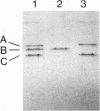
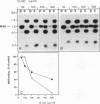
Abstract
Chromatin in eukaryotic nuclei is thought to be partitioned into functional loop domains that are generated by the binding of defined DNA sequences, named MARs (matrix attachment regions), to the nuclear matrix. We have previously identified B-type lamins as MAR-binding matrix components (M. E. E. Ludérus, A. de Graaf, E. Mattia, J. L. den Blaauwen, M. A. Grande, L. de Jong, and R. van Driel, Cell 70:949-959, 1992). Here we show that A-type lamins and the structurally related proteins desmin and NuMA also specifically bind MARs in vitro. We studied the interaction between MARs and lamin polymers in molecular detail and found that the interaction is saturable, of high affinity, and evolutionarily conserved. Competition studies revealed the existence of two different types of interaction related to different structural features of MARs: one involving the minor groove of double-stranded MAR DNA and one involving single-stranded regions. We obtained similar results for the interaction of MARs with intact nuclear matrices from rat liver. A model in which the interaction of nuclear matrix proteins with single-stranded MAR regions serves to stabilize the transcriptionally active state of chromatin is discussed.
Full text
PDF

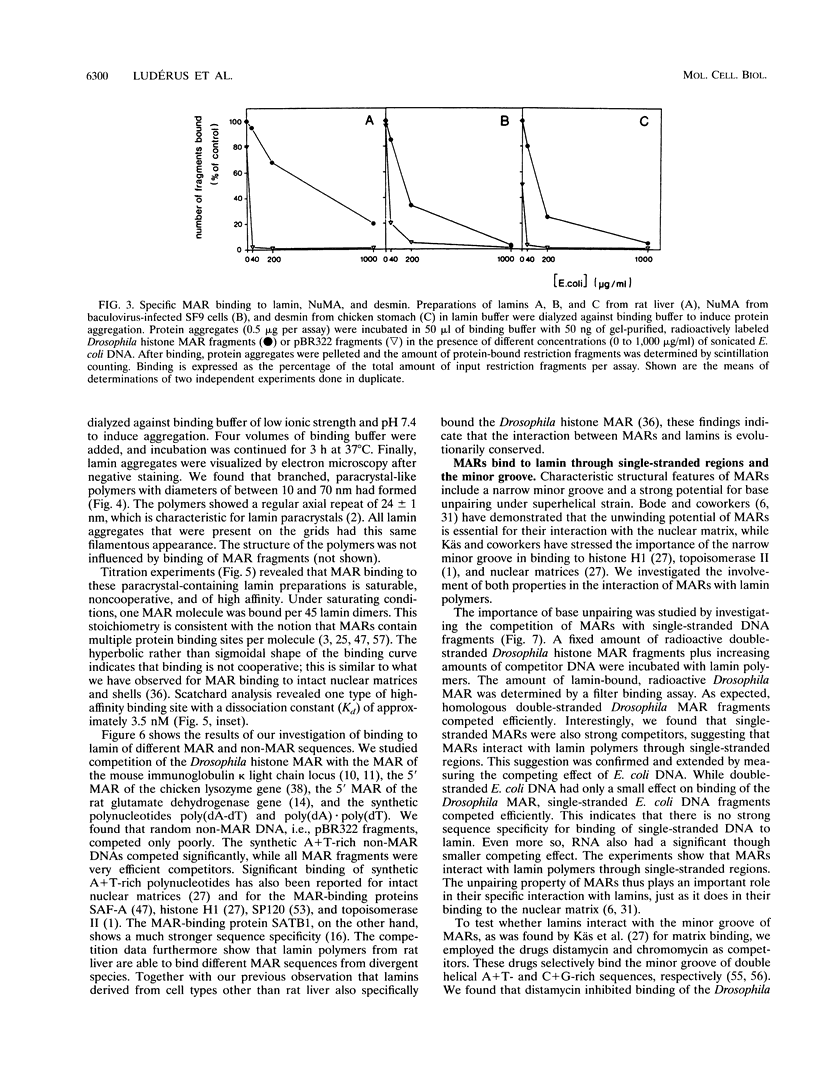
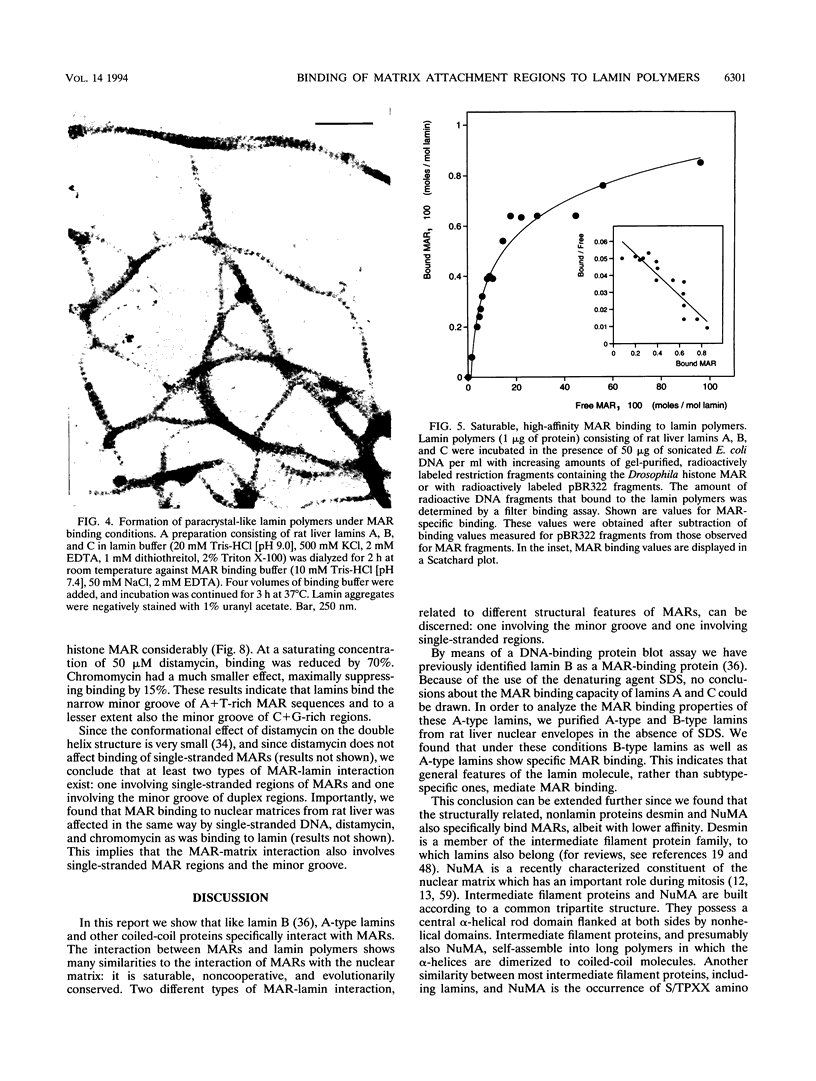
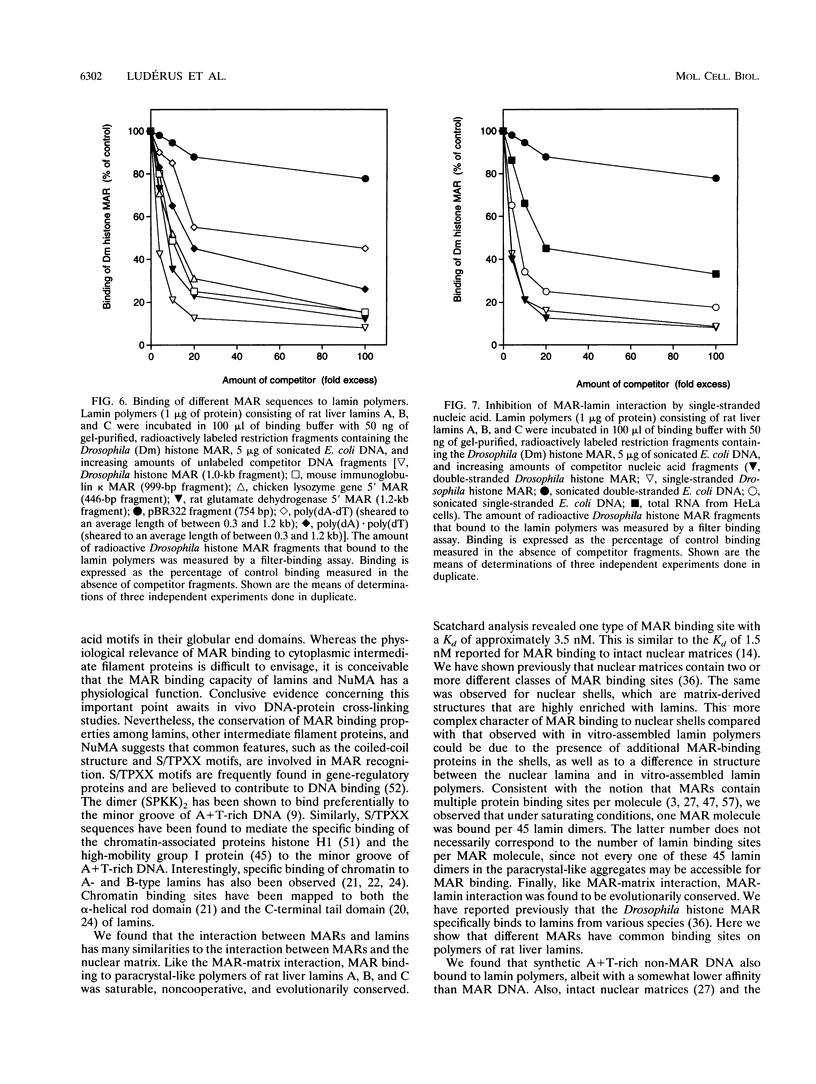
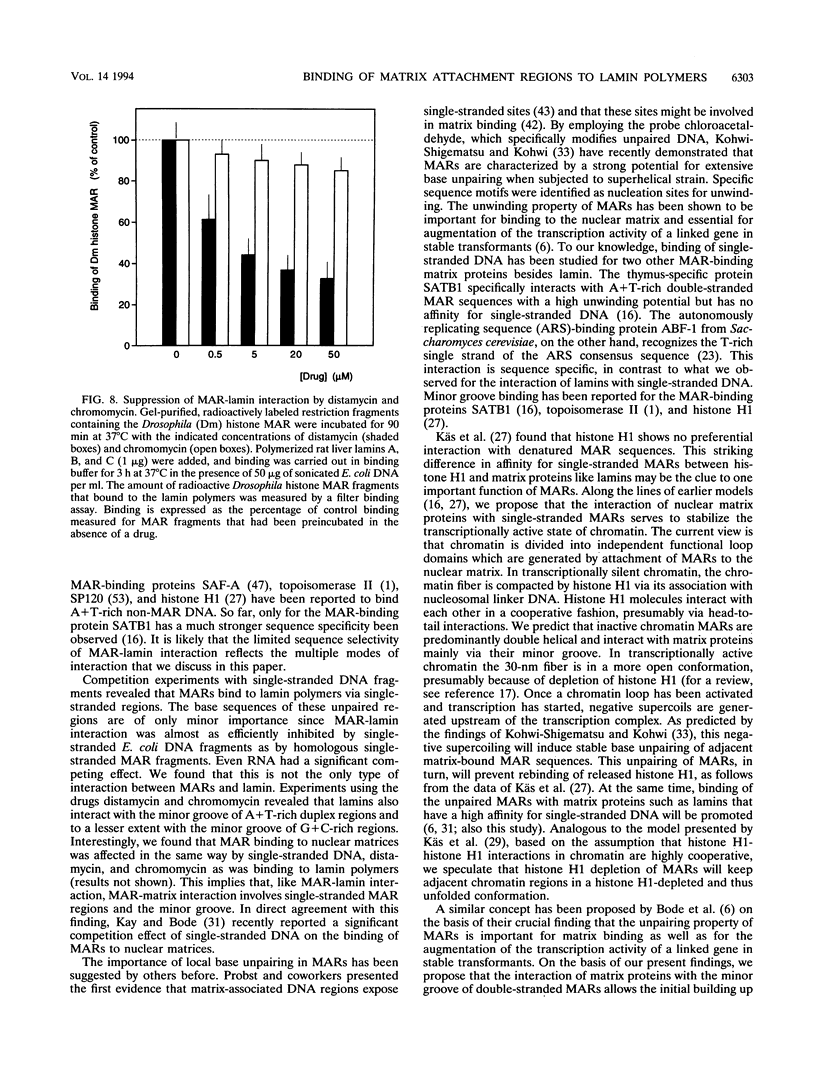


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi Y., Käs E., Laemmli U. K. Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. EMBO J. 1989 Dec 20;8(13):3997–4006. doi: 10.1002/j.1460-2075.1989.tb08582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi U., Cohn J., Buhle L., Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 1986 Oct 9;323(6088):560–564. doi: 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- Amati B., Pick L., Laroche T., Gasser S. M. Nuclear scaffold attachment stimulates, but is not essential for ARS activity in Saccharomyces cerevisiae: analysis of the Drosophila ftz SAR. EMBO J. 1990 Dec;9(12):4007–4016. doi: 10.1002/j.1460-2075.1990.tb07622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney R., Coffey D. S. Identification of a nuclear protein matrix. Biochem Biophys Res Commun. 1974 Oct 23;60(4):1410–1417. doi: 10.1016/0006-291x(74)90355-6. [DOI] [PubMed] [Google Scholar]
- Blasquez V. C., Xu M., Moses S. C., Garrard W. T. Immunoglobulin kappa gene expression after stable integration. I. Role of the intronic MAR and enhancer in plasmacytoma cells. J Biol Chem. 1989 Dec 15;264(35):21183–21189. [PubMed] [Google Scholar]
- Bode J., Kohwi Y., Dickinson L., Joh T., Klehr D., Mielke C., Kohwi-Shigematsu T. Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science. 1992 Jan 10;255(5041):195–197. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- Bonifer C., Hecht A., Saueressig H., Winter D. M., Sippel A. E. Dynamic chromatin: the regulatory domain organization of eukaryotic gene loci. J Cell Biochem. 1991 Oct;47(2):99–108. doi: 10.1002/jcb.240470203. [DOI] [PubMed] [Google Scholar]
- Bonifer C., Vidal M., Grosveld F., Sippel A. E. Tissue specific and position independent expression of the complete gene domain for chicken lysozyme in transgenic mice. EMBO J. 1990 Sep;9(9):2843–2848. doi: 10.1002/j.1460-2075.1990.tb07473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill M. E., Suzuki M. 'SPKK' motifs prefer to bind to DNA at A/T-rich sites. EMBO J. 1989 Dec 20;8(13):4189–4195. doi: 10.1002/j.1460-2075.1989.tb08604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986 Jan 31;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage sites appear to be evolutionarily conserved. FEBS Lett. 1986 Aug 11;204(1):5–7. doi: 10.1016/0014-5793(86)81377-1. [DOI] [PubMed] [Google Scholar]
- Compton D. A., Cleveland D. W. NuMA is required for the proper completion of mitosis. J Cell Biol. 1993 Feb;120(4):947–957. doi: 10.1083/jcb.120.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton D. A., Szilak I., Cleveland D. W. Primary structure of NuMA, an intranuclear protein that defines a novel pathway for segregation of proteins at mitosis. J Cell Biol. 1992 Mar;116(6):1395–1408. doi: 10.1083/jcb.116.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A. T., Ludérus M. E., Lamers W. H. Identification and analysis of a matrix-attachment region 5' of the rat glutamate-dehydrogenase-encoding gene. Eur J Biochem. 1993 Aug 1;215(3):777–785. doi: 10.1111/j.1432-1033.1993.tb18092.x. [DOI] [PubMed] [Google Scholar]
- Dickinson L. A., Joh T., Kohwi Y., Kohwi-Shigematsu T. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 1992 Aug 21;70(4):631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Laemmli U. K. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986 Aug 15;46(4):521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- Gerace L., Burke B. Functional organization of the nuclear envelope. Annu Rev Cell Biol. 1988;4:335–374. doi: 10.1146/annurev.cb.04.110188.002003. [DOI] [PubMed] [Google Scholar]
- Gerace L., Foisner R. Integral membrane proteins and dynamic organization of the nuclear envelope. Trends Cell Biol. 1994 Apr;4(4):127–131. doi: 10.1016/0962-8924(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Glass C. A., Glass J. R., Taniura H., Hasel K. W., Blevitt J. M., Gerace L. The alpha-helical rod domain of human lamins A and C contains a chromatin binding site. EMBO J. 1993 Nov;12(11):4413–4424. doi: 10.1002/j.1460-2075.1993.tb06126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass J. R., Gerace L. Lamins A and C bind and assemble at the surface of mitotic chromosomes. J Cell Biol. 1990 Sep;111(3):1047–1057. doi: 10.1083/jcb.111.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J. F., Gasser S. M. Identification and purification of a protein that binds the yeast ARS consensus sequence. Cell. 1991 Mar 8;64(5):951–960. doi: 10.1016/0092-8674(91)90319-t. [DOI] [PubMed] [Google Scholar]
- Homberger H. P. Bent DNA is a structural feature of scaffold-attached regions in Drosophila melanogaster interphase nuclei. Chromosoma. 1989 Aug;98(2):99–104. doi: 10.1007/BF00291044. [DOI] [PubMed] [Google Scholar]
- Höger T. H., Krohne G., Kleinschmidt J. A. Interaction of Xenopus lamins A and LII with chromatin in vitro mediated by a sequence element in the carboxyterminal domain. Exp Cell Res. 1991 Dec;197(2):280–289. doi: 10.1016/0014-4827(91)90434-v. [DOI] [PubMed] [Google Scholar]
- Jarman A. P., Higgs D. R. Nuclear scaffold attachment sites in the human globin gene complexes. EMBO J. 1988 Nov;7(11):3337–3344. doi: 10.1002/j.1460-2075.1988.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann S. H., Shaper J. H. A subset of non-histone nuclear proteins reversibly stabilized by the sulfhydryl cross-linking reagent tetrathionate. Polypeptides of the internal nuclear matrix. Exp Cell Res. 1984 Dec;155(2):477–495. doi: 10.1016/0014-4827(84)90208-8. [DOI] [PubMed] [Google Scholar]
- Kay V., Bode J. Binding specificity of a nuclear scaffold: supercoiled, single-stranded, and scaffold-attached-region DNA. Biochemistry. 1994 Jan 11;33(1):367–374. doi: 10.1021/bi00167a047. [DOI] [PubMed] [Google Scholar]
- Klehr D., Maass K., Bode J. Scaffold-attached regions from the human interferon beta domain can be used to enhance the stable expression of genes under the control of various promoters. Biochemistry. 1991 Feb 5;30(5):1264–1270. doi: 10.1021/bi00219a015. [DOI] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T., Kohwi Y. Torsional stress stabilizes extended base unpairing in suppressor sites flanking immunoglobulin heavy chain enhancer. Biochemistry. 1990 Oct 16;29(41):9551–9560. doi: 10.1021/bi00493a009. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käs E., Izaurralde E., Laemmli U. K. Specific inhibition of DNA binding to nuclear scaffolds and histone H1 by distamycin. The role of oligo(dA).oligo(dT) tracts. J Mol Biol. 1989 Dec 5;210(3):587–599. doi: 10.1016/0022-2836(89)90134-4. [DOI] [PubMed] [Google Scholar]
- Käs E., Laemmli U. K. In vivo topoisomerase II cleavage of the Drosophila histone and satellite III repeats: DNA sequence and structural characteristics. EMBO J. 1992 Feb;11(2):705–716. doi: 10.1002/j.1460-2075.1992.tb05103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käs E., Poljak L., Adachi Y., Laemmli U. K. A model for chromatin opening: stimulation of topoisomerase II and restriction enzyme cleavage of chromatin by distamycin. EMBO J. 1993 Jan;12(1):115–126. doi: 10.1002/j.1460-2075.1993.tb05637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Wilson B., Fortier C. The limits of the DNase I-sensitive domain of the human apolipoprotein B gene coincide with the locations of chromosomal anchorage loops and define the 5' and 3' boundaries of the gene. J Biol Chem. 1989 Dec 15;264(35):21196–21204. [PubMed] [Google Scholar]
- Loc P. V., Strätling W. H. The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO J. 1988 Mar;7(3):655–664. doi: 10.1002/j.1460-2075.1988.tb02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludérus M. E., de Graaf A., Mattia E., den Blaauwen J. L., Grande M. A., de Jong L., van Driel R. Binding of matrix attachment regions to lamin B1. Cell. 1992 Sep 18;70(6):949–959. doi: 10.1016/0092-8674(92)90245-8. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Mirault M. E., Laemmli U. K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984 Nov;39(1):223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Nelson H. C., Finch J. T., Luisi B. F., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987 Nov 19;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- Phi-Van L., von Kries J. P., Ostertag W., Strätling W. H. The chicken lysozyme 5' matrix attachment region increases transcription from a heterologous promoter in heterologous cells and dampens position effects on the expression of transfected genes. Mol Cell Biol. 1990 May;10(5):2302–2307. doi: 10.1128/mcb.10.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst H., Herzog R. DNA regions associated with the nuclear matrix of Ehrlich ascites cells expose single-stranded sites after deproteinization. Eur J Biochem. 1985 Jan 2;146(1):167–171. doi: 10.1111/j.1432-1033.1985.tb08634.x. [DOI] [PubMed] [Google Scholar]
- Probst H., Hofstaetter T., Jenke H. S., Gentner P. R., Müller-Scholz D. Metabolism and non-random occurrence of nonnascent short chains in the DNA of Ehrlich ascites cells. Biochim Biophys Acta. 1983 Jun 24;740(2):200–211. doi: 10.1016/0167-4781(83)90078-7. [DOI] [PubMed] [Google Scholar]
- Razin S. V., Vassetzky Y. S., Hancock R. Nuclear matrix attachment regions and topoisomerase II binding and reaction sites in the vicinity of a chicken DNA replication origin. Biochem Biophys Res Commun. 1991 May 31;177(1):265–270. doi: 10.1016/0006-291x(91)91977-k. [DOI] [PubMed] [Google Scholar]
- Reeves R., Langan T. A., Nissen M. S. Phosphorylation of the DNA-binding domain of nonhistone high-mobility group I protein by cdc2 kinase: reduction of binding affinity. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1671–1675. doi: 10.1073/pnas.88.5.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberge M., Gasser S. M. DNA loops: structural and functional properties of scaffold-attached regions. Mol Microbiol. 1992 Feb;6(4):419–423. doi: 10.1111/j.1365-2958.1992.tb01485.x. [DOI] [PubMed] [Google Scholar]
- Romig H., Fackelmayer F. O., Renz A., Ramsperger U., Richter A. Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. EMBO J. 1992 Sep;11(9):3431–3440. doi: 10.1002/j.1460-2075.1992.tb05422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. Intermediate filaments: structure, assembly and molecular interactions. Curr Opin Cell Biol. 1990 Feb;2(1):91–100. doi: 10.1016/s0955-0674(05)80037-7. [DOI] [PubMed] [Google Scholar]
- Stief A., Winter D. M., Strätling W. H., Sippel A. E. A nuclear DNA attachment element mediates elevated and position-independent gene activity. Nature. 1989 Sep 28;341(6240):343–345. doi: 10.1038/341343a0. [DOI] [PubMed] [Google Scholar]
- Suzuki M. SPKK, a new nucleic acid-binding unit of protein found in histone. EMBO J. 1989 Mar;8(3):797–804. doi: 10.1002/j.1460-2075.1989.tb03440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M. SPXX, a frequent sequence motif in gene regulatory proteins. J Mol Biol. 1989 May 5;207(1):61–84. doi: 10.1016/0022-2836(89)90441-5. [DOI] [PubMed] [Google Scholar]
- Tsutsui K., Tsutsui K., Okada S., Watarai S., Seki S., Yasuda T., Shohmori T. Identification and characterization of a nuclear scaffold protein that binds the matrix attachment region DNA. J Biol Chem. 1993 Jun 15;268(17):12886–12894. [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Chromomycin, mithramycin, and olivomycin binding sites on heterogeneous deoxyribonucleic acid. Footprinting with (methidiumpropyl-EDTA)iron(II). Biochemistry. 1983 May 10;22(10):2373–2377. doi: 10.1021/bi00279a011. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Hammer R. E., Blasquez V. C., Jones S. L., Garrard W. T. Immunoglobulin kappa gene expression after stable integration. II. Role of the intronic MAR and enhancer in transgenic mice. J Biol Chem. 1989 Dec 15;264(35):21190–21195. [PubMed] [Google Scholar]
- Yang C. H., Lambie E. J., Snyder M. NuMA: an unusually long coiled-coil related protein in the mammalian nucleus. J Cell Biol. 1992 Mar;116(6):1303–1317. doi: 10.1083/jcb.116.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong L., van Driel R., Stuurman N., Meijne A. M., van Renswoude J. Principles of nuclear organization. Cell Biol Int Rep. 1990 Dec;14(12):1051–1074. doi: 10.1016/0309-1651(90)90014-p. [DOI] [PubMed] [Google Scholar]
- van Driel R., Humbel B., de Jong L. The nucleus: a black box being opened. J Cell Biochem. 1991 Dec;47(4):311–316. doi: 10.1002/jcb.240470405. [DOI] [PubMed] [Google Scholar]
- von Kries J. P., Buhrmester H., Strätling W. H. A matrix/scaffold attachment region binding protein: identification, purification, and mode of binding. Cell. 1991 Jan 11;64(1):123–135. doi: 10.1016/0092-8674(91)90214-j. [DOI] [PubMed] [Google Scholar]