Abstract
Synthetic equivalents of phosphoprotein-specific antibodies would be valuable reagents for biological research, since these antibodies can often be difficult to produce. Protein phosphorylation is thought to result in significant conformational changes in most substrate proteins. Therefore, one approach might be to simply screen combinatorial libraries for ligands to the phosphorylated state in the hope of isolating a ligand that binds to a pocket created by the conformational shift. In this study, we probe this strategy by screening a peptoid library for ligands to the phosphorylated form of the Brd4 transcriptional coactivator protein. We find that peptoids with high selectivity for binding to phopshorylated form of Brd4 can indeed be isolated in this screen. Moreover, these ligands do not bind promiscuously to other phospho-proteins. However, attempts to employ these reagents as antibody substitutes in an immunoaffinity purification-like application showed that they do not perform as well as bona fide antibodies and that significant optimization will be required. This study highlights the potential and current limitations of a naïve library screening strategy for phosphoprotein-specific antibody surrogates.
Protein phosphorylation is one of the most common post-translational modifications in eukaryotic cells and plays a central role in signaling. Therefore, antibodies capable of recognizing the phosphorylated form of a protein are valuable reagents for biomedical research. These phosphorylation state-specific antibodies (PSSAs) are almost always produced by immunization of animals with synthetic phosphopeptides. However, the success rate in the generation of good PSSAs is poor due to the low immunogenicity of the phosphate group, the possibility for physiological cleavage of the group during the immunization process and other reasons1,2. In addition, it can be the case that the resultant antibodies do not work well in immunoprecipitation experiments due to incomplete exposure of the antigen in the native protein3. Because of these limitations, it would be useful to develop inexpensive and easy-to-make synthetic compounds that would act as PSSA surrogates, that is, bind tightly and selectively to a particular phospho-form of a given protein. Since it is quite difficult to identify small molecules that bind tightly to linear peptide epitopes, it is not possible to take an approach strictly parallel to the production of PSSAs. An alternative would be to rely on the fact that many protein phosphorylation events result in significant conformational changes in the protein, which might create small molecule binding pockets that are absent in the unmodified protein. There is precedent for this view in the recent demonstration that small molecule inhibitors can bind selectively to either the active or inactive forms of protein kinases. For example, Ranjitkar et al. has characterized small-molecule inhibitors that target an inactive conformation of protein kinases4.
To test this idea, we conducted a screen of a peptoid library against the phosphorylated form of the phosphorylation-dependent interaction domain (PDID) of the Brd4 transcriptional coactivator (Figure 1). We and others have demonstrated that libraries of peptoids5 (oligo-N-substituted glycines) are rich sources of protein-binding ligands 6-10. Brd4 is a double bromodomain-containing protein that is used as a cellular adaptor by some animal and human papillomaviruses (HPV) for anchoring viral genomes to mitotic chromosomes. Mammalian Brd4 plays a crucial role in cell growth, and is involved in cell cycle control, DNA replication and many other cellular processes11. There are two domains that are phosphorylated by the kinase CK2. One is Brd4 (598-785), and the other is Brd4 (287-530), which is also known as the PDID (Figure 1)12,13. When expressed in bacculovirus-infected insect cells, this protein is produced in a mulitply phosphorylated form12. Recently, Filippakopoulous et al identified a cell-permeable small molecule (JQ1) as a selective and potent inhibitor of recombinant Brd4 protein derived from bacteria14. In this work, we report the first example of a peptidomimetic compound selectively recognizes the phosphorylated domain, PDID, of Brd4 protein derived from insect cells.
Figure 1. Background of Bromodomain4 (Brd4) and PDID domain.
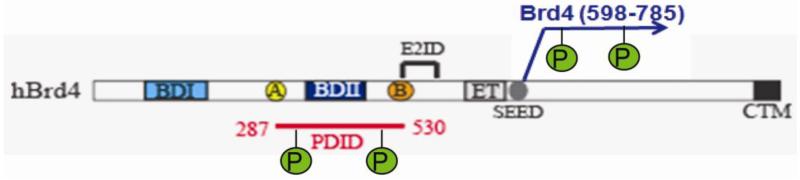
Both Brd4 (287-530), named as PDID (phosphorylation-dependent intereaction domain) and Brd4 (598-785) are highly phosphorylated by CK2.
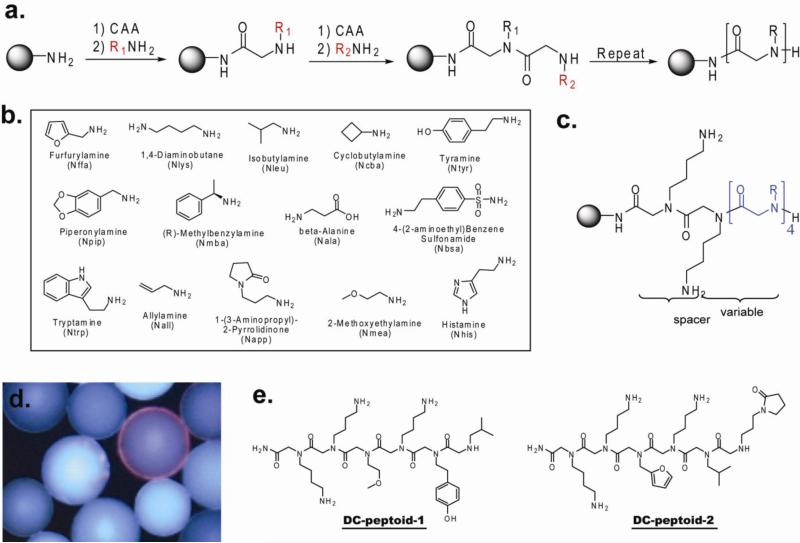
A “one bead one compound” (OBOC) peptoid library with a theoretical diversity of 144 (38, 416) compounds was synthesized using microwave-assisted sub-monomer peptoid synthesis15 on Tentagel beads using the “split and pool” strategy (Figure 2a) 5. The fourteen amines shown in Figure 2b were employed as sub-monomers. These amines were chosen so that the library would include a variety of different functional groups. Two C-terminal Nlys (diaminobutane) residues, which are positively charged at neural pH, were present in every sequence (Figure 2c) to facilitate display of the compound in aqueous solution.
Figure 2. Peptoid library construction and screening.
(a) Synthesis scheme for peptoid library. (CAA = activated ester of chloroacetic acid). (b) Amine monomers used for the sub-monomer synthesis-based library construction15. (c) General structure of the peptoids in the synthesized library. (d) A fluorescence micrograph of a single bead displaying a red halo that was picked as a hit in the screening experiment. (e) Structures of DC-peptoid 1 and DC-peptoid 2, putative phospho-PDID-binding peptoids, as determined by Edman degradation.
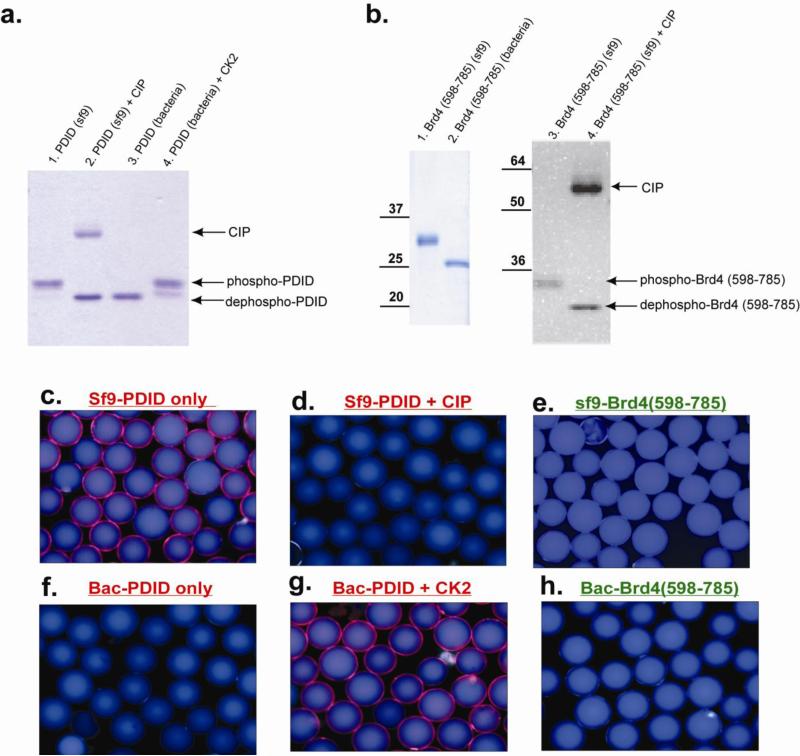
For screening, approximately 150, 000 peptoid-displaying Tentagel beads were blocked with 4% BSA and then exposed to Flag-tagged phospho-PDID (500nM) in the presence of 4% BSA. After incubation, the beads were washed thoroughly and probed with anti-Flag monoclonal antibody and a secondary anti-IgG antibody conjugated with red-emitting quantum dots (Qdots). The beads were then visualized under a fluorescent microscope, and those that displayed a red halo were isolated manually using a micropipette (16 beads total; a representative picture is shown in Figure 2d). The isolated beads were stripped of protein with 1% SDS and then re-probed with anti-Flag antibody and Qdot-conjugated secondary antibody to identify compounds that bind directly to primary or secondary antibodies rather than the phospho-PDID. Only two beads (0.0013% of the library) did not bind primary or secondary antibodies directly and thus presumably displayed phos-PDID ligands. These beads were subjected to Edman degradation, and the sequence of each peptoid was identified. The hits were named DC-peptoid 1 and DC-peptoid 2, respectively (Figure 2e). Although the number of beads employed in the screen should have provided an almost 4-fold coverage of the theoretical diversity of the library, multiple copies of each hit were not isolated, as might have been expected. It is not clear why this was the case. Both compounds were re-synthesized on TentaGel macrobeads and incubated with different forms of the protein. Phospho-PDID protein purified from insect sf9 cells was dephosphorylated with calf intestinal alkaline phosphatase (CIP) (Figure 3a, lane 1&2), and non-phosphorylated PDID protein purified from bacterial cells was phosphorylated by casein kinase 2 (CK2) (Figure 3a, lane 3&4). The different forms of PDID were incubated with bead-displayed DC-peptoid 1 and DC-peptoid 2, followed by anti-FLAG monoclonal antibody and Qdot-655-conjugated secondary antibody incubation using the same conditions as described for the library screening experiment. The hit peptoids, when incubated with phospho-PDID protein (Figure 3c, Supplementary Figure 1a), emitted red-fluorescence, confirming them as true phospho-PDID binders. Very little bead-bound fluorescence was observed when the phospho-PDID was dephosphorylated with CIP prior to incubation (Figure 3d, Supplementary Figure 1b). In the reciprocal experiment, no signal was observed when the peptoids were incubated with unmodified PDID (Figure 3f, Supplementary Figure 1d), yet red fluorescence was detected when the bacterially expressed protein was phosphorylated by CK2 (Figure 3g, Supplementary Figure 1e). These results demonstrate that the hit peptoids isolated from an unbiased library can bind to the phosphorylated PDID, but not the non-phosphorylated form of the protein.
Figure 3. The bindings between hit peptoid and PDIDs treated with different enzymes.
(a) In vitro kinase assay and dephosphorylation assay in PDID domain. (b) In vitro dephosphorylation assay in Brd4 (598-795). On-bead binding assay between immobilized DC-peptoid 1 and (c) phos-PDID only, (d) phos-PDID + CIP, (e) phos-Brd4 (598-785), (f) PDID, (g) PDID + CK2, (h) Brd4 (598-785).
To test whether the hit peptoids are general serine-, threonine-phosphorylated protein binders, another phosphorylated domain of Brd4 protein, which includes amino acids (598-785), was examined in the on-bead binding assay. Brd4 (598-785) has a comparable molecular weight and similar number of CK2 phosphorylation sites as phospho-PDID13. The same protocol was applied to phospho-Brd4 (598-785) and Brd4 (598-785) with both hit peptoids (Figure 3b). DC-peptoid 1 showed no detectable binding to either phosphorylated or non-phosphorylated Brd4 (595-785) (Figure 3e & 3h). A very weak signal was observed when DC-peptoid 2 was incubated with phos-Brd4 (598-785) (Supplementary Figure 1c), suggesting it might bind to phospho-Brd4 (598-785) with an affinity much lower than with phospho-PDID. This result suggests that these peptoids are selective ligands for phospho-PDID, rather than acting as general binders to any phospho-Serine/Threonine-containing proteins. Since DC-peptoid 1 seemed to have a higher level of selectivity for phospho-PDID than DC-peptoid 2, subsequent characterization experiments were focused on this compound.
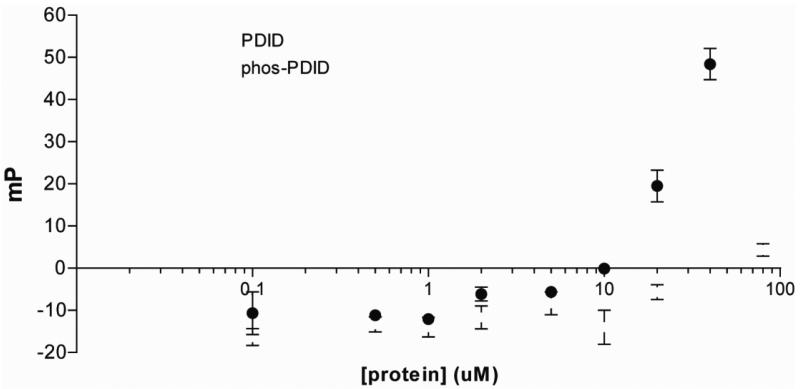
The binding selectivity and affinity of the DC-peptoid 1 for phospho-PDID was then measured in the solution phase. Fluoresence polarization experiments, in which a constant level of fluorescein-labeled, soluble DC-peptoid 1 was titrated with increasing amounts of either phospho-PDID, or unmodified PDID, were employed to measure the KD of the peptoid-protein complexes. The KD of the phospho-PDID-DC-peptoid 1 complex could not be determined precisely, since saturation was not observed at the highest concentration of protein achievable, but the data (Figure 4) suggest that the value is likely in the 50-100 μM range, which, not surprisingly, is far weaker than the affinity of an antibody for its target protein. Binding of the peptoid to the unmodified PDID protein was at least 100 times weaker (Figure 4).
Figure 4.
Fluoresence polarization of peptoid binding to PDID & phos-PDID.
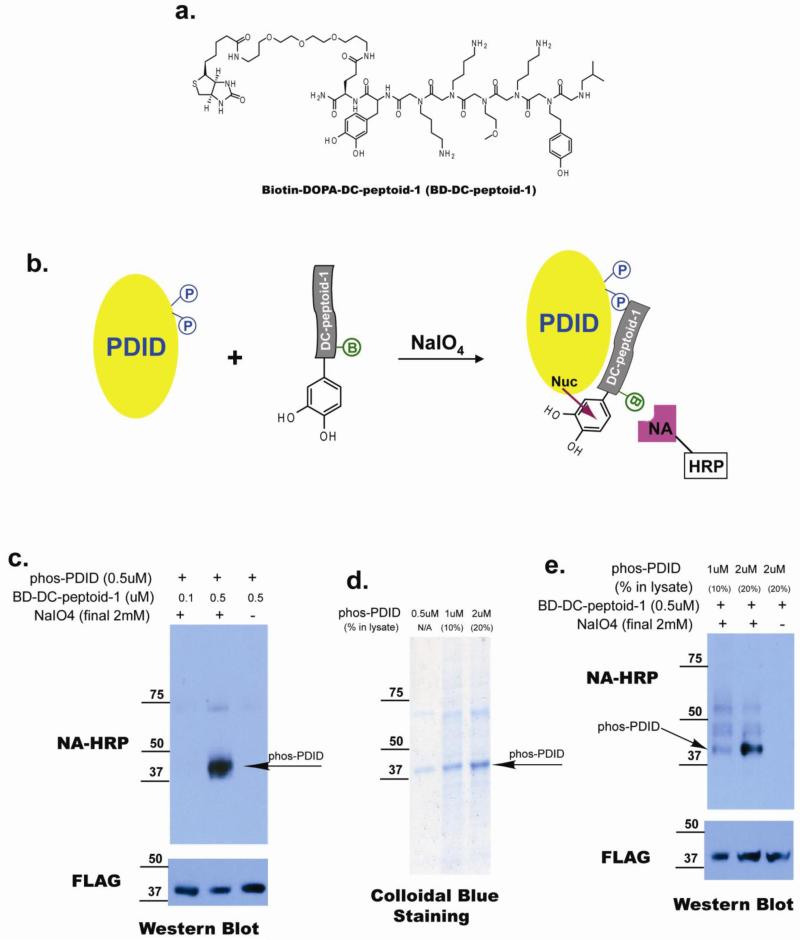
To further evaluate the binding of soluble DC-peptoid 1 to phospho-PDID protein, we constructed a hybrid molecule containing DC-peptoid 1, DOPA and biotin (BD-DC-peptoid 1; Figure 5a). We have shown previously that, when oxidized with sodium periodate, 3,4-Dihydroxylphenylalanine (DOPA)-containing small molecules will cross-link efficiently to nearby proteins via nucleophilic attack of an electron-rich protein residue on the resultant ortho-quinone (Figure 5b)16,17. The products can then be detected on gels by probing for the biotin tag. Unlike most chemical cross-linking methods, DOPA cross-linking results in little or no non-specific products even in complex protein mixtures18 because sulfhydryl groups in the buffer quench the reactive intermediate efficiently if it is not in intimate contact with a partner protein at the time of its generation. BD-DC-peptoid 1 (either 0.1 or 0.5 μM) was incubated with 0.5 μM immunopurified Flag-tagged phos-PDID. After addition of sodium periodate and a brief incubation, the reaction was quenched. Cross-linked products were separated by SDS-PAGE, transferred to PVDF membrane and probed with Neutravidin (NA)-HRP to detect biotin-containing cross-linked products. A band corresponding to phospho-PDID (~40kD) was observed in the presence of 0.5 μM peptoid and 2mM NaIO4 (Figure 5c, lane 2). No signal was detected when the peptoid concentration was reduced to 0.1 μM or when NaIO4 was absent (Figure 5c, lane 1&3).
Figure 5. Binding affinity selectivity of soluble DC-peptoid 1, and its capacity to recognize its target protein in the context of whole cell lysate.
(a) Structure of biotinylated DC-peptoid-1 conjugated with DOPA crosslinker (BD-DC-peptoid-1). (b) Cartoon illustrating mechanism of action of Biotin-DOPA conjugated DC-peptoid 1. B=Biotin, NA-HRP=Neutravidin horse radish peroxidase. (c) Biotin-DOPA-DC-peptoid-1 can cross-link with phos-PDID in the solution, and the product can be detected by Neutra-Avidin HRP. (d) Gel electrophoresis of phos-PDID in the absence or presence of crude extract. (f) Biotin-DOPA-DC-peptoid-1 can cross-link with phos-PDID when crude extract is present.
Having established that BD-DC-peptoid-1 can be cross-linked to phospho-PDID using this chemistry, we next examined the cross-linking reaction in the context of a whole cell lysate. If highly efficient and selective cross-linking of the peptoid to the phospho-PDID protein were observed, then the result would be the equivalent of a Western blot using a phospho-specific antibody. Sf9 cell lysates doped with purified phospho-PDID to a level of either 10% or 20% of the total protein (see Figure 5d, lanes 2 and 3) were used for the cross-linking experiment. When the reaction products were separated by SDS-PAGE and blotted with NA-HRP, a major band corresponding to the size of phospho-PDID (~40kD) was indeed detected. However, three other bands resulting from cross-linking of BD-DC-peptoid-1 to other proteins in the lysate were also observed at significant levels (Figure 5e, lane 1&2). Again, when NaIO4 was omitted from the reaction, no cross-linked products were observed (Figure 5e, lane 3).
These results demonstrate that the soluble BD-DC-peptoid-1 reagent can selectively detect phospho-PDID protein in the context of whole cell lysate, but that the binding selectivity is not sufficiently good to “light up” only a single protein in the lysate, even when that protein is a substantial fraction of the total protein.
Phospho-specific antibodies can be employed as affinity reagents for the purification of their cognate protein from crude extracts. To begin to explore the utility of DC-peptoid 1 as a potential affinity reagent for phospho-PDID, this compound was re-synthesized on Rink Amide beads with a cysteine reside at the C-terminus of the peptoid to facilitate compound quantification and immobilization on agarose gels. The HPLC purified Cys-DC-peptoid 1 was quantified using Elman's reagent and coupled to SulfoLink Coupling Gel (Pierce) following the manufacturer's protocols. L-cysteine was also conjugated to the Sulfolink agarose beads to serve as a negative control. An equal amount of DC-peptoid 1- or L-Cysteine-coated resin (~6 μl) were incubated with a lysate prepared from either sf9 cells that express phospho-PDID, or bacterial cells that express non modified-PDID respectively (Supplementary Figure 2a). After removing the supernatant (unbound proteins) and washing, bound proteins were stripped from the beads with the same volume of 1% SDS and the proteins were analyzed by SDS-PAGE and Western blotting using an anti-Flag antibody that recognizes both the phosphorylated and non-phosphorylated forms of the protein. As expected, control beads that are not conjugated with a peptoid ligand do not bind to the phospho-PDID (Supplementary Figure 2b, lane 2). Cys-DC-peptoid 1-conjugated agarose beads capture phospho-PDID protein from the whole insect cell lysate with a recovery efficiency of about 20% under these conditions (Supplementary Figure 2b, lane 3), but showed no binding to non-phosphorylated-PDID protein in the presence of bacterial cell lysate (Supplementary Figure 2b, lane 6), demonstrating the high selectivity of DC-peptoid 1 towards phospho-PDID in the context of a whole cell lysate. However, as shown in the Colloidal blue staining gel, there are bands other than the anticipated phos-PDID that are also precipitated by DC-peptoid-1-conjugated beads (Supplementary Figure 2c, lane 3&6). The cross-reactivity with other components in the crude extract and the relatively low recovery rate (~20%) are the challenges faced by the first peptoid ligand selective for a phosphor-protein. Structure activity relationship studies are currently under way to optimize the structure of the compound to yield better binding affinity for its target phosphorylated protein.
In conclusion, we have demonstrated that a highly phosphor-selective ligand for modified Brd4 can be identified from a modest-sized peptidomimetic library. In this case, no counter-screen against unmodified protein was necessary, but could be carried out easily if were. Not surprisingly for a primary hit from such a modest library, the affinity of the peptoids for phospho-PDID was much weaker than that evinced by a good antibody and significant optimization will be required for such molecules to be of practical use in biology. Nonetheless, this study does demonstrate the feasibility of isolation phospho-protein-selective ligands from modest combinatorial libraries.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by a contract from the National Heart, Lung, and Blood Institute (NO1-HV-28185) for the UT Southwestern Center for Proteomics Research
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPORTING INFORMATION. Two figures, as well as detailed experimental protocols are available in the supporting information section.
REFERENCES
- 1.Czernik AJ, Girault JA, Nairn AC, Chen J, Snyder G, Kebabian J, Greengard P. Methods Enzymol. 1991;201:264–83. doi: 10.1016/0076-6879(91)01025-w. [DOI] [PubMed] [Google Scholar]
- 2.Heffetz D, Fridkin M, Zick Y. Eur J Biochem. 1989;182:343–8. doi: 10.1111/j.1432-1033.1989.tb14836.x. [DOI] [PubMed] [Google Scholar]
- 3.Paradela A, Albar JP. J Proteome Res. 2008;7:1809–18. doi: 10.1021/pr7006544. [DOI] [PubMed] [Google Scholar]
- 4.Ranjitkar P, Brock AM, Maly DJ. Chem Biol. 17:195–206. doi: 10.1016/j.chembiol.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simon RJ, Kania RS, Zuckermann RN, Huebner VD, Jewell DA, Banville S, Ng S, Wang L, Rosenberg S, Marlowe CK, et al. Proc Natl Acad Sci U S A. 1992;89:9367–71. doi: 10.1073/pnas.89.20.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Udugamasooriya DG, Dineen SP, Brekken RA, Kodadek T. J Am Chem Soc. 2008;130:5744–52. doi: 10.1021/ja711193x. [DOI] [PubMed] [Google Scholar]
- 7.Xiao X, Yu P, Lim HS, Sikder D, Kodadek T. Angew Chem Int Ed Engl. 2007;46:2865–8. doi: 10.1002/anie.200604485. [DOI] [PubMed] [Google Scholar]
- 8.Lim HS, Archer CT, Kodadek T. J Am Chem Soc. 2007;129:7750–1. doi: 10.1021/ja072027p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alluri P, Liu B, Yu P, Xiao X, Kodadek T. Mol Biosyst. 2006;2:568–79. doi: 10.1039/b608924k. [DOI] [PubMed] [Google Scholar]
- 10.Simpson LS, Burdine L, Dutta AK, Feranchak AP, Kodadek T. J Am Chem Soc. 2009 doi: 10.1021/ja900852k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu SY, Chiang CM. J Biol Chem. 2007;282:13141–5. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 12.Lee AY, Chiang CM. J Biol Chem. 2009;284:2778–86. doi: 10.1074/jbc.M805835200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawa C, Nedea E, Krogan N, Wada T, Handa H, Greenblatt J, Buratowski S. Mol Cell Biol. 2004;24:4734–42. doi: 10.1128/MCB.24.11.4734-4742.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Nature. 468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olivos HJ, Alluri PG, Reddy MM, Salony D, Kodadek T. Org Lett. 2002;4:4057–9. doi: 10.1021/ol0267578. [DOI] [PubMed] [Google Scholar]
- 16.Burdine L, Gillette TG, Lin HJ, Kodadek T. J Am Chem Soc. 2004;126:11442–3. doi: 10.1021/ja045982c. [DOI] [PubMed] [Google Scholar]
- 17.Liu B, Burdine L, Kodadek T. J Am Chem Soc. 2006;128:15228–35. doi: 10.1021/ja065794h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li WW, Heinze J, Haehnel W. J Am Chem Soc. 2005;127:6140–1. doi: 10.1021/ja050974x. [DOI] [PubMed] [Google Scholar]
- 19.Alluri PG, Reddy MM, Bachhawat-Sikder K, Olivos HJ, Kodadek T. Journal of the American Chemical Society. 2003;125:13995–4004. doi: 10.1021/ja036417x. [DOI] [PubMed] [Google Scholar]
- 20.Chiang CM, Ge H, Wang Z, Hoffmann A, Roeder RG. EMBO J. 1993;12:2749–62. doi: 10.1002/j.1460-2075.1993.tb05936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu SY, Thomas MC, Hou SY, Likhite V, Chiang CM. J Biol Chem. 1999;274:23480–90. doi: 10.1074/jbc.274.33.23480. [DOI] [PubMed] [Google Scholar]
- 22.Chiang CM, Roeder RG. Pept Res. 1993;6:62–4. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.