Summary
M. tuberculosis generally reside in phagosomes within human macrophages that resist maturation and acidification, but exhibit significant heterogeneity. In this study we have constructed an IPTG inducible-GFP expression system in M. tuberculosis to assess the relationship between the metabolic status of M. tuberculosis and the degree of phagosomal maturation. Using these recombinant bacteria, we have found that, in human macrophages, M. tuberculosis that respond to IPTG with expression of GFP fluorescence, and hence are metabolically active, reside in non-acidified phagosomes that have not fused with Texas Red dextran prelabeled lysosomes. In contrast, M. tuberculosis that fail to express GFP in response to IPTG, and hence are metabolically inactive, reside within acidified phagosomes that have fused with Texas Red dextran labeled lysosomes. These studies demonstrate that metabolic activity of M. tuberculosis correlates strongly with phagosomal maturation and that the inducible GFP expression system is useful for assessing metabolic activity of intracellular M. tuberculosis.
Keywords: tuberculosis, CD63, LAMP, phagolysosome, immunofluorescence
Introduction
Mycobacterium tuberculosis, the etiological agent of tuberculosis, is a facultative intracellular bacterium. In human macrophages, M. tuberculosis has been shown to reside in a membrane-bound phagosomal compartment that resists fusion with lysosomes and is only mildly acidified (Armstrong & Hart, 1971, Clemens, 1996, Clemens & Horwitz, 1995, Fratti et al., 2000, Malik et al., 2000, Via, 1997, Xu et al., 1994). In previous studies, using the cryosection immunogold technique, we have found that the M. tuberculosis phagosome exhibits delayed clearance of MHC class I molecules, and relatively weak staining for lysosomal membrane glycoproteins, CD63, LAMP-1, and LAMP-2, and lysosomal acid protease, cathepsin D (Clemens, 1996, Clemens & Horwitz, 1995, Clemens & Horwitz, 1996, Clemens et al., 2000b, Clemens et al., 2000a). Studies by other investigators have also demonstrated that M. tuberculosis and other mycobacterial species reside in phagosomes that are less mature and less fusiogenic with lysosomes than phagosomes containing inert particles (Russell, 1994, Sturgill-Koszycki et al., 1994, Xu et al., 1994). These results are consistent with the hypothesis that M. tuberculosis retards the maturation of its phagosome along the endolysosomal pathway and resides in a compartment that has not matured fully to that of a phagolysosome (Clemens & Horwitz, 1995).
Although the majority of M. tuberculosis reside in immature phagosomes, M. tuberculosis phagosomes within a host cell consistently exhibit different degrees of maturation as manifest by heterogeneity in the intensity of their staining for endosomal and lysosomal markers, and in the extent to which they have fused with exogenously added tracers that traffic through the endosomal-lysosomal pathway. We hypothesize that the metabolic status of M. tuberculosis correlates with the degree of phagosomal maturation.
We describe in this study the construction of an inducible GFP expression system and the utility of such a system in reporting the metabolic status of intracellular M. tuberculosis in human macrophages. Using this reporter system, we demonstrate that metabolically active M. tuberculosis reside in less mature and less acidified phagosomes than metabolically inactive M. tuberculosis.
Results
Establishment of a two-plasmid based inducible-expression system for mycobacteria
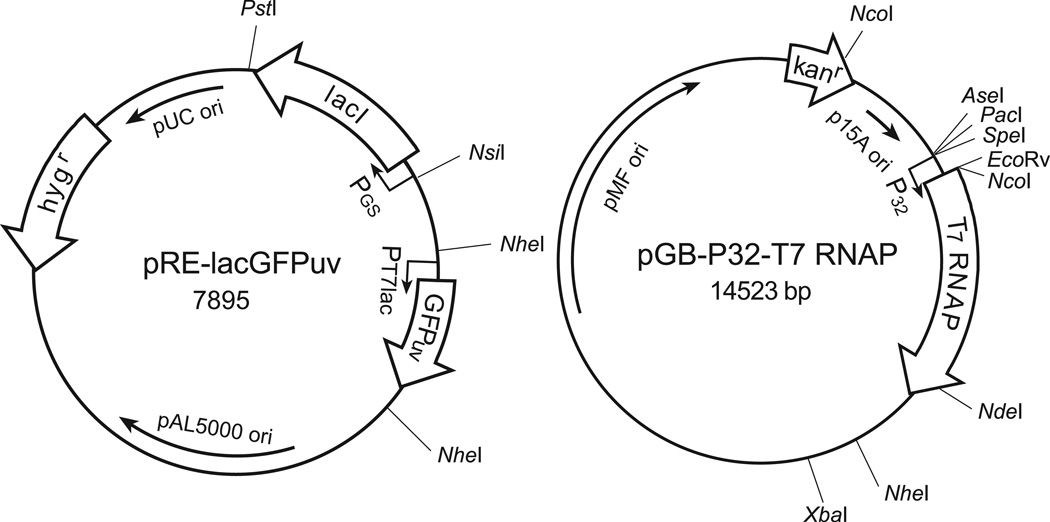
We adopted the T7lac repressor/operator system for expressing recombinant proteins in Escherichia coli to generate an inducible system with which to control gene expression in mycobacteria during extracellular growth in liquid medium as well as during intracellular growth in human macrophages. For its wide application in bioimaging, we selected the green fluorescent protein gene as a reporter gene and placed it under the transcriptional control of PT7lac, an IPTG inducible promoter. In the absence of an inducer, gfp gene expression is suppressed by the repressor LacIq. The Lac repressor is produced constitutively under the transcriptional control of the glnA1 promoter of M. tuberculosis. Both the inducible gfp and the lac repressor gene cassettes were cloned into pRE1, a pAL5000 derivative, to generate the plasmid construct pRE-lacGFP (Fig. 1). We constructed pGB-T7 RNAP by placing the T7 RNA polymerase gene under the control of the promoter for the 32-kDa mycolyl transferase gene of M. tuberculosis on pGB9.2, a pMF1 derivative (Harth et al., 2004). Addition of IPTG to the growing culture of mycobacteria carrying the two plasmids displaces the Lac repressor from the T7lac promoter region and allows the T7 RNA polymerase to initiate transcription of gfp. As a result, the bacterium becomes green fluorescent. The two plasmids, pAL5000 and pMF1, are compatible and each has been estimated to be maintained at approximately 5 and 1.3 copies per bacterium (Bachrach et al., 2000, Stover et al., 1991). To establish an expression system in mycobacteria that met the dual goals of low background in the absence of an inducer and strong green fluorescence upon induction, we placed the lac repressor and gfp gene cassettes on a pAL5000 replicon based multicopy plasmid and the T7 RNA polymerase gene cassette on a pMF1 replicon based single copy plasmid. To demonstrate the utility of such an inducible expression system, we introduced it into M. tuberculosis (Mtb-iGFP) for characterization in liquid culture and cell culture. We refer to this strain as Mtb-P32-iGFP or simply Mtb-iGFP.
Fig. 1. A Two-plasmid based inducible GFP expression system for mycobacteria.
The gene cassettes for the UV optimized gfp and the lacI repressor are located on the first plasmid construct pRE-lacGFPuv. The T7 RNA polymerase gene cassette is located on the second plasmid construct pGB-T7 RNAP. Upon IPTG induction, mycobacteria carrying both plasmids express green fluorescent protein and become green fluorescent. PGS, promoter for M. tuberculosis glnA1 gene. P32, promoter for the M. tuberculosis 32-kDa mycolyl transferase gene. hygr, hygromycin resistant gene. kanr, kanamycin resistant gene. Some of the useful restriction sites on the plasmids are shown.
In vitro growth and IPTG-induction of Mtb-iGFP
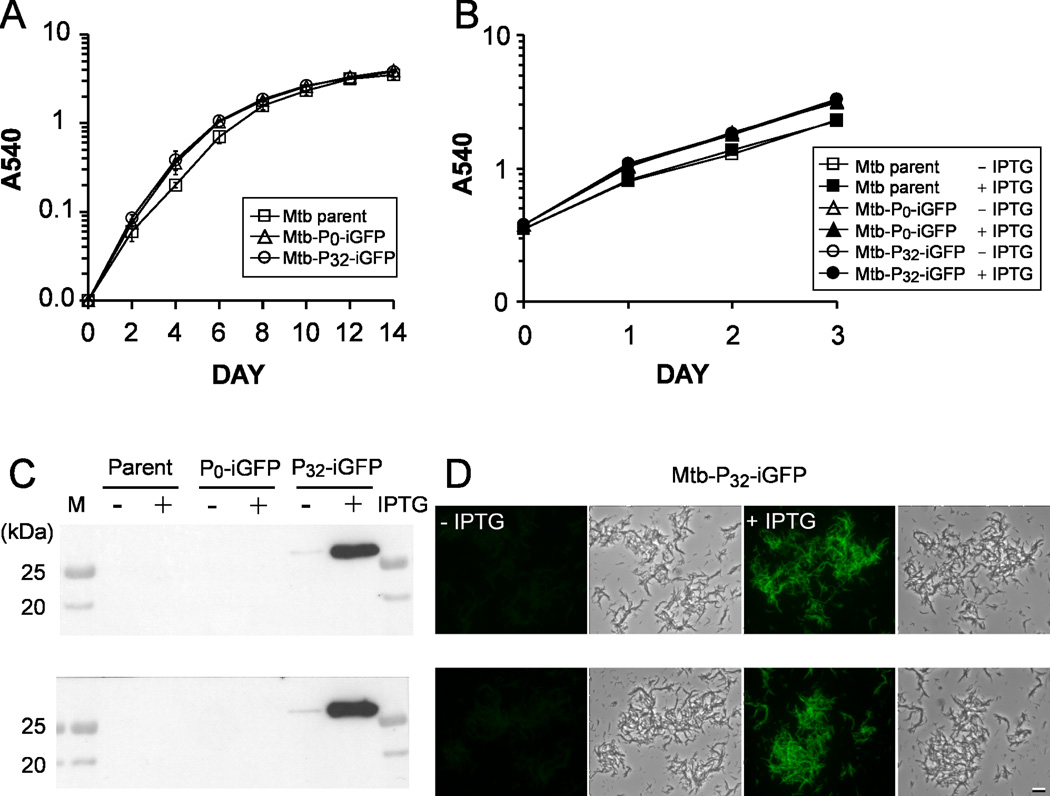
In liquid culture medium, the Mtb-iGFP strain grew at a rate that was indistinguishable from either the M. tuberculosis parent or Mtb-P0-iGFP, a control strain possessing the two plasmids but lacking a promoter for the T7 RNA polymerase gene cassette. Cultures of the three strains started at an optical density of 0.01 reached an O.D. of approximately 1.0 at 1 week and almost 4.0 at 2 weeks (Fig. 2A). Furthermore, the Mtb-iGFP strain grew at the same rate with and without IPTG induction (Fig. 2B).
Fig. 2. Growth and GFP expression of M. tuberculosis strains in broth.
A. Growth rates of M. tuberculosis strains in the absence of IPTG by monitoring optical density of the cultures over a 2-week period.
B. Growth rates of M. tuberculosis parental, Mtb-P0-iGFP and Mtb-P32-iGFP (Mtb-iGFP) strains in the presence or absence of 1 mM IPTG.
Data represent the means and standard deviations for two independent experiments. Growth curves of each strain in the presence or absence of IPTG were superimposable.
C. Assessment of GFP expression by immunoblot analysis of equal amounts of protein (20 µg) from bacterial lysates obtained from M. tuberculosis cultures incubated with or without IPTG (1 mM) for 1 day (upper) or for 3 days (lower).
D. Fluorescence microscopy examination of M. tuberculosis cultures with or without IPTG induction for 1 day (with a 1.5 second constant exposure time) (upper) or for 3 days (with a 1.0 second constant exposure time) (lower). Size bar, 10 microns.
Western blot analysis of bacterial cultures showed that GFP was detectable in Mtb-iGFP one day after IPTG induction (Fig. 2C, upper panel). The intensity of the GFP detected by Western immunoblotting from Mtb-iGFP after one day induction was more than 40 fold greater than that observed without induction as measured by densitometry and quantification using NIH image software. The expression of GFP polypeptide by Mtb-iGFP after 1 day of induction (Fig. 2C, upper panel) was almost as great as after 3 days of induction. (Fig. 2C, lower panel). Fluorescence microscopy revealed that, after IPTG induction for 1 day, Mtb-iGFP were intensely green fluorescent, whereas the un-induced bacteria were only dimly fluorescent (Fig. 2D, upper panel). After IPTG induction for 3 days, green fluorescence from individual Mtb-iGFP bacteria was even brighter while the fluorescence of un-induced bacteria remained very low (Fig. 2D, lower panel). Newly synthesized GFP polypeptides must mature to acquire fluorescence properties (Tsien, 1998). The further increase in fluorescence intensity from 1 day to 3 days with IPTG induction is likely due to an increased number of mature GFP molecules in the bacterium. Taken together, these results indicate that the growth rate of Mtb-iGFP and parental strains are similar in broth culture and expression of the inducible GFP does not impair growth. Moreover, the expression system exhibits the desired properties of a low background expression in the absence of an inducer and readily detectable target gene expression after one day induction, a time period that represents approximately one multiplication for a slow growing mycobacterium such as M. tuberculosis. We have tested the induction of GFP at three concentrations of IPTG (1, 2.5 and 5 mM) and found that the higher concentrations of IPTG did not result in GFP production during a 3-day induction period (Fig. S2). In this study, we chose to use 1 mM IPTG for induction because the intensity of green fluorescence emitted from the intracellular bacteria induced at this concentration was sufficiently bright for microscopic analysis.
Intracellular growth and IPTG induction of Mtb-iGFP
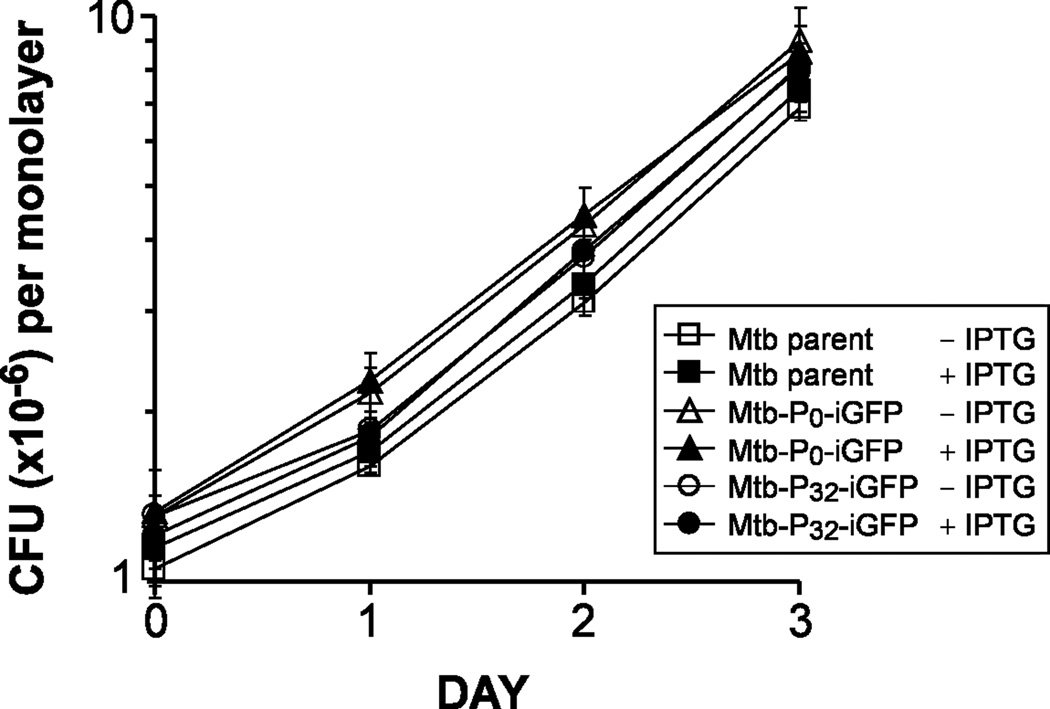
To determine whether IPTG induction of GFP expression in M. tuberculosis affects the growth of the bacteria in macrophages, we infected adherent monolayers of THP-1 cells with Mtb-iGFP for 2 h in the absence of IPTG, washed the macrophages to remove extracellular bacteria, and added fresh medium with or without 1 mM IPTG. The growth of the Mtb-iGFP in the macrophages over a 3-day period was followed by lysing the monolayers at sequential times after infection and enumerating the number of CFU in the monolayers by plating serial dilutions of the lysate on 7H11 agar plates. All strains exhibited similar growth rates in THP-1 cells in the absence or presence of IPTG induction. (Fig. 3).
Fig. 3. Growth of M. tuberculosis strains in THP-1 cells.
Growth rates of M. tuberculosis parental, Mtb-P0-iGFP and Mtb-P32-iGFP (Mtb-iGFP) strains in the presence or absence of 1 mM IPTG.
Data represent the means and standard deviations of two independent experiments.
Metabolically active M. tuberculosis do not colocalize with the lysosomal marker Texas Red dextran, whereas killed and metabolically inactive M. tuberculosis do colocalize with Texas Red dextran
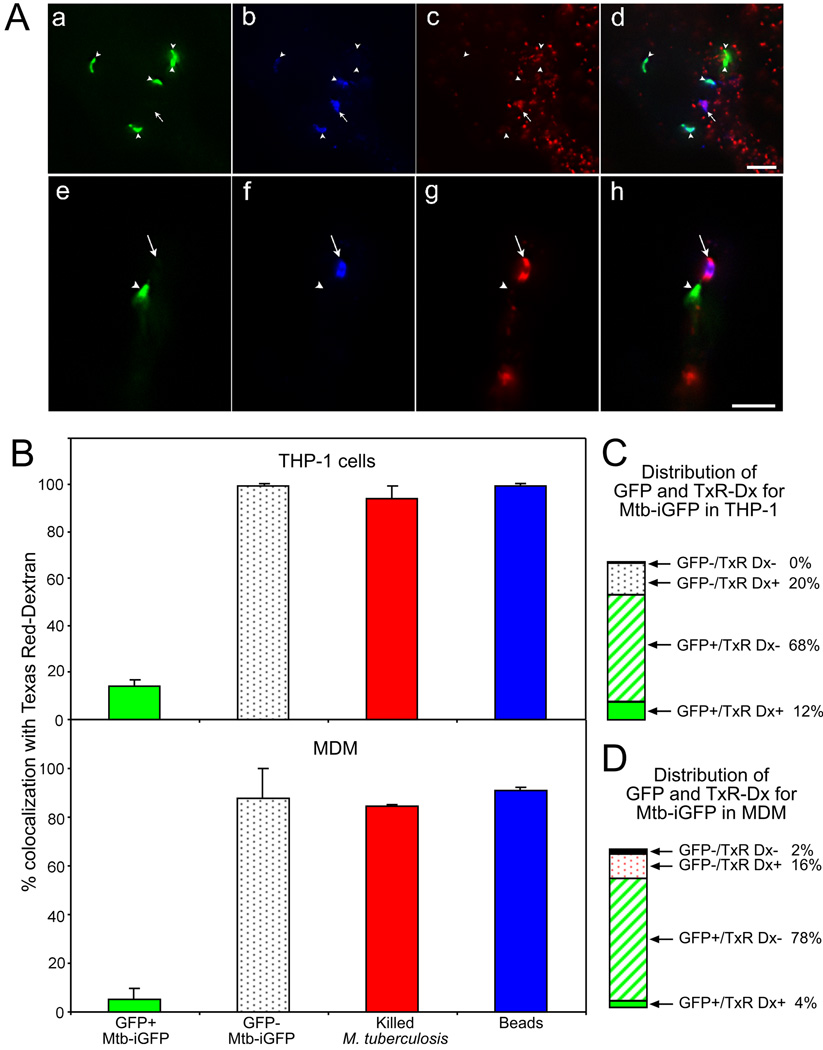
The maturational status of a phagosome correlates with its capacity to fuse with secondary lysosomes (Desjardins, 1995, Desjardins et al., 1994, Pitt et al., 1992), and phagolysosomal compartments are capable of homotypic fusion with lysosomes. M. tuberculosis phagosomes have consistently been reported to reside within less fusiogenic compartments (Armstrong & Hart, 1971, Clemens & Horwitz, 1995, Crowle et al., 1991, Goren et al., 1976, Malik et al., 2000, Malik et al., 2001, Xu et al., 1994). Nevertheless, some “live” M. tuberculosis (i.e. freshly harvested and not deliberately killed) are typically observed within fused compartments. To assess the degree to which resistance to fusion with exogenously labeled lysosomes correlates with metabolic activity, we labeled lysosomes in Mtb-iGFP infected macrophages by overnight incubation with Texas Red dextran in the presence or absence of 1mM IPTG (Fig. 4A). With the relatively low multiplicity of infection and short time course of infection (48 h) used in these experiments, we observed that the infected macrophages were well spread and did not show any evidence of cytotoxicity (Fig. S4). In the absence of IPTG, the Mtb-iGFP exhibited no green fluorescence, consistent with our observations in broth culture (Fig.2 and Figs. S1 and S3). Our original infecting inoculum contains both metabolically active and metabolically inactive bacteria. After 1 day of IPTG induction in broth culture, we observe that approximately 80% of the Mtb-iGFP exhibit green fluorescence. This percentage is in good agreement with the level of viability that we have reported previously in our infecting inoculum, as assessed by comparing bacterial CFUs on agar plates and particle counts obtained using a Petroff-Hausser chamber (Clemens & Horwitz, 1995). We observed that at 2 days after infection and IPTG induction, 80 – 90% of the Mtb-iGFP (identified by rabbit anti-LAM immunofluorescence staining) exhibited fluorescence indicative of GFP expression. Thus, approximately 10 – 20% of bacteria that stained by anti-LAM immunofluorescence were metabolically inactive and failed to express GFP in response to IPTG. We observed that whereas only 14% and 5% of the metabolically active Mtb-iGFP colocalized with Texas Red dextran in THP-1 cells and MDM, respectively, 100% and 88% of the metabolically inactive (non-GFP expressing) Mtb-iGFP colocalized with Texas Red dextran in THP-1 cells and MDM, respectively (Fig. 4B). Thus, colocalization with Texas Red dextran correlates extremely well with metabolic inactivity of the M. tuberculosis. Interestingly, a subpopulation of approximately 10% of the GFP-positive bacteria were only weakly stained by the rabbit anti-LAM antibody, and this subpopulation of GFP-expressing Mtb-iGFP did not colocalize with Texas Red dextran.
Fig. 4. Metabolically inactive Mtb-iGFP fuse with Texas Red dextran prelabelled lysosomes, whereas metabolically active Mtb-iGFP do not.
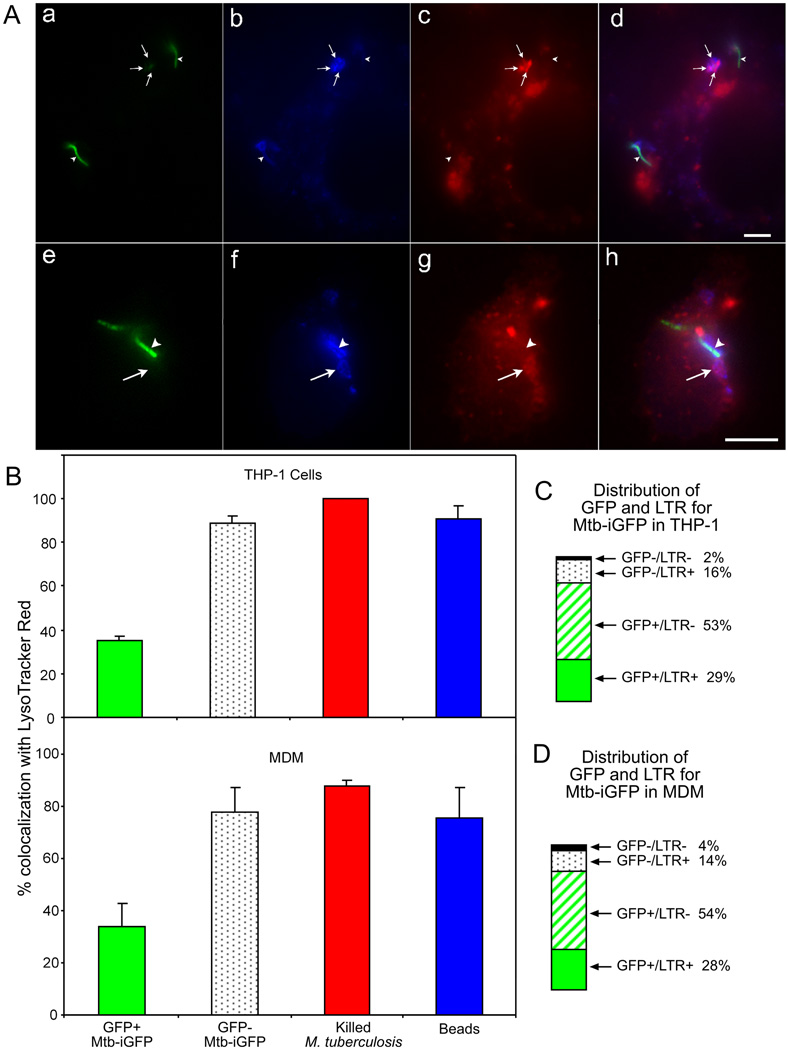
A. Epifluorescence microscopy was used to assess the extent of colocalization of Mtb-iGFP or formalin-killed M. tb.-GFP with Texas Red (TxR)-dextran in THP-1 cells (a – d) and MDM (e – h). Metabolically active Mtb-iGFP were distinguished from metabolically inactive Mtb-iGFP by their green fluorescence protein expression (a, e) and Mtb-iGFP, independent of metabolic status, were visualized by staining with amino methyl coumarin (AMC)-labeled anti-LAM antibody (b, f). Metabolically inactive Mtb-iGFP, but not metabolically active Mtb-iGFP, colocalized uniformly with TxR-dextran (c, g), Merged color images are shown on the right (d, h). Arrowheads indicate the metabolically active Mtb-iGFP and arrows indicate metabolically inactive Mtb-iGFP. Size bars are 5 microns.
B. Quantitative assessment of Texas Red dextran colocalization with metabolically active (GFP-positive) and inactive (GFP-negative) Mtb-iGFP, formalin-killed M. tb.-GFP and latex beads at 48 h post-infection and post-IPTG induction in THP-1 cells (top) and MDM (bottom). The experiment was performed twice with similar results. Values shown are means and standard deviations of duplicate determinations of at least 40 bacteria or beads.
C and D. Percentage of GFP+ and GFP− Mtb-iGFP found in Texas Red dextran positive and negative compartments in THP-1 cells (C) and MDM (D). The sum of all 4 compartments is 100%
Metabolically active M. tuberculosis associate weakly with LysoTracker red, whereas metabolically inactive M. tuberculosis colocalize strongly with LysoTracker red
It has been reported in multiple studies by several different groups that several different species of mycobacteria (including M. tuberculosis, M. bovis BCG, and M. avium) resist acidification of their phagosomes (Crowle et al., 1991, Mwandumba et al., 2004, Oh & Straubinger, 1996, Sturgill-Koszycki et al., 1994, Via et al., 1998, Xu et al., 1994). We employed the lysosomotropic fluorescent dye LysoTracker red to assess the degree to which metabolic activity of M. tuberculosis correlates with phagosomal acidification (Fig. 5A). Whereas killed M. tuberculosis and metabolically inactive Mtb-iGFP showed very high levels (80 – 100%) of colocalization with LysoTracker red, metabolically active Mtb-iGFP showed a much lower level (30%) of colocalization (Fig. 5B), indicating that metabolically active, but not metabolically inactive, M. tuberculosis resist acidification of their phagosomes.
Fig. 5. Metabolically inactive Mtb-iGFP colocalize with LysoTracker red (DND-99), whereas metabolically active Mtb-iGFP do not.
A. Epifluorescence microscopy was used to assess colocalization of Mtb-iGFP with LysoTracker red (DND-99) in THP-1 cells (a – d) and human monocyte-derived macrophages (e – h). As described above, GFP expression (green) is used to identify metabolically active bacteria (a, e); LAM immunofluorescence (blue) is used to identify the M. tuberculosis (b, f); and LysoTracker red (c, g) is used to identify acidified compartments (c, g). Merged color images are shown in panels (d) and (h). Whereas metabolically active bacteria (arrowheads) do not colocalize with LysoTracker red, the metabolically inactive bacteria (arrows) do colocalize with LysoTracker red. Size bars are 5 microns.
B. Quantitative assessment of LysoTracker red colocalization with metabolically active (GFP-positive) and inactive (GFP-negative) Mtb-iGFP, formalin-killed M. tb.-GFP and latex beads at 48 h post-infection and post-IPTG induction in THP-1 cells (top) and MDM (bottom) . Beads, killed M. tb.-GFP, and metabolically inactive Mtb-iGFP colocalize with LysoTracker red more frequently than metabolically active Mtb-iGFP (strong GFP expression). The experiment was performed twice with similar results. Values shown are means and standard deviations of duplicate determinations for at least 40 bacteria or beads.
C and D. Percentage of GFP+ and GFP− Mtb-iGFP found in LysoTracker red positive and negative compartments in THP-1 cells (C) and MDM (D). The sum of all 4 compartments is 100%.
Metabolically active and inactive M. tuberculosis reside in CD63 positive compartments at two days after infection
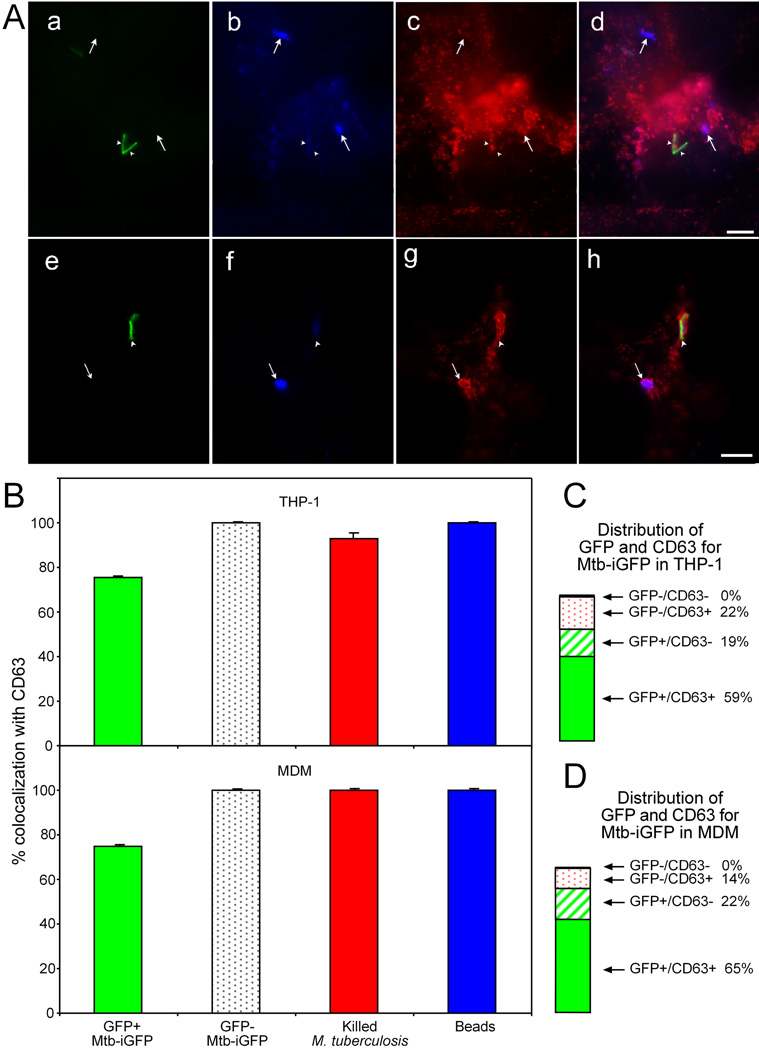
We have previously documented heterogeneity in the degree of staining of the M. tuberculosis phagosome for lysosomal membrane glycoproteins, including CD63, by quantitative cryosection immunogold electron microscopy (Clemens, 1996, Clemens & Horwitz, 1995). Thus, while some M. tuberculosis phagosomes have little or no staining for CD63, other M. tuberculosis phagosomes, even in the same cells, have much more intense staining. This is not attributable to morphological disruption of some of the phagosomes (van der Wel et al., 2007) because the heterogeneity is apparent at 48 h post-infection, a time at which we consistently observe all M. tuberculosis residing within a phagosomal compartment with a distinct membrane bilayer. In contrast to phagosomes containing live M. tuberculosis, we consistently observe more intense and more uniform CD63 immunogold staining and immunofluorescence associated with killed M. tuberculosis and latex beads. To determine if the CD63 staining correlates with metabolic activity, we examined both the magnitude of GFP-induced expression by Mtb-iGFP and the magnitude of CD63 immunofluorescence on individual bacteria in human THP-1 cells and MDM 48 h after infection (Fig. 6A). Whereas 100% of the inert particles (latex beads and killed M. tuberculosis) and the non-GFP expressing Mtb-iGFP colocalized with CD63, a significantly lower percentage (76 ± 0.8%) of the GFP-expressing Mtb-iGFP colocalized with CD63 (Fig. 6B). Moreover, the majority of Mtb-iGFP expressing GFP had less intense CD63 staining than did bacteria that failed to express GFP, although we observed occasional Mtb-iGFP bacteria expressing GFP (metabolically active) in intensely positive CD63 compartments (Fig. 6A). These results held true both for PMA-differentiated THP-1 macrophages and for MDM. As noted above, approximately 10% of the GFP-positive Mtb-iGFP were only weakly stained by the rabbit anti-LAM antibody. This subpopulation of LAM-negative GFP-positive Mtb-iGFP did not differ from the GFP-positive bacteria with respect to degree of colocalization with CD63. Thus, whereas colocalization with Texas Red dextran correlated extremely well with metabolic inactivity of the M. tuberculosis, CD63 immunofluorescence is a relatively poor indicator of the metabolic activity of the bacteria, since the majority of both live and dead M. tuberculosis reside in CD63 positive compartments.
Fig. 6. Colocalization of metabolically active and metabolically inactive Mtb-iGFP with the lysosomal membrane glycoprotein, CD63.
A. Epifluorescence microscopy was used to assess colocalization of Mtb-iGFP with CD63 in THP-1 cells (a – d) and human monocyte-derived macrophages (e – h) infected with Mtb-iGFP. Metabolic activity of Mtb-iGFP was assessed by green fluorescence protein expression (a, e). Mtb-iGFP, independent of metabolic status, were visualized by staining with AMC-labeled anti-lipoarabinomannan (LAM) antibody (b, f) and CD63 was visualized by staining with Texas red labeled anti-CD63 antibody (c, g). The merged color images are shown on the right (d, h). Metabolically inactive Mtb-iGFP (failed to express GFP after IPTG induction) consistently colocalized with CD63 labeled with Texas red, both in THP-1 cells (a–d) and MDM (e–h). The majority of metabolically active Mtb-iGFP (intense green fluorescence after IPTG induction) also colocalized with CD63. White arrowheads indicate the metabolically active bacteria. Arrows indicate the metabolically inactive bacteria. Note that the lower arrowhead in panel (a) is associated with a metabolically active bacterium with negligible CD63 immunofluorescence (lower arrowhead in panel (c)). Even some bacteria with relatively intense CD63 fluorescence rimming the phagosome (upper arrowhead, in (c) and arrowhead in (g) are associated with metabolically active (GFP-positive) bacteria (corresponding arrowheads in (a) and (e)). Size bars are 5 microns.
B. Quantitative assessment of CD63 colocalization with metabolically active (GFP-positive) and inactive (GFP-negative) Mtb-iGFP, formalin-killed M. tb.-GFP and latex beads at 48 h post-infection and post-IPTG induction in THP-1 cells (top) and MDM (bottom). The experiment was performed twice with similar results. Values shown are means and standard deviations for duplicate determinations for at least 40 bacteria or beads.
C and D. Percentage of GFP+ and GFP− Mtb-iGFP found in CD63 positive and negative compartments in THP-1 cells (C) and MDM (D). The sum of all 4 compartments is 100%.
Clumps of Mtb-iGFP exhibit metabolic activity despite residence in acidified phagolysosomes
Although we seek to minimize the presence of clumps of mycobacteria from the initial infecting inoculum, a small percentage of the bacteria are nevertheless found as clumps. We have previously reported that clumps of M. tuberculosis traffick to phagolysosomes (Clemens & Horwitz, 1995). As expected, we observe that clumps of metabolically inactive Mtb-iGFP (i.e. that fail to exhibit green fluorescence after IPTG induction) uniformly colocalize with CD63, LysoTracker red, and Texas Red dextran. In the current study, we also examined the compartment occupied by clumps of metabolically active Mtb-iGFP. We defined metabolically active clumps as tightly clustered groups of at least 8 M. tuberculosis, with at least 3/4ths of the bacteria in the clump exhibiting green fluorescence 48 h after induction with IPTG. We observed that the majority of these clumps of metabolically active Mtb-iGFP colocalized with CD63 in both THP-1 cells and MDM (86% ± 13% and 95% ± 6%, respectively) and with LysoTracker red (83% ± 4% and 90% ± 0.3%, respectively). Many of the clumps of metabolically active Mtb-iGFP in THP-1 cells and MDM also colocalized with Texas Red dextran (48% ± 3% and 75% ± 7%, respectively). These data indicate that the clumps of metabolically active M. tuberculosis traffic differently and show less resistance to fusion with lysosomes and less resistance to acidification than do individual metabolically active M. tuberculosis.
Discussion
We have previously demonstrated the practical use of two compatible plasmids for stable and simultaneous expression of multiple extracellular M. tuberculosis proteins for recombinant BCG vaccine development (Harth et al., 2004). We report in this study incorporation of the recombinant E. coli T7lac repressor/operator inducible system into the two-plasmid based mycobacteria expression system. We have generated an M. tuberculosis strain that responds to IPTG induction in human macrophages in such a way that it allows us to distinguish intracellular bacteria that are metabolically active from those that are metabolically inactive based on the intensity of bacterial green fluorescence after IPTG induction. Upon induction, new protein synthesis must occur to turn an intracellular bacterium from non-green fluorescent to green fluorescent. This phenotypic change requires a functional and efficiently coordinated transcription and translation machinery in the bacterium. Not only do transcription and translation require the active participation of hundreds of protein and RNA molecules (Szaflarski & Nierhaus, 2007), but the process also requires energy (ATP, GTP) and metabolites (nucleotides, amino acids, NADH). Thus, M. tuberculosis bacteria that do not engage in nutrient acquisition or energy production are unable to respond to IPTG induction by initiating transcription and translation. An absent or weak fluorescent signal in Mtb-iGFP can not be attributed to degradation of GFP by proteases in metabolically active bacteria because GFP is extremely stable in mycobacteria [(Blokpoel et al., 2003) and Fig. S3]. Thus, M. tuberculosis whose GFP expression is negative in the presence of the IPTG signal are bacteria that lack the metabolic capacity to respond to IPTG induction, rather than metabolically active bacteria that have subsequently degraded their GFP.
Failure of Mtb-iGFP to respond to IPTG is not attributable to failure of IPTG to access the intracellular compartment in which the Mtb-iGFP reside. IPTG has been shown to access multiple intracellular compartments. For example, Dargelos et al. have shown that IPTG can access the nucleus of muscle cells (Dargelos et al., 2002), and Dancz et al. placed lysteriolysin under the control of IPTG and demonstrated that IPTG could access Listeria monocytogenes trapped in macrophage phagosomes (Dancz et al., 2002). The data presented in our manuscript demonstrate that IPTG accesses Mtb-iGFP within LAMP+, Texas red dextran negative, non-acidified compartments. In addition, as demonstrated in our analysis of clumps of mycobacteria, IPTG accesses acidified, Texas red dextran positive compartments. Thus, failure of Mtb-iGFP to respond to IPTG is not attributable to lack of access of IPTG to the bacteria.
We observe that the majority of both metabolically active and inactive M. tuberculosis colocalize with the lysosomal membrane glycoprotein CD63, although the intensity and uniformity of staining is greater for the metabolically inactive M. tuberculosis. On the other hand, the extent of colocalization of the M. tuberculosis phagosome with exogenously added Texas Red dextran correlates well with metabolic state, with resistance to fusion with Texas Red dextran serving as a reliable marker of metabolically active M. tuberculosis. CD63 is present on both late endosomes and lysosomes and it can be present on phagosomes of varying degrees of maturation and fusiogenicity. These data demonstrate the utility of the Mtb-iGFP in probing the correlation between M. tuberculosis metabolic activity and its intracellular niche in vitro. As it is practicable to administer IPTG to animals in vivo, the Mtb-iGFP construct that we describe may prove useful for studies investigating the metabolic viability of M. tuberculosis in animal models as well as for in vitro studies.
Our data show that the metabolic status of M. tuberculosis correlates with the degree of phagosomal maturation. Maturation of a phagosome to a phagolysosome is not the result of an all or nothing fusion event, but instead occurs along a spectrum and requires multiple fusion events (Desjardins, 1995). The presence of lysosomal membrane glycoproteins on a compartment is not synonymous with full phagolysosomal maturation. Although the M. avium, M. bovis BCG, and M. tuberculosis phagosomes do acquire limited amounts of lysosome associated membrane glycoproteins (Clemens & Horwitz, 1995, Deretic et al., 1997, Russell et al., 1996), the compartments occupied by these bacteria differ markedly from fully mature phagolysosomal compartments. Our data demonstrate that metabolically active M. tuberculosis that are capable of GFP-expression at 48 h are less fusiogenic with lysosomes than are metabolically inactive M. tuberculosis that fail to express GFP in response to IPTG.
We have observed that a sub-population of GFP positive Mtb-iGFP stain weakly by anti-LAM antibody and that these Mtb-iGFP reside in Texas Red dextran negative, LysoTracker red negative compartments. This raises the concern that there might also be a population of Mtb-iGFP that are LAM−/GFP− and thus completely undetectable by immunofluorescence in these experiments. However, since we observe an inverse correlation between GFP fluorescence intensity and LAM fluorescence intensity, this would be contrary to the pattern that we observe. We believe that the inverse relationship between the intensity of the GFP and LAM signals may be due to a change in LAM molecular structure that accompanies intracellular growth of the bacteria within macrophages. While it is conceivable that some dead bacteria may be degraded to the point that they have lost LAM immunoreactivity, in our immunogold electron microscopy studies (Clemens, 1996, Clemens & Horwitz, 1995, Clemens & Horwitz, 1996, Clemens et al., 2000b, Clemens et al., 2000a), we have not observed any morphologically discernible M. tuberculosis that are not stained by the rabbit polyclonal anti-LAM antibody used in this study. Indeed, the bacteria that are the most severely disrupted morphologically typically have the strongest LAM immunogold staining, in agreement with the inverse correlation that we observe between GFP and LAM fluorescence intensity by fluorescence microscopy in this study.
It has been reported that M. tuberculosis is able to disrupt its phagosome and to translocate into the host cell cytoplasm (McDonough et al., 1993, Myrvik et al., 1984, van der Wel et al., 2007). We have examined our M. tuberculosis infected macrophages for this phenomenon and have observed that more than 90% of the mycobacteria reside within phagosomes with readily discernible membrane bilayers in the first 3 days post-infection. However, we do observe loss of phagosomal membrane by approximately 25% of the M. tuberculosis by 5 days post-infection. Furthermore, we have also demonstrated by microinjection studies, that M. tuberculosis is not accessible to the cytoplasm in the first 2 days post-infection (Clemens et al., 2002). Thus, the heterogeneity in phagosomal maturation during the first 2 – 3 days post-infection is not attributable to escape of M. tuberculosis into the cytoplasm.
The heterogeneity in the intensity of immunogold staining for endosomal and lysosomal markers that we consistently observed in our prior studies can be explained, in part, by differences in the metabolic status of the bacteria in the population studied. We previously suspected that clumps of mycobacteria were dead based on the fact that they usually stained intensely for CD63 and Texas Red dextran. However, in the present studies, we observe clumps of Mtb-iGFP expressing intense GFP fluorescence despite residence in compartments rich in CD63 and Texas Red dextran, indicating that clumps of M. tuberculosis can retain metabolic viability despite residence within phagolysosomal-like compartments. Our observation in the present study is at 48 h after infection. It is possible that with additional time after infection, these clumps of bacteria may either become metabolically inactive, i.e. lose their capacity to express GFP in response to IPTG, or lose their colocalization with lysosomal markers.
Our studies have shown a correlation between M. tuberculosis metabolic activity and phagosomal phenotype. Our experiments have not addressed the issue of causality, i.e. whether metabolic inactivity was the cause of accelerated phagosomal maturation or whether residence in a phagolysosome causes the M. tuberculosis to become metabolically inactive. The observation that killed M. tuberculosis reside in acidified compartments that fuse with Texas Red dextran labeled lysosomes suggests that metabolic activity is essential for retarding phagosomal maturation. However, additional studies, such as following individual bacteria over time in live imaging studies, are needed to determine whether metabolic inactivity precedes or follows residence in a phagosolysosome.
Experimental Procedures
Bacterial strains and growth conditions
M. tuberculosis Erdman strain (35801; American Type Culture Collection, VA) was used for introducing plasmid constructs made in this study. M. tuberculosis strains were grown on Middlebrook 7H11 agar at 37°C, 5% CO2-95% air atmosphere. Bacterial strains were stored at −80°C in Middlebrook 7H9 medium with 10% OADC enrichment and 20% glycerol. For broth culture, M. tuberculosis strains were grown in 7H9 medium containing 2% glucose and 0.01% Tyloxapol (Sigma-Aldrich, St. Louis, MO) at 37°C, 5% CO2-95% air atmosphere. E. coli DH5á used for molecular cloning was grown in Luria-Bertani (LB) medium at 37°C with rotation at 250 rpm. Antibiotics were used at the following concentrations: carbenicillin, 100 µg ml−1; kanamycin, 10 µg ml−1; hygromycin, 250 µg ml−1 (for E. coli) and 50 µg ml−1 (for M. tuberculosis). Prior to use of bacteria for infection of human macrophages, bacterial aggregates were dispersed by sonication of the bacteria in a water bath sonicator (Astrason Scientific) for 8 periods of 15 sec, with cooling of the suspension in an ice bath for 5 sec between sonications. Residual aggregates were removed by centrifugation at 200 g for 10 min at 4°C. The pellet of aggregated bacteria was discarded and the supernate suspension centrifuged again under the same conditions, and the process repeated a total of three times.
Construction of an inducible expression system for mycobacteria
The inducible expression system is composed of two plasmid constructs, pRE-lacGFP and pGB-T7 RNA polymerase. The plasmid pRE-lacGFP was constructed by placing two gene cassettes, PT7-lacGFPuv and PGS-lacI, into pRE1, an E. coli-mycobacteria shuttle plasmid modified from pNBV1 (Howard et al., 1995). The first gene cassette, PT7-lacGFPuv, was constructed by PCR amplification of the UV-optimized gfp coding sequence from plasmid pGFPuv and cloned into pET15b, immediate downstream of PT7lac, in between NdeI and BamHI restriction sites to generate the plasmid, pET15b-GFPuv. Primer pET15-148F and primer pET15-561R were used next to amplify the gene cassette, PT7-lacGFPuv, by PCR from the plasmid pET15b-GFPuv. The 1-kb PCR product was digested with NheI and ligated into the pRE1 vector digested with the same enzyme to generate an intermediate plasmid pRE-GFP. For the second gene cassette, PGS-lacI, primer lacI-F and primer lacImyc-R were used to amplify the coding sequence for lacIq from pET15b. The 1.1-kb PCR product was treated with PstI and cloned into pZErO-2 in between EcoRv and PstI. A 325 bp promoter region for the glutamine synthetase (glnA1) gene of M. tuberculosis was amplified from pNBV1-MtbGS with the primer pair PGS-F1 and PGS-R. The PCR product (PGS) was cloned into pZErO-lacI, between XbaI and NdeI restriction sites, preceding the lacI gene to generate the intermediate plasmid construct pZErO-PGSlacI. Primer PGS-F2 and primer PGS-R were then used to amplify the entire PGS-lacI cassette from pZErO-PGSlacI and cloned into the NsiI and PstI sites on pRE-GFPuv to generate the plasmid pRE-lacGFPuv.
The second plasmid of the inducible system, pGB-T7 RNA polymerase was constructed by amplifying the promoter region for the 32-kDa mycolyl transferase gene of M. tuberculosis using primer P32-F and P32-R. The 0.4-kb PCR product was cloned into pGB9.2 between SpeI and EcoRv sites to generate the intermediate plasmid, pGB-P32. Primer mycT7 pol-F and primer T7 pol-R were used to amplify T7 RNA polymerase from pAR1173. The 2.7-kb PCR product was digested with EcoRv and AseI and ligated with pGB-P32 digested with EcoRv and NdeI to generate pGB-T7 RNA polymerase (pGB-T7 RNAP).
Generation of the inducible GFP strain of M. tuberculosis
Plasmid constructs pRE-lacGFP and pGB-T7 RNA polymerase were introduced into M. tuberculosis by two separate electroporations. In brief, M. tuberculosis was electroporated with plasmid pRE-lacGFP and selected on 7H11 plates with hygromycin. Once the pRE-lacGFP harboring M. tuberculosis strain was established, it was electroporated with the second plasmid pGB-T7 RNA polymerase and selected on 7H11 plates with hygromycin and kanamycin to generate the M. tuberculosis-iGFP strain.
For electroporation, M. tuberculosis in 10% glycerol (100 µl) was mixed with plasmid DNA (1 µg) and pulsed electrically in a 0.1-cm cuvette (Bio-Rad, Hercules, CA) at 1.25 kV, 25 µF, 1000 Ω. The bacteria were transferred to a tube with 0.9 ml 7H9 medium containing 10% OADC enrichment and incubated at 37°C, 5% CO2-95% air atmosphere overnight before being plated on selective media.
Preparation of formalin-killed GFP-M. tuberculosis
GFP-expressing M. tuberculosis prepared as previously described (Clemens et al., 2002) were plated on 7H11 agar plates containing hygromycin, incubated at 37°C, 5% CO2-95% air, for 10 – 14 days, scraped into Dulbecco’s phosphate buffered saline (PBS, Irvine Scientific Co.), and aggregates removed by water bath sonication and low speed centrifugation as described above. Paraformaldehyde was added to the bacterial suspension to a final concentration of 4% paraformaldehyde. After 30 min at room temperature, the formalin-killed bacteria were washed three times in PBS by centrifugation at 10,000 g for 10 min. Because washing by centrifugation induces aggregation, bacterial aggregates were removed by centrifugation at 200 g for 10 min immediately prior to use of the formalin-killed bacteria in an infection experiment.
Evaluation of growth rates of recombinant M. tuberculosis in 7H9 medium
We inoculated strains of M. tuberculosis into 7H9 medium containing 2% glucose and 0.01% Tyloxapol at an optical density (540 nm) of 0.01. We maintained the cultures at 37°C, 5% CO2-95% air atmosphere and measured the optical density at 540 nm of 1 ml aliquots of the culture every other day for a period of two weeks using a spectrophotometer (Ultraspec III, Amersham Pharmacia, Piscataway, NJ). To evaluate the impact of IPTG on the growth rate of parental and recombinant strains of M. tuberculosis, we initiated the bacterial cultures as described above, and when the optical density reached a value of approximately 0.4, we subdivided the cultures into equal aliquots, continued the incubation with or without 1 mM IPTG, and measured the optical density of the cultures daily for the next 3 days.
Analysis of GFP expression in M. tuberculosis cultures with or without IPTG induction
M. tuberculosis cultures in 7H9 medium containing 2% glucose and 0.01% Tyloxapol with or without IPTG were pelleted by centrifugation at 3,500 g, 4°C for 30 min, resuspended in 1 ml of PBS with protease inhibitors (1:1000 dilution, Novagen, La Jolla, CA) and sonicated on ice with a probe tip sonicator (model W-375; Heat Systems Ultrasonics, Plainview, N.Y.) at 50% duty cycle for a duration of 1 min three times with a 3 min rest on ice between sonications to lyse the bacteria. Sonication was done inside a sealed container within a biosafety cabinet. Sonicated samples were centrifuged at 10,000 g for 10 min in an aerosol-tight microcentrifuge within a biohazard hood and the resulting supernate was passed through an Acrodisc syringe filter with 0.8/0.2 µm Supor membrane (Pall Corporation, Ann Arbor, MI). Protein concentration of the filtered samples were determined by bicinchonic acid method (Pierce, Rockford, IL) according to the manufacturer’s instructions.
The bacterial protein samples were separated by SDS-polyacrylamide gel electrophoresis and electrophoretically transferred onto a nitrocellulose membrane. For detection of green fluorescent protein (GFP), the membrane blot was incubated with a rabbit anti-GFP antibody (Assay Design, Ann Arbor, MI) at a dilution of 1:10,000 followed by a peroxidase conjugated goat anti-rabbit antibody (Sigma). The membrane was developed with SuperSignal West Pico chemiluminescent substrate (Pierce) and the signal from immunoreactive protein bands detected by exposing the membrane to X-ray film.
For detection of bacterial green fluorescence, M. tuberculosis bacteria grown in culture medium with or without IPTG induction for 1 to 3 days were harvested by centrifugation in an aerosol-tight microcentrifuge as described above. Bacterial pellets were resuspended in 3.7% formaldehyde and epifluorescence examined with an Eclipse TE2000-S microscope equipped with an X-Cite 120 light source (Nikon) and images acquired with a SPOT RT-KE monochrome camera and SPOT software (Diagnostic Instruments, Sterling Heights, MI).
THP-1 cells
The human monocytic cell line, THP-1 (ATCC, TIB 202), was grown in RPMI-1640 (Mediatech, Herndon, VA) supplemented with 2 mM glutamine, 10% heat-inactivated fetal bovine serum (HI-FBS), and penicillin-streptomycin (1000 IU ml−1 and 100 µg ml−1, respectively). Prior to use in an infection experiment, the THP-1 cells were added to glass coverslips in 2 cm2 tissue culture wells (2 × 105 cells/cm2) and differentiated with phorbol 12-myristate 13-acetate (PMA, 100 nM) in RPMI-1640 with 10% heat-inactivated fetal bovine serum for 3 days at 37°C in air containing 5% CO2.
Human peripheral blood mononuclear cells
Heparinized blood from normal blood donors was diluted 1:1 with 0.9% saline, and the mononuclear cell fraction obtained by centrifugation at 800 g for 30 min at 24°C over a Ficoll-sodium diatrizoate solution (Ficoll-Paque, Pharmacia Fine Chemicals, Inc.). The layer containing the mononuclear cell fraction was removed, diluted 1:1 with RPMI-1640, and the mononuclear cells collected by centrifugation at 400 g for 10 min at 4°C. The mononuclear cells were washed twice by centrifugation at 115 g for 10 min at 4°C, resuspended in RPMI-1640, counted in a hemocytometer (Clay Adams Div., Becton Dickinson and Co.), and added to glass coverslips in 2 cm2 tissue culture wells (1.5 × 106 cells/coverslip in 0.5 ml of culture medium) in RPMI-1640 containing 15% autologous serum. Cells were allowed to adhere to the coverslips for 90 min at 37°C, 5% CO2, washed twice with RPMI-1640, and incubated for 2 days in fresh culture medium with 15% autologous serum prior to use in an infection experiment. The participation of normal human blood donors in our research was approved by the UCLA Institutional Review Board.
Intracellular growth of recombinant M. tuberculosis in THP-1 cells
PMA differentiated THP-1 cells were infected with M. tuberculosis-iGFP strains at the ratio of 24 bacteria per macrophage in RPMI containing 20% human serum type AB for 2 h at 37°C, 5% CO2. Extracellular bacteria were removed by washing extensively and fresh medium with or without IPTG (1 mM) was added to the monolayer. Under these conditions, 10 – 20% of the infecting inocula were internalized, yielding an average of 2–4 bacteria per macrophage. Infected monolayers were incubated for 2 h to 3 days, lysed with 0.1% SDS, serially diluted in 7H9 medium with 10% OADC enrichment and 0.05% Tween 80, plated on 7H11 agar, and numbers of colony forming units (CFU) enumerated after a 2-week incubation.
Evaluation of GFP-expression and maturational state of M. tuberculosis phagosomes in macrophages by fluorescence microscopy
Monolayers of differentiated THP-1 cells or human monocytes on coverslips were co-incubated for 90 min with M. tuberculosis-iGFP or with formalin-killed GFP-M. tuberculosis and fluorescent blue latex beads (1 µ diameter, Polysciences, at a 4000-fold dilution of a stock suspension of 10% solids) in RPMI-1640 containing 10% fresh AB-serum (THP-1 cells) or autologous serum (monocytes) as described above. Monolayers were washed with culture medium, incubated in fresh medium with or without 1 mM IPTG at 37°C for 48 hours, fixed in 4% paraformaldehyde in 0.075 M sodium phosphate buffer, pH 7.4, for 30 min at room temperature, permeablized in 0.1% saponin in PBS containing 10 mM glycine, and incubated with 5% goat serum in PBS with 1% BSA to block non-specific staining. Coverslips were stained with mouse antibody to human CD63 (5 µg ml−1 IgG, Becton Dickinson) diluted in the same buffer overnight at 4°C. Coverslips were washed with PBS and incubated with Texas red-conjugated goat anti-mouse IgG, diluted 1:50 in 5% goat serum, 1% BSA in PBS) for 90 min at room temperature. The coverslips were washed in PBS, incubated with in rabbit antibody to lipoarabinomannan (1:1000 dilution), washed in PBS, and stained with amino methyl coumarin (AMC)-conjugated goat-anti-rabbit IgG (Sigma Chemical Company, 1:50 dilution), post-fixed in 2% paraformaldehyde in PBS, incubated with 2.5 µM DAPI in PBS for 15 min at room temperature to label host nuclei, washed in PBS, blotted, mounted with Prolong Antifade mounting medium and viewed by epifluorescence microscopy as described above or by confocal scanning microscopy with a Leica TCS-SP confocal and 2-photon microscope and Leica confocal software. Texas red, Oregon green, and DAPI fluorochromes were excited with confocal krypton and argon lasers and 2-photon scanning with a short-pulsed titanium:sapphire laser, respectively.
Labelling of lysosomal compartments with Texas Red dextran
Lysosomal compartments were labeled by incubating the infected monolayers overnight in culture medium containing 50 µg ml−1 lysine-fixable Texas red-conjugated dextran, 70,000 kDa (Molecular Probes, Eugene, OR) prior to fixation, permeabilization, and staining with rabbit anti-LAM and AMC-conjugated goat anti-rabbit as described above.
Labelling of acidified compartments with LysoTracker red DND-99
Acidifed compartments were identified by incubating infected monolayers with 50 nM LysoTracker red DND-99 (Lepperdinger et al., 1998, Wubbolts et al., 1996) in RPMI containing 10% serum for 2 h at 37°C prior to fixation, permeabilization, and immunostaining for LAM as described above.
Supplementary Material
Table 1.
Strains, plasmids and oligonucleotide primers
| Strain, plasmid, or primers |
Description | Source or reference |
|---|---|---|
| M. tuberculosis | ||
| Erdman | Parental strain | ATCC |
| Mtb-P0-iGFP | Erdman strain carrying pRE-lacGFPuv and pGB-P0-T7 RNAP | This work |
| Mtb-P32-iGFP | Erdman strain carrying pRE-lacGFPuv and pGB-P32-T7 RNAP | This work |
| E. coli | ||
| DH5α | Plasmid construction and cloning | BRL |
| Plasmidsa | ||
| pET15b | Ampr, source of lacIq repressor and T7lac promoter | Novagen |
| pGFPuv | Ampr, source of gfp | Clontech |
| pAR1173 | Ampr, Tetr, source of T7 RNA polymerase | ATCC |
| pNBV1-MtbGS | Hygr, source of glnA1 promoter | (Tullius et al., 2003) |
| pZErO-2 | Kanr, cloning vector for PCR products | Invitrogen |
| pRE1 | Hygr, a derivate of the E. coli-mycobacteria shuttle plasmid pNBV1 | Tullius and Horwitz unpublished work |
| pGB9.2 | Kanr, a low-copy-number E. coli-mycobacteria shuttle plasmid | (Harth et al., 2004) |
| pET15-GFPuv | Ampr, UV-optimized gfp in pET15b | This work |
| pRE-lacGFPuv | Hygr, lacI and gfp gene cassettes in pRE1 | This work |
| pGB-P0-T7 RNAP | Kanr, a promoterless T7 RNA polymerase gene cassette in pGB9.2 | This work |
| pGB-P32-T7 RNAP | Kanr, pGB9.2 with T7 RNA polymerase cassette | This work |
| Primersb | ||
| pET15-148F | 5’-AGCTTTGGCTAGCAGGCTTGGTTATGCCGGTACTG-3’ | |
| pET15-561R | 5’-GACTAGTGCTAGCGCCAGCAACCGCACCTGTGGC-3’ | |
| GFP-F | 5’-GGAATTCTCATGAAGTAAAGGAGAAGAACTTTTCACTGGA-3’ | |
| GFP-R | 5’-CGGGATCCTCATTTGTAGAGCTCATCCATGCCATGTG-3’ | |
| lacI-F | 5’-GGAATTCCATATGAAGCCGGTCACGTTGTACGATGTCGCA-3’ | |
| lacImyc-R | 5’-GATCCTGCAGTCACATGGCGCTGTTCAGGTCCTCTTCGGAG ATGAGCTTCTGCCCGCTTTCCAGTCGGGAAAC-3’ | |
| PGS-F1 | 5’-GGATCTAGAGCGATCAGCCAGTCGATCAGCAGAGCC-3’ | |
| PGS-F2 | 5’-GGAATGCATGCGATCAGCCAGTCGATCAGCAGAGCC-3’ | |
| PGS-R | 5’-GGAATTCCATATGAATGCTCCTTTACTGTTCCGCGG-3’ | |
| mycT7 pol-F | 5’-ACCGATATCCATGGCCTCGATGCAGAAGCTGATCTCGGAGG AGGACCTGAACACGATTAACATCGCTAAGAACGAATTC-3’ | |
| T7 pol-R | 5’-TCCGATTAATGGATCCTTACGCGAACGCGAAGTCCGACTCT AAG-3’ | |
| P32-F | 5’-GACTAGTGATCACCCTGCACCGATTCCTCTC-3’ | |
| P32-R | 5’-ACCGATATCTTCCCTCATCCTCATCTTCAACGCATCCATGC-3’ | |
Ampr, Hygr, Kanr, and Tetr indicate resistance to ampicillin, hygromycin, kanamycin and tetracycline, respectively.
Underlined nucleotides indicate restriction enzyme recognition sequence on the primer that was used in cloning.
Acknowledgements
This work was supported by grant HL077000 from the National Institutes of Health.
References
- Armstrong JA, Hart PD. Response of cultured macrophages to Mycobacterium tuberculosis with observations on fusion of lysosomes with phagosomes. J. Exp. Med. 1971;134:713–740. doi: 10.1084/jem.134.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrach G, Colston MJ, Bercovier H, Bar-Nir D, Anderson C, Papavinasasundaram KG. A new single-copy mycobacterial plasmid, pMF1, from Mycobacterium fortuitum which is compatible with the pAL5000 replicon. Microbiology. 2000;146:297–303. doi: 10.1099/00221287-146-2-297. [DOI] [PubMed] [Google Scholar]
- Blokpoel MC, O'Toole R, Smeulders MJ, Williams HD. Development and application of unstable GFP variants to kinetic studies of mycobacterial gene expression. J Microbiol Methods. 2003;54:203–211. doi: 10.1016/s0167-7012(03)00044-7. [DOI] [PubMed] [Google Scholar]
- Clemens DL. Characterization of the Mycobacterium tuberculosis phagosome. Trends Microbiol. 1996;4:113–118. doi: 10.1016/0966-842X(96)81528-9. [DOI] [PubMed] [Google Scholar]
- Clemens DL, Horwitz MA. Characterization of the Mycobacterium tuberculosis phagosome and evidence that phagosomal maturation is inhibited. J Exp Med. 1995;181:257–270. doi: 10.1084/jem.181.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Horwitz MA. The Mycobacterium tuberculosis phagosome interacts with early endosomes and is accessible to exogenously administered transferrin. J Exp Med. 1996;184:1349–1355. doi: 10.1084/jem.184.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Lee BY, Horwitz MA. Deviant expression of Rab5 on phagosomes containing the intracellular pathogens Mycobacterium tuberculosis and Legionella pneumophila is associated with altered phagosomal fate. Infect Immun. 2000a;68:2671–2684. doi: 10.1128/iai.68.5.2671-2684.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Lee BY, Horwitz MA. Mycobacterium tuberculosis and Legionella pneumophila phagosomes exhibit arrested maturation despite acquisition of Rab7. Infect Immun. 2000b;68:5154–5166. doi: 10.1128/iai.68.9.5154-5166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Lee BY, Horwitz MA. The Mycobacterium tuberculosis phagosome in human macrophages is isolated from the host cell cytoplasm. Infect Immun. 2002;70:5800–5807. doi: 10.1128/IAI.70.10.5800-5807.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowle AJ, Dahl R, Ross E, May MH. Evidence that vesicles containing living, virulent Mycobacterium tuberculosis or Mycobacterium avium in cultured human macrophages are not acidic. Infect Immun. 1991;59:1823–1831. doi: 10.1128/iai.59.5.1823-1831.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancz CE, Haraga A, Portnoy DA, Higgins DE. Inducible control of virulence gene expression in Listeria monocytogenes: temporal requirement of listeriolysin O during intracellular infection. J Bacteriol. 2002;184:5935–5945. doi: 10.1128/JB.184.21.5935-5945.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dargelos E, Moyen C, Dedieu S, Veschambre P, Poussard S, Vuillier-Devillers K, Brustis JJ, Cottin P. Development of an inducible system to assess p94 (CAPN3) function in cultured muscle cells. J Biotechnol. 2002;96:271–279. doi: 10.1016/s0168-1656(02)00052-4. [DOI] [PubMed] [Google Scholar]
- Deretic V, Via LE, Fratti RA, Deretic D. Mycobacterial phagosome maturation, rab proteins, and intracellular trafficking. Electrophoresis. 1997;18:2542–2547. doi: 10.1002/elps.1150181409. [DOI] [PubMed] [Google Scholar]
- Desjardins M. Biogenesis of phagolysosomes: the 'kiss and run' hypothesis. Trends Cell Biol. 1995;5:183–186. doi: 10.1016/s0962-8924(00)88989-8. [DOI] [PubMed] [Google Scholar]
- Desjardins M, Huber LA, Parton RG, Griffiths G. Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J Cell Biol. 1994;124:677–688. doi: 10.1083/jcb.124.5.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratti RA, Vergne I, Chua J, Skidmore J, Deretic V. Regulators of membrane trafficking and Mycobacterium tuberculosis phagosome maturation block. Electrophoresis. 2000;21:3378–3385. doi: 10.1002/1522-2683(20001001)21:16<3378::AID-ELPS3378>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Goren MB, Hart PD, Young MR, Armstrong JA. Prevention of phagosome-lysosome fusion in cultured macrophages by sulfatides of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 1976;73:2510–2514. doi: 10.1073/pnas.73.7.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harth G, Maslesa-Galic S, Horwitz MA. A two-plasmid system for stable, selective-pressure-independent expression of multiple extracellular proteins in mycobacteria. Microbiology. 2004;150:2143–2151. doi: 10.1099/mic.0.27113-0. [DOI] [PubMed] [Google Scholar]
- Howard NS, Gomez JE, Ko C, Bishai WR. Color selection with a hygromycin-resistance-based Escherichia coli-mycobacterial shuttle vector. Gene. 1995;166:181–182. doi: 10.1016/0378-1119(95)00597-x. [DOI] [PubMed] [Google Scholar]
- Lepperdinger G, Strobl B, Kreil G. HYAL2, a human gene expressed in many cells, encodes a lysosomal hyaluronidase with a novel type of specificity. J. Biol. Chem. 1998;273:22466–22470. doi: 10.1074/jbc.273.35.22466. [DOI] [PubMed] [Google Scholar]
- Malik ZA, Denning GM, Kusner DJ. Inhibition of Ca(2+) signaling by Mycobacterium tuberculosis is associated with reduced phagosome-lysosome fusion and increased survival within human macrophages. J Exp Med. 2000;191:287–302. doi: 10.1084/jem.191.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik ZA, Iyer SS, Kusner DJ. Mycobacterium tuberculosis phagosomes exhibit altered calmodulin-dependent signal transduction: contribution to inhibition of phagosome-lysosome fusion and intracellular survival in human macrophages. J Immunol. 2001;166:3392–3401. doi: 10.4049/jimmunol.166.5.3392. [DOI] [PubMed] [Google Scholar]
- McDonough KA, Kress Y, Bloom BR. Pathogenesis of tuberculosis: interaction of Mycobacterium tuberculosis with macrophages. Infect Immun. 1993;61:2763–2773. doi: 10.1128/iai.61.7.2763-2773.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwandumba HC, Russell DG, Nyirenda MH, Anderson J, White SA, Molyneux ME, Squire SB. Mycobacterium tuberculosis resides in nonacidified vacuoles in endocytically competent alveolar macrophages from patients with tuberculosis and HIV infection. J Immunol. 2004;172:4592–4598. doi: 10.4049/jimmunol.172.7.4592. [DOI] [PubMed] [Google Scholar]
- Myrvik QN, Leake ES, Wright MJ. Disruption of phagosomal membranes of normal alveolar macrophages by the H37Rv strain of Mycobacterium tuberculosis A correlate of virulence. Am Rev Respir Dis. 1984;129:322–328. [PubMed] [Google Scholar]
- Oh YK, Straubinger RM. Intracellular fate of Mycobacterium avium: use of dual-label spectrofluorometry to investigate the influence of bacterial viability and opsonization on phagosomal pH and phagosome-lysosome interaction. Infect Immun. 1996;64:319–325. doi: 10.1128/iai.64.1.319-325.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt A, Mayorga LS, Schwartz AL, Stahl PD. Transport of phagosomal components to an endosomal compartment. J Biol Chem. 1992;267:126–132. [PubMed] [Google Scholar]
- Russell DG. Immunoelectron microscopy of endosomal trafficking in macrophages infected with microbial pathogens. Methods Cell Biol. 1994;45:277–288. doi: 10.1016/s0091-679x(08)61857-9. [DOI] [PubMed] [Google Scholar]
- Russell DG, Dant J, Sturgill-Koszycki S. Mycobacterium avium- and Mycobacterium tuberculosis-containing vacuoles are dynamic, fusion-competent vesicles that are accessible to glycosphingolipids from the host cell plasmalemma. J Immunol. 1996;156:4764–4773. [PubMed] [Google Scholar]
- Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF, Snapper SB, Barletta RG, Jacobs WR, Jr, Bloom BR. New use of BCG for recombinant vaccines. Nature. 1991;351:458–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- Sturgill-Koszycki S, Schlesinger PH, Chakraborty P, Haddix PL, Collins HL, Fok AK, Allen RD, Gluck SL, Heuser J, Russell DG. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science. 1994;263:678–681. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
- Szaflarski W, Nierhaus KH. Question 7: optimized energy consumption for protein synthesis. Orig Life Evol Biosph. 2007;37:423–428. doi: 10.1007/s11084-007-9091-4. [DOI] [PubMed] [Google Scholar]
- Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- Tullius MV, Harth G, Horwitz MA. Glutamine synthetase (GlnA1) is essential for growth of Mycobacterium tuberculosis in human macrophages and in guinea pigs. Infect. Immun. 2003;71:3927–3936. doi: 10.1128/IAI.71.7.3927-3936.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, Brenner M, Peters PJ. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- Via LE, Deretic D, Ulmer RJ, Hibler NS, Huber LA, Deretic V. Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J. Biol. Chem. 1997;272:13326–13331. doi: 10.1074/jbc.272.20.13326. [DOI] [PubMed] [Google Scholar]
- Via LE, Fratti RA, McFalone M, Pagan-Ramos E, Deretic D, Deretic V. Effects of cytokines on mycobacterial phagosome maturation. J Cell Sci. 1998;111(Pt 7):897–905. doi: 10.1242/jcs.111.7.897. [DOI] [PubMed] [Google Scholar]
- Wubbolts R, Fernandez-Borja M, Oomen L, Verwoerd D, Janssen H, Calafat J, Tulp A, Dusseljee S, Neefjes J. Direct vesicular transport of MHC class II molecules from lysosomal structures to the cell surface. J. Cell Biol. 1996;135:611–622. doi: 10.1083/jcb.135.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Cooper A, Sturgill-Koszycki S, van Heyningen T, Chatterjee D, Orme I, Allen P, Russell DG. Intracellular trafficking in Mycobacterium tuberculosis and Mycobacterium avium-infected macrophages. J Immunol. 1994;153:2568–2578. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.