) and treZ (PA14_36580), the predicted treYZ operon contains glgA (PA14_36570), malQ (PA14_36590), hypothetical gene (PA14_36620) and glgX (PA14_36630). glgA, malQ, glgX encode enzymes with a putative role in α-1,4-linked glucan synthesis (glgA) and degradation (malQ, glgX), that could serve as precursors for trehalose synthesis. In addition to treS, the treS operon contains a predicted α-amylase (PA14_36740), and glgB (PA14_36710), a predicted α-1,4-branching enzyme (Figure 1).
Figure 1. Annotation of a 42.23 kb region of the P. aeruginosa PA14 genome encoding 38 genes (PA14_36375–36830) and schematic representation of transposon and deletion mutants used in this study.
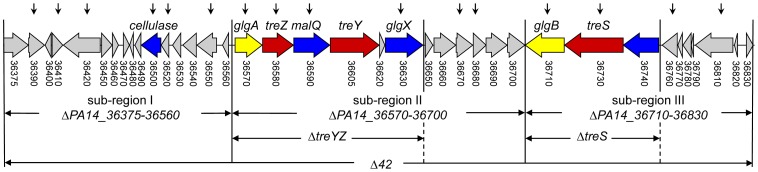
Colors depict trehalose biosynthetic genes (red), glucanolytic genes (blue) and glucan synthesis genes (yellow). Numbers below the genes correspond to PA14 gene locus tags (http://ausubellab.mgh.harvard.edu/pa14sequencing). On the top of the Figure, vertical arrows indicate the positions of MAR2xT7 transposon insertions [16], and on the bottom, horizontal arrows denote the extent of in-frame deletion mutants.
The 42.23 kb PA14_36375–36830 region containing 38 genes is highly conserved among several sequenced P. aeruginosa strains that were examined and the treYZ and treS operons are conserved among pseudomonads in general (Table S2).
We utilized a previously constructed non-redundant PA14 transposon insertion mutant library [16] to determine whether particular PA14 genes in the 38-gene region promote pathogenesis in Arabidopsis. Among 16 transposon insertions in 16 different genes that were available in the library, two were significantly attenuated in virulence. These mutants, with insertions in glgA and treZ, exhibited a decrease in virulence of 20 and 16 fold, respectively, as measured by in planta growth (Table S1). glgA and treZ are the first two genes in the treYZ operon, pointing to an important role for trehalose in the infectious process.
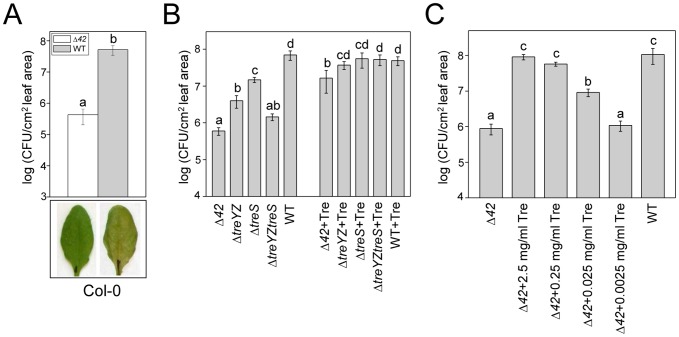
To further investigate whether the trehalose operons and/or other genes in the 38-gene cluster are required for virulence, we constructed an in-frame deletion of the entire 42.23 kb region (referred to hereafter as Δ42) by homologous recombination. In contrast to insertions in glgA and treZ, which exhibited at most a 20 fold decrease in growth compared to wild-type, the Δ42 mutant exhibited severe attenuation in virulence, affecting growth of PA14 infiltrated into Arabidopsis leaves about 120 fold and preventing the appearance of pathogenic symptoms (Figure 2A). Similar results were obtained with four independently constructed Δ42 mutants (data not shown), demonstrating that the non-pathogenic phenotype was caused by the deletion of the 42.23 kb region. Importantly, the Δ42 mutant does not appear to be slow growing or to be generally deficient in a variety of phenotypes associated with virulence in P. aeruginosa. The Δ42 deletion mutant was not auxotrophic, grew at the same rate as wild-type PA14 in a variety of minimal and rich media, and had no observable phenotypes with respect to the production of pyocyanin (Figure S3), motility, or biofilm formation (Table S3), and similar results were obtained with a second independently-constructed Δ42 mutant (Figure S3; Table S3). Because independently-constructed Δ42 mutants exhibited the same phenotypes, one of the Δ42 mutants was chosen for subsequent experiments.
Figure 2. Attenuation of Δ42 and trehalose biosynthetic mutants in Arabidopsis leaves and suppression of attenuation of trehalose mutants with exogenous trehalose.
(A) Columbia (Col-0) ecotype plants were infiltrated with PA14 wild-type or Δ42. Leaves were harvested 3 days after infiltration and bacterial counts determined as described in Materials and Methods. Representative photographs of infected leaves were taken 3 days after infiltration. Four independently constructed Δ42 mutants exhibited the same dramatic non-pathogenic phenotype in Arabidopsis leaves. The experiment was repeated more than three times. (B, C) Before the infiltration of leaves with PA14 wild-type or PA14-derived deletion mutants, trehalose (Tre) at the indicated concentrations was added to bacterial suspensions. In panel B, the trehalose concentration was 2.5 mg/ml. The leaves were harvested 3 days post-infiltration (dpi) and bacterial counts determined as described in Materials and Methods. Data represent the mean of bacterial titers ± SE of six leaf disks excised from 6 leaves of 3 plants. Letters above bars denote statistically significant differences (P<0.05, Fisher's PLSD test). See Figure 1 for a description of the mutants. The experiments in (B) and (C) were repeated four times and two times, respectively.
We next constructed several smaller deletions within the 42 kb region to determine which of the 38 encoded genes are primarily responsible for the severe avirulent phenotype of Δ42: ΔPA14_36375-36560 (sub-region I) contains a deletion of the cellulase/peptidase operon and several adjacent genes, and ΔPA14_36570-36700 (sub-region II) and ΔPA14_36710-36830 (sub-region III) contain deletions of the treYZ and treS genes, respectively, including some neighboring genes (Figure 1).
Deletion of sub-region I that includes the putative cellulase/peptidase gene had a modest 3.3 fold reduction in virulence. In contrast, deletion of sub-region II that contains the treYZ operon had a much more significant effect on virulence (28.7 fold decrease in growth; Figure S4), whereas deletion of sub-region III that contains the treS operon caused a 5.9 fold decrease in growth (Figure S4). These experiments suggested that the treYZ and treS operons play a significant role in PA14 pathogenesis in Arabidopsis.
To corroborate the involvement of the trehalose genes in plant pathogenesis we constructed ΔPA14_36570-36630 (ΔtreYZ) and ΔPA14_36710-36740 (ΔtreS) containing deletions of only the two putative operons containing the treYZ and treS genes, respectively, and ΔPA14_36570-36630;PA14_36710-36740 (ΔtreYZΔtreS) containing deletions of both of the trehalose biosynthetic operons (Figure 1). Deleting either the putative treYZ or the treS operons (Figure 2B) had approximately the same effects as deleting the more extensive corresponding subregions II or III, respectively (Figure S4), and deleting both trehalose operons resulted in an approximately 50 fold decrease in virulence compared to the approximate 120 fold decrease observed with the Δ42 mutant (Figure 2B). These data show that the treYZ and treS operons play a key role in pathogenesis in Arabidopsis leaves, but that genes in the 42 kb region in addition to those involved in trehalose biosynthesis also play a role in plant pathogenesis.
Further evidence suggesting an important role for trehalose biosynthesis in plant pathogenesis was obtained by measuring the levels of trehalose synthesized in vitro by PA14 wild-type and trehalose biosynthetic mutants. While wild-type PA14 synthesized readily detectable levels of trehalose, there was approximately 50% less trehalose in the ΔtreS mutant, and there were undetectable levels of trehalose in the glgA, treZ, ΔtreYZ, ΔtreYZΔtreS, and Δ42 mutants (Table 1). These data show that the treYZ and treS operons encode enzymes involved in trehalose biosynthesis. These data also suggest that treS operon may be dependent on treYZ for trehalose production, as reported previously [17]. When we compared the levels of trehalose synthesized in vitro (Table 1) and the extent of growth of the various strains in Arabidopsis leaves (Figure 2B; Table S1), we found an excellent positive correlation coefficient (R2 = 0.87).
Table 1. Trehalose levels in the trehalose mutants.
| Strains | Trehalose (mg/ml) |
| 1. PA14_36570::MAR2xT7 (glgA) | not detected |
| 2. PA14_36580::MAR2xT7 (treZ) | not detected |
| 3. ΔPA14_36570-36630 (ΔtreYZ) | not detected |
| 4. ΔPA14_36710-36740 (ΔtreS) | 0.212±0.015 |
| 5. ΔPA14_36570-36630;PA14_36710-36740 (ΔtreYZtreS) | not detected |
| 6. ΔPA14_36375-36830 (Δ42) | not detected |
| 7. WT | 0.444±0.003 |
P. aeruginosa strains were grown at 37°C in MinA medium supplemented with 0.5 M NaCl. Trehalose was extracted and quantified enzymatically as described in Materials and Methods. See Figure 1 for a description of the mutants. Data represent the mean ± SE of two replicate samples and are representative of at least three independent experiments.
Importantly, we found that co-infiltration of the PA14 trehalose mutants and pure trehalose essentially completely suppressed the avirulent phenotypes of the ΔtreYZ, ΔtreS, and ΔtreYZΔtreS mutants and mostly suppressed the phenotype of the Δ42 mutant (Figures 2B and 2C). However, 0.25 mg/ml trehalose also rescued the Δ42 mutant almost as well as 2.5 mg/ml, and 0.025 mg/ml trehalose partially suppressed the growth defect of the Δ42 mutant (Figure 2C). These data indicated a requirement for trehalose for PA14 virulence in planta, potentially at physiologically relevant concentrations.
In summary, the data in this section shows that the ΔtreYZ, ΔtreS, and ΔtreYZΔtreS mutants are less virulent in planta, that they either synthesize undetectable (ΔtreYZ and ΔtreYZΔtreS) or reduced (ΔtreS) levels of trehalose, that their level of virulence positively correlates with the level of trehalose they synthesize, and that their reduced virulence phenotype can be suppressed by exogenous trehalose. These data demonstrate that the virulence deficient phenotypes of the ΔtreYZ, ΔtreS, and ΔtreYZΔtreS mutants are a consequence of the inability of these strains to synthesize trehalose, thereby correlating the genotype of these mutants with their avirulent phenotypes.
Trehalose biosynthetic genes are not required for virulence in metazoans
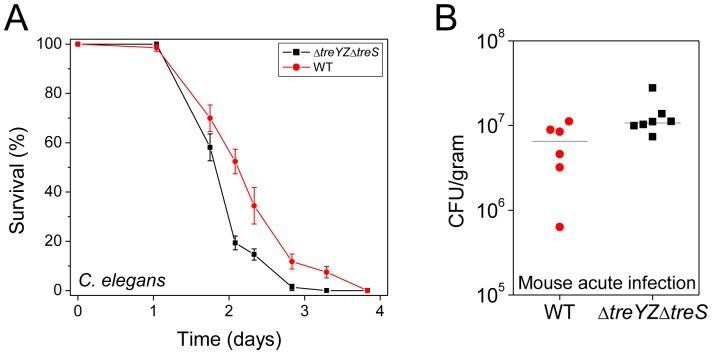
As described in the Introduction, PA14 infection models have previously been established in C. elegans [3], D. melanogaster [18], and mice [19], [20], as well as in other metazoans. Interestingly, the ΔtreYZΔtreS double trehalose mutant was not less virulent in a C. elegans killing model or in a murine acute pneumonia model. In fact, the ΔtreYZΔtreS appeared to be slightly more virulent in the metazoan hosts (Figure 3). Similar results were obtained with the Δ42 mutant in these two models as well as in a D. melanogaster ingestion model and in a chronic oropharyngeal colonization model in transgenic mutant mice lacking the cystic fibrosis transmembrane conductance regulator protein (see Materials and Methods for the mutant description)(Figure S5). These data suggest that trehalose appears to be specifically required for plant but not for metazoan pathogenesis. In the sections that follow, we considered several hypotheses concerning the role of trehalose in promoting the virulence of P. aeruginosa during the infectious process in plants but not in animals.
Figure 3. The ΔtreYZΔtreS mutant is more virulent than wild-type PA14 in nematodes and mice.
(A) C. elegans are more susceptible to killing by ΔtreYZΔtreS than PA14 wild-type (P<0.0001). Mutant fer15;fem1 C. elegans animals were exposed to P. aeruginosa strains and survival was determined as described in Materials and Methods. Data at each time point correspond to the average of three plates per strain, each with approximately 40 animals per plate, and are representative of two independent experiments. (B) The Δ42 mutant is more virulent than wild-type PA14 in a murine acute lung infection model. See Materials and Methods for details of infection protocol. The median CFU/gram of lung tissue of mice infected with ΔtreYZΔtreS is 2-fold higher than with wild-type PA14 18 hours post intranasal infection (P<0.05, Mann-Whitney U test). Data are representative of two independent experiments.
Specific Arabidopsis cell wall mutants suppress the phenotype of PA14 trehalose mutants
Since a major difference between plant and animals cells is the plant cellulosic cell wall, we reasoned that trehalose may function in a process that involves the plant cell wall. Because PA14 infection in Arabidopsis leaves causes extensive degradation of mesophyll cell walls [11], we first investigated the possibility that trehalose enhances the activity of cell wall degrading enzymes (CWDEs). We tested whether trehalose enhanced the activity of a variety of commercial CWDEs to hydrolyze partially purified Arabidopsis cell walls in vitro to generate reducing sugars, which were measured using the Somogyi-Nelson assay [21], [22]. However, we were not able to conclusively demonstrate that trehalose enhanced the activity of the CWDEs tested (data not shown).
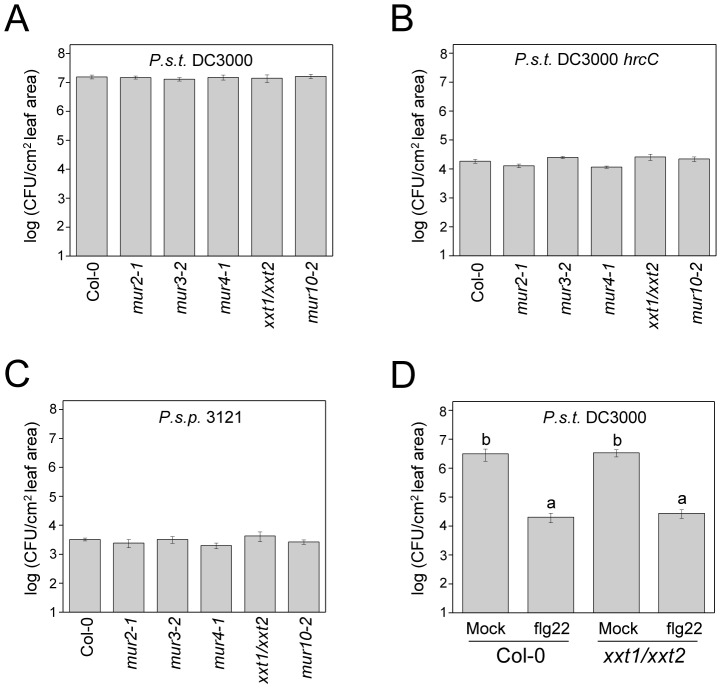
We next reasoned that if trehalose interacts with the plant cell wall, specific Arabidopsis cell wall mutants might suppress the phenotype of the ΔtreYZΔtreS mutant. We tested the growth of wild-type PA14 and the ΔtreYZΔtreS mutant in several Arabidopsis cell wall mutants involved in xyloglucan (mur2-1, mur3-2, xxt1/xxt2), arabinose (mur4-1), or cellulose (mur10-2) synthesis. Remarkably, the ΔtreYZΔtreS mutant grew to the same titer as wild-type PA14 in an xxt1/xxt2 double mutant that completely lacks xyloglucan in its cell walls and in a mur4-1 mutant that has decreased levels of arabinose in pectins, xylans, and xyloglucans [23] (Figure 4). Similar results were obtained with the Δ42 mutant; i.e., the Arabidopsis xxt1/xxt2 mutant completely suppressed and the mur4-1 mutant mostly suppressed the avirulent phenotype of the Δ42 mutant (Figure S6).
Figure 4. The in planta growth defect of the PA14 ΔtreYZΔtreS mutant in Arabidopsis is suppressed by cell wall mutations.
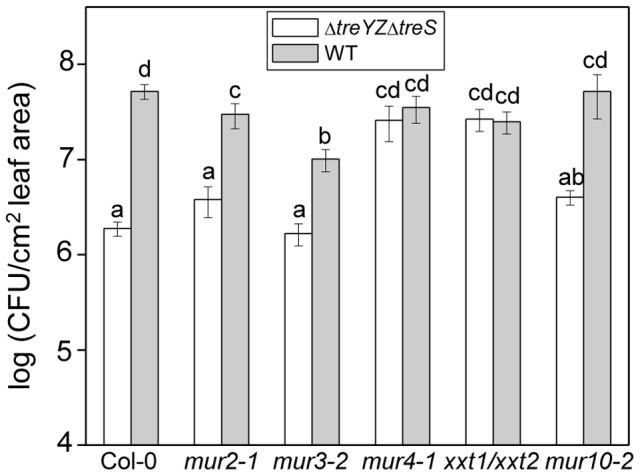
Growth of PA14 wild-type and the ΔtreYZΔtreS mutant 3 days post infiltration in Arabidopsis cell wall mutants mur2-1, mur3-2, mur4-1, mur10-2 and xxt1/xxt2. Data represent the mean of bacterial titers ± SE of six leaf disks excised from 6 leaves of 3 plants. Letters above bars denote statistically significant differences (P<0.05, Fisher's PLSD test). The experiments were repeated at least two times.
We ruled out the possibility that the Arabidopsis cell wall mutants suppress the avirulent phenotype of the PA14 trehalose mutants simply because they are generally more susceptible to pathogen attack. As shown in Figure 5A, the cell wall mutants did not exhibit enhanced susceptibility to the P. syringae pv. tomato strain DC3000, a well-studied bona fide Arabidopsis pathogen. The Arabidopsis cell wall mutants were also not more susceptible to a DC3000 hrcC mutant (Figure 5B), which is greatly impaired in virulence, or to the bean pathogen P. syringae pv. phaseolicola strain 3121 (Figure 5C), which is not normally pathogenic in Arabidopsis. Consistent with these data, we also showed that the xxt1/xxt2 mutant, which exhibits the most severe cell wall defect of the Arabidopsis mutants tested, mounts a normal defense response when challenged with the flagellin peptide flg22 (Figure 5D). Flg22 elicits so-called “pattern triggered immunity” in Arabidopsis. When Arabidopsis leaves are pre-infiltrated with flg22, flg22 exerts a protective effect against subsequent infection with P. syringae DC3000 [24]. As shown in Figure 5D, flg22 elicits the same level of protection against P. syringae DC3000 in xxt1xxt2 plants as in wild-type plants.
Figure 5. Arabidopsis cell wall mutants are not more susceptible to virulent or non-pathogenic P. syringae strains and the xxt1/xxt2 mutant mounts an effective innate immune response.
Growth of (A) P. syringae pv. tomato strain DC3000 (B) P. syringae pv. tomato strain DC3000 hrcC (C) P. syringae pv. phaseolicola strain 3121 three days post infiltration in Arabidopsis cell wall mutants mur2-1, mur3-2, mur4-1, mur10-2 and xxt1/xxt2 and Col-0 wild-type and (D) P. syringae pv. tomato strain DC3000 three days post infiltration of Col-0 and xxt1/xxt2 mutant preinfiltrated with 1 µM flg22 for 24 hours. Data represent the mean of bacterial titers ± SE of six leaf disks excised from 6 leaves of 3 plants. Letters above bars denote statistically significant differences (P<0.05, Fisher's PLSD test). Absence of letters indicates no statistically significant differences. The experiments were repeated at least two times.
Ammonium and nitrate but not glucose or sucrose suppress the phenotype of trehalose mutants
As described in the Introduction, because bacterial plant pathogens primarily replicate in the intercellular spaces in a leaf, they need to acquire nutrients from plant mesophyll cells. We therefore tested whether trehalose may be involved in the acquisition of a variety of nutrient sources including carbon, nitrogen, sulfur and phosphorous. If this were the case, we reasoned that co-infiltration of particular nutrients with the ΔtreYZΔtreS or the Δ42 mutant would suppress their non-pathogenic phenotypes.
Co-infiltration of the ΔtreYZΔtreS double mutant with glucose (Figure 6) or co-infiltration of the Δ42 mutant with glucose or sucrose (Figure S7A) did not rescue the attenuated phenotype in the Arabidopsis leaf assay. These experiments showed that the ΔtreYZΔtreS or the Δ42 mutant is not limited by carbon. The fact that trehalose but not glucose or sucrose suppressed the phenotype of the Δ42 mutant also shows that the putative cellulase/peptidase and other hypothetical glucanolytic enzymes encoded in the 38 gene region deleted in the Δ42 mutant do not play a critical role in supplying a carbon source to PA14.
Figure 6. The in planta growth defect of the ΔtreYZΔtreS double mutant is suppressed by ammonium or nitrate ions.
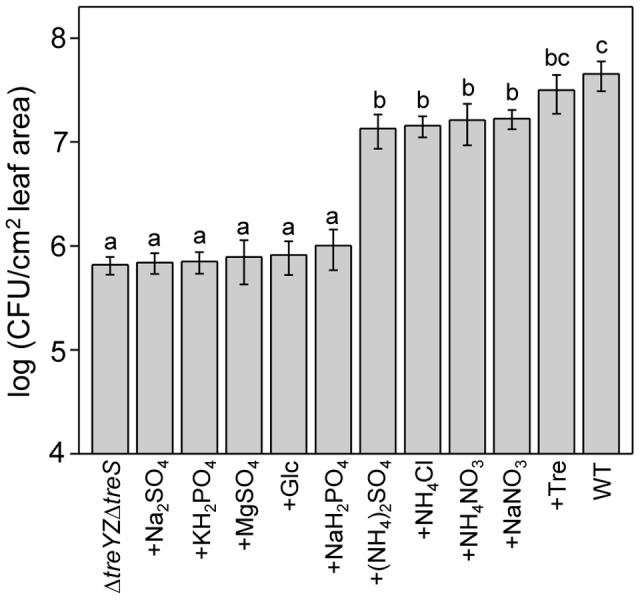
Leaves of four-week-old Arabidopsis Col-0 plants were infiltrated with PA14 wild-type or with Δ42 co-inoculated with various solutions of phosphate, sulfate, nitrate, or ammonium salts at 1 mM. Suppression of the growth defect of Δ42 with 2.5 mg/ml trehalose (Tre) and 1.25 mg/ml glucose (Glc) were tested as positive and negative controls, respectively. Data represent the mean of bacterial titers ± SE of six leaf disks excised from 6 leaves of 3 plants. Letters above bars denote statistically significant differences (P<0.05, Fisher's PLSD test). The experiments were repeated at least two times.
We also entertained the possibility that PA14 could accumulate trehalose as a storage sugar, analogous to glycogen or starch, and then hydrolyze trehalose using the enzyme trehalase (PA14_33450, treA) and utilize the resulting glucose as a carbon source, thereby promoting virulence. We ruled out this possibility, however, by showing that co-infiltration of a double Δ42treA::MAR2xT7 mutant (which cannot metabolize trehalose) with trehalose rescued the non-pathogenic phenotype similarly as co-infiltration of the Δ42 mutant with trehalose (Figure S7B). We also confirmed that the Δ42treA::MAR2xT7 cannot metabolize trehalose and utilize it as a carbon source (see Materials and Methods).
Finally, we tested various salts to determine whether they would suppress the phenotypes of the ΔtreYZΔtreS (Figure 6) or the Δ42 mutant (Figure S8). Interestingly, ammonium and nitrate ions almost completely suppressed the lack of growth phenotype of the ΔtreYZΔtreS (Figure 6) or the Δ42 mutant (Figure S8), whereas sulfates and phosphates did not have a significant effect.
The data in this section suggest that trehalose enhances access to nitrogen sources during an Arabidopsis infection. An alternative model is that ammonium nitrate (as well as trehalose) suppresses the avirulent phenotype of the PA14 trehalose mutants by suppressing the plant defense response. To test this possibility, we tested whether infiltration of leaves with trehalose or ammonium nitrate resulted in enhanced susceptibility to P. syringae DC3000 (Figure 7A), the DC3000 hrcC mutant (Figure 7B), or P. syringae pv. phaseolicola strain 3121 (Figure 7C); however, neither trehalose nor ammonium nitrate increased the susceptibility to any of these strains. Moreover, infiltration of trehalose or ammonium nitrate did not block the ability of flg22 to elicit protection against infection by P. syringae DC3000 (Figure 7A).
Figure 7. Trehalose or ammonium nitrate does not suppress the Arabidopsis flg22-mediated defense response and does not make Arabidopsis more susceptible to non-pathogenic P. syringae strains.
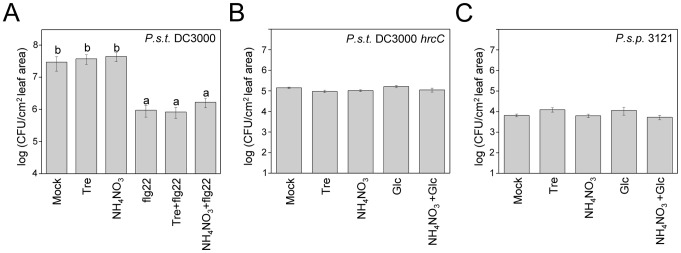
(A) Growth of P. syringae pv. tomato strain DC3000 three days post infiltration of Col-0 plants pretreated for 24 hours with 1 µM flg22, 1 mM trehalose, 1 mM ammonium nitrate individually or with a mixture of flg22 with trehalose or ammonium nitrate. (B) P. syringae pv. tomato strain DC3000 hrcC or (C) P. syringae pv. phaseolicola strain 3121 were co-inoculated with 1 mM trehalose or 1 mM ammonium nitrate and bacterial counts determined 3 days post infiltration. Glucose or glucose plus ammonium nitrate were included as controls. Data represent the mean of bacterial titers ± SE of six leaf disks excised from 6 leaves of 3 plants. Letters above bars denote statistically significant differences (P<0.05, Fisher's PLSD test). Absence of letters indicates no statistically significant differences. The experiments were repeated at least two times.
Trehalose does not appear to function as a stress-response molecule either in vivo or in vitro
Trehalose is well-studied as a so-called compatible solute, which is defined as a molecule that functions as an osmolyte and helps an organism survive osmotic stress. We therefore tested whether other di- and trisaccharide compatible solutes would suppress the avirulent phenotype of the Δ42 mutant. Indeed, as shown in Figure S9, both maltose and maltotriose functioned similarly to trehalose in allowing the Δ42 mutant to grow in planta, albeit somewhat less efficiently than did trehalose.
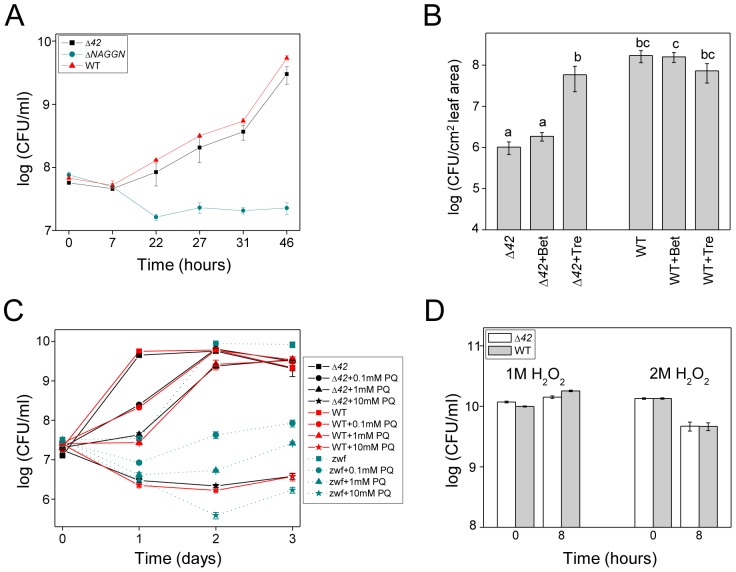
Given these results, we next considered the hypothesis that trehalose enhances the virulence of PA14 by ameliorating a variety of environmental stresses [17], [25], [26]. However, the Δ42 mutant was not more susceptible than wild-type PA14 to osmotic stress in response to 0.5 M NaCl (Figure 8A). As a positive control for the osmotic stress experiment, we constructed an in-frame deletion of a predicted (http://www.pseudomonas.com) three-gene operon (PA14_19350-19370) responsible for the synthesis of a major organic osmoprotectant in P. aeruginosa, N-acetylglutaminylglutamine amide (NAGGN) [27]. As expected, the ΔPA14_19350-19370 mutant (ΔNAGGN) was more susceptible to 0.5 M NaCl than wild-type PA14 or the Δ42 mutant (Figure 8A).
Figure 8. The Δ42 mutant is not more susceptible to osmotic or oxidative stress.
(A) Growth of Δ42, ΔNAGGN and PA14 wild-type under osmotic stress in vitro. Cells were grown at 37°C in MinA medium supplemented with 0.5 M NaCl. Data represent the mean ± SE of 3 replicates. (B) The in planta growth defect of Δ42 is suppressed by trehalose but not betaine. See Figure 2 for experimental details. Data represent the mean ± SE of six replicate samples. (C) In vitro survival of PA14 wild-type, Δ42, and a PA14 zwf::MAR2xT7 mutant cultured for three days in MinA medium supplemented with various concentrations of paraquat (PQ). (D) Survival of PA14 wild-type and Δ42 in LB medium containing 1 M or 2 M hydrogen peroxide added directly to overnight cultures grown for 14 h (inoculum, zero time point on x axis). Cells were further incubated for 8 h at 37°C. 3 M H2O2 was a lethal dose. All experiments in Figure 8 were repeated at least two times.
We further tested whether trehalose functions to protect PA14 from osmotic stress in vitro by comparing its ability to enhance growth in minimal medium supplemented with 0.5 M NaCl compared to the well-studied osmoprotectant molecule betaine [27]. In vitro, betaine rescued the growth of PA14, Δ42, and the ΔNAGGN mutant in 0.5 M NaCl whereas trehalose had no effect (Figure S10A). We also tested whether betaine would rescue the Δ42 mutant for in planta growth, similarly to trehalose. However, as shown in Figure 8B, betaine had no significant effect in rescuing Δ42 growth in planta, showing that the ability of trehalose to rescue Δ42 in planta is not likely due to the fact that it is functioning to protect Δ42 from osmotic stress. In contrast to Δ42, the ΔNAGGN mutant, which is very susceptible to osmotic stress in vitro, had no significant impairment in growth in planta (Table S1). These data show that the Δ42 mutant is not highly susceptible to osmotic stress and that trehalose does not play a major role as an osmoprotectant in PA14.
As an alternative to functioning as an osmoprotectant, we investigated whether trehalose protects PA14 from reactive oxygen-mediated stress generated as a consequence of the plant innate immune response. However, we found no significant difference between the Δ42 mutant and wild-type PA14 with respect to tolerance to paraquat or hydrogen peroxide (Figures 8C and 8D, respectively). Because a P. aeruginosa zwf mutant has been reported to be hyper-sensitive to paraquat-mediated killing [28], we also tested a PA14 zwf::MAR2xT7 mutant [16] as a positive control for determining the sensitivity of PA14 and Δ42 to paraquat. As shown in Figure 8C, the zwf mutant exhibited enhanced susceptibility to paraquat in vitro, but did not exhibit an impaired growth phenotype in planta (Table S1). These data show that it is unlikely that trehalose functions to protect PA14 from oxidative stress.
In addition to oxidative and osmotic stress, we also tested whether the Δ42 mutant is susceptible to pH or temperature stress, displayed a defect in biofilm formation under osmotic stress, or was deficient in the generation of persister cells in the presence of antibiotics. However, wild-type PA14 and the Δ42 mutant were indistinguishable in all of these tests (Figure S10B–E).
Trehalose functions extracellularly
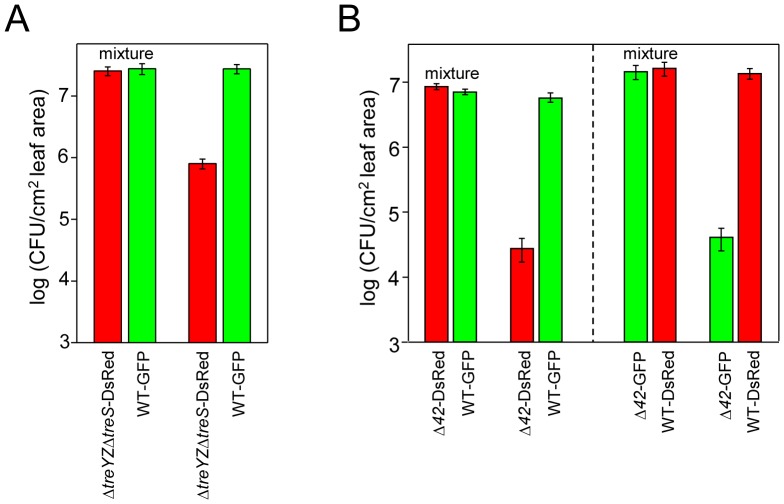
The data in the previous section suggest that trehalose does not function intracellularly to protect PA14 from a variety of stresses during free-living growth. To provide evidence that trehalose functions extracellularly, we tested whether wild-type PA14 “complements” the growth defect of PA14 trehalose mutants in planta. Specifically, we co-inoculated Arabidopsis leaves with equal mixtures of wild-type PA14 and the ΔtreYZΔtreS double trehalose mutant carrying plasmids that express GFP or DsRed, respectively (Figure 9A). Dramatically, co-inoculation of wild-type PA14 with ΔtreYZΔtreS completely rescued the growth defect of ΔtreYZΔtreS (Figure 9A), strongly suggesting that trehalose is most likely acting extracellularly and not internally within PA14 cells. Similar results were obtained when PA14 expressing GFP was mixed with the Δ42 mutant expressing DsRed (Figure 9B, left panel). In this latter experiment, to make sure that the expression of red or green fluorescent protein does not affect bacterial strain viability, we also carried out an experiment in which the plasmids expressing fluorescent proteins were switched in wild-type PA14 and the Δ42 mutant and obtained the same result (Figure 9B, right panel).
Figure 9. PA14 trehalose mutants are rescued in trans in planta by wild-type PA14.
(A) Four-week-old Arabidopsis Col-0 plants were inoculated with a 1∶1 mixture of PA14 wild-type carrying pSMC2 (GFP) and the ΔtreYZΔtreS mutant carrying pAA100 (DsRed) at a total concentration of 3.5×102 CFU/cm2 leaf area. As a control the strains were inoculated individually. (B) In the left panel, four-week-old Arabidopsis Col-0 plants were inoculated with a 1∶1 mixture of PA14 wild-type carrying pSMC2 (GFP) and the Δ42 mutant carrying pAA100 (DsRed) at a total concentration of 6×102 CFU/cm2 leaf area. As a control the strains were inoculated individually. In the right panel, PA14 wild-type carried pAA100 and the Δ42 mutant carried pSMC2. Three days post infiltration, leaves were harvested and CFU determined by counting colonies using a Zeiss Stemi SV6 dissecting microscope fitted with a dual GFP/RFP filter. Data represent the mean of bacterial titers ± SE of six leaf disks excised from 6 leaves of 3 plants and are representative of three independent experiments.
Discussion
In this study, we report that synthesis of the disaccharide trehalose by the multi-host opportunistic pathogen P. aeruginosa is required for plant pathogenesis, but not for pathogenesis in at least three metazoan hosts, mice, D. melanogaster or C. elegans. Trehalose has been extensively characterized as a stress-response molecule that protects cells from osmotic, oxidative, and other environmental stresses. Surprisingly, however, our data suggest that trehalose does not function internally in P. aeruginosa to alleviate a variety of stresses that P. aeruginosa might encounter in its interaction with a plant host. Trehalose also does not function as a major osmoprotectant molecule for P. aeruginosa. Instead, because nitrate and ammonium ions, but not glucose, sucrose, or betaine, suppress the non-pathogenic phenotype of trehalose mutants in planta, we propose that trehalose may function to promote the acquisition of nitrogen-containing nutrients, thereby allowing P. aeruginosa to replicate in the intercellular spaces in a leaf. Moreover, because Arabidopsis cell wall mutants also suppress the non-pathogenic phenotype of the trehalose mutants, it is possible that the plant cell wall normally functions directly or indirectly as a barrier to block nutrient uptake by extracellular bacteria.
Trehalose as a virulence factor for plant and animal pathogens
An important result from this work is that in contrast to many other bacteria and fungi, trehalose appears to have very little effect on protecting P. aeruginosa PA14 from a variety of diverse stresses, including osmotic, oxidative, pH, antibiotic, and temperature stress, and yet trehalose mutants are highly impaired in virulence in Arabidopsis. Instead of trehalose, our data show that N-acetylglutaminylglutamine amide (NAGGN) and glycine-betaine appear to be the primary stress response molecules in P. aeruginosa, in agreement with published data showing that osmotically stressed P. aeruginosa cultures accumulate NAGGN and glycine-betaine [27], [29]. Specifically, we found that a ΔNAGGN mutant was highly impaired in growth under osmotic stress and that exogenously added glycine-betaine, but not trehalose, protected Δ42, ΔNAGGN, and wild-type PA14 from osmotic stress (Figures 8A and S10A). Importantly, however, even though glycine-betaine is a potent stress protection molecule in vitro, it did not rescue the Δ42 mutant in vivo (Figure 8B). Conversely, the ΔNAGGN or a zwf mutant, which are highly susceptible to osmotic or oxidative stress, respectively, were not impaired in plant infection (Table S1). These data suggest that trehalose does not play a role as a stress protection molecule in P. aeruginosa during plant infection.
Does trehalose function as a virulence factor for other bacterial phytopathogens in addition to P. aeruginosa? As shown in Table S2, the treYZ and treS trehalose biosynthetic operons are highly conserved among pseudomonads, including P. syringae, but it is not known whether trehalose functions as a virulence factor in these species. A recent study showed that deletion of P. syringae trehalose biosynthetic genes resulted in lowered fitness on the surface of plant leaves, but whether this was due to reduced virulence or increased susceptibility to hyperosmotic stress is not known [17].
What is the explanation for our observation that P. aeruginosa does not require trehalose for pathogenesis in at least three diverse metazoan hosts (mice, insects, and nematodes), and in fact may be a detriment for infection? In contrast to plants, mammals do not synthesize trehalose [30], and it is likely that trehalose, which is a stable and non-reactive molecule, has little effect on mammalian cells, at least at relatively modest concentrations. In the case of insects, trehalose is a major component of the hemolymph. Trehalose is also synthesized by C. elegans, where it accumulates during the formation of desiccation-resistant dauer larvae [31] and exogenous trehalose promotes C. elegans longevity [32]. Thus in the case of flies and worms, trehalose is beneficial and it appears unlikely that the additional levels of trehalose that are synthesized by P. aeruginosa would have a significant physiological affect. The apparent hypervirulence of the trehalose mutants in metazoan models of infection may simply be the result of increased fitness of the strain, which conserves energy by not synthesizing trehalose.
Role of plant cell walls in PA14 infection
Because Arabidopsis cell wall mutants suppress the non-pathogenic phenotype of trehalose mutants, it seems likely that the virulence-enhancing role of trehalose is mediated through the plant cell wall. Can we attribute the lack of a particular plant cell wall polymer as playing a key role in the suppression of the non-pathogenic phenotype of the trehalose mutants? As shown in Figures 4 and S7, several Arabidopsis mutants that we tested either completely (xxt1/xxt2 and mur4-1) or partially (mur2-1, mur3-2, and mur10-2) suppressed the phenotype of the trehalose mutants. A common feature of all of the cell wall mutants that we tested (Figures 4 and S6) is that they exhibit alterations in xyloglucan, the most abundant hemicellulose in the walls of dicotyledonous plants. The xxt1/xxt2 mutant completely lacks xyloglucan [33], mur2-1 and mur3-2 display altered side chains in xyloglucan [34], [35], mur4-1 has decreased levels of arabinose in xyloglucan [23], and mur10-2 exhibits alterations in xyloglucan remodeling throughout the plant [36]. Interestingly, wild-type PA14 grew significantly less in mur3-2 than in Col-0 plants, showing that mur3-2 is more resistant to PA14 than wild-type plants. Thus, the fact that the trehalose mutants grew to the same extent in mur3-2 as in Col-0 (Figures 4 and S6) suggests that mur3-2 also partially suppresses its growth defect. These data suggest that xyloglucan may be a key component of the cell wall that affects the virulence of P. aeruginosa.
At the mechanistic level, it is not necessarily the case that the rigid plant cell wall is functioning, for example, simply as a physical barrier that blocks the ability of P. aeruginosa to extract nutrients from the cytoplasm of mesophyll cells. If the primary role of trehalose is to facilitate nutrient uptake, the source of the nutrients could be the apoplastic fluid or even components of the cell wall itself, such as specific cell-wall associated proteins.
Importantly, the enhanced susceptibility of the Arabidopsis cell wall mutants to the P. aeruginosa trehalose mutants is not simply a consequence of enhanced susceptibility to pathogens in general or the inability of the cell wall mutants to elicit an effective defense response. As shown in Figure 5, the cell wall mutants are not more susceptible to virulent or non-pathogenic P. syringae strains and appear to mount an effective innate immune response when challenged with the flagellin peptide flg22.
How does trehalose promote P. aeruginosa virulence?
Specialized bacterial foliar phytopathogens primarily replicate in the intercellular spaces between mesophyll cells. It is poorly understood which plant-derived nutrients are critical for bacterial growth in this environment as well as the mechanisms utilized by pathogens to obtain nutrients from their hosts. The majority of these pathogens utilize type III effectors not only to suppress the host innate immune response [37], [38] but also to extract nutrients [10] from mesophyll cells. Interestingly, however, the P. aeruginosa type III secretion system is not necessary for pathogenesis in plants [6] and it seems unlikely that a broad host range pathogen such as P. aeruginosa would encode host-specific effectors that subvert the Arabidopsis sugar export system analogously to the Xanthomonas effectors that activate glucose efflux in mesophyll cells [10]. Indeed, it also seems highly unlikely that any particular P. aeruginosa strain has extensively co-evolved with any particular host [4], [9]. Instead, our finding that P. aeruginosa utilizes trehalose as a major virulence factor for plant pathogenesis is consistent with our studies with P. aeruginosa as a C. elegans pathogen, which have shown that the majority of virulence related factors required to infect nematodes correspond to genes that encode conserved global transcriptional regulators or “house-keeping” genes that encode enzymes involved in conserved metabolic processes [4], [9]. Trehalose biosynthesis is highly conserved. All pseudomonads (Table S2) and at least 30% of sequenced prokaryotic genomes encode presumptive trehalose biosynthetic enzymes (J. Urbach and F. Ausubel, unpublished data). It appears that P. aeruginosa has capitalized on what is mostly likely an ancient biosynthetic pathway to promote plant pathogenesis.
How does trehalose promote pathogenesis in an Arabidopsis leaf? Trehalose can serve as a carbon and energy source for growth of many bacteria and fungi including P. aeruginosa [25], [39]. However, we have shown that sucrose and glucose do not suppress the phenotype of P. aeruginosa trehalose mutants and co-inoculation of a double Δ42treA::MAR2xT7 mutant (which cannot metabolize trehalose) with trehalose rescues the non-pathogenic phenotype similarly as co-inoculation of the Δ42 mutant with trehalose (Figure S6). These data suggest that either the level of carbon is not limiting or that trehalose is not involved in carbon acquisition. In addition, experiments designed to determine whether trehalose promotes activity of CWDEs failed to provide evidence that trehalose plays a significant role in cell wall degradation, with the caveats, however, that the experiments we carried out were done in vitro with commercial CWDEs and that in our particular assay a relatively low level of trehalose – enhanced hydrolysis would have not been detected.
Does trehalose function as a general toxin to disrupt host cellular processes? A number of studies have shown that exogenously applied trehalose can have a major negative impact on seedling growth and development [40]–[42]. On the other hand, the concentrations of trehalose used in these seedling experiments (from 30 mM to 100 mM) were significantly higher than the levels that would be expected to be encountered under natural conditions. By way of contrast, in our experiments we used mature four-week old plants and substantially lower concentrations of trehalose (most often 1 mM). In mature plants, trehalose did not have a toxic effect as evidenced by the lack of any visible symptoms following trehalose (1 mM) infiltration (data not shown). Importantly, in our experiments, trehalose concentrations as low as 0.74 mM largely suppressed the non-pathogenic phenotype of the Δ42 mutant and 0.074 mM had a significant effect (Figure 2C). Taken together, our data indicate that trehalose does not have a toxic effect on mature plants in the P. aeruginosa - plant infection model.
Does trehalose upregulate PA14 virulence genes expression? If so, it would have to specifically upregulate genes required for plant pathogenesis because as shown in Figures 3 and S5, PA14 trehalose mutants are not less virulent in nematodes, flies, or mice. However, we do not favor this explanation. As shown in Figures 6, S8, and S9, nitrate, ammonium, maltose and maltotriose functioned similarly to trehalose in suppressing the inability of the PA14 trehalose mutants to grow in planta. It seemly highly unlikely that all three sugars as well as nitrate and ammonium would function similarly to each other as signaling molecules.
Since the non-pathogenic phenotype of P. aeruginosa trehalose mutants can also be suppressed by ammonium nitrate, we propose that trehalose promotes the acquisition of nitrogenous compounds and that nitrogen is limiting in the intercellular environment. The intercellular spaces in leaves are mostly filled with air [43] and very little is known about the mechanisms that plant pathogens utilize to obtain nutrients in this dry environment. Nitrogen limitation during P. aeruginosa infection in plants has been reported previously [44], [45]. One way that trehalose could promote nitrogen acquisition is by modulating host nitrogen metabolism, thereby diverting nitrogen-containing compounds to invading P. aeruginosa cells. Several in planta studies have shown that trehalose-6-phosphate (T6P) plays a key role in the regulation of carbon and nitrogen metabolism [41], [46], [47] and is associated with altered cell wall structure and starch accumulation [48]–[50]. In our study, preliminary transcriptional profiling analysis has shown that infiltration of trehalose into Arabidopsis leaves at a concentration that is effective in rescuing the trehalose mutants (1 mM) has only a very modest effect on Arabidopsis gene expression (S. Djonovic and F. Ausubel, unpublished data). In addition, we showed that trehalose or ammonium nitrate does not modulate plant defense responses, since infiltration of ammonium nitrate or trehalose into Arabidopsis leaves did not make them more susceptible or resistant to virulent or non-pathogenic P. syringae strains or interfere with their ability to mount an effective innate immune response when challenged with the flagellin peptide flg22 (Figure 7). Finally, another way that P. aeruginosa could use trehalose to promote nitrogen acquisition is by generating a high local concentration of trehalose to create an osmotic gradient that causes an efflux of nitrogen containing nutrients from neighboring plant cells, perhaps in conjunction with P. aeruginosa-encoded pore forming toxins. The data in Figure 9, which shows that trehalose functions externally to P. aeruginosa, is consistent with these proposed models.
Conclusions
We have found that P. aeruginosa-synthesized trehalose plays a key role as a virulence factor during infection of plant leaves. Although the mechanistic details remain to be elucidated, our data suggest that a role of trehalose during the infectious process involves the procurement of nitrogen-containing molecules. In contrast to specialized plant pathogens that utilize highly evolved Type III virulence effectors to promote virulence, the multi-host opportunistic pathogen P. aeruginosa, which is not likely to have co-evolved with particular plant hosts, appears to have repurposed a highly conserved anabolic pathway (trehalose biosynthesis) as a potent virulence factor.
Materials and Methods
Ethics statement
Experiments with mice were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The animal protocol was approved by the Harvard Medical Area Institutional Animal Care and Use Committee (Permit Number: 404). All efforts were made to minimize suffering.
Bacterial strains and media
P. aeruginosa strain UCBPP-PA14 [1], P. syringae pv. tomato strain DC3000 [51], and P. syringae pv. phaseolicola strain 3121 [52] have been described. A nonpolar hrcC mutant of P. syringae strain DC3000 (CUCPB5112) was obtained from A. Collmer and B. Kvitklo, Cornell University. Escherichia coli strain SM10 λpir was used for triparental mating [53]. Strains were routinely maintained at 37°C on Luria-Bertani (LB) agar plates or cultured in LB broth supplemented with appropriate antibiotics as needed. The concentrations of antibiotics were: ampicillin or carbenicillin, 50 µg/ml for E. coli or 300 µg/ml for P. aeruginosa; and rifampicin 100 µg/ml. Minimal medium (M63) or modified minimal A medium (MinA) that contained glucose (0.3%) [39] were also used for the growth of P. aeruginosa.
Generation of in-frame PA14 deletion mutants
The ΔPA14_36375-36830 deletion mutant (Δ42) was constructed using a 2.25 kb sequence containing regions immediately flanking the deleted region that was generated by a standard 3-step PCR protocol using FastStart Taq DNA Polymerase (Roche, Germany) and cloned into the KpnI and BamHI sites of pEX18Ap [54] creating plasmid pEX18PA14_36375-36830Δ1. The resulting plasmid was used to introduce the deleted PA14_36375-36830 region into the wild-type PA14 genome by homologous recombination [53]. Similar strategies were used to construct other deletion mutants. For ΔPA14_36375-36560, ΔPA14_36570-36630, ΔPA14_36570-36700, ΔPA14_36710-36740, ΔPA14_36710-36830, and ΔPA14_19350-19370, 12.64-, 10.58-, 16.28-, 7.50-, 12.81-, and 4.78 kb wild-type sequences were deleted by recombination using 1.30, 1.27, 1.06, 1.28, 1.30, and 1.26 kb fragments, respectively, containing the relevant flanking sequences. A double mutant lacking both trehalose operons was constructed by recombining the deleted treS operon in pEX18PA14_36710-36830Δ1 into the ΔPA14_36570-36700 (treYZ) mutant background. A double Δ42treA mutant was constructed by recombining the Δ42 deletion in pEX18PA14_36375-830Δ1 into a treA::MR2xT7 transposon insertion mutant [16]. We confirmed that treA::MR2xT7 mutant could not grow when provided trehalose as sole carbon source in diluted LB and that treA::MR2x7 could not hydrolyze trehalose to glucose using the Somogyi-Nelson assay [21], [22]. All deletion mutants were confirmed by PCR analysis and sequencing.
Generation of fluorescently labeled bacterial strains
PA14 wild-type and the Δ42 mutant were transformed with pSMC2 carrying green fluorescent protein (GFP) [55]. To construct strains expressing red fluorescent protein (RFP), a variant of DsRed2, DsRed.T3(DNT), from Vibrio fischeri [56] was transferred (on a 719 bp SphI – XbaI fragment from pVSV208) into the SphI – XbaI sites of pUCP19 [57] generating pUCP19/DsRed.T3(DNT), which was designated pAA100. pAA100 was transformed into PA14 wild-type and Δ42 by electroporation.
Motility and growth assays
Twitching and swimming motility assays were performed as previously described [58]. To compare growth rates of wild-type and mutants, the cultures were grown at 37°C overnight in LB, centrifuged, washed and resuspended into minimal medium (M63). Bacterial growth was monitored in vitro by plating and counting CFU/ml at 3-, 6- and 9- hour time points. Growth rate (h−1) was calculated using the equation for exponential growth: μ = (lnN 1−lnN 0)/(t 1−t 0), where N 0 and N 1 equal bacterial abundance (CFU/ml) at the beginning (t 0) and end (t 1) of the exponential growth phase. Each experiment was repeated at least twice with similar results.
Plant material and growth of plants
Arabidopsis ecotypes Columbia (Col-0) was obtained from the Arabidopsis Biological Resource Center (Columbus, OH). Plants were grown on 30-mm Jiffy-7 peat pellets (Jiffy Products, Shippagan, New Brunswick, Canada) in a Conviron E7/2 chamber (Winnipeg, Manitoba, Canada) set at a 23°C/20°C day/night regime with a 12-h photoperiod at a light intensity of 100 µE m−2 s−1 and 60% relative humidity. Arabidopsis cell wall mutants were obtained from the Arabidopsis Biological Resource Center: mur2-1 (AT2G03220; CS8565), mur3-2 (AT2G20370, CS8567), mur4-1 (at1g30620, CS8568), mur10-2 (at5g17420, CS8578), and an xxt1/xxt2 double T-DNA insertion line (at3g6272; SALK_119658C/at4g02500; SALK_1013080) as previously published [33].
Arabidopsis pathogenicity assays
Plant infection assays were carried out as previously described [1] with some modifications. P. aeruginosa strains were grown in LB medium overnight, subcultured and grown to an OD600 of 2.5. Cells were centrifuged, washed and resuspended in 10 mM MgSO4. Leaves of four-week old plants were inoculated with a 1×105 CFU/ml suspension of PA14 wild-type or various PA14 mutants, which corresponds to 1×103 CFU/cm2 leaf area. Infected plants were incubated in a growth chamber at 28°C with a 12-h photoperiod at a light intensity of 60 µE m−2s−1 and 90% relative humidity. Six to eight leaves were harvested from three to four plants for CFU determination. Each experiment was repeated at least two to four times with similar results. Co-inoculation of bacteria with betaine, trehalose or sucrose (Sigma, St. Louis, MO) was performed as described above. Before inoculation of leaves, betaine, trehalose, glucose, or sucrose was added to bacterial suspensions at the indicated concentrations, or various phosphate, sulfate, nitrate, or ammonium salts were added at 1 mM. Trehalose was initially added at 2.5 mg/ml, but after we carried out dose response curves and found that 0.25 mg/ml (0.74 mM) was an effective concentration, subsequent experiments were carried out using 1 mM trehalose.
Infection assays with P. syringae strains were performed the same way as with P. aeruginosa with a few exceptions. The temperature in the growth chamber was 22°C and bacterial strains were cultured in King's B medium (protease peptone, 10 mg/ml; glycerol, 15 mg/ml; K2HPO4, 1.5 mg/ml; MgSO4, 5 mM, pH 7.0) until late logarithmic phase. Elicitation assays were performed by infiltration of leaves with 1 µM flg22, 1 mM trehalose, 1 mM glucose or 1 mM ammonium nitrate (or in combinations) 24 hours prior to bacterial inoculation.
Metazoan pathogenicity assays
C. elegans slow killing assays were performed as previously described [3]. Briefly, PA14, Δ42, or ΔtreYZΔtreS mutants were grown overnight in LB and 10 µl of each liquid culture was spread onto 3 SK plates (modified NGM medium; [3]). The plates were incubated at 37°C for 24 hours and then at 25°C for 20–24 hours. 35–45 fer-15;fem-1 sterile L4 nematodes were picked to the SK plates seeded with PA14, Δ42, or ΔtreYZΔtreS and the plates were incubated at 25°C. Live and dead animals were counted daily for approximately 8 days. A worm was scored dead when it no longer responded to touch.
Infection survival assays in D. melanogaster were conducted with D. melanogaster strains w[118] (Bloomington stock #6326) or Oregon R, which were grown under non-crowded conditions on standard cornmeal-molasses medium. Fly husbandry and infections were carried out at 25°C, 70% humidity, 12 hours light cycle. For infections assays, P. aeruginosa was grown aerated at 37°C in LB medium containing 50 µg/ml rifampicin, and subcultured to an OD600 = 2.3–2.5. The bacterial culture was diluted to a final concentration 80% LB, 4% sucrose, 50 µg/ml rifampicin and 3×108 CFU/ml and 7 ml of infection mixture was pipetted onto sterilized cotton balls at the bottom of clean, empty fly vials. 25 male flies, 4 days old, were added and their survival monitored several times a day.
Mouse experiments complied with institutional and federal guidelines regarding the use of animals in research. For the acute pneumonia model, a modified version of a previously described method of intranasal inoculation of anesthetized mice was utilized [19]. Briefly, 6- to 8-week-old female C3H/HeN mice (Harlan) were sedated with ketamine and xylazine and then 10 µl of a bacterial suspension was applied to each nostril. Bacterial suspensions were prepared in PBS (OD600 = 0.5) after overnight growth of frozen stock on TSA. Doses were determined by serial dilution and plating on MacConkey agar (1.5×107 CFU/20 µl for PA14, 1.4×107 CFU/20 µl for Δ42). After 18 hr, mice were euthanized with carbon dioxide and then lungs and spleens were removed, weighed, and homogenized in 1 ml of 1% proteose peptone in water. Viable counts were determined by serial dilution and plating.
For the chronic oropharyngeal colonization model in transgenic CF mice, we utilized mouse strain Cftr tm1Unc-TgN(FABP-CFTR) (denoted FABP-CFTR), which has a stop codon in the murine cftr gene (S489X) but also expresses human CFTR in the gut epithelium due to transgenic introduction of human Cftr under the control of the fatty acid binding protein (FABP) promoter [59]. These FABP-CFTR mice have been bred into the FVB/N genetic background (breeding pairs were initially provided by Dr. J. Whitsett, University of Cincinnati). These FABP-CFTR mice are susceptible to chronic oropharyngeal colonization with P. aeruginosa after exposure in the drinking water [20]. To establish colonization, age- and gender-matched mice were given oral levofloxacin in their drinking water for 5 days, followed by gentamicin for 2 days, followed by bacteria (either PA14 or the Δ42 mutant) suspended in water at 107 CFU/ml. Bacterial levels in the drinking water were unchanged at the end of 7-day exposure. Throat cultures were then taken every 1–2 weeks using a swab inserted into the oropharynx of mice anesthetized with isofluorane. The swab was placed in 1 ml tryptic soy broth and incubated at 37°C for 3 hours. Next, 1 ml of nitrofurantoin (2 mg/ml) was added to suppress the growth of any contaminating Enterobacter spp., which can interfere with detection of P. aeruginosa. The culture was incubated overnight at 37°C and then subcultured overnight on cetrimide agar. All mice in both groups (n = 9 for the Δ42 mutant, n = 8 for PA14 WT) had positive throat cultures after colonization. Mice were then followed for survival.
Bioinformatic analysis of PA14_36375-36830 genes
To assign putative functions to genes within the block of PA14_36375 through PA14_36830, each protein in the 42 kb cluster was used as a query in a BLAST or PSI-BLAST homology search. In the process of assigning putative functions, several types of information were taken into account: homologous proteins with experimentally assigned function; homologous proteins with computationally predicted function; matches to HMMs from conserved domain databases; and the genomic/operon context of close homologs.
The protein and nucleotide sequences of prokaryotic genomes were obtained from NCBI (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/). Additionally, two P. aeruginosa genomes (P. aeruginosa 2192; P. aeruginosa C3719) were obtained from the Broad Institute of Harvard and MIT (http://www.broadinstitute.org/annotation/genome/pseudomonas_group/MultiHome.html).
To identify orthologs to PA14_36375-36830 genes, two criteria were used. First, putative ortholog pairs were required to be reciprocal best hits, with an e-value less than or equal to 0.0001 for best hits of the PA14 proteins against compared protein sets, and an e-value less than or equal to 0.001 for reciprocal best hits against the PA14 protein set. Secondly, the putative orthologs were required to align for at least 80 percent of their lengths and have less than a 20% difference in protein sequence lengths, thereby conserving overall domain structure. Of these constraints, the e-value and sequence length constraints are very permissive, whereas the requirement for alignment length is stringent.
Trehalose quantification assay
P. aeruginosa strains were grown at 37°C in MinA medium with 0.5 M NaCl to an early stationary phase. Trehalose was extracted from a 19 ml culture by pelleting the cells, resuspending in 0.5 ml water, and heating at 95°C for 20 min [60]. The concentration of trehalose in the supernatants was determined using an enzymatic assay by converting trehalose to glucose with trehalase and then measuring the glucose using a trehalose assay kit (Megazyme International Ireland Limited). The pre-existing glucose in each sample was determined in a control reaction without trehalase and subtracted from the total glucose. The experiment was repeated at least twice with similar results.
Stress response, resistance and biofilm assays
Osmotic stress sensitivity: P. aeruginosa was grown at 37°C in MinA containing 17 mM glucose [39], washed and subcultured into MinA containing 0.5 M NaCl. Bacterial growth was monitored by plating CFU. Persistence assay: This assay was performed as previously described [61]. Briefly, persisters were determined by exposure of stationary cultures to antibiotics at concentrations exceeding the corresponding bacterial minimal inhibitory concentrations (MICs). The antibiotics were used at the following concentrations: 6 µg/ml tobramycin, 2 µg/ml ciprofloxacin and 3 mg/ml carbenicillin. Oxidative stress resistance: Hydrogen peroxide was added directly to an overnight culture grown for 14 h in LB medium and the cells were incubated for 8 h at 37°C. The following concentrations of hydrogen peroxide were used: 1 M (non-lethal), 2 M (sub-lethal), and 3 M (lethal dose). To test sensitivity to paraquat (Sigma, 856177), overnight MinA cultures were diluted 100 fold in MinA containing different amounts of paraquat (0.1, 1 and 10 mM) and cultured for three days. pH stress: Cultures were grown to stationary phase in LB medium that had been titrated with HCl to pH 4, 5, 6, or 7. Thermotolerance: Small volumes of stationary phase cells were heated in Eppendorf tubes in a heating block at different temperatures and incubation times, then rapidly diluted and plated. Biofilm formation: Biofilm attachment assays were performed using wild-type PA14, Δ42 and ΔPA14_19350-19370 (ΔNAGGN) cultures grown in 96-well polyvinylchloride (PVC) plates as described previously [62]. Overnight cultures were diluted 1/100 in MinA medium or MinA medium supplemented with 0.5 M or 0.75 M NaCl. Aliquots of 100 µL were dispensed into the wells of PVC microtiter plates and incubated at 37°C. Attachment was detected by staining with 1% crystal violet dissolved in water. Dye not associated with bacteria was removed by rinsing with water. Bacteria-associated dye was solubilized using 95% ethanol and absorbance was determined at 550 nm. Each experiment was repeated at least twice with similar results.
Statistical analysis
Statistical analyses in animal experiments were performed using GraphPad Prism 5 software (La Jolla, CA) and a log rank (Mantel-Cox) test to assess the significance of differential survival, and a Mann-Whitney U non-parametric test for significance of CFU data, which were not normally distributed. Statistics in all other experiments was performed using analysis of variance (ANOVA) and a Fisher's PLSD test (Statview v. 5.0.1, SAS Institute, Cary, NC).
Supporting Information
Expression of PA14_36500 encoding a putative cellulase/peptidase in infected Arabidopsis leaves. Semiquantitative RT-PCR was carried out as described in Materials and Methods. (A) PA14_36500 transcript levels on various days post infiltration (dpi) with PA14 wild-type. (B) PA14_36500 expression in wild-type PA14 and a PA14 ΔmvfR mutant 2 days post-infiltration. P. aeruginosa PA14 ribosomal protein L21 (rplU) was used as a control for equal amounts of cDNA. The experiment was repeated at least two times with similar results.
(TIF)
The 42 kb cluster encodes two independent pathways for trehalose biosynthesis. In the top pathway, gene product 36605 (a putative maltooligosyltrehalose synthase) alters the regiochemistry of the terminal sugar linkage from alpha-1,4 to alpha-1,1; the terminal disaccharide is subsequently cleaved by gene product 36580 (a putative maltooligosyltrehalose trehalohydrolase), releasing trehalose. In the bottom pathway, gene product 36740 (a putative alpha-amylase) cleaves the terminal disaccharide of the alpha-1,4-glucan, releasing maltose. The alpha-1,4 linkage of the maltose disaccharide is then isomerized to alpha-1,1 by gene product 36730 (a putative trehalose synthase), yielding trehalose.
(TIF)
Pyocyanin production by Δ42 and PA14 wild-type. Bacterial strains were streaked onto Pseudomonas agar P to assess pyocyanin production (see Materials and Methods) and incubated 20 h at 37°C. Sectors: Δ42-1 and Δ42-2 (two independent Δ42 deletion constructs); phzM (phzM::MAR2xT7, negative control: a pyocyanin-defective mutant); WT (PA14 wild-type). Characteristic blue-green color indicates that the strain is proficient in pyocyanin production.
(TIF)
Growth of P. aeruginosa Δ42, three sub-region in-frame deletion mutants, and PA14 wild-type in Arabidopsis Col-0 leaves. Plants were inoculated and incubated as described in Materials and Methods. The leaves were harvested 3 days post-inoculation. Data represent the mean of bacterial titers ± SE of six leaf disks excised from 6 leaves of 3 plants. Different letters above bars denote statistically significant differences (P<0.05, Fisher's PLSD test). See Figure 1 for a description of the mutants.
(TIF)
The Δ42 mutant is more virulent than wild-type PA14 in nematodes, insects, and mice. (A) C. elegans are more sensitive to killing by the Δ42 mutant than PA14 wild-type (P<0.004). Mutant fer15;fem1 C. elegans animals were exposed to P. aeruginosa strains and survival was determined as described in Materials and Methods. Data at each time point correspond to the average of three plates per strain, each with approximately 40 animals per plate, and are representative of two independent experiments. (B) D. melanogaster infected with Δ42 die faster than flies infected with PA14 wild-type (P<0.03). D. melanogaster strain Oregon R was infected with P. aeruginosa and approximately 25 flies per vial were scored several times a day for survival throughout the time course of infection. Data are representative of four independent experiments carried out with two different D. melanogaster lines (Oregon R and w[118]). (C) The Δ42 mutant is more virulent in a murine acute lung infection model. See Materials and Methods for details of infection protocol. CFU/gram of lung tissue of mice infected with Δ42 mutant is 3.8-fold higher than with wild-type PA14 18 hours post intranasal infection (P<0.01, Mann-Whitney U test). Data are representative of two independent experiments. (D) FABP-CFTR transgenic mice are more susceptible to killing by Δ42 mutant than by PA14 wild-type after oropharyngeal colonization (P<0.04, log rank test). All mice in both groups (n = 9 for Δ42; n = 8 for PA14 WT) had positive throat cultures for the duration of the experiment after initial colonization by exposure to bacteria in drinking water for one week.
(TIF)
The in planta growth defect of the Δ42 mutant is suppressed by Arabidopsis cell wall mutants. Growth of PA14 wild-type or Δ42 3 days post infiltration in Arabidopsis cell wall mutants mur2-1, mur3-2, mur4-1, mur10-2 and xxt1/xxt2. Data represent the mean of bacterial titers ± SE of six leaf disks excised from 6 leaves of 3 plants. Letters above bars denote statistically significant differences (P<0.05, Fisher's PLSD test). The experiments were repeated at least two times.
(TIF)
The growth of the Δ42 mutant in planta is not limited by lack of a carbon source. (A) The growth of the Δ42 mutant in planta is not suppressed by 1.25 mg/ml glucose or 2.5 mg/ml sucrose. (B) Suppression of the growth defect of Δ42treA::MAR2xT7 with trehalose in Arabidopsis leaves. Plants were inoculated and incubated as described in Materials and Methods and leaves were harvested 3 days post-inoculation. In (A) and (B), data represent the mean of bacterial titers ± SE of six leaf disks excised from 6 leaves of 3 plants. Different letters above bars denote statistically significant differences (P<0.05, Fisher's PLSD test). The experiments were repeated at least two times.
(TIF)
The in planta growth defect of the Δ42 mutant is suppressed by ammonium or nitrate ions. Leaves of four-week-old Arabidopsis Col-0 plants were infiltrated with PA14 wild-type or Δ42 as described in Materials and Methods except that the infiltration solution contained various phosphate, sulfate, nitrate, or ammonium salts at 1 mM. Suppression of the growth defect of Δ42 with 2.5 mg/ml trehalose (Tre) and 1.25 mg/ml glucose (Glc) were tested as positive and negative controls, respectively. Data represent the mean of bacterial titers ± SE of six leaf disks excised from 6 leaves of 3 plants. Letters above bars denote statistically significant differences (P<0.05, Fisher's PLSD test). The experiments were repeated at least two times.
(TIF)
Suppression of attenuation of Δ42 with exogenous maltose and maltotriose. Col-0 plants were infiltrated with PA14 wild-type or Δ42 co-inoculated with maltose (M) or malotriose (MT) at the indicated concentrations. Leaves were harvested 3 days post-infiltration. Data represent the mean of bacterial titers ± SE of six leaf disks excised from 6 leaves of 3 plants. Letters above bars denote statistically significant differences (P<0.05, Fisher's PLSD test). The experiment was repeated at least two times.
(TIF)
Response of Δ42 to various stress conditions. (A) Rescue of Δ42, ΔNAGGN and PA14 wild-type with betaine but not trehalose under osmotic stress in vitro. Cells were grown at 37°C in MinA medium with 0.5 M NaCl (squares), or with 0.5 M NaCl+1 mM betaine (Bet) (circles) or 1 mM trehalose (Tre) (triangles). Data represent the mean ± SE of 3 replicates. (B) Thermotolerance. Survival of stationary phase bacteria after a 30 minute exposure to 53°C. Temperatures below 53°C were non-lethal and above 56°C were 100% lethal. (C) Biofilm attachment under osmotic stress. Overnight cultures were diluted 1/100 in MinA medium supplemented with 0.5 or 0.75 M NaCl. Attachment assays were performed as described in Materials and Methods. (D) Growth under pH stress. Cultures were grown to stationary phase in LB medium adjusted to pH 4, 5, 6, or 7. (E) Persistence assay. Persisters were determined by exposure of stationary cultures (inoculum, time point zero on x axis) to 6 µg/ml tobramycin, 2 µg/ml ciprofloxacin or 3 mg/ml carbenicillin. The assay was performed as described in Materials and Methods. Based on analysis of variance (ANOVA) and Fisher's PLSD test (P<0.05), there was no significant differences between Δ42 and PA14 wild-type in any of the assays (A–E).
(TIF)
Growth of P. aeruginosa mutants in Arabidopsis Col-0 leaves. Plants were inoculated and incubated as described in Materials and Methods and leaves were harvested 3 days post-inoculation. Data represent the mean of bacterial titers ± SE of six leaf disks excised from 6 leaves of 3 plants. Different superscript letters denote statistically significant differences (P<0.05, Fisher's PLSD test).
(DOC)
Predicted functions and orthologs of PA14_36375-36830 genes across sequenced Pseudomonas genomes. Putative functions of PA14_36375-36830 genes were assigned as described in Materials and Methods. Genes of the reference taxon PA14 are indicated in the left column (PA14 loci), and different pseudomonad genomes are indicated on the top row. The presence of an ortholog is indicated by a checkmark. Boxes of identical hue indicate that the genes are contiguous in a particular genome, with the lighter shades on the top strand, and darker shades on the bottom strand. Checks in white boxes indicate an ortholog that is not contiguous with other PA14_36375-36830 block orthologs.
(PDF)
Growth, biofilm formation and motility of Δ42 and PA14 wild-type. Growth rate (h−1) of Pseudomonas strains in minimal media (M63) was calculated by the equation for exponential growth (see Materials and Methods). Biofilm formation was measured as attachment to polyvinylchloride plates in absorbance units (OD550; see Materials and Methods). Swimming and twitching motility are represented as a radius of a halo in cm (see Materials and Methods). Two independent Δ42 deletion constructs were tested. Data represent the mean ± SE. Based on analysis of variance (ANOVA) and Fisher's PLSD test (P<0.05), there was no significant differences between Δ42 mutant and PA14 wild-type in any of the assays.
(DOC)
Acknowledgments
We thank M. Kesarwani, R. Hazan and L. Rahme for helpful discussions and for sharing transcriptional profiling data prior to publication, A. Adonizio for constructing pAA100, N. Clay for providing seeds of Arabidopsis cell wall mutants, and S. Yu for helping with C. elegans assays.
Funding Statement
This work was funded by an MBRC Tosteson Fellowship awarded to SD, an Opportunity Award from the Program for Breakthrough Biomedical Research and NIH grant DP2 OD007290 awarded to MAF, NIH grant R01 HL092515 awarded to GPP, and NIH grants R37 GM48707 and R01 AI085581 and by NSF grant MCB-0519898 awarded to FMA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, et al. (1995) Common virulence factors for bacterial pathogenicity in plants and animals. Science 268: 1899–1902. [DOI] [PubMed] [Google Scholar]
- 2. Jander G, Rahme LG, Ausubel FM (2000) Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. Journal of bacteriology 182: 3843–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tan MW, Mahajan-Miklos S, Ausubel FM (1999) Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proceedings of the National Academy of Sciences of the United States of America 96: 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee DG, Urbach JM, Wu G, Liberati NT, Feinbaum RL, et al. (2006) Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol 7: R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM (1999) Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96: 47–56. [DOI] [PubMed] [Google Scholar]
- 6. Miyata S, Casey M, Frank DW, Ausubel FM, Drenkard E (2003) Use of the Galleria mellonella caterpillar as a model host to study the role of the type III secretion system in Pseudomonas aeruginosa pathogenesis. Infect Immun 71: 2404–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rahme LG, Tan MW, Le L, Wong SM, Tompkins RG, et al. (1997) Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc Natl Acad Sci U S A 94: 13245–13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mathee K, Narasimhan G, Valdes C, Qiu X, Matewish JM, et al. (2008) Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci U S A 105: 3100–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Feinbaum RL, Urbach JM, Liberati NT, Djonovic S, Adonizio A, et al. (2012) Genome-Wide Identification of Pseudomonas aeruginosa Virulence-Related Genes Using a Caenorhabditis elegans Infection Model. PLoS Pathog 8: e1002813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, et al. (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468: 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Plotnikova JM, Rahme LG, Ausubel FM (2000) Pathogenesis of the human opportunistic pathogen Pseudomonas aeruginosa PA14 in Arabidopsis. Plant Physiology 124: 1766–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Waite RD, Paccanaro A, Papakonstantinopoulou A, Hurst JM, Saqi M, et al. (2006) Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles. BMC Genomics 7: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deziel E, Gopalan S, Tampakaki AP, Lepine F, Padfield KE, et al. (2005) The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol Microbiol 55: 998–1014. [DOI] [PubMed] [Google Scholar]
- 14. Maruta K, Mitsuzumi H, Nakada T, Kubota M, Chaen H, et al. (1996) Cloning and sequencing of a cluster of genes encoding novel enzymes of trehalose biosynthesis from thermophilic archaebacterium Sulfolobus acidocaldarius . Biochim Biophys Acta 1291: 177–181. [DOI] [PubMed] [Google Scholar]
- 15. Nishimoto T, Nakano M, Nakada T, Chaen H, Fukuda S, et al. (1996) Purification and properties of a novel enzyme, trehalose synthase, from Pimelobacter sp. R48. Biosci Biotechnol Biochem 60: 640–644. [DOI] [PubMed] [Google Scholar]
- 16. Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, et al. (2006) An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103: 2833–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Freeman BC, Chen C, Beattie GA (2010) Identification of the trehalose biosynthetic loci of Pseudomonas syringae and their contribution to fitness in the phyllosphere. Environ microbiol 12: 1486–1497. [DOI] [PubMed] [Google Scholar]
- 18. Limmer S, Haller S, Drenkard E, Lee J, Yu S, et al. (2011) Pseudomonas aeruginosa RhlR is required to neutralize the cellular immune response in a Drosophila melanogaster oral infection model. Proc Natl Acad Sci U S A 108: 17378–17383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Allewelt M, Coleman FT, Grout M, Priebe GP, Pier GB (2000) Acquisition of expression of the Pseudomonas aeruginosa ExoU cytotoxin leads to increased bacterial virulence in a murine model of acute pneumonia and systemic spread. Infect Immun 68: 3998–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Coleman FT, Mueschenborn S, Meluleni G, Ray C, Carey VJ, et al. (2003) Hypersusceptibility of cystic fibrosis mice to chronic Pseudomonas aeruginosa oropharyngeal colonization and lung infection. Proc Natl Acad Sci U S A 100: 1949–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nelson NJ (1957) Colorimetric analysis of sugars. Methods Enzymol 3: 85–86. [Google Scholar]
- 22. Somogyi M (1952) Notes on sugar determination. J Biol Chem 195: 19–23. [PubMed] [Google Scholar]
- 23. Burget EG, Reiter WD (1999) The mur4 mutant of Arabidopsis is partially defective in the de novo synthesis of uridine diphospho L-arabinose. Plant Physiol 121: 383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, et al. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767. [DOI] [PubMed] [Google Scholar]
- 25. Arguelles JC (2000) Physiological roles of trehalose in bacteria and yeasts: a comparative analysis. Arch Microbiol 174: 217–224. [DOI] [PubMed] [Google Scholar]
- 26. Iturriaga G, Suarez R, Nova-Franco B (2009) Trehalose metabolism: from osmoprotection to signaling. Int J Mol Sci 10: 3793–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. D'Souza-Ault MR, Smith LT, Smith GM (1993) Roles of N-acetylglutaminylglutamine amide and glycine betaine in adaptation of Pseudomonas aeruginosa to osmotic stress. Appl Environ Microbiol 59: 473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ma JF, Hager PW, Howell ML, Phibbs PV, Hassett DJ (1998) Cloning and characterization of the Pseudomonas aeruginosa zwf gene encoding glucose-6-phosphate dehydrogenase, an enzyme important in resistance to methyl viologen (paraquat). J Bacteriol 180: 1741–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Behrends V, Ryall B, Wang X, Bundy JG, Williams HD (2010) Metabolic profiling of Pseudomonas aeruginosa demonstrates that the anti-sigma factor MucA modulates osmotic stress tolerance. Mol Biosyst 6: 562–569. [DOI] [PubMed] [Google Scholar]
- 30. Elbein AD, Pan YT, Pastuszak I, Carroll D (2003) New insights on trehalose: a multifunctional molecule. Glycobiology 13: 17R–27R. [DOI] [PubMed] [Google Scholar]
- 31. Erkut C, Penkov S, Khesbak H, Vorkel D, Verbavatz JM, et al. (2011) Trehalose renders the dauer larva of Caenorhabditis elegans resistant to extreme desiccation. Curr Biol 21: 1331–1336. [DOI] [PubMed] [Google Scholar]
- 32. Honda Y, Tanaka M, Honda S (2010) Trehalose extends longevity in the nematode Caenorhabditis elegans . Aging Cell 9: 558–569. [DOI] [PubMed] [Google Scholar]
- 33. Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, et al. (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20: 1519–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, et al. (2002) The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc Natl Acad Sci U S A 99: 3340–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Madson M, Dunand C, Li X, Verma R, Vanzin GF, et al. (2003) The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15: 1662–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bosca S, Barton CJ, Taylor NG, Ryden P, Neumetzler L, et al. (2006) Interactions between MUR10/CesA7-dependent secondary cellulose biosynthesis and primary cell wall structure. Plant Physiol 142: 1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. He P, Shan L, Lin NC, Martin GB, Kemmerling B, et al. (2006) Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125: 563–575. [DOI] [PubMed] [Google Scholar]
- 38. Li X, Lin H, Zhang W, Zou Y, Zhang J, et al. (2005) Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc Natl Acad Sci U S A 102: 12990–12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Aspedon A, Palmer K, Whiteley M (2006) Microarray analysis of the osmotic stress response in Pseudomonas aeruginosa . J Bacteriol 188: 2721–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gravot A, Grillet L, Wagner G, Jubault M, Lariagon C, et al. (2011) Genetic and physiological analysis of the relationship between partial resistance to clubroot and tolerance to trehalose in Arabidopsis thaliana . New Phytol 191: 1083–1094. [DOI] [PubMed] [Google Scholar]
- 41. Schluepmann H, van Dijken A, Aghdasi M, Wobbes B, Paul M, et al. (2004) Trehalose mediated growth inhibition of Arabidopsis seedlings is due to trehalose-6-phosphate accumulation. Plant Physiol 135: 879–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bae H, Herman E, Bailey B, Bae H, Sicher R (2005) Exogenous trehalose alters Arabidopsis transcripts involved in cell wall modification, abiotic stress, nitrogen metabolism, and plant defense. Physiologia Plantarum 125: 114–126. [Google Scholar]
- 43. Sifton HB (1945) Air-space tissue in plants. Bot Rev 11: 108–143. [Google Scholar]
- 44. Bielecki P, Puchalka J, Wos-Oxley ML, Loessner H, Glik J, et al. (2011) In-vivo expression profiling of Pseudomonas aeruginosa infections reveals niche-specific and strain-independent transcriptional programs. PLoS One 6: e24235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Weir TL, Stull VJ, Badri D, Trunck LA, Schweizer HP, et al. (2008) Global gene expression profiles suggest an important role for nutrient acquisition in early pathogenesis in a plant model of Pseudomonas aeruginosa infection. Appl Environ Microbiol 74: 5784–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang R, Okamoto M, Xing X, Crawford NM (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ramon M, Rolland F (2007) Plant development: introducing trehalose metabolism. Trends Plant Sci 12: 185–188. [DOI] [PubMed] [Google Scholar]
- 48. Wingler A, Fritzius T, Wiemken A, Boller T, Aeschbacher RA (2000) Trehalose induces the ADP-glucose pyrophosphorylase gene, ApL3, and starch synthesis in Arabidopsis. Plant Physiol 124: 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Best M, Koenig K, McDonald K, Schueller M, Rogers A, et al. (2011) Inhibition of trehalose breakdown increases new carbon partitioning into cellulosic biomass in Nicotiana tabacum . Carbohydr Res 346: 595–601. [DOI] [PubMed] [Google Scholar]
- 50. Gomez LD, Baud S, Gilday A, Li Y, Graham IA (2006) Delayed embryo development in the ARABIDOPSIS TREHALOSE-6-PHOSPHATE SYNTHASE 1 mutant is associated with altered cell wall structure, decreased cell division and starch accumulation. Plant J 46: 69–84. [DOI] [PubMed] [Google Scholar]
- 51. Whalen MC, Innes RW, Bent AF, Staskawicz BJ (1991) Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lindgren PB, Peet RC, Panopoulos NJ (1986) Gene cluster of Pseudomonas syringae pv. “phaseolicola” controls pathogenicity of bean plants and hypersensitivity of nonhost plants. J Bacteriol 168: 512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Donnenberg MS, Kaper JB (1991) Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun 59: 4310–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Prentki P, Krisch HM (1984) In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29: 303–313. [DOI] [PubMed] [Google Scholar]
- 55. Bloemberg GV, O'Toole GA, Lugtenberg BJ, Kolter R (1997) Green fluorescent protein as a marker for Pseudomonas spp. . Appl Environ Microbiol 63: 4543–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV (2006) New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol 72: 802–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. West SE, Schweizer HP, Dall C, Sample AK, Runyen-Janecky LJ (1994) Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa . Gene 148: 81–86. [DOI] [PubMed] [Google Scholar]
- 58. Darzins A (1993) The pilG gene product, required for Pseudomonas aeruginosa pilus production and twitching motility, is homologous to the enteric, single-domain response regulator CheY. Journal of Bacteriology 175: 5934–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhou L, Dey CR, Wert SE, DuVall MD, Frizzell RA, et al. (1994) Correction of lethal intestinal defect in a mouse model of cystic fibrosis by human CFTR. Science 266: 1705–1708. [DOI] [PubMed] [Google Scholar]
- 60. Kandror O, DeLeon A, Goldberg AL (2002) Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc Natl Acad Sci U S A 99: 9727–9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Moker N, Dean CR, Tao J (2010) Pseudomonas aeruginosa increases formation of multidrug-tolerant persister cells in response to quorum-sensing signaling molecules. Journal of Bacteriology 192: 1946–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. O'Toole GA, Kolter R (1998) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Molecular Microbiology 28: 449–461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of PA14_36500 encoding a putative cellulase/peptidase in infected Arabidopsis leaves. Semiquantitative RT-PCR was carried out as described in Materials and Methods. (A) PA14_36500 transcript levels on various days post infiltration (dpi) with PA14 wild-type. (B) PA14_36500 expression in wild-type PA14 and a PA14 ΔmvfR mutant 2 days post-infiltration. P. aeruginosa PA14 ribosomal protein L21 (rplU) was used as a control for equal amounts of cDNA. The experiment was repeated at least two times with similar results.
(TIF)
The 42 kb cluster encodes two independent pathways for trehalose biosynthesis. In the top pathway, gene product 36605 (a putative maltooligosyltrehalose synthase) alters the regiochemistry of the terminal sugar linkage from alpha-1,4 to alpha-1,1; the terminal disaccharide is subsequently cleaved by gene product 36580 (a putative maltooligosyltrehalose trehalohydrolase), releasing trehalose. In the bottom pathway, gene product 36740 (a putative alpha-amylase) cleaves the terminal disaccharide of the alpha-1,4-glucan, releasing maltose. The alpha-1,4 linkage of the maltose disaccharide is then isomerized to alpha-1,1 by gene product 36730 (a putative trehalose synthase), yielding trehalose.
(TIF)
Pyocyanin production by Δ42 and PA14 wild-type. Bacterial strains were streaked onto Pseudomonas agar P to assess pyocyanin production (see Materials and Methods) and incubated 20 h at 37°C. Sectors: Δ42-1 and Δ42-2 (two independent Δ42 deletion constructs); phzM (phzM::MAR2xT7, negative control: a pyocyanin-defective mutant); WT (PA14 wild-type). Characteristic blue-green color indicates that the strain is proficient in pyocyanin production.
(TIF)
Growth of P. aeruginosa Δ42, three sub-region in-frame deletion mutants, and PA14 wild-type in Arabidopsis Col-0 leaves. Plants were inoculated and incubated as described in Materials and Methods. The leaves were harvested 3 days post-inoculation. Data represent the mean of bacterial titers ± SE of six leaf disks excised from 6 leaves of 3 plants. Different letters above bars denote statistically significant differences (P<0.05, Fisher's PLSD test). See Figure 1 for a description of the mutants.
(TIF)
The Δ42 mutant is more virulent than wild-type PA14 in nematodes, insects, and mice. (A) C. elegans are more sensitive to killing by the Δ42 mutant than PA14 wild-type (P<0.004). Mutant fer15;fem1 C. elegans animals were exposed to P. aeruginosa strains and survival was determined as described in Materials and Methods. Data at each time point correspond to the average of three plates per strain, each with approximately 40 animals per plate, and are representative of two independent experiments. (B) D. melanogaster infected with Δ42 die faster than flies infected with PA14 wild-type (P<0.03). D. melanogaster strain Oregon R was infected with P. aeruginosa and approximately 25 flies per vial were scored several times a day for survival throughout the time course of infection. Data are representative of four independent experiments carried out with two different D. melanogaster lines (Oregon R and w[118]). (C) The Δ42 mutant is more virulent in a murine acute lung infection model. See Materials and Methods for details of infection protocol. CFU/gram of lung tissue of mice infected with Δ42 mutant is 3.8-fold higher than with wild-type PA14 18 hours post intranasal infection (P<0.01, Mann-Whitney U test). Data are representative of two independent experiments. (D) FABP-CFTR transgenic mice are more susceptible to killing by Δ42 mutant than by PA14 wild-type after oropharyngeal colonization (P<0.04, log rank test). All mice in both groups (n = 9 for Δ42; n = 8 for PA14 WT) had positive throat cultures for the duration of the experiment after initial colonization by exposure to bacteria in drinking water for one week.
(TIF)
The in planta growth defect of the Δ42 mutant is suppressed by Arabidopsis cell wall mutants. Growth of PA14 wild-type or Δ42 3 days post infiltration in Arabidopsis cell wall mutants mur2-1, mur3-2, mur4-1, mur10-2 and xxt1/xxt2. Data represent the mean of bacterial titers ± SE of six leaf disks excised from 6 leaves of 3 plants. Letters above bars denote statistically significant differences (P<0.05, Fisher's PLSD test). The experiments were repeated at least two times.
(TIF)
The growth of the Δ42 mutant in planta is not limited by lack of a carbon source. (A) The growth of the Δ42 mutant in planta is not suppressed by 1.25 mg/ml glucose or 2.5 mg/ml sucrose. (B) Suppression of the growth defect of Δ42treA::MAR2xT7 with trehalose in Arabidopsis leaves. Plants were inoculated and incubated as described in Materials and Methods and leaves were harvested 3 days post-inoculation. In (A) and (B), data represent the mean of bacterial titers ± SE of six leaf disks excised from 6 leaves of 3 plants. Different letters above bars denote statistically significant differences (P<0.05, Fisher's PLSD test). The experiments were repeated at least two times.
(TIF)
The in planta growth defect of the Δ42 mutant is suppressed by ammonium or nitrate ions. Leaves of four-week-old Arabidopsis Col-0 plants were infiltrated with PA14 wild-type or Δ42 as described in Materials and Methods except that the infiltration solution contained various phosphate, sulfate, nitrate, or ammonium salts at 1 mM. Suppression of the growth defect of Δ42 with 2.5 mg/ml trehalose (Tre) and 1.25 mg/ml glucose (Glc) were tested as positive and negative controls, respectively. Data represent the mean of bacterial titers ± SE of six leaf disks excised from 6 leaves of 3 plants. Letters above bars denote statistically significant differences (P<0.05, Fisher's PLSD test). The experiments were repeated at least two times.
(TIF)
Suppression of attenuation of Δ42 with exogenous maltose and maltotriose. Col-0 plants were infiltrated with PA14 wild-type or Δ42 co-inoculated with maltose (M) or malotriose (MT) at the indicated concentrations. Leaves were harvested 3 days post-infiltration. Data represent the mean of bacterial titers ± SE of six leaf disks excised from 6 leaves of 3 plants. Letters above bars denote statistically significant differences (P<0.05, Fisher's PLSD test). The experiment was repeated at least two times.
(TIF)
Response of Δ42 to various stress conditions. (A) Rescue of Δ42, ΔNAGGN and PA14 wild-type with betaine but not trehalose under osmotic stress in vitro. Cells were grown at 37°C in MinA medium with 0.5 M NaCl (squares), or with 0.5 M NaCl+1 mM betaine (Bet) (circles) or 1 mM trehalose (Tre) (triangles). Data represent the mean ± SE of 3 replicates. (B) Thermotolerance. Survival of stationary phase bacteria after a 30 minute exposure to 53°C. Temperatures below 53°C were non-lethal and above 56°C were 100% lethal. (C) Biofilm attachment under osmotic stress. Overnight cultures were diluted 1/100 in MinA medium supplemented with 0.5 or 0.75 M NaCl. Attachment assays were performed as described in Materials and Methods. (D) Growth under pH stress. Cultures were grown to stationary phase in LB medium adjusted to pH 4, 5, 6, or 7. (E) Persistence assay. Persisters were determined by exposure of stationary cultures (inoculum, time point zero on x axis) to 6 µg/ml tobramycin, 2 µg/ml ciprofloxacin or 3 mg/ml carbenicillin. The assay was performed as described in Materials and Methods. Based on analysis of variance (ANOVA) and Fisher's PLSD test (P<0.05), there was no significant differences between Δ42 and PA14 wild-type in any of the assays (A–E).
(TIF)
Growth of P. aeruginosa mutants in Arabidopsis Col-0 leaves. Plants were inoculated and incubated as described in Materials and Methods and leaves were harvested 3 days post-inoculation. Data represent the mean of bacterial titers ± SE of six leaf disks excised from 6 leaves of 3 plants. Different superscript letters denote statistically significant differences (P<0.05, Fisher's PLSD test).
(DOC)
Predicted functions and orthologs of PA14_36375-36830 genes across sequenced Pseudomonas genomes. Putative functions of PA14_36375-36830 genes were assigned as described in Materials and Methods. Genes of the reference taxon PA14 are indicated in the left column (PA14 loci), and different pseudomonad genomes are indicated on the top row. The presence of an ortholog is indicated by a checkmark. Boxes of identical hue indicate that the genes are contiguous in a particular genome, with the lighter shades on the top strand, and darker shades on the bottom strand. Checks in white boxes indicate an ortholog that is not contiguous with other PA14_36375-36830 block orthologs.
(PDF)
Growth, biofilm formation and motility of Δ42 and PA14 wild-type. Growth rate (h−1) of Pseudomonas strains in minimal media (M63) was calculated by the equation for exponential growth (see Materials and Methods). Biofilm formation was measured as attachment to polyvinylchloride plates in absorbance units (OD550; see Materials and Methods). Swimming and twitching motility are represented as a radius of a halo in cm (see Materials and Methods). Two independent Δ42 deletion constructs were tested. Data represent the mean ± SE. Based on analysis of variance (ANOVA) and Fisher's PLSD test (P<0.05), there was no significant differences between Δ42 mutant and PA14 wild-type in any of the assays.
(DOC)