Background: The metabolism of host lipids is central to the survival of intracellular Mtb.
Results: Degradation of cholesterol generates propionyl-CoA, which is, in part, detoxified through incorporation into methyl-branched lipids.
Conclusion: The balance of acetyl-CoA versus propionyl-CoA concentrations in Mtb impacts detoxification of propionyl-CoA.
Significance: The ability of Mtb to utilize host-derived lipids effectively is key to its success as a pathogen.
Keywords: Cholesterol, Cholesterol Metabolism, Lipid Metabolism, Mycobacterium tuberculosis, Tuberculosis, Propionate, Propionyl-CoA
Abstract
Recent data indicate that the nutrients available to Mycobacterium tuberculosis (Mtb) inside its host cell are restricted in their diversity. Fatty acids and cholesterol appear more favored; however, their degradation can result in certain metabolic stresses. Their breakdown can generate propionyl-CoA, which gives rise to potentially toxic intermediates. Detoxification of propionyl-CoA relies on the activity of the methylcitrate cycle, the methylmalonyl pathway, or incorporation of the propionyl-CoA into methyl-branched lipids in the cell wall. The current work explores carbon flux through these pathways, focusing primarily on those pathways responsible for the incorporation of propionyl-CoA into virulence-associated cell wall lipids. Exploiting both genetic and biochemical rescue, we demonstrate that these metabolic pressures are experienced by Mtb inside its host macrophage and that the bacterium accesses host fatty acid stores. The metabolism of these host lipids expands the acetyl-CoA pool and alleviates the pressure from propionyl-CoA. These data have major implications for our appreciation of central metabolism of Mtb during the course of infection.
Introduction
There is a growing body of literature indicating that Mycobacterium tuberculosis (Mtb),2 a facultative intracellular pathogen, utilizes a restricted set of host-derived nutrients to persist within its host cell. Most notably, host lipids appear to be the primary carbon source for Mtb in vivo or in infected macrophages in culture. Evidence dates back to the early observations of Bloch and Segal (1), who reported the preferential utilization of fatty acids as carbon source by Mtb harvested from the lungs of infected mice. Following the sequencing of the Mtb genome, Cole et al. (2) noted the relative “overrepresentation” of genes predicted to be involved in the processing and degradation of fatty acids. Moreover, genes involved in routing the products of fatty acid degradation through pathways such as the glyoxylate shunt and gluconeogenesis strongly influence the bacterium's phenotype in a range of different infection models (3–6). Finally, cholesterol is known to be prominent among the host lipids that are required for the growth and persistence of Mtb in mice (7), and mutants defective in either uptake or processing of cholesterol exhibit severe defects in intracellular growth and survival (8–11).
The reliance on host-derived nutrients such as cholesterol as a major carbon source comes at a cost to Mtb. In addition to the degradation of the A, B, C, and D, rings of cholesterol, the acyl side chain is predicted to be degraded by rounds of β-oxidation, which will give rise to propionyl-CoA (9, 12, 13). Mtb is exquisitely sensitive to increases in the propionyl-CoA pool, and the bacterium has three different means of metabolizing this precursor of potentially toxic metabolite(s) (14–16). ICL1 functions both as an isocitrate lyase in the glyoxylate shunt and as a methylisocitrate lyase that catalyzes the last reaction of the methylcitrate cycle (MCC), which converts propionyl-CoA to succinate and pyruvate that feeds into the TCA cycle (5, 14). Alternatively, propionyl-CoA carboxylase can generate methylmalonyl-CoA that can enter the vitamin B12-dependent methylmalonyl pathway (MMP), leading to the production of succinyl-CoA (15). Finally, and of considerable significance to infection, these three-carbon intermediates in the form of methylmalonyl-CoA may be used as building blocks for the bacterium's cell wall lipids (17–19). The complex lipids of the Mtb cell wall form an impressive hydrophobic barrier around the bacterium and are also known modulators of host cell function, acting as highly potent virulence factors (20). These bioactive lipids can be esterified with up to five multiple methyl-branched (MB) long chain fatty acids, which provides an effective “sink” for excess propionyl-CoA. In Mtb, these MB lipids include the phthiocerol dimycocerosates (PDIM) and the trehalose ester families, including sulfolipid-1 (SL-1), diacyltrehalose, triacyltrehalose, and polyacyltrehalose (21). These Mtb lipids are synthesized by individual polyketide synthase complexes from malonyl-CoA and methylmalonyl-CoA (MM-CoA), which originate from acetyl-CoA and propionyl-CoA, respectively. Thus, the three-carbon metabolite propionyl-CoA plays a significant role in the generation of cell wall lipids known to be required for the modulation of the host at level of both the host cell and the infected tissue (22–27).
Cholesterol is the best characterized source of three-carbon metabolites in Mtb. Additionally, degradation of uneven chain length fatty acids or MB amino acids will all generate propionyl-CoA; therefore, the interplay between the assimilatory pathways (MCC and MMP) and the biosynthetic incorporation of MM-CoA into MB cell wall lipids is critical to minimizing the potential toxicity of these metabolites while maximizing their “usefulness” for either energy production or building blocks for the bacterium's cell wall lipids. In this current study, we selectively manipulated carbon flux through the three different propionyl-CoA processing pathways to elucidate both the genetic and biochemical linkages between the pathways as well as the factors that promote incorporation of three-carbon intermediates into the cell wall lipids of the bacterium. Moreover, we demonstrate through biochemical rescue that Mtb can access and metabolize the lipid stores in its host cell to shunt propionyl-CoA through favored, non-intoxicating routes of degradation or synthesis. Finally, the accumulation or sequestration of host lipids and their utilization by Mtb are part of the metabolic reprogramming of the host cell that has been documented both in cell culture models and in human tuberculosis granulomas (25, 28–31). The current study further elaborates the tight interplay between the lipid metabolism of both host and pathogen during tuberculosis infection.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions
Mtb strains were maintained in Middlebrook 7H9 medium supplemented with 0.2% glycerol, 10% OADC (0.5 g/liter oleic acid, 50 g/liter albumin, 20 g/liter dextrose, and 0.04 g/liter catalase), and 0.05% tyloxapol. Kanamycin (20 μg/ml) or hygromycin (50 μg/ml) were used where necessary. For growth on defined carbon sources, strains were grown without shaking in minimal medium (0.5 g/liter asparagine, 1.0 g/liter KH2PO4, 2.5 g/liter Na2HPO4, 50 mg/liter ferric ammonium citrate, 0.5 g/liter MgSO4·7H2O, 0.5 mg/liter CaCl2, 0.1 mg/liter ZnSO4, 10 mm glycerol, and 0.05% tyloxapol) (7) containing propionate or fatty acids (Sigma). Prior to addition, fatty acids having longer chain length than butyric acid (C4) were dissolved to 100 mm in a solution of tyloxapol/ethanol (1:1) at 80 °C for 30 min. To overcome the poor solubility of long chain fatty acids (C10–C24), a prewarmed 100 mm stock solution of each long chain fatty acid was added to the medium to a final concentration of 0.05 mm. To activate the MMP, vitamin B12 (VitB12) was added to a concentration of 10 μg/ml. Bacterial growth was monitored by measuring optical density at 600 nm.
Mutant Strains
The acs:Tn mutant (Tn insertion at 866 bp), the prpD:Tn mutant (Tn insertion at 643 bp), and the prpC:Tn mutant (Tn insertion at 1019 bp) were constructed using MycomarT7 in H37Rv in a Δicl1 background. To construct a PDIM (or SL-1)-deficient mutant in the Δicl1 background, an internal 1066-bp fragment of ppsD (Rv2934) and an internal 1044-bp fragment of pks2 (Rv3825) were PCR-amplified and cloned into the plasmid pMV307 (KmR), a derivative of pMV306 without the int gene, which was used to construct the ppsD knock-out mutant by single crossover. Then plasmid DNA was UV-irradiated and electroporated into the Δicl1 strain. Transformants were selected on kanamycin and confirmed by PCR. To quantify survival of Mtb in macrophage infection experiments, we transformed the relevant H37Rv strains with pVV16-smyc′::mCherry expressing the fluorescent protein mCherry driven by the smyc promoter (32).
Pyruvate Quantification
For pyruvate quantification, the Δicl1 mutant was grown in the minimal medium supplemented with 10 mm glycerol up to A600 = 1.0. 100 ml was pelleted and resuspended in 10 ml of minimal medium containing 10 mm propionate and 10 mm glycerol for 48 h. The pyruvate released into the medium was quantified using a pyruvate quantification kit (Sigma).
Metabolic Labeling and Lipid Analysis
Metabolic radiolabeling of mycobacterial cell wall lipids was performed as described (32). Briefly, bacterial strains were cultured in the 20-ml minimal medium with 4 μCi of [1-14C]sodium propionate, 4 μCi of [1-14C]stearic acid, or 4 μCi of [1,2-14C]sodium acetate for 2 weeks. Cultures were centrifuged and washed in PBS, and the cell wall lipids were extracted twice with CHCl3/CH3OH (2:1, v/v). After determination of 14C incorporation by a scintillation counter, 10,000 cpm from each lipid extract was loaded onto a TLC plate and resolved by running twice in a petroleum ether/ethyl acetate (98:2, v/v) solvent system. Radiolabeled lipids were visualized by autoradiography with a PhosphorImager, and the ratio of total PDIM (A and B), to TAG was quantified (GeneSnap, Syngene).
For intracellular labeling, lipid droplets were induced in macrophages (2 × 108) by the addition of 400 μm oleate and 20 μCi of [1-14C]oleic acid, 20 μCi [1-14C]stearic acid, or [1-14C]sodium propionate for 24 h. The cells were washed and infected with either Δicl1 or Erdman (MOI = 10:1) for 4 h at 37 °C. After 4 days, macrophages were washed and harvested in PBS. Lipids from intracellular Mtb lipids were isolated as detailed by Singh et al. (33), whereby infected macrophages were washed twice with methanol, followed by extraction of mycobacterial lipids with CHCl3/CH3OH (2:1, v/v) for 48 h.
In order to track the fate of the double-labeled stearic acid, [1-14C,9,10-3H]stearic acid (PerkinElmer Life Sciences) was used. To isolate labeled PDIM, lipid extracts were spotted on TLC and run twice in a petroleum ether/ethyl acetate (98:2, v/v) solvent system. The PDIM was visualized by iodine vapor and scraped from the plates, and lipids were extracted in petroleum ether. The relative ratio of 3H and 14C in the isolated PDIM was determined by a scintillation counter from two biological replicates analyzed in triplicate.
Lipid Droplet Induction and Macrophage Infection
C57BL/6 mouse bone marrow-derived macrophages were cultured in DMEM containing 15% L929-conditioned medium, 10% fetal bovine serum, and 2 mm glutamate for 7 days. Lipid droplet induction was performed as described (34). Briefly, 400 μm oleate, conjugated to defatted BSA (1:5), was added to the macrophage medium. To monitor lipid droplet formation, these macrophages were transferred to 24 wells containing sterile coverslips and incubated for up to 5 days in the presence of 400 μm oleate. At each time point, cells were fixed in 4% paraformaldehyde. Then lipid droplets were stained with BODIPY 493/503.
To assess the impact of lipid droplet formation on the survival of the Δicl1 mutant, lipid droplets were induced in macrophages by incubation with 400 μm oleate for 24 h prior to infecting the cells with Δicl1 (MOI = 10) for 4 h at 37 °C. The cultures were washed extensively, and the intracellular bacterial load was assessed over a period of 8–9 days. The bacterial load was quantified by lysing the macrophages; plating the lysate on Middlebook 7H10 medium supplemented with 0.2% glycerol, 10% OADC; and counting cfu. Alternatively, the infection was performed in 96-well plates at comparable MOI with Δicl1 (smyc′::mCherry), and the mCherry fluorescence was measured by a PerkinElmer Life Sciences EnVision plate reader over a period of 9 days. Cultures were manipulated through the addition of vitamin B12 to 50 μg/ml and maintained throughout the infection.
Construction of Transposon Mutant Libraries
Transposon libraries were made by using the Transposon donor phagemid φMycoMarT7 in the Δicl1 H37Rv background as described (35). Briefly, for transduction, mycobacterial cultures grown to an A600 of 0.8–1.0 in liquid culture were washed twice with MP buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 10 mm MgSO4, 2 mm CaCl2) and concentrated in one-tenth of the original culture volume in MP buffer. Then cells were infected with 1 × 1010 phage/ml of bacterial culture for 4 h at 37 °C. The mixture was plated on 7H10 plus OADC and 20 μg/ml kanamycin, and libraries were collected by scraping colonies from plates. Each library consisted of at least 1 × 105 independent mutants.
Transposon Site Hybridization (TraSH)
Approximately 2 × 106 cfu from an aliquot of the library were inoculated into minimal media containing either 10 mm glycerol or 10 mm glycerol with 0.05 mm propionate and 0.05 mm stearic acid. To limit the overselection of fast growers, the cultures were only allowed to grow up to an OD of 0.3. Then the control pool and the propionate/fatty acid rescue pool were compared (two biological replicates and two technical replicates), using TraSH (35). Briefly, genomic DNA from each pool was partially digested with HinPI followed by MspI. 0.5–2-kb fragments were purified and ligated to asymmetric adaptors, and transposon chromosome junctions were amplified using PCR. A custom-designed, high density microarray was used to identify the insertion sites. This array, synthesized by Agilent Technologies, consisted of 60-mer oligonucleotides every 350 bp of the Mtb genome. We calculated that this oligonucleotide density would allow size-selected (200–500-bp) labeled probes to hybridize to at least one oligonucleotide and therefore provide sufficient coverage to identify the majority of insertion sites. Mutants that were significantly overrepresented or underrepresented represented after the screen were defined using the following criteria: arbitrary fluorescence intensity >300 in one of the channels, fluorescence ratio >3, and t test p value <0.05 (GeneSpring GX, Agilent).
RESULTS
The Relative, but Not the Absolute, Abundance of Three-carbon versus Two-carbon Metabolites Impacts Mtb Fitness
To elucidate the pressures induced by increased concentrations of three-carbon intermediates, we developed approaches to experimentally modulate the concentration of propionyl-CoA and the bacterium's ability to process the metabolite. Propionyl-CoA, a high energy metabolite, is a key precursor to several cell wall lipids in Mtb (36); however, the buildup of propionyl-CoA is potentially toxic to the cell. Propionyl-CoA may be metabolized through either the MCC or the MMP; however, transcriptional profiling of Mtb in macrophages indicates that propionyl-CoA is metabolized predominantly through the MCC, leading to the production of pyruvate and succinate (37). Additionally, the MCC genes are transcriptionally induced and key MCC metabolites accumulate when Mtb is grown in the presence of cholesterol, indicating enhanced flux of propionyl-CoA carbons through the MCC (9). ICL1 (Rv0467) functions as a 2-methylisocitrate lyase and catalyzes that last reaction in the MCC; thus, this enzyme is critical in relieving the potential toxicity of propionyl-CoA (6).
It had been shown previously that mutants defective in expression of ICL1 are unable to grow on propionate as a sole carbon source (5, 38). We modified this approach to demonstrate that the growth of a Δicl1 mutant was normal in minimal medium with 10 mm glycerol but was inhibited strongly by the addition of increasing concentrations of propionate (Fig. 1A). The growth of wild-type H37Rv was unimpaired under these conditions. This demonstrated that propionate or its products were intoxicating the Δicl1 mutant even in the presence of an alternate carbon source, suggesting strongly that a functional MCC is critical to alleviating the increased propionyl-CoA concentration. Savvi et al. (15) had shown that the MMP offered an alternate route for propionyl-CoA degradation but that this pathway was dependent on the activity of a MM-CoA synthase, which requires VitB12 as a co-factor. Although Mtb does possess the genes required for synthesis of VitB12, it is probably not produced under normal in vitro culture conditions. We found that the addition of VitB12 to the culture medium containing the Δicl1 mutant rescued the mutant from propionate-mediated intoxication (Fig. 1A), thus further confirming the results of Savvi et al. (15) and demonstrating that both MCC and MMP function to detoxify propionyl-CoA rather than render it usable as an alternative carbon source.
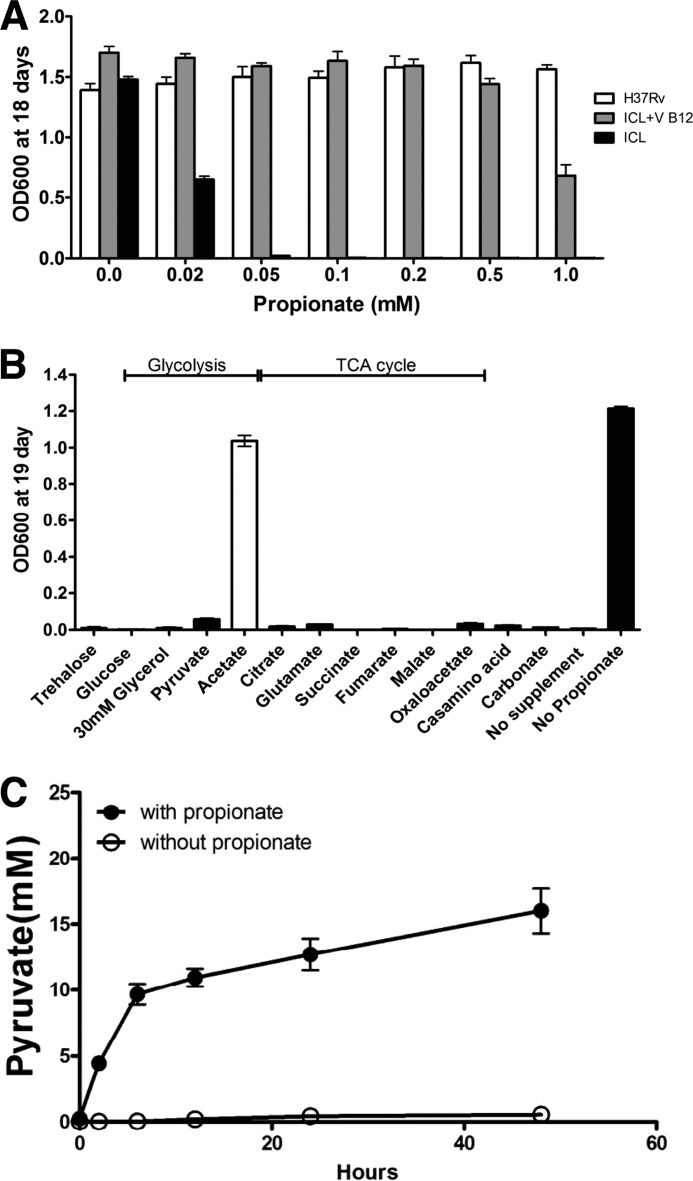
FIGURE 1.
Mitigation of propionate toxicity in a Δicl1 mutant strain by acetate and VitB12. The Δicl1 mutant strain of Mtb grows poorly in the presence of propionate, but growth can be rescued by the addition of either VitB12 or acetate. A, bacterial growth, measured by absorbance at 600 nm, was determined for Mtb H37Rv and Δicl strain grown for 18 days in minimal medium containing 10 mm glycerol as the primary carbon source and propionate across the range from 0.02 to 1 mm. Vitamin B12 at a concentration of 10 μg/ml reversed the propionate toxicity observed in the Δicl1 mutant strain. B, propionate toxicity is rescued in the Δicl1 mutant strain with acetate. Of the central metabolic intermediates tested, only acetate (4 mm) was able to rescue growth of the Δicl1 mutant strain in the presence of propionate (0.05 mm). C, pyruvate accumulates and is released from the Δicl1 mutant when grown in the presence of propionate over the course of 48 h. Data are representative of three experiments. Error bars, Standard Deviation.
Earlier studies on propionate metabolism in Aspergillus nidulans suggested that a paucity of intracellular acetyl-CoA relative to the high concentration of propionyl-CoA directly impacted growth and that propionyl-CoA inhibited pyruvate dehydrogenase (PDH) activity (39). To confirm whether or not a similar mechanism was operating in Mtb, we tested growth of the Δicl1 mutant on minimal medium supplemented with 0.05 mm propionate and a range of carbon intermediates from the TCA cycle and glycolysis. Only acetate supplementation rescued the propionate toxicity (Fig. 1B), and, although we cannot formally prove that Mtb has access to all of these intermediates, the data are consistent with the previous study with Aspergillus. When grown on glycerol as the main carbon source, much of the acetyl-CoA in Mtb is generated by PDH from pyruvate, and to test whether propionate addition impairs PDH activity, we monitored the accumulation of extracellular pyruvate in the presence and absence of exogenous propionate in Δicl1 mutant cultures. As hypothesized, pyruvate rapidly accumulated in the medium in the presence of propionate (Fig. 1C), but pyruvate was almost undetectable in the control culture. To further confirm this phenotype, we grew the Δicl1 mutant on minimal medium supplemented with 0.05 mm propionate and acetate in increasing concentrations from 0.5 to 4.0 mm, illustrated in (Fig. 2A). Interestingly, compared with the control culture with no propionate, the bacterial growth rates were similar when the acetate concentration exceeded 4 mm. These findings indicate that propionyl-CoA or its products impair the production of acetyl-CoA and that the addition of increasing concentrations of acetate relieves this toxicity, which implies that it is the relative balance of two- and three-carbon intermediates that is critical for bacterial fitness.
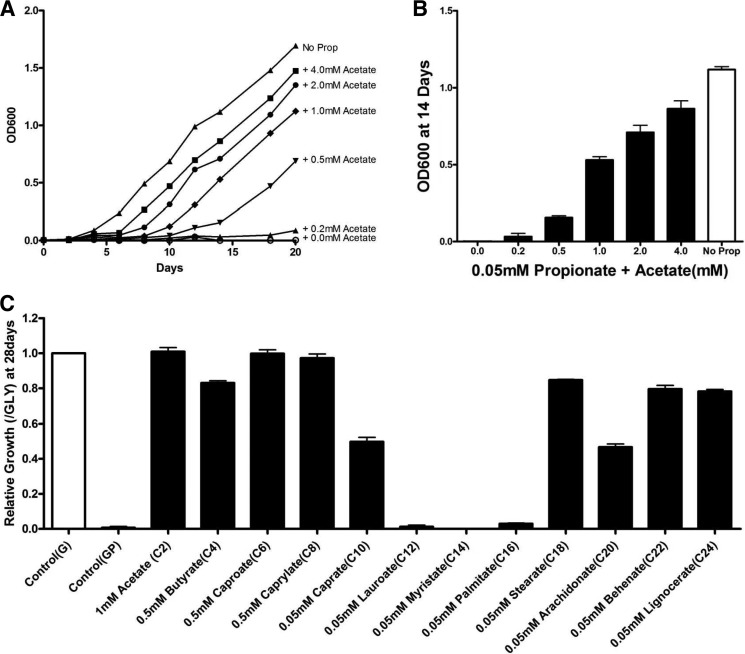
FIGURE 2.
Fatty acids of differing chain lengths rescue the Δicl1 strain from propionate toxicity. Acetate, short chain fatty acids, and long chain fatty acids similar to those found in the host macrophage can rescue propionate toxicity in Δicl1 Mtb. A and B, the C2 compound, acetate, rescues propionate toxicity in Δicl1 Mtb in a dose-dependent manner. Bacteria were grown for 18 days in medium containing 10 mm glycerol (G) and 0.05 mm propionate supplemented with acetate across the range from 0.05 to 4 mm. C, short to mid-length and long chain fatty acids also rescue propionate toxicity in Δicl1 Mtb. Bacteria were grown for 18 days in medium containing 10 mm glycerol and 0.05 mm propionate (GP) supplemented with saturated even chain length fatty acids (C2–C24) at the following concentrations. Acetate (C2) was added at 1 mm, short chain fatty acids (C4–C8) at 0.5 mm, and intermediate and long chain fatty acids (C10–C24) at 0.05 mm. Bacterial growth was measured by absorbance at 600 nm, and results are representative of three replicates. Error bars, Standard Deviation.
To extend this observation to more physiologically relevant substrates, we examined the ability of longer chain fatty acids to rescue growth of the Δicl1 mutant in propionate-containing medium. We focused on even chain length fatty acids because odd chain fatty acids and MB fatty acids would also yield propionyl-CoA that would elevate propionyl-CoA levels in the bacterium. First, we measured the growth of WT Mtb in glycerol-containing media in the presence of saturated even chain fatty acids, from acetic acid (C2) to lignoceric acid (C24). As noted previously (40), free fatty acids of intermediate chain length (C10–C16) were toxic to Mtb, but the bacterium grew well on either short or longer chain length fatty acids. Next, the fatty acids that supported WT Mtb growth were tested for their ability to rescue the Δicl1 mutant from propionate-mediated toxicity. Growth was restored to the Δicl1 mutant through supplementing the propionate- and glycerol-containing media with the majority of the fatty acids, except for C12 and C14, suggesting that the acetyl-CoA pool may also be expanded though β-oxidation of long chain fatty acids (Fig. 2B). Interestingly, the shorter chain fatty acid (C2–C8) supplementation abrogated the propionate-mediated toxicity efficiently, whereas long chain fatty acids (C18–C24) appeared to sustain growth in propionate following a lag period. Together, these observations support the contention that it is the relative abundance of two- versus three-carbon intermediates that plays an important role in protecting Mtb from the potential toxicity of propionyl-CoA.
The Methyl-branched Lipid, PDIM, Acts as a Sink for Propionyl-CoA
The third proposed route of detoxification of propionyl-CoA is through its incorporation into the MB lipids of cell wall lipids. To synthesize MB lipids, Mtb would require fatty acids to act as primers for the polyketide synthase-catalyzed addition of MM-CoA from propionyl-CoA (41). In a typical mycocerosic acid in PDIM, there is a 4:1 molar ratio of MM-CoA to acyl primer required for synthesis. We therefore hypothesized that under our defined growth conditions, the Δicl1 mutant grown in minimal medium containing propionate and glycerol in the absence of VitB12 is dependent upon the biosynthesis of MB lipids as a means of detoxifying excess propionyl-CoA. If correct, then provision of acetate or longer chain fatty acids would facilitate synthesis of fatty acid-AMP precursors that would act as acceptors for methylmalonyl-CoA and enable Mtb to incorporate excess propionate into cell wall lipids, such as PDIM. This hypothesis would predict that radiolabel in the form of 14C in acetate, long chain fatty acids, or propionate should all be incorporated into PDIM.
To follow the flow of carbon into PDIM, we metabolically labeled Mtb with [14C]propionate, [14C]acetate, and [14C]stearic acid. We used both the Δicl1 mutant and wild-type Erdman strains. Unlike H37Rv, which is the parental strain of the Δicl1 mutant and has a defective icl2 gene (38), Erdman is a high PDIM-producing strain with an intact icl2 gene (6). The peripheral cell wall lipids were isolated and analyzed by thin layer chromatography (TLC). As shown in Fig. 3, 14C label was incorporated into PDIM in both the Δicl1 mutant and the WT strain from either [14C]acetate or [14C]stearic acid. The amount of radiolabel incorporated into PDIM from [14C]acetate, and [14C]stearic acid was enhanced in the Δicl1 mutant in the presence of 0.05 mm unlabeled propionate. In the WT strain, which has an intact MCC, 14C label from [14C]propionate was incorporated into both TAG and PDIM in the presence of 0.05 mm unlabeled propionate. Significantly, in the absence of unlabeled propionate, the [14C]propionate tracer was found only in PDIM.
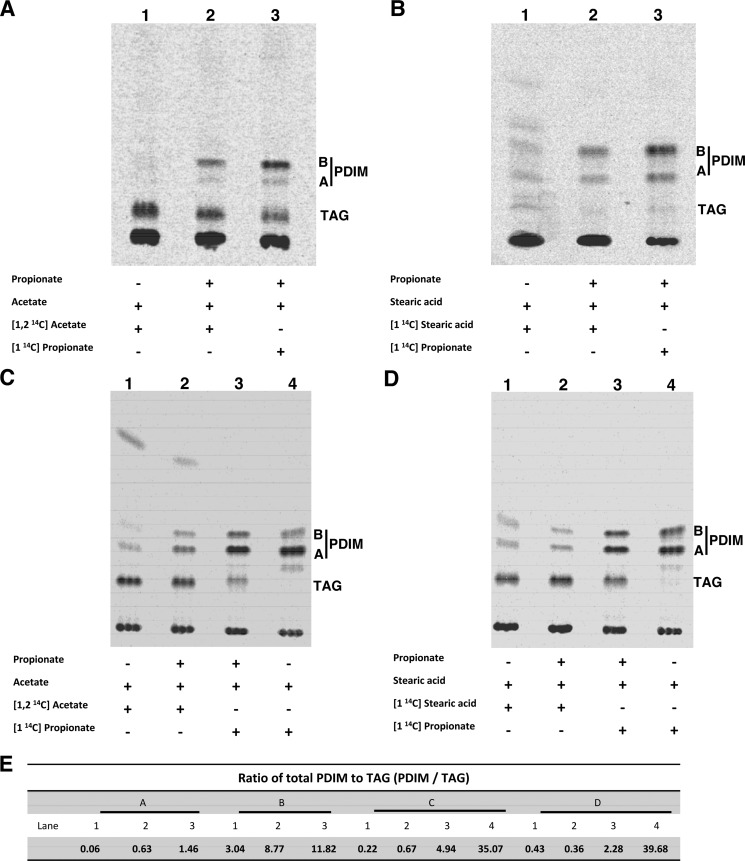
FIGURE 3.
Fatty acids rescue propionate toxicity in Mtb through incorporation into methyl-branched lipids, such as PDIM, in both the Δicl1 mutant strain and wild-type Mtb. TLC analysis of the cell lipids indicates that radiolabel derived from [1-14C]propionate, [1,2-14C]acetate, or [1-14C]stearic acid is incorporated into the cell wall lipid PDIM. 14C incorporation into PDIM is enhanced by 0.05 mm propionate but is a constitutive process in both the Δicl1 mutant strain and in the high PDIM-producing wild-type Erdman strain. Bacteria were grown in glycerol medium containing propionate, acetate, and stearic acid as indicated. A, lipids from the Δicl1 mutant incubated with [1,2-14C]acetate in the absence of propionate (lane 1), incubated with [1,2-14C] acetate in the presence of 0.05 mm unlabeled propionate (lane 2), or incubated with [1-14C]propionate in the presence of 0.05 mm unlabeled propionate (lane 3). C, lipids from Erdman. B, the experiment was repeated using the long chain fatty acid, stearic acid (C18), instead of acetate (C2). Shown are lipids from the Δicl1 mutant incubated with [1-14C] stearic acid in the absence of propionate (lane 1), with [1-14C]stearic acid in the presence of 0.05 mm unlabeled propionate (lane 2), or with [1-14C]propionate in the presence of 0.05 mm unlabeled propionate (lane 3). D, lipids from Erdman. E, ratio of PDIM to TAG. The identity of the PDIM and TAG bands had been established previously by mass spectrometry (32). PDIM A, phthiocerol dimycocerosate A; PDIM B, phthiodiolone dimycocerosate.
These data indicate that, at least in a high PDIM-producing strain of Mtb, under these in vitro conditions, flux of propionyl-CoA into PDIM is the preferred route even in the presence of an intact MCC. These data demonstrate that utilization of both long chain fatty acids and the de novo synthesis of fatty acids from acetate may facilitate the formation of the fatty acid-AMP primers to support the incorporation of propionyl-CoA into the MB cell wall lipid PDIM.
Stearic Acid Is Incorporated into Mtb PDIM without Degradation through β-Oxidation
The preceding result does not discriminate between the direct incorporation of stearic acid into PDIM as an acyl-primer and the incorporation of acetyl-CoA, which would be released by β-oxidation of the stearic acid. To determine whether intact stearic acid is used as a fatty acid primer for PDIM biosynthesis, we metabolically labeled WT Erdman and the Δicl1 mutant with double isotope-labeled stearic acid ([9,10-3H,1-14C]stearic acid) in the presence of propionate. If the stearic acid were catabolized by β-oxidation, the 1-14C radiolabel would be released as [14C]acetyl-CoA, which can enter the central metabolism or be used as a substrate by the FAS1 system and incorporated into n-fatty acids. Therefore, we predict that if the stearic acid were β-oxidized, the ratio of 3H to 14C would change, indicating a loss of 14C from stearic acid. However, we found that the 3H/14C ratio within PDIM was virtually identical to that of the [9,10-3H,1-14C]stearic acid standard (Table 1). This result demonstrates that long chain fatty acids, such as stearic acid, can bypass catabolic β-oxidation and serve as a primer for direct incorporation into PDIM.
TABLE 1.
Stearic acid is incorporated into Mtb PDIM intact, without β-oxidation
Double-labeled [9,10-3H,1-14C]stearic acid was used for metabolic labeling of the Δicl1 or Erdman strain in minimal medium containing glycerol, 0.05 mm propionate, and 0.05 mm stearic acid. After 2 weeks of incubation, Mtb wall surface lipids were extracted, and PDIM was purified from the PDIM bands by TLC. The ratio of 3H and 14C radioactivity (cpm) was determined for both the starting material and the extracted PDIM bands. Average and S.D. values were calculated from three technical replicates from each experiment. Expt., experiment.
| [9,10-3H,1-14C]Stearic acid dual isotope labeling |
||||
|---|---|---|---|---|
|
3H/14C cpm ratios of H37Rv Δicl |
3H/14C cpm ratios of Erdman |
|||
| Expt. 1 | Expt. 2 | Expt. 1 | Expt. 2 | |
| Total PDIM | 0.2962 ± 0.007 | 0.3087 ± 0.012 | 0.2784 ± 0.001 | 0.2821 ± 0.004 |
| [9,10-3H,1-14C]Stearic acid | 0.2932 ± 0.005 | 0.3130 ± 0.003 | 0.2898 ± 0.002 | 0.2966 ± 0.002 |
Intracellular Mtb Can Exploit Host Lipid Stores to Alleviate Propionate-mediated Stress
Mtb mutants defective in cholesterol utilization or in the expression of ICL1 exhibit reduced survival in macrophages and mice, and this defect is exacerbated further by activation of the host macrophages or the development of an effective immune response (5, 7, 14). The similarity in phenotype between Mtb mutants defective in cholesterol utilization and mutants defective in expression of ICL1 is intriguing because it could result from related metabolic defects. Mtb mutants defective in utilization of cholesterol will starve if dependent on the sterol as a carbon source, whereas mutants defective in ICL1 expression will be vulnerable to propionate toxicity should they metabolize cholesterol. Daniel et al. (28) reported that intracellular Mtb can access and metabolize triacylglycerol from lipid droplet stores in its host cell. Recently, we confirmed this observation and demonstrated that the increased accumulation of lipid in infected macrophages correlates with reduced turnover of host lipids (31). These observations suggest that it is feasible to establish whether or not Mtb experiences propionyl-CoA toxicity under the physiological conditions within its host cell through manipulation of host lipid stores to provide metabolic rescue. To test this hypothesis and to determine the physiological significance of propionyl-CoA toxicity during infection, we induced lipid droplet formation in macrophages through the addition of 400 μm oleate to the culture medium (34). The resultant lipid droplets, detected by BODIPY 493/503, saturated within 1 day of culture (Fig. 4). These macrophages and control, untreated macrophages were subsequently infected with the Δicl1 mutant (containing smyc′::mCherry) (MOI = 10), and bacterial growth was monitored in parallel by both cfu counts and the fluorescent signal from mCherry. As reported previously (34), the growth of the Δicl1 mutant was severely impaired in the control, untreated macrophages. In contrast, growth of the Δicl1 mutant was rescued in the macrophages preloaded with lipid droplets induced by the addition of oleate. Moreover, the addition of VitB12 to the culture medium was also capable of restoring intracellular growth to the Δicl1 mutant. The trends were comparable in both the cfu and the fluorescence readouts (Fig. 4), although the cfu count probably underestimates bacterial viability due to stress inflicted during bacterial isolation, whereas the fluorescence readout probably overestimates viability due to persistent signal from dying bacteria.
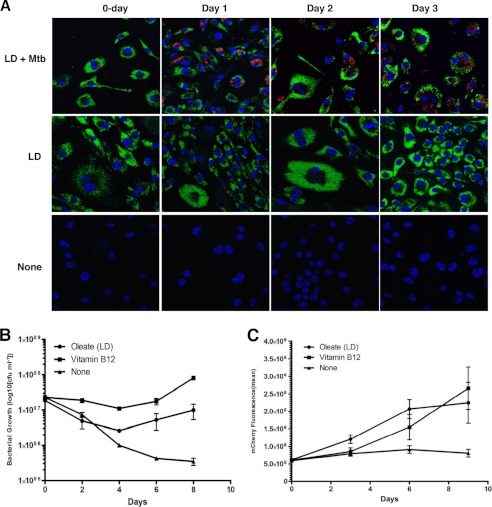
FIGURE 4.
Intracellular growth is restored to the Δicl1 Mtb through the induction of oleate-containing lipid droplets in the infected cell. The survival of the Δicl1 mutant is impaired in macrophages; however, growth could be restored by preloading the host cells with exogenous oleate or by adding VitB12 to the cell culture medium to facilitate operation of the MMP. A, induction of macrophage lipids droplets, detected with BODIPY 493/503 (green), Δicl1 strain expressing pVV16-mCherry (smyc′::mCherry) (red), and nuclei (blue) in oleate-loaded macrophages versus untreated macrophages over the duration of the experimental period. B, bacterial cfu counts were determined at 2-day intervals across an 8-day period for the Δicl1 Mtb in untreated, control macrophages (triangle), in lipid droplet-containing, oleate-loaded macrophages (circle), and in macrophage supplemented by the addition of VitB12 to the medium (square). C, the experiment was repeated with a Δicl1 strain expressing pVV16-mCherry (smyc′::mCherry), and bacterial proliferation was quantified by measuring bacterial mCherry fluorescence across an 8-day infection period. Both methods demonstrated the enhanced growth and survival of the Δicl1 mutant in the presence of either oleate-induced droplets or exogenous VitB12. Error bars, Standard Deviation values from three representative replicates.
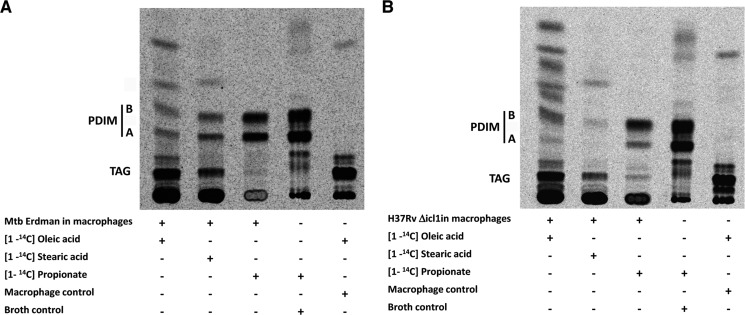
In order to demonstrate the incorporation of host-derived fatty acids into PDIM in both the Δicl1 mutant and the WT Erdman strain, we metabolically labeled macrophage lipid droplets with [14C]propionate, [14C]oleate, and [14C]stearic acid prior to infection with Mtb. Subsequent analysis of Mtb cell wall lipids postinfection revealed that 14C label from the [14C]propionate, [14C]oleate, and [14C]stearic acid was incorporated into PDIM (Fig. 5), providing biochemical confirmation of the intracellular growth phenotype shown in Fig. 4. These data confirm and extend three issues that are critical to our understanding of the physiology of Mtb within its host macrophage. First, propionyl-CoA metabolism and the relative balance of two- and three-carbon intermediates represent a potential problem for Mtb within the environment of the host macrophage. Second, the routing of propionyl-CoA through MM-CoA and into MB cell wall lipids, such as PDIM, is active in intracellular Mtb, most notably in the WT Erdman strain. Finally, the balance of lipids in the lipid droplets in the host macrophage may have considerable bearing on the fitness and growth potential of the bacterium within its host cell.
FIGURE 5.
Mtb inside lipid droplet-loaded macrophage incorporates the host-derived fatty acids into PDIM. Radiolabeled fatty acid precursors are incorporated into PDIM of both the WT Mtb (A) and the Δicl1 Mtb (B) when grown in lipid droplet-containing macrophages. Lipid droplets were induced in macrophages with excess oleic acid containing tracer amounts of [1-14C]oleic acid (lane 1), [1-14C]stearic acid (lane 2), or [1-14C]propionate (lane 3) for 24 h. The lipid-loaded macrophages were infected with either WT Mtb or Δicl1 Mtb at an MOI of 5:1 for 4 h. At 5 days postinfection, Mtb cell wall lipids were extracted from macrophage and analyzed by TLC. Control labeling experiments were run with WT Mtb grown in broth culture labeled with [1-14C]propionate (lane 4) and macrophages alone labeled with [1-14C]oleic acid (lane 5). The identity of the PDIM species has been established previously by mass spectrometry (32). PDIM A, phthiocerol dimycocerosate A; PDIM B, phthiodiolone dimycocerosate.
Of specific significance to this last point is the capacity of Mtb to induce lipid droplet formation in host macrophages in culture and within the macrophages in the human tuberculosis granuloma (25, 29). Isolation and analysis of the lipids from within the caseum of human tuberculosis granulomas revealed abundant cholesterol, cholesterol esters, and triacylglycerol (25). This may represent a “well balanced” diet for Mtb because it contains a mix of lipids and sterols that would generate appropriate levels of two- and three-carbon products.
Genetic Profiling of Propionate and Stearic Acid (C18:0) Metabolism
The above data argue that the pathways of lipid degradation and cell wall synthesis play an important role in the modulation of the relative levels of three-carbon intermediates, such as propionyl-CoA, and that this regulation is critical to the survival of intracellular Mtb. These data were generated from a very targeted approach that artificially controlled carbon flux through the MCC and the MMP and into MB cell wall lipids. The simplicity of the resultant phenotypes is, however, somewhat misleading because the bacterium undoubtedly possesses complex feedback loops and regulatory circuits to control both upstream and downstream pathways of central carbon metabolism and biosynthetic pathways. To expand our study and incorporate a more holistic understanding of other bacterial gene products that impact propionyl-CoA metabolism, we exploited our experimental system to construct a forward genetic screen to identify other genes implicated in this metabolic process.
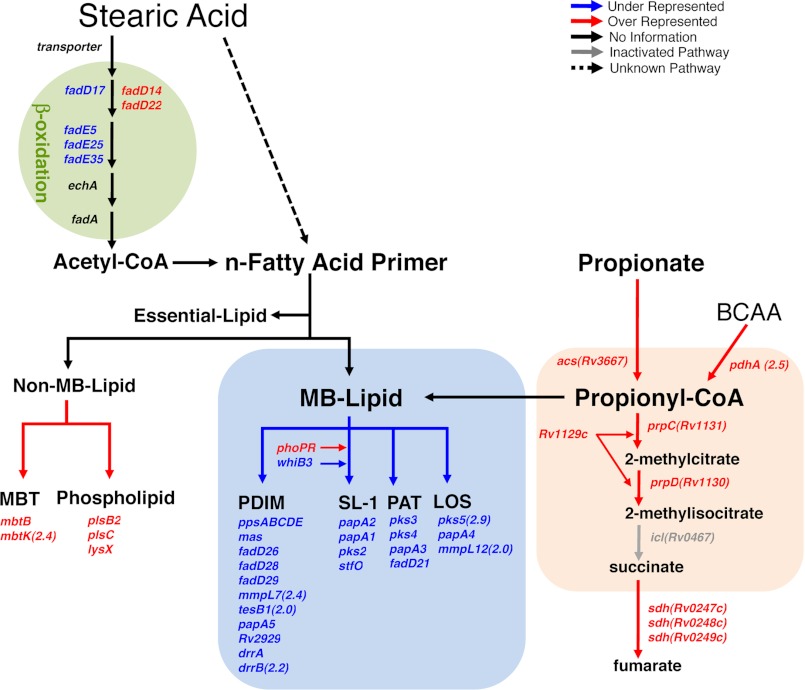
To identify such loci, we designed a TraSH screen to reveal mutants that were incapable of detoxifying propionyl-CoA and that were unable to grow in the presence of propionate when provided long chain fatty acids as a means of rescue. We focused on a saturated C18 fatty acid, stearic acid, to provide metabolic rescue because we have shown that stearic acid is an effective primer for synthesis of PDIM and is known to be one of the more abundant fatty acid species in the granulomatous lesion of tuberculosis as well as in the mammalian host (25, 28). We constructed a 105 cfu library in H37Rv Δicl1 using the mariner transposon (MycomarT7) (35). This library was grown in minimal medium containing 10 mm glycerol (input pool) versus minimal medium containing 10 mm glycerol, 0.05 mm propionate, and 0.05 mm stearic acid (output pool). To identify genes that impact the metabolism of stearic acid or propionate, we compared the input and output pools of mutants to quantify those mutations that were under- or overrepresented following selection. The TraSH analysis revealed 116 overrepresented mutations and 72 underrepresented mutations that satisfied our statistical cut-off (3-fold change, p < 0.05, intensity >300) (supplemental Tables S1 and S2). These genes are arranged diagrammatically in the context of their annotation and assignment to known metabolic pathways (Fig. 6). We hypothesized that those mutants lost during selection will include genes that are required for stearic acid uptake and rescue from propionyl-CoA-mediated toxicity, whereas those mutants enriched in the screen would include genes that mitigate propionyl-CoA-mediated toxicity and facilitate growth. Not surprisingly, the most dominant themes to emerge from the screen are loci involved in MB lipid biosynthesis, the MCC, and β-oxidation (Fig. 6).
FIGURE 6.
Phenotypic TraSH screen identifies genes involved in propionate utilization and toxicification. Graphic illustration of the differentially represented genes in the context of the metabolic pathways most relevant to propionate utilization and detoxification. Genes required for Δicl1 Mtb growth in the presence propionate and long chain fatty acids as a means of propionate toxicity rescue are indicated in blue type. Genes that, when mutated, enhance Δicl1 Mtb growth in the presence of propionate and long chain fatty acids are indicated in red type. The differentially represented genes are shown in relation to the relevant carbon metabolic pathways: synthesis of non-essential MB lipids, synthesis of non-MB lipids, the MMC, and genes involved in the β-oxidation breakdown of long chain fatty acids. All genes selected were >3-fold overrepresented or >3-fold underrepresented with p < 0.05. The full list of overrepresented and underrepresented genes is provided in supplemental Tables S1 and S2. BCAA, branched chain amino acids; PAT, polyacyltrehalose; LOS, lipooligosaccharide.
Prominent among underrepresented mutations were those genes involved in the biosynthesis of MB lipids: locus Rv2930–Rv2950 for PDIM synthesis, Rv3820–Rv3825c for SL-1 synthesis, Rv1180–Rv1185 for polyacyltrehalose and diacyltrehalose synthesis, and Rv1527c/Pks5–Rv1528c/PapA4, which are related to mycobacterial lipooligosaccharide biosynthetic genes in Mycobacterium marinum and Mycobacterium canetti (42). Although Mtb is not reported to make lipooligosaccharide, pks5 has been implicated in mycocerosoic acid synthesis, and mutants defective in expression of pks5 have a marked growth defect in mice (43). These data provide independent verification that the biosynthesis of MB lipids provides a sink for excess propionyl-CoA and are consistent with the observation that increases in propionate pools enhance the biosynthesis of PDIM and SL-1 or longer MB lipids (17). In addition, mutants of a transcriptional regulator, phoP (phoP 10.1X, p = 0.00007; phoR 2.2X, p = 0.003), were also enriched following selection. It has been shown previously that phoP mutants exhibit a marked enhancement in the production of PDIM and other mycocerosates (32, 44), which is consistent with a previous report that documented that PhoP mutants exhibited enhanced resistance to both propionate toxicity and 3-nitropropionate, a known inhibitor of ICL activity (45).
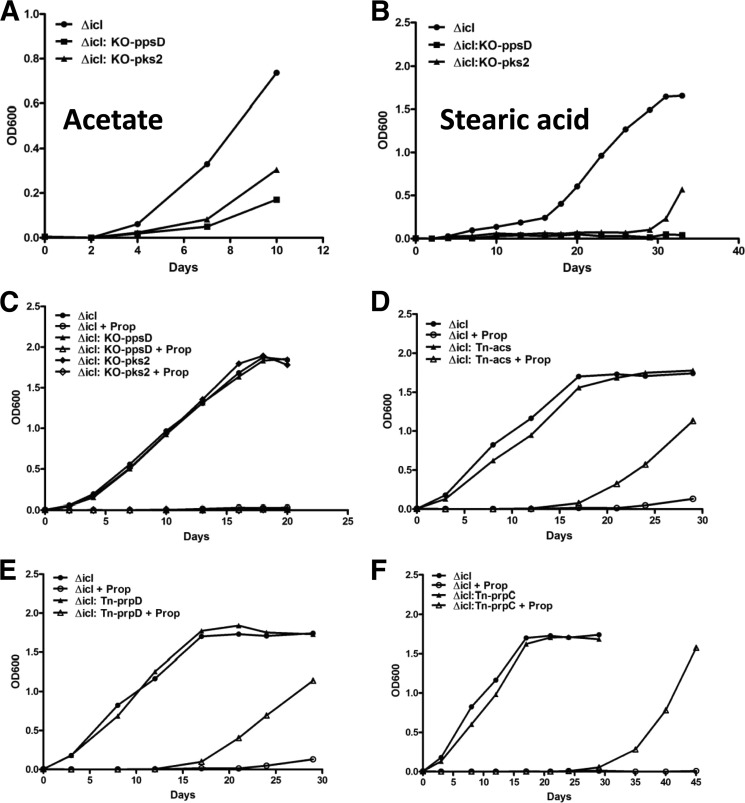
To verify the TraSH data, we generated and determined the phenotype of two double mutants: Δicl1:KO-ppsD (PDIM synthesis) and Δicl1:KO-pks2 (SL-1 synthesis). Although both mutants grew normally on control medium, neither the PDIM-deficient nor the SL-1-deficient double mutants could grow in propionate-containing medium supplemented by either acetate or stearic acid (Fig. 7, A–C), whereas the parent Δicl1 mutant grew in propionate supplemented with either acetate or stearic acid. These results both validate the genetic screen and provide biochemical verification of the phenotypes of the double mutants.
FIGURE 7.
Validation of the genes implicated in propionate utilization and detoxification in Δicl Mtb. Genes identified by the TraSH screen were validated through the generation of double mutants and characterized by growth phenotype. A–C, genes negatively selected for involved in methyl branch-containing lipid biosynthesis. Growth of the Δicl1 parental strain and Δicl1:KO-ppsD (PDIM biosynthesis) and Δicl1:KO-pks2 (SL-1 biosynthesis) double mutant strains was assessed in minimal media containing glycerol, propionate, and 1 mm acetate (A) or 0.05 mm stearic acid (B) or without any fatty acid supplement (C). In each instance, the mutants exhibited defective growth in fatty acid with propionate (A and B) yet grew normally in minimal medium with glycerol (C). D–F, genes positively selected as being involved in propionate utilization and the generation of toxic intermediates. Shown is growth of the Δicl1 parental strain Δicl1:Tn acs (Rv3667) (D), Δicl1:Tn prpD (Rv1130) (E), or Δicl1:Tn prpC (Rv1131) (F) in minimal media containing glycerol and 0.05 mm propionate (D and E) or 0.1 mm propionate (F). These mutants all grew normally in minimal medium, and each of the latter two mutations conferred partial resistance to propionate toxicity, consistent with their enrichment under propionate selection. Results are representative of three experiments.
Among those mutations that were overrepresented or positively selected in the screen, the most striking were those genes implicated in the metabolism of propionate through MCC (Fig. 6). The screen was conducted in a Δicl1 mutant background; thus, the MCC was inactivated at the 2-methylisocitrate to succinate and pyruvate step. This suggests strongly that the accumulation of all three upstream intermediates (propionyl-CoA, 2-methylcitrate, and 2-methylisocitrate) is, individually or collectively, toxic to Mtb. The inactivation of Rv1129c, Rv1130/prpD, or Rv1131/prpC resulted in marked enhancement of growth, which implies that 2-methycitrate and/or 2-methylisocitrate are more toxic than propionyl-CoA (Rv1129c (103.7X, p = 0.003), Rv1130 (136.8X, p = 0.0002), Rv1131 (15.6X, p = 0.01), and Rv3667 (17.7X, p = 0.04)). Finally, we also found that a mutation in Rv2497c (pdhA) encoding a subunit of the branched chain keto acid dehydrogenase (46) was advantageous to growth. It is therefore possible that a significant fraction of propionyl-CoA in Mtb may be produced via catabolism of MB amino acids, such as valine and isoleucine, in addition to the breakdown of cholesterol.
To validate those loci identified as overrepresented in the TraSH screen, we examined the phenotypes of the double knock-out mutants, Δicl1:Tn::Rv3667 (an acetyl-CoA synthase), Δicl1:Tn::prpC (citrate synthase, GLTA1), and Δicl1:Tn::prpD (methylcitrate synthase). We included Rv3667 in this analysis because, although it is annotated as an acetyl-CoA synthase, it was strongly selected for in the TraSH screen (enriched almost 18-fold) and may function as a propionyl-CoA synthetase, required for the conversion of propionate to propionyl-CoA. All three double mutant strains exhibited resistance to propionate toxicity (0.05 and 0.01 mm) and were able to grow to varying degrees in propionate-containing medium, whereas the growth of the Δicl1 single mutant strain was strongly inhibited (Fig. 7, D–F). The prolonged lag period observed in the growth of these mutants is not due to suppressor mutations because similar lag periods are observed upon the passing of the mutants into fresh culture medium containing propionate. Again, these data provide biochemical verification of the phenotypes of the double mutants.
The above themes have been selected because they validate the TraSH screen and the selection pressure applied to the mutant pools; however, there are other, less obvious sets of genetic loci that have been enriched or depleted from the mutant pool. Notably, several fadD and fadE genes appeared as underrepresented, suggesting that these may be involved in stearic acid catabolism and expansion of the acetyl-CoA pool, in addition to the role of stearic acid as an acyl primer. This finding is particularly interesting given the high redundancy of genes thought to be involved in β-oxidation of fatty acids (2) because it implies that there may be at least a measure of substrate specificity within this pathway. Finally, we found that mutations in genes involved in the synthesis of certain non-MB lipids, such as mycobactin (mbtB and mbtK) (47) and phospholipid (lysX and plsC) (48), conferred a growth advantage, which was unexpected. However, Mtb requires C16–C20 fatty acids (or C12 in mycobactin) to synthesize phospholipids and mycobactin (49) as well as MB lipids, such as PDIM (36). Therefore, because biosynthesis pathways of non-MB lipids and MB lipids probably compete for these fatty acid primers, the reduced carbon flow into biosynthesis of non-MB lipids may lead to the increased availability of fatty acid primers that can be redirected into MB lipids, thus enhancing detoxification.
DISCUSSION
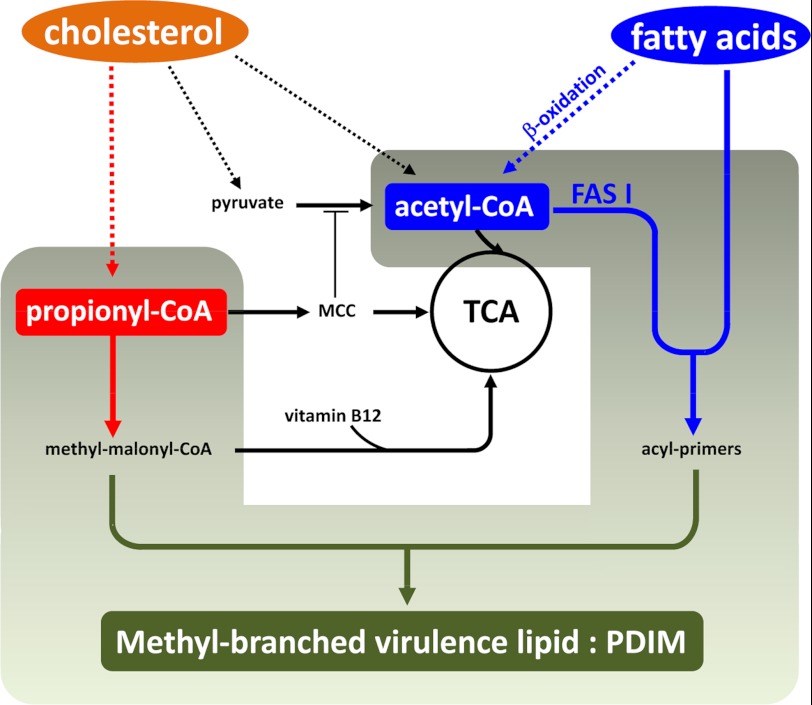
The ability of Mtb to utilize cholesterol as a carbon source during the course of infection has been shown to be critical to its success as a pathogen (7). However, the degradation of cholesterol expands the propionyl-CoA pool with potentially toxic consequences (15, 16). In the current study, we performed genetic and biochemical manipulation of the MCC and MMP to reveal the metabolic requirements for incorporation of propionyl-CoA into PDIM and other virulence-associated lipids of the bacterial cell wall. Moreover, we showed that both the Δicl1 mutant and WT Erdman Mtb can access and utilize fatty acids from host lipid stores to provide primers for assembly of the MB precursors of these virulence lipids. These data demonstrate that the metabolic pressures that we generated artificially in culture have validity within a host cell infection model. Moreover, the unbiased genetic screen to identify mutants that are impaired in, or protected from, propionate intoxication revealed both known genes and novel pathways that control both propionate metabolism and the generation of acyl primers for the synthesis of cell wall lipids (Fig. 8).
FIGURE 8.
Model of assimilation of host lipids and fatty acids into methyl-branched Mtb virulence lipids. Mtb has access to both cholesterol and fatty acids from the macrophage host cell. Degradation of cholesterol expands the pool of propionyl-CoA, which can be used to fuel central metabolism and biosynthetic pathways. Accumulation of propionyl-CoA and/or products of the MCC may limit the generation of acetyl-CoA from pyruvate by inhibiting PDH activity. Inhibition of PDH activity places more pressure on the acetyl-CoA pool, which is utilized as malonyl-CoA for the assembly of Mtb cell wall lipids. Additionally, MB cell wall lipids can be built using n-acyl primers generated either from de novo synthesis or from preformed long chain fatty acids imported by the bacterium. The integration of these two- and three-carbon metabolic pathways is clearly critical to the growth of Mtb and its success within its host cell environment.
The “choices” open to Mtb are both catabolic, through the degradation of propionyl-CoA by the MCC and MMP and the incorporation of its products into the TCA cycle, and anabolic, through the generation of malonyl-CoA primers to act as acceptors of propionyl-CoA in the synthesis of MB lipids. In the WT Erdman strain, we observed the incorporation of propionate into PDIM in both the presence and absence of exogenous propionate and the incorporation of propionate into TAG only when exogenous propionate was present. This suggests, first, that Mtb routes propionate into MB lipids by preference, and second, because the generation of TAG from propionate requires the MCC, this indicates that the MCC is activated in response to additional pressure from propionate. The “opening” of the MCC has the potential to generate more acetyl-CoA, which in turn could enhance PDIM production. This hypothesis is consistent with the observations of Jain et al. (17), who observed that increased levels of methylmalonyl-CoA lead to increased synthesis of PDIM and SL-1, and furthermore, impairment in synthesis of these lipids leads to reduced virulence in mice. Finally, transcriptional profiling of Mtb in macrophages over an extended period of infection demonstrated that the genes encoding all of the steps of the MCC were highly up-regulated throughout the duration of the infection (37). These data argue strongly that the tightly coordinated interplay between the MCC, MMP, and MB lipid synthesis is important to Mtb and may represent an “Achilles' heel” in the metabolic network in a bacterium that is highly evolved to survive and persist in its human host.
Acknowledgment
We thank Linda Bennett for technical support throughout this study.
This work was supported, in whole or in part, by National Institutes of Health Grants AI 067027, AI080651, and HL055936 (to D. G. R.).

This article contains supplemental Tables S1 and S2.
- Mtb
- M. tuberculosis
- MB
- methyl-branched
- MCC
- methylcitrate cycle
- MM-CoA
- methylmalonyl-CoA
- MMP
- methylmalonyl pathway
- PDH
- pyruvate dehydrogenase
- PDIM
- phthiocerol dimycocerocates
- SL-1
- sulfolipid-1
- TAG
- triacylglycerol
- TCA
- tricarboxylic acid
- VitB12
- vitamin B12
- MOI
- multiplicity of infection
- TraSH
- transposon site hybridization.
REFERENCES
- 1. Bloch H., Segal W. (1956) Biochemical differentiation of Mycobacterium tuberculosis grown in vivo and in vitro. J. Bacteriol. 72, 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 3. Liu K., Yu J., Russell D. G. (2003) pckA-deficient Mycobacterium bovis BCG shows attenuated virulence in mice and in macrophages. Microbiology 149, 1829–1835 [DOI] [PubMed] [Google Scholar]
- 4. Marrero J., Rhee K. Y., Schnappinger D., Pethe K., Ehrt S. (2010) Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc. Natl. Acad. Sci. U.S.A. 107, 9819–9824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McKinney J. D., Höner zu Bentrup K., Muñoz-Elías E. J., Miczak A., Chen B., Chan W. T., Swenson D., Sacchettini J. C., Jacobs W. R., Jr., Russell D. G. (2000) Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406, 735–738 [DOI] [PubMed] [Google Scholar]
- 6. Muñoz-Elías E. J., McKinney J. D. (2005) Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 11, 638–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pandey A. K., Sassetti C. M. (2008) Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. U.S.A. 105, 4376–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chang J. C., Miner M. D., Pandey A. K., Gill W. P., Harik N. S., Sassetti C. M., Sherman D. R. (2009) igr genes and Mycobacterium tuberculosis cholesterol metabolism. J. Bacteriol. 191, 5232–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Griffin J. E., Pandey A. K., Gilmore S. A., Mizrahi V., McKinney J. D., Bertozzi C. R., Sassetti C. M. (2012) Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem. Biol. 19, 218–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miner M. D., Chang J. C., Pandey A. K., Sassetti C. M., Sherman D. R. (2009) Role of cholesterol in Mycobacterium tuberculosis infection. Indian J. Exp. Biol. 47, 407–411 [PubMed] [Google Scholar]
- 11. Van der Geize R., Yam K., Heuser T., Wilbrink M. H., Hara H., Anderton M. C., Sim E., Dijkhuizen L., Davies J. E., Mohn W. W., Eltis L. D. (2007) A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. U.S.A. 104, 1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang X., Nesbitt N. M., Dubnau E., Smith I., Sampson N. S. (2009) Cholesterol metabolism increases the metabolic pool of propionate in Mycobacterium tuberculosis. Biochemistry 48, 3819–3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thomas S. T., VanderVen B. C., Sherman D. R., Russell D. G., Sampson N. S. (2011) Pathway profiling in Mycobacterium tuberculosis. Elucidation of cholesterol-derived catabolite and enzymes that catalyze its metabolism. J. Biol. Chem. 286, 43668–43678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Muñoz-Elías E. J., Upton A. M., Cherian J., McKinney J. D. (2006) Role of the methylcitrate cycle in Mycobacterium tuberculosis metabolism, intracellular growth, and virulence. Mol. Microbiol. 60, 1109–1122 [DOI] [PubMed] [Google Scholar]
- 15. Savvi S., Warner D. F., Kana B. D., McKinney J. D., Mizrahi V., Dawes S. S. (2008) Functional characterization of a vitamin B12-dependent methylmalonyl pathway in Mycobacterium tuberculosis. Implications for propionate metabolism during growth on fatty acids. J. Bacteriol. 190, 3886–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Upton A. M., McKinney J. D. (2007) Role of the methylcitrate cycle in propionate metabolism and detoxification in Mycobacterium smegmatis. Microbiology 153, 3973–3982 [DOI] [PubMed] [Google Scholar]
- 17. Jain M., Petzold C. J., Schelle M. W., Leavell M. D., Mougous J. D., Bertozzi C. R., Leary J. A., Cox J. S. (2007) Lipidomics reveals control of Mycobacterium tuberculosis virulence lipids via metabolic coupling. Proc. Natl. Acad. Sci. U.S.A. 104, 5133–5138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Russell D. G., VanderVen B. C., Lee W., Abramovitch R. B., Kim M. J., Homolka S., Niemann S., Rohde K. H. (2010) Mycobacterium tuberculosis wears what it eats. Cell Host Microbe 8, 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rainwater D. L., Kolattukudy P. E. (1982) Isolation and characterization of acyl coenzyme A carboxylases from Mycobacterium tuberculosis and Mycobacterium bovis, which produce multiple methyl-branched mycocerosic acids. J. Bacteriol. 151, 905–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cox J. S., Chen B., McNeil M., Jacobs W. R., Jr. (1999) Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402, 79–83 [DOI] [PubMed] [Google Scholar]
- 21. Kaur D., Guerin M. E., Skovierová H., Brennan P. J., Jackson M. (2009) Chapter 2. Biogenesis of the cell wall and other glycoconjugates of Mycobacterium tuberculosis. Adv. Appl. Microbiol. 69, 23–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Beatty W. L., Rhoades E. R., Ullrich H. J., Chatterjee D., Heuser J. E., Russell D. G. (2000) Trafficking and release of mycobacterial lipids from infected macrophages. Traffic 1, 235–247 [DOI] [PubMed] [Google Scholar]
- 23. Beatty W. L., Ullrich H. J., Russell D. G. (2001) Mycobacterial surface moieties are released from infected macrophages by a constitutive exocytic event. Eur. J. Cell Biol. 80, 31–40 [DOI] [PubMed] [Google Scholar]
- 24. Geisel R. E., Sakamoto K., Russell D. G., Rhoades E. R. (2005) In vivo activity of released cell wall lipids of Mycobacterium bovis bacillus Calmette-Guerin is due principally to trehalose mycolates. J. Immunol. 174, 5007–5015 [DOI] [PubMed] [Google Scholar]
- 25. Kim M. J., Wainwright H. C., Locketz M., Bekker L. G., Walther G. B., Dittrich C., Visser A., Wang W., Hsu F. F., Wiehart U., Tsenova L., Kaplan G., Russell D. G. (2010) Caseation of human tuberculosis granulomas correlates with elevated host lipid metabolism. EMBO Mol. Med. 2, 258–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rhoades E., Hsu F., Torrelles J. B., Turk J., Chatterjee D., Russell D. G. (2003) Identification and macrophage-activating activity of glycolipids released from intracellular Mycobacterium bovis BCG. Mol. Microbiol. 48, 875–888 [DOI] [PubMed] [Google Scholar]
- 27. Rhoades E. R., Geisel R. E., Butcher B. A., McDonough S., Russell D. G. (2005) Cell wall lipids from Mycobacterium bovis BCG are inflammatory when inoculated within a gel matrix. Characterization of a new model of the granulomatous response to mycobacterial components. Tuberculosis 85, 159–176 [DOI] [PubMed] [Google Scholar]
- 28. Daniel J., Maamar H., Deb C., Sirakova T. D., Kolattukudy P. E. (2011) Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog. 7, e1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peyron P., Vaubourgeix J., Poquet Y., Levillain F., Botanch C., Bardou F., Daffé M., Emile J. F., Marchou B., Cardona P. J., de Chastellier C., Altare F. (2008) Foamy macrophages from tuberculous patients' granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS Pathog. 4, e1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Russell D. G., Cardona P. J., Kim M. J., Allain S., Altare F. (2009) Foamy macrophages and the progression of the human tuberculosis granuloma. Nat. Immunol. 10, 943–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Podinovskaia M., Lee W., Caldwell S., Russell D. G. (2013) Infection of macrophages with Mycobacterium tuberculosis induces global modifications to phagosomal function. Cell. Microbiol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Abramovitch R. B., Rohde K. H., Hsu F. F., Russell D. G. (2011) aprABC. A Mycobacterium tuberculosis complex-specific locus that modulates pH-driven adaptation to the macrophage phagosome. Mol. Microbiol. 80, 678–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Singh A., Crossman D. K., Mai D., Guidry L., Voskuil M. I., Renfrow M. B., Steyn A. J. (2009) Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 5, e1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Listenberger L. L., Brown D. A. (2007) Fluorescent detection of lipid droplets and associated proteins. Curr. Protoc. Cell Biol. Chapter 24, Unit 24.2 [DOI] [PubMed] [Google Scholar]
- 35. Murry J. P., Sassetti C. M., Lane J. M., Xie Z., Rubin E. J. (2008) Transposon site hybridization in Mycobacterium tuberculosis. Methods Mol. Biol. 416, 45–59 [DOI] [PubMed] [Google Scholar]
- 36. Minnikin D. E., Kremer L., Dover L. G., Besra G. S. (2002) The methyl-branched fortifications of Mycobacterium tuberculosis. Chem. Biol. 9, 545–553 [DOI] [PubMed] [Google Scholar]
- 37. Rohde K. H., Veiga D. F., Caldwell S., Balázsi G., Russell D. G. (2012) Linking the transcriptional profiles and the physiological states of Mycobacterium tuberculosis during an extended intracellular infection. PLoS Pathog. 8, e1002769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Höner Zu Bentrup K., Miczak A., Swenson D. L., Russell D. G. (1999) Characterization of activity and expression of isocitrate lyase in Mycobacterium avium and Mycobacterium tuberculosis. J. Bacteriol. 181, 7161–7167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brock M., Buckel W. (2004) On the mechanism of action of the antifungal agent propionate. Eur. J. Biochem. 271, 3227–3241 [DOI] [PubMed] [Google Scholar]
- 40. Dubos R. J. (1950) The effect of organic acids on mammalian tubercle bacilli. J. Exp. Med. 92, 319–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Trivedi O. A., Arora P., Vats A., Ansari M. Z., Tickoo R., Sridharan V., Mohanty D., Gokhale R. S. (2005) Dissecting the mechanism and assembly of a complex virulence mycobacterial lipid. Mol. Cell 17, 631–643 [DOI] [PubMed] [Google Scholar]
- 42. Rombouts Y., Alibaud L., Carrère-Kremer S., Maes E., Tokarski C., Elass E., Kremer L., Guérardel Y. (2011) Fatty acyl chains of Mycobacterium marinum lipooligosaccharides. Structure, localization, and acylation by PapA4 (MMAR_2343) protein. J. Biol. Chem. 286, 33678–33688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rousseau C., Sirakova T. D., Dubey V. S., Bordat Y., Kolattukudy P. E., Gicquel B., Jackson M. (2003) Virulence attenuation of two Mas-like polyketide synthase mutants of Mycobacterium tuberculosis. Microbiology 149, 1837–1847 [DOI] [PubMed] [Google Scholar]
- 44. Walters S. B., Dubnau E., Kolesnikova I., Laval F., Daffe M., Smith I. (2006) The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol. Microbiol. 60, 312–330 [DOI] [PubMed] [Google Scholar]
- 45. Gonzalo-Asensio J., Mostowy S., Harders-Westerveen J., Huygen K., Hernández-Pando R., Thole J., Behr M., Gicquel B., Martín C. (2008) PhoP. A missing piece in the intricate puzzle of Mycobacterium tuberculosis virulence. PLoS One 3, e3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Venugopal A., Bryk R., Shi S., Rhee K., Rath P., Schnappinger D., Ehrt S., Nathan C. (2011) Virulence of Mycobacterium tuberculosis depends on lipoamide dehydrogenase, a member of three multienzyme complexes. Cell Host Microbe 9, 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Krithika R., Marathe U., Saxena P., Ansari M. Z., Mohanty D., Gokhale R. S. (2006) A genetic locus required for iron acquisition in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 2069–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Maloney E., Lun S., Stankowska D., Guo H., Rajagoapalan M., Bishai W. R., Madiraju M. V. (2011) Alterations in phospholipid catabolism in Mycobacterium tuberculosis lysX mutant. Front. Microbiol. 2, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bhatt A., Molle V., Besra G. S., Jacobs W. R., Jr., Kremer L. (2007) The Mycobacterium tuberculosis FAS-II condensing enzymes. Their role in mycolic acid biosynthesis, acid-fastness, pathogenesis and in future drug development. Mol. Microbiol. 64, 1442–1454 [DOI] [PubMed] [Google Scholar]