Abstract
Introduction
Because the receptor for Parvovirus B19 (B19V) is on erythrocytes, we investigated B19V distribution in blood by in-vitro spiking experiments and evaluated viral compartmentalization and persistence in natural infection.
Methods
Two whole blood protocols (ultracentrifugation and a rapid RBC lysis/removal protocol) were evaluated using quantitative real-time PCR. Whole blood (WB) was spiked with known concentrations of B19V and recovery in various blood fractions was determined. The rapid RBC lysis/removal protocol was then used to compare B19V concentrations in 104 paired whole blood and plasma samples collected longitudinally from 43 B19V infected donors with frozen specimens in the REDS Allogeneic Donor and Recipient Repository (RADAR).
Results
In B19V spiking experiments, ~one-third of viral DNA was recovered in plasma and two-thirds was loosely bound to erythrocytes. In the IgM positive stage of infection in blood donors when plasma B19V DNA concentrations were > 100 IU/mL, median DNA concentrations were ~30-fold higher in WB than in plasma. In contrast, when IgM was absent and when the B19V DNA concentration was lower, the median whole blood to plasma ratio was ~1. Analysis of longitudinal samples demonstrated persistent detection of B19V in WB but declining ratios of WB/plasma B19V with declining plasma VL levels and loss of IgM-reactivity.
Conclusions
The WB/plasma B19V DNA ratio varies by stage of infection. Further study is required to determine if this is related to the presence of circulating DNA-positive erythrocytes derived from B19V infected erythroblasts, B19V-specific IgM mediated binding of virus to cells, or other factors.
Introduction
The understanding of the natural history of Parvovirus B19 Virus (B19V) infection is that in most immunocompetent individuals (e.g. blood donors) viremia occurs approximately one week post infection and persists at high-titers in plasma for approximately five days.1 IgM antibody develops at about 12 days post-infection and IgG antibody follows within days coinciding with precipitous declines in plasma viremia levels. Subsequently, plasma viremia disappears generally within weeks, IgM antibody becomes undetectable after several months (although this precise duration is unknown), whereas IgG antibody persists long-term and is thought to convey immunity to reinfection. Recently it has become established that a variation of this natural history occurs in people in whom chronic persistent B19V infection occurs; this is characterized by low plasma levels of B19V DNA persisting for more than six months in conjunction with IgG antibody.2-7
The receptor for B19V on bone marrow erythrocyte progenitor cells is the P blood group antigen.8,9 This receptor is also present at high concentrations on mature circulating erythrocytes in almost all individuals, with the exception of rare persons with the null p phenotype. Binding of B19V to mature erythrocytes is known to occur and has been exploited in development of red cell B19V antigen agglutination assays.10 Thus, it is theoretically possible that a substantial proportion of B19V in blood is adsorbed to or persists within erythrocytes from the infected erythroblast phase, and that B19V DNA concentrations will consequently differ in plasma and cellular blood compartments. Also, it is unknown if the partitioning of B19V between plasma and cellular blood compartments varies during different stages of infection, possibly due to the effect of IgM and IgG antibodies on B19V particles enhancing or blocking binding to one or more cellular blood elements (e.g. erythrocytes, leukocytes, or platelets).
B19V contamination of plasma derivatives has led to widespread adoption of B19V DNA screening of source and recovered plasma donations to interdict high-titer viremic units prior to pooling and fractionation.11-13 Transfusion-transmitted B19V infection from blood component transfusion occurs infrequently but has been documented in several case reports, including a recent case in the U.S.14 Although screening of whole blood components intended for individual patient transfusions is not currently routinely performed (except in Germany, Austria, and Japan, where this screening is performed on plasma and targets units with high B19V DNA concentration), the issue of compartmentalization of B19V in blood could be important if policies evolve toward further testing.
This study’s major objective was to establish the relative concentrations of B19V DNA in plasma versus whole blood and to determine if this “compartmentalization” varies in different stages of infection. To accomplish this, we developed procedures to apply a sensitive B19V PCR assay to whole blood samples. This involved a series of in-vitro spiking experiments to establish that 1) B19 viral standards contained intact viral particles that could be pelleted by our ultracentrifugation protocol; 2) spiking high titer B19V standards into fresh and frozen whole blood to establish the partitioning of exogenously spiked B19V in various blood compartments; and 3) development of a special reagent to overcome the inability of the ultracentrifugation-target capture (TC) protocol (as demonstrated in our experiments) to reliably recover and detect low levels of B19V DNA in whole blood. Following this in-vitro experimental work, we then used our standard plasma TC-PCR and this novel whole blood protocol to test serial samples from donors with previously documented B19 plasma viremia whose samples were stored in the REDS Allogeneic Donor and Recipient repository (RADAR). As a secondary objective, we also evaluated the rate of B19V DNA persistence in those B19V DNA positive donors who had a repository donation that was given at least six months before or subsequent to their DNA positive donation.
Methods
Standard B19V assays
Plasma B19V PCR assay
This assay has previously been described in detail 15,16. It is based on a magnetic-bead B19V DNA capture step followed by a TaqMan real-time PCR assay targeting the VP1 region of the genotype 1 B19V genome. Assay sensitivity was established as a 50% LOD of 1.6 IU per mL (95% CI: 1.2-2.1) and a 95% LOD of 16.5 IU per mL (95% CI: 10.6 – 33.9). When run against a standard curve, the assay can also be used to quantify B19V DNA with the lower limit of quantitation at 20 IU/mL. Alternately, in in-vitro spiking studies a difference in cycle threshold (i.e., the cycle at which DNA was initially detected) was used to compare relative quantities of B19V DNA in different blood compartments.
B19V antibody testing
Specimens were tested for the presence of B19V IgG and IgM antibodies against a recombinant VP2 protein with FDA licensed test kits (Biotrin, Dublin, Ireland). Due to sample volume considerations, testing was performed in singlicate (rather than in duplicate as stated in the package insert) by accessing a 0.25-mL subaliquot of plasma from the RADAR repository. If results fell into the equivocal zone, the assay was repeated in singlicate and the repeat result was taken as the overall final result for the specimen. 15,16
Protocol for development of whole blood PCR assays and for in vitro spiking studies to compare levels of B19V in plasma versus whole blood compartments
B19V standard and spiked controls
We used a genotype-1 B19V standard from the Center of Biologics Evaluation and Research (CBER, Rockville, MD) to prepare our spiking B19V preparation. The CBER B-19 positive standard 17 was from a window period plasma donation.. We generated plasma and whole blood spiked controls at serial two-fold dilutions, with concentrations equal to 1000 IU/mL, 500 IU/mL, 250 IU/mL, 125 IU/mL, 62.5 IU/mL and 31.25 IU/mL. These spiked standards were aliquoted and frozen at −80C. Unspiked samples were also prepared as negative controls. These standards were used in both the ultracentrifugation and the HemoBind™ Whole Blood Processing protocols. In order to make a relevant and equitable comparison all the whole blood and plasma assays used in this study were designed to assay 0.5mL per reaction, as was used in our previous study.
Validation of the ultracentrifugation protocol using plasma controls
Plasma spiking standards, as described above, were ultra-centrifuged for 2 hours (4°C) at 50,377xg (Sorvall Stratos) to pellet intact viral particles, and thereby confirm that B19V DNA in the CBER standard was virion-associated and that the ultracentrifugation protocol could efficiently recover all B19V in the standard. The supernatant was removed and the pellet digested with proteinase K (20mg/mL) overnight; these samples were processed by B19V target-capture and amplified using our real-time PCR protocol. The results from the ultracentrifugation protocol were compared to the results generated using our standard assay procedure applied to corresponding spiked plasma preparations which were not subjected to ultracentrifugation and pellet extraction.
Ultracentrifugation protocol for frozen whole blood
Fresh whole blood, spiked with B19V to achieve a concentration of 100,000 IU/mL (derived from a high titer plasma donation that tested negative for B19 IgM and IgG antibodies by the Biotrin assay), was incubated at room temperature for one hour and then aliquoted and frozen at −80°C. The frozen blood was thawed and mixed with an equal volume of RBC saponin lysis solution (0.4% Saponin in 0.5% NaCl (pH 7.4) in order to completely lyse residual RBC in the already hemolyzed thawed frozen whole blood. The preparation was ultra-centrifuged at 50,377xg for 2 hours. The supernatant was transferred to a second tube, leaving the degraded RBC membranes and viral particles in a pellet in the primary tube. The pellet was subjected to protein digestion by adding 200uL of an equal part of Solution A (0.1M KCl, 0.01M Tris Base pH 8.3, 0.0025M MgCl26H2O) and Solution B (10 mM Tris, pH8.3, 2.5 mM MgCl26H2O, 1% Tween-20, 1% NP40) mixed with 1.25 uL of proteinase K (20mg/mL). Protein digestion was performed at 60°C, for 2 hours, vortexing vigorously every 15 minutes. Both pellets and supernatants were tested for B19V DNA by target-capture and real time PCR amplification.
Separation of whole blood into compartments after incubation with B19V
Plasma containing B19 virions was spiked into freshly drawn whole blood to achieve a concentration of 100,000 IU/mL followed by incubation at room temperature for 24 hours. After incubation, the whole blood was centrifuged at 833xg for 5 minutes to separate the plasma from the packed red cell (pRBC) compartment, which includes the buffy coat layer. The plasma collected at this point served as reference data for other compartments.
In a “without wash” experiment, after removal of plasma, the pRBCs were not washed and were directly subjected to RBC lysis followed by ultracentrifugation to generate a pellet composed of viral particles, RBC membranes and leukocytes/platelets. In the “wash” experiment, after removal of plasma, the remaining pRBCs with buffy coat (leukocytes/platelets) were washed twice with PBS to ensure that viral particles that were not tightly bound to cells were washed away before proceeding with RBC lysis using a Saponin RBC lysis solution. After lysis, the preparation was centrifuged at 4000xg for 5 minutes, generating a hemolyzed RBC supernatant and a pellet of leukocytes and large platelets. The supernatant, containing lysed RBC membranes and presumably bound viral particles, was collected into a tube and subsequently ultra-centrifuged. The pellet was washed again to ensure no residual lysis solution remained. The RBC supernatant and the wash solutions were ultra-centrifuged at 50,377xg (Sorvall Stratos) for 2 hours to bring down viral particles, RBC membranes, and viral particles bound to the membranes. The pellet and the supernatant gathered after ultracentrifugation were collected for testing. The RBC membrane pellets from the ultracentrifugation and the leukocyte/platelet pellets were subjected to protein digestion. B19V DNA was extracted from all compartments using target-capture and amplified using real-time PCR.
Novel RBC Lysis/Removal Protocol for Whole Blood Preparations
To address the problem of processing whole blood for detection of both plasma and cell-associated nucleic acids without use of ultracentrifugation, we developed a new method for rapid RBC lysis and removal of hemoglobin and other possible inhibitors of target capture and PCR processing (HemoBind™ Whole Blood Processing Protocol, US patent application pending; http://www.faqs.org/patents/app/20100092980). An equal volume of 6 molar guanidine-HCL-0.2M EDTA lysis buffer (pH 8.0) was added to 0.5mL of thawed frozen whole blood. The sample was vortexed and incubated at room temperature while gently mixing in a rocker. The sample was pulse spun at high speed in a microcentrifuge to pellet particulate material; all cell and plasma derived nucleic acids would be expected to remain in the guanidine lysate supernatant. One milliliter of HemoBind™ buffer was added to 1mL of the lysate supernatant, along with 20uL of proteinase K (20mg/mL). The mixture was vortexed and incubated at 60°C for 30 minutes, and then clarified of precipitates by centrifugation in a high speed microcentrifuge for one minute. The supernatant was transferred to a clean mirocentrifuge tube and heated at 100°C for 5 minutes. The preparation was microcentrifuged at high speed for one minute and the clear supernatant was transferred to the ten-tube units used in the Procleix™ target-capture system. The protocol for plasma target-capture was followed. As with the plasma protocol, the WB samples were amplified with internal controls. Results from samples with positive internal controls were deemed valid. Valid results with less or equal to cycle threshold of 40 were considered positive and those with amplifications greater than cycle threshold of 40 were considered negative.
Selection of paired plasma and whole blood samples from the RADAR repository
The National Heart, Lung, and Blood Institute (NHLBI) Retrovirus Epidemiology Donor Study Allogeneic Donor and Recipient (RADAR) repository was established to investigate possible transfusion-transmitted infections and has been described in detail elsewhere.18 Repository specimens were collected from 2000 through 2003 by blood centers and selected hospitals at seven geographically dispersed US locations. All enrolled subjects gave informed consent for frozen storage of their specimens and for subsequent testing of these specimens for possible transfusion transmissible infections. There was no restriction on how many times a repeat donor could be recruited to give a repository sample.
The donor portion of the repository consists of 13,201 donation specimens (given by 12,408 distinct donors) that were transfused to enrolled recipients and 99,906 donation specimens (contributed by 84,339 donors) from donations that were not transfused to enrolled RADAR recipients. Repository specimens consisted of two frozen 1.8 mL plasma aliquots and a 1.5 mL sample of frozen whole blood.
In previous studies, B19V DNA and antibody testing had been performed on plasma samples from 17,549 donor repository samples (5,020 from donors without enrolled recipients and 12,529 from donors with enrolled recipients).15,16 As previously reported, this testing identified 149 samples from 147 donors that tested positive for B19V DNA in plasma. For this study, we searched through the RADAR repository records to determine whether any of the donors with positive PCR results had made additional repository donations which had not yet been evaluated for B19V DNA. We found that of the 149 PCR positive donors, 43 had samples from more than one blood donation included in the repository. In total, there were 137 donations given by these 43 donors. The index samples (i.e., those previously selected for B19V analysis), the immediate previous and subsequent donations, and the first and last donation in donors with greater than 3 donations from each of these 43 donors were selected for this study; additional intervening donations were also tested in selected cases. Thus, a total of 104 donations were tested by plasma B19V PCR, whole blood B19V PCR using the Hemobind™ protocol, and B19V IgM and IgG antibodies. Quantitative results from plasma and whole blood PCR assays were obtained as two separate replicate determinations with results reported as the average of these results. Personnel performing the study testing were kept blinded as to the identities of the paired whole blood and plasma samples. For the 43 index donations, we used the plasma B19V PCR and antibody results from previous studies rather than performing these tests again due to limitations in sample volumes. We constructed a ratio of B19V DNA concentration in whole blood divided by B19V DNA concentration in plasma. Samples with positive PCR results that were below the quantitative detection limit of 20 IU/mL of B19V DNA were assigned a value of 10 IU/mL.
Donation/ donor classification
A donor was classified as having chronic persistent infection if two consecutive samples separated by 6 or more months were B19V DNA positive. A donor was classified as recently infected if a donation was IgM antibody positive or if a positive DNA result occurred in a donation given subsequent to a donation that tested B19V DNA, IgM and IgG negative. A donor was classified as a remote infection if neither of the above criteria applied and if the DNA positive donation was also IgG positive.
Statistical analysis
B19V DNA concentrations in paired whole blood and plasma samples were compared using the sign test. The median (and 95% confidence interval) ratio of B19V DNA in whole blood and plasma was determined. A non-parametric regression of B19V DNA in whole blood and plasma was performed.
Results
Recovery of B19V DNA from spiked plasma and whole blood by ultracentrifugation
Quantitative standard curve plasma controls from a previous study were used to validate an ultracentrifugation protocol designed to recover B19 viral particles from plasma and whole blood preparations. Spiked plasma controls were ultra-centrifuged and resulting pellets (which should contain all intact viral particles but not soluble DNA) and supernatant were each tested to evaluate the efficacy of viral DNA recovery. Depending on the dilution assayed, the cycle threshold of the amplification curve derived from the pellet was 6 to 16 cycles earlier than for the supernatant. Since each PCR cycle threshold (CT) difference corresponds to a 2-fold difference in starting concentration of the target nucleic acid, these ΔCT results indicate that the B19 virion-particle associated DNA recovery using the ultracentrifugation protocol was >98% (i.e., only 1/26 to 1/216 of B19V DNA remained in the supernatant), thereby establishing the ability to recover virtually all B19 viral particles from plasma samples spiked with B19V standards. Of note, the CT values observed following amplification of ultracentrifuge pellets derived from each concentration of spiked virus were similar to those of our previous study, in which the spiked plasma samples were directly subjected to the same target-capture and real time PCR assay (Table 1 and reference 16).
Table 1.
Recovery of spiked B-19V DNA spiked into plasma using the ultracentrifugation protocol
| B19V DNA spike (IU /mL) |
Pellet Cτ | Supernatant Cτ | ΔCτ | %Recovery in pellet |
|---|---|---|---|---|
| 1000 | 30.5 | 37 | 6.5 | 98.9 |
| 500 | 31.5 | 41 | 9.5 | 99.8 |
| 250 | 32.5 | 40 | 7.5 | 99.5 |
| 125 | 33 | 39 | 6 | 98.4 |
| 62.5 | 34 | 50 | 16 | 100 |
Frozen whole blood samples spiked with 100,000 IU/mL of B19 virions were then tested to determine the recovery of B19V DNA by the ultracentrifugation protocol. The CT values of real-time PCR amplifications were compared between the ultracentrifuge pellets and supernatants. In four experiments, the ΔCT between the pellet (CT = 22, 26, 24, 20) and supernatants (CT = 28, 33, 50, 30) were 6, 7, 16, and 10, respectively, indicating B19 viral/DNA recoveries of >98%, similar to the results with comparably spiked and processed plasma controls.
B19V DNA recovery from blood compartments following in vitro spiking of whole blood
We incubated fresh whole blood with high-titer B19V positive plasma from a donor in the acute pre-seroconversion phase of infection to achieve a final concentration of 100,000 IU/mL whole blood, thereby simulating a blood specimen acutely infected with B19V. After incubation for 24 hours, whole blood was processed into three compartments: plasma, RBCs and leukocytes/platelets. The B19V DNA levels were quantified in each of these compartments using sample processing, ultracentrifugation and target-capture real-time PCR protocols detailed in the methods section.
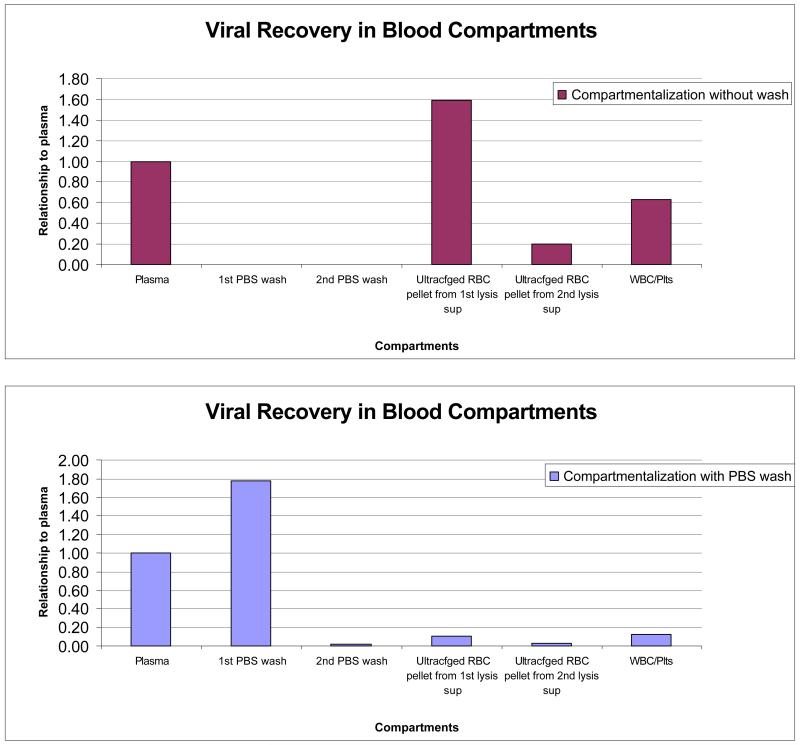
The results are presented in Figure 1. Plasma was used as the reference compartment, and hence data from plasma were plotted as 1.0 in both Figures 1A and 1B, with levels of B19V DNA derived from the other compartments plotted as ratios to the corresponding plasma data. The data in Figure 1A were generated without a wash step, and therefore do not include results for “1st PBS wash” and “2nd PBS wash”. The concentration of B19V DNA derived from the RBC and leukocyte/platelets compartments was more than twice the level derived from plasma; in other words, the plasma B19V DNA concentration was less than a third of the total B19 virus/DNA that had been spiked into whole blood.
Figure 1.
Distribution of B19V DNA in compartments of whole blood samples following incubation with B19 virus stock at 100,000 IU/mL. The relative concentrations of recovered B19V DNA are plotted on the y-axis using the concentration in the plasma sample as a reference with a designated value of 1.00. The x-axis shows the different compartments prepared from the whole blood samples. Less than a third of total B19V DNA was present in plasma.
The data in Figure 1B were generated using a slightly modified protocol incorporating washing of cellular (RBCs and leukocyte/platelets) preparations before proceeding with lysis of RBCs, pelleting, and B19V DNA extraction. In this protocol, the cells were washed twice with PBS and the amplification results of washing solution specimens were plotted as “1st PBS wash” and “2nd PBS wash”. Added together, these 2 washes yielded about twice the B19V DNA as did plasma. After washing twice with PBS, very little B19V DNA was recovered in pellets derived from the RBC and leukocyte/platelet compartments. This indicates that following in vitro spiking, approximately two-thirds of B19 virus/DNA is associated with cellular constituents and one-third with plasma, similar to the results derived with the ultracentrifugation protocol using spiked whole blood preparations, but that this binding is weak and reversible.
Validation of HemoBind ™/target-capture protocol
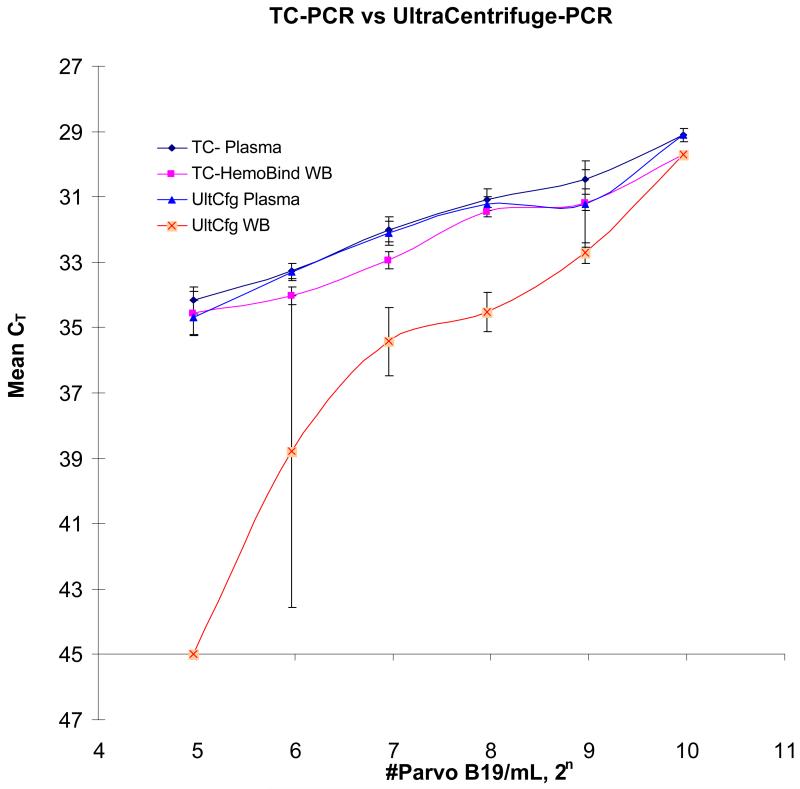
For the in-vitro spiking experiments reported above, high concentrations of virus (100,000 IU/mL) were spiked into whole blood samples to show compartmentalization of the virus. However, this input level is much higher than the concentrations of B19V that we have observed in RADAR donation samples, 15,16 and also beyond the limits of our standard curve. When the ultracentrifugation protocol was applied to whole blood spiked with the standard curve controls, the recovery at 1000IU/mL was the same for plasma and whole blood, but the recovery in whole blood samples decreased and became variable with decreasing viral input compared to the plasma samples (Figure 2). The whole blood spiked with the 62.5 IU/mL control yielded poor and erratic recovery of virus (standard deviation +/−4.8 cycles), while the whole blood spiked with 31.25 IU/mL control yielded no detectable B19V DNA following ultracentrifugation and TC-PCR. Because the ultracentrifugation protocol had low viral recovery at the low end of the whole blood standard curve, we created a new protocol using HemoBind™ to process whole blood samples to enhance the sensitivity of the assay.
Figure 2.
Viral Recovery from Ultracentrifugation and HemoBind™ Target Capture Protocols The y-axis shows mean CTs (n=4) with +/−1STD error bars of real-time PCR amplification. The B19 viral input is plotted on the x-axis in base 2 log format since PCR amplicons theoretically double in quantity per amplification cycle. The control standards are serial two fold dilutions (1000, 500, 250, 125, 62.5 and 31.25 IU/mL). TC-HemoBind™ WB refers to the protocol using HemoBind™, target-capture and real-time PCR amplification. TC-Plasma refers to the protocol using target-capture and real-time PCR amplification for plasma. UltCfg-Plasma and UltCfg-WB refer to protocols using ultracentrifugation and real-time PCR amplification.
Frozen whole blood standard controls (spiked and non-spiked) were processed using HemoBind™ to eliminate cellular debris and potential inhibitors of target capture and real-time PCR. After HemoBind™ was added into frozen whole blood and a clarified solution obtained, the samples were processed in parallel with plasma standard controls. In Figure 2, the CT of real-time PCR amplifications were compared between HemoBind™ processed whole blood samples and the corresponding plasma samples processed using the standard protocol. The slopes for the three procedures [target-capture/real-time PCR on plasma (TC-RT plasma), the HemoBind™ /target-capture real-time PCR on whole blood (HB-TC-RT WB), and the ultra-centrifuged/real-time PCR (UltCfg-RT) on plasma] were −0.99, −0.98, and −1.00, respectively, very close to the theoretical slope of −1.00. The intercept of TC-RT plasma and HB-TC-RT WB were 39.1 and 39.6, respectively. The similar slopes and intercepts indicate minimal difference between the two protocols, with differential CT’s of only ~0.5 cycles.
Comparison of B19V DNA in paired whole blood and plasma samples from blood donors
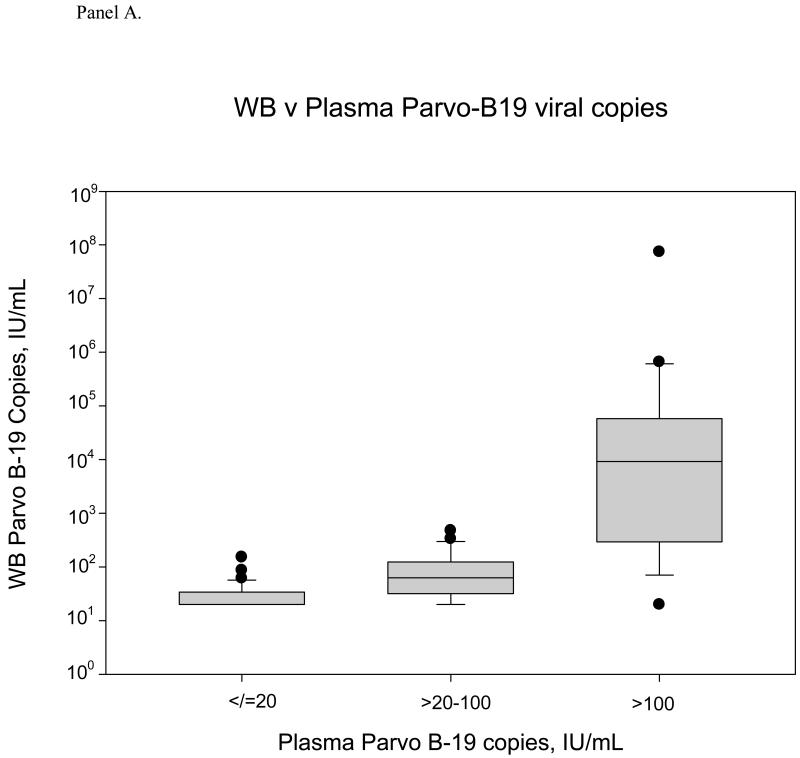
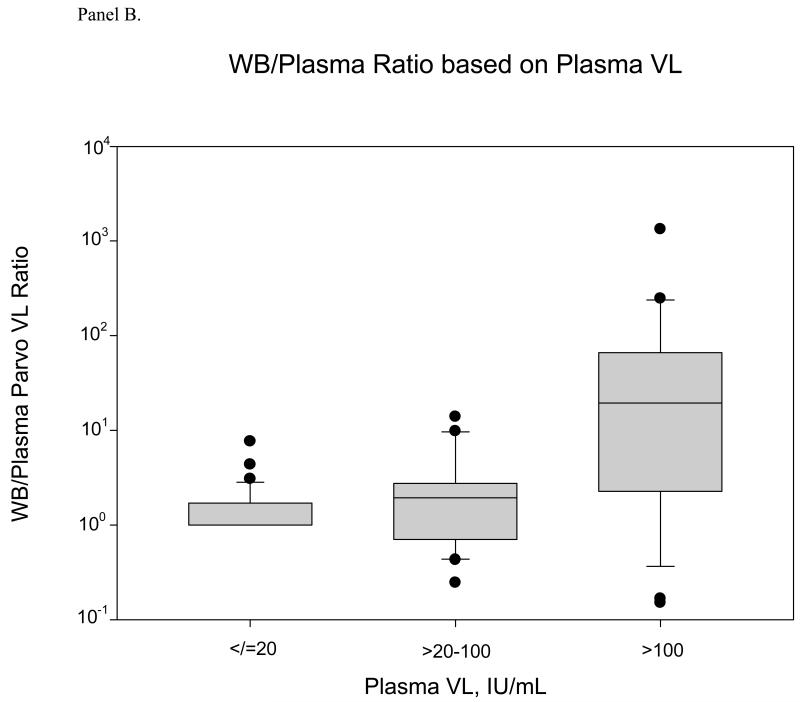
Figure 3 summarizes the whole blood relative to plasma concentrations of B19V for all 104 samples with positive B19V DNA results. Concentrations in whole blood samples tended to be higher than concentrations in plasma samples (p=0.0005, sign test). For the total sample set, whole blood samples yielded 1.9-fold higher B19V DNA concentrations than their paired plasma samples (median WB/plasma ratio 1.9, 95% CI 1.2, 4.2). This ratio varied throughout the range of plasma B19V DNA load; for plasma samples with B19V DNA concentrations >100 IU/mL, this median ratio was 19.5, whereas the ratio was only 1.9 for samples with plasma B19V DNA concentrations of >20-100 IU/mL. While the WB/plasma ratio for the complete dataset is statistically significantly greater than 1, the ratio varies in samples with much higher ratios observed among donor specimens with plasma high B19V DNA levels (p=0.01 by non-parametric regression goodness of fit test).
Figure 3.
Panel A. Box and whisker plot of Parvo B-19 viral loads in the whole blood compartment compared to corresponding plasma viral loads. When plasma VL is positive but <20 IU/mL (n=58), the mean VL in whole blood is 27.54 IU/mL (median is 20). In the mid range of VL (>20-100 IU/mL, n=24), the mean whole blood VL is 100.63 IU/mL (median: 63.2). At the high end, when the plasma viral load is >100 IU/mL (n=22), the mean whole blood VL is 3.48×106 IU/mL (median: 9199).
Panel B. WB-to-plasma VL ratio relative to plasma viral load. At the low end of the plasma viral load distribution, the WB/Plasma ratio ranges from 0-7.66, with a median of 1 and mean of 1.38. In the mid range (>20-100 IU/mL, n=24), the WB/Plasma ratio range is 0.25-13.90, with a median of 1.95 and mean of 2.83. When the plasma VL is greater than 100 IU/mL, the WB/Plasma ratio range is from 0.15-1332, with a median of 19.47 and mean of 103.52.
B19V DNA whole blood/ plasma ratios at different infection stages
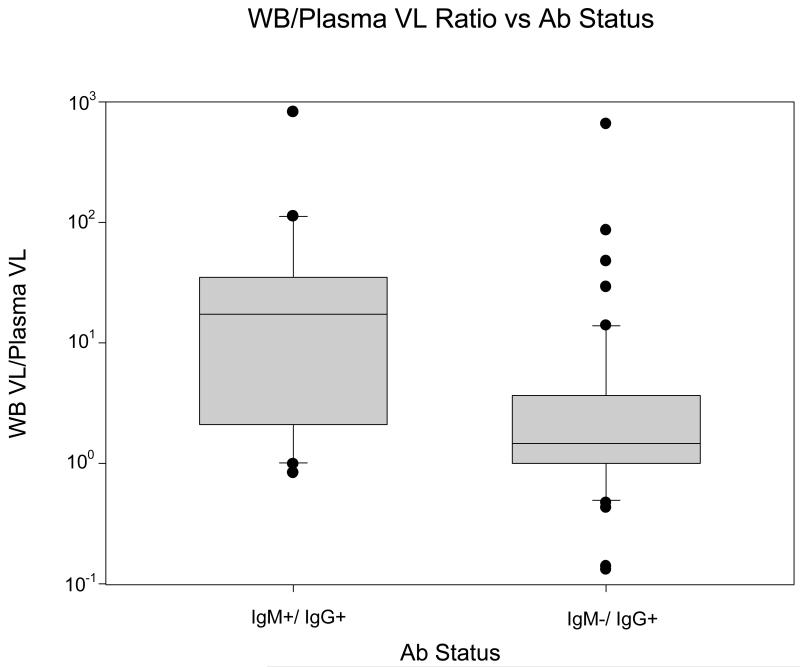
In Figure 4, B19V DNA whole blood to plasma ratios for 74 blood donations with quantitative (e.g. non-zero) whole blood and plasma DNA results were analyzed relative to the IgM and IgG antibody status of the donation, which reflects the stage of B19V infection. For 20 IgM positive, IgG positive recent infections, the mean ratio was 66.1, with a median of 17.4 and a range of 0.8 to 824.3. For 54 remote infections (IgM negative, IgG positive), this mean ratio was 15.4 with a median of 1.5 and a range of 0.06 to 657.0. The difference in whole blood/plasma B19V DNA ratios between these two stages of infection was statistically significant (p =<0.001, Mann-Whitney Rank Sum Test).
Figure 4.
Box and whisker plot of the B19V DNA ratios of whole blood samples to plasma samples based on IgM and IgG status, IgM+/IgG+ (n=20) and IgM-/IgG+ (n=54). The B19V DNA ratios are plotted on the y-axis and the serological status is plotted on the x-axis. [p>0.001, Wilcoxon Rank Sum Test].
Table 2 provides a composite analysis of B19V DNA whole blood to plasma ratios classified by both plasma DNA concentration and IgM status. The highest median ratio (29.7) was found in IgM positive donations with B19V DNA > 100 IU/mL.
Table 2.
Whole blood to plasma ratios for units with quantitative B19V DNA results > 20 IU/mL
| Plasma DNA concentration |
Whole blood/plasma DNA ratio | |||||
|---|---|---|---|---|---|---|
| IgM Pos | IgM Neg | |||||
| n | Mean | Median | n | Mean | Median | |
| >20-100 | 6 | 4.43 | 2.15 | 18 | 2.19 | 1.06 |
| >100 Total* | 14 20 | 137.3 97.4 | 29.7 13.9 | 8 26 | 44.4 15.2 | 2.64 1.82 |
data for 58 IgM donations with DNA results that were negative or < 20 IU/mL are not included
Stage of B19V infection and persistence in blood donors
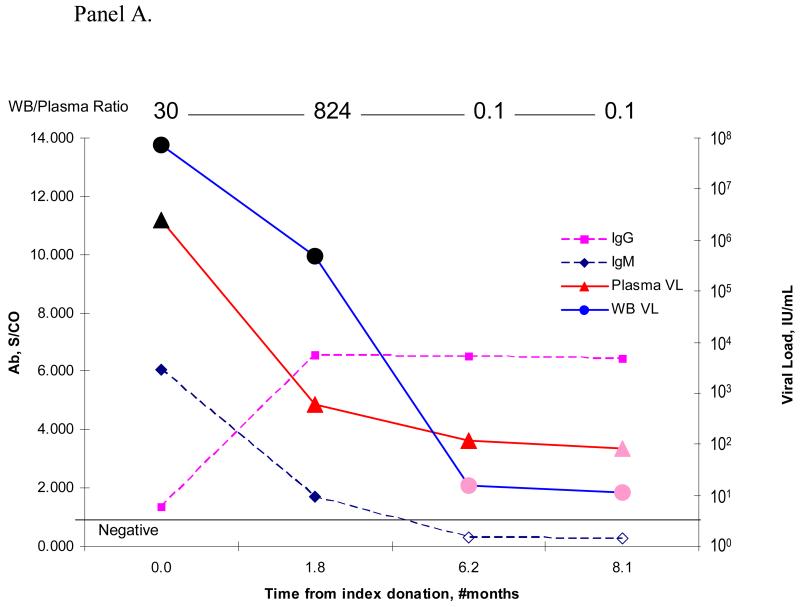
Figure 5 plots data for the donors with multiple donations from which paired whole blood and plasma samples were available in the RADAR repository. Panel A presents detailed quantitative DNA and antibody results from one representative B19 viremic blood donor with recent infection (IgM+) that evolved to chronic persistent infection. This donor had repository samples from donations made during both the early and later stages of B19V infection as evidenced by serial donation samples with decreasing IgM reactivity in parallel with increasing IgG seroreactivity. Note that the B19V DNA loads of whole blood samples at the early stage of infection were significantly higher than those of their paired plasma samples, as indicated by the whole blood/plasma B19V DNA ratios at the top of the figure. In contrast, at the later stages of infection, the whole blood and plasma B19V DNA levels were almost equal.
Figure 5.
B19V distributions in blood compartments and persistence in infected RADAR donors.
Panel A. This figure shows the B19V DNA concentration and serological status over time for a blood donor with chronic persistent B19V infection. IgM and IgG S/C ratio is plotted on the left y-axis, while the corresponding plasma and whole blood B19V DNA concentrations in IU/mL are plotted on the right y-axis. The timing of the donation events is plotted on the x-axis. The WB/Plasma ratio is indicated above the corresponding donation.
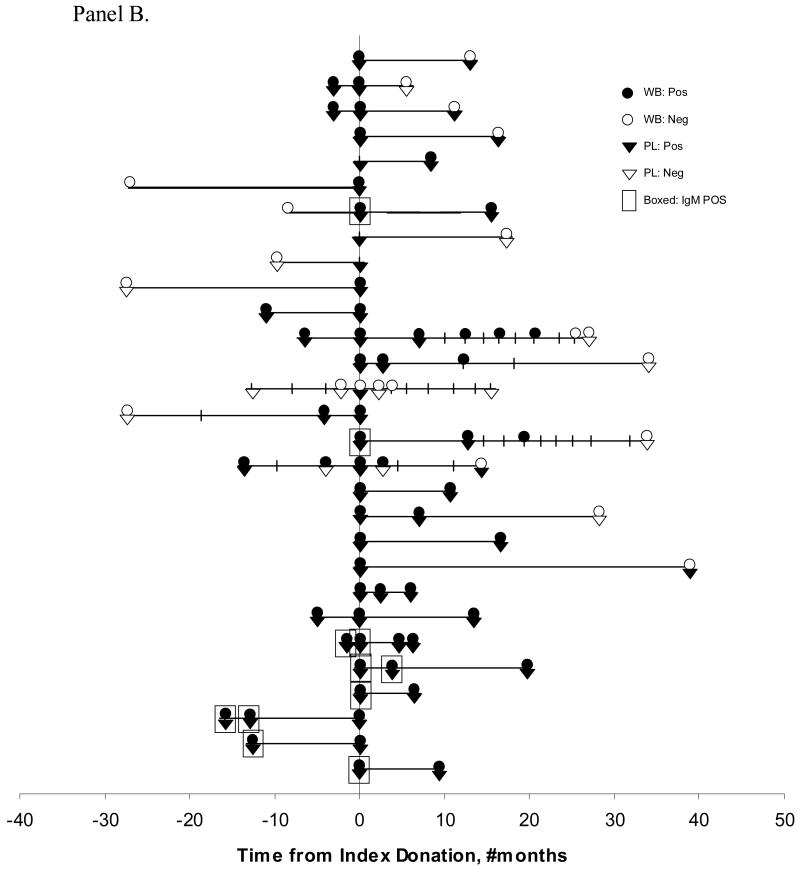
Panel B. Quantitative B19V DNA results for all tested donations from 29 donors with at least one donation at a >6 month interval. The x-axis represents time from index donation with the index donation plotted as time 0. Additional donations going backwards or forward from a positive index donation were tested by quantitative PCR, performed on both whole blood (●) and plasma samples (▼). The closed symbols denote samples with detectable B19V DNA results and the empty symbols denote negative B19V DNA results. IgM positive donations are indicated by boxed symbols.
Panel B shows results for 29 donors whose donations were separated by at least six months. Of these, 25 made at least one donation at a >6 month interval from their initial DNA positive donation. Based on plasma B19V DNA detection, 22 (88%) showed chronic persistent infection; this percentage fell to 72% (18 donors) with the whole blood assay. Five of the persistently infected donors had additional test results at >2 years at which time four donors were B19V DNA negative on both their plasma and whole blood samples. All donors with detectable IgM showed loss of the antibody over time. The 14 additional donors (not shown in Panel B), with less than 6 months of follow-up showed an even higher rate of persistence over the short time frame in which they were evaluated..
Four donors in this study were documented to have incident infection, characterized by a donation that was negative for B19V DNA and antibody followed by a sample that was DNA positive, IgG positive, and in 3 of 4 cases, IgM positive (Figure 5B, and not shown). An additional 12 donors had recent infection based on the detection of IgM antibody in the initial PCR positive donation (8 cases shown in Figure 5B; 4 cases not shown). Three donors with B19V IgG antibody but without detectable B19V DNA on their initial donation showed detectable low level B19V DNA on their follow-up donation; this may represent either intermittent low level viremia or a limitation of our assay in consistently detecting such low B19V DNA levels (Figure 5B, and not shown). Finally, we found one antibody negative donor with a low level of B19V DNA in both plasma (132 IU/mL) and whole blood (15 IU/mL); on a subsequent donation given almost 6 months later, this donor tested B19V DNA negative but did not demonstrate antibody seroconversion (case not plotted in Figure 5B).
Discussion
Because B19V is known to infect and replicate in erythroblasts in the bone marrow and because mature erythrocytes contain the B19V receptor (i.e., the P antigen) on their cell surface,19 we speculated that B19V may be within or bound to circulating erythroid cells and hence viral DNA concentrations might differ in whole blood and plasma. In addition, previous work with other viral agents had demonstrated that for some viruses that exhibit significant plasma viremia (e.g., HIV, HCV, WNV, Dengue), a substantial proportion of viral nucleic acid in blood is found to be cell-associated when appropriate whole blood based NAT is performed.[18-23] Potential mechanisms to explain this finding are the presence of intracellular viral nucleic acid due to active replication in cellular compartments (e.g. HIV DNA and RNA in CD4+ T cells) and/or binding of viral particles to cellular constituents of blood, including platelets (for HIV and Dengue)19-22 and erythrocytes (for HIV and WNV).23,24
Prior to evaluating cases of natural B19V infection, we began our study by evaluating different laboratory protocols for B19V whole blood nucleic acid testing. Whole blood NAT assays are challenging due to the fact that hemoglobin and other cell-derived constituents inhibit nucleic acid amplification.25 This applies both to PCR assays (which are inhibited by as little as 1.3 micrograms/mL of hemoglobin) and to the transcription mediated amplification assay method (TMA) commercially developed by Gen-Probe (San Diego, CA). In this latter assay, whole blood inhibition occurs both at the target capture and amplification steps.26,27 Although a 5-10-fold dilution of whole blood has been demonstrated to overcome this interference, this dilution results in a loss of assay sensitivity.26,27
In this study we evaluated whether ultracentrifugation of blood lysates could quantitatively recover viral particles and overcome these inhibitory effects. We demonstrated that our ultracentrifugation protocol was effective at recovering B19V when high concentrations of B19V were spiked into plasma or whole blood. Using this protocol, we established that less than a third of B19V spiked into whole-blood was present in plasma, with the remainder in the cellular compartments and primarily in the erythrocyte compartment. When this cellular compartment was subjected to serial low speed centrifugation and washing steps, most of the initially bound virus was present in the eluate rather than retained in the cellular preparation. We speculate that the explanation for these results is loose binding of spiked B19V to erythrocytes, either through weak binding to the P blood group receptor site or through non-specific binding. These findings are not inconsistent with development of B19 antigen agglutination assays employed in donor screening in Japan, given that those assays employ optimized conditions to maximize binding and they require >1010 B19 particles/mL of plasma for positive results.28,29
The above spiking and recovery studies were performed using a high concentration of B19V. Further experiments demonstrated that the ultracentrifugation protocol was not efficient in recovering B19V when lower concentrations of virion particles were spiked into whole blood. We speculate that the difference in the protocol efficiency at high and low viral spiking concentrations was due to the need to have sufficient concentrations of virus to create ultracentrifugation pellets that could withstand subsequent washing and recovery procedures. Use of unrelated carrier virus particles or microbeads to facilitate B19V recovery at low spiking levels could potentially overcome the sensitivity problem of the ultracentrifugation protocol (as discussed in reference 20).
As part of our ongoing nucleic acid testing research, we developed a new whole blood processing protocol using a novel buffer to precipitate and eliminate the hemoglobin and other proteins which inhibit target capture and PCR or TMA amplification. This unique buffer and associated protocol is termed “HemoBind™” (patent pending), and delivers viral nucleic acids in a supernatant in a sufficiently pure form so that there is no inhibition of either the target capture procedure or subsequent amplification steps. Through spiking experiments, we demonstrated that combining HemoBind™ with target-capture and real-time PCR amplification yielded a quantitative B19V DNA assay that was efficient and reproducible in recovering B19V DNA spiked into whole blood over a wide spectrum of spiking concentrations.
We applied the HemoBind™ target-capture real-time PCR assay to paired whole blood and plasma samples from serial donations of RADAR donors who had been shown to have detectable plasma B19V DNA. During early stages of B19V infection when IgM was present and when B19V DNA concentrations were higher, we demonstrated that the median whole blood B19V DNA concentration was approximately 30 times higher than in plasma, and in some samples the whole blood to plasma ratio was > 1000. In contrast, when IgM was absent later in infection and when B19V DNA concentrations were lower, the median whole blood to plasma ratio varied only by a factor of 1.06. Possible explanations for these results include: B19V is preferentially bound to erythrocytes when it is present in IgM immune complexes, more B19V is bound at higher plasma concentration due to steric effects on receptor mediated binding, or that B19V DNA is present at high levels within a subset of peripheral blood erythrocytes in the earlier as compared to the later stages of B19V infection. This last hypothesis is intriguing given that B19V propagates in erythroblasts and the period of high level cell-associated B19 viremia corresponds roughly to the 120 day survival period of red cells in peripheral blood, and hence RBC derived from infected erythroblasts could circulate for several months harboring B19V and account for the differential portioning of DNA during the convalescent phase of infection. Further experiments that will be needed to investigate these possible explanations would require access to fresh blood specimens from acutely infected subjects to allow purification of intact cell subsets by centrifugation and elutriation or flow cytometry-based sorting, coupled with quantitative B19V detection analyses.
Although B19V infection had been classically considered to result in an acute transient infection, recent studies have established that chronic asymptomatic as well as symptomatic persistent B19V infections occur in a proportion of infected persons. 2-7 Such infections are characterized by prolonged periods with low levels of B19V DNA in blood of IgG seropositive donors and patients. The rate of occurrence and determinants of persistent B19V infections are poorly understood. A few patients have been reported to have asymptomatic persistent B19 viremia for up to 3-5 years using very highly sensitive PCR assays. Prolonged persistence of low-level plasma viremia has also been observed in follow-up studies of healthy donors. 2-7 In addition, recent studies indicate that B19V DNA can persist in solid tissues for years or even decades following clearance of circulating viremia and seroconversion.5
In this study, using a highly sensitive B19V DNA assay, we demonstrated that persistent B19V infection occurred in 88% of evaluable blood donors who had given at least one sample >6 months after plasma B19V DNA was initially detectable. B19V DNA levels either decreased from higher levels or remained at consistently low-levels in these donors. These findings are similar to those in blood donor studies reported recently from Germany and Japan. 6-7 In addition, we found that B19V DNA persistence could also be detected by our whole blood assay, but without incremental detection relative to plasma; this is explained by the low titers of virus and absence of IgM during the persistent stage of infection.
While almost all donors demonstrated findings consistent with the known natural history of B19V infection, we did identify one donor with unusual results. B19V DNA was detectable at low levels in the absence of IgM and IgG antibody, which is contrary to the usual high viral loads (>106 IU/mL) that are detectable prior to antibody seroconversion. In addition, this donor did not seroconvert to IgG on a follow-up sample as would be expected. It is possible that this donor had a transient B19V infection (as indicated by the low DNA concentration) but for unknown reasons did not develop antibody to the VP2 antigen that is detectable by the antibody kits used in this study. We do not think the results can be explained by laboratory error since the positive index donation DNA results were obtained on both plasma and whole blood samples from this donation.
Since B19V is only rarely transmitted by blood transfusion, our finding of differential (high) whole blood to plasma concentrations in the early IgM+ stages of infection have limited if any significance for blood safety. However, we believe that the general issue of differential levels of whole blood versus plasma nucleic acid concentrations for viruses and other blood borne pathogens is important, and encourage further development of sample preparative methods to enable performance of nucleic acid testing on whole blood specimens. For transfusion-transmissible viruses with infectious window periods not currently detected by NAT screening, it is possible (though speculative) that whole blood nucleic acid testing could achieve greater sensitivity than plasma testing, and thereby offer a tool to decrease residual risk. Whole blood methods could also enable detection of persistent viral nucleic acids in convalescent or low-level persistent stages of infection (e.g., “occult HBV” and “elite controller HIV” infections), similar to our findings of more efficient detection of B19V during the convalescent IgM+ phase of infection and the reported more efficient detection of WNV in whole blood relative to plasma in asymptomatic donors.24 Also, adoption of high-throughput sample processing to allow use of whole blood in NAT assay systems would open the door to detection of cell associated viruses and of parasitic and other non-viral agents, including those that are internal to erythrocytes (e.g., malaria or babesia parasites) or that partition with leukocyte fractions (e.g., T cruzi).
Acknowledgments
We thank Hailing Yan of CBER/FDA for performing some of the anti-B19V assays, and Dr Venkatakrishna Shyamala, formerly of Chiron Corporation, for research and development on the prototype assay and for further assay validation work.
This work was supported by NHLBI contracts N01-HB-47175 and -57181 The authors report no conflict of interest.
References
- 1.Corcoran A, Doyle S. Advances in the biology, diagnosis and host-pathogen interactions of parvovirus B19. J Med Microbiol. 2004;53:459–75. doi: 10.1099/jmm.0.05485-0. [DOI] [PubMed] [Google Scholar]
- 2.Cassinotti P, Siegl G. Quantitative evidence for persistence of human parvovirus B19 DNA in an immunocompetent individual. Eur J Clin Microbiol Infect Dis. 2000;19:886–7. doi: 10.1007/s100960000384. [DOI] [PubMed] [Google Scholar]
- 3.Lefrere JJ, Servant-Delmas A, Candotti D, et al. Persistent B19 infection in immunocompetent individuals: implications for transfusion safety. Blood. 2005;106:2890–5. doi: 10.1182/blood-2005-03-1053. [DOI] [PubMed] [Google Scholar]
- 4.Musiani M, Manaresi E, Gallinella G, et al. Recurrent erythema in patients with long-term parvovirus B19 infection. Clin Infect Dis. 2005;40:e117–9. doi: 10.1086/430442. [DOI] [PubMed] [Google Scholar]
- 5.Norja P, Hokynar K, Aaltonen LM, et al. Bioportfolio: lifelong persistence of variant and prototypic erythrovirus DNA genomes in human tissue. Proc Natl Acad Sci U S A. 2006;103:7450–3. doi: 10.1073/pnas.0602259103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsukura H, Shibata S, Tani Y, et al. Persistent infection by human parvovirus B19 in qualified blood donors. Transfusion. 2008;48:1036–7. doi: 10.1111/j.1537-2995.2008.01704.x. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt M, Themann A, Drexler C, et al. Blood donor screening for parvovirus B19 in Germany and Austria. Transfusion. 2007;47:1775–82. doi: 10.1111/j.1537-2995.2007.01443.x. [DOI] [PubMed] [Google Scholar]
- 8.Brown KE, Hibbs JR, Gallinella G, et al. Resistance to parvovirus B19 infection due to lack of virus receptor (erythrocyte P antigen) N Engl J Med. 1994;330:1192–6. doi: 10.1056/NEJM199404283301704. [DOI] [PubMed] [Google Scholar]
- 9.Mortimer PP, Humphries RK, Moore JG, et al. A human parvovirus-like virus inhibits haematopoietic colony formation in vitro. Nature. 1983;302:426–9. doi: 10.1038/302426a0. [DOI] [PubMed] [Google Scholar]
- 10.Hitzler WE, Runkel S. Prevalence of human parvovirus B19 in blood donors as determined by a haemagglutination assay and verified by the polymerase chain reaction. Vox Sang. 2002;82:18–23. doi: 10.1046/j.0042-9007.2001.00124.x. [DOI] [PubMed] [Google Scholar]
- 11.Brown KE, Simmonds P. Parvoviruses and blood transfusion. Transfusion. 2007;47:1745–50. doi: 10.1111/j.1537-2995.2007.01459.x. [DOI] [PubMed] [Google Scholar]
- 12.Wu CG, Mason B, Jong J, et al. Parvovirus B19 transmission by a high-purity factor VIII concentrate. Transfusion. 2005;45:1003–10. doi: 10.1111/j.1537-2995.2005.04387.x. [DOI] [PubMed] [Google Scholar]
- 13.Brown KE, Young NS, Alving BM, Barbosa LH. Parvovirus B19: implications for transfusion medicine. Summary of a workshop. Transfusion. 2001;41:130–5. doi: 10.1046/j.1537-2995.2001.41010130.x. [DOI] [PubMed] [Google Scholar]
- 14.Yu MY, Alter HJ, Virata-Theimer ML, et al. Parvovirus B19 infection transmitted by transfusion of red blood cells confirmed by molecular analysis of linked donor and recipient samples. Transfusion. doi: 10.1111/j.1537-2995.2010.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleinman SH, Glynn SA, Lee TH, et al. A linked donor-recipient study to evaluate parvovirus B19 transmission by blood component transfusion. Blood. 2009;114:3677–83. doi: 10.1182/blood-2009-06-225706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleinman SH, Glynn SA, Lee TH, et al. Prevalence and quantitation of parvovirus B19 DNA levels in blood donors with a sensitive polymerase chain reaction screening assay. Transfusion. 2007;47:1756–64. doi: 10.1111/j.1537-2995.2007.01341.x. [DOI] [PubMed] [Google Scholar]
- 17.Saldanha J, Lelie N, Yu MW, Heath A. Establishment of the first World Health Organization International Standard for human parvovirus B19 DNA nucleic acid amplification techniques. Vox Sang. 2002;82:24–31. doi: 10.1046/j.1423-0410.2002.00132.x. [DOI] [PubMed] [Google Scholar]
- 18.Kleinman SH, Glynn SA, Higgins MJ, et al. The RADAR repository: a resource for studies of infectious agents and their transmissibility by transfusion. Transfusion. 2005;45:1073–83. doi: 10.1111/j.1537-2995.2005.00171.x. [DOI] [PubMed] [Google Scholar]
- 19.Brown KE, Anderson SM, Young NS. Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science. 1993;262:114–7. doi: 10.1126/science.8211117. [DOI] [PubMed] [Google Scholar]
- 20.Lee TH, Stromberg RR, Heitman JW, et al. Distribution of HIV type 1 (HIV-1) in blood components: detection and significance of high levels of HIV-1 associated with platelets. Transfusion. 1998;38:580–8. doi: 10.1046/j.1537-2995.1998.38698326338.x. [DOI] [PubMed] [Google Scholar]
- 21.Lee TH, Stromberg RR, Henrard D, Busch MP. Effect of platelet-associated virus on assays of HIV-1 in plasma. Science. 1993;262:1585–6. doi: 10.1126/science.8248811. [DOI] [PubMed] [Google Scholar]
- 22.Wang S, He R, Patarapotikul J, et al. Antibody-enhanced binding of dengue-2 virus to human platelets. Virology. 1995;213:254–7. doi: 10.1006/viro.1995.1567. [DOI] [PubMed] [Google Scholar]
- 23.Beck Z, Brown BK, Wieczorek L, et al. Human erythrocytes selectively bind and enrich infectious HIV-1 virions. PLoS One. 2009;4:e8297. doi: 10.1371/journal.pone.0008297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rios M, Daniel S, Chancey C, et al. West Nile virus adheres to human red blood cells in whole blood. Clin Infect Dis. 2007;45:181–6. doi: 10.1086/518850. [DOI] [PubMed] [Google Scholar]
- 25.Al-Soud WA, Radstrom P. Purification and characterization of PCR-inhibitory components in blood cells. J Clin Microbiol. 2001;39:485–93. doi: 10.1128/JCM.39.2.485-493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nugent CT, Dockter J, Bernardin F, et al. Detection of HIV-1 in alternative specimen types using the APTIMA HIV-1 RNA Qualitative Assay. J Virol Methods. 2009;159:10–4. doi: 10.1016/j.jviromet.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tobler LH, Dockter J, Stramer SL, et al. Use of the Procleix HIV-1 and HCV discriminatory assays to detect HIV and HCV RNA in whole blood. Transfusion. 2002;42:1525–7. doi: 10.1046/j.1537-2995.2002.00266.x. [DOI] [PubMed] [Google Scholar]
- 28.Sato H, Takakura F, Kojima E, et al. Screening of blood donors for human parvovirus B19. Lancet. 1995;346:1237–8. doi: 10.1016/s0140-6736(95)92950-9. [DOI] [PubMed] [Google Scholar]
- 29.Wakamatsu C, Takakura F, Kojima E, et al. Screening of blood donors for human parvovirus B19 and characterization of the results. Vox Sang. 1999;76:14–21. doi: 10.1159/000031014. [DOI] [PubMed] [Google Scholar]