Abstract
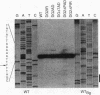
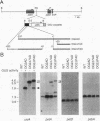
Complex processing of primary transcripts occurs during the expression of higher-plant chloroplast genes. In Chlamydomonas reinhardtii, most chloroplast genes appear to possess their own promoters, rather than being transcribed as part of multicistronic operons. By generating specific deletion mutants, we show that petD, which encodes subunit IV of the cytochrome b6/f complex, has an RNA processing site that is required for accumulation of monocistronic petD mRNA in petD promoter deletion mutants; in such mutants, transcription of petD originates from the upstream petA promoter. The 5' ends of transcripts initiated at the petD promoter are probably also generated by processing, since the 5' end of monocistronic petD mRNA is the same in wild-type strains as it is in the petD promoter mutants. The location and function of the processing site were further examined by inserting petD-uidA fusion genes into the chloroplast genome (uidA is an Escherichia coli gene that encodes beta-glucuronidase). When a promoterless petD-uidA fusion gene was inserted downstream of petA, a monocistronic uidA transcript accumulated, which was apparently initiated at the petA promoter and was processed at a site corresponding precisely to the petD mRNA 5' end. When a construct including only sequences downstream of +25 relative to the mature mRNA 5' end was inserted into the same site, a dicistronic petA-uidA transcript accumulated but no monocistronic uidA transcript could be detected, suggesting that a processing site lies at least partially within the region from -1 to +25. Beta-glucuronidase activity was not detected in transformants that accumulated only the dicistronic petA-uidA transcript, suggesting that the first 25 bp of the 5' untranslated region are required for translation initiation. One explanation for this translational defect is that Chlamydomonas chloroplasts cannot translate the second coding region of some dicistronic messages.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barkan A. Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 1988 Sep;7(9):2637–2644. doi: 10.1002/j.1460-2075.1988.tb03116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D. C., Rogers S. A., Chen L. J., Orozco E. M., Jr A primary transcript in spinach chloroplasts that completely lacks a 5' untranslated leader region. Plant Mol Biol. 1990 Jul;15(1):111–119. doi: 10.1007/BF00017728. [DOI] [PubMed] [Google Scholar]
- Blowers A. D., Ellmore G. S., Klein U., Bogorad L. Transcriptional analysis of endogenous and foreign genes in chloroplast transformants of Chlamydomonas. Plant Cell. 1990 Nov;2(11):1059–1070. doi: 10.1105/tpc.2.11.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton J. E., Gillham N. W., Harris E. H., Hosler J. P., Johnson A. M., Jones A. R., Randolph-Anderson B. L., Robertson D., Klein T. M., Shark K. B. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 1988 Jun 10;240(4858):1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- Cannistraro V. J., Subbarao M. N., Kennell D. Specific endonucleolytic cleavage sites for decay of Escherichia coli mRNA. J Mol Biol. 1986 Nov 20;192(2):257–274. doi: 10.1016/0022-2836(86)90363-3. [DOI] [PubMed] [Google Scholar]
- Deutscher M. P. Ribonuclease multiplicity, diversity, and complexity. J Biol Chem. 1993 Jun 25;268(18):13011–13014. [PubMed] [Google Scholar]
- Hanley-Bowdoin L., Orozco E. M., Jr, Chua N. H. In vitro synthesis and processing of a maize chloroplast transcript encoded by the ribulose 1,5-bisphosphate carboxylase large subunit gene. Mol Cell Biol. 1985 Oct;5(10):2733–2745. doi: 10.1128/mcb.5.10.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G. S., Mason J. G., Holton T. A., Koller B., Cox G. B., Whitfeld P. R., Bottomley W. A gene cluster in the spinach and pea chloroplast genomes encoding one CF1 and three CF0 subunits of the H+-ATP synthase complex and the ribosomal protein S2. J Mol Biol. 1987 Jul 20;196(2):283–298. doi: 10.1016/0022-2836(87)90690-5. [DOI] [PubMed] [Google Scholar]
- Jefferson R. A., Kavanagh T. A., Bevan M. W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987 Dec 20;6(13):3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallas T., Spiller S., Malkin R. Characterization of two operons encoding the cytochrome b6-f complex of the cyanobacterium Nostoc PCC 7906. Highly conserved sequences but different gene organization than in chloroplasts. J Biol Chem. 1988 Oct 5;263(28):14334–14342. [PubMed] [Google Scholar]
- Kindle K. L., Richards K. L., Stern D. B. Engineering the chloroplast genome: techniques and capabilities for chloroplast transformation in Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1721–1725. doi: 10.1073/pnas.88.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy D., Zhang W., Cao J., Wu R. Isolation of an efficient actin promoter for use in rice transformation. Plant Cell. 1990 Feb;2(2):163–171. doi: 10.1105/tpc.2.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B. J., Schottel J. L. Characterization of cat messenger RNA decay suggests that turnover occurs by endonucleolytic cleavage in a 3' to 5' direction. Mol Microbiol. 1992 May;6(9):1095–1104. doi: 10.1111/j.1365-2958.1992.tb01547.x. [DOI] [PubMed] [Google Scholar]
- Reinbothe S., Reinbothe C., Heintzen C., Seidenbecher C., Parthier B. A methyl jasmonate-induced shift in the length of the 5' untranslated region impairs translation of the plastid rbcL transcript in barley. EMBO J. 1993 Apr;12(4):1505–1512. doi: 10.1002/j.1460-2075.1993.tb05794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W., Kindle K. L., Stern D. B. In vivo analysis of Chlamydomonas chloroplast petD gene expression using stable transformation of beta-glucuronidase translational fusions. Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):497–501. doi: 10.1073/pnas.90.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W., Sturm N. R., Kindle K. L., Stern D. B. petD mRNA maturation in Chlamydomonas reinhardtii chloroplasts: role of 5' endonucleolytic processing. Mol Cell Biol. 1994 Sep;14(9):6180–6186. doi: 10.1128/mcb.14.9.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd H. S., Boynton J. E., Gillham N. W. Mutations in nine chloroplast loci of Chlamydomonas affecting different photosynthetic functions. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1353–1357. doi: 10.1073/pnas.76.3.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Gruissem W. Control of plastid gene expression: 3' inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell. 1987 Dec 24;51(6):1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- Stern D. B., Kindle K. L. 3'end maturation of the Chlamydomonas reinhardtii chloroplast atpB mRNA is a two-step process. Mol Cell Biol. 1993 Apr;13(4):2277–2285. doi: 10.1128/mcb.13.4.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Radwanski E. R., Kindle K. L. A 3' stem/loop structure of the Chlamydomonas chloroplast atpB gene regulates mRNA accumulation in vivo. Plant Cell. 1991 Mar;3(3):285–297. doi: 10.1105/tpc.3.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura M. The chloroplast genome. Plant Mol Biol. 1992 May;19(1):149–168. doi: 10.1007/BF00015612. [DOI] [PubMed] [Google Scholar]
- Verwoerd T. C., Dekker B. M., Hoekema A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989 Mar 25;17(6):2362–2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff P., Herrmann R. G. Complex RNA maturation in chloroplasts. The psbB operon from spinach. Eur J Biochem. 1988 Feb 1;171(3):551–564. doi: 10.1111/j.1432-1033.1988.tb13824.x. [DOI] [PubMed] [Google Scholar]