Summary
Caspases, a family of apoptotic proteases, are increasingly recognized as being extensively phosphorylated, usually leading to inactivation. To date, no structural mechanism for phosphorylation-based caspase inactivation is available, although this information may be key to achieving caspase-specific inhibition. Caspase-6 has recently been implicated in neurodegenerative conditions including Huntington's and Alzheimer's diseases. A full understanding of caspase-6 regulation is crucial to caspase-6-specific inhibition. Caspase-6 is phosphorylated by ARK5 kinase at serine 257 leading to suppression of cell death via caspase-6 inhibition. Our structure of the fully inactive phosphomimetic S257D reveals that phosphorylation results in a steric clash with P201 in the L2′ loop. Removal of the proline side chain alleviates the clash resulting in nearly wild-type activity levels. This phosphomimetic-mediated steric clash causes misalignment of the substrate-binding groove, preventing substrate binding. Substrate-binding loop misalignment appears to be a widely used regulatory strategy among caspases and may present a new paradigm for caspase-specific control.
Introduction
Caspases are a family of cysteine proteases with exquisite specificity for cleaving after aspartate residues. Caspases were originally identified for orchestrating the complex process of apoptotic programmed cell death. Caspases have since been found to play a variety of roles in inflammation, differentiation, and development. Apoptotic caspases are categorized as initiator (upstream, caspase-2, -8, and -9) or executioner (downstream, caspase-3, -6 and -7). Among the apoptotic caspases, categorization of caspase-6 was historically the most ambiguous. Caspase-6 is weakly apoptotic, although overexpression of caspase-6 does result in apoptosis (Suzuki et al., 2004) similarly to caspase-3 and -7, which are mutually homologous to caspase-6 (∼37% sequence identity). Caspase-6 has been reported to act upstream of the initiator caspase-8 (Cowling and Downward, 2002) and to be activated by the inflammatory caspase-1 (Guo et al., 2006), which would set it outside of the traditional executioner caspase category. Together, these observations suggest that caspase-6 functions in a diversity of cellular processes, rendering categorization of caspase-6 complex and ambiguous.
Caspase-6 is now known to play important roles in several neurodegenerative diseases. Caspase-6 cleaves amyloid precursor protein at position D664 leading to production of the toxic APP-C31 fragment. Mice in which this caspase-6 cleavage site is blocked are protected from development of the symptoms of Alzheimer's disease (Galvan et al., 2006), suggesting a causal role for caspase activity. Likewise, cleavage of the polyglutamine-expanded Huntingtin protein at a site recognized by caspase-6 (D586), but not by caspase-3, is required for the development of the behavioral and neuropathological features of Huntington's disease. Substitution of the caspase-6 site in mice provides protection from the neural dysfunction, suggesting a causal relationship between caspase-6 activity and Huntington's disease (Graham et al., 2006). Thus, significant interest exists in full elucidation of the caspase-6 structure, function, and particularly its regulation, which together give rise to both apoptotic and neurodegenerative roles.
Due to their cell-death inducing potential, the activities of all apoptotic caspases are tightly regulated. The regulation of each caspase is necessarily unique so that each can perform its independent and nonredundant cellular roles. The most prevalent mechanisms of caspase regulation include zymogen activation, binding of inhibitor of apoptosis protein (IAP) family members, and posttranslational modification. The intersection of these regulatory pathways may offer especially good therapeutic targets.
Many proteases are controlled via zymogen activation, in which proteolytic processing results in the generation of an active protease. Caspase zymogens (also called procaspases) are maintained in a full-length, unprocessed, and inactive form prior to induction of apoptosis or other activating cellular stimuli. Processing by an upstream caspase removes a prodomain and cleaves an intersubunit linker. The mature, active caspase is a dimer of dimers, composed of two large and two small subunits. Processing of executioner caspases renders them active and competent to cleave cellular targets, resulting in apoptosis. Cleavage of the intersubunit linker produces two nascent loops that participate in a four-loop substrate-binding groove bundle. These loops are known to be extremely mobile. In a number of caspase structures these loops become disordered in the absence of substrate. Locking one of these loops (L2′) into a zymogen-like (down) conformation results in allosteric inhibition (Hardy et al., 2004; Hardy and Wells, 2009) and can be used to exogenously inactivate caspase-7 (Witkowski and Hardy, 2011). When L2′ is in the down conformation it does not support the L2 loop and as a consequence none of the loops in the substrate-binding groove bundle (L2, L3, L4, or L2′) attain the proper conformation. In caspase-6 the loops are capable of a strand-to-helix transition that results in a unique conformation (Baumgartner et al., 2009; Vaidya et al., 2011), which can not be obtained by any other caspase (Vaidya and Hardy, 2011). Caspase-6 is also the only caspase for which autoactivation has been confirmed and a mechanism of autoactivation has been elucidated (Wang et al., 2010). In terms of both zymogen activation and loop control, caspase-6 is distinct from all other caspases.
Apoptotic caspases are generally regulated by the IAP family of inhibitors. The most well studied, XIAP, inhibits caspase-3 and -7, caspase-6's closest homologs, by binding to the active site (Chai et al., 2001; Scott et al., 2005). XIAP also inhibits caspase-9, but does so by blocking self dimerization rather than active-site binding (Shiozaki et al., 2003). Curiously, caspase-6 is the only apoptotic executioner caspase resistant to XIAP-mediated inhibition at either the active site or the dimer interface (Deveraux et al., 1997; Stennicke et al., 2002). This could be because caspase-6 exists in the extended helical conformation or because the intersubunit linker blocks XIAP active site binding.
Another mechanism of caspase regulation is via posttranslational modification. Phosphorylation is emerging as the most critical and prevalent form of posttranslational modification in caspases (for review see Kurokawa and Kornbluth, 2009). Phosphorylation was first reported to inactivate caspase-9 (Cardone et al., 1998), but has now been discovered to regulate caspase-2, -3, -6, -7, -8, and -9, although the structural details of how phosphorylation events activate or inactivate caspases remain to be discovered. Only a single phosphorylation site has been reported for caspase-6. Caspase-6 is phosphorylated at S257 by the kinase ARK5 (Suzuki et al., 2004). ARK5-mediated phosphorylation results in inactivation of caspase-6, which is sufficient to suppress Akt-dependent cell death mediated by the death receptor complex. This single phosphorylation site in caspase-6 may present an opportunity for understanding phosphorylation-mediated inhibition of caspases generally.
Phosphorylation occurs in all structural domains of caspases and typically results in caspase inactivation. Phosphorylation of the sites of zymogen processing blocks activity by preventing formation of the active, cleaved caspase (Duncan et al., 2011). To date no structural information is available to explain how phosphorylation of other caspase domains results in inactivation. It remains to be determined whether similar mechanisms of phosphorylation-based inhibition will emerge or whether phosphorylation at each site functions using independent mechanisms. In this study we uncover the structural mechanism by which caspase-6 phosphorylation results in inhibition.
Results
S257 Phosphomimetics Inactivate Caspase-6
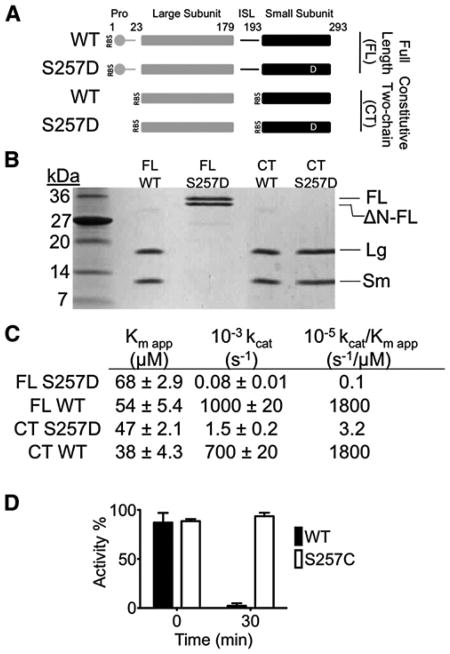
Phosphorylation of capase-6 at residue serine 257 is reported to inactivate the protein (Suzuki et al., 2004). To study this effect we made a phosphomimetic version of caspase-6 in which serine 257 is substituted by aspartate (S257D, Figure 1A). Substitution of serine by aspartate or glutamate often mimics the effects of serine phosphorylation due to the introduction of a negative charge equidistant from the protein backbone. Caspase-6 is the only caspase for which a molecular mechanism of autoactivation in cis has been reported (Klaiman et al., 2009), although other caspases are activated during overexpression in Escherichia coli, presumably by cleavage in trans. Expression of full-length (FL) caspase-6 results in self-processing, and formation of two cleaved fragments, the large and small subunits that together form active caspase-6 (Figure 1B). Cleavage of the intersubunit linker, which generates the two-chain form, is essential for caspase-6 activity (Vaidya et al., 2011). As expected for an uncleaved zymogen, FL S257D was inactive relative to the mature wild-type protein (Figure 1C). Two other S257 variants were also tested for autoactivation to address whether substitution at the S257 site in general negatively impacts caspase-6's autoactivating ability. The inactivating S257W substitution also prevented autoactivation, but the S257C substitution, which has no influence on catalytic efficiency underwent autoactivation like wild-type (Figure S1A available online), showing that the ability to autoactivate is a reflection of catalytic competence. The fact that S257D prevents caspase-6 autoactivation suggests that phosphorylation of S257 is sufficient to either prevent catalytic activity or to block zymogen autoactivation.
Figure 1. S257D Is a Robust Caspase-6 Phosphomimetic.
(A) Expression constructs showing accessible cleavage sites (gap) in full-length (FL) or constitutive two-chain (CT) caspase-6. Protein domains include the prodomain (gray circle), large subunit (gray bar), intersubunit linker (ISL, black line), and small subunit (black bar). The phosphomimetic substitution serine to aspartate is depicted as a white D. Residue numbers for each domain are indicated. RBS indicates ribosome binding sites for initiation of translation in the CT constructs.
(B) Cleavage patterns in E. coli expression constructs. Full-length (FL) wild-type (WT) caspase-6 is self-cleaved to generate large (Lg) and small (Sm) subunits. The phosphomimetic S257D blocks zymogen processing resulting in FL and N-terminal pro-domain deleted (ΔN-FL) proteins. The constitutive two-chain (CT) variant expresses the large subunit residues 1–179 independently of the small subunit, which is expressed from an introduced ribosome-binding site and a start codon at residue 193. The identity of these bands was confirmed by mass spectrometry.
(C) Kinetics parameters of caspase-6 variants. Kinetic parameters Km and kcat were fit from substrate titrations measured from independent duplicate dilutions of substrate on 3 independent days.
(D) The effect of in vitro caspase-6 phosphorylation by the ARK5 kinase on caspase-6 activity was measured from independent duplicate dilutions on 3 independent days; error bars represent the standard deviation of those measurements.
See Figure S1.
To test whether phosphorylation of S257 inactivates mature two-chain caspase-6 or simply blocks zymogen processing, we introduced S257D into a constitutively two-chain (CT) caspase-6 expression construct (Vaidya et al., 2011) (Figures 1A and 1B). Two-chain S257D caspase-6 is dramatically inactivated, with a catalytic efficiency nearly three orders of magnitude lower than wild-type caspase-6 (Figure 1C). To ensure that enzyme concentrations and thus kcat were accurately measured, the concentration of caspase-6 variants were measured by active-site titration (Figure S1B). In all cases the concentration determined by active-site titration agreed well with the concentration based on absorbance at 280 nm. To assess the similarity between S257D and phosphorylated caspase-6, we incubated WT caspase-6 or the S257C variant with the kinase ARK5. Although ARK5 is capable of fully inactivating WT, caspase-6, it has no effect on the unphosphorylatable S257C variant (Figure 1D). Thus, the S257D variant is a reliable surrogate for phosphorylation at S257. This result further suggests that phosphorylation of S257 is capable of catalytically inactivating both the zymogen and the mature (cleaved) forms of caspase-6.
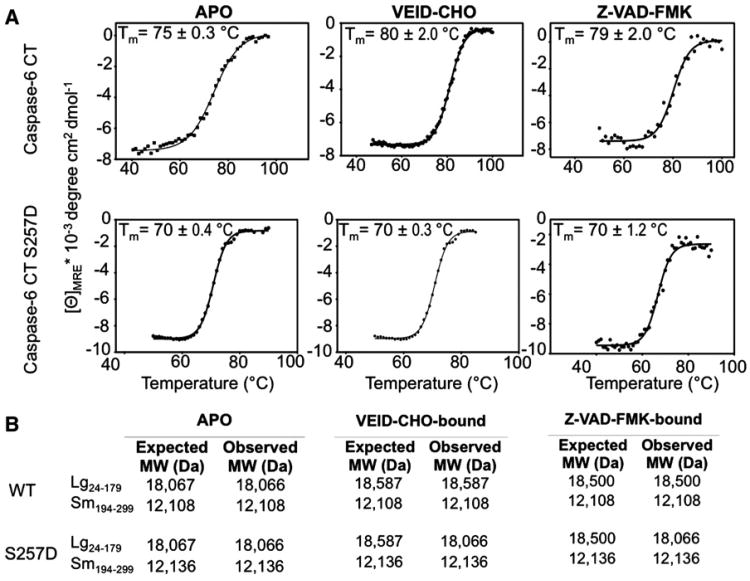
Loss of catalytic ability could result from either disruption of the catalytic machinery or loss of the ability to bind substrate. Binding of a substrate-like ligand (VEID-aldehyde) to caspase-6 results in a shift in the thermal stability of the liganded protein (Vaidya et al., 2011). Wild-type caspase-6 is stabilized by 3°C in the presence of the substrate-like ligands (Figure 2A). The S257D variant is slightly destabilized relative to wild-type caspase-6, but is still stable to well above human growth temperatures. In S257D caspase-6 no increase in thermal stability is observed in the presence of the substrate-like ligand, suggesting that it is incapable of binding substrate. The same conclusion can be drawn from samples analyzed by mass spectrometry (Figure 2B). The active-site adducts of VEID-aldehyde (VEID-CHO) or the more general caspase inhibitor z-VAD-FMK, which both bind covalently to the catalytic cysteine in the large subunit were each observed for wild-type caspase-6 but not for S257D caspase-6 indicating that S257D prevents binding of substrate. Together the substrate binding data suggest that S257 phosphorylation inactivates caspase-6 by preventing substrate binding.
Figure 2. Circular Dichroism and Mass Spectrometry Indicate Only Wild-Type Caspase-6 Can Bind Ligand.
(A) Thermal denaturation profiles monitored by circular dichroism of unliganded (APO) and active- site ligand-bound (VEID-CHO-bound) or (Z-VAD- FMK-bound) caspase-6 are shown as a function of temperature. Each condition was measured in three independently prepared samples on 3 different days, which were used to calculate average and standard deviations, with representative curves shown. The increase in the thermal melting temperature (Tm) indicates that wild-type but not S257D binds active-site ligand.
(B) The listed molecular weight observed by mass spectrometry after incubation with the covalent binding inhibitors Ac-VEID-CHO or Z-VAD-FMK further indicates that wild-type, but not S257D, binds ligand at the active site cysteine, C163, which is in the large subunit.
See Figure S1
Classically, prevention of substrate binding should be expected to influence Km. We fit kinetic substrate titrations for WT and S257D caspase-6 data using Michaelis-Menten conventions, recognizing that the caspase-6 catalytic mechanism (Scheme S1) is more complex than can be described by simple Michaelis-Menten assumptions wherein Km is often viewed as a surrogate for substrate binding affinity. Due to the fact that caspase-6 does not function by a standard Michaelis-Menten mechanism, we report kcat and Km app. Given the complexity of the caspase-6 reaction mechanism (Scheme S1) we focus on overall catalytic properties reflected in kcat/Km app and refrain from interpreting individual changes for kcat or Km app from a mechanistic viewpoint.
Phosphomimetics Alter Accessibility of the Caspase-6 Intersubunit Linker Suggesting an Altered Substrate-Binding Groove
One mechanism by which substrate binding could be prevented by phosphorylation is through disruption of the flexible loops that form the caspase-6 substrate-binding groove. In the crystal structure, the caspase-6 zymogen binds its own intersubunit linker within the substrate-binding groove (Wang et al., 2010). This self-binding allows autoactivation at the D193 site but also prevents caspase-3 from accessing and cleaving D193 cleavage site. Caspase-6 is efficiently processed by caspase-3 at the available D179 site and can subsequently be processed at the D193 site. Measuring susceptibility of caspase-6 to cleavage by caspases at the intersubunit linker reports the status of the substrate binding-grooves in wild-type and S257D caspase-6. If the substrate-binding groove of S257D is in the same conformation as the C163S zymogen, we would expect cleavage of the two versions of caspase-6 to occur at identical rates. If the substrate-binding groove is ordered, the D193 site is expected to be protected from caspase-3 cleavage. Changes in the substrate-binding groove are expected to alter the accessibility of the linker to cleavage.
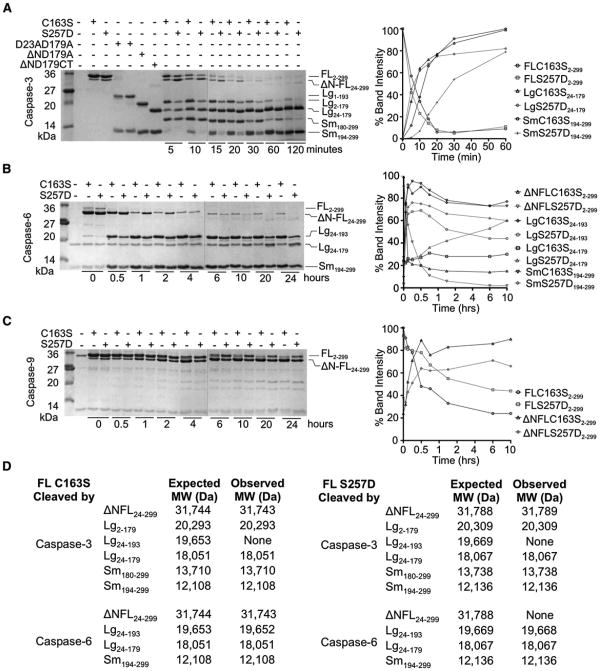
To test this accessibility of the D193 cleavage site, we examined the ability of caspase-3 to cleave zymogen caspase-6 (C163S) or S257D. The cleavage products were assessed by comparison to caspase-6 processing variants of known composition (D23A D179A, ΔND179A, and ΔND179CT) (Vaidya et al., 2011) (Figure 3A) and were unambiguously identified by mass spectrometry (Figure 3D). The procaspase-6 zymogen (C163S), which is catalytically inactive is incapable of self-processing and is therefore a useful surrogate for assessing cleavage of caspase-6 by other caspases. Caspase-6 C163S was first cleaved by caspase-3 at D179, generating Lg24–179 and Sm180–299 (Figure 3A). The large subunit is then further processed to remove the prodomain (Lg24–179) and finally at the D193 site in the intersubunit linker resulting in the final products Lg24–179 and Sm194–299. This pattern is altered in both order and kinetics for caspase-3 cleavage of S257D. S257D is first principally cleaved at the D23 site removing the prodomain and subsequently cleaved at the D179 site (Figures 3A and 3D). As expected, cleavage by caspase-3 did not result in activation although S257D attained the mature, processed form (Figure S2). The order of cleavage by caspase-3 was confirmed with the D179A/S257D variant (Figures S3A and S3C). As is clearly evident at time points >30 min, cleavage of the D193 site is much slower in S257D phosphomimetic than in the C163S zymogen. These differences in order and kinetics of cleavage by caspase-3 suggest that the substrate-binding groove of S257D differs structurally from that of active caspase-6, which has the same loop conformation as the zymogen.
Figure 3. S257D Cleavage Patterns Suggest a Substrate-Binding Groove that Is Ordered Differently from Wild-Type Caspase-6.
(A–C) One percent active caspase-3 (A), 10% caspase-6 (B), or 40% caspase-9 (C) were used. The ratio of active caspase to inactive substrate was selected so that the same number of units of active caspase-3, -6 or -9 were present in all reactions. The dotted vertical lines indicate the edges of two gels used for analysis. Gel bands quantified as disappearance of uncleaved full-length procaspase-6 C163S, full-length caspase-6 S257D, or full-length procaspase-7 C186A were set to 100% or as appearance of cleaved products. Although all bands were measured, only quantification of the large and small subunits are shown, for clarity.
(A) Caspase-3-mediated cleavage of caspase-6 C163S zymogen or S257D phosphomimetic. Control constructs expressing defined domains D23A/D179A, ΔN D179A, and ΔN D179 CT are presented for comparison of the electrophoretic mobility of caspase-6 fragments produced at the indicated time points. Fragments resulting from caspase-3-mediated cleavage at the 20 min time point were assessed by mass spectrometry to identify precise cleavage sites. Full length (FL), FL lacking the N-terminal prodomain (ΔN-FL), large (Lg), and small subunits (Sm) with the amino acids present in those bands (subscripts) are labeled. (B) Caspase-6-mediated cleavage of caspase-6 C163S zymogen or S257D phosphomimetic are shown at the indicated time points. Fragments resulting from caspase-3-mediated cleavage at the 2 hr time point were assessed by mass spectrometry to identify precise cleavage sites. Fragment labels are as in (A). (C) Caspase-9-mediated cleavage of caspase-6 C163S zymogen or S257D phosphomimetic at the indicated time points. Fragment labels are as in (A).
(D) Masses of caspase-6 fragments produced by active caspase-3 or active caspase-6 analyzed by mass spectrometry. Expected molecular weights (MW) were calculated using the average isotopic masses of the amino acids translated from the DNA sequences of the expression constructs. The N-terminal methionine was cleaved during bacterial expression, resulting in the presence of Lg2-179. The observed MW were measured after 20 min of cleavage by caspase-3 or 2 hr of cleavage by caspase-6.
When catalytically active caspase-6 was incubated with the zymogen C163S or with phosphomimetic S257D, differences in patterns of cleavage by active caspase-6 were also observed (Figure 3B). In both C163S and S257D cleavage first occurred at the prodomain D23 site and second at the D193 site generating Lg24–193 and Sm194–299. For the C163S zymogen, subsequent cleavage at D179 was much slower than cleavage of S257D at D179 in which more Lg24–179 accumulated over the course of the experiment. This was also confirmed with the D179A/S257D variant (Figure S3). Thus the accessibility of the intersubunit linker to caspase-6 also appears to be altered for S257D further suggesting changes to the substrate-binding loops in the phosphomimetic variant.
Caspase-6 has been reported to be a poor substrate of caspase-9, (Srinivasula et al., 1996) although caspase-9 can efficiently cleave caspase-7, a canonical caspase-9 substrate (Figure S4). We reasoned that because phosphorylation alters the substrate-binding grooves, it might also alter the availability of caspase-6 to caspase-9 cleavage. For both the C163S and S257D versions of caspase-6 the only prominent cleavage site was the D23 prodomain site. Cleavage of zymogen C163S at D23 was more rapid than S257D. Cleavage at the intersubunit linker was very slow, however, C163S was cleaved faster than S257D. For example, at 0.5 hr, 20% more C163S was cleaved than S257D (Figure 3C). Nevertheless, caspase-9-mediated cleavage of S257D also differs from zymogen C163S further underscoring the observation that the substrate-binding groove of caspase-6 S257D is organized differently than the caspase-6 zymogen.
S257 Phosphomimetic Has a Canonical Caspase Fold, but Misaligned Substrate-Binding Groove
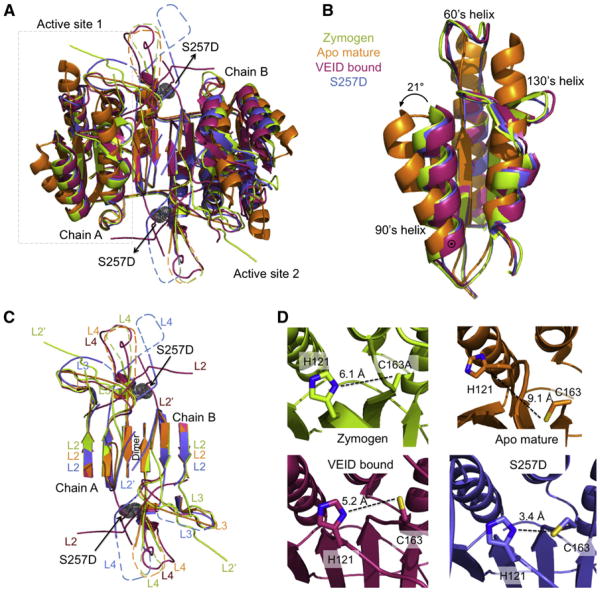
To determine the status of the substrate-binding groove, we solved the crystal structure of S257D at 2.87 Å resolution. This structure had eight monomers (here a monomer comprises one large and one small subunit so there are four full dimeric caspase-6s with eight active sites present) in the asymmetric unit. The structure refined to Rcryst/Rfree to 21.5%/25.5% (Table 1). The four dimers in the asymmetric unit are identical, each representing one functional biological unit. S257D shows the canonical executioner caspase dimer of dimers, each composed of a two large and two small subunits. The caspase-6 structures in the zymogen form, apo mature, VEID-bound, and our phosphomimetic S257D variant all exhibit the same core fold (Figure 4A). Our phosphomimetic variant structure is consistent with the overall fold of all caspase-6 structures with the exception of the 60's, 90's and 130's helices elongation from the mature form but similar to the zymogen and VEID-bound form (Figure 4B). Structurally our phosphomimetic variant S257D is located at the base of loop 4 at the dimer interface. There are several differences around the active site and within the substrate-binding loops that are unique to our phosphomimetic variant compared with the other caspase-6 structures (Figure 4C). Specifically, loops L3 and L4 are in a different conformation than other caspase-6 structures and loop L2′ is folded over the dimer interface as seen in the apo, mature structures. The folded conformation of the L2′ loop has also been observed in both the mature and allosterically inactivated caspase-7 structures. This down conformation does not support the formation of the substrate-binding groove loop bundle or allow proper ordering of the L2 loop. The L2 loop is disordered and cannot be seen in the S257D structure due to the down conformation of the L2′ loop. Three of the four substrate-binding groove loops are misaligned relative to the active conformation observed when the substrate mimic VEID is bound to caspase-6 in the active conformation.
Table 1. Crystallographic Data Collection and Statistics.
| Data Collection Statistics | |
| Wavelength (Å) | 1.54 |
| Diffraction resolution (Å) | 50.0–2.8 (2.9–2.8) |
| Measured reflections (n) | 175,642 |
| Unique reflections | 47,774 |
| Completeness (%) | 90.5 (80.6) |
| Redundancy | 3.7 |
| I/σ(I) | 27.1/12.6 (2.2) |
| Rsym | 0.134 (0.312) |
| Space group | P21 |
| a (Å) | 81.7 |
| b (Å) | 163.7 |
| c (Å) | 89.0 |
| α = γ (°) | 90.0 |
| β (°) | 94.2 |
| Refinement Statistics | |
| No. of atoms | 13,944 |
| No. waters | 47 |
| Rwork/Rfree (%) | 21.5/25.6 |
| RMSD bond length (Å) | 0.005 |
| RMSD bond angle (°) | 0.753 |
| Average B-factor (Å2) | 45.5 |
| Ramachandran Plot | |
| Core (%) | 93.8 |
| Allowed (%) | 6.2 |
| Disallowed (%) | 0 |
Values in parentheses are for the highest resolution bin. RMSD, root-mean-square deviation.
Figure 4. Caspase-6 S257D Maintains the Canonical Core Structure with Misaligned Loops.
(A) Superposition of dimeric caspase-6 structures highlights conformational changes between various states: the caspase-6 zymogen (3NR2, green), apo mature (3K7E, orange), VEID-bound (3OD5, red), and S257D (3S8E, blue). The boxed region indicates the substrate-binding region shown in (B). S257D is shown as gray dots.
(B) Only apo, mature caspase-6 attains the extended helical state of the 60's and 130's helices. Superposition of caspase-6 structures allows comparison the 60's, 90's and 130's helices of the caspase-6 structures (coloring as in A). ⊙ Denotes the hinge around which the 90's helix pivots by 21° transitioning from the apo, mature state to all other states.
(C) The substrate-binding groove loops differ dramatically between various caspase-6 conformational states. Superposition of caspase-6 loops L2, L3, L4 from chain A of the dimer and L2′ from chain B of the dimer show significantly different conformations in S257D than in all other caspase-6 states. Dashed loops represent disordered regions not observed in the crystal structures. Coloring is as in (A).
(D) Active site geometry and cysteine-histidine dyad distances in various states of caspase-6. Distances between the catalytic cysteine (C163, sticks) and catalytic histidine (H121, sticks) are drawn (dashed black line). Coloring is as in (A).
Loop Conformation More Critical for Activity than Active-Site Geometry
Activity of a caspase or any enzyme requires that the enzyme be able to bind substrate in the appropriate orientation to facilitate the chemical reaction and that the amino acid side chains involved in catalysis be properly positioned. The mechanism of caspase catalysis has not been well studied and remains under debate. We expect that caspases share a common mechanism with other cysteine and serine proteases, in which deprotonation of the catalytic thiol by the histidine in the catalytic dyad precedes nucleophilic attack. An alternative mechanism has been also suggested that does not involve catalytic thiol deprotonation (Fuentes-Prior and Salvesen, 2004). To date, no caspase structures have been observed with the catalytic machinery optimally positioned to carry out proton abstraction preceding the hydrolysis of the peptide bond. All zymogen structures have been obtained using variants in which the catalytic nucleophile has been removed by substitution of the cysteine with alanine or serine, making observation of the catalytically relevant geometry impossible. The majority of caspase crystal structures have focused on peptide-based inhibitors, which bind covalently to the active site. Covalent modification of the active-site cysteine positions the catalytic nucleophile (C163) over 5 Å from the catalytic base (H121), too far for efficient proton abstraction. The mature apo conformation is unique to caspase-6 and inactivates by holding the catalytic dyad over 9 Å apart in a catalytically incompetent conformation. S257D is profoundly inactive (Figure 1B) but is the first structure, to our knowledge, to show reasonable geometry for catalysis (Figure 4D). Detailed calculations suggest that the optimal heavy atom distance for a Cys-his dyad in a cysteine protease is 3.5 Å (Podobnik et al., 1997). The active site dyad of S257D is the only structure that shows optimal geometry and distance for a proton abstraction mechanism (3.5 Å). This alignment of the catalytic machinery is closer than has been observed in any caspase-6 structure (Figure 4D). Given this finding, the conformation of the loops seems to be more critical for activity than the geometry of the catalytic site. Even when the catalytic dyad is properly positioned, misalignment of the substrate-binding groove loops is sufficient to fully inactivate caspase-6.
Phosphorylation Mediates Inhibition by Steric Clash
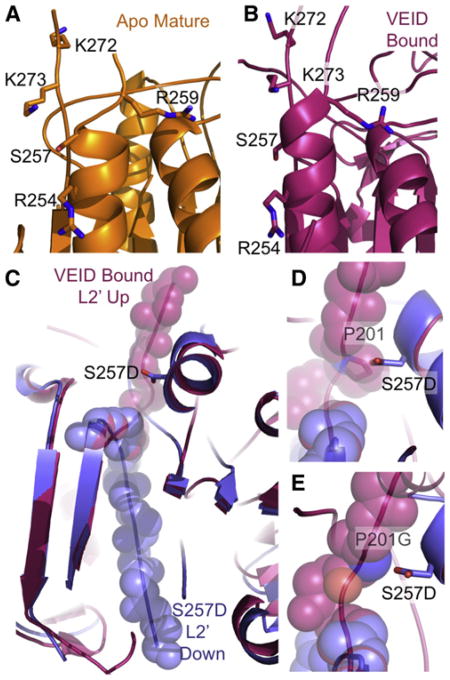
Phosphorylation changes the targeted substrate protein in two ways: alteration of the electrostatic state and addition of steric bulk. In structures of other phosphorylated proteins, the addition of negatively charged phosphates mediate electrostatic interactions with the backbone or with neighboring amino acids (Hurley et al., 1990; Johnson, 1992; Johnson and Barford, 1994). Phosphorylation of other proteins results in disordering or reordering elements of secondary structure due to steric clash. We sought to determine which of these factors played a role in the inhibition of caspase-6 by phosphorylation at S257. In the structures of two inhibited forms of caspase-6, the apo mature form (Figure 5A) and the VEID-aldehyde-bound form (Figure 5B) positively charged residues are clustered in the vicinity of S257. The role of electrostatic interactions in S257D were interrogated by changing the positively charged neighboring residues R254, R259, K272, and K273 to alanine or a negatively-charged amino acid. If electrostatic interactions play a role in inhibition of S257D, these secondary substitutions would be expected to lessen the impact of the phosphomimetic S257D substitution, which decreases the activity of caspase-6 by nearly three orders of magnitude (Table 2). Of the double variants, only the R254D substitution showed any impact on the kinetics of catalysis. Reversing the charge in R254D returned one order of magnitude in activity, although no direct interaction between these residues was observed in the S257D structure. Together this suggests that perhaps long-range electrostatic interactions may play a role in S257D inactivation.
Figure 5. S257 Phosphorylation Blocks Caspase-6 Activity through Steric Clash with P201.
(A and B) S257 is surrounded by positively charged residues (R254, R259, K272, and K273) in both the apo, mature (A), and in the VEID-bound (B) structures. These positively-charged residues could participate in electrostatic interactions with the negatively charged phosphate bound to S257.
(C) The L2′ loop is held in the up (active) conformation when caspase-6 (red) is bound by VEID, a substrate mimic, but is observed in the down (inactive) conformation in the S257D structure (blue).
(D) Proline 201 in the L2′ loop in the up conformation in the VEID-bound structure (red) sterically clashes with the superimposed S257D phosphomimetic (blue). This steric clash prevents S257D from attaining the active conformation of the L2′ loop.
(E) The substitution P201G modeled here is predicted to alleviate the steric clash between S257D and P201 in the active (VEID-bound, red) conformation.
See Figure S5.
Table 2. Kinetics Parameters for Caspase-6 Variants Demonstrate that Inhibition Is the Result of Steric Clash between S257D and P201.
| Km app(μM) | 10−3 × kcat(S−1) | 10−5 × kcat/Km app(S−1/μM) | |
|---|---|---|---|
| WT | 38 ± 4.3 | 700 ± 20 | 1,800 |
| S257D | 47 ± 2.1 | 1.5 ± 0.2 | 3.2 |
| S257D/R254D | 54 ± 3.9 | 17 ± 1 | 32 |
| S257D/R254A | 37 ± 1.2 | 2.3 ± 0.1 | 6.2 |
| S257D/R259E | 43 ± 2.1 | 4.5 ± 0.3 | 10 |
| S257D/R259A | 28 ± 0.2 | 2.1 ± 0.2 | 7.5 |
| S257D/K272A | 41 ± 0.2 | 2.3 ± 0.1 | 5.6 |
| S257D/K273A | 37 ± 3.6 | 5.1 ± 0.1 | 13 |
| S257C | 39 ± 1.8 | 700 ± 30 | 1,800 |
| S257N | 72 ± 4.0 | 3.5 ± 0.1 | 4.9 |
| S257W | 31 ± 2.8 | 0.62 ± 0.03 | 2.0 |
| S257D/P201G | 35 ± 3.0 | 100 ± 30 | 290 |
Substrate titrations to assess Km app and kcat were measured from independent duplicate dilutions of substrate on 3 separate days.
Given that removal of positive charges was insufficient to fully reverse the effects of the phosphomimetic S257D substitution, we interrogated the role of steric clash in the mechanism of S257D inhibition (Table 2). Superposition of the S275D structure with the VEID-bound structure suggests that S257D sterically clashes with proline at position 201 in the L2′ loop (Figures 5C and 5D). Analysis of the superimposed structures suggested that this clash might be sufficient to prevent the up (active) conformation of the L2′ loop, which is critical for substrate binding. The substitution S257C, which does not change the side-chain size, leads to a fully active enzyme. S257N, which is isosteric (same size, but uncharged), with S257D also results in the same three orders of magnitude inhibition as S257D. Introducing a larger residue, S257W, results in a more severely impacted enzyme, which is catalytically crippled by four orders of magnitude. These S257 substitutions suggest that the greater the steric clash, the more severe the impact. The superposition of S257D with VEID-bound caspase suggests a direct clash between S257D and P201. Removal of the proline side chain by the P201G substitution in the presence of S257D (modeled Figure 5E) results in recovery of nearly full enzyme activity (over two orders magnitude of activity, Table 2). Thus removal of the P201 side chain prevents the inhibition that is introduced by the phosphomimetic S257D indicating that this single steric clash is the predominant factor in phosphorylation-mediated inhibition of caspase-6, whereas long-range electrostatics play a minor role.
Discussion
Our results suggest that phosphorylation of caspase-6 S257 can block both apoptotic and neurodegenerative functions. Until recently, a compelling question about the role of caspase-6 in neurodegeneration was how caspase-6 could be activated in the nucleus to cleave lamin A or in the cytosol to cleave APP or huntingtin proteins without inducing apoptosis. The recent crystal structure of zymogen caspase-6 clearly demonstrates how caspase-6 bound to its own intersubunit linker can prevent processing by caspase-3 or -9 and can autoprocess and thus autoactivate (Wang et al., 2010). Autoprocessing of caspase-6 does not result in apoptosis (Klaiman et al., 2009). Our results suggest that phosphorylation at S257 can prevent caspase-6 autoprocessing. Thus any nonapoptotic caspase-6-dependant activity that relies on autoprocessing, such as cleavage of APP or huntingtin proteins will be blocked by S257 phosphorylation. The activity of cleaved (activated) caspase-6, which is responsible for the apoptotic role of caspase-6 is also blocked by S257 phosphorylation. Thus phosphorylation of active caspase-6 by ARK5 or other kinases will also block both the apoptotic activity of cleaved caspase-6 and the neurodegenerative activities of autoprocessed caspase-6.
Cleavage patterns by caspase-3 and -6 as well as the structure of S257 reveal that the substrate-binding groove loops of caspase-6 are misaligned by phosphorylation. In other caspases, the conformation of the substrate-binding groove loops are also critical for function. In both caspase-1 (Scheer et al., 2006) and caspase-7 (Hardy et al., 2004; Hardy and Wells, 2009) binding of allosteric inhibitors at the dimer interface alter the conformation of the substrate-binding groove loops, preventing proper formation of the substrate-binding groove and inhibiting enzymatic activity. In caspase-7, locking one of the active-site loops into a different conformation can inactivate the protein (Witkowski and Hardy, 2009, 2011). In S257D caspase-6, the geometry of the catalytic dyad is more optimal than has ever been observed in any caspase structure, yet S257D is profoundly inactive. Proteolytic activity comprises at least three necessary components: substrate-binding, optimal positioning of the catalytic nucleophile relative to the labile peptide bond, and the presence of an oxyanion hole to stabilize the transition state. None of these components are sufficient on their own. The fact that phosphorylation of S257 alters the substrate-binding loops without any negative impact on the geometry of the catalytic dyad (Figure 4D) or the oxyanion hole (Figure S5) supports the idea that the conformation of the binding loops is at least as important, if not more important than appropriate geometry at the active site.
Loop Misalignment Predicted for Other Caspases
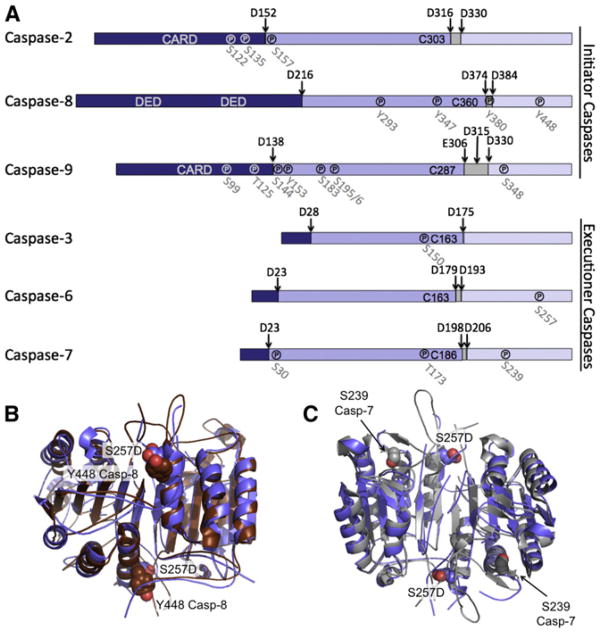
Phosphorylation of S257 disrupts the loops that form the substrate-binding groove and inactivates caspase-6. We predict that this mechanism of caspase regulation is present in other caspases as well. Both initiator and executioner caspases are extensively regulated by phosphorylation and the majority of these phosphorylation events result in inactivation of the caspase (for review see Kurokawa and Kornbluth [2009] and López-Otín and Hunter [2010]). The most prevalent sites for phosphorylation cluster in the large and small subunits (Figure 6A). In caspase-8 only tyrosine phosphorylation has been observed whereas all other caspases show serine/threonine phosphorylation. We predict that phosphorylation of Y448, which inactivates capase-8 (Jia et al., 2008) at the structurally homologous site functions by a similar mechanism of loop misalignment. Y448 in caspase-8 sits immediately adjacent to S257 in caspase-6 (Figure 6B). We predict that Y448 phosphorylation would likewise prevent the proper ordering of the L2′ loop and thus prevent substrate binding leading to the observed loss in activity. In caspase-7, S239 phosphorylation also leads to inactivation (Li et al., 2011). S239 sits at the bottom of the L3 loop, which forms the base of the substrate-binding cleft (Figure 6C). Phosphorylation of this residue is predicted to prevent the L3 loop from adopting its proper folded and flattened conformation, likewise misaligning the substrate-binding groove. Our analysis of the locations of caspase phosphorylation sites suggests that loop misalignment may be one of the predominant mechanisms of caspase inactivation by phosphorylation. Although blocking access of upstream caspases to the cleavage sites and preventing zymogen processing is also a possibility, this may be a relatively rarer mechanism as to date it has only been observed for caspase-3 (Duncan et al., 2011). Based on the locations of known sites of caspase phosphorylation, misalignment of the substrate-binding groove loops appears to be a general phenomenon across the family of apoptotic caspases.
Figure 6. Phosphorylation Is Predicted to Misalign Active Site Loops in Many Caspases.
(A) Both initiator and executioner caspases are phosphorylated. Phosphorylation (℗) occurs at indicated sites in all domains of caspases: prodomain (dark blue), large subunit (medium blue), linker (gray), and small subunit (light blue). Interdomain cleavage sites are indicated by arrows. Phosphorylation typically results in inactivation.
(B) In caspase-8 Y448 phosphorylation leads to inactivation. Structural alignment of caspase-6 (blue) with caspase-8 (brown) shows capase-8 Y448 (brown spheres) is immediately adjacent to S257 in caspase-6 (blue spheres). Y448 phosphorylation is therefore predicted to lead to inhibition through a similar loop-misalignment mechanism.
(C) In caspase-7 phosphorylation of S239 results in inactivation. Structural alignment of caspase-6 (blue) with caspase-7 (gray) shows caspase-7 S239 (gray spheres) sitting at the base of the L3 loop. S239 phosphorylation is thus predicted to disrupt the folded state of L3 and prevent substrate binding.
The location of the phosphorylation within the caspase dictates relevant mechanisms of dephosphorylation. If phosphorylation is in the prodomain or intersubunit linker (e.g., phosphorylation of S135 in the caspase-8 prodomain or T125 in the caspase-9 prodomain), inhibition will be relieved by zymogen activation, which removes these regions. S257 phosphorylation is in the small subunit, which forms the core of folded caspase-6, so dephosphorylation must require a phosphatase counteracting ARK5 function. This phosphatase has not yet been identified. Activating ARK5 or suppressing the counteracting phosphatase represent two more potential points of intervention for treating caspase-6-mediated neurodegeneration.
It is striking to us that steric clash between a small number of atoms on S257 and the side chain of P201 is sufficient to lead to full enzyme inactivation. Addition of a phosphate to serine increases steric bulk by just four additional heavy atoms and changes serine from neutral to doubly negatively charged phosphoserine. Phosphorylation sometimes leads to ordering of regions of the phosphoprotein through electrostatic interactions, particularly with arginine residues (e.g., glycogen phosphorylase) (Barford et al., 1991; Johnson, 1992; Johnson and Barford, 1994). On the other hand, steric clash of the phosphate leading to disorder is an equally or more common structural phenomenon induced by phosphorylation. In isocitrate dehydrogenase the added phosphate sterically occludes and electrostatically excludes the negatively charged isocitrate substrate from the active site (Hurley et al., 1990). In two related enzymes, α-ketoacid dehydrogenase and pyruvate dehydrogenase, phosphorylation leads to steric clash, which in turn disorders a loop and prevents binding of a partner protein (Kato et al., 2008; Wynn et al., 2004). Similarly, the steric clash we observe between phospho-S257 and P201 likewise prevents ordering of the substrate-binding groove loops that in turn prevents binding substrate. Thus the mechanism we observe for inhibition of caspase-6 appears to be utilized across many families of proteins.
We now recognize that the region around S257 is a very susceptible site. The clash of a phosphate with just three side-chain atoms is sufficient to misalign an entire substrate-binding groove. Clashes of just a few atoms, especially in small drug-like molecules have been observed to propagate to large changes in structure and function of PTP-1B (Hansen et al., 2005), Factor VIIa (Dennis et al., 2001; Roberge et al., 2001), glycogen phosphorylase (Zographos et al., 1997), and many other proteins. These examples demonstrate that small molecules are able to perform similar functions to that of phosphorylation at S257 in caspase-6, namely inducing conformational changes in proteins. Thus, our results suggest that any protein or small molecule that locks caspase-6 into the S257 phosphorylated conformation would potentially be useful for treatment of neurodegenerative diseases in which caspase-6 plays a role.
Experimental Procedures
Experimental procedures for all aspects of this article are included as Supplemental Experimental Procedures. These procedures include: generation of caspase-6 variants, caspase-6 expression and purification, caspase-3 and -7 expression and purification, caspase-9 expression and purification, activity assays, in vitro caspase-6 phosphorylation by ARK5, proteolytic cleavage by active caspase-3, -6, or -9, mass spectrometry, stability measurements by circular dichroism, crystallization and data collection, and structure determination.
Supplementary Material
Acknowledgments
We thank Kristen Huber for providing caspase-7 C186A and caspase-9, Stephen Eyles for assistance with mass spectral data obtained at the University of Massachusetts Mass Spectrometry Facility, which is supported, in part, by the National Science Foundation. This work was supported by the National Institutes of Health (GM80532). E.M.V-D. was supported by the National Science Foundation (S21000025700000 and DGE-0504485).
Footnotes
Accession Numbers: Coordinates and structure factors have been deposited in the Protein Data Bank as PDB ID 3S8E.
Supplemental Information: Supplemental Information includes five figures, Supplemental Experimental Procedures, and one scheme and can be found with this article online at doi:10.1016/j.str.2012.02.003.
References
- Barford D, Hu SH, Johnson LN. Structural mechanism for glycogen phosphorylase control by phosphorylation and AMP. J Mol Biol. 1991;218:233–260. doi: 10.1016/0022-2836(91)90887-c. [DOI] [PubMed] [Google Scholar]
- Baumgartner R, Meder G, Briand C, Decock A, D'arcy A, Hassiepen U, Morse R, Renatus M. The crystal structure of caspase-6, a selective effector of axonal degeneration. Biochem J. 2009;423:429–439. doi: 10.1042/BJ20090540. [DOI] [PubMed] [Google Scholar]
- Cardone MH, Roy N, Stennicke HR, Salvesen GS, Franke TF, Stanbridge E, Frisch S, Reed JC. Regulation of cell death protease caspase-9 by phosphorylation. Science. 1998;282:1318–1321. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- Chai J, Shiozaki E, Srinivasula SM, Wu Q, Datta P, Alnemri ES, Shi Y. Structural basis of caspase-7 inhibition by XIAP. Cell. 2001;104:769–780. doi: 10.1016/s0092-8674(01)00272-0. [DOI] [PubMed] [Google Scholar]
- Cowling V, Downward J. Caspase-6 is the direct activator of caspase-8 in the cytochrome c-induced apoptosis pathway: absolute requirement for removal of caspase-6 prodomain. Cell Death Differ. 2002;9:1046–1056. doi: 10.1038/sj.cdd.4401065. [DOI] [PubMed] [Google Scholar]
- Dennis MS, Roberge M, Quan C, Lazarus RA. Selection and characterization of a new class of peptide exosite inhibitors of coagulation factor VIIa. Biochemistry. 2001;40:9513–9521. doi: 10.1021/bi010591l. [DOI] [PubMed] [Google Scholar]
- Deveraux QL, Takahashi R, Salvesen GS, Reed JC. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- Duncan JS, Turowec JP, Duncan KE, Vilk G, Wu C, Lüscher B, Li SS, Gloor GB, Litchfield DW. A peptide-based target screen implicates the protein kinase CK2 in the global regulation of caspase signaling Sci. Signal. 2011;4:ra30. doi: 10.1126/scisignal.2001682. [DOI] [PubMed] [Google Scholar]
- Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J. 2004;384:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan V, Gorostiza OF, Banwait S, Ataie M, Logvinova AV, Sitaraman S, Carlson E, Sagi SA, Chevallier N, Jin K. Reversal of Alzheimer's-like pathology and behavior in human APP transgenic mice by mutation of Asp664. Proc Natl Acad Sci USA. 2006;103:7130–7135. doi: 10.1073/pnas.0509695103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, Pearson J, Shehadeh J, Bertram L, Murphy Z, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Guo H, Pétrin D, Zhang Y, Bergeron C, Goodyer CG, LeBlanc AC. Caspase-1 activation of caspase-6 in human apoptotic neurons. Cell Death Differ. 2006;13:285–292. doi: 10.1038/sj.cdd.4401753. [DOI] [PubMed] [Google Scholar]
- Hansen SK, Cancilla MT, Shiau TP, Kung J, Chen T, Erlanson DA. Allosteric inhibition of PTP1B activity by selective modification of a non-active site cysteine residue. Biochemistry. 2005;44:7704–7712. doi: 10.1021/bi047417s. [DOI] [PubMed] [Google Scholar]
- Hardy JA, Lam J, Nguyen JT, O'Brien T, Wells JA. Discovery of an allosteric site in the caspases. Proc Natl Acad Sci USA. 2004;101:12461–12466. doi: 10.1073/pnas.0404781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy JA, Wells JA. Dissecting an allosteric switch in caspase-7 using chemical and mutational probes. J Biol Chem. 2009;284:26063–26069. doi: 10.1074/jbc.M109.001826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Dean AM, Sohl JL, Koshland DE, Jr, Stroud RM. Regulation of an enzyme by phosphorylation at the active site. Science. 1990;249:1012–1016. doi: 10.1126/science.2204109. [DOI] [PubMed] [Google Scholar]
- Jia SH, Parodo J, Kapus A, Rotstein OD, Marshall JC. Dynamic regulation of neutrophil survival through tyrosine phosphorylation or dephosphorylation of caspase-8. J Biol Chem. 2008;283:5402–5413. doi: 10.1074/jbc.M706462200. [DOI] [PubMed] [Google Scholar]
- Johnson LN. Glycogen phosphorylase: control by phosphorylation and allosteric effectors. FASEB J. 1992;6:2274–2282. doi: 10.1096/fasebj.6.6.1544539. [DOI] [PubMed] [Google Scholar]
- Johnson LN, Barford D. Electrostatic effects in the control of glycogen phosphorylase by phosphorylation. Protein Sci. 1994;3:1726–1730. doi: 10.1002/pro.5560031011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Wynn RM, Chuang JL, Tso SC, Machius M, Li J, Chuang DT. Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops. Structure. 2008;16:1849–1859. doi: 10.1016/j.str.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaiman G, Champagne N, LeBlanc AC. Self-activation of Caspase-6 in vitro and in vivo: Caspase-6 activation does not induce cell death in HEK293T cells. Biochim Biophys Acta. 2009;1793:592–601. doi: 10.1016/j.bbamcr.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Kurokawa M, Kornbluth S. Caspases and kinases in a death grip. Cell. 2009;138:838–854. doi: 10.1016/j.cell.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wen W, Liu K, Zhu F, Malakhova M, Peng C, Li T, Kim HG, Ma W, Cho YY, et al. Phosphorylation of caspase-7 by p21-activated protein kinase (PAK)2 inhibits chemotherapeutic drugs-induced apoptosis of breast cancer cell lines. J Biol Chem. 2011;286:22291–22299. doi: 10.1074/jbc.M111.236596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C, Hunter T. The regulatory crosstalk between kinases and proteases in cancer. Nat Rev Cancer. 2010;10:278–292. doi: 10.1038/nrc2823. [DOI] [PubMed] [Google Scholar]
- Podobnik M, Kuhelj R, Turk V, Turk D. Crystal structure of the wild-type human procathepsin B at 2.5 A resolution reveals the native active site of a papain-like cysteine protease zymogen. J Mol Biol. 1997;271:774–788. doi: 10.1006/jmbi.1997.1218. [DOI] [PubMed] [Google Scholar]
- Roberge M, Santell L, Dennis MS, Eigenbrot C, Dwyer MA, Lazarus RA. A novel exosite on coagulation factor VIIa and its molecular interactions with a new class of peptide inhibitors. Biochemistry. 2001;40:9522–9531. doi: 10.1021/bi010592d. [DOI] [PubMed] [Google Scholar]
- Scheer JM, Romanowski MJ, Wells JA. A common allosteric site and mechanism in caspases. Proc Natl Acad Sci USA. 2006;103:7595–7600. doi: 10.1073/pnas.0602571103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott FL, Denault JB, Riedl SJ, Shin H, Renatus M, Salvesen GS. XIAP inhibits caspase-3 and -7 using two binding sites: evolutionarily conserved mechanism of IAPs. EMBO J. 2005;24:645–655. doi: 10.1038/sj.emboj.7600544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Alnemri ES, Fairman R, Shi Y. Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell. 2003;11:519–527. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Fernandes-Alnemri T, Zangrilli J, Robertson N, Armstrong RC, Wang L, Trapani JA, Tomaselli KJ, Litwack G, Alnemri ES. The Ced-3/interleukin 1beta converting enzyme-like homolog Mch6 and the lamin-cleaving enzyme Mch2alpha are substrates for the apoptotic mediator CPP32. J Biol Chem. 1996;271:27099–27106. doi: 10.1074/jbc.271.43.27099. [DOI] [PubMed] [Google Scholar]
- Stennicke HR, Ryan CA, Salvesen GS. Reprieval from execution: the molecular basis of caspase inhibition. Trends Biochem Sci. 2002;27:94–101. doi: 10.1016/s0968-0004(01)02045-x. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Kusakai G, Kishimoto A, Shimojo Y, Miyamoto S, Ogura T, Ochiai A, Esumi H. Regulation of caspase-6 and FLIP by the AMPK family member ARK5. Oncogene. 2004;23:7067–7075. doi: 10.1038/sj.onc.1207963. [DOI] [PubMed] [Google Scholar]
- Vaidya S, Hardy JA. Caspase-6 latent state stability relies on helical propensity. Biochemistry. 2011;50:3282–3287. doi: 10.1021/bi2001664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya S, Velázquez-Delgado EM, Abbruzzese G, Hardy JA. Substrate-induced conformational changes occur in all cleaved forms of caspase-6. J Mol Biol. 2011;406:75–91. doi: 10.1016/j.jmb.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Cao Q, Liu X, Wang KT, Mi W, Zhang Y, Li LF, LeBlanc AC, Su XD. Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation. EMBO Rep. 2010;11:841–847. doi: 10.1038/embor.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski WA, Hardy JA. L2′ loop is critical for caspase-7 active site formation. Protein Sci. 2009;18:1459–1468. doi: 10.1002/pro.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski WA, Hardy JA. A designed redox-controlled caspase. Protein Sci. 2011;20:1421–1431. doi: 10.1002/pro.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn RM, Kato M, Machius M, Chuang JL, Li J, Tomchick DR, Chuang DT. Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation. Structure. 2004;12:2185–2196. doi: 10.1016/j.str.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Zographos SE, Oikonomakos NG, Tsitsanou KE, Leonidas DD, Chrysina ED, Skamnaki VT, Bischoff H, Goldmann S, Watson KA, Johnson LN. The structure of glycogen phosphorylase b with an alkyldihydropyridine-dicarboxylic acid compound, a novel and potent inhibitor. Structure. 1997;5:1413–1425. doi: 10.1016/s0969-2126(97)00292-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.