Abstract
Diffusion tensor imaging (DTI) is a useful tool for studying anatomy and pathology in the rodent central nervous system (CNS).The unique tissue contrasts provided by DTI are well suited for monitoring disease progression, studying brain development, and characterizing anatomical phenotypes. Recent technical developments have vastly improved the speed and resolution of rodent DTI. Ongoing research efforts exploring the microstructural basis of DTI signals have provided useful insights into its capabilities to delineate brain structures and detect neuropathology. Significant progress has also been made in combining DTI results with data acquired using other imaging modalities to enhance our understanding of the rodent CNS.
Keywords: diffusion tensor imaging, rodent, brain, spinal cord, neuroanatomy
Introduction
The laboratory mouse and rat are widely used in neuroscience research to investigate the mechanisms that control neuronal structures, functions, and disease processes. The ever-increasing sophistication in research that involves mutant mouse strains or rodent models of neurological disease demands an array of techniques to capture various aspects of the rodent central nervous system (CNS). Histology has been the standard method by which to examine the cellular and molecular aspects of the rodent CNS and its disease processes, but has several limitations. Histology is highly invasive and often focuses on predefined anatomical regions of interest at a single time point, which is limiting if the phenotype or pathology of interest evolves over time or extends beyond the pre-defined regions. Thus, imaging techniques capable of in vivo monitoring, with whole-brain or spinal cord coverage, could bring new perspectives and extend our knowledge of the rodent CNS.
Among the imaging modalities available, diffusion tensor magnetic resonance imaging (DT-MRI), or simply diffusion tensor imaging (DTI) [1], is a unique technique for visualizing structural organization and pathology in the rodent CNS (Figure 1). By measuring 3D diffusion of water molecules in tissues and fitting the results into a tensor model, DTI provides several image contrasts that carry a wealth of neuroanatomical information (Box 1). Details about the concept, implementation, and biomedical applications of DTI can be found in several excellent reviews and books [2–7]. With its unique tissue contrasts and its non-invasive nature for examinations with whole-brain or spinal cord coverage, DTI is well suited for imaging the rodent CNS, especially white-matter structures, in a macroscopic fashion that is complementary to histology.
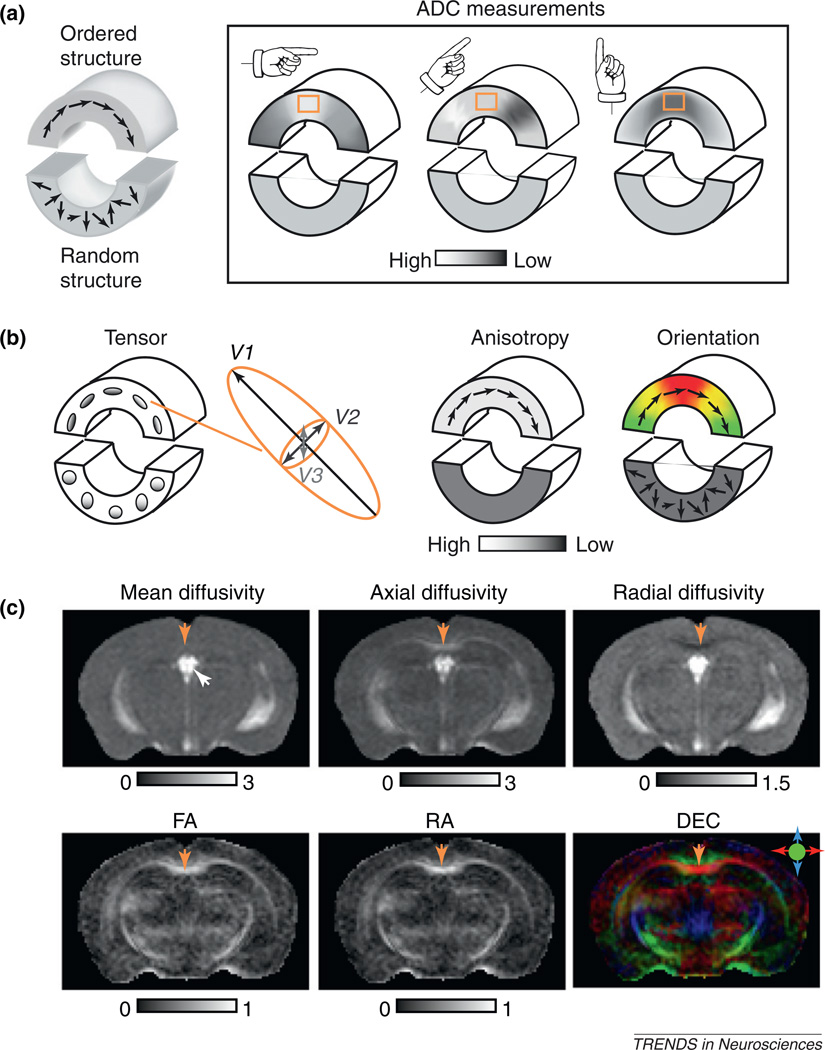
Figure 1.
From diffusion anisotropy to diffusion tensor. (a) A 2D diagram illustrating diffusion anisotropy [39]. The phantom in the diagram consists of an upper part that contains ordered structures (e.g. a bundle of axons or well-organized neurons) with coherent tissue orientations (represented by the arrows) and a lower part that contains random structures without coherent orientations. When measuring ADC along three different orientations, ADC measurements in the upper part show orientation dependence (i.e. anisotropic diffusion). For example, ADC values measured in the orange box change from high to low when the measuring orientation changes from horizontal to vertical. In comparison, ADC measured in the lower part does not show this orientation dependence. (b) Diffusion tensors visualized as ellipsoids and rendered as anisotropy and orientation-encoded color maps. The size, shape, and orientation of each ellipsoid are dictated by the eigenvalues and eigenvectors of the corresponding diffusion tensor. In the anisotropy map, the upper part of the phantom has higher diffusion anisotropy than the lower part. When diffusion anisotropy is high, the long axes of the diffusion tensor often coincide with the underlying structural orientation and can be visualized using a direction-encoded color map (DEC). In this 2D example, regions with structural orientation running horizontally are red and those running vertically are green. Transition areas become yellow, a mixture of red and green. Note that the technique cannot distinguish afferent and efferent axonal tracts. (c) DTI contrasts in the mouse brain. In the mean diffusivity image, the cerebral spinal fluid in the ventricle (indicated by the white arrow) appears bright while the mouse brain parenchyma appears rather homogeneous. The axial diffusivity and radial diffusivity images show more contrasts between gray- and white-matter structures than the mean diffusivity image. The CC (indicated by yellow arrows) has higher axial diffusivity values and lower radial diffusivity values than the neighboring cortical regions. In the FA and RA images, major white-matter structures, such as the CC, can easily be distinguished from gray-matter structures by their high anisotropy values. In the DEC image, major white-matter tracts are accentuated by their high anisotropy (reflected by their intensities) and color-coded orientations. The color scheme is: red, left–right; green, rostral–caudal; and blue, superior–inferior. The CC appears red in the DEC image because most fibers in the CC are arranged along the left–right orientation. The unit for mean, axial, and radial diffusivity is µm2/ms. FA and RA are unitless. DTI images adapted, with permission, from [16,34].
Box 1. Measuring and visualizing diffusion tensor in the mouse brain using MRI.
DTI uses the process of water molecule diffusion as a probe to infer tissue microstructure and organization. By applying a pair of diffusion-sensitizing magnetic field gradients (or simply diffusion gradients) along an arbitrary orientation, the ADC along that orientation can be determined based on the amount of MR signal attenuations due to diffusion. Basically, we compare MR images acquired without diffusion gradients (non-diffusion-weighted images) to images acquired with diffusion gradients applied along the selected orientation (diffusion-weighted images, with one or more diffusion weightings or b values). MR signals are fitted to a mono-exponential model S(b) = S0e−b·ADC to estimate the ADC. S(b) and S0 are the amplitude of the MR signals with and without diffusion gradients, respectively, and b is calculated based on the magnitude and timing of the diffusion gradient [4]. For anisotropic diffusion (Figure 1a), ADC measured along one orientation is apparently not adequate to represent the orientation-dependent property, and a diffusion tensor based on a 3D Gaussian model for water molecule diffusion can be used instead. In DTI, MR signals acquired with diffusion gradients applied along at least six non-collinear, non-coplanar directions are compared to MR signals acquired without diffusion gradients, and a diffusion tensor is calculated from these signals using multivariate regression [71]. It is necessary to note that different experimental parameters, such as choices of b and timing of the diffusion gradients, may affect the estimated ADCs and diffusion tensors [72–74].
A diffusion tensor can be visualized using an ellipsoid (Figure 1b) for which the length of the three axes is proportional to the three eigenvalues of the diffusion tensor (λ1, λ2, and λ3 in descending order), and the three orthogonal axes coincide with the three eigenvectors (V1, V2, and V3). Because it is difficult to visualize and comprehend diffusion tensors directly, several tensor-derived scalar indices are frequently used to generate grayscale images for visualization and quantification (Figure 1c). These indices include mean diffusivity (MD), axial diffusivity (λ‖), radial diffusivity (λ⊥), fractional anisotropy (FA), and relative anisotropy (RA) [75]:
Mean diffusivity is useful for detecting stroke injuries in the brain [76]. The use of axial and radial diffusivity is discussed in this article. Both FA and RA are commonly used measures of diffusion anisotropy, with FA offering better noise characteristics compared to RA [77]. The orientation information encoded in diffusion tensor can be visualized using a direction-encoded color map (DEC; Figure 1c) [78]. In a DEC image, red, green, and blue colors represent left–right, anterior–posterior, and superior–inferior orientations, respectively, with the overall intensity proportional to diffusion anisotropy (FA is commonly used here).
Early applications of DTI in the rodent CNS date back more than a decade ago [8,9] and its development since then has been driven primarily by needs arising from neuroscience research (e.g. the need to examine anatomical phenotypes in mutant mouse brains or neuropathology in various rodent models of disease). With an increasing number of small-animal magnetic resonance (MR) scanners installed in research centers and the continuing advances in MR instruments and techniques, rodent DTI has become a practical and useful tool for the neuroscience community, as evidenced by the large number of related studies in the last few years. Here, we review these recent findings and discuss the current technical capabilities of rodent DTI, as well as the potential pitfalls and limitations of DTI in detecting rodent neuropathology. We also discuss studies that have used DTI to study brain development and phenotypes.
Recent technical developments in rodent DTI
For DTI, speed and spatial resolution have been the two main limiting factors that prevented its widespread application to rodent models. Rodent DTI requires much higher spatial resolution (0.1–0.2 mm per pixel) compared to clinical DTI (1–2 mm per pixel) for reliable delineation of miniature structures in the rodent CNS. This requirement puts rodent DTI at a significant signal-to-noise ratio disadvantage, which, together with prolonged acquisition and sensitivity to subject motion, makes in vivo DTI of the rodent CNS quite challenging. Recent advances in small-animal MRI systems (e.g. high-sensitivity imaging coils, high-performance gradient systems, and implementation of fast echo planar imaging sequences for small-animal imaging on high-field systems [10,11]) have greatly improved the speed and spatial resolution of in vivo rodent DTI. Using current state-of-the-art small-animal instruments, in vivo DTI of the mouse brain can be acquired at a resolution of 0.156 mm × 0.156 mm for a 0.5-mm slice thickness within 90 min [12]. At this resolution, several small white-matter tracts in the mouse brain (e.g. the stria terminalis, which were previously elusive in in vivo DTI) can be reliably delineated, along with large white-matter structures such as the corpus callosum (Figure 2a). The 3D trajectories of these structures can also be reconstructed [12]. Compared to the rodent brain, DTI of the rodent spinal cord is more challenging because of its proximity to the sources of cardiac and respiratory motion. Using a surface radiofrequency coil and a custom-made animal holder, a recent study demonstrated in vivo DTI of the mouse cervical spinal cord at a resolution of 0.117 mm × 0.059 mm with a slice thickness of 0.5 mm within 1 h [13]. These technical developments have enhanced our ability to non-invasively examine white-matter structures and injuries in the rodent CNS.
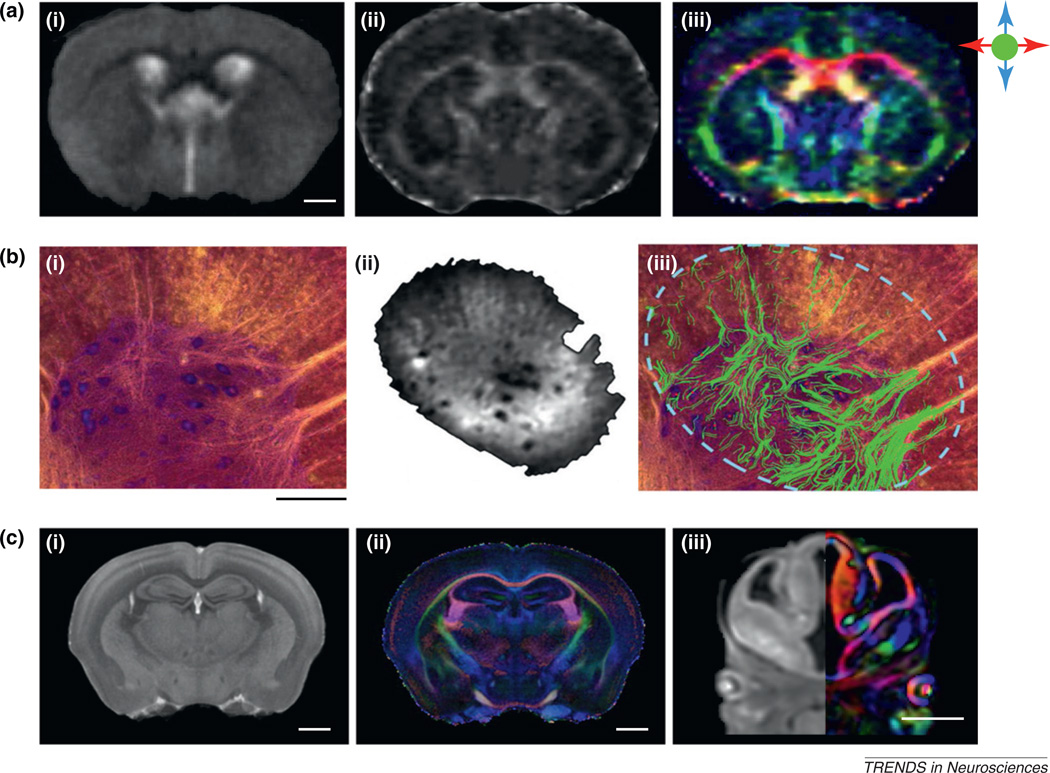
Figure 2.
Recent efforts to achieve high-resolution DTI of the rodent CNS. (a) In vivo (i) T2-weighted, (ii) fractional anisotropy (FA), and (iii) direction-encoded color map (DEC) images of an adult C57BL/6J mouse brain with a resolution of 156 µm × 156 µm × 500 µm [12]. The color scheme is: red, left–right; green, rostral–caudal; and blue, superior–inferior. The scale bar corresponds to 1 mm. (b) (i) A Nissl-stained section and (ii) a diffusion-weighted image of a section of the ventral horn of rat cervical spinal cord [15]. The MR image was acquired at a resolution of 15.6 µm × 15.6 µm × 25 µm. (iii) DTI-based fiber tracking results (green fibers) overlaid on the Nissl-stained section [15]. The scale bar corresponds to 250 µm. (c) Ex vivo diffusion-weighted and DEC images of (i) an adult C57BL/6J mouse brain [60] and (iii) an E12 CD1 mouse embryo [16], acquired at an isotropic resolution of 55 µm per voxel. Also shown are average T2* and DEC images of adult C57BL/6J mouse brains (n = 8) at an isotropic resolution of 43 µm per voxel [60]. The color scheme is identical to that for (a). The scale bars correspond to 1 mm. Reproduced, with permission, from [12] (a), [15] (b), [16] (c iii), and [60] (c i, ii).
Although in vivo DTI allows monitoring of white-matter structures in live animals, most studies that demand spatial resolution higher than 0.1 mm must still resort to ex vivo DTI of post mortem specimens. For ex vivo DTI, the available resolution is primarily dictated by the sensitivity and stability of imaging instruments. The theoretical resolution limit of ex vivo DTI is believed to be approximately 0.005–0.01 mm, depending on the apparent diffusion coefficient (ADC) (see Glossary) of water molecules in tissue specimens and imaging parameters [14]. A specially designed high-sensitivity coil was recently used to acquire ultra-high-resolution DTI data from a section of rat spinal cord at an in-plane resolution of 0.016 mm × 0.016 mm (Figure 2b) [15]. At this resolution, the diffusion MRI data could be readily co-registered and compared side by side with histological images, and small white-matter tracts in the ventral horn of the rat spinal cord could be reconstructed from the DTI data in exquisite detail. Although these images were acquired from a thin tissue section rather than from the entire brain or spinal cord, they nonetheless demonstrated that such resolution is within the reach of DTI and the level of detail that high-resolution DTI can reveal. Two recent studies reported ex vivo DTI of the adult and embryonic mouse brain at isotropic resolution of 0.04–0.05 mm (Figure 2c) [16,17]. The rich anatomical details revealed in these high-resolution images allow delineation of white-matter tracts previously only visible on histological images. For example, small axonal bundles in the adult striatum and thalamus and early axonal tracts in the embryonic mouse brains were visualized.
DTI of rodent neuropathology
Using DTI to study white matter injury
DTI is increasingly used to examine neuropathology in rodent models of disease such as multiple sclerosis [18], stroke [19–22], amyotrophic lateral sclerosis [23,24], traumatic brain injury (TBI), and spinal cord injury [25,26], so an understanding of the relationships between DTI-based imaging markers and specific pathology is imperative. Extensive research in the last few years has greatly advanced our knowledge in this area. It was previously shown that the two most frequently used DTI-derived markers, mean diffusivity (MD) and fractional anisotropy (FA) (Box 1), have limited sensitivity and specificity. For example, either axonal injury or myelin injury may alter FA values in white-matter structures. On the basis of experiments in rodent models, several reports demonstrated that axial and radial diffusivities were able to separate axonal and myelin injuries, with a decrease in axial diffusivity indicating potential axonal injury and an increase in radial diffusivity indicating potential demyelination or dysmyelination [27–29]. In subsequent years, the relationships between axial and radial diffusivities and axon and myelin injuries have been scrutinized in additional rodent models with unique pathological features. In a study of Wallerian degeneration in the mouse optic nerve and optic tract after retinal ischemia, in vivo DTI revealed an early decrease in axial diffusivity at 3 days after ischemia, followed by an increase in radial diffusivity 6 days later [30]. DTI of the same injury model in slow Wallerian degeneration (Wlds) mutant mice showed similar but delayed changes in axial and radial diffusivities in the optic nerve of mutant compared to wild-type mice [31]. These observations correlated well with histological findings and other results regarding the timing of Wallerian degeneration in wild-type and Wlds mice.
The relationships, however, became less straightforward in complex models. In the mouse cuprizone model of demyelination, in vivo DTI showed increased radial diffusivity in the corpus callosum of treated mice during chronic demyelination (after 6 weeks of cuprizone treatment), which returned to baseline levels after re-myelination, as expected [32]. What is interesting is that at 4 weeks after the start of cuprizone treatment, histology showed severe demyelination and inflammation, whereas radial diffusivity measured in vivo showed no significant difference from the control mice [33,34]. It is now assumed that the presence of inflammation, with microglia and macro-phage activation, cell infiltration, and vasogenic edema, could be one of the confounding factors that reduce the sensitivity of radial diffusivity to demyelination in this model [35].
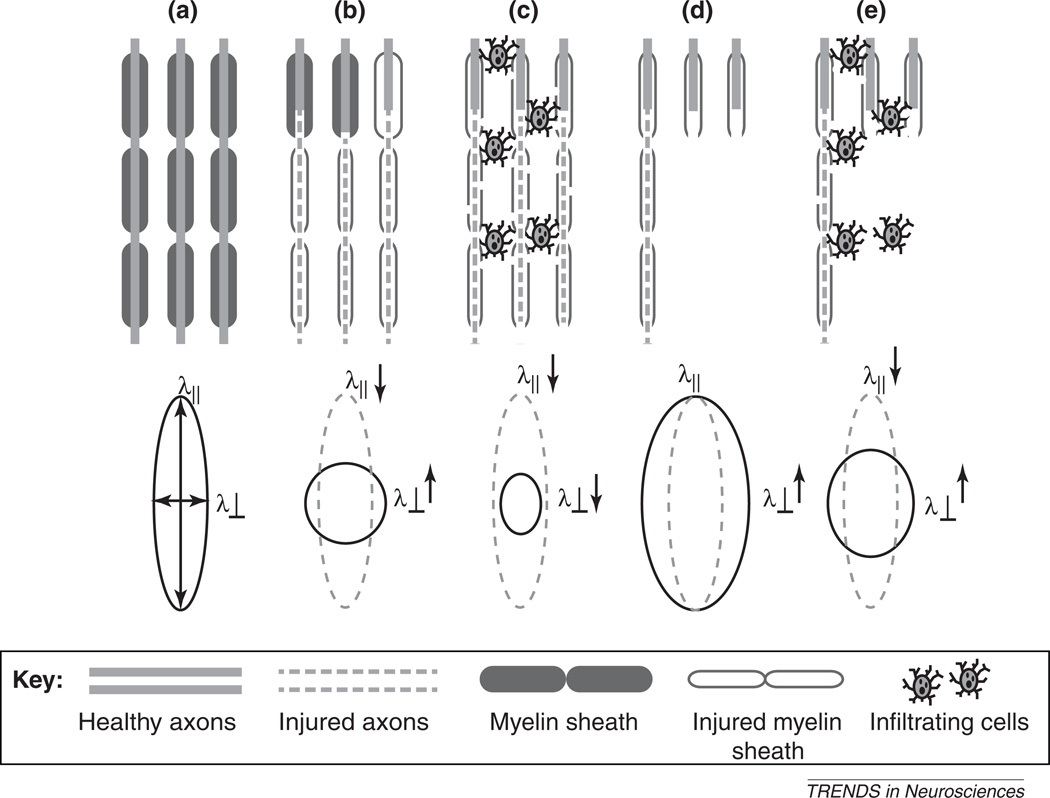
To understand such findings, it is important to realize that DTI signals reflect the average diffusion properties of tissues within a voxel that encompasses thousands of cell bodies, axons, and myelin sheaths. Inflammation and subsequent microstructural changes will inevitably affect the microstructural composition of a voxel, and subsequently its diffusion signals. Several simplified scenarios, in which cell infiltration in white-matter regions might influence the diffusion anisotropy and diffusivity measurements, have been suggested (Figure 3) [35]. In the case of classic Wallerian degeneration, axon and myelin injury during the early stages (Figure 3b) will lead to decreases in axial diffusivity and increases in radial diffusivity, as described in an optic nerve model [30] and a rat spinal-cord axotomy model [36]. As the axon and myelin debris are gradually removed in the later stages (Figure 3d), both axial and radial diffusivities will increase, which may be the mechanism behind the so-called pseudo-normalization of axial diffusivity in the sub-acute stages reported in TBI models [37]. When inflammation is an additional factor (Figure 3c,e), the presence of infiltrating cells could suppress signal increases in axial and radial diffusivities, and, as a result, decrease the sensitivity of these DTI measurements to actual axon and myelin injuries [35]. These reports demonstrate that DTI measurements alone are not sufficient to tackle complex pathology. To resolve this issue, diffusion MRI models more sophisticated than the diffusion tensor model [35] need to be developed. Alternatively, additional MRI contrasts, such as T1, T2, and magnetization transfer MRI [38], should be included in the study design.
Figure 3.
Schematic diagrams illustrating how complex neuropathology can affect DTI results. (a) Normal myelinated axons and the corresponding diffusion tensor and axial and radial diffusivity (λ‖ and λ⊥, respectively). The diffusion tensor is represented by the ellipsoid here, and is shown in subsequent plots as a grey ellipsoid with dashed lines. (b,c) Axon and myelin injury with and without cell infiltration. (d,e) Axon and myelin injury with axonal loss, with and without cell infiltration. Adapted, with permission, from [35].
Using DTI to study gray-matter alterations
Although white-matter injury has been the focus of the majority of studies that employed DTI, a few studies that used DTI to examine gray-matter injury have been reported. Several rodent gray-matter structures (e.g. the hippocampus, cerebellum, and embryonic or neonatal cortex) contain highly organized axons or cell bodies. These unique structural organizations can be recognized on DTI by their distinct patterns of diffusion anisotropy [39,40], and, when disrupted by injury, may be detected by DTI. In rats treated with kainic acid and pilocarpine to induce status epilepticus, FA values measured in the dentate gyrus correlated with the density of mossy fibers and myelinated axons [41]. In neonatal rat brains with hypoxic ischemic injury, disrupted radial organization in the affected cortical area could be visualized by DTI [42]. For other gray-matter regions that lack highly organized cellular structures, it is often a challenge to pin down the exact histological substrate of DTI readouts. For example, increases in FA in rat cortex observed after TBI have been linked to white-matter reorganization [43], gliosis, and other factors [44] whose contributions are difficult to evaluate.
Relationships between DTI signals and histological markers
In the last few years, several studies have used histology–MRI co-registration to systematically examine the relationships between DTI signals and histological markers in a pixel-by-pixel fashion. For complex disease models, comparison of MRI and DTI data from different animals using conventional approaches based on manually defined regions of interest is often difficult because the location and degree of axon and myelin injuries vary from one animal to another and it is unrealistic to define lesions based solely on MRI and DTI data. By normalizing maps of DTI parameters and histological images to a common space, the relationships between DTI parameters and histological markers of axonal and myelin damage can be examined throughout entire images with lesions and normal regions easily defined in histology. Such an approach was demonstrated recently in a mouse chronic experimental autoimmune encephalomyelitis (EAE) model (Figure 4) [18]. After histology–MRI co-registration, pixel-by-pixel analysis between DTI parameters and histological markers showed significant correlation between axial diffusivity and axonal injury, as measured by neurofilament staining. Pixel-by-pixel analysis between DTI parameters and clinical scores also showed significant correlation between axial diffusivity and behavioral scores. By contrast, radial diffusivity showed large variations in spatial patterns and was not significantly correlated with histological markers, probably because of inflammation in this model. Normalized histological sections showed strong correlation between inflammation, demyelination, and axonal damage in spinal-cord white matter, confirming the complex pathology in this model. The potential impact of this and similar studies on our understanding of complex disease models can hardly be overestimated, because direct, across-the-board comparisons of co-registered histological and MRI and DTI data will inevitably contribute to more complete knowledge about the relationships between histological and MRI and DTI measurements.
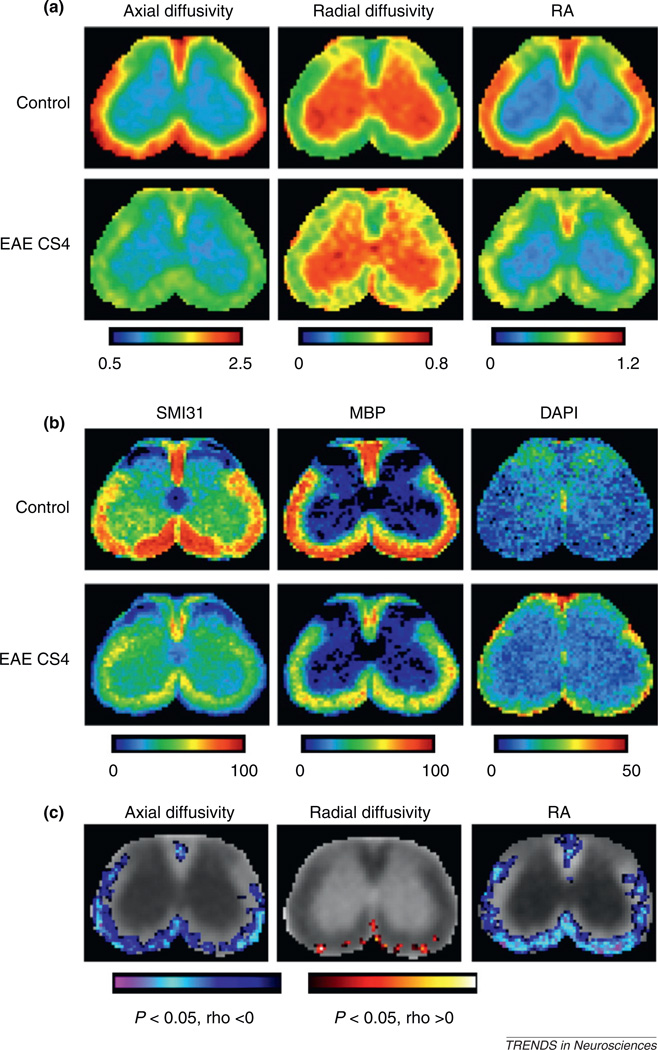
Figure 4.
Co-registration of histological and DTI images allows detailed analysis of the relationships between histological and DTI markers. (a) Average maps of axial diffusivity, radial diffusivity, and relative anisotropy (RA) of the lumbar spinal cord from control mice and mice with severe EAE (clinical score of 4, CS4) [18]. The maps show reduced axial diffusivity and RA values in spinal-cord white matter, especially in the ventrolateral part, of mice with severe EAE compared to control mice. The maps also show increased radial diffusivity values, but with a sporadic pattern. The unit for axial and radial diffusivity is µm2/ms. The color code in these images indicates RA and diffusivity values in the spinal cord, with red indicating the highest levels of RA and diffusivity. (b) Average histological images stained for phosphorylated neurofilament (SMI31), myelin basic protein (MBP), and 4′,6-diamidino-2-phenylindole (DAPI) from control mice and mice with severe EAE at the same lumbar location as shown in (a) [18]. These maps reveal patterns of axonal injury, myelin loss, and inflammatory cells in spinal-cord white matter of mice with severe EAE. The color code in these images indicates the area fraction of positive staining, with red indicating the highest levels of staining. (c) Pixel-by-pixel correlations between DTI markers and clinical scores. In most white-matter regions of the mouse lumbar spinal cord, axial diffusivity and RA were significantly correlated with clinical score, whereas radial diffusivity was not [18]. Adapted, with permission, from [18].
DTI of brain development and its use as an anatomical reference tool
Applications of DTI during brain development
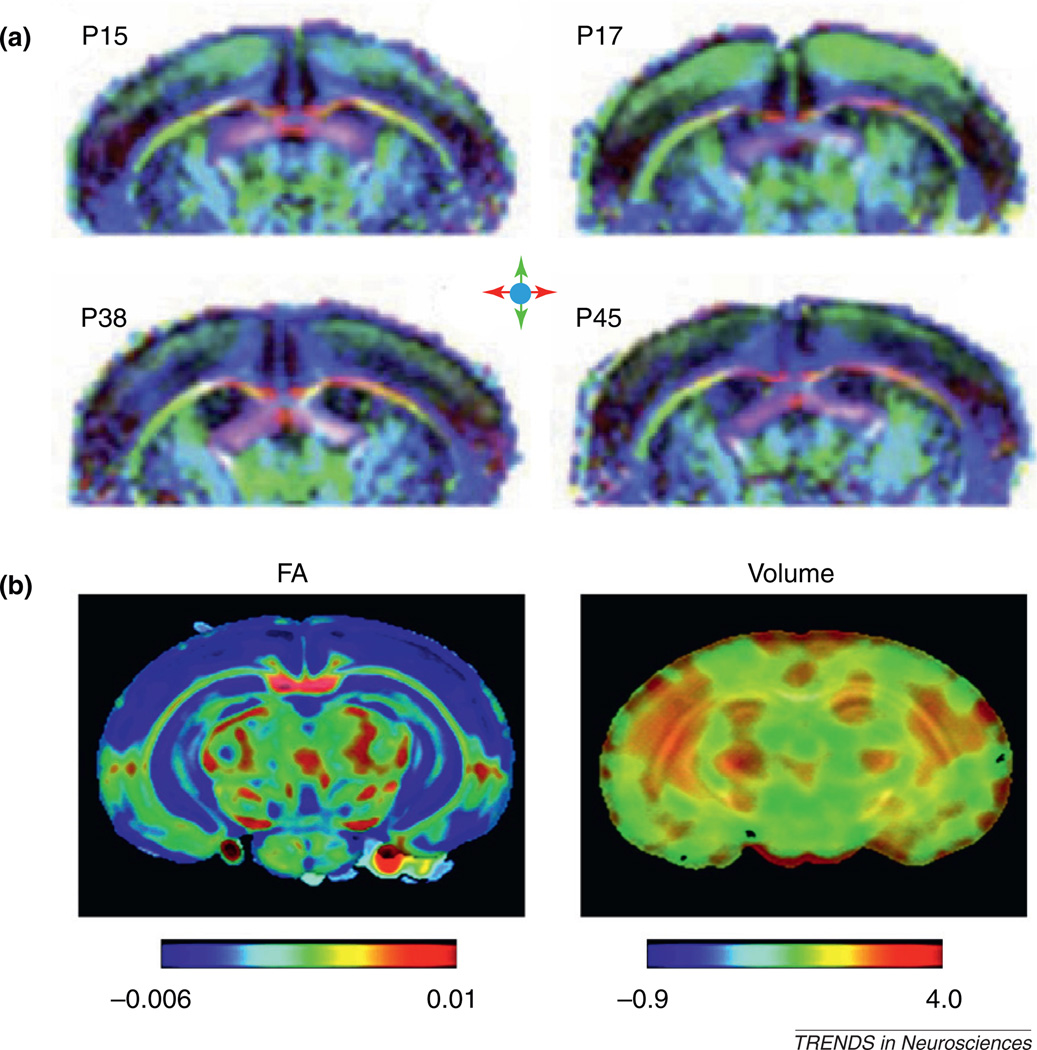
Anatomical changes during brain development and their control mechanisms have been under active investigation. The superb tissue contrasts provided by DTI in embryonic and neonatal rodent brains allow tracking of brain structures and their growth at different developmental stages and may also reveal microstructural changes associated with maturation [39,45]. Several groups measured time-related changes in DTI measurements during postnatal brain development (Figure 5a) [46–48] and showed that in major white-matter structures, ADC values decrease with time, whereas FA values increase with time [47]. A recent study compared histological and DTI data from the post-natal rat corpus callosum and reported that changes in FA correlated strongly with myelination [48]. For gray-matter structures, such as the cortex, both ADC and FA values decrease with time. Another recent study examined the spatiotemporal patterns of FA changes in postnatal rat cortex and showed that FA changes in the cingulate, insular, visual–auditory, and sensory–motor cortices followed slightly different trajectories [49]. The investigators hypothesized that the different trajectories might reflect differences in the degree of structural complexity during development. Detailed spatiotemporal maps of changes in FA, ADC, and local tissue volume in the developing mouse brain based on high-resolution ex vivo DTI data have also been reported [50]. These maps were generated using advanced computational techniques to normalize brain images to a common template, from which time-related changes in MR parameters could be mapped throughout development and volumetric growth could be quantified (Figure 5b) [50].
Figure 5.
DTI of the developing mouse brain. (a) In vivo diffusion tensor direction-encoded color map images of mouse brains on postnatal days 15, 17, 38, and 45, with apparent changes visible in the cerebral cortex. The color schemes are: red, left–right; green, rostral–caudal; and blue, superior–inferior. (b) Time-related changes in FA and local tissue volume in the developing mouse brain. Red indicates an increase in FA values or local tissue volume, and blue indicates a decrease in FA values or local tissue volume over development. The results show widespread decreases in FA in the cortex and hippocampus and increases in tissue volumes mostly in the lateral part of the brain during postnatal development. Adapted, with permission, from [47] (a) and [50] (b).
DTI allows easy reconstruction of the 3D trajectories of major white-matter tracts, as well as visualization of the spatial relationships between white-matter tracts and neighboring structures, so it is useful for screening abnormal white-matter tracts in mutant mouse brains [51,52]. The 3D information provided by DTI may also be useful for guiding subsequent histological examinations. A recent study used DTI in tandem with DiI tracing to investigate Probst bundles in Netrin-1- and DCC-deficient mutant mouse strains, which lack a properly formed corpus callosum [53]. Using DTI-based tractography, the extent of Probst bundles and their spatial relationship with nearby cortical regions were visualized in 3D. The results revealed that Probst bundles consist of axons that had originated from parts of the parietal cortex, in addition to the frontal lobe shown by previous studies using DiI tracing. It should be noted that DTI-based tractography cannot resolve crossing axons, for which more sophisticated diffusion MRI techniques are required. More information on this topic can be found in a recent article in which the authors systematically compared several tractography approaches based on data acquired from post mortem mouse brains using DTI and other diffusion MRI techniques [54].
Advanced imaging analysis techniques
Although gross anatomical changes in the rodent CNS can be detected in DTI data by visual inspection or tractography, the ability to locate subtle structural changes will require advanced image analysis techniques. One such technique is voxel-based analysis (VBA), which involves voxel-wise cross-subject analysis in images that have been spatially normalized to a common space [55]. The advantage of VBA is that it can examine the entire brain in an automated fashion, without requiring a priori knowledge of the locations of structural changes. A recent study applied VBA to examine ex vivo DTI data for a transgenic mouse model of early-onset dystonia, and the results showed abnormal FA values in the cerebellothalamic and thalamocortical pathways [56]. Another study applied VBA and in vivo DTI to examine structural remodeling of the rat hippocampus and reported significant changes in mean diffusivity in the rat hippocampus after 2 h of repeated water maze tasks [57]. VBA, however, demands accurate spatial normalization of subject images, which may be difficult to achieve when severe injury or tissue deformation is present, such as in rodent models of stroke. For such models, tract-based spatial statistics (TBSS) [58] can assist in localizing changes in major white-matter tracts. TBSS has been applied to evaluate potential recovery of injured rat corticospinal tracts after ischemic stroke and revealed significant increases in FA values in the ipsilesional cerebral peduncle and internal capsule at 70 days after stroke [59].
Digital atlases of the rodent brain
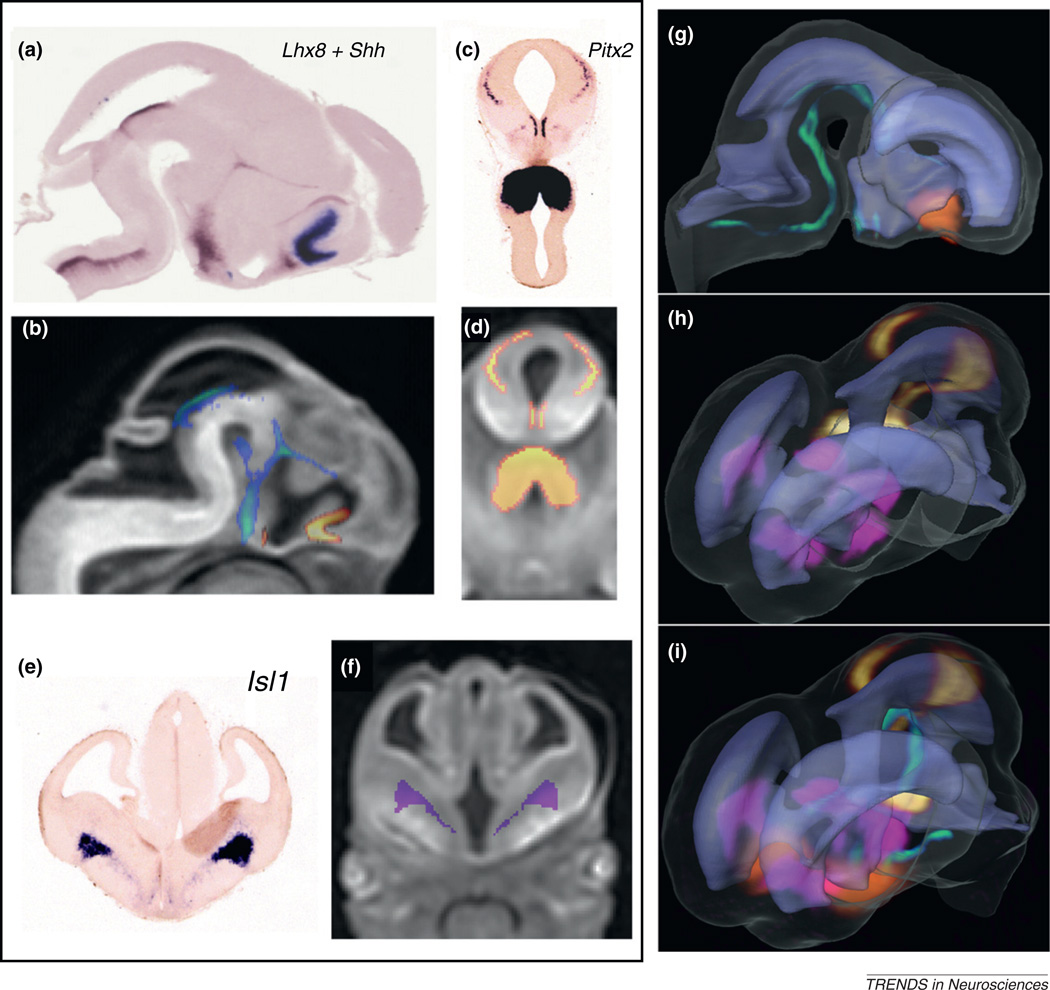
With high-resolution ex vivo DTI data, several digitized rodent brain atlases have been established to visualize 3D brain structures, to guide stereotaxic operations, and to serve as a common framework for data integration and analysis [60–63]. The 3D visualization of brain anatomy by DTI also lends itself to mapping and integration of experimental data from different modalities, such as gene and proteomic expression patterns [64,65]. Studies based on high-throughput techniques, such as in situ hybridization (ISH), contribute an increasing wealth of snapshot data on the expression of individual genes at specific locations in the developing mouse brain. However, a comprehensive understanding of the regionalized activation patterns of multiple genes and their spatiotemporal relations with the evolving embryonic neuroanatomy requires integration of these data to a common anatomical framework [66–68]. High-resolution DTI in the embryonic stages offers one such anatomical framework by which to register expression data from histological sectioning of multiple specimens [16,69]. Figure 6 illustrates 3D registration and visualization of gene expression patterns from serial ISH sections to µDTI images of an embryonic (E12) mouse brain. Serial ISH sections from E12 brains examined for the expression of several homeobox transcription factors and the sonic hedgehog homolog (Shh) gene were mapped to MR images using intensity- and landmark-based nonlinear image-matching techniques [16]. From the original data sets for 2D ISH sections, it was difficult to determine the spatial relations or potential overlaps between the expression sites of different genes (Figure 6a,c,e). Mapping of the expression profiles of multiple genes from different specimens and sectioning orientations facilitated simultaneous visualization of the 3D expression patterns and relative spatial distributions of these genes within the E12 mouse brain (Figure 6g–i). In addition, the mapping of gene expression data to DTI images can provide complementary information for a better understanding of embryonic CNS development, with delineation of anatomical structures from DTI and developmental boundaries that are defined by spatially restricted patterns of gene expression. DTI of the rodent brain can thus be used as an anatomical reference for mapping and interpretation of histological data, gene expression patterns, and proteomic profiles. Hence, it will be an important modality in ongoing efforts to create multi-modal digital atlases of the developing mouse brain [68].
Figure 6.
Mapping of gene expression data to 3D DTI images in an E12 mouse embryo. Expression data for (a) LIM homeobox 8 (Lhx8) and Shh from serial sagittal ISH sections and (c) pituitary homeobox 2 (Pitx2) and (e) LIM homeobox 1 (Isl1) from serial coronal sections were mapped to (b,d,f) MR images using intensity- and landmark-based registration [16]. (g,h,i) Mapping of the expression data allowed visualization of the 3D expression patterns of Lhx8 (orange in g and i), Shh (green in g and i), Pitx2 (yellow in h and i), and Isl1 (pink in h and i), and their relative spatial distributions in the embryonic brain in relation to anatomical structures such as the ventricles (rendered in light purple). Adapted, with permission, from [16].
Concluding remarks and future perspectives
In summary, DTI is a useful tool for monitoring disease progression, studying brain development, and characterizing anatomical phenotypes. Recent technical developments in DTI allow the rodent CNS to be examined in greater detail. We expect that future technical advances in imaging instruments will continue to push the resolution and speed limits of DTI, especially in vivo rodent DTI, to extract more information from the rodent CNS. For example, a recent report demonstrated that the newly available cryogenic probe can significantly improve the resolution and speed of mouse brain MRI [70]. It should be noted that DTI is based on a simplified model, and complex tissue microstructure or pathology often diminishes the value of DTI (Box 2). Integration of DTI with other imaging modalities may help to resolve such complex cases and further enhance our ability to examine various aspects of the rodent CNS.
Box 2. Outstanding questions.
Although major gray- and white-matter structures in the rodent brain have been delineated with DTI [60,61], systematic efforts to identify and delineate small structures in the rodent brain (e.g. subregions in the thalamus and transient structures during embryonic and neonatal stages) are still lacking. Fully segmented and annotated high-resolution maps or atlases of the rodent brain are essential for DTI application in phenotyping and developmental studies, as well as for detection of brain injuries.
A variety of microstructural changes happen during the repair and reorganizational stages that follow brain injury. Can DTI, by itself or in combination with other imaging modalities, detect and quantify such changes? If so, what are the contrast mechanisms that allow DTI to detect such changes?
Acknowledgments
The authors are supported by National Institutes of Health grants R21NS059529 (J.Z.), R21NS065306 (J.Z.), R01NS070909 (J.Z.), R01AG20012 (S.M.), and R01EB003543 (S.M.). We thank Mary McAllister (Johns Hopkins University) for her editorial assistance.
Glossary
- Apparent diffusion coefficient (ADC)
water diffusion in tissue is hindered or restricted by many structural barriers, such as cell membranes and myelin sheaths. As a result, the mean square distance traveled by water molecules in unit time, or diffusivity, depends on its environment and is sensitive to changes in tissue microstructure. ADC is equivalent to the diffusivity when the underlying diffusion process is Gaussian. In normal tissue, the ADC value is approximately 1 × 10–3 mm2/s.
- Diffusion anisotropy
in biological tissues, diffusivity or ADC values may vary depending on the direction of the diffusion-sensitizing magnetic field gradients (see Box 1 for details). This phenomenon is called anisotropic diffusion and is the source of the unique tissue contrasts that make DTI useful.
- Diffusion tensor
the 3 × 3 matrix used to characterize the 3D properties of the diffusion of water molecules based on a Gaussian model. From each diffusion tensor, three pairs of eigenvalues and eigenvectors can be calculated using matrix diagonalization. The eigenvector that corresponds to the largest eigenvalue is called the primary eigenvector, whose orientation often coincides with that of the axon fibers in most white-matter tracts. The model has its own limitations and cannot resolve complex tissue microstructures such as crossing fibers.
- DTI tractography
technique for reconstructing the 3D trajectories of major white-matter tracts based on diffusion tensor data. DTI tractography is useful for visualizing 3D trajectories of white-matter tracts in normal and diseased brains. There are several ways to perform DTI tractography, including deterministic and probabilistic methods.
References
- 1.Basser PJ, et al. MR diffusion tensor spectroscopy and imaging. Biophys. J. 1994;66:259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le Bihan D. Looking into the functional architecture of the brain with diffusion MRI. Nat. Rev. Neurosci. 2003;4:469–480. doi: 10.1038/nrn1119. [DOI] [PubMed] [Google Scholar]
- 3.Mori S, Zhang J. Principles of diffusion tensor imaging and its applications to basic neuroscience research. Neuron. 2006;51:527–539. doi: 10.1016/j.neuron.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Basser PJ, Jones DK. Diffusion-tensor MRI: theory, experimental design and data analysis - a technical review. NMR Biomed. 2002;15:456–467. doi: 10.1002/nbm.783. [DOI] [PubMed] [Google Scholar]
- 5.Jones DK. Diffusion MRI: Theory, Methods, and Applications. 1st edn. Oxford University Press; 2000. [Google Scholar]
- 6.Mori S. Introduction to Diffusion Tensor Imaging. Elsevier; 2007. [Google Scholar]
- 7.Johansen-Berg H, Behrens TEJ, editors. Diffusion MRI: From Quantitative Measurement to In Vivo Neuroanatomy. 1st edn. Academic Press; 2009. [Google Scholar]
- 8.Inglis BA, et al. Diffusion anisotropy in excised normal rat spinal cord measured by NMR microscopy. Magn. Reson. Imaging. 1997;15:441–450. doi: 10.1016/s0730-725x(96)00242-1. [DOI] [PubMed] [Google Scholar]
- 9.Gulani V, et al. A multiple echo pulse sequence for diffusion tensor imaging and its application in excised rat spinal cords. Magn. Reson. Med. 1997;38:868–873. doi: 10.1002/mrm.1910380603. [DOI] [PubMed] [Google Scholar]
- 10.van de Looij Y, et al. Diffusion tensor echo planar imaging using surface coil transceiver with a semiadiabatic RF pulse sequence at 14.1T. Magn. Reson. Med. 2011;65:732–737. doi: 10.1002/mrm.22656. [DOI] [PubMed] [Google Scholar]
- 11.Asanuma T, et al. Diffusion tensor imaging and fiber tractography of C6 rat glioma. J. Magn. Reson. Imaging. 2008;28:566–573. doi: 10.1002/jmri.21473. [DOI] [PubMed] [Google Scholar]
- 12.Harsan LA, et al. In vivo diffusion tensor magnetic resonance imaging and fiber tracking of the mouse brain. NMR Biomed. 2010;23:884–896. doi: 10.1002/nbm.1496. [DOI] [PubMed] [Google Scholar]
- 13.Kim JH, et al. Diffusion tensor imaging of mouse brain stem and cervical spinal cord. J. Neurosci. Methods. 2009;176:186–191. doi: 10.1016/j.jneumeth.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Callagan PT. Principles of Nuclear Magnetic Resonance Microscopy. Oxford University Press; 1991. [Google Scholar]
- 15.Flint JJ, et al. Cellular-level diffusion tensor microscopy and fiber tracking in mammalian nervous tissue with direct histological correlation. Neuroimage. 2010;52:556–561. doi: 10.1016/j.neuroimage.2010.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aggarwal M, et al. Three-dimensional diffusion tensor microimaging for anatomical characterization of the mouse brain. Magn. Reson. Med. 2010;64:249–261. doi: 10.1002/mrm.22426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Y, Johnson GA. Microscopic diffusion tensor imaging of the mouse brain. Neuroimage. 2010;50:465–471. doi: 10.1016/j.neuroimage.2009.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Budde MD, et al. Axial diffusivity is the primary correlate of axonal injury in the experimental autoimmune encephalomyelitis spinal cord: a quantitative pixelwise analysis. J. Neurosci. 2009;29:2805–2813. doi: 10.1523/JNEUROSCI.4605-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan KC, et al. Late measures of microstructural alterations in severe neonatal hypoxic-ischemic encephalopathy by MR diffusion tensor imaging. Int. J. Dev. Neurosci. 2009;27:607–615. doi: 10.1016/j.ijdevneu.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 20.Wang S, et al. Characterization of white matter injury in a hypoxic-ischemic neonatal rat model by diffusion tensor MRI. Stroke. 2008;39:2348–2353. doi: 10.1161/STROKEAHA.107.509927. [DOI] [PubMed] [Google Scholar]
- 21.Stone BS, et al. Delayed neural network degeneration after neonatal hypoxia-ischemia. Ann. Neurol. 2008;64:535–546. doi: 10.1002/ana.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Zijden JP, et al. Longitudinal in vivo MRI of alterations in perilesional tissue after transient ischemic stroke in rats. Exp. Neurol. 2008;212:207–212. doi: 10.1016/j.expneurol.2008.03.027. [DOI] [PubMed] [Google Scholar]
- 23.Underwood CK, et al. Non-invasive diffusion tensor imaging detects white matter degeneration in the spinal cord of a mouse model of amyotrophic lateral sclerosis. Neuroimage. 2011;55:455–461. doi: 10.1016/j.neuroimage.2010.12.044. [DOI] [PubMed] [Google Scholar]
- 24.Kim JH, et al. Noninvasive detection of brainstem and spinal cord axonal degeneration in an amyotrophic lateral sclerosis mouse model. NMR Biomed. 2011;24:163–169. doi: 10.1002/nbm.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JH, et al. Diffusion tensor imaging at 3 hours after traumatic spinal cord injury predicts long-term locomotor recovery. J. Neurotrauma. 2010;27:587–598. doi: 10.1089/neu.2009.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mac Donald CL, et al. Diffusion tensor imaging reliably detects experimental traumatic axonal injury and indicates approximate time of injury. J. Neurosci. 2007;27:11869–11876. doi: 10.1523/JNEUROSCI.3647-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song SK, et al. Diffusion tensor imaging detects and differentiates axon and myelin degeneration in mouse optic nerve after retinal ischemia. Neuroimage. 2003;20:1714–1722. doi: 10.1016/j.neuroimage.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 28.Song SK, et al. Dysmyelination revealed through MRI as increased radial (but unchanged axial) diffusion of water. Neuroimage. 2002;17:1429–1436. doi: 10.1006/nimg.2002.1267. [DOI] [PubMed] [Google Scholar]
- 29.Song SK, et al. Demyelination increases radial diffusivity in corpus callosum of mouse brain. Neuroimage. 2005;26:132–140. doi: 10.1016/j.neuroimage.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 30.Sun SW, et al. Evolving Wallerian degeneration after transient retinal ischemia in mice characterized by diffusion tensor imaging. Neuroimage. 2008;40:1–10. doi: 10.1016/j.neuroimage.2007.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie M, et al. Delayed axonal degeneration in slow Wallerian degeneration mutant mice detected using diffusion tensor imaging. Neuroscience. 2011;197:339–347. doi: 10.1016/j.neuroscience.2011.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun SW, et al. Noninvasive detection of cuprizone induced axonal damage and demyelination in the mouse corpus callosum. Magn. Reson. Med. 2006;55:302–308. doi: 10.1002/mrm.20774. [DOI] [PubMed] [Google Scholar]
- 33.Xie M, et al. Rostrocaudal analysis of corpus callosum demyelination and axon damage across disease stages refines diffusion tensor imaging correlations with pathological features. J. Neuropathol. Exp. Neurol. 2010;69:704–716. doi: 10.1097/NEN.0b013e3181e3de90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, et al. In vivo and ex vivo diffusion tensor imaging of cuprizone-induced demyelination in the mouse corpus callosum. Magn. Reson. Med. 2012;67:750–759. doi: 10.1002/mrm.23032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, et al. Quantification of increased cellularity during inflammatory demyelination. Brain. 2011;134:3590–3601. doi: 10.1093/brain/awr307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, et al. Diffusion tensor magnetic resonance imaging of Wallerian degeneration in rat spinal cord after dorsal root axotomy. J. Neurosci. 2009;29:3160–3171. doi: 10.1523/JNEUROSCI.3941-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mac Donald CL, et al. Detection of traumatic axonal injury with diffusion tensor imaging in a mouse model of traumatic brain injury. Exp. Neurol. 2007;205:116–131. doi: 10.1016/j.expneurol.2007.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merkler D, et al. Multicontrast MRI of remyelination in the central nervous system. NMR Biomed. 2005;18:395–403. doi: 10.1002/nbm.972. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, et al. Three-dimensional anatomical characterization of the developing mouse brain by diffusion tensor microimaging. Neuroimage. 2003;20:1639–1648. doi: 10.1016/s1053-8119(03)00410-5. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J, et al. Three dimensional diffusion tensor magnetic resonance micro-imaging of adult mouse brain and hippocampus. Neuroimage. 2002;15:892–901. doi: 10.1006/nimg.2001.1012. [DOI] [PubMed] [Google Scholar]
- 41.Laitinen T, et al. Diffusion tensor MRI of axonal plasticity in the rat hippocampus. Neuroimage. 2010;51:521–530. doi: 10.1016/j.neuroimage.2010.02.077. [DOI] [PubMed] [Google Scholar]
- 42.Sizonenko SV, et al. Developmental changes and injury induced disruption of the radial organization of the cortex in the immature rat brain revealed by in vivo diffusion tensor MRI. Cereb. Cortex. 2007;17:2609–2617. doi: 10.1093/cercor/bhl168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang Q, et al. MRI evaluation of axonal reorganization after bone marrow stromal cell treatment of traumatic brain injury. NMR Biomed. 2011;24:1119–1128. doi: 10.1002/nbm.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Budde MD, et al. The contribution of gliosis to diffusion tensor anisotropy and tractography following traumatic brain injury: validation in the rat using Fourier analysis of stained tissue sections. Brain. 2011;134:2248–2260. doi: 10.1093/brain/awr161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, et al. Mapping postnatal mouse brain development with diffusion tensor microimaging. Neuroimage. 2005;26:1042–1051. doi: 10.1016/j.neuroimage.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 46.Bockhorst KH, et al. Early postnatal development of rat brain: in vivo diffusion tensor imaging. J. Neurosci. Res. 2008;86:1520–1528. doi: 10.1002/jnr.21607. [DOI] [PubMed] [Google Scholar]
- 47.Chahboune H, et al. Neurodevelopment of C57B/L6 mouse brain assessed byin vivo diffusion tensor imaging. NMR Biomed. 2007;20:375–382. doi: 10.1002/nbm.1130. [DOI] [PubMed] [Google Scholar]
- 48.Jito J, et al. Maturational changes in diffusion anisotropy in the rat corpus callosum: comparison with quantitative histological evaluation. J. Magn. Reson. Imaging. 2008;28:847–854. doi: 10.1002/jmri.21496. [DOI] [PubMed] [Google Scholar]
- 49.Huang H, et al. Quantitative cortical mapping of fractional anisotropy in developing rat brains. J. Neurosci. 2008;28:1427–1433. doi: 10.1523/JNEUROSCI.3194-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baloch S, et al. Quantification of brain maturation and growth patterns in C57BL/6J mice via computational neuroanatomy of diffusion tensor images. Cereb. Cortex. 2009;19:675–687. doi: 10.1093/cercor/bhn112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J, et al. Magnetic resonance diffusion tensor microimaging reveals a role for Bcl-x in brain development and homeostasis. J. Neurosci. 2005;25:1881–1888. doi: 10.1523/JNEUROSCI.4129-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y, et al. Axonal growth and guidance defects in Frizzled3. knock-out mice: a comparison of diffusion tensor magnetic resonance imaging, neurofilament staining, and genetically directed cell labeling. J. Neurosci. 2006;26:355–364. doi: 10.1523/JNEUROSCI.3221-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ren T, et al. Diffusion tensor magnetic resonance imaging and tract-tracing analysis of Probst bundle structure in Netrin1- and DCC-deficient mice. J. Neurosci. 2007;27:10345–10349. doi: 10.1523/JNEUROSCI.2787-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moldrich RX, et al. Comparative mouse brain tractography of diffusion magnetic resonance imaging. Neuroimage. 2010;51:1027–1036. doi: 10.1016/j.neuroimage.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashburner J, Friston KJ. Voxel-based morphometry -the methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 56.Ulug AM, et al. Cerebellothalamocortical pathway abnormalities in torsinA DYT1 knock-in mice. Proc. Natl. Acad. SciU.SA. 2011;108:6638–6643. doi: 10.1073/pnas.1016445108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sagi Y, et al. Learning in the fast lane: new insights into neuroplasticity. Neuron. 2012;73:1195–1203. doi: 10.1016/j.neuron.2012.01.025. [DOI] [PubMed] [Google Scholar]
- 58.Smith SM, et al. Acquisition and voxelwise analysis of multi-subject diffusion data with tract-based spatial statistics. Nat. Protoc. 2007;2:499–503. doi: 10.1038/nprot.2007.45. [DOI] [PubMed] [Google Scholar]
- 59.van Meer MP, et al. Extent of bilateral neuronal network reorganization and functional recovery in relation to stroke severity. J. Neurosci. 2012;32:4495–4507. doi: 10.1523/JNEUROSCI.3662-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang Y, Johnson GA. Microscopic diffusion tensor atlas of the mouse brain. Neuroimage. 2011;56:1235–1243. doi: 10.1016/j.neuroimage.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chuang N, et al. An MRI-based atlas and database of the developing mouse brain. Neuroimage. 2011;54:80–89. doi: 10.1016/j.neuroimage.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aggarwal M, et al. Magnetic resonance imaging and micro-computed tomography combined atlas of developing and adult mouse brains for stereotaxic surgery. Neuroscience. 2009;162:1339–1350. doi: 10.1016/j.neuroscience.2009.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chugh BP, et al. Measurement of cerebral blood volume in mouse brain regions using micro-computed tomography. Neuroimage. 2009;47:1312–1318. doi: 10.1016/j.neuroimage.2009.03.083. [DOI] [PubMed] [Google Scholar]
- 64.MacKenzie-Graham A, et al. The informatics of a C57BL/6J mouse brain atlas. Neuroinformatics. 2003;1:397–410. doi: 10.1385/NI:1:4:397. [DOI] [PubMed] [Google Scholar]
- 65.MacKenzie-Graham A, et al. A multimodal, multidimensional atlas of the C57BL/6J mouse brain. J. Anat. 2004;204:93–102. doi: 10.1111/j.1469-7580.2004.00264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 67.Streicher J, Muller GB. 3D modelling of gene expression patterns. Trends Biotechnol. 2001;19:145–148. doi: 10.1016/s0167-7799(00)01571-7. [DOI] [PubMed] [Google Scholar]
- 68.Hawrylycz M, et al. Digital atlasing and standardization in the mouse brain. PLoS Comput. Biol. 2011;7:e1001065. doi: 10.1371/journal.pcbi.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dhenain M, et al. Three-dimensional digital mouse atlas using high-resolution MRI. Dev. Biol. 2001;232:458–470. doi: 10.1006/dbio.2001.0189. [DOI] [PubMed] [Google Scholar]
- 70.Baltes C, et al. Micro MRI of the mouse brain using a novel 400 MHz cryogenic quadrature RF probe. NMR Biomed. 2009;22:834–842. doi: 10.1002/nbm.1396. [DOI] [PubMed] [Google Scholar]
- 71.Basser PJ, et al. Estimation of the effective self-diffusion tensor from the NMR spin echo. J. Magn. Reson. Ser. B. 1994;103:247–254. doi: 10.1006/jmrb.1994.1037. [DOI] [PubMed] [Google Scholar]
- 72.Hui ES, et al. B-value dependence of DTI quantitation and sensitivity in detecting neural tissue changes. Neuroimage. 2010;49:2366–2374. doi: 10.1016/j.neuroimage.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 73.Kim S, et al. Dependence on diffusion time of apparent diffusion tensor of ex vivo calf tongue and heart. Magn. Reson. Med. 2005;54:1387–1396. doi: 10.1002/mrm.20676. [DOI] [PubMed] [Google Scholar]
- 74.Clark CA, et al. Diffusion time dependence of the apparent diffusion tensor in healthy human brain and white matter disease. Magn. Reson. Med. 2001;45:1126–1129. doi: 10.1002/mrm.1149. [DOI] [PubMed] [Google Scholar]
- 75.Basser PJ, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. J. Magn. Reson. Ser. B. 1996;111:209–219. doi: 10.1006/jmrb.1996.0086. [DOI] [PubMed] [Google Scholar]
- 76.Sotak CH. The role of diffusion tensor imaging in the evaluation of ischemic brain injury - a review. NMR Biomed. 2002;15:561–569. doi: 10.1002/nbm.786. [DOI] [PubMed] [Google Scholar]
- 77.Hasan KM, et al. Does fractional anisotropy have better noise immunity characteristics than relative anisotropy in diffusion tensor MRI? An analytical approach. Magn. Reson. Med. 2004;51:413–417. doi: 10.1002/mrm.10682. [DOI] [PubMed] [Google Scholar]
- 78.Pajevic S, Pierpaoli C. Color schemes to represent the orientation of anisotropic tissues from diffusion tensor data: 1999 [PubMed] [Google Scholar]