Abstract
A heme-containing transmembrane ferric reductase domain (FRD) is found in bacterial and eukaryotic protein families, including ferric reductases (FRE), and NADPH oxidases (NOX). The aim of this study was to understand the phylogeny of the FRD superfamily. Bacteria contain FRD proteins consisting only of the ferric reductase domain, such as YedZ and short bFRE proteins. Full length FRE and NOX enzymes are mostly found in eukaryotic cells and all possess a dehydrogenase domain, allowing them to catalyze electron transfer from cytosolic NADPH to extracellular metal ions (FRE) or oxygen (NOX). Metazoa possess YedZ-related STEAP proteins, possibly derived from bacteria through horizontal gene transfer. Phylogenetic analyses suggests that FRE enzymes appeared early in evolution, followed by a transition towards EF-hand containing NOX enzymes (NOX5- and DUOX-like). An ancestral gene of the NOX(1-4) family probably lost the EF-hands and new regulatory mechanisms of increasing complexity evolved in this clade. Two signature motifs were identified: NOX enzymes are distinguished from FRE enzymes through a four amino acid motif spanning from transmembrane domain 3 (TM3) to TM4, and YedZ/STEAP proteins are identified by the replacement of the first canonical heme-spanning histidine by a highly conserved arginine. The FRD superfamily most likely originated in bacteria.
Introduction
All aerobic living organisms face a dilemma when confronted with the need to assimilate the essential element iron. Indeed, iron is the second most abundant metal on earth, yet the primary form found in the environment is the water insoluble and metabolically inactive ferric ion (Fe3+) [1]. The introduction and accumulation of dioxygen, into the ancient oceans and atmosphere, by Cyanobacteria completely changed the earth’s initial reductive environment by gradually causing it to become oxidative. As a result, the absorption of bioactive and water soluble ferrous ion (Fe2+) became a challenge for all forms of life and left a great impact on evolution [2]. One solution to the dilemma was the emergence of ferric reductases (FRE), which transfer electrons from cytosolic NADPH to extracellular ferric ions to generate the reduced form of ferrous ions, which can then be transported across the plasma membrane by specific iron transporters [3], [4].
Ferric reductases (FRE) and NADPH oxidases (NOX) are homologs [5]. Indeed, three canonical domains are commonly shared by both protein families: a heme-containing 6 transmembrane (6TM) ferric reductase domain and the two C-terminal cytoplasmic FAD-binding and NADPH-binding domains [6]. This common organization most probably reflects the fact that they catalyze similar reactions: Fe3++e− = Fe2+ (ferric reductase) and O2+ e− = O2 − (NADPH oxidase).
NOXs transfer electrons to oxygen to produce short-lived superoxide which is the primary reactive oxygen species (ROS), which is then transformed into various other ROS, such as hydrogen peroxide, hypochlorite or ozone [7]. ROS can also be generated as a byproduct in aerobic metabolisms, typically by mitochondria, peroxisomes, chloroplasts, or cytochrome p-450. In contrast, NOXs are devoted to the generation of biologically functional ROS, which play important roles in innate immunity [8], inter/intra-cellular signaling [9], morphogenesis and development [10], [11]. The various physiological and pathophysiological roles of NOX enzymes have been intensively studied and reviewed [7], [12].
Detailed bioinformatics analyses highlighted both the gene phylogeny and the structure of family members [13]–[16]. It was thus shown that ferric reductase domain (FRD) superfamily members exist in a wide variety of organisms, and many species carry multiple gene copies [15]. What is more, structural models have been developed for differing family members and a large number of conserved positions were identified [14]. Various studies inferred the evolutionary relationships of ROS-generating NADPH oxidase families [13]–[15]. Finally, homologs that possess only the conserved ferric reductase domain have been identified in bacteria (YedZ) and eukaryotes (STEAP; Six Transmembrane Epithelial Antigen of Prostate) [16].
In the present study, we characterized the evolutionary history of the FRD superfamily. By adding homologs of species from deep-branching nodes of the species tree, we showed that families and subfamilies emerged earlier than had been previously thought. The gene phylogeny is discussed in the context of structural and functional features. Findings to be highlighted include 1) conserved residues predicted to be crucial for NADPH-oxidases; 2) the probable lateral inheritance of the metazoan STEAP family from bacteria and the synchronous loss of the ancient ferric reductases in this clade; 3) the emergence of the NOX family from EF-hand containing superfamily members; 4) the origin of the FRD superfamily from a bacterial homolog that consists solely of a ferric reductase domain.
Materials and Methods
Data Collection
Eukaryotic homologs of the FRD superfamily were obtained from UniProtKB (release 2011_07) [17] [18], by searching – by way of cross-references – for three conserved domains predicted by the Pfam database [19]: the ferric reductase-like transmembrane component (PF01794), the FAD-binding domain (PF08022) and the NADPH-binding domain (PF08030). Further family members were identified in UniProtKB using Blast [20] [21]. The dataset was then complemented with homologs predicted by Ensembl (release 63, 30 June 2011), EnsemblMetazoa (release 10, July 2011), EnsemblPlant (release 10, July 2011), EnsemblFungi (release 10, July 2011), and EnsemblProtist (release 10, July 2011). 47 eukaryotic species were selected as representatives of their corresponding taxonomic groups. Finally, homologs from species of deep-branching nodes of the species tree were added. In order to root the tree, we retrieved homologs from UniProtKB with a conserved ferric reductase domain from complete bacterial proteomes. 29 homologs – including long and short homologs from 16 representative bacteria – were selected for further analysis. The dataset is available in the File S1. For the analysis of the full superfamily phylogeny we retrieved the 2876 predicted ferric reductase domains from Pfam (release 26.0, November 2011).
Phylogenetic Analysis
Sequences were aligned using MAFFT (version 6) [22] with parameter settings optimized for data with multiple conserved domains and long gaps (E-INS-i) and scoring matrix JTT200 [23]. The gap opening penalty (1.80) and the offset value (0.1) were set above the default. The multiple sequence alignment (MSA) was inspected and edited with JalView (version 2.6.1) [24]. Sequences with long gaps were either replaced by more appropriate isoforms or removed from the alignment. The non-homologous N-terminus of the amino acid sequences was trimmed to the beginning of the conserved ferric reductase domain. From the obtained sequences we constructed a second dataset which included the NOX family members and four members of the DUOX family as an outgroup. Both datasets were realigned as described above. From both datasets we constructed different data models from the conserved regions of the alignments. Following this strategy, we tried to obtain phylogenetic signals at different levels of depth in the gene history. The accuracy of the alignments was evaluated with GUIDANCE (version 1.1) [25] and unreliable columns were removed manually, mostly according to the GUIDANCE score. The best-fit models of evolution were determined with ProtTest (version 3.2) [26] ML phylogenies were calculated for all data models with PhyML 3.0 [27] under the LG amino acid substitution model [28], using eight rate categories which approximated a gamma distribution; the alpha parameter and the proportion of invariable sites were estimated from the dataset. The likelihood was maximized by optimizing the tree topology and branch lengths. The degree of support for internal branches was assessed by the approximate likelihood-ratio test based on the non-parametric Shimodaira-Hasegawa-like procedure (SH-aLRT) as implemented in PhyML.
For the analysis of the superfamily, the sequence redundancy of the predicted ferric reductase domains was reduced to 90% using Jalview. Truncated and dubious sequences were removed and sequences were realigned with MAFFT, using settings optimized for data with one conserved domain and long gaps (L-INS-i) and the scoring matrix JTT200. No other default parameter values were changed. The alignment was edited again with Jalview. This time, the data model was selected in a less stringent way to keep a maximum number of positions for the analysis. Sequence redundancies above 82% were removed from the data model. An ML tree was constructed with PhyML using the same evolutionary model as described above. Phylogenetic trees were inspected and drawn using Archaeopteryx (http://www.phylosoft.org/archaeopteryx).
Protein Sequence Analysis
All sequences were searched for possible transit peptides (SignalP [29], TargetP [30]), transmembrane domains (TMHMM [31], Phobius [32], MEMSAT [33]), homologous, biased and functional regions (InterPro [34], Pfam [35]), and post-translational modifications (NMT [36]). To avoid over-prediction, we considered positive results only, if 1) a region or site was shown to exist or was analyzed in depth in a previous study for at least one family member; 2) predictors had a high specificity or a region or site is predicted by more than one predictor, or 3) the hit was conserved for the majority of members within a clade.
For the identification of conserved sites within transmembrane domains and conserved adjacent regions, representative homologs of the NOX family (NOX1-4 and co-orthologs from Fungi, Amoebozoa and Naegleria gruberi), NOX-EF group (EF-hand(s) containing members of the NOX group), ppFRE group (protist and plant homologs of the FRE group), and fuFRE (fungal homologs of the FRE group) families, as well as the bacterial short and long forms and the STEAP family were obtained from the UniProt server and previous datasets. Sequences of each family were aligned using MAFFT. The alignments were then inspected manually and sequences that possessed atypical insertions within the transmembrane domains were removed from the MSA. The monophyletic group of YedZ-like proteins was removed from the MSA of bacterial short forms; in the following the group was treated as a group on its own. Well-aligned regions – including the predicted transmembrane domains (TM3–TM5) – were extracted from the alignments. Finally, sequence conservation logos were constructed from the eight MSAs using the WebLogo server (weblogo.threeplusone.com). The alignments used for this purpose included 443 (bacterial short forms), 107 (bacterial long forms), 75 (STEAP), 22 (bacterial short forms similar to bacterial long forms), 123 (fuFRE), 54 (ppFRE), 66 (NOX-EF), and 54 (NOX) sequences, respectively.
The analyses were performed on the Vital-IT High Performance Computing Center (www.vital-it.ch), mafft.cbrc.jp/alignment/server (MAFFT), guidance.tau.ac.il (GUIDANCE), darwin.uvigo.es/software/prottest.html (ProtTest), www.phylogeny.fr [37] (PhyML), www.cbs.dtu.dk/services (SignalP, TargetP, TMHMM), phobius.sbc.su.se (Phobius), pfam.sanger.ac.uk (Pfam), www.ebi.ac.uk/interpro (InterPro), weblogo.berkeley.edu (WebLogo) and local computers.
Collection of Functional Knowledge
A systematic literature search on experimentally characterized homologs was conducted for NOX homologs and for selected members of the FRE group using PubMed/MEDLINE at www.ncbi.nlm.nih.gov/pubmed and in the reference section of relevant UniProtKB entries.
Results and Discussion
Phylogeny of the FRD Superfamily
At the highest level of structural organization, members of the FRD superfamily are characterized by a common transmembrane ferric reductase domain (Pfam: PF01794) with structural similarity to cytochrome b of the mitochondrial bc(1) complex. Our analysis initially focused on eukaryotic homologs, whose superfamily members also possess a dehydrogenase module consisting of an FAD-binding domain (Pfam: PF08022) and an NADPH-binding domain (Pfam: PF08030), both oriented towards the cytoplasm. Conserved blocks of these homologous regions account for the data model used in the phylogenetic analyses of the eukaryotic genes; the phylogeny of the FRD superfamily is inferred based on the homologous regions of the ferric reductase domain (Figure 1). The many lineage-specific radiations in the eukaryotic clades throughout evolution are striking (Figure 2, and File S2). Hence, most of the analyzed species possess more than one gene copy of this superfamily. However, organisms do not systematically contain FRD homologs. For 33 out of 263 complete eukaryotic proteomes in UniProtKB (release 2011_12), no homologs with a ferric reductase domain have been predicted until now. These species belong to the taxonomic groups Alveolata (18), Diplomonadida (3), Euglenozoa (2), Microsporidia (4), Parabasalia (1), stramenopiles (1), Platyhelminthes (2) and Arthropoda (2). For bacteria, FRD homologs are annotated in only 37% of all complete proteomes.
Figure 1. Maximum likelihood phylogeny of the FRD superfamily.
A. Phylogram of the NOX family rooted to DUOX genes (outgroup not shown). The tree topology suggests lineage-specific gene duplications in all major taxonomic clades. The NOX1-3 and NOX4 subfamilies possibly diverged before the emergence of metazoans. B. Phylogeny of eukaryotic gene families of the FRE group and the NOX group. According to this model, the DUOX family and NOX family form sister clades, but not the EF-hands containing protein families NOX5 and DUOX. C. Phylogenetic tree of the FRD superfamily. The tree topology proposes that the metazoan STEAP family (red) emerges from the bacterial clade at the base of the YedZ family. The gene AM1_3152 from the cyanobacterium Acaryochloris marina (strain MBIC 11017) (UniProtKB: B0CEP3) was probably obtained from an ancestral gene of the eukaryotic NOX5 family. Explanation: The names of gene families and gene groups are indicated with curly brackets. Branch colors correspond to those of the listed taxonomic groups.
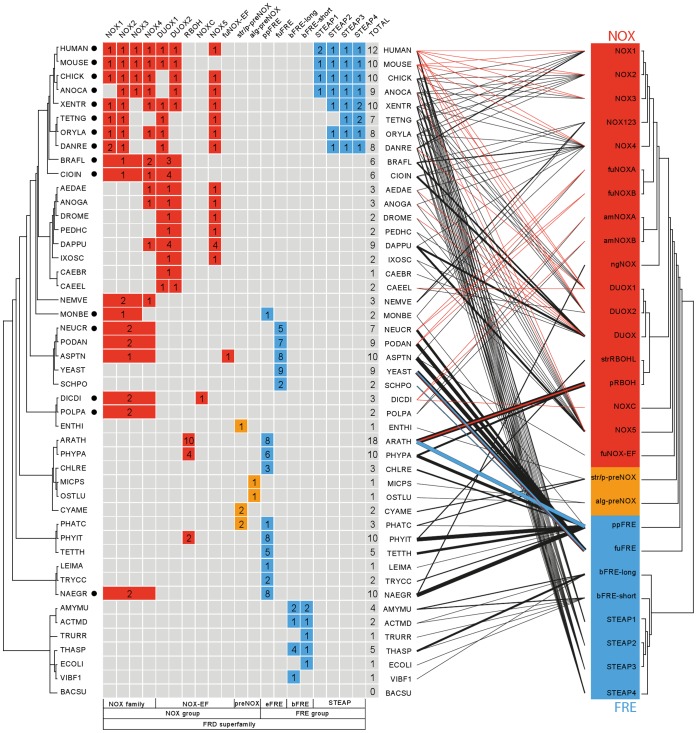
Figure 2. Phyletic profile and molecular function of the FRD superfamily.
On the left-hand side, phyletic profile for 47 species: gene copy numbers are plotted in accordance with the species phylogeny (left) and gene families: The number of NOX homologs of a species is given in red cells, FRE homologs in blue cells, and preNOX in orange ones. Some cells are merged according to the family hierarchy. The number of predicted homologs is given in the last column of the phyletic profile. On the right-hand side, gene copies are represented by lines which link the corresponding species and protein families; the thickness of these lines indicates the number of gene copies. Colored lines flag experimentally confirmed gene functions: red = ROS-generating NADPH oxidase activity; blue = metalloreductase activity. Black circles mark species that possess p22phox homologs.
The phylogenetic tree of the FRD superfamily suggests three major protein groups. Clade 1 includes homologs of the ROS-generating protein families (NOX group) as well as homologs of ferric reductases (FRE) predominantly from plants and protists (ppFRE). This group is of special interest in this study, because – based on functionally characterized family members – we presume that the functional shift from a metalloreductase to a ROS-generating protein took place within this branch in an ancestral gene of very early eukaryotes. The phylogeny of the ROS-generating NADPH oxidases (NOX1-5, DUOX, RBOH, preNOX) was analyzed previously [13]–[15]. In agreement with these results are the sub-familial relationships within the NOX family (Figure 1A). The sea anemone Nematostella vectensis currently represents the most basal metazoan, and its proteome contains three gene copies, i.e. two ortholog to the NOX1-3 subfamily and one ortholog to the NOX4 subfamily. There is even some evidence that the divergence of these two subfamilies took place prior to the emergence of animal – NOX homologs of the choanoflagellates Monosiga brevicollis and Salpingoeca sp. (strain ATCC 50818) cluster steadily with the NOX1-3 clade. Furthermore, the NOX family includes the independently duplicated genes from fungi, Amoebozoa and Naegleria gruberi. The presence of these co-orthologs reveals that a gene of the NOX family was probably already present in very early eukaryotes.
What is more challenging is the interpretation of the evolution of the EF-hand containing protein group (NOX-EF, including NOX5, NOXC, DUOX, and RBOH families). It has been suggested that the EF-hand domain was acquired only once [38]; but, to our knowledge, none of the phylogenetic studies so far have achieved a tree topology that supports a monophyletic origin of NOX-EF prior to the divergence from the NOX family. In our analyses, alternative interfamilial relationships are predicted for the EF-hand containing members when datasets, analysis methods, or analysis parameters are changed (File S3). One explanation for the observed inconsistent topology could be multiple gene duplications that happened close together in time, leaving no or only marginal traces of common evolution in these clades. Because results indicate that the NOX family emerged from the NOX-EF group, we hypothesize that the EF-hand was lost in an ancestral gene of the NOX family.
At the base of clade 1, metalloreductases predominantly from protists and plants (ppFRE) diverge (Figure 1B). Minor clades in between ppFRE and the NOX group are hereafter named preNOX (Figure 1).
Clade 2 includes solely fungal metalloreductases (fuFRE), and the topological placement of this branch outside other eukaryotic metalloreductases is notable (Figure 1B). Our analysis includes genes from five representative fungal proteomes, which possess between two and nine homologs. A more comprehensive analysis of this clade – with homologs from 29 eumycotal proteomes – classified members of the FRE group into 24 families [39].
So far, no metazoan eFRE orthologs have been identified. Indeed, animals possess enzymes with the conserved ferric reductase domain and a ferric reductase activity, but this family (STEAP) is probably xenolog to eFRE (Figures 1C and Figure S4-4 in File S1).
In order to root the tree, we examined FRD homologs from prokaryotes (clade 3). This clade is composed of bacterial ferric reductases (bFRE) including the bacterial YedZ family and the eukaryotic STEAP family. Bacterial members can be roughly classified into short and long forms; the short forms consist basically of the ferric reductase domain, and the long forms of all three domains conserved in the eukaryotic families (Figure 3). According to Pfam cross-references in UniProtKB (release 2011_12), short forms are about 5.5 times more frequent than the long form. Furthermore, some bacterial proteomes seem to contain only a long form, others only a short form, while some have both forms or none at all (Figure 2). Out of 1471 complete bacterial proteomes provided by UniProtKB, only 543 species possess at least one predicted homolog (Figure S4-1 in File S4). Yet we observed no conspicuous correlation between the bacterial taxonomy and the occurrence of homologs (Figure S4-2 in File S4). During the analysis, we noticed only a few likely cases of HGT from eukaryotes to bacteria. One example is the gene AM1_3152 from the cyanobacterium Acaryochloris marina (strain MBIC 11017) (UniProtKB: B0CEP3), which could have been obtained from an ancestral gene of the NOX5 family (Figure 1C). UniProtKB includes four FRE family members from the archaeal domain. A phylogenetic analysis of a prokaryotic dataset reveals that the archaeal genes were probably derived by HGT from bacteria in three independent evolutionary events (Figure S4-3 in File S4).
Figure 3. Domain architecture of FRD superfamily members.
Models of domain architectures are mapped to the phylogenetic gene trees of bacterial (A) and eukaryotic (B) FRD homologs. Tree branch colors correspond to the color code of the models (see highlight color of model identifiers). The three conserved domains of the ‘eukaryotic structural core’ are colored, and other predicted domains are given in black. Domain forms indicate their function; rounded rectangle = binding of electron donor/hydrogen acceptor: FAD-binding, NADPH-binding (M3), FMN (M4); triangle = electron transfer agent: Ferredoxin/Fer2 (M8), Rieske (M4), DOMON (M10), peroxidase-like domain (M15); circle = regulation of enzyme activity: EF-hands (M14–M16); hexagon = protein-protein interaction: NADPH-oxidase-like domain (M16), SH3 (M17); ellipse = transport of small solutes: MSF (M6).
Modularity and Functional Diversification
The basic structural component of this superfamily is the transmembrane ferric reductase domain that is present in all the analyzed superfamily members. This domain includes four canonical conserved heme-coordinating histidines in transmembrane domain 3 (TM3) and transmembrane domain 5 (TM5). Their positions in the human NOX2 sequence are 101 and 115 (TM3), and 209 and 222 (TM5). The heme close to the cytoplasm is expected to bind to His-101 and His-209, while the heme close to the cell surface is expected to bind to His-115 and His-222. Most of the bFRD clade homologs consist solely of this domain, but seven distinct modules have been identified, which contribute to the extension of this simple ‘bacterial structural core’ (Figure 3A, model M7). According to Pfam cross-references in UniProtKB, only about 12% of the bacterial homologs have a predicted architecture similar to eukaryotic metal reductases and NADPH oxidases, comprising the transmembrane ferric reductase domain followed by the cytoplasmic FAD-binding and NADPH-binding domains (Figure 3A, model M1). All in all, nine variations of this ‘eukaryotic structural core’ (model M1) were predicted for sequences of our dataset (Figure 3B). Interestingly, additional functional components are generally fused to the N-terminal of the conserved triad. The two C-terminal domains form an elaborated functional unit, in which the NADPH-binding domain provides the enzyme with a readily available electron donor, thereby enhancing its efficiency. Meanwhile the FAD-binding region optimizes the energetic profile of the electron transport chain. This module with redox and electron transfer properties is beneficial to many redox systems, and thus represents an abundant structural compound found in the vicinity of oxidoreductase domains, either as a module in multi-domain enzymes or as a subunit of protein complexes. This structure is often referred to as dehydrogenase domain or dehydrogenase module. While analyzing FRD superfamily members and other unrelated proteins with a dehydrogenase module, transmembrane domain predictors often reported a positive hit in between the two cofactor-binding domains. In all logic, such a protein structure would contradict the module’s biological function. An inspection of known structures (PDB: 3a1f, 1gvh) revealed a hydrophobic region located in the center of the NADPH-binding domain, spanning the C-terminus of a parallel beta-sheet, a short loop opposite the cofactor-binding site and the N-terminal of a proximate alpha-helix. Based on this analysis, both the existence of a transmembrane domain and a hairpin-like structure in between the neighboring domains is therefore unlikely.
In the following sections, the different domain architectures are discussed along with examples of well-studied proteins (Figure 3). An overview of the molecular function of characterized family members is given in Figure 2, and a description of the experimentally confirmed protein functions is available in Table 1.
Table 1. Biological functions of eukaryotic FRD superfamily members from published experiments.
| Protein, species, UniProtKB identifier | Biological Function | References |
| Mammalian NOX1-4 | ||
| NOX1 human: Q9Y5S8, mouse: Q8CIZ9 | Signaling (e.g. smooth muscle proliferation, angiogenesis) | [72]–[74] |
| NOX2 human: P04839, mouse: Q61093 | Host defense; signaling to limit inflammation and immune activation | [7] [75]–[78] |
| NOX3 human: Q9HBY0, mouse: Q672J9 | Signaling and/or biosynthesis in the inner ear (otoconia formation) | [79] [80] |
| NOX4 rat: Q924V1, mouse: Q9JHI8 | Signaling (e.g. myofibroblast differentiation, hypoxia response) | [81]–[83] |
| Fungal NoxA/B | ||
| NoxA Epichloe festucae: Q2PEP0 | Maintain mutualistic status with host plant | [84] |
| NoxA Botryotinia fuckeliana: B0BES1 | Maintain pathogenicity; develop penetration structure to infect host plant | [85] |
| NoxA Emericella nidulans: Q8J0N4 | Sexual development | [86] |
| NoxA Magnaporthe grisea: A6ZIB7 | Maintain pathogenicity; develop penetration structure to infect host plant | [87] |
| NoxA Neurospora crassa: Q7RW00 | Female sexual structure formation; asexual development and hyphal growth | [66] |
| NoxA Podospora anserine: B2AA06 | Develop penetration structure to infect host plant; degrade host plant cellulose; fruiting body differentiation | [88] [89] |
| NoxB B. fuckeliana: B0BES2 | Maintain pathogenicity; colonize in host plant | [85] |
| NoxB M. grisea: A6ZIB8 | Maintain pathogenicity; develop penetration structure to infect host plant | [87] |
| NoxB N. crassa: A7UW98 | Spores germination | [66] |
| NoxB P. anserine: B2AL10 | Ascospore germination; develop penetration structure to infect host plant; degrade host plant cellulose | [88] [89] |
| Amoebozoan NOX homologs | ||
| NoxA D. discoideum: Q9XYS3 | Development and spore formation | [10] |
| NoxB D. discoideum: Q86GL4 | Development and spore formation | [10] |
| NOX-EF | ||
| DUOX1 human: Q9NRD9, mouse: A2AQ92 | Thyroid hormone synthesis; mucosal host defense; signaling (e.g. urothelium mechanosensing) | [90]–[96] |
| DUOX2 human: Q9NRD8, mouse: A2AQ99 | Thyroid hormone synthesis; mucosal host defense | [90]–[94] [96] [97] |
| DUOX Danio rerio: F1QVF2 | Signaling (chemotaxis, wound healing) | [98] |
| DUOX Aedes aegypti: Q171Q3 | Innate immunity; intestinal host defense | [99] |
| DUOX Anopheles gambiae: Q7Q147 | Midgut nitration and apoptosis during invasion of Plasmodium berghei | [100] |
| DUOX D. melanogaster: Q9VQH2 | Innate immunity; signaling (Ca2+ channel); protein cross-linking for wing stabilization;epidermal wound healing | [11] [101]–[103] |
| DUOX1 Caenorhabditis elegans: O61213 | Innate immunity; host defense; protein cross-linking in cuticular extracellular matrix | [104]–[106] |
| DUOX Lytechinus variegatus: Q5XMJ0 | Protein cross-linking in fertilization envelope | [107] |
| RBOHC Arabidopsis thaliana: O81210 | Signaling (Ca2+ channel); root cell elongation | [108] |
| RBOHD A. thaliana: Q9FIJ0, RBOHF:A. thaliana: O48538 | Host-pathogen interaction; signaling (e.g. ROS as second messengers in abscisic acidsignaling in guard cells) | [109]–[112] |
| NOX5 human: Q96PH1 | Signaling (e.g. prostate cancer cells, spermatocytes, marginal B lymphocytes) | [113]–[116] |
| NOX5 A. gambiae: Q7PNG0 | Midgut epithelial nitration and innate immunity | [117] |
| NOXC D. discoideum: Q54F44 | Development and spore formation | [10] |
| Plant eFRE homologs | ||
| FRO2 A. thaliana: P92949 | Fe3+ reduction/acquisition in root surface; iron and copper homeostasis; chilling stresstolerance (block ROS signaling during chilling) | [3] [118]–[120] |
| FRO3 A. thaliana: F4I4K7 | Fe3+ reduction/acquisition in root vascular cylinder and shoots (mitochondria); iron and copper homeostasis | [120] [121] |
| FRO4 A. thaliana: Q8W110 | Iron and copper homeostasis | [122] |
| FRO5 A. thaliana: Q9FLW2 | Iron and copper homeostasis in root | [120] [122] |
| FRO6 A. thaliana: Q8RWS6 | Fe3+ reduction in shoots (chloroplast); iron and copper homeostasis | [120] [121] [123] |
| FRO7 A. thaliana: Q3KTM0 | Fe3+ reduction/acquisition in chloroplast for photosynthesis; iron and copper homeostasis | [48] [120] |
| FRO8 A. thaliana: Q8VY13 | Iron and copper homeostasis in leaves | [120] [121] |
| Fungal eFRE homologs | ||
| FRE1 S. cerevisiae: P32791 | Fe3+ and Cu2+ reduction/acquisition; iron and copper homeostasis | [44] [124] [125] |
| FRE2 S. cerevisiae: P36033 | Fe3+ and Cu2+ reduction/acquisition; iron and copper homeostasis | [124] [126] |
| FRE3 S. cerevisiae: Q08905 | Fe3+ reduction; iron homeostasis | [46] [124] |
| FRE4 S. cerevisiae: P53746 | Fe3+ reduction; iron homeostasis | [46] [124] |
| FRE5 S. cerevisiae: Q08908 | Iron homeostasis | [46] |
| FRE6 S. cerevisiae: Q12473 | Export iron and copper from vacuole; iron and copper homeostasis | [46] [127] [128] |
| FRE7 S. cerevisiae: Q12333 | Copper homeostasis | [46] |
| AIM14 S. cerevisiae: P53109 | ROS generation; apoptosis; actin cable formation | [47] |
| FRP1 S. pombe: Q04800 | Fe3+ reduction/acquisition; iron homeostasis | [129] |
bFRD: Extensions to a Simple Module
Typical representatives of the ‘one-domain’ homologs from bacteria are members of the YedZ family found in Proteobacteria, which are involved in redox regulation and transmembrane electron transfer [40]. The E.coli homolog of this family has been shown to form an oxidoreductase complex with the soluble catalytic molybdoenzyme YedY [41]. Out of 1,755 currently available bacterial homologs in UniProtKB, YedZ is – to our knowledge – the only characterized family member. bFRD homologs are probably involved in many different biological processes, as can be inferred from the diverse fusion proteins. Six modular extensions to the bacterial structural core have been predicted for members of the bacterial clade and, as a matter of fact, all these domains are related to functions of electron transport and/or exchange. Structural modifications are also observed in homologs of the eukaryotic STEAP family that emerged from the bacterial FRE branch. Like its bacterial homologs, STEAP1 proteins consist only of a ferric reductase domain but its paralogs, STEAP2-4, possess additionally an NADPH-binding domain (‘NADP oxidoreductase coenzyme F420-dependent’ domain; Pfam: PF03807) in their N-terminal region. Such a domain architecture has not been predicted for bacterial metal reductases. STEAP family members have been shown to possess both ferric reductase and cupric reductase activities [42] [43]. Thus even members of the FRD superfamily, which do not have the three canonical domains, appear to be involved in the same molecular function, but probably using different reaction mechanisms. We compared the conserved amino acids found in the transmembrane domains of both the short and the long forms of bFRD. This revealed that most short forms possess only two (STEAP2-4: His-115, His-222) or three (YedZ, STEAP1: His-115, His-209, His-222) of the conserved histidines and thus probably bind only the surface-proximal heme (Figure 4). Experimental evidence for functional electron transport through the membrane via a single heme is scarce but some evidence has been reported for YedZ [41]. Instead of the cytoplasm-proximal heme-binding histidines we found a well-conserved arginine (TM3) and glutamine (TM5). In contrast, the long forms possess all four conserved histidines. The few short forms that contain all four conserved histidines are found in the direct vicinity of the long forms in our phylogenetic tree. This finding supports the tree topology in that STEAP is more close related to the bacterial short form, and bacterial long forms are more closely related to eukaryotic homologs.
Figure 4. Sequence conservation logos and the proposed structure of the ferric reductase domain of protein groups from the FRD superfamily.
A. Transmembrane domains TM3 to TM5 - as predicted for human cytochrome b-245 heavy chain (NOX2) - are indicated by gray rectangles. The cladogram indicates the phylogenetic relationship of the analyzed homologous groups. In the conservation logos, the height of the stacks indicates sequence conservation; the width of the stacks is proportional to the fraction of amino acids, thus narrowed within gapped regions. B. Proposed structure of ferric reductase domain with conserved amino acid residues corresponding to the annotation in figure 4A.
Most structural modifications are found in orthologs of closely related species, thus indicating that variations of the basic core are generally not persistent in bacteria, a fact which is typical for the evolution of prokaryotic proteins. Hence, the enduring bacterial ‘long form’ might be more efficient than functional equivalents or could have a unique function.
eFRE: Variations in ‘Transmembrane’
Most, if not all, fungal metalloreductases (fuFRE) contain an additional predicted transmembrane domain that precedes the three conserved domains of the eukaryotic structural core. The nine Saccharomyces cerevisiae homologs are among the best-studied examples of this group. As they are important components of the high affinity uptake system for iron and copper ions, FRE1 and FRE2 are involved in both ferric and cupric ion reduction. There is evidence that FRE3-6 homologs are specific iron reductases, whereas the FRE7 homolog is a specific copper reductase [44]–[46]. A recent study shows that yeast YNO1 (AIM14) - one of the 9 yeast FREs - generates ROS and affects both apoptosis and actin cable formation [47]. The function of the N-terminally fused transmembrane region is unknown.
In some ways similar to the fungal metalloreductases, the majority of protist and plant homologs (ppFRE) in this group bear predicted transmembrane regions in addition to those of the ferric reductase domain. Two or six transmembrane domains are found in the N-terminal of these homologs, but additional transmembrane helices are also predicted close to the C-terminus, within the NADPH-binding domain in clade members from Viridiplantae and stramenopiles. A well-studied example is the tissue-specific expression of the eight paralogs of the ferric chelate reductase (FRO) family from Arabidopsis thaliana [3]. Three of these homologs are located in the membrane of organelles: FRO7 in chloroplasts, FRO3 and FRO8 in mitochondria [48] [49]. Nevertheless, all these gene copies emerge from the eukaryotic clade and it is therefore unlikely that they originate from an organelle genome.
Besides additional transmembrane domains, a single clade of the eFRE group has acquired a new structural module. Members of a subfamily – including seven paralogs from the Heterolobosea N. gruberi and homologs from the stramenopiles Albugo laibachii (UniProtKB: F0X089) and Phytophthora infestans (UniProtKB: D0MUZ3) – contain one or two extracellular DOMON domains (Pfam: PF03351). The DOMON domain was predicted to possess a hydrophobic pocket that binds heme and sugar [50] [51]. It has also been suggested that it participates in an electron-transfer system [50]. A signal peptide was predicted only for a homolog presenting two DOMON domains.
Note that no FRE-EF forms were observed in bacteria and eukaryotes, suggesting that the regulatory EF hand domains were acquired by superoxide-producing NOX enzymes.
preNOX and NOX-EF: Emergence of ROS Generation and Activity Regulation
At the base of the NOX group, homologs from Rhodophyta, stramenopiles and Viridiplantae contain four predicted additional transmembrane helices between the FAD-binding and NADPH-binding domains. One study revealed a potential ROS-generating NADPH oxidase activity for the homolog of the red algae Chondrus crispus, [52]. This experimental evidence is interesting, because with the occurrence of EF-hands at the N-terminus, such homologs are known or expected to produce ROS for specific biological functions. The algae, however, diverged before this domain was acquired.
The functional shift from metalloreductases to ROS-generating NADPH oxidases probably occurred early on in the NOX group and, to some extent, seems to be linked to the occurrence of EF-hands in the N-terminus. The binding of Ca2+ to EF-hands can cause conformational changes linked to regulatory functions, as for instance in calmodulin [53]. A need for regulated ROS production is plausible because of the toxicity of the products, as illustrated by their function in oxidative innate immune defense. At least one EF-hand was predicted for all members of the NOX-EF group. As the biologically active structure is generally a pair of EF-hands, and not all predicted EF-hands have the capacity to bind Ca2+, representative family members were inspected manually (Cox JA, personal communication). All the studied homologs possessed at least two EF-hands, of which at least one was canonical and hence assumed to be Ca2+-binding. Fungal members of the paraphyletic group of NOX-EF have a single Ca2+-binding EF-hand motif. They seem to exhibit ROS-generating NADPH oxidase activity and have been suggested to be regulated by Ca2+ [15] [54] [55]. The metazoan NOX5 family members contain four EF-hands, and studies in human and mouse confirmed Ca2+-regulated ROS-generation [56] [57].
A comparison of the eFRE metal reductases with expected NADPH oxidases indicated two positions that are conserved in members of the latter group: a histidine in addition to the two heme-binding histidines in transmembrane domain 3 (TM3), and a conserved threonine in transmembrane domain 4 (TM4) (Figure 4). In accordance with their sequence position in human NOX2, we refer to these sites as His-119 and Thr-178. His-119 is located within the alpha-helix close to the cell surface. Well-studied heme-binding proteins involved in oxygen-transport are hemoglobin and myoglobin. In both molecules, dioxygen-binding is stabilized by a hydrogen bond to a conserved histidine residue distal to the heme-binding sites [58]. We propose that a similar stabilizing mechanism involving His-119 could apply to the ROS-generating homologs of the superfamily, as this position is conserved in the vast majority of the analyzed homologs of the NOX group. Exceptions were found in species with multiple NOX inparalogs, and each of these species’ proteomes presented at least one gene copy that possessed the conserved histidine (His-119). One of these exceptions is NOXC of D. discoideum. Preliminary studies using a NOXC knockout strain showed reduced levels of stimulated ROS generation in vegetative and starved cells (Zhang X, Soldati T, unpublished data). We therefore assume, that the second conserved residue, Thr-178, is equally important for NADPH oxidase activity, but a gating function seems more likely than oxygen-binding. According to an analysis of the functional roles of catalytic residues [59], threonine has been found to predominantly perform stabilizing functions. Of note is a conserved glycine residue (Gly-179) adjacent to the conserved threonine, which is actually not specific to NOX homologs, but which forms a conserved dipeptide in the probable ROS-generating proteins. Natural variants in human NOX2 have been identified in patients with inheritable disorders related to innate immunity: mutations in His-119 as well as in Gly-179 lead to a chronic granulomatous disease (CGD) phenotype [60] [61], and mutations in Thr-178 to Mendelian susceptibility to mycobacterial disease (MSMD) syndrome phenotype [62]. In summary, four conserved positions are probably indicative in distinguishing ROS-generating homologs of the NOX group from presumptive metalloreductases: His-101, His-119, Thr-178 and Gly-179.
These findings are key in identifying the point of functional divergence in evolution. The preNOX group diverges at the very base of the NOX group, and includes genes from stramenopiles, red algae as well as green algae and fungi. Except for the fungal genes, all these members lack His-119; green algae even lack the second heme-binding histidine in TM3, but all homologs possess the conserved dipeptide. Hence, the homologs from stramenopiles and rhodophytes – but not the genes of the chlorophytes analyzed here – are likely to possess a ROS-generating activity that is not regulated by Ca2+. All other homologs of the NOX-EF group possess two or more EF-hands. In addition, two other structural components are found in members of this group: a domain of unknown function that encompasses the N-terminus of the NADPH oxidase (NADPH-oxidase Ox; Pfam: PF08414) and the heme-containing peroxidase-like domain (Pfam: PF03098). These two additional domains are the hallmark of two separate NOX families. The cytoplasmic NADPH-oxidase domain is attached to the N-terminus of the EF-hands in the RBOH family of land plants. Interestingly, closely related homologs from stramenopiles (RBOH-like) seem to possess EF-hands, but show no predicted domains at the N-terminus. Members of the DUOX family comprise up to three predicted EF-hand motifs, and a further transmembrane domain links to an extracellular peroxidase-like domain. The predicted signal peptide might be required to translocate the peroxidase-like domain to the extracellular space. Members from both families have been biochemically characterized as ROS-generating and Ca2+-regulated enzymes, strongly suggesting a conserved regulation pattern within the NOX-EF group (see Table 1).
The NOX Family: ROS Generation and Increasing Complexity of Regulation
Analyses of the extremely divergent domain architectures suggest that two distinct series of events appear to have led to an increased complexity of regulation of ferric reductase domain enzymes. First, as detailed in previous sections, during the early evolution of the superfamily, NOX enzymes acquired longer N-termini with various regulatory and functional domains. Second, the N-terminus was lost subsequent to the gene duplication event that gave rise to the emergence of the NOX1-4 family, which includes the meatozoan NOX1-4 homologs, and the independently duplicated co-orthologs of fungi (NOXA, NOXB), Amoebozoa (NOXA, NOXB) and Naegleria. The loss of the EF hand-containing N-termini occurred concomitantly with the emergence of separate cytosolic and membrane bound regulatory subunits. An advantage of multi-domain proteins is the colinear expression of functional units that are essential for a specific cellular task, bypassing the need for coordinated expression of separate gene products. However, a more fine-grained regulation is possible when multiple differentially expressed and regulated subunits assemble into a functional complex.
Developmental steps to a higher regulation complexity from NOX4, to NOX3, NOX1 and NOX2 can be traced in the NOX family. Of all these, the best studied NOX ortholog is NOX2 and its regulatory network has shown to be particularly complex. Indeed, its fine regulation requires an interaction both with the membrane component p22phox and the cytosolic components p47phox, p67phox, and p40phox. Finally, it also associates with a Rac GTPase that is responsible for the activation of the protein complex [15]. As a paradigm for the regulation of this class of enzymes, in the resting state of human NOX2, the proline rich region (PRR) at the N-terminus of p47phox interacts with the N-terminal SH3 domain of p67phox, while the PB1 domains of p67phox and p40phox interact with each other. Upon stimulation, GTP-bound Rac interacts with the N-terminal region of p67phox. Due to conformational changes in these cytosolic components, the SH3 domain of p47phox is able to interact with the N-terminal PRR of p22phox, which will further interact with NOX2 and activate its enzymatic function [15]. Only four regulators are needed for the function of NOX1: namely p22phox, the p47phox homolog NOXO1 (NOX1 organizer), the p67phox homolog NOXA1 (NOX1 activator), and Rac [15]. It is remarkable that most of these subunits are paralogs of the NOX2 regulators, stressing the importance of differential regulation. In contrast to this, the organization around NOX3 and NOX4 seems to be less tight. Indeed, NOX3 activity is up-regulated either by NOXO1 or by NOXA1, but only p22phox is required for ROS production [63]. NOX4, the most distant paralog to NOX2, requires only p22phox for ROS generation and seems to function independently of further regulators [15] [64].
Precursors of the domains that interact with these essential regulatory subunits can already be found in the NOX homologs of unicellular organisms. The two orthologs of Heterolobosea N. gruberi, for instance, contain an SH3 domain (Pfam: PF00018) within their C-terminal NADPH binding domain. Usually, the SH3 domain mediates protein complex assembly via binding to proline rich regions (PRR) of binding partners [65]. Thus, the SH3 domain found within the NADPH-binding domain of N. gruberi NOX homologs suggests that some unknown PRR-containing protein partner is involved in their regulation [15]. Interestingly, an SH3 domain has been identified in the human p67phox, but, alternatively, the homolog of the social amoeba Dictyostelium discoideum carries a functionally analogous WW domain for protein-protein interactions (Pfam: PF00397) [10]. Furthermore, the N-terminus of both homologs consist of four TPR repeats, (Pfam: PF00515), a feature also conserved in fungal NOX regulators (NOXR) found in Pezizomycotina, including Aspergillus nidulans, Neurospora crassa and Epichloe festucae [66] [67]. This conserved region mediates interaction with small Rac GTPases, thereby linking these NOX enzymes directly to the regulation by Rho family members [68]. Although it is still not clear which of the many Rac homologs might be involved in the regulation of D. discoideum NOXA and NOXB, Rac1A, -B and -C show the greatest homology with Rac2 in human neutrophils. In addition, all of the Rac2 residues at the interface with p67phox are conserved, suggesting that a similar activation process might also have occurred in D. discoideum [10].
Functional Congruence in Domain Composition, Sequence Divergence at Functional Sites
Domain architectures evolve at different modes in prokaryotes and eukaryotes and our results are characteristic for the two superkingdoms. Prokaryotic proteins generally possess a less complex domain composition than eukaryotes, but undergo fusion and fission events more frequently. Hence, proteins with a more complex domain organization are found in small clades (Figure 3). In contrast, eukaryotic proteins tend to be composed of more domains, but structural alterations can occur stepwise and at a low rate. Thus, to some extent it is possible to trace back evolution based on the continuous increase of structural complexity in proteins from related species. In the case of the FRD superfamily, this leads to a higher structural diversity in eukaryotic homologs than prokaryotic ones. Despite the differing mode of evolution regarding the domain architecture, congruent evolution can be observed with respect to the function of domains that enlarge the bacterial and eukaryotic structural core. First of all, it is likely that the bacterial long forms and the equivalent eukaryotic structural core result from independent fusion events. Furthermore, four distinct domains serve as electron transfer agents (Ferredoxin (Fer2), Rieske, DOMON, peroxidase-like domain) and two distinct domains assist protein-protein interactions (NADPH-oxidase-like domain, SH3). Thus, the reductive function of the ferric reductase domain was most probably conserved over more than two billion years. This assumption is supported by the conservation of the surface-proximal heme-binding histidines in TM3 and TM5. Finally it is notable that domains with equivalent functions can occur on either side of the membrane.
At the sequence level, divergent evolution can be observed at functional sites, which are assumed to reflect changes in substrate selectivity and possibly the reaction mechanism. The functional shift from metal reductase to NADPH oxidase activity probably occurred early on in evolution at the base of the NOX clade. Four amino acids have been identified to distinguish metalloreductases from ROS-generating NADPH oxidases. The replacement of the first canonical heme-spanning histidine by a highly conserved arginine in members of the prokaryotic YedZ family and the eukaryotic STEAP family may indicate a change in the reaction mechanism.
Origin of the FRD Superfamily
The ferric reductase domain may well have emerged from cytochrome b. The phylogeny of the FAD-binding domain suggests the fusion of a ferric reductase domain to a dehydrogenase module (File S5). Based on the likely absence of ancient ferric reductase domain gene copies in archaeal genomes, it seems reasonable to expect a bacterial origin of the ferric reductase domain. However, as shown above, only about 37% of all bacterial genomes possess a homolog of the FRD superfamily. Supporting arguments for a bacterial origin of this superfamily are – in particular – the presence of homologs in a broad variety of taxonomic groups compounded by the fact that the ferric reductase activity is only essential for obligate aerobic species that have no access to ferrous ion resources and possess no non-reductive iron uptake system. Furthermore, as a typical operational gene, ferric reductase is not only transferred more easily between bacteria [69], but is also likely to descend from bacteria early on in eukaryotic evolution [69]–[71]. Our phylogenetic superfamily tree shows a bacterial and a eukaryotic clade, suggesting a unique event of ancient gene transfer between the superkingdoms. The model is summarized in Figure 5.
Figure 5. A model of the evolutionary history of the FRD superfamily.
The ancestral system may have used reduced quinol to produce soluble ferrous ions and progressed into a highly regulated system that generates immunologically potent ROS by using NADPH as electron source.
Conclusions
This study reveals that the ferric reductase domain superfamily probably originated from a short bacterial homolog that consisted solely of a ferric reductase domain. During evolution these ‘one-domain’ homologs were extended by different modules that either participate in redox systems or are regulatory components. The C-terminal fusion of an FAD- and NADPH-binding domain probably resulted in the core of the long homologs, which are mostly found in eukaryotes. Phylogenetic analysis reveals that the highly diverse number of gene copies per species is derived by extensive lineage-specific gene gain and gene loss throughout evolution. The functional shift from metal reductase to NADPH oxidase activity probably occurred early in evolution at the base of the NOX clade. The generation of potentially toxic ROS is accompanied by increased complexity of regulatory systems. One such regulatory component is p22phox, which appears with the emergence of the NOX family; its origin is still unknown.
Four amino acids have been identified to distinguish metalloreductases from ROS-generating NADPH oxidases. We believe that it might be possible to detect further sites for the construction of even more specific function predictors based on the comparison of clade-specific conservation signatures. Finally, we plan to use conservation signatures for the revision of unresolved nodes. Preliminary results suggest taking the option of classifying the superfamily into three main groups, namely NOX, FRE and YedZ/STEAP. These and other related issues will be the subject of future interesting studies.
Supporting Information
Sequence data and domain predictions.
(XLSX)
Maximum likelihood phylogeny of eukaryotic gene families of the FRE group and the NOX group. The phylogenetic tree is a detailed presentation of Figure 1B. Tree branches are colored according to the taxonomic classification of the species.
(TIF)
Exploration of the tree space. Phylogenies of the FRD superfamily from multiple analyses. Gene families are colored in the phylogenetic tree and family names are given. Branch support values of major internal nodes are indicated.
(PDF)
Analysis of bacterial FRD superfamily members.
(PDF)
Evolution of the FRD superfamily from the view of its core domains.
(PDF)
Acknowledgments
The authors wish to thank Jos A. Cox for his help in the analysis of EF-hand domains. A special thanks to Nicolas Salamin for feedback on the manuscript, and to Vivienne Baillie Gerritsen for proofreading it. The authors are grateful to two anonymous reviewers for their valuable comments and suggestions to improve the quality of the study.
Contributions
Conceived and designed the experiments: KHK TS BB. Performed the experiments: XZ BB. Analyzed the data: XZ BB TS KHK IX. Wrote the paper: BB XZ KHK TS IX.
Funding Statement
This work was supported in part by the Swiss Federal Government through the Federal Office of Education and Science (BB) and by a Swiss National Science Foundation ProDoc Research Module, PDFMP3_127302 (TS and KHK). Funding for open access charge: Swiss Institute of Bioinformatics. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kosman DJ (2010) Redox cycling in iron uptake, efflux, and trafficking. The Journal of biological chemistry 285: 26729–26735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Crichton RR, Pierre JL (2001) Old iron, young copper: from Mars to Venus. Biometals: an international journal on the role of metal ions in biology, biochemistry, and medicine 14: 99–112. [DOI] [PubMed] [Google Scholar]
- 3. Vasconcelos M, Eckert H, Arahana V, Graef G, Grusak MA, et al. (2006) Molecular and phenotypic characterization of transgenic soybean expressing the Arabidopsis ferric chelate reductase gene, FRO2. Planta 224: 1116–1128. [DOI] [PubMed] [Google Scholar]
- 4. Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, et al. (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. The Plant cell 14: 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shatwell KP, Dancis A, Cross AR, Klausner RD, Segal AW (1996) The FRE1 ferric reductase of Saccharomyces cerevisiae is a cytochrome b similar to that of NADPH oxidase. The Journal of biological chemistry 271: 14240–14244. [DOI] [PubMed] [Google Scholar]
- 6. Finegold AA, Shatwell KP, Segal AW, Klausner RD, Dancis A (1996) Intramembrane bis-heme motif for transmembrane electron transport conserved in a yeast iron reductase and the human NADPH oxidase. The Journal of biological chemistry 271: 31021–31024. [DOI] [PubMed] [Google Scholar]
- 7. Bedard K, Krause KH (2007) The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiological reviews 87: 245–313. [DOI] [PubMed] [Google Scholar]
- 8. Brown GD (2011) Innate antifungal immunity: the key role of phagocytes. Annual review of immunology 29: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Groeger G, Quiney C, Cotter TG (2009) Hydrogen peroxide as a cell-survival signaling molecule. Antioxidants & redox signaling 11: 2655–2671. [DOI] [PubMed] [Google Scholar]
- 10. Lardy B, Bof M, Aubry L, Paclet MH, Morel F, et al. (2005) NADPH oxidase homologs are required for normal cell differentiation and morphogenesis in Dictyostelium discoideum. Biochim Biophys Acta 1744: 199–212. [DOI] [PubMed] [Google Scholar]
- 11. Anh NT, Nishitani M, Harada S, Yamaguchi M, Kamei K (2011) Essential role of Duox in stabilization of Drosophila wing. The Journal of biological chemistry 286: 33244–33251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Touyz RM, Briones AM, Sedeek M, Burger D, Montezano AC (2011) NOX isoforms and reactive oxygen species in vascular health. Molecular interventions 11: 27–35. [DOI] [PubMed] [Google Scholar]
- 13. Boeckmann B, Robinson-Rechavi M, Xenarios I, Dessimoz C (2011) Conceptual framework and pilot study to benchmark phylogenomic databases based on reference gene trees. Brief Bioinform 12: 423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kawahara T, Quinn MT, Lambeth JD (2007) Molecular evolution of the reactive oxygen-generating NADPH oxidase (Nox/Duox) family of enzymes. BMC Evol Biol 7: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sumimoto H (2008) Structure, regulation and evolution of Nox-family NADPH oxidases that produce reactive oxygen species. The FEBS journal 275: 3249–3277. [DOI] [PubMed] [Google Scholar]
- 16. Sanchez-Pulido L, Rojas AM, Valencia A, Martinez AC, Andrade MA (2004) ACRATA: a novel electron transfer domain associated to apoptosis and cancer. BMC Cancer 4: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, et al. (2005) The Universal Protein Resource (UniProt). Nucleic acids research 33: D154–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Magrane M, Consortium U (2011) UniProt Knowledgebase: a hub of integrated protein data. Database: the journal of biological databases and curation 2011: bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Finn RD, Mistry J, Tate J, Coggill P, Heger A, et al. (2010) The Pfam protein families database. Nucleic acids research 38: D211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, et al. (2008) NCBI BLAST: a better web interface. Nucleic acids research 36: W5–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dereeper A, Audic S, Claverie JM, Blanc G (2010) BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC evolutionary biology 10: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Katoh K, Toh H (2008) Recent developments in the MAFFT multiple sequence alignment program. Briefings in bioinformatics 9: 286–298. [DOI] [PubMed] [Google Scholar]
- 23. Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282. [DOI] [PubMed] [Google Scholar]
- 24. Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Penn O, Privman E, Ashkenazy H, Landan G, Graur D, et al. (2010) GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Res 38: W23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105. [DOI] [PubMed] [Google Scholar]
- 27. Guindon S, Delsuc F, Dufayard JF, Gascuel O (2009) Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol 537: 113–137. [DOI] [PubMed] [Google Scholar]
- 28. Le SQ, Gascuel O (2008) An improved general amino acid replacement matrix. Mol Biol Evol 25: 1307–1320. [DOI] [PubMed] [Google Scholar]
- 29. Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- 30. Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016. [DOI] [PubMed] [Google Scholar]
- 31. Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580. [DOI] [PubMed] [Google Scholar]
- 32. Kall L, Krogh A, Sonnhammer EL (2007) Advantages of combined transmembrane topology and signal peptide prediction–the Phobius web server. Nucleic Acids Res 35: W429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jones DT (2007) Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics 23: 538–544. [DOI] [PubMed] [Google Scholar]
- 34. Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, et al. (2012) InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res 40: D306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, et al. (2012) The Pfam protein families database. Nucleic Acids Res 40: D290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Eisenhaber F, Eisenhaber B, Kubina W, Maurer-Stroh S, Neuberger G, et al. (2003) Prediction of lipid posttranslational modifications and localization signals from protein sequences: big-Pi, NMT and PTS1. Nucleic Acids Res 31: 3631–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bedard K, Lardy B, Krause KH (2007) NOX family NADPH oxidases: not just in mammals. Biochimie 89: 1107–1112. [DOI] [PubMed] [Google Scholar]
- 39. Grissa I, Bidard F, Grognet P, Grossetete S, Silar P (2010) The Nox/Ferric reductase/Ferric reductase-like families of Eumycetes. Fungal Biol 114: 766–777. [DOI] [PubMed] [Google Scholar]
- 40. von Rozycki T, Yen MR, Lende EE, Saier MH Jr (2004) The YedZ family: possible heme binding proteins that can be fused to transporters and electron carriers. Journal of molecular microbiology and biotechnology 8: 129–140. [DOI] [PubMed] [Google Scholar]
- 41. Brokx SJ, Rothery RA, Zhang G, Ng DP, Weiner JH (2005) Characterization of an Escherichia coli sulfite oxidase homologue reveals the role of a conserved active site cysteine in assembly and function. Biochemistry 44: 10339–10348. [DOI] [PubMed] [Google Scholar]
- 42. Ohgami RS, Campagna DR, McDonald A, Fleming MD (2006) The Steap proteins are metalloreductases. Blood 108: 1388–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Knutson MD (2007) Steap proteins: implications for iron and copper metabolism. Nutr Rev 65: 335–340. [DOI] [PubMed] [Google Scholar]
- 44. Dancis A, Roman DG, Anderson GJ, Hinnebusch AG, Klausner RD (1992) Ferric reductase of Saccharomyces cerevisiae: molecular characterization, role in iron uptake, and transcriptional control by iron. Proceedings of the National Academy of Sciences of the United States of America 89: 3869–3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stearman R, Yuan DS, Yamaguchi-Iwai Y, Klausner RD, Dancis A (1996) A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271: 1552–1557. [DOI] [PubMed] [Google Scholar]
- 46. Martins LJ, Jensen LT, Simon JR, Keller GL, Winge DR (1998) Metalloregulation of FRE1 and FRE2 homologs in Saccharomyces cerevisiae. The Journal of biological chemistry 273: 23716–23721. [DOI] [PubMed] [Google Scholar]
- 47. Rinnerthaler M, Buttner S, Laun P, Heeren G, Felder TK, et al. (2012) Yno1p/Aim14p, a NADPH-oxidase ortholog, controls extramitochondrial reactive oxygen species generation, apoptosis, and actin cable formation in yeast. Proc Natl Acad Sci U S A 109: 8658–8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jeong J, Cohu C, Kerkeb L, Pilon M, Connolly EL, et al. (2008) Chloroplast Fe(III) chelate reductase activity is essential for seedling viability under iron limiting conditions. Proceedings of the National Academy of Sciences of the United States of America 105: 10619–10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jeong J, Connolly EL (2009) Iron uptake mechanisms in plants: Functions of the FRO family of ferric reductases. Plant Science 176: 709–714. [Google Scholar]
- 50. Iyer LM, Anantharaman V, Aravind L (2007) The DOMON domains are involved in heme and sugar recognition. Bioinformatics 23: 2660–2664. [DOI] [PubMed] [Google Scholar]
- 51. Aravind L (2001) DOMON: an ancient extracellular domain in dopamine beta-monooxygenase and other proteins. Trends in biochemical sciences 26: 524–526. [DOI] [PubMed] [Google Scholar]
- 52. Herve C, Tonon T, Collen J, Corre E, Boyen C (2006) NADPH oxidases in Eukaryotes: red algae provide new hints! Current genetics. 49: 190–204. [DOI] [PubMed] [Google Scholar]
- 53. Tripathi S, Portman JJ (2008) Inherent flexibility and protein function: The open/closed conformational transition in the N-terminal domain of calmodulin. J Chem Phys 128: 205104. [DOI] [PubMed] [Google Scholar]
- 54. Kawahara T, Quinn MT, Lambeth JD (2007) Molecular evolution of the reactive oxygen-generating NADPH oxidase (Nox/Duox) family of enzymes. BMC evolutionary biology 7: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takemoto D, Tanaka A, Scott B (2007) NADPH oxidases in fungi: diverse roles of reactive oxygen species in fungal cellular differentiation. Fungal genetics and biology: FG & B 44: 1065–1076. [DOI] [PubMed] [Google Scholar]
- 56. Tirone F, Cox JA (2007) NADPH oxidase 5 (NOX5) interacts with and is regulated by calmodulin. FEBS Lett 581: 1202–1208. [DOI] [PubMed] [Google Scholar]
- 57. Banfi B, Tirone F, Durussel I, Knisz J, Moskwa P, et al. (2004) Mechanism of Ca2+ activation of the NADPH oxidase 5 (NOX5). J Biol Chem 279: 18583–18591. [DOI] [PubMed] [Google Scholar]
- 58. Olson JS, Mathews AJ, Rohlfs RJ, Springer BA, Egeberg KD, et al. (1988) The role of the distal histidine in myoglobin and haemoglobin. Nature 336: 265–266. [DOI] [PubMed] [Google Scholar]
- 59. Holliday GL, Mitchell JB, Thornton JM (2009) Understanding the functional roles of amino acid residues in enzyme catalysis. J Mol Biol 390: 560–577. [DOI] [PubMed] [Google Scholar]
- 60. Rae J, Newburger PE, Dinauer MC, Noack D, Hopkins PJ, et al. (1998) X-Linked chronic granulomatous disease: mutations in the CYBB gene encoding the gp91-phox component of respiratory-burst oxidase. Am J Hum Genet 62: 1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tsuda M, Kaneda M, Sakiyama T, Inana I, Owada M, et al. (1998) A novel mutation at a probable heme-binding ligand in neutrophil cytochrome b558 in atypical X-linked chronic granulomatous disease. Hum Genet 103: 377–381. [DOI] [PubMed] [Google Scholar]
- 62. Bustamante J, Arias AA, Vogt G, Picard C, Galicia LB, et al. (2011) Germline CYBB mutations that selectively affect macrophages in kindreds with X-linked predisposition to tuberculous mycobacterial disease. Nat Immunol 12: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ueno N, Takeya R, Miyano K, Kikuchi H, Sumimoto H (2005) The NADPH oxidase Nox3 constitutively produces superoxide in a p22phox-dependent manner: its regulation by oxidase organizers and activators. J Biol Chem 280: 23328–23339. [DOI] [PubMed] [Google Scholar]
- 64. Martyn KD, Frederick LM, von Loehneysen K, Dinauer MC, Knaus UG (2006) Functional analysis of Nox4 reveals unique characteristics compared to other NADPH oxidases. Cell Signal 18: 69–82. [DOI] [PubMed] [Google Scholar]
- 65. Mayer BJ (2001) SH3 domains: complexity in moderation. Journal of cell science 114: 1253–1263. [DOI] [PubMed] [Google Scholar]
- 66. Cano-Dominguez N, Alvarez-Delfin K, Hansberg W, Aguirre J (2008) NADPH oxidases NOX-1 and NOX-2 require the regulatory subunit NOR-1 to control cell differentiation and growth in Neurospora crassa. Eukaryotic cell 7: 1352–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Takemoto D, Tanaka A, Scott B (2006) A p67Phox-like regulator is recruited to control hyphal branching in a fungal-grass mutualistic symbiosis. The Plant cell 18: 2807–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lapouge K, Smith SJ, Walker PA, Gamblin SJ, Smerdon SJ, et al. (2000) Structure of the TPR domain of p67phox in complex with Rac.GTP. Molecular cell 6: 899–907. [DOI] [PubMed] [Google Scholar]
- 69. Jain R, Rivera MC, Lake JA (1999) Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci U S A 96: 3801–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yutin N, Makarova KS, Mekhedov SL, Wolf YI, Koonin EV (2008) The deep archaeal roots of eukaryotes. Mol Biol Evol 25: 1619–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Makarova KS, Wolf YI, Mekhedov SL, Mirkin BG, Koonin EV (2005) Ancestral paralogs and pseudoparalogs and their role in the emergence of the eukaryotic cell. Nucleic Acids Res 33: 4626–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Matsuno K, Yamada H, Iwata K, Jin D, Katsuyama M, et al. (2005) Nox1 is involved in angiotensin II-mediated hypertension: a study in Nox1-deficient mice. Circulation 112: 2677–2685. [DOI] [PubMed] [Google Scholar]
- 73. Dikalova A, Clempus R, Lassegue B, Cheng G, McCoy J, et al. (2005) Nox1 overexpression potentiates angiotensin II-induced hypertension and vascular smooth muscle hypertrophy in transgenic mice. Circulation 112: 2668–2676. [DOI] [PubMed] [Google Scholar]
- 74. Garrido-Urbani S, Jemelin S, Deffert C, Carnesecchi S, Basset O, et al. (2011) Targeting vascular NADPH oxidase 1 blocks tumor angiogenesis through a PPARalpha mediated mechanism. PLoS One 6: e14665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kumatori A, Faizunnessa NN, Suzuki S, Moriuchi T, Kurozumi H, et al. (1998) Nonhomologous recombination between the cytochrome b558 heavy chain gene (CYBB) and LINE-1 causes an X-linked chronic granulomatous disease. Genomics 53: 123–128. [DOI] [PubMed] [Google Scholar]
- 76. Price MO, Atkinson SJ, Knaus UG, Dinauer MC (2002) Rac activation induces NADPH oxidase activity in transgenic COSphox cells, and the level of superoxide production is exchange factor-dependent. The Journal of biological chemistry 277: 19220–19228. [DOI] [PubMed] [Google Scholar]
- 77. Babior BM (2000) The NADPH oxidase of endothelial cells. IUBMB life 50: 267–269. [DOI] [PubMed] [Google Scholar]
- 78. Jendrysik MA, Vasilevsky S, Yi L, Wood A, Zhu N, et al. (2011) NADPH oxidase-2 derived ROS dictates murine DC cytokine-mediated cell fate decisions during CD4 T helper-cell commitment. PLoS One 6: e28198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Paffenholz R, Bergstrom RA, Pasutto F, Wabnitz P, Munroe RJ, et al. (2004) Vestibular defects in head-tilt mice result from mutations in Nox3, encoding an NADPH oxidase. Genes & development 18: 486–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Banfi B, Malgrange B, Knisz J, Steger K, Dubois-Dauphin M, et al. (2004) NOX3, a superoxide-generating NADPH oxidase of the inner ear. The Journal of biological chemistry 279: 46065–46072. [DOI] [PubMed] [Google Scholar]
- 81. Paravicini TM, Chrissobolis S, Drummond GR, Sobey CG (2004) Increased NADPH-oxidase activity and Nox4 expression during chronic hypertension is associated with enhanced cerebral vasodilatation to NADPH in vivo. Stroke; a journal of cerebral circulation 35: 584–589. [DOI] [PubMed] [Google Scholar]
- 82. Kim KS, Choi HW, Yoon HE, Kim IY (2010) Reactive oxygen species generated by NADPH oxidase 2 and 4 are required for chondrogenic differentiation. The Journal of biological chemistry 285: 40294–40302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Cucoranu I, Clempus R, Dikalova A, Phelan PJ, Ariyan S, et al. (2005) NAD(P)H oxidase 4 mediates transforming growth factor-beta1-induced differentiation of cardiac fibroblasts into myofibroblasts. Circulation research 97: 900–907. [DOI] [PubMed] [Google Scholar]
- 84. Tanaka A, Christensen MJ, Takemoto D, Park P, Scott B (2006) Reactive oxygen species play a role in regulating a fungus-perennial ryegrass mutualistic interaction. The Plant cell 18: 1052–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Segmuller N, Kokkelink L, Giesbert S, Odinius D, van Kan J, et al. (2008) NADPH oxidases are involved in differentiation and pathogenicity in Botrytis cinerea. Molecular plant-microbe interactions : MPMI 21: 808–819. [DOI] [PubMed] [Google Scholar]
- 86. Lara-Ortiz T, Riveros-Rosas H, Aguirre J (2003) Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Molecular microbiology 50: 1241–1255. [DOI] [PubMed] [Google Scholar]
- 87. Egan MJ, Wang ZY, Jones MA, Smirnoff N, Talbot NJ (2007) Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proceedings of the National Academy of Sciences of the United States of America 104: 11772–11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Malagnac F, Lalucque H, Lepere G, Silar P (2004) Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal genetics and biology: FG & B 41: 982–997. [DOI] [PubMed] [Google Scholar]
- 89. Brun S, Malagnac F, Bidard F, Lalucque H, Silar P (2009) Functions and regulation of the Nox family in the filamentous fungus Podospora anserina: a new role in cellulose degradation. Molecular microbiology 74: 480–496. [DOI] [PubMed] [Google Scholar]
- 90. Song Y, Driessens N, Costa M, De Deken X, Detours V, et al. (2007) Roles of hydrogen peroxide in thyroid physiology and disease. The Journal of clinical endocrinology and metabolism 92: 3764–3773. [DOI] [PubMed] [Google Scholar]
- 91. Geiszt M, Witta J, Baffi J, Lekstrom K, Leto TL (2003) Dual oxidases represent novel hydrogen peroxide sources supporting mucosal surface host defense. The FASEB journal: official publication of the Federation of American Societies for Experimental Biology 17: 1502–1504. [DOI] [PubMed] [Google Scholar]
- 92. Ohye H, Sugawara M (2010) Dual oxidase, hydrogen peroxide and thyroid diseases. Experimental biology and medicine 235: 424–433. [DOI] [PubMed] [Google Scholar]
- 93. De Deken X, Wang D, Many MC, Costagliola S, Libert F, et al. (2000) Cloning of two human thyroid cDNAs encoding new members of the NADPH oxidase family. The Journal of biological chemistry 275: 23227–23233. [DOI] [PubMed] [Google Scholar]
- 94. Cardoso LC, Martins DC, Figueiredo MD, Rosenthal D, Vaisman M, et al. (2001) Ca(2+)/nicotinamide adenine dinucleotide phosphate-dependent H(2)O(2) generation is inhibited by iodide in human thyroids. The Journal of clinical endocrinology and metabolism 86: 4339–4343. [DOI] [PubMed] [Google Scholar]
- 95. Donko A, Ruisanchez E, Orient A, Enyedi B, Kapui R, et al. (2010) Urothelial cells produce hydrogen peroxide through the activation of Duox1. Free radical biology & medicine 49: 2040–2048. [DOI] [PubMed] [Google Scholar]
- 96. Grasberger H, De Deken X, Mayo OB, Raad H, Weiss M, et al. (2012) Mice deficient in dual oxidase maturation factors are severely hypothyroid. Molecular endocrinology 26: 481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Moreno JC, Visser TJ (2007) New phenotypes in thyroid dyshormonogenesis: hypothyroidism due to DUOX2 mutations. Endocrine development 10: 99–117. [DOI] [PubMed] [Google Scholar]
- 98. Niethammer P, Grabher C, Look AT, Mitchison TJ (2009) A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature 459: 996–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kumar S, Molina-Cruz A, Gupta L, Rodrigues J, Barillas-Mury C (2010) A peroxidase/dual oxidase system modulates midgut epithelial immunity in Anopheles gambiae. Science 327: 1644–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kumar S, Gupta L, Han YS, Barillas-Mury C (2004) Inducible peroxidases mediate nitration of anopheles midgut cells undergoing apoptosis in response to Plasmodium invasion. The Journal of biological chemistry 279: 53475–53482. [DOI] [PubMed] [Google Scholar]
- 101. Ritsick DR, Edens WA, Finnerty V, Lambeth JD (2007) Nox regulation of smooth muscle contraction. Free radical biology & medicine 43: 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Ha EM, Oh CT, Bae YS, Lee WJ (2005) A direct role for dual oxidase in Drosophila gut immunity. Science 310: 847–850. [DOI] [PubMed] [Google Scholar]
- 103. Juarez MT, Patterson RA, Sandoval-Guillen E, McGinnis W (2011) Duox, Flotillin-2, and Src42A are required to activate or delimit the spread of the transcriptional response to epidermal wounds in Drosophila. PLoS Genet 7: e1002424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Meitzler JL, Brandman R, Ortiz de Montellano PR (2010) Perturbed heme binding is responsible for the blistering phenotype associated with mutations in the Caenorhabditis elegans dual oxidase 1 (DUOX1) peroxidase domain. The Journal of biological chemistry 285: 40991–41000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Edens WA, Sharling L, Cheng G, Shapira R, Kinkade JM, et al. (2001) Tyrosine cross-linking of extracellular matrix is catalyzed by Duox, a multidomain oxidase/peroxidase with homology to the phagocyte oxidase subunit gp91phox. The Journal of cell biology 154: 879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Chavez V, Mohri-Shiomi A, Garsin DA (2009) Ce-Duox1/BLI-3 generates reactive oxygen species as a protective innate immune mechanism in Caenorhabditis elegans. Infection and immunity 77: 4983–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wong JL, Creton R, Wessel GM (2004) The oxidative burst at fertilization is dependent upon activation of the dual oxidase Udx1. Developmental cell 7: 801–814. [DOI] [PubMed] [Google Scholar]
- 108. Foreman J, Demidchik V, Bothwell JH, Mylona P, Miedema H, et al. (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446. [DOI] [PubMed] [Google Scholar]
- 109. Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, et al. (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. The EMBO journal 22: 2623–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Fagard M, Dellagi A, Roux C, Perino C, Rigault M, et al. (2007) Arabidopsis thaliana expresses multiple lines of defense to counterattack Erwinia chrysanthemi. Molecular plant-microbe interactions: MPMI 20: 794–805. [DOI] [PubMed] [Google Scholar]
- 111. Aguirre J, Lambeth JD (2010) Nox enzymes from fungus to fly to fish and what they tell us about Nox function in mammals. Free radical biology & medicine 49: 1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Torres MA, Dangl JL, Jones JD (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proceedings of the National Academy of Sciences of the United States of America 99: 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Brar SS, Corbin Z, Kennedy TP, Hemendinger R, Thornton L, et al. (2003) NOX5 NAD(P)H oxidase regulates growth and apoptosis in DU 145 prostate cancer cells. American journal of physiology Cell physiology 285: C353–369. [DOI] [PubMed] [Google Scholar]
- 114. Hong J, Behar J, Wands J, Resnick M, Wang LJ, et al. (2010) Bile acid reflux contributes to development of esophageal adenocarcinoma via activation of phosphatidylinositol-specific phospholipase Cgamma2 and NADPH oxidase NOX5-S. Cancer research 70: 1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Banfi B, Molnar G, Maturana A, Steger K, Hegedus B, et al. (2001) A Ca(2+)-activated NADPH oxidase in testis, spleen, and lymph nodes. The Journal of biological chemistry 276: 37594–37601. [DOI] [PubMed] [Google Scholar]
- 116. Zhou X, Li D, Resnick MB, Behar J, Wands J, et al. (2011) Signaling in H2O2-induced increase in cell proliferation in Barrett’s esophageal adenocarcinoma cells. The Journal of pharmacology and experimental therapeutics 339: 218–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Oliveira Gde A, Lieberman J, Barillas-Mury C (2012) Epithelial nitration by a peroxidase/NOX5 system mediates mosquito antiplasmodial immunity. Science 335: 856–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697. [DOI] [PubMed] [Google Scholar]
- 119. Einset J, Winge P, Bones AM, Connolly EL (2008) The FRO2 ferric reductase is required for glycine betaine’s effect on chilling tolerance in Arabidopsis roots. Physiologia plantarum 134: 334–341. [DOI] [PubMed] [Google Scholar]
- 120. Mukherjee I, Campbell NH, Ash JS, Connolly EL (2006) Expression profiling of the Arabidopsis ferric chelate reductase (FRO) gene family reveals differential regulation by iron and copper. Planta 223: 1178–1190. [DOI] [PubMed] [Google Scholar]
- 121. Wu H, Li L, Du J, Yuan Y, Cheng X, et al. (2005) Molecular and biochemical characterization of the Fe(III) chelate reductase gene family in Arabidopsis thaliana. Plant & cell physiology 46: 1505–1514. [DOI] [PubMed] [Google Scholar]
- 122. Bernal M, Casero D, Singh V, Wilson GT, Grande A, et al. (2012) Transcriptome Sequencing Identifies SPL7-Regulated Copper Acquisition Genes FRO4/FRO5 and the Copper Dependence of Iron Homeostasis in Arabidopsis. The Plant cell 24: 738–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Feng H, An F, Zhang S, Ji Z, Ling HQ, et al. (2006) Light-regulated, tissue-specific, and cell differentiation-specific expression of the Arabidopsis Fe(III)-chelate reductase gene AtFRO6. Plant physiology 140: 1345–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Yun CW, Bauler M, Moore RE, Klebba PE, Philpott CC (2001) The role of the FRE family of plasma membrane reductases in the uptake of siderophore-iron in Saccharomyces cerevisiae. The Journal of biological chemistry 276: 10218–10223. [DOI] [PubMed] [Google Scholar]
- 125. Hassett R, Kosman DJ (1995) Evidence for Cu(II) reduction as a component of copper uptake by Saccharomyces cerevisiae. The Journal of biological chemistry 270: 128–134. [DOI] [PubMed] [Google Scholar]
- 126. Georgatsou E, Mavrogiannis LA, Fragiadakis GS, Alexandraki D (1997) The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. The Journal of biological chemistry 272: 13786–13792. [DOI] [PubMed] [Google Scholar]
- 127. Singh A, Kaur N, Kosman DJ (2007) The metalloreductase Fre6p in Fe-efflux from the yeast vacuole. The Journal of biological chemistry 282: 28619–28626. [DOI] [PubMed] [Google Scholar]
- 128. Rees EM, Thiele DJ (2007) Identification of a vacuole-associated metalloreductase and its role in Ctr2-mediated intracellular copper mobilization. The Journal of biological chemistry 282: 21629–21638. [DOI] [PubMed] [Google Scholar]
- 129. Roman DG, Dancis A, Anderson GJ, Klausner RD (1993) The fission yeast ferric reductase gene frp1+ is required for ferric iron uptake and encodes a protein that is homologous to the gp91-phox subunit of the human NADPH phagocyte oxidoreductase. Molecular and cellular biology 13: 4342–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence data and domain predictions.
(XLSX)
Maximum likelihood phylogeny of eukaryotic gene families of the FRE group and the NOX group. The phylogenetic tree is a detailed presentation of Figure 1B. Tree branches are colored according to the taxonomic classification of the species.
(TIF)
Exploration of the tree space. Phylogenies of the FRD superfamily from multiple analyses. Gene families are colored in the phylogenetic tree and family names are given. Branch support values of major internal nodes are indicated.
(PDF)
Analysis of bacterial FRD superfamily members.
(PDF)
Evolution of the FRD superfamily from the view of its core domains.
(PDF)