Abstract
Protein-protein interactions (PPIs) control the assembly of multi-protein complexes and, thus, these contacts have enormous potential as drug targets. However, the field has produced a mix of both exciting success stories and frustrating challenges. Here, we review known examples and explore how the physical features of a PPI, such as its affinity, hotspots, off-rates, buried surface area and topology, may influence the chances of success in finding inhibitors. This analysis suggests that concise, tight binding PPIs are most amenable to inhibition. However, it is also clear that emerging technical methods are expanding the repertoire of “druggable” protein contacts and increasing the odds against difficult targets. In particular, natural product-like compound libraries, high throughput screens specifically designed for PPIs and approaches that favor discovery of allosteric inhibitors appear to be attractive routes. The first group of PPI inhibitors has entered clinical trials, further motivating the need to understand the challenges and opportunities in pursuing these types of targets.
Keywords: high throughput screening, allostery, multi-protein complexes, Hsp70, Hsp90, fragment based drug discovery, natural products, protein-protein interactions
Introduction
As revealed by proteomic and two-hybrid strategies, protein-protein interactions (PPIs) are extensive and ubiquitous in biology (1, 2). PPIs hold together the key multi-protein complexes of the cell, guide subcellular trafficking and form the backbone of its major signaling pathways (3–6). Accordingly, many PPIs are also potential therapeutic targets in disease (7, 8) and inhibiting PPIs has become an increasingly attractive goal for both drug discovery and the generation of new research probes (9).
The last twenty years have witnessed considerable progress in the area of small molecule-based PPI inhibitors, with an explosion of literature reports and multiple PPI inhibitors entering clinical trials (10, 11). For example, a 2012 search of PubChem for projects involving “protein-protein” interactions reveals more than 800 results. In these projects, research groups are employing a wide array of technologies and often these methods are particularly suited for PPIs. At the same time, structural and computational biologists are becoming more adept at predicting PPIs and many groups are studying these interfaces to identify common topological features suitable for binding to inhibitors (12–15). Clearly, this field is maturing rapidly and “tricks” are being developed to overcome the historical challenges associated with targeting PPIs. In other words, the prevailing attitude has changed considerably in the last two decades and PPIs are no longer considered uniformly “undruggable”. In this review, we retrospectively analyze a subset of successful cases to explore what lessons can be learned and we analyze the frontiers of PPI research, where inhibitors are still difficult to identify.
Challenges of Targeting Protein-Protein Interactions
Despite advances, small molecule inhibitors of PPIs remain a relatively daunting challenge, an idea clearly articulated in a number of recent reviews (16–23). Informal polls of colleagues in the field suggest that successes are equally balanced with frustrations. There are several, well-described factors that contribute to this issue. Firstly, proteins that interact with other proteins typically do so using relatively large contact surfaces (1500 to 3000 Å2) (3). This value is much larger than the average size of the contact area between a small molecule and protein target, which is estimated to be between 300 to 1000 Å2 (24). This larger surface creates problems because molecules that target PPIs through competitive binding must typically have a high molecular weight to overcome the distributed free energy (ΔG) of the larger contact surface (16). Accordingly, the resulting compounds may have difficulty fitting within the reported limits on the size of orally available drugs, as summarized by the Rule of Five (RO5) (25). RO5 violations may shelve traditional drug leads, however a strict adherence to the RO5 for PPI programs seems likely to prove detrimental. In fact, the chemical properties of successful PPI antagonists have steadily shifted away from this benchmark (10). Still, the sheer size of many PPIs poses an unavoidable challenge. One possible solution to this problem is that 'hotspots' within some PPI sites create a scenario in which a handful of amino acids contribute a disproportionate amount of the binding ΔG (26, 27). Thus, targeting these specific regions typically has a more dramatic influence on the overall affinity and effectiveness of a small molecule. One of the earliest strategies to take advantage of this idea was the tethering method explored by Wells and colleagues (26). In this approach, compound fragments are covalently directed to hotspots to maximize the chances of developing potent PPI inhibitors. More recently, fragment-based screening by mass spectrometry, crystallography and nuclear magnetic resonance (NMR) have become next-generation platforms for this type of discovery (28). NMR, in particular, has proven a powerful method for finding building blocks that bind to topologically or energetically interesting sites on protein targets.
Another major issue related to PPI inhibition is that, compared to more traditional targets of antagonism, PPI partners often lack substantial grooves or deep pockets at their interacting surfaces (29). For example, the Arora group reported an interesting study in which they analyzed PPI structures from the Protein Data Base (PDB), focusing on those PPIs involving α-helical interactions. They categorize these PPIs into examples with well-defined clefts and those with extended, flat interfaces and use this information to suggest that PPIs with shallow surfaces are less likely to be readily inhibited (30).
Regardless of the type of contact, screens for chemical PPI inhibitors often produce hits that are large, with complex topology, especially compared to traditional inhibitors (16, 25, 31). These features can create synthetic challenges, in addition to possible issues with pharmacokinetics and solubility. As such, many groups have explored ways of improving chemical libraries to maximize the topological complexity of the library members and enrich for PPI inhibitors. The goal in these approaches, such as diversity-oriented synthesis and others (32, 33), is to produce compounds able to match the topology of PPIs. These challenges have also sparked revitalized interest in natural products as another rich source of potential PPI inhibitors, given the high average complexity of these molecules (34). Similarly, peptidomimetics, cyclic peptides and stapled peptides are being increasingly used to inhibit PPIs (35, 36). These ligands mimic natural protein-protein contacts by presenting multiple amino acid side chains from architecturally complex cores (37–41). Finally, small molecule-protein hybrids have been developed to artificially increase apparent molecular mass and target the most difficult PPIs (42, 43).
A related challenge to overcome in the search for PPI inhibitors is that a single contact surface on a protein can often have multiple binding partners (44) and, moreover, these partners can exchange in complex, dynamic equilibria (45). Thus, PPI inhibitors may have to contend with multiple protein competitors in the cell and, conversely, a single inhibitor may simultaneously disrupt multiple contacts, which could have unintended consequences on biology. Multi-protein complexes are often combinatorially assembled from a selection of possible subunits in the cell. For example, certain chromatin-remodeling complexes and chaperone systems are built from components that bind in a mutually exclusive way (46, 47), allowing creation of complexes with distinct functions. This is the biological complexity that must be overcome when considering which PPI to target and what to expect from successful inhibition.
Classes of Protein-Protein Interactions Inhibited by Small Molecules
The first literary example of inhibiting a PPI came from a peptide mimic against the ribonucleotide reductase (RR) of herpes simplex virus type 1 (HSV1) in 1986. Two independent groups showed that a nonapeptide representing the C-terminus of an essential subunit in the HSV1 RR holoenzyme was sufficient to inhibit RR activity (48, 49). Since then, the field has produced a large and ever increasing number of successful inhibitors (16, 17, 23). With that growing list of examples, can we begin to “bin” PPIs into categories that are predictive of their relative chances of success? Similarly, can this retrospective analysis reveal common topological features that make PPIs more or less challenging?
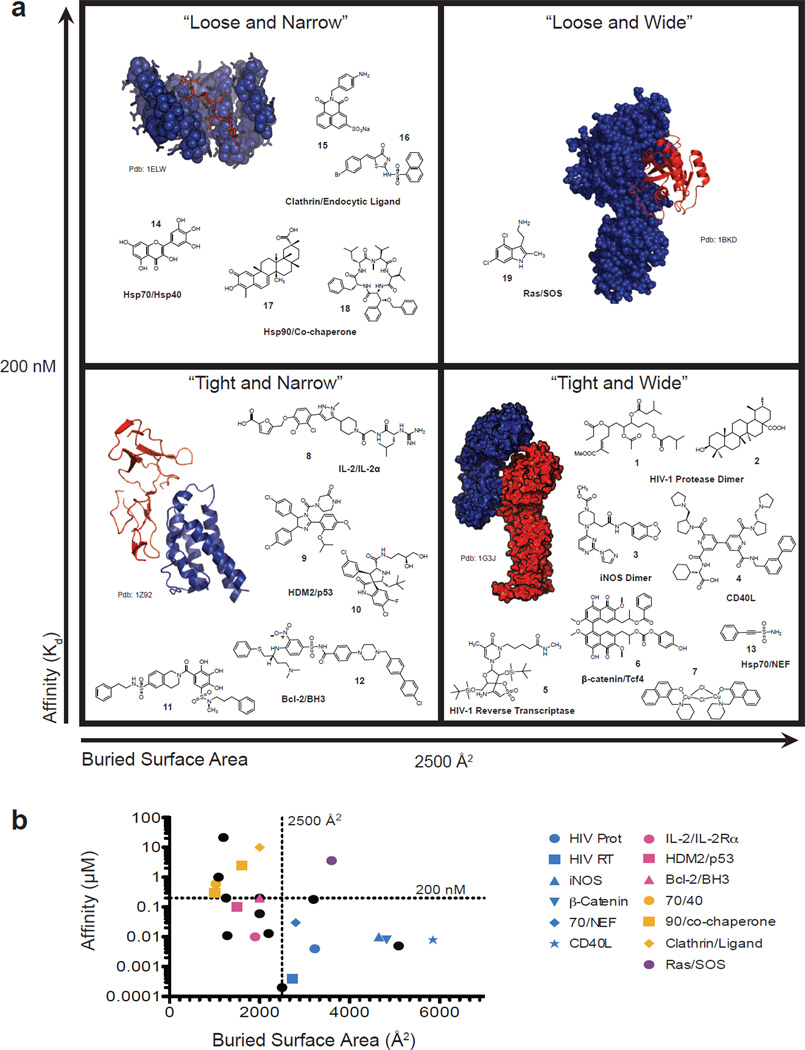
As one way to approach these questions, we compiled a list of PPIs with known inhibitors from the 2P2I (50) and TIMBAL (51) databases, as well as from recent examples in the literature (Table 1). For each of these “inhibit-able” PPIs, we measured the surface area that is buried in the protein-protein contact, based on available crystal structures (Fig 1a), and we looked up the reported affinity (KD) of the interaction. Plotting each PPI according to these two physical properties provided an overview of the types of interactions that have been reported to be inhibited by small molecules (Fig. 1b). We were struck by how this analysis seemed to create four general categories of PPIs and we colloquially termed these quadrants: “Tight and Narrow”, “Tight and Wide”, “Loose and Narrow” and “Loose and Wide” (Fig. 1a). In these arbitrary categories, narrow was defined as less than 2500 Å2, while wide was greater than 2500 Å2. Likewise, the affinity of the contact was separated into tight (Kd less than 200 nM) or loose (Kd greater than 200 nM). From this analysis, it was clear that there are relatively few examples of inhibitors in the “Loose and Wide” category, while inhibitors were clustered in the “Tight and Narrow” and “Weak and Narrow” categories. To see if specific types of chemical structures tended to cluster in these categories, we examined 19 published inhibitors (Fig. 1a). However, other than a previously observed tendency towards high molecular mass (16), we did not note any obvious consensus. In the following sections, we further discuss PPI inhibitors in the context of this quadrant nomenclature to ask what lessons might be gleaned.
Table 1.
Features of select PPIs and their inhibitors.
| Interaction | Buried surface area (Å2) |
Complex affinity (µM) |
Inhibitor affinity (µM)a |
References |
|---|---|---|---|---|
| Tight and Wide | ||||
| HIV-1 Protease | 3224 | 0.004 | 2–3 | (66, 67, 70, 71) |
| HIV-1 Reverse Transcriptase | 2730 | 0.0004 | 0.56 | (68, 69, 73) |
| iNOS | 4650 | 0.01 | 0.0022 | (79, 147, 148) |
| β-catenin/Tcf4 | 4820 | 0.008 | 3–5 | (74–76, 149) |
| Hsp70/NEF | 2800 | 0.03 | ND | (120) |
| CD40L | 5850 | 0.008 | 25 | (83) |
| RGS4/Gα | 5090 | 0.005 | 4.2 | (90, 164) |
| TNFα | 2500 | 0.0002 | 22 | (84, 168, 169) |
| cMyc/Max | 3200 | 0.18 | 30 | (82, 177, 178) |
| Tight and Narrow | ||||
| IL-2/IL-2Rα | 1900 | 0.01 | 0.06 | (53, 150, 151) |
| HDM2/p53 (peptide) | 1498 | 0.1 | 0.005–0.067 | (56, 58, 152–154) |
| Bcl-2/BH3 (peptide) | <2500b | 0.2 | 0.0006–0.11 | (60, 155, 156) |
| HPV E1/E2 | ~2000c | 0.06 | 0.1 | (157, 158) |
| S100B/p53 | 1041 | 0.2 | 1 | (165–167) |
| XIAP/Caspase 9 | 2200 | 0.013 | 0.01 | (171–173) |
| LEDGF/Integrase | 1280 | 0.011 | 1 | (174–176) |
| CD28/B7.1 | 1255 | 0.2 | 0.004 | (179, 180) |
| Loose and Narrow | ||||
| Hsp70/Hsp40 | 1028 | 0.6 | ND | (118, 124) |
| Hsp90/TPR | 1000 | 0.3 | 0.4 | (121, 122, 140) |
| Hsp90/cdc37 | 1600 | 2.5 | ND | (135, 161) |
| Clathrin/Endolytic Ligand | <2500b | 1–100 | 12–18 | (99, 159, 160) |
| ZipA/FtsZ (peptide) | 1197 | 21.6 | 12 | (162, 163) |
| UL30/UL42 | 1087 | 1 | 12 | (170) |
| Loose and Wide | ||||
| Ras/SOS | 3600 | 3.6 | ND | (103, 104, 106) |
PPIs in the table represent all interactions contained in both the 2P2I (Ref. 50) and TIMBAL (Ref. 51) databases, as well as a select set of recent literary examples.
When direct binding data was not available, Ki or IC50 values were used instead.
The absolute value for surface area has not been described for these interactions. The values in the table are predictions based off of known structural information.
Predicted value (Ref. 157).
Figure 1. Categorization of protein-protein interactions (PPIs) and their inhibitors.
PPIs with known inhibitors were from obtained from the 2P2I (Ref. 10) and TIMBAL (Ref. 50) databases and recent literature. These PPIs were then categorized by the affinity (KD) of the protein-protein interaction and the buried surface area from co-crystal structures (see Table 1). These values were used to arbitrarily categorize the “inhibit-able” PPIs into four quadrants. To illustrate the types of proteins in each category, structures of representative PPIs from each class are shown, with each partner depicted in either blue or red. For “Tight and Wide” the interaction is between the armadillo repeat region of β-catenin and the catenin binding domain of Xenopus TCF3 (PDB: 1G3J). For “Tight and Narrow” the interaction is between IL-2 and the IL-2α Receptor (PDB: 1Z92). For “Loose and Narrow” the interaction is between TPR1 and a C-terminal peptide of Hsc70 (PDB: 1ELW) and for “Loose and Wide” the interaction is between Ras and SOS (PDB: 1BKD). Also shown are 19 representative chemical structures of the PPI inhibitors, illustrating the lack of consensus in molecular weight, shape or other characteristics.
Protein-Protein Interactions "Tight and Narrow"
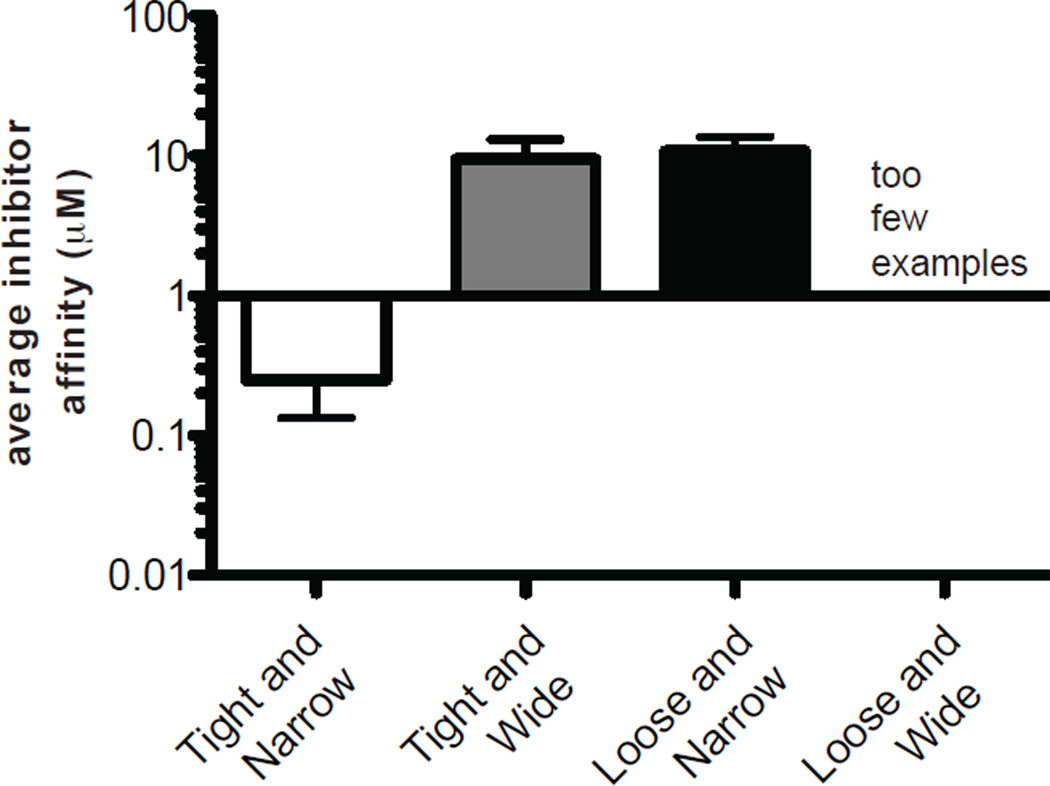
To date, this category of PPI has proven most amenable to inhibition, producing the most potent inhibitors and a large number of examples (Fig. 2). The reasons for this relative success could be due to the fact that the physical features shared by members of this class are most similar to traditional, enzyme targets. These PPIs are those with high affinity encompassed in a relatively small surface area. They also typically have deep pockets that are engaged by less than five major amino acids that contribute a majority of the binding ΔG. Because of these concise features, some of the strategies used in typical drug discovery campaigns, such as high throughput screening (HTS) and structure-based design, can be readily employed to target these PPIs. However, a number of PPI-specific methods have also been developed and some of those methods have subsequently been used to tackle more challenging targets.
Figure 2. Average potency of inhibitors in each class of protein-protein interaction.
The most potent inhibitors published for each interaction listed in Table 1 were averaged within the four PPI categories. The category of “Loose and Wide” did not have enough examples to be included.
In 1997, a group at Hoffmann-La Roche screened a series of acylphenylalanine derivatives intending to competitively inhibit the interaction between interleukin 2 (IL-2) and the IL-2α receptor (52). Their mid-micromolar inhibitor was developed by researchers at Sunesis Pharmaceuticals into the mid-nanomolar antagonist SP4206, Compound 8, (Ki= 60 nM; Fig. 1). The Sunesis group utilized structural information provided by NMR, combined with tethering, to incrementally build this molecule (53). An instructive idea that arose from that work came from the fact that the structural characterization of these compounds was originally based on their direct binding to IL-2. It was only after the structure of the IL-2/IL2Rα complex was solved that it became apparent that SP4206 bound to IL-2 and stabilized a conformation that was less competent to bind the IL-2Rα. Moreover, SP4206 induced this conformational change at the PPI surface by interacting with the same residues that were shown by alanine scanning to encompass a disproportionate amount of binding ΔG for the IL-2/IL-2Rα interaction (e.g. a “hotspot”) (54). Thus, this early example of a successful PPI inhibitor uncovered a number of principles that became repetitive themes in other systems; namely, compound-induced conformational change and hotspot binding.
Small-molecule inhibitors of the PPI between p53 and mouse double minute 2 (MDM2) were identified based on the results of a high throughput screen. As was observed in the case of IL2, these cis-imidazolines, termed nutlins, were shown to occupy the same binding pocket on MDM2 that is critical for binding to p53 (55). Nutlin-3, Compound 9 (Fig. 1), was shown to have mid-nanomolar (~70 nM) and enantioselective activity towards the p53-MDM2 complex, leading to an accumulation of p53 and subsequent tumor suppression (56). Nutlin-3 is currently in phase I clinical trial for the treatment of retinoblastoma, illustrating the promise of PPI inhibitors as drugs and solidifying the idea that surface mimicry and hotspot binding are key tools for targeting this class of PPI. The concept of mimicking the natural interactions was also used in a parallel strategy to inhibit p53-MDM2. This strategy was inspired by the natural product spiro(oxindole-3,3'-pyrrilodine) scaffold, which mimics the indole ring of Trp23 in p53 that binds to a deep, hydrophobic cavity in MDM2 (57). This rational-design approach, coupled with medicinal chemistry efforts yielded MI-63, which was further developed to MI-219, Compound 10 (Fig. 1) to improve its pharmacokinetic profile. MI-219 shows low nanomolar (~5 nM) inhibition of complex formation with sub-micromolar (0.4 to 0.8 µM) IC50 values for tumor growth inhibition (58). The Wang group has pioneered additional rational design approaches in which they start with the structure of the PPI, perform alanine scans to identify possible hotspots and then design peptidomimetics and synthetic scaffolds that are intended to disrupt critical contacts (57, 59). These examples are clear cases in which the structure of the PPI can be used to launch inhibitor programs.
Another key lesson is illustrated by the work of Abbott investigators in their search for inhibitors of B-cell lymphoma 2 (Bcl-2). Fesik and colleagues employed fragment-based screening by NMR, followed by extensive SAR by NMR to develop ABT-737, Compound 12 (Fig. 1), which binds the anti-apoptotic molecules Bcl-XL, Bcl-2, and Bcl-W and prevents their association with pro-apoptotic proteins BAD and BAX (Ki <1 nM) (60). This compound, and its orally bioavailable derivative ABT-263, shows anti-proliferative activity against a number of cancer cell lines, as well as anti-tumor activity in xenograft animal models (61). ABT-263 is currently in phase I/II trial as a single agent for relapsed or refractory lymphoid malignancies, and in phase II trial for lymphatic leukemia in combination with the antibody therapeutic rituximab. This work was some of the first to document how NMR could be used as a primary discovery tool for identifying and elaborating drug leads, and the first to do so using a fragment-based approach (62). More broadly, NMR-based design of PPI inhibitors, often combined with some form of HTS, has been particularly successful in this category of interactions, as illustrated by examples in the Runx1-CBFβ (63) and MLL (64).
Protein-Protein Interactions "Tight and Wide"
Some PPIs involve extensive and often convoluted or discontinuous interaction surfaces, creating contacts with large buried contact areas and tight affinities. These features can create special difficulties in developing small-molecule inhibitors because of the slow off rates and the large surfaces to overcome. Still, a number of successful examples have been reported and a review of these cases suggests some methodologies with potentially far-reaching utility.
Of the 15 enzymes encoded in the human immunodeficiency virus (HIV) genome, three are essential homo- or pseudo-dimers (65). Two of these proteins, HIV-1 protease (HIVp) and reverse transcriptase (RT) have been successfully targeted with small-molecule inhibitors. The HIVp dimer has an interacting face with over 3,000 Å2 of buried surface area (66) and a Kd value in the low nanomolar range (67). Similarly, the HIV-1 RT multimer interface buries 2,730 Å2 (68) with a Kd of 400 pM (69). In the late 1990s, two groups identified HIVp dimerization inhibitors by screening natural products (Compounds 1 & 2; Fig. 1) (70, 71). Likewise, exploration of non-nucleoside inhibitors of HIV-1 RT revealed Compound 5 (Fig. 1), which was subsequently shown to have anti-dimerization activity (72, 73). These findings support the idea that topologically complex natural products are suitable scaffolds for inhibiting even complex and large PPIs. To further exemplify this idea, the interaction surface between β-catenin and Tcf/LEF family members is also particularly large (> 3000 Å2), making it another difficult target (74). Yet, Compound 6 (Fig. 1) was identified in a natural product screen that relied on measuring binding of Tcf4 to β-catenin (75). However, natural products are not exclusive in their ability to inhibit these types of interactions. Recently, an in silico screen revealed Compound 7 (Fig. 1), a simpler structure that is predicted to bind to a hotspot region and inhibit the β-catenin-Tcf4 interaction (76). Additional structural information will be needed to fully understand the binding mode and mechanistic basis for these activities, but the findings suggest that topologically complex chemical libraries may be good starting points for identifying PPI inhibitors.
The inducible isoform of nitric oxide synthase (iNOS) has been implicated in inflammatory and autoimmune diseases (77), while the endothelial isoform plays a vital role in vascular homeostasis (78). In an effort to identify selective inhibitors of iNOS, McMillan et al. (2000) conducted an in vitro screen against iNOS enzyme activity using a compound library based off of a pyrimidine-imidazole core (79). This screen resulted in no active compounds, but a cell-based screen against nitric oxide production yielded the potent inhibitor Compound 3 (Fig. 1). The compound was shown to have a high affinity for the iNOS monomer and, upon binding, it allosterically inhibited subsequent dimerization, explaining why the original in vitro screen against the pre-formed iNOS dimer produced no inhibitors. This example nicely illustrates a growing realization that allostery and allosteric mechanisms are powerful tools in targeting larger PPIs (12, 80). Moreover, this study illustrates how dynamics can have a large impact on success in HTS-based PPI campaigns. Only when iNOS was allowed to sample both monomeric and dimeric structures were allosteric inhibitors uncovered.
Other interesting examples of this class are found in inhibitors of the c-Myc/Max dimer, the CD40 ligand (CD40L) trimer, and the eukaryotic translation initiation factors (eIF) eIF4E and eIF4G. A hurdle to small-molecule inhibition of c-Myc/Max dimerization is the vast increase in dimer stabilization in the presence of DNA (81). To circumvent this issue, the Berg group developed a clever HTS approach in which c-Myc/Max binding to a fluorophore-labeled oligonucleotide was measured, revealing Mycros 1 and 2 as micromolar inhibitors (82). This study nicely leveraged known biophysical features of the PPI to design a screen especially targeting a key aspect of the complex. Similarly, identifying inhibitors of CD40L trimerization are also difficult, given the tight affinity of this complex. Yet, a direct binding assay was used to produce BIO8898, Compound 4 (Fig. 1), a small molecule that populates a deep, allosteric pocket between two subunits of the trimer (83). Binding by BIO8898 distorts the interface enough to prevent binding of CD40L to the CD40 receptor. This is a more subtle form of inhibition than seen in previous studies of the homologous TNFα, where an inhibitor was found that completely ejected one subunit from the trimer (84). These examples suggest that even challenging PPIs can sometimes be amenable to HTS approaches, using methodologies, such as AlphaLisa, fluorescence and lumiscence complementation, ELISA, SPR and FRET (85–88), and the resulting compounds, if the screen is designed carefully, can access unexpected and interesting molecular mechanisms. This concept is further re-enforced by work from the Wagner group, in which they developed a high-throughput fluorescence polarization screen against the initiation factor eIF4E using a peptide from the binding motif of eIF4G (89). This screen revealed the inhibitor 4EGI-1 that disrupts full-length eIF4G binding, and interestingly, clears the path for natural modulators of translation (4E-BP) to interact and thereby inhibit translation. Together, these examples demonstrate the diversity of solutions to the problem of blocking large, tight interactions.
As illustrated by the examples above, this class of PPIs has been successfully targeted and the resulting compounds access interesting mechanisms. Another example was provided by the Neubig group, in which they used a flow cytometry protein interaction assay (FCPIA) to target regulators of G-protein signaling (RGS). They identified CCG-4986, which inhibits the RGS4-Gα PPI by covalent modification of a cysteine residue adjacent to the interaction surface (90). This example highlights a growing resurgence of covalent modifiers as probes and drugs (91). Covalent modifiers might be particularly attractive for PPIs because irreversible binding can be used to overcome problems of weak interactions and shallow binding sites.
Amyloids are ordered protein aggregates defined by a characteristic appearance by electron microscopy, affinity for the dye, congo red, and large contact interfaces between monomers (92). The interface challenge is exacerbated by the repetitive structure of amyloids, involving thousands of monomer interactions and thousands of cumulative Å2 of buried surface area. Amyloids underlie a number of neurodegenerative disorders and other diseases of protein misfolding (93), so inhibitors of amyloid PPIs are of medical interest. Numerous small molecules with tight affinity for amyloids have been described, some based on synthetic scaffolds and others based on peptidomimetics (92, 94, 95). Some of these molecules have even advanced to clinical trials in Alzheimer’s disease. Interestingly, these compounds typically have good Kd values, yet their ability to block PPIs between amyloid-forming monomers (IC50) is typically 10- to 1000-fold worse. The disconnect between these values is thought to arise from compound binding being insufficient to fully block the large amyloid surfaces, which often lack clear “hotspots”. In 2004, it was discovered that hybrids between congo red and the FK506-binding protein (FKBP) created bifunctional inhibitors that better matched the size and complexity of the amyloid surface, producing inhibitors with nanomolar IC50 values (42). Interestingly, increasing the size of the FKBP portion enhanced the apparent IC50 of the hybrids, suggesting that larger surfaces were more effective inhibitors (96). Another clever solution to this problem can be found in anti-amyloid strategies using compounds that dissolve pre-formed aggregates by allostery (97). Thus, even for some of the most extreme PPIs, allostery and other mechanisms can be used to inhibit their formation.
Protein-Protein Interactions "Loose and Narrow"
PPIs within this category are characterized by weak (Kd > 200 nM) binding but relatively small contact areas. Because these interactions are typically transient, it is not unusual for the surfaces to be shared by multiple partners. These hurdles, often coupled to a lack of structural data and relatively shallow binding pockets, make these interactions especially challenging targets (Fig. 2). However, in one example of a successful approach, the N-terminal β-propeller domain (TD) of the clathrin heavy chain was targeted. Clathrin heavy chain serves as a central interacting hub for accessory proteins in the endocytic pathway (98). Two molecules termed pitstops, one from a naphthalimide core Compound 15 and the other from rhodanine Compound 16 (Fig. 1) were identified in an ELISA-based high-throughput screen (99). These compounds were shown to compete with accessory proteins for binding to a common site on the clathrin TD, limiting endocytosis and thereby inhibiting viral entry. Despite this success, general strategies for targeting this class are not yet clear.
In another interesting example that illustrates the challenges in this type of PPI, the Mapp group identify compounds that inhibit transcriptional activation within the activator complex (100). These authors recognized that a conserved structural element of natural activation domains is that they are amphipathic. They displayed polar functionality from an isoxazolidine core and, indeed, found that the only apparent requirement for creating artificial activators was that the molecule needed to be amphipathic (101, 102). This relatively loose structural constraint suggests that the strategies for optimizing inhibitors of this type of PPI will be substantially different than for other types of targets.
Protein-Protein Interactions “Loose and Wide”
At the extreme end of PPIs are the contacts defined by large surface areas and weak affinities. To our knowledge, few potent inhibitors of contacts within this category have been described and these targets remain a particularly challenging area. Yet, very recent evidence suggests that inhibition at this level is possible. The interaction between the small GTP-binding protein Ras and its guanidine nucleotide exchange factor (GEF) SOS (Son of Sevenless) spans approximately 3600 Å2 (103) and the catalytic domains bind with an affinity in the low micromolar range (104), placing this PPI squarely within the “loose and wide” category. Recent work has produced a stapled peptide (105) and a small molecule (Compound 19; Fig. 1) (106) capable of inhibiting this interaction both in vitro and in cells. Other biological examples of these PPIs are plentiful in the literature, especially in the area of GPCR clustering, cell-cell interactions and carbohydrate-protein interactions (107), creating a need for PPI inhibitors.
Advancing protein-protein interaction inhibitors in difficult systems
As evident from visually placing PPIs into quadrants (Fig. 1), some systems, especially weaker interactions and those that make contacts over a wide area, remain notoriously resistant to inhibition. This challenge is further evident by the large differences in the average potency values for inhibitors targeting the different classes. On average, compounds that inhibit “Tight and Narrow” PPIs have 10-fold better potency than those targeting “Loose and Narrow” contacts (Fig. 2). Many of these resistant systems have commonalities among them, such as limited structural information, transient and weak contacts and promiscuous binding interfaces. To further illustrate these ideas and highlight methodologies developed to specifically address these challenges, we focus on the heat shock protein 70 (Hsp70) and heat shock protein 90 (Hsp90) systems for further discussion. Hsp70 and Hsp90 are molecular chaperones that each form multi-protein complexes with important roles in protein folding and stabilization. Moreover, there is compelling evidence, in both cases, to suggest that targeting PPIs in the Hsp70 and Hsp90 complexes may be an effective therapeutic strategy in cancer and neurodegeneration (108–112). These systems also provide a convenient model for these discussions because the PPIs inherent in Hsp70 and Hsp90 complexes provide examples of nearly every type of PPI category.
Heat shock protein 70
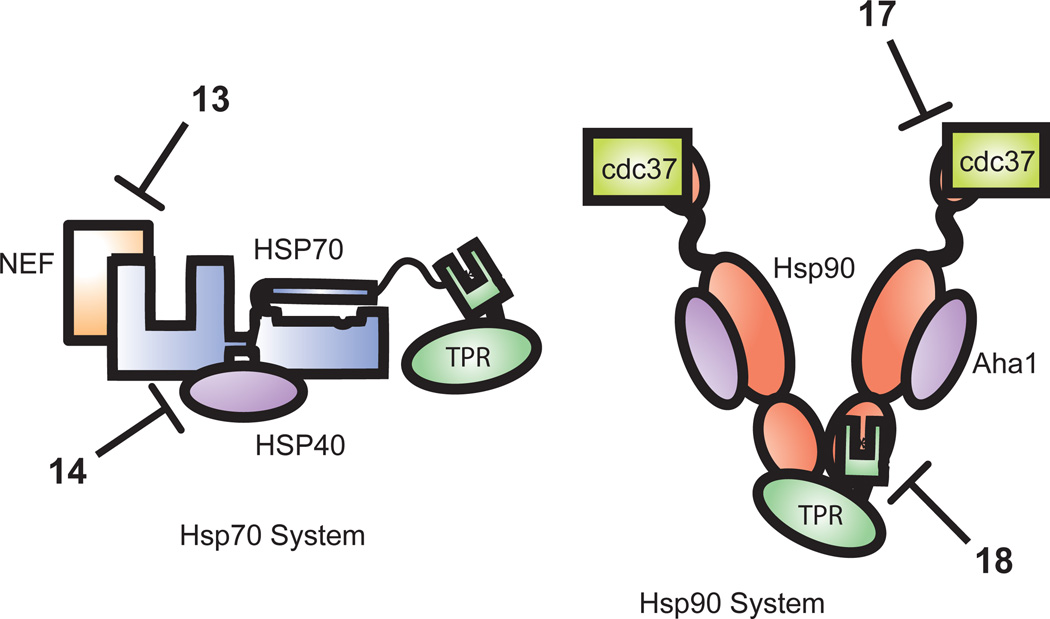
Hsp70 plays important roles in normal protein homeostasis and it is implicated in several disease states, such as cancer, neurodegeneration, and amyloidosis (108, 109, 113, 114). The protein consists of two domains, a nucleotide binding domain (NBD) that hydrolyzes ATP and a substrate-binding domain (SBD) that binds to exposed hydrophobic regions of polypeptides. NMR-based fragment screens conducted by the biotechnology company, Vernalis, have shown that the ATP-binding site of Hsp70 is not particularly amenable to discovery of selective or potent inhibitors (115). Thus, the PPIs between Hsp70 and its numerous co-chaperones have become attractive alternatives (108, 109, 116). There are three main classes of proteins that bind to Hsp70s. The Hsp40 (or DnaJ) superfamily is characterized by a conserved J-domain that binds to Hsp70 and stimulates its ATPase activity (117). This stimulation of ATP turnover promotes tight binding of substrates in the SBD via a conformational change. Nucleotide exchange factors (NEFs) bind Hsp70 and facilitate ADP release, helping to release substrates. And, finally, a family of tritetracopeptide repeat (TPR) domain-containing co-chaperones binds to the SBD, helping to arbitrate the fate of Hsp70-bound substrates (114). Thus, either promoting or inhibiting PPIs between Hsp70 and its co-chaperones can modulate the biology of the system (108, 109).
The J domain of the prokaryotic Hsp40, DnaJ, binds to the Hsp70, DnaK, across a largely polar interface between the NBD and SBD (118). The Kd of this interaction is weak (> 1 µM) and a structure of the auxilin J-domain fused to mammalian Hsp70 suggests a relatively modest interaction surface (1028 Å2) (119). Co-structures of Hsp70 and NEFs suggest a larger (2800 Å2) interaction, with much higher affinity (30 nM) (120). Finally, the TPR domain interaction with Hsp70 is approximately 1 µM (121) and occurs over an area of 1330 Å2, based on a crystal structure of a representative TPR domain with the C-terminus of Hsp70 (122). Based on genetic studies, each of these interfaces is attractive as a therapeutic target.
Our group has become particularly interested in targeting the Hsp70-Hsp40 interaction because of the unusually weak affinity between these partners and the importance of the contact in chaperone biology (117). Recent work by the Zuiderweg group has shown that the prokaryotic Hsp70-Hsp40 contact is largely polar, with a complex and shallow topology (118). We originally considered it unlikely that a screen for direct (e.g. competitive) inhibitors of the direct PPI would be fruitful, given the weak binding of the two partners. Accordingly, we pursued a different strategy, termed “gray box screening”. In this method, the ATPase activity of Hsp70 is stimulated by reconstituting its complex with an Hsp40 in vitro. Any compounds that disrupt Hsp40-stimulated ATP turnover would be identified as a “hit” in the screen. This approach is termed gray box screening because it has some features in common with “black box” screens, in which whole cells or animals are used as the target. In cell-based screens, the physiological relevance of the platform is high, but target identification is a challenging task. In the gray box approach, some of the natural complexity of the system is mimicked by reconstitution of the multi-protein system. This approach has been used to identify a number of inhibitors of the Hsp70-Hsp40 complex, some of which bind directly at the PPI interface (123) and others that bind distal, allosteric sites (124). For example, the flavonoid myricetin Compound 14 (Fig. 1) was found to bind an unanticipated site in the NBD, about 30 Å from the Hsp70-Hsp40 interface, trapping a conformation that is not able to interact with Hsp40 (124). Interestingly, the binding site for myricetin is not apparent in the crystal structures of the Hsp70 NBD, suggesting that dynamic movements in this region are required to open the compound-binding site (125). Because this screening approach is amenable to HTS in low volume, large numbers of compounds can be screened (126, 127). This strategy might be applicable in other systems involving weak interactions, especially those in which non-enzyme partners allosterically modify the activity of a core enzyme component.
The tighter Hsp70-NEF interaction has been targeted using a “black box” high throughput screen. From a cell-based assay against p53-mediated apoptosis, PES Compound 13 (Fig. 1) was identified as a small-molecule that decreases tumor cell viability (128). In follow-up studies, PES appears to block the binding of Hsp70 to the M isoform of bcl-2 associated athanogene 1 (BAG-1), a NEF for Hsp70. These findings (along with the nutlin work described above) suggest that phenotypic screens can sometimes reveal PPI inhibitors, even if the target PPI is relatively large. One power of these methods is that the target PPI is allowed to undergo its natural dynamics, often providing unanticipated mechanisms of inhibition. The challenge is that the mechanism of inhibition is not clear until follow-up studies are performed.
Heat shock protein 90
The Hsp70 and Hsp90 systems are linked through a shared TPR-domain co-chaperone, HOP (Hsp70-Hsp90 organizing protein) (129). And like Hsp70, Hsp90 is an abundant molecular chaperone that relies on a network of these co-chaperones for its activity (130). Under both stress and normal conditions, Hsp90 regulates the stability and maturation of over 200 client proteins, many of which either harbor mutations or are over-expressed in cancers (131). In fact, inhibitors of the ATPase activity of Hsp90, which bind to the N-terminal ATP-binding pocket, have been extensively explored and some of these have advanced to clinical trials as anti-cancer agents (132). These inhibitors bind classically defined pockets and do not appear to directly impact co-chaperone binding. However, one drawback of these molecules is that they elicit a heat shock response, through activation of heat shock factor 1 (HSF1) (133). This cytoprotective response has the potential to undermine the anti-proliferative effects of Hsp90 inhibition. These issues have driven interest in targeting the C-terminal ATP-binding site (134) and, importantly for this review, PPIs between Hsp90 and its co-chaperones.
Hsp90 interacts with the important co-chaperones Aha1, cdc37, p23 and a number of TPR-domain proteins (129). These interactions tune the ATPase activity of Hsp90 and these PPIs are being recognized as potential drug targets (112). The TPR-Hsp90 interaction surface resembles that in the TPR-Hsp70 system, being relatively weak but narrow. Cdc37 interacts with relatively poor affinity (2.5 µM) (135) to Hsp90, burying 1600 Å2 of solvent exposed surface area, while Aha1 binds across a very large, polar surface of Hsp90 (136) with moderate affinity (0.6 µM) (137). Thus, like the Hsp70 system, this multi-protein complex has a wide range of affinities and surface areas.
An investigation into how the natural product, celastrol Compound 17 (Fig. 1) inhibited Hsp90 and elicited a heat-shock response initially revealed that the compound reduced the interaction between Hsp90 and the cancer associated co-chaperone cdc37 (138). Upon further analysis, it was revealed that celastrol covalently binds to the Hsp90 co-chaperone, cdc37 (139). Recently, the molecule Compound 18 (Fig. 1) based off of another natural product, Sansalvamide A, was reported to bind to Hsp90 and inhibit the interaction of TPR domain-containing co-chaperones (140). Other inhibitors of the Hsp90-TPR interaction have also been identified by HTS approaches (140, 141). Thus, like in the Hsp70 system, “biology-driven” HTS was employed as a successful strategy to identify PPIs inhibitors in the Hsp90 system and natural products were common hits.
Conclusions
Protein-protein interactions are the “glue” that drives biology. To take advantage of potential therapeutic and emerging research opportunities, a growing urgency has emerged around inhibiting PPIs. In addition, a number of successes in the field of chemical inhibitors of PPIs, including initiation of multiple clinical trials, have provided a strong motivator for continued experimental focus.
What can be learned from analyzing prior successes and failures in targeting PPIs with small molecules? One over-whelming observation is that PPI inhibitors are not as hard to find as one might expect. Many straightforward HTS methods have successfully produced micromolar and nanomolar inhibitors, especially of concise (e.g. “Tight and Narrow”) PPIs. Also, many of the most successful PPI inhibitors have taken advantage of hotspots that effectively reduce large, flat surfaces to more manageable targets. Another common solution is found in compounds that bind allosteric sites to modify PPIs, as was the case with the CD40L inhibitor and myricetin in the Hsp70 system. These compounds utilized well-defined pockets to enact global changes at either adjacent (in the case of CD40L) or distal (in the case of Hsp70) PPI contacts. Allostery can work over substantial distances (142), further suggesting that even topologically complex surfaces can be impacted. Also, allosteric sites can be versatile tools, sometimes allowing switching between agonism and antagonism (123, 143).
What methods are best for identifying PPI inhibitors? The answer to this question appears to be dependent on the type of PPI, the specific biological goals and other factors. Rational design approaches have succeeded in cases of both large surface areas (as is the case with stapled peptides) and concise PPIs (as was seen with inhibitors of p53-MDM2). NMR is likely to continue to be a powerful method for discovery of PPI inhibitors because it combines structural insights with fragment-based approaches (as was seen in the case of Bcl-2). Finally, unbiased HTS methods, especially gray box screening and cell-based methods, have proven surprisingly fruitful in the search for PPI inhibitors. These methods seem particularly attractive in systems involving large contact surfaces, owing to their propensity to find unanticipated allosteric sites.
What do these studies mean for understanding basic biology? Many, if not all, biological processes are dependent on the function of multi-protein complexes (144–146). In a post-genomic world, chemical biologists and biologists are increasingly focusing on PPIs as key regulatory hubs. Thus, the development of research probes that target these interactions is important for understanding the logic of biological networks. It seems likely that substantial insights will emerge from efforts to create new solutions to the problem of inhibiting PPIs.
Figure 3. Inhibition of protein-protein interactions in the chaperone complexes.
Complexes between Hsp70 and Hsp90 and their co-chaperones are shown. PES 13 inhibits the Hsp70/Bag1 interaction, myricetin 14 inhibits the Hsp70/Hsp40 interaction, celastrol 17 inhibits Hsp90/cdc37, and San A 18 inhibits the Hsp90/TPR interactions. Abbreviations: NEF, nucleotide exchange factor; TPR, tetratricopeptide repeat domain-containing co-chaperone; San A, sansalvamide A.
Acknowledgements
We apologize that many outstanding examples of PPI inhibitors were not able to be included due to space constraints. Our work on PPI inhibitors is funded by the NIH (NS059690) and NSF (MCB-0844512). M.C.S. is funded by a training grant from the NIH (AG000114) and a Rackham Merit Fellowship. The authors also thank J. A. Townes and members of the Gestwicki group for helpful conversations.
References
- 1.Stelzl U, et al. A Human Protein-Protein Interaction Network: A Resource for Annotating the Proteome. Cell. 2005;122(6):957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 2.Rual J-F, et al. Towards a Proteome-Scale Map of the Human Protein-Protein Interaction Network. Nature. 2005;437(7062):1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 3.Jones S, Thornton JM. Principles of Protein-Protein Interactions. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(1):13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gavin AC, et al. Functional Organization of the Yeast Proteome by Systematic Analysis of Protein Complexes. Nature. 2002;415(6868):141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 5.Ryan DP, Matthews JM. Protein-Protein Interactions in Human Disease. Current Opinion in Structural Biology. 2005;15(4):441–446. doi: 10.1016/j.sbi.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Kuriyan J, Eisenberg D. The Origin of Protein Interactions and Allostery in Colocalization. Nature. 2007;450(7172):983–990. doi: 10.1038/nature06524. [DOI] [PubMed] [Google Scholar]
- 7.Balch WE, Yates JR. Application of Mass Spectrometry to Study Proteomics and Interactomics in Cystic Fibrosis. Cystic Fibrosis: Diagnosis and Protocols, Vol Ii: Methods and Resources to Understand Cystic Fibrosis. 2011;742:227–247. doi: 10.1007/978-1-61779-120-8_14. [DOI] [PubMed] [Google Scholar]
- 8.Vidal M, Cusick M, Barabasi A-L. Interactome Networks and Human Disease. Cell. 2011;144(6):986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arkin M. Protein-Protein Interactions and Cancer: Small Molecules Going in for the Kill. Current Opinion in Chemical Biology. 2005;9(3):317–324. doi: 10.1016/j.cbpa.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Morelli X, Bourgeas R, Roche P. Chemical and Structural Lessons from Recent Successes in Protein-Protein Interaction Inhibition (2p2i) Current Opinion in Chemical Biology. 2011;15(4):475–481. doi: 10.1016/j.cbpa.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 11.Thangudu RR, et al. Modulating Protein-Protein Interactions with Small Molecules: The Importance of Binding Hotspots. Journal of Molecular Biology. 2012;415(2):443–453. doi: 10.1016/j.jmb.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reynolds K, McLaughlin R, Ranganathan R. Hot Spots for Allosteric Regulation on Protein Surfaces. Cell. 2011;147(7):1564–1575. doi: 10.1016/j.cell.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang C-Y, Wang S. Computational Analysis of Protein Hotspots. ACS Medicinal Chemistry Letters. 2010;1(3):125–129. doi: 10.1021/ml100026a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geppert T, et al. Context-Based Identification of Protein-Protein Interfaces And "Hot-Spot" Residues. Chemistry & Biology. 2011;18(3):344–353. doi: 10.1016/j.chembiol.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Reichmann D, et al. The Modular Architecture of Protein-Protein Binding Interfaces. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(1):57–62. doi: 10.1073/pnas.0407280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wells JA, McClendon CL. Reaching for High-Hanging Fruit in Drug Discovery at Protein-Protein Interfaces. Nature. 2007;450(7172):1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 17.Keskin O, et al. Principles of Protein-Protein Interactions: What Are the Preferred Ways for Proteins to Interact? Chemical Reviews. 2008;108(4):1225–1244. doi: 10.1021/cr040409x. [DOI] [PubMed] [Google Scholar]
- 18.Berg T. Modulation of Protein–Protein Interactions with Small Organic Molecules. Angewandte Chemie International Edition. 2003;42(22):2462–2481. doi: 10.1002/anie.200200558. [DOI] [PubMed] [Google Scholar]
- 19.Veselovsky AV, et al. Protein–Protein Interactions: Mechanisms and Modification by Drugs. Journal of Molecular Recognition. 2002;15(6):405–422. doi: 10.1002/jmr.597. [DOI] [PubMed] [Google Scholar]
- 20.Gordo S, Giralt E. Knitting and Untying the Protein Network: Modulation of Protein Ensembles as a Therapeutic Strategy. Protein Science. 2009;18(3):481–493. doi: 10.1002/pro.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meireles LMC, Mustata G. Discovery of Modulators of Protein-Protein Interactions: Current Approaches and Limitations. Current Topics in Medicinal Chemistry. 2011;11(3):248–257. doi: 10.2174/156802611794072632. [DOI] [PubMed] [Google Scholar]
- 22.Toogood PL. Inhibition of Protein-Protein Association by Small Molecules: Approaches and Progress. Journal of Medicinal Chemistry. 2002;45(8):1543–1558. doi: 10.1021/jm010468s. [DOI] [PubMed] [Google Scholar]
- 23.Arkin MR, Whitty A. The Road Less Traveled: Modulating Signal Transduction Enzymes by Inhibiting Their Protein-Protein Interactions. Current Opinion in Chemical Biology. 2009;13(3):284–290. doi: 10.1016/j.cbpa.2009.05.125. [DOI] [PubMed] [Google Scholar]
- 24.Cheng AC, et al. Structure-Based Maximal Affinity Model Predicts Small-Molecule Druggability. Nature Biotechnology. 2007;25(1):71–75. doi: 10.1038/nbt1273. [DOI] [PubMed] [Google Scholar]
- 25.Lipinski CA, et al. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Advanced Drug Delivery Reviews. 2001;46(1–3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 26.Clackson T, Wells J. A Hot Spot of Binding Energy in a Hormone-Receptor Interface. Science. 1995;267(5196):383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 27.Erlanson DA, Wells JA, Braisted AC. Tethering: Fragment-Based Drug Discovery. Annual Review of Biophysics and Biomolecular Structure. 2004;33(1):199–223. doi: 10.1146/annurev.biophys.33.110502.140409. [DOI] [PubMed] [Google Scholar]
- 28.Valkov E, et al. Fragment-Based Drug Discovery and X-Ray Crystallography. Heidelberg: Springer Berlin; 2012. Targeting Protein–Protein Interactions and Fragment-Based Drug Discovery; pp. 145–179. [DOI] [PubMed] [Google Scholar]
- 29.Hopkins AL, Groom CR. The Druggable Genome. Nature Reviews Drug Discovery. 2002;1(9):727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 30.Jochim AL, Arora PS. Systematic Analysis of Helical Protein Interfaces Reveals Targets for Synthetic Inhibitors. Acs Chemical Biology. 2010;5(10):919–923. doi: 10.1021/cb1001747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walters WP, Murcko A, Murcko MA. Recognizing Molecules with Drug-Like Properties. Current Opinion in Chemical Biology. 1999;3(4):384–387. doi: 10.1016/s1367-5931(99)80058-1. [DOI] [PubMed] [Google Scholar]
- 32.Schreiber SL. Target-Oriented and Diversity-Oriented Organic Synthesis in Drug Discovery. Science. 2000;287(5460):1964–1969. doi: 10.1126/science.287.5460.1964. [DOI] [PubMed] [Google Scholar]
- 33.Xu Y, et al. A Credit-Card Library Approach for Disrupting Protein-Protein Interactions. Bioorganic & Medicinal Chemistry. 2006;14(8):2660–2673. doi: 10.1016/j.bmc.2005.11.052. [DOI] [PubMed] [Google Scholar]
- 34.Reayi A, Arya P. Natural Product-Like Chemical Space: Search for Chemical Dissectors of Macromolecular Interactions. Current Opinion in Chemical Biology. 2005;9(3):240–247. doi: 10.1016/j.cbpa.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Woodman R, et al. Design and Validation of a Neutral Protein Scaffold for the Presentation of Peptide Aptamers. Journal of Molecular Biology. 2005;352(5):1118–1133. doi: 10.1016/j.jmb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 36.Kritzer JA, et al. Miniature Protein Inhibitors of the P53–Hdm2 Interaction. ChemBioChem. 2006;7(1):29–31. doi: 10.1002/cbic.200500324. [DOI] [PubMed] [Google Scholar]
- 37.Verdine GL, et al. Methods in Enzymology. Academic Press; 2012. Chapter One - Stapled Peptides for Intracellular Drug Targets; pp. 3–33. [DOI] [PubMed] [Google Scholar]
- 38.Walensky LD, et al. Activation of Apoptosis in Vivo by a Hydrocarbon-Stapled Bh3 Helix. Science. 2004;305(5689):1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, et al. A Cell-Penetrating Helical Peptide as a Potential Hiv-1 Inhibitor. Journal of Molecular Biology. 2008;378(3):565–580. doi: 10.1016/j.jmb.2008.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horswill AR, Savinov SN, Benkovic SJ. A Systematic Method for Identifying Small-Molecule Modulators of Protein-Protein Interactions. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(44):15591–15596. doi: 10.1073/pnas.0406999101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruvo M, et al. Branched Peptides for the Modulation of Protein-Protein Interactions: More Arms Are Better Than One? Current Medicinal Chemistry. 2011;18(16):2429–2437. doi: 10.2174/092986711795843191. [DOI] [PubMed] [Google Scholar]
- 42.Gestwicki JE, Crabtree GR, Graef IA. Harnessing Chaperones to Generate Small-Molecule Inhibitors of Amyloid B Aggregation. Science. 2004;306(5697):865–869. doi: 10.1126/science.1101262. [DOI] [PubMed] [Google Scholar]
- 43.Gestwicki JE, Marinec PS. Chemical Control over Protein-Protein Interactions: Beyond Inhibitors. Combinatorial Chemistry & High Throughput Screening. 2007;10(8):667–675. doi: 10.2174/138620707782507296. [DOI] [PubMed] [Google Scholar]
- 44.DeLano WL, et al. Convergent Solutions to Binding at a Protein-Protein Interface. Science. 2000;287(5456):1279–1283. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- 45.Sprinzak E, Altuvia Y, Margalit H. Characterization and Prediction of Protein-Protein Interactions within and between Complexes. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(40):14718–14723. doi: 10.1073/pnas.0603352103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lessard JA, Crabtree GR. Chromatin Regulatory Mechanisms in Pluripotency. In: Schekman RGLLR, editor. Annual Review of Cell and Developmental Biology. Vol 26. 2010. pp. 503–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hohfeld J, Cyr DM, Patterson C. From the Cradle to the Grave: Molecular Chaperones That May Choose between Folding and Degradation. Embo Reports. 2001;2(10):885–890. doi: 10.1093/embo-reports/kve206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen EA, et al. Specific-Inhibition of Herpesvirus Ribonucleotide Reductase by a Nonapeptide Derived from the Carboxy Terminus of Subunit-2. Nature. 1986;321(6068):441–443. doi: 10.1038/321441a0. [DOI] [PubMed] [Google Scholar]
- 49.Dutia BM, et al. Specific-Inhibition of Herpesvirus Ribonucleotide Reductase by Synthetic Peptides. Nature. 1986;321(6068):439–441. doi: 10.1038/321439a0. [DOI] [PubMed] [Google Scholar]
- 50.Bourgeas R, et al. Atomic Analysis of Protein-Protein Interfaces with Known Inhibitors: The 2p2i Database. Plos One. 2010;5(3) doi: 10.1371/journal.pone.0009598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Higueruelo AP, et al. Atomic Interactions and Profile of Small Molecules Disrupting Protein–Protein Interfaces: The Timbal Database. Chemical Biology & Drug Design. 2009;74(5):457–467. doi: 10.1111/j.1747-0285.2009.00889.x. [DOI] [PubMed] [Google Scholar]
- 52.Tilley JW, et al. Identification of a Small Molecule Inhibitor of the Il-2/Il-2rα Receptor Interaction Which Binds to Il-2. Journal of the American Chemical Society. 1997;119(32):7589–7590. [Google Scholar]
- 53.Raimundo BC, et al. Integrating Fragment Assembly and Biophysical Methods in the Chemical Advancement of Small-Molecule Antagonists of Il-2: An Approach for Inhibiting Protein-Protein Interactions. Journal of Medicinal Chemistry. 2004;47(12):3111–3130. doi: 10.1021/jm049967u. [DOI] [PubMed] [Google Scholar]
- 54.Thanos CD, DeLano WL, Wells JA. Hot-Spot Mimicry of a Cytokine Receptor by a Small Molecule. Proceedings of the National Academy of Sciences. 2006;103(42):15422–15427. doi: 10.1073/pnas.0607058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fry DC, et al. Nmr Structure of a Complex between Mdm2 and a Small Molecule Inhibitor. Journal of Biomolecular NMR. 2004;30(2):163–173. doi: 10.1023/B:JNMR.0000048856.84603.9b. [DOI] [PubMed] [Google Scholar]
- 56.Vassilev LT, et al. In Vivo Activation of the P53 Pathway by Small-Molecule Antagonists of Mdm2. Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 57.Ding K, et al. Structure-Based Design of Potent Non-Peptide Mdm2 Inhibitors. Journal of the American Chemical Society. 2005;127(29):10130–10131. doi: 10.1021/ja051147z. [DOI] [PubMed] [Google Scholar]
- 58.Shangary S, et al. Temporal Activation of P53 by a Specific Mdm2 Inhibitor Is Selectively Toxic to Tumors and Leads to Complete Tumor Growth Inhibition. Proceedings of the National Academy of Sciences. 2008;105(10):3933–3938. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang GP, et al. Structure-Based Design of Potent Small-Molecule Inhibitors of Anti-Apoptotic Bcl-2 Proteins. Journal of Medicinal Chemistry. 2006;49(21):6139–6142. doi: 10.1021/jm060460o. [DOI] [PubMed] [Google Scholar]
- 60.Bruncko M, et al. Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-Xl. Journal of Medicinal Chemistry. 2007;50(4):641–662. doi: 10.1021/jm061152t. [DOI] [PubMed] [Google Scholar]
- 61.Oltersdorf T, et al. An Inhibitor of Bcl-2 Family Proteins Induces Regression of Solid Tumours. Nature. 2005;435(7042):677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 62.Murray CW, Rees DC. The Rise of Fragment-Based Drug Discovery. Nat Chem. 2009;1(3):187–192. doi: 10.1038/nchem.217. [DOI] [PubMed] [Google Scholar]
- 63.Gorczynski MJ, et al. Allosteric Inhibition of the Protein-Protein Interaction between the Leukemia-Associated Proteins Runx1 and Cbf Beta. Chemistry & Biology. 2007;14(10):1186–1197. doi: 10.1016/j.chembiol.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 64.Grembecka J, et al. Menin-Mll Inhibitors Reverse Oncogenic Activity of Mll Fusion Proteins in Leukemia. Nature Chemical Biology. 2012;8(3):277–284. doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frankel AD, Young JAT. Hiv-1: Fifteen Proteins and an Rna. Annual Review of Biochemistry. 1998;67(1):1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- 66.Weber IT. Comparison of the Crystal-Structures and Intersubunit Interactions of Human Immunodeficiency and Rous-Sarcoma Virus Proteases. Journal of Biological Chemistry. 1990;265(18):10492–10496. [PubMed] [Google Scholar]
- 67.Zhang ZY, et al. Dissociative Inhibition of Dimeric Enzymes - Kinetic Characterization of the Inhibition of Hiv-1 Protease by Its Cooh-Terminal Tetrapeptide. Journal of Biological Chemistry. 1991;266(24):15591–15594. [PubMed] [Google Scholar]
- 68.Ding J, et al. Buried Surface Analysis of Hiv-1 Reverse Transcriptase P66/P51 Heterodimer and Its Interaction with Dsdna Template/Primer. Journal of Molecular Recognition. 1994;7(2):157–161. doi: 10.1002/jmr.300070212. [DOI] [PubMed] [Google Scholar]
- 69.Divita G, Restle T, Goody RS. Characterization of the Dimerization Process of Hiv-1 Reverse-Transcriptase Heterodimer Using Intrinsic Protein Fluorescence. Febs Letters. 1993;324(2):153–158. doi: 10.1016/0014-5793(93)81383-b. [DOI] [PubMed] [Google Scholar]
- 70.Fan X, Flentke GR, Rich DH. Inhibition of Hiv-1 Protease by a Subunit of Didemnaketal A. Journal of the American Chemical Society. 1998;120(34):8893–8894. [Google Scholar]
- 71.Quere L, Wenger T, Schramm HJ. Triterpenes as Potential Dimerization Inhibitors of Hiv-1 Protease. Biochemical and Biophysical Research Communications. 1996;227(2):484–488. doi: 10.1006/bbrc.1996.1533. [DOI] [PubMed] [Google Scholar]
- 72.Balzarini J, et al. 2',5'-Bis-O-(Tert-Butyldimethylsilyl)-3'-Spiro-5"-(4"-Amino-1",2"-Oxathiole-2",2'-Dioxide)Pyrimidine (Tsao) Nucleoside Analogues: Highlyselective Inhibitors of Human Immunodeficiency Virus Type 1 That Are Targeted at the Viral Reverse Transcriptase. Proceedings of the National Academy of Sciences. 1992;89(10):4392–4396. doi: 10.1073/pnas.89.10.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bonache Ma-C, et al. Improving the Antiviral Efficacy and Selectivity of Hiv-1 Reverse Transcriptase Inhibitor Tsao-T by the Introduction of Functional Groups at the N-3 Position. Journal of Medicinal Chemistry. 2005;48(21):6653–6660. doi: 10.1021/jm050437n. [DOI] [PubMed] [Google Scholar]
- 74.Graham TA, et al. Crystal Structure of a B-Catenin/Tcf Complex. Cell. 2000;103(6):885–896. doi: 10.1016/s0092-8674(00)00192-6. [DOI] [PubMed] [Google Scholar]
- 75.Lepourcelet M, et al. Small-Molecule Antagonists of the Oncogenic Tcf/B-Catenin Protein Complex. Cancer Cell. 2004;5(1):91–102. doi: 10.1016/s1535-6108(03)00334-9. [DOI] [PubMed] [Google Scholar]
- 76.Tian W, et al. Structure-Based Discovery of a Novel Inhibitor Targeting the B-Catenin/Tcf4 Interaction. Biochemistry. 2012;51(2):724–731. doi: 10.1021/bi201428h. [DOI] [PubMed] [Google Scholar]
- 77.Amin AR, Abramson SB. The Role of Nitric Oxide in Articular Cartilage Breakdown in Osteoarthritis. Current Opinion in Rheumatology. 1998;10(3):263–268. doi: 10.1097/00002281-199805000-00018. [DOI] [PubMed] [Google Scholar]
- 78.Nathan C, Xie QW. Nitric-Oxide Synthases - Roles, Tolls, and Controls. Cell. 1994;78(6):915–918. doi: 10.1016/0092-8674(94)90266-6. [DOI] [PubMed] [Google Scholar]
- 79.McMillan K, et al. Allosteric Inhibitors of Inducible Nitric Oxide Synthase Dimerization Discovered Via Combinatorial Chemistry. Proceedings of the National Academy of Sciences. 2000;97(4):1506–1511. doi: 10.1073/pnas.97.4.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee GM, Craik CS. Trapping Moving Targets with Small Molecules. Science. 2009;324(5924):213–215. doi: 10.1126/science.1169378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fieber W, et al. Structure, Function, and Dynamics of the Dimerization and DNA-Binding Domain of Oncogenic Transcription Factor V-Myc. Journal of Molecular Biology. 2001;307(5):1395–1410. doi: 10.1006/jmbi.2001.4537. [DOI] [PubMed] [Google Scholar]
- 82.Kiessling A, et al. Selective Inhibition of C-Myc/Max Dimerization and DNA Binding by Small Molecules. Chemistry & Biology. 2006;13(7):745–751. doi: 10.1016/j.chembiol.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 83.Silvian LF, et al. Small Molecule Inhibition of the Tnf Family Cytokine Cd40 Ligand through a Subunit Fracture Mechanism. Acs Chemical Biology. 2011;6(6):636–647. doi: 10.1021/cb2000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.He MM, et al. Small-Molecule Inhibition of Tnf-Alpha. Science. 2005;310(5750):1022–1025. doi: 10.1126/science.1116304. [DOI] [PubMed] [Google Scholar]
- 85.Magliery TJ, et al. Detecting Protein-Protein Interactions with a Green Fluorescent Protein Fragment Reassembly Trap: Scope and Mechanism. Journal of the American Chemical Society. 2005;127(1):146–157. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- 86.Liu B, et al. Label Transfer Chemistry for the Characterization of Protein-Protein Interactions. Journal of the American Chemical Society. 2007;129(41):12348–12349. doi: 10.1021/ja072904r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heeres JT, et al. Identifying Modulators of Protein-Protein Interactions Using Photonic Crystal Biosensors. Journal of the American Chemical Society. 2009;131(51):18202–18203. doi: 10.1021/ja907066r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Porter JR, et al. A General and Rapid Cell-Free Approach for the Interrogation of Protein-Protein, Protein-DNA, and Protein-Rna Interactions and Their Antagonists Utilizing Split-Protein Reporters. Journal of the American Chemical Society. 2008;130(20):6488–6497. doi: 10.1021/ja7114579. [DOI] [PubMed] [Google Scholar]
- 89.Moerke NJ, et al. Small-Molecule Inhibition of the Interaction between the Translation Initiation Factors Eif4e and Eif4g. Cell. 2007;128(2):257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 90.Roman DL, et al. Identification of Small-Molecule Inhibitors of Rgs4 Using a High-Throughput Flow Cytometry Protein Interaction Assay. Molecular Pharmacology. 2007;71(1):169–175. doi: 10.1124/mol.106.028670. [DOI] [PubMed] [Google Scholar]
- 91.Potashman MH, Duggan ME. Covalent Modifiers: An Orthogonal Approach to Drug Design. Journal of Medicinal Chemistry. 2009;52(5):1231–1246. doi: 10.1021/jm8008597. [DOI] [PubMed] [Google Scholar]
- 92.Reinke AA, Gestwicki JE. Insight into Amyloid Structure Using Chemical Probes. Chemical Biology & Drug Design. 2011;77(6):399–411. doi: 10.1111/j.1747-0285.2011.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Eisenberg D, Jucker M. The Amyloid State of Proteins in Human Diseases. Cell. 2012;148(6):1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Findeis MA. Approaches to Discovery and Characterization of Inhibitors of Amyloid Beta-Peptide Polymerization. Biochimica Et Biophysica Acta-Molecular Basis of Disease. 2000;1502(1):76–84. doi: 10.1016/s0925-4439(00)00034-x. [DOI] [PubMed] [Google Scholar]
- 95.Lee VMY. Amyloid Binding Ligands as Alzheimer's Disease Therapies. Neurobiology of Aging. 2002;23(6):1039–1042. doi: 10.1016/s0197-4580(02)00121-5. [DOI] [PubMed] [Google Scholar]
- 96.Bose M, et al. 'Nature-Inspired' Drug-Protein Complexes as Inhibitors of a Beta Aggregation. Biochemical Society Transactions. 2005;33:543–547. doi: 10.1042/BST0330543. [DOI] [PubMed] [Google Scholar]
- 97.Roberts BE, et al. A Synergistic Small-Molecule Combination Directly Eradicates Diverse Prion Strain Structures. Nat Chem Biol. 2009;5(12):936–946. doi: 10.1038/nchembio.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schmid EM, McMahon HT. Integrating Molecular and Network Biology to Decode Endocytosis. Nature. 2007;448(7156):883–888. doi: 10.1038/nature06031. [DOI] [PubMed] [Google Scholar]
- 99.von Kleist L, et al. Role of the Clathrin Terminal Domain in Regulating Coated Pit Dynamics Revealed by Small Molecule Inhibition. Cell. 2011;146(3):471–484. doi: 10.1016/j.cell.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 100.Lee LW, Mapp AK. Transcriptional Switches: Chemical Approaches to Gene Regulation. Journal of Biological Chemistry. 2010;285(15):11033–11038. doi: 10.1074/jbc.R109.075044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Buhrlage SJ, et al. Amphipathic Small Molecules Mimic the Binding Mode and Function of Endogenous Transcription Factors. Acs Chemical Biology. 2009;4(5):335–344. doi: 10.1021/cb900028j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Casey RJ, et al. Expanding the Repertoire of Small Molecule Transcriptional Activation Domains. Bioorganic & Medicinal Chemistry. 2009;17(3):1034–1043. doi: 10.1016/j.bmc.2008.02.045. [DOI] [PubMed] [Google Scholar]
- 103.Boriack-Sjodin PA, et al. The Structural Basis of the Activation of Ras by Sos. Nature. 1998;394(6691):337–343. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- 104.Sondermann H, et al. Structural Analysis of Autoinhibition in the Ras Activator Son of Sevenless. Cell. 2004;119(3):393–405. doi: 10.1016/j.cell.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 105.Patgiri A, et al. An Orthosteric Inhibitor of the Ras-Sos Interaction. Nat Chem Biol. 2011;7(9):585–587. doi: 10.1038/nchembio.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Maurer T, et al. Small-Molecule Ligands Bind to a Distinct Pocket in Ras and Inhibit Sos-Mediated Nucleotide Exchange Activity. Proceedings of the National Academy of Sciences. 2012;109(14):5299–5304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kiessling LL, Gestwicki JE, Strong LE. Synthetic Multivalent Ligands as Probes of Signal Transduction. Angewandte Chemie International Edition. 2006;45(15):2348–2368. doi: 10.1002/anie.200502794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Evans CG, Chang L, Gestwicki JE. Heat Shock Protein 70 (Hsp70) as an Emerging Drug Target. Journal of Medicinal Chemistry. 2010;53(12):4585–4602. doi: 10.1021/jm100054f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Patury S, Miyata Y, Gestwicki JE. Pharmacological Targeting of the Hsp70 Chaperone. Current Topics in Medicinal Chemistry. 2009;9(15):1337–1351. doi: 10.2174/156802609789895674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Brodsky JL, Chiosis G. Hsp70 Molecular Chaperones: Emerging Roles in Human Disease and Identification of Small Molecule Modulators. Current Topics in Medicinal Chemistry. 2006;6(11):1215–1225. doi: 10.2174/156802606777811997. [DOI] [PubMed] [Google Scholar]
- 111.Brandt GEL, Blagg BSJ. Alternate Strategies of Hsp90 Modulation for the Treatment of Cancer and Other Diseases. Current Topics in Medicinal Chemistry. 2009;9(15):1447–1461. doi: 10.2174/156802609789895683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Powers MV, Workman P. Inhibitors of the Heat Shock Response: Biology and Pharmacology. Febs Letters. 2007;581(19):3758–3769. doi: 10.1016/j.febslet.2007.05.040. [DOI] [PubMed] [Google Scholar]
- 113.Hartl FU, Hayer-Hartl M. Converging Concepts of Protein Folding in Vitro and in Vivo. Nat Struct Mol Biol. 2009;16(6):574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- 114.Meimaridou E, Gooljar SB, Chapple JP. From Hatching to Dispatching: The Multiple Cellular Roles of the Hsp70 Molecular Chaperone Machinery. Journal of Molecular Endocrinology. 2009;42(1–2):1–9. doi: 10.1677/JME-08-0116. [DOI] [PubMed] [Google Scholar]
- 115.Massey AJ. Atpases as Drug Targets: Insights from Heat Shock Proteins 70 and 90. Journal of Medicinal Chemistry. 2010;53(20):7280–7286. doi: 10.1021/jm100342z. [DOI] [PubMed] [Google Scholar]
- 116.Miyata Y, et al. Molecular Chaperones and Regulation of Tau Quality Control: Strategies for Drug Discovery in Tauopathies. Future Medicinal Chemistry. 2011;3(12):1523–1537. doi: 10.4155/fmc.11.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kampinga HH, Craig EA. The Hsp70 Chaperone Machinery: J Proteins as Drivers of Functional Specificity. Nature Reviews Molecular Cell Biology. 2010;11(8):579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ahmad A, et al. Heat Shock Protein 70 Kda Chaperone/Dnaj Cochaperone Complex Employs an Unusual Dynamic Interface. Proceedings of the National Academy of Sciences. 2011;108(47):18966–18971. doi: 10.1073/pnas.1111220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jiang J, et al. Structural Basis of J Cochaperone Binding and Regulation of Hsp70. Molecular cell. 2007;28(3):422–433. doi: 10.1016/j.molcel.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Harrison CJ, et al. Crystal Structure of the Nucleotide Exchange Factor Grpe Bound to the Atpase Domain of the Molecular Chaperone Dnak. Science. 1997;276(5311):431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- 121.Schmid AB, et al. The Architecture of Functional Modules in the Hsp90 Co-Chaperone Sti1/Hop. EMBO J. 2012;31(6):1506–1517. doi: 10.1038/emboj.2011.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Scheufler C, et al. Structure of Tpr Domain-Peptide Complexes: Critical Elements in the Assembly of the Hsp70–Hsp90 Multichaperone Machine. Cell. 2000;101(2):199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 123.Wisen S, et al. Binding of a Small Molecule at a Protein-Protein Interface Regulates the Chaperone Activity of Hsp70-Hsp40. Acs Chemical Biology. 2010;5(6):611–622. doi: 10.1021/cb1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chang L, et al. Chemical Screens against a Reconstituted Multiprotein Complex: Myricetin Blocks Dnaj Regulation of Dnak through an Allosteric Mechanism. Chemistry & Biology. 2011;18(2):210–221. doi: 10.1016/j.chembiol.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bhattacharya A, et al. Allostery in Hsp70 Chaperones Is Transduced by Subdomain Rotations. Journal of Molecular Biology. 2009;388(3):475–490. doi: 10.1016/j.jmb.2009.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chang L, et al. High-Throughput Screen for Small Molecules That Modulate the Atpase Activity of the Molecular Chaperone Dnak. Analytical Biochemistry. 2008;372(2):167–176. doi: 10.1016/j.ab.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 127.Miyata Y, et al. High-Throughput Screen for Escherichia Coli Heat Shock Protein 70 (Hsp70/Dnak): Atpase Assay in Low Volume by Exploiting Energy Transfer. Journal of Biomolecular Screening. 2010;15(10):1211–1219. doi: 10.1177/1087057110380571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Leu JIJ, et al. A Small Molecule Inhibitor of Inducible Heat Shock Protein 70. Molecular cell. 2009;36(1):15–27. doi: 10.1016/j.molcel.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li J, Soroka J, Buchner J. The Hsp90 Chaperone Machinery: Conformational Dynamics and Regulation by Co-Chaperones. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2012;1823(3):624–635. doi: 10.1016/j.bbamcr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 130.Kamal A, Boehm MF, Burrows FJ. Therapeutic and Diagnostic Implications of Hsp90 Activation. Trends in molecular medicine. 2004;10(6):283–290. doi: 10.1016/j.molmed.2004.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Whitesell L, Lindquist SL. Hsp90 and the Chaperoning of Cancer. Nat Rev Cancer. 2005;5(10):761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 132.Porter JR, Fritz CC, Depew KM. Discovery and Development of Hsp90 Inhibitors: A Promising Pathway for Cancer Therapy. Current Opinion in Chemical Biology. 2010;14(3):412–420. doi: 10.1016/j.cbpa.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 133.Bagatell R, et al. Induction of a Heat Shock Factor 1-Dependent Stress Response Alters the Cytotoxic Activity of Hsp90-Binding Agents. Clinical Cancer Research. 2000;6(8):3312–3318. [PubMed] [Google Scholar]
- 134.Donnelly A, Blagg BSJ. Novobiocin and Additional Inhibitors of the Hsp90 C-Terminal Nucleotide-Binding Pocket. Current Medicinal Chemistry. 2008;15(26):2702–2717. doi: 10.2174/092986708786242895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sreeramulu S, et al. The Human Cdc37.Hsp90 Complex Studied by Heteronuclear Nmr Spectroscopy. Journal of Biological Chemistry. 2009;284(6):3885–3896. doi: 10.1074/jbc.M806715200. [DOI] [PubMed] [Google Scholar]
- 136.Meyer P, et al. Structural Basis for Recruitment of the Atpase Activator Aha1 to the Hsp90 Chaperone Machinery. EMBO J. 2004;23(3):511–519. doi: 10.1038/sj.emboj.7600060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Siligardi G, et al. Co-Chaperone Regulation of Conformational Switching in the Hsp90 Atpase Cycle. Journal of Biological Chemistry. 2004;279(50):51989–51998. doi: 10.1074/jbc.M410562200. [DOI] [PubMed] [Google Scholar]
- 138.Zhang T, et al. Characterization of Celastrol to Inhibit Hsp90 and Cdc37 Interaction. Journal of Biological Chemistry. 2009;284(51):35381–35389. doi: 10.1074/jbc.M109.051532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sreeramulu S, et al. Molecular Mechanism of Inhibition of the Human Protein Complex Hsp90–Cdc37, a Kinome Chaperone–Cochaperone, by Triterpene Celastrol. Angewandte Chemie International Edition. 2009;48(32):5853–5855. doi: 10.1002/anie.200900929. [DOI] [PubMed] [Google Scholar]
- 140.Ardi VC, et al. Macrocycles That Inhibit the Binding between Heat Shock Protein 90 and Tpr-Containing Proteins. Acs Chemical Biology. 2011;6(12):1357–1366. doi: 10.1021/cb200203m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yi F, Regan L. A Novel Class of Small Molecule Inhibitors of Hsp90. Acs Chemical Biology. 2008;3(10):645–654. doi: 10.1021/cb800162x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gandhi PS, et al. Structural Identification of the Pathway of Long-Range Communication in an Allosteric Enzyme. Proceedings of the National Academy of Sciences. 2008;105(6):1832–1837. doi: 10.1073/pnas.0710894105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Motlagh HN, Hilser VJ. Agonism/Antagonism Switching in Allosteric Ensembles. Proceedings of the National Academy of Sciences. 2012;109(11):4134–4139. doi: 10.1073/pnas.1120519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chari A, Fischer U. Cellular Strategies for the Assembly of Molecular Machines. Trends in Biochemical Sciences. 2010;35(12):676–683. doi: 10.1016/j.tibs.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 145.Peterson-Kaufman KJ, et al. Nucleating the Assembly of Macromolecular Complexes. ChemBioChem. 2010;11(14):1955–1962. doi: 10.1002/cbic.201000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Good MC, Zalatan JG, Lim WA. Scaffold Proteins: Hubs for Controlling the Flow of Cellular Information. Science. 2011;332(6030):680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cardinale D, et al. Homodimeric Enzymes as Drug Targets. Current Medicinal Chemistry. 2010;17(9):826–846. doi: 10.2174/092986710790712156. [DOI] [PubMed] [Google Scholar]
- 148.Panda K, et al. Distinct Dimer Interaction and Regulation in Nitric-Oxide Synthase Types I, Ii, and Iii. Journal of Biological Chemistry. 2002;277(34):31020–31030. doi: 10.1074/jbc.M203749200. [DOI] [PubMed] [Google Scholar]
- 149.Knapp S, et al. Thermodynamics of the High-Affinity Interaction of Tcf4 with B-Catenin. Journal of Molecular Biology. 2001;306(5):1179–1189. doi: 10.1006/jmbi.2001.4463. [DOI] [PubMed] [Google Scholar]
- 150.Rickert M, et al. The Structure of Interleukin-2 Complexed with Its Alpha Receptor. Science. 2005;308(5727):1477–1480. doi: 10.1126/science.1109745. [DOI] [PubMed] [Google Scholar]
- 151.Braisted AC, et al. Discovery of a Potent Small Molecule Il-2 Inhibitor through Fragment Assembly. Journal of the American Chemical Society. 2003;125(13):3714–3715. doi: 10.1021/ja034247i. [DOI] [PubMed] [Google Scholar]
- 152.Yu GW, et al. The Central Region of Hdm2 Provides a Second Binding Site for P53. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(5):1227–1232. doi: 10.1073/pnas.0510343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kussie PH, et al. Structure of the Mdm2 Oncoprotein Bound to the P53 Tumor Suppressor Transactivation Domain. Science. 1996;274(5289):948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 154.Grasberger BL, et al. Discovery and Cocrystal Structure of Benzodiazepinedione Hdm2 Antagonists That Activate P53 in Cells. Journal of Medicinal Chemistry. 2005;48(4):909–912. doi: 10.1021/jm049137g. [DOI] [PubMed] [Google Scholar]
- 155.Sattler M, et al. Structure of Bcl-X(L)-Bak Peptide Complex: Recognition between Regulators of Apoptosis. Science. 1997;275(5302):983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 156.Tang G, et al. Pyrogallol-Based Molecules as Potent Inhibitors of the Antiapoptotic Bcl-2 Proteins. Journal of Medicinal Chemistry. 2007;50(8):1723–1726. doi: 10.1021/jm061400l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Abbate EA, Berger JM, Botchan MR. The X-Ray Structure of the Papillomavirus Helicase in Complex with Its Molecular Matchmaker E2. Genes & Development. 2004;18(16):1981–1996. doi: 10.1101/gad.1220104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang Y, et al. Crystal Structure of the E2 Transactivation Domain of Human Papillomavirus Type 11 Bound to a Protein Interaction Inhibitor. Journal of Biological Chemistry. 2004;279(8):6976–6985. doi: 10.1074/jbc.M311376200. [DOI] [PubMed] [Google Scholar]
- 159.Edeling MA, Smith C, Owen D. Life of a Clathrin Coat: Insights from Clathrin and Ap Structures. Nat Rev Mol Cell Biol. 2006;7(1):32–44. doi: 10.1038/nrm1786. [DOI] [PubMed] [Google Scholar]
- 160.Miele AE, et al. Two Distinct Interaction Motifs in Amphiphysin Bind Two Independent Sites on the Clathrin Terminal Domain [Beta]-Propeller. Nat Struct Mol Biol. 2004;11(3):242–248. doi: 10.1038/nsmb736. [DOI] [PubMed] [Google Scholar]
- 161.Siligardi G, et al. Regulation of Hsp90 Atpase Activity by the Co-Chaperone Cdc37p/P50 Cdc37. Journal of Biological Chemistry. 2002;277(23):20151–20159. doi: 10.1074/jbc.M201287200. [DOI] [PubMed] [Google Scholar]
- 162.Mosyak L, et al. The Bacterial Cell-Division Protein Zipa and Its Interaction with an Ftsz Fragment Revealed by X-Ray Crystallography. EMBO J. 2000;19(13):3179–3191. doi: 10.1093/emboj/19.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rush TS, et al. A Shape-Based 3-D Scaffold Hopping Method and Its Application to a Bacterial Protein, Àíprotein Interaction. Journal of Medicinal Chemistry. 2005;48(5):1489–1495. doi: 10.1021/jm040163o. [DOI] [PubMed] [Google Scholar]
- 164.Tesmer JJG, et al. Structure of Rgs4 Bound to Alf4-Activated Gia1: Stabilization of the Transition State for Gtp Hydrolysis. Cell. 1997;89(2):251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- 165.Fernandez-Fernandez MR, Veprintsev DB, Fersht AR. Proteins of the S100 Family Regulate the Oligomerization of P53 Tumor Suppressor. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(13):4735–4740. doi: 10.1073/pnas.0501459102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Rust RR, Baldisseri DM, Weber DJ. Structure of the Negative Regulatory Domain of P53 Bound to S100b(Bβ) Nat Struct Mol Biol. 2000;7(7):570–574. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- 167.Markowitz J, et al. Identification and Characterization of Small Molecule Inhibitors of the Calcium-Dependent S100b-P53 Tumor Suppressor Interaction. Journal of Medicinal Chemistry. 2004;47(21):5085–5093. doi: 10.1021/jm0497038. [DOI] [PubMed] [Google Scholar]
- 168.Mukai Y, et al. Solution of the Structure of the Tnf-Tnfr2 Complex. Sci. Signal. 2010;3(148) doi: 10.1126/scisignal.2000954. ra83-. [DOI] [PubMed] [Google Scholar]
- 169.Aggarwal BB, Eessalu TE, Hass PE. Characterization of Receptors for Human Tumour Necrosis Factor and Their Regulation by Γ-Interferon. Nature. 1985;318(6047):665–667. doi: 10.1038/318665a0. [DOI] [PubMed] [Google Scholar]
- 170.Pilger BD, Cui C, Coen DM. Identification of a Small Molecule That Inhibits Herpes Simplex Virus DNA Polymerase Subunit Interactions and Viral Replication. Chemistry & Biology. 2004;11(5):647–654. doi: 10.1016/j.chembiol.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 171.Vucic D, et al. Engineering Ml-Iap to Produce an Extraordinarily Potent Caspase 9 Inhibitor: Implications for Smac-Dependent Anti-Apoptotic Activity of Ml-Iap. Biochemical Journal. 2005;385:11–20. doi: 10.1042/BJ20041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shiozaki EN, et al. Mechanism of Xiap-Mediated Inhibition of Caspase-9. Molecular Cell. 2003;11(2):519–527. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 173.Oost TK, et al. Discovery of Potent Antagonists of the Antiapoptotic Protein Xiap for the Treatment of Cancer. Journal of Medicinal Chemistry. 2004;47(18):4417–4426. doi: 10.1021/jm040037k. [DOI] [PubMed] [Google Scholar]
- 174.Cherepanov P, et al. Structural Basis for the Recognition between Hiv-1 Integrase and Transcriptional Coactivator P75. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(48):17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tsiang M, et al. Affinities between the Binding Partners of the Hiv-1 Integrase Dimer-Lens Epithelium-Derived Growth Factor (in Dimer-Ledgf) Complex. Journal of Biological Chemistry. 2009;284(48):33580–33599. doi: 10.1074/jbc.M109.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Christ F, et al. Rational Design of Small-Molecule Inhibitors of the Ledgf/P75-Integrase Interaction and Hiv Replication. Nat Chem Biol. 2010;6(6):442–448. doi: 10.1038/nchembio.370. [DOI] [PubMed] [Google Scholar]
- 177.Nair SK, Burley SK. X-Ray Structures of Myc-Max and Mad-Max Recognizing DNA: Molecular Bases of Regulation by Proto-Oncogenic Transcription Factors. Cell. 2003;112(2):193–205. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- 178.Hu J, Banerjee A, Goss DJ. Assembly of B/Hlh/Z Proteins C-Myc, Max, Mad1 with Cognate DNA: Importance of Protein-Protein and Protein-DNA Interactions. Biochemistry. 2005;44(35):11855–11863. doi: 10.1021/bi050206i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Stamper CC, et al. Crystal Structure of the B7-1/Ctla-4 Complex That Inhibits Human Immune Responses. Nature. 2001;410(6828):608–611. doi: 10.1038/35069118. [DOI] [PubMed] [Google Scholar]
- 180.Erbe DV, et al. Small Molecule Ligands Define a Binding Site on the Immune Regulatory Protein B7.1. Journal of Biological Chemistry. 2002;277(9):7363–7368. doi: 10.1074/jbc.M110162200. [DOI] [PubMed] [Google Scholar]
Further Reading
- Bourgeas R, et al. Atomic Analysis of Protein-Protein Interfaces with Known Inhibitors: The 2p2i Database. PLoS ONE. 2010;5(3):e9598. doi: 10.1371/journal.pone.0009598. http://2p2idb.cnrs-mrs.fr/ This article and accompanying website detail the creation of a database compiling structural information of PPIs with known inhibitors.
- Higueruelo AP, et al. Atomic Interactions and Profile of Small Molecules Disrupting Protein–Protein Interfaces: The Timbal Database. Chemical Biology & Drug Design. 2009;74(5):457–467. doi: 10.1111/j.1747-0285.2009.00889.x. http://www-cryst.bioc.cam.ac.uk/databases/timbal. This article and accompanying website describe a database of the molecular properties of PPIs with known inhibitors.
- Drewry DH, Macarron R. Enhancements of Screening Collections to Address Areas of Unmet Medical Need: An Industry Perspective. Current Opinion in Chemical Biology. 2010;14(3):289–298. doi: 10.1016/j.cbpa.2010.03.024. This review further describes the need to develop and maintain appropriate tools to successfully target historically “undruggable” targets like PPIs.
- Uversky VN. Intrinsically Disordered Proteins and Novel Strategies for Drug Discovery. Expert Opinion on Drug Discovery. 2012;7(6):475–488. doi: 10.1517/17460441.2012.686489. This comprehensive review describes efforts in targeting intrinsically disordered proteins. This class of proteins share many similar characteristics with PPIs, and have proven to be similarly intractable for drug development.
- Jubb H, et al. Structural Biology and Drug Discovery for Protein-Protein Interactions. Trends in Pharmacological Sciences. 2012;33(5):241–248. doi: 10.1016/j.tips.2012.03.006. This recent review describes methods for developing small molecule modulators of PPIs with available structural information. The authors focus on fragment-based approaches that have become valuable tools for discovering de novo binding sites at these interaction surfaces.
- Verdine GL, et al. Methods in Enzymology. Academic Press; 2012. Chapter One - Stapled Peptides for Intracellular Drug Targets; pp. 3–33. This recent chapter describes the dense body of work related to developing cell permeable, stapled peptides as inhibitors of intracellular PPIs; a topic that was regrettably outside the scope of the current review.