Background: Sarcolipin and phospholamban, the regulators of SERCA, are differentially expressed in muscle.
Results: Only sarcolipin binds to SERCA in the presence of Ca2+ and interacts with SERCA throughout the kinetic cycle.
Conclusion: Sarcolipin alone promotes uncoupling of the SERCA pump leading to increased heat production.
Significance: Sarcolipin-mediated regulation of SERCA plays an important role in muscle-based thermogenesis.
Keywords: Calcium ATPase, Calcium Transport, Mutagenesis, Protein Cross-linking, Sarcoplasmic Reticulum (SR), Phospholamban, Sarcolipin
Abstract
Sarco(endo)plasmic reticulum Ca2+ATPase (SERCA) pump activity is modulated by phospholamban (PLB) and sarcolipin (SLN) in cardiac and skeletal muscle. Recent data suggest that SLN could play a role in muscle thermogenesis by promoting uncoupling of the SERCA pump (Lee, A.G. (2002) Curr. Opin. Struct. Biol. 12, 547–554 and Bal, N. C., Maurya, S. K., Sopariwala, D. H., Sahoo, S. K., Gupta, S. C., Shaikh, S. A., Pant, M., Rowland, L. A., Bombardier, E., Goonasekera, S. A., Tupling, A. R., Molkentin, J. D., and Periasamy, M. (2012) Nat. Med. 18, 1575–1579), but the mechanistic details are unknown. To better define how binding of SLN to SERCA promotes uncoupling of SERCA, we compared SLN and SERCA1 interaction with that of PLB in detail. The homo-bifunctional cross-linker (1,6-bismaleimidohexane) was employed to detect dynamic protein interaction during the SERCA cycle. Our studies reveal that SLN differs significantly from PLB: 1) SLN primarily affects the Vmax of SERCA-mediated Ca2+ uptake but not the pump affinity for Ca2+; 2) SLN can bind to SERCA in the presence of high Ca2+, but PLB can only interact to the ATP-bound Ca2+-free E2 state; and 3) unlike PLB, SLN interacts with SERCA throughout the kinetic cycle and promotes uncoupling of the SERCA pump. Using SERCA transmembrane mutants, we additionally show that PLB and SLN can bind to the same groove but interact with a different set of residues on SERCA. These data collectively suggest that SLN is functionally distinct from PLB; its ability to interact with SERCA in the presence of Ca2+ causes uncoupling of the SERCA pump and increased heat production.
Introduction
The sarco(endo)plasmic reticulum Ca2+ATPase (SERCA)3 is primarily responsible for maintaining low cytosolic and high luminal Ca2+ in the sarcoplasmic reticulum (SR) of muscle by coupling energy from ATP hydrolysis to transport Ca2+ (1, 55). SERCA pump activity is modulated by phospholamban (PLB) and sarcolipin (SLN) in cardiac and skeletal muscle (2). Although PLB and SLN have been considered to be homologous proteins, there are several distinct features that have been overlooked. First, the pattern of expression of these proteins is very distinct. In mammals, PLB is expressed in cardiac muscle and to a lesser extent in slow twitch skeletal muscles, whereas SLN is predominantly expressed in skeletal muscles and its expression in the heart is restricted to atria (3, 4). Interestingly, SLN expression is severalfold higher in fast and slow twitch skeletal muscles of larger mammals when compared with rodents (3). Secondly, they bear key structural differences both at the N terminus and at the C terminus with limited sequence similarity in the transmembrane region (5–9). The transmembrane helix of SLN is only 19 amino acids (aa) long, whereas that of PLB is 30 aa long with 21 aa within the membrane and 9 aa protruding out into the cytosol (supplemental Fig. 1) (8). The cytosolic portion of PLB has an additional helix, whereas the unstructured cytosolic SLN region allows flexibility. Further, only SLN has a luminal segment (5 aa) that protrudes into the SR lumen, whereas PLB does not.
These distinct properties of SLN and PLB suggest that they play unique roles in cardiac and skeletal muscle physiology. The function of PLB has been well studied; PLB in the dephosphorylated state is known to decrease the apparent affinity of Ca2+ without any effect on the Vmax of SERCA (10, 11). The inhibitory effect of PLB on SERCA is abolished at high Ca2+ and PLB phosphorylation at Ser-16 or Thr-17 by protein kinase A and Ca2+/calmodulin-dependent protein kinase II (CaMKII) as a result of β-adrenergic stimulation (8, 10, 11). The interaction between PLB and SERCA has been characterized extensively by chemical cross-linking and co-immunoprecipitation experiments (12–20). Recent studies have shown that the presence of Ca2+ influences SERCA-PLB as well as SERCA-SLN interactions. PLB binds most favorably to the Ca2+-free E2 state of SERCA in the presence of ATP, but cannot functionally interact with SERCA at high Ca2+. These studies concluded that PLB and Ca2+ binding to SERCA are mutually exclusive (15, 18, 19, 21). In contrast, the mechanism of SLN inhibition of SERCA is poorly understood. Previous studies have suggested that SLN can affect the affinity and/or the Vmax of SERCA pump (22–25).
It is currently unknown whether SLN binds to SERCA in the same region as PLB and whether SLN binding to SERCA has a different functional outcome when compared with PLB (12, 26, 27). Mall et al. (28) showed that SLN binding to SERCA promotes uncoupling of the SERCA pump and slippage of Ca2+ into the cytoplasm instead of the SR lumen. These studies also suggested that SLN binding with SERCA can increase ATP hydrolysis and heat production and therefore could contribute to muscle thermogenesis (29, 30). We further tested this idea in vivo by generating an SLN−/− mouse model (22, 31). Our results showed that SLN is essential for thermogenesis in muscle and that mice lacking SLN develop hypothermia when exposed to acute cold. Moreover SLN−/− mice became significantly obese when fed on high fat diet, whereas WT mice were less obese and significantly up-regulated SLN expression (31). These data suggested that the muscle-based SLN-SERCA interaction contributes to heat production and energy expenditure.
A major goal of this study was to investigate how SLN binding with SERCA contributes to muscle thermogenesis. In the current study, we investigated whether PLB plays a role in muscle thermogenesis using PLB−/− mice. Our results show that PLB was not essential for thermogenesis. By comparing SLN binding with SERCA with that of PLB-SERCA, we demonstrate that SLN-SERCA interaction is unique; it can bind to SERCA in the presence of high Ca2+ and interact with SERCA throughout the kinetic cycle, which may facilitate uncoupling of SERCA. Using mutagenesis, we show that SLN binds with a different set of residues on SERCA when compared with PLB. These findings provide new insight into the molecular basis of SLN-SERCA interaction and highlight that SLN alone is responsible for muscle thermogenesis.
EXPERIMENTAL PROCEDURES
Materials
The cross-linking reagents, 1,6-bismaleimidohexane (BMH) and dibromobimane, were purchased from Pierce (Thermo Scientific). Thapsigargin (TG), sodium orthovanadate, AlCl3, and KOH were purchased from Sigma. Lipofectamine and DMEM were obtained from Invitrogen. [45Ca]Cl2 was obtained from PerkinElmer Life Sciences.
Mouse Models and Acute Cold Challenge Experiments
PLB−/− mice were a kind gift from Litsa Kranias, University of Cincinnati. SLN−/− mice have been generated previously (22). Both PLB−/− and SLN−/− mice were bred (C57BL/6J genetic background) and housed at ambient temperature. The study protocol was approved by the Ohio State University Institutional Animal Care and Use Committee (OSU-IACUC). Acute cold exposure of mice to 4 °C was performed in the Comprehensive Lab Animal Monitoring System (CLAMS) setup as described before (31).
Mutagenesis and Expression of SLN, PLB, and SERCA in HEK293 Cells
The rat SERCA1 cDNA sequence, which has 99% homology with mouse SERCA1, was cloned into the pcDNA3.1 (+) vector. Mouse SLN and PLB cDNAs were PCR-amplified and cloned into pcDNA3.1 (+) vector. Desired mutagenesis of rat SERCA1, mouse SLN, and mouse PLB were done using the QuikChangeTM site-directed mutagenesis kit (Agilent Technologies). All cDNA clones and mutated constructs were confirmed by direct sequencing. HEK293 cells were co-transfected with SERCA and SLN or PLB construct cDNAs using Lipofectamine 2000. Co-expression was done at 1:2 ratios of SERCA and SLN or PLB. 48 h after transfection, cells were harvested in PBS, and pellet was stored at −80 °C after flash freezing in liquid nitrogen. Microsomes from transfected cells were prepared as described previously (32). Briefly, cells were resuspended in a hypotonic solution containing 10 mm Tris·HCl (pH 7.5) and 0.5 mm MgCl2 for 20 min. Protease inhibitor was added, and cells were homogenized by 30 strokes in a Dounce homogenizer on ice. Homogenates were diluted by an equal volume of 10 mm Tris·HCl (pH 7.5), 0.5 m sucrose, 300 mm KCl. The cell extracts were centrifuged at 10,000 × g to pellet cell debris. The supernatants were diluted with KCl to a final concentration of 0.6 m and centrifuged at 100,000 × g for 1 h at 4 °C. The pellet was resuspended in storage buffer containing 10 mm MOPS (pH 7.0) and 10% sucrose and stored at −80 °C in small aliquots.
Chemical Cross-linking of SLN to SERCA
Chemical cross-linking of proteins was performed by homo-bifunctional sulfhydryl cross-linker BMH (18). SERCA and SLN or PLB were cross-linked in cross-linking buffer containing 40 mm MOPS (pH 7.0), 3.2 mm MgCl2, 75 mm KCl, and 1 mm EGTA. 15 μg of microsomes was mixed in cross-linking buffer, and 3 mm ATP was added followed by the addition of 0.1 mm cross-linker to start cross-linking. The reaction was incubated for 1 h at 25 °C. The reaction was stopped by the addition of SDS-PAGE sample-loading buffer containing 100 mm dithiothreitol. Specific cross-linking of SLN to SERCA interaction shows a 113-kDa band probed with anti-SLN antibody, whereas PLB interaction with SERCA shows a 116-kDa band probed with anti-PLB antibody.
Effect of Ca2+ and TG on SERCA and SLN Interaction
The effect of Ca2+ on SLN binding was assessed by the addition of increasing concentrations of Ca2+ in the presence of ATP and cross-linker as described previously (31). The effect of TG, a SERCA inhibitor, was studied by adding TG (0–10 μm) to the reaction mixture without Ca2+, in the presence of ATP before the addition of cross-linker. The reactions were stopped by adding SDS sample buffer and analyzed as described previously.
SERCA-mediated Ca2+ Uptake and ATP Hydrolysis Assays
To determine the inhibitory effect of SLN on SERCA-mediated Ca2+ transport, oxalate-supported Ca2+ uptake assay was performed (33). SERCA1 was co-transfected with pcDNA3.1, SLN, or E7C-SLN in HEK cells. Briefly, HEK homogenates were incubated in buffer containing 20 mm MOPS (pH 7.0), 5 mm MgCl2, 100 mm KCl, 5 mm NaN3, 5 mm ATP, 5 mm K+-oxalate, and 0.5 mm EGTA. Different concentrations of CaCl2 were added to obtain desired free Ca2+ levels as determined by the MAXCHELATOR program. Samples were incubated in reaction buffer, and aliquots were collected at different time intervals and filtered through 0.45-μm filters. The filters were washed by wash solution, and bound radioactive Ca2+ was measured by scintillation counting. ATPase activities were measured using the BIOMOL green phosphate assay (28, 34). Microsomes were incubated in the Ca2+ uptake buffer containing 15–20 μg of protein in the presence or absence of 5 μm ionophore (A23187). The reaction was initiated by the addition of CaCl2, and samples were collected at different time intervals. The amount of Pi release was calculated as nmol Pi/mg/min. Control reactions were carried out in the presence of TG.
Cross-linking of SLN to the Various Kinetic States of SERCA
The major intermediates of SERCA kinetic steps and their stable analogs were produced by incubating microsomes in cross-linking reaction buffer with different analogs for 45 min at 25 °C before the addition of BMH (35, 36). The E2 state was obtained by incubating the microsomes without ATP. E1·Ca2 and E1PCa2 reaction tubes contained 100 μm free Ca2+ in the absence and presence, respectively, of 3 mm ATP. E1·AlFx·ADP complex, an E1PCa2·ADP analog, was obtained by incubating microsomes with 50 μm AlCl3, 3 mm KF, and 3 mm ADP. E2·AlF4−, the transition state analog of the E2P hydrolysis, was produced by the addition of 50 μm AlCl3 and 3 mm KF to cross-linking buffer. E2Vi, the E2P analog, was produced by preincubating microsomes with 0.1 mm orthovanadate. Chemical cross-linking was done by adding 0.1 mm BMH at 25 °C for 1 h. Cross-linked samples were analyzed by SDS-PAGE and immunoblotted with anti-SLN antibody or anti-PLB antibody.
RESULTS
PLB Does Not Play a Role in Muscle Thermogenesis
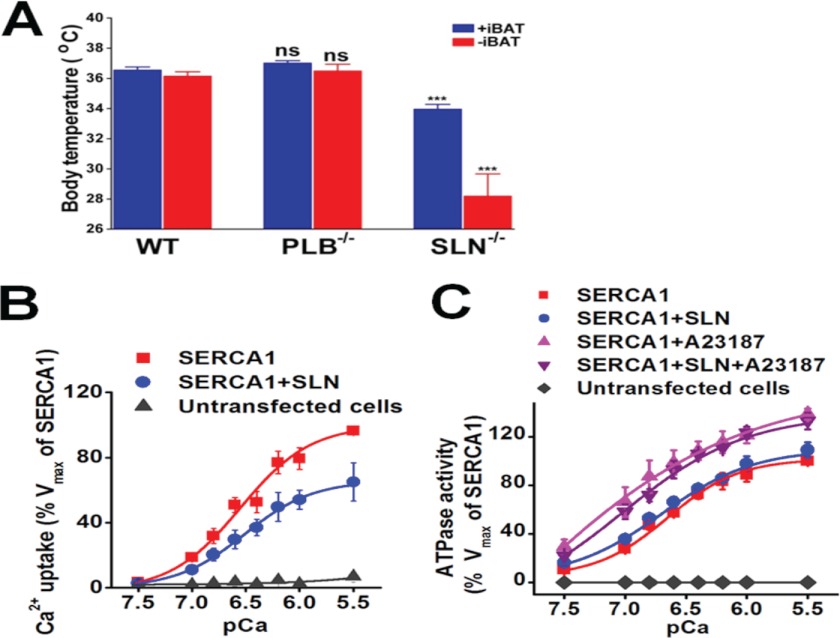
PLB and SLN are key regulators of the SERCA pump, but it is currently unknown whether PLB is also important for thermogenesis as found for SLN (31). Therefore, in this study, PLB−/− and SLN−/− mice were challenged to acute cold (4 °C), and their core body temperature (Tc) was followed for a period of 8 h. To minimize contribution from brown adipose tissue (BAT), we also surgically removed interscapular BAT (iBAT) from one set of mice. Results showed that after 8 h of cold exposure, SLN−/− mice (with iBAT) had a reduced Tc (34.0 ± 0.3 °C). Moreover iBAT-ablated SLN−/− mice showed a further decrease in Tc (28.2 ± 1.5 °C) and developed severe hypothermia, as reported previously (31). However, both PLB−/− and WT mice (with or without iBAT) showed similar heat generation capacity during cold challenge and were able to maintain optimal Tc at ∼37 °C, suggesting that the absence of PLB does not affect thermogenesis. These data clearly demonstrate that PLB is not involved in heat generation and that SLN alone is responsible for muscle thermogenesis (Fig. 1A).
FIGURE 1.
SLN alone is responsible for muscle thermogenesis. A, maintenance of Tc in WT, PLB−/−, and SLN−/− mice with or without iBAT exposed to 4 °C for a period of 8 h. Data are mean ± S.E. of 4–10 animals; ***, p < 0.001 versus WT; ns, not significant as analyzed by Student's t test. B, Ca2+ dependence of Ca2+ uptake was measured in samples expressing SERCA1 alone or with SLN. Untransfected HEK cells were used as control. The values are given as mean of the percentage of SERCA1 mean ± S.E. (n = 3–5 samples). C, the effect of SLN on ATP hydrolysis in the absence and presence of ionophore A23187. The values are given as mean of the percentage of SERCA1 mean ± S.E. (n = 3 samples).
SLN Decreases SERCA Ca2+ Uptake (Vmax) but Does Not Affect ATP Hydrolysis
The finding that SLN alone is responsible for muscle thermogenesis prompted us to further define how SLN interacts with SERCA and modulates pump activity in detail. Previous studies have reported opposing results; some studies showed that SLN causes a decrease in Ca2+ affinity, whereas others showed an increase or decrease in Vmax of Ca2+ uptake (13, 22–25, 27, 28, 37). To further characterize the effect of SLN on SERCA, both Ca2+ uptake and ATP hydrolysis assays were performed using microsomes from HEK cells transfected with SERCA1 alone or with SLN. The expression level of SERCA and SLN was verified by immunoblotting using specific antibodies (supplemental Fig. 2). Oxalate-supported Ca2+ uptake assay showed that the presence of SLN did not affect the apparent Ca2+ affinity of SERCA1 (EC50 SERCA = 6.5 ± 0.1; EC50 SERCA + SLN = 6.4 ± 0.09 mean ± S.E.). In contrast, SLN inhibited the Vmax of the SERCA pump at all Ca2+ concentrations (Fig. 1B). Our result showed a 33% decrease in maximal Ca2+ uptake, and this finding that SLN primarily inhibits the Vmax of SERCA is very similar to previously reported results in mouse hearts deficient in SLN and in studies of rat slow twitch muscle with SLN gene transfer (22, 24). Next, we determined whether SLN had an effect on ATP hydrolysis by measuring the ATPase activity at different Ca2+concentrations (Fig. 1C). This was also tested both in the presence and in the absence of Ca2+ ionophore (A23187) because the addition of ionophore dissipates the Ca2+ concentration gradient across the SR membrane and abolishes the back inhibition of SERCA activity (38). Quantitation of phosphate release showed that there was no significant difference in ATP hydrolysis between samples expressing SERCA alone or with SLN (Fig. 1C). As expected, the addition of ionophore increased the ATPase activity in both samples; however, the presence of SLN did not further modify ATPase activity.
SLN Binds to SERCA in the Same Groove as PLB
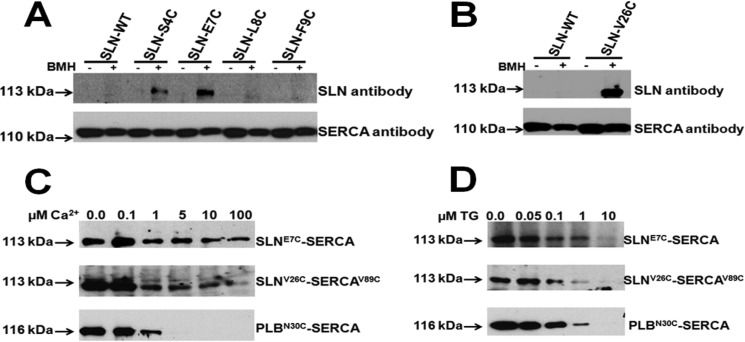
Next, we sought to map the site of SLN binding on SERCA. SLN and SERCA1 were expressed in HEK cells, and protein-protein interaction was studied using a 10 Å chemical cross-linker. The structural basis for PLB and SERCA interaction has been well characterized in vitro using different length chemical cross-linkers (15, 16, 18, 19). These studies have shown that residue Asn-30 of PLB resides less than a distance of 10 Å from cysteine 318 of SERCA2, when the proteins functionally interact (15, 18). To identify the SLN residues that lie at/or within 10 Å from Cys-318 of SERCA, selected amino acids in SLN were mutated to cysteine. The SLN mutants studied include S4C-SLN (corresponding to N27C-PLB), E7C-SLN (corresponding to N30C-PLB), L8C-SLN and F9C-SLN (Fig. 2A), and V26C-SLN (corresponding to V49C of PLB that cross-links with V89C-SERCA) (15). The binding between SERCA1 and SLN was studied with homo-bifunctional cross-linking reagent BMH (Fig. 2, A and B), and the cross-linked SLN-SERCA complex was detected by immunoblotting with anti-SLN antibody. Data showed that residues S4C and E7C in SLN could be specifically cross-linked to Cys-318 of SERCA1, whereas L8C and F9C in SLN could not (Fig. 2A), indicating that Ser-4 and Glu-7 of SLN lie at/within 10 Å from Cys-318 of SERCA. Similarly the C-terminal V26C-SLN showed specific cross-linking with V89C-SERCA1 in the presence of BMH, suggesting that Val-26 of SLN and Val-89 of SERCA are located within 10 Å distance (Fig. 2B). These results suggest that SLN and PLB bind to the same groove of SERCA formed by TMs M2, M4, M6, and M9 (13).
FIGURE 2.
SLN interacts with SERCA1, and its interaction with SERCA is decreased under increasing Ca2+ but abolished by TG. A, cross-linking of SERCA WT (C318) with SLN N-terminal cysteinized mutants (S4C, E7C, L8C, and F9C). B, cross-linking of V89C-SERCA with C-terminal cysteinized SLN mutant V26C-SLN. C, the effect of increasing Ca2+ on SLN-SERCA and PLB-SERCA interaction. E7C-SLN (upper panel) and N30C-PLB (lower panel) were cross-linked with SERCA, and V26C-SLN (middle panel) was cross-linked with V89C-SERCA by adding BMH in the presence of increasing concentrations of Ca2+. D, the effect of increasing TG on SLN-SERCA and PLB-SERCA interaction. E7C-SLN (upper panel), V26C-SLN (Middle panel) or N30C-PLB (lower panel). In all panels, the 113-kDa band refers to SLN cross-linking to SERCA, and the 116-kDa band refers to PLB cross-linking to SERCA1.
SLN Interacts with SERCA in the Presence of High Ca2+, but Its Interaction Is Abolished by Thapsigargin
It is well known that Ca2+ modulates the activity of SERCA, and at high Ca2+, SERCA is maximally activated. We therefore studied the effect of Ca2+ on SERCA-SLN binding and compared how SLN binding differs from that of PLB-SERCA at similar Ca2+ concentrations. SLN can be maximally cross-linked to SERCA in the absence of Ca2+. Interestingly, as shown in Fig. 2C, SLN continues to interact with SERCA even at high Ca2+ (100 μm), but at a reduced level, both at the N terminus (E7C) and at the C terminus (V26C) cross-linking sites. In contrast, PLB binding with SERCA is abolished at Ca2+ concentrations above 1 μm (Fig. 2C). These data suggest that unlike PLB, SLN is able to bind to SERCA even in the presence of high Ca2+ (concentrations ranging from 5 to 100 μm), which is an important distinction between these two molecules. To prove that the binding between SLN and SERCA is dynamic and requires an active SERCA pump, we investigated the effect of TG, a known inhibitor, which forms an irreversible and nonphysiological complex with SERCA (39, 40). We examined the effect of increasing concentrations of TG (1–10 μm) and found that at a higher concentration (10 μm), TG completely abolished the binding between SERCA and SLN at both the N terminus and the C terminus (Fig. 2D). TG also had a similar inhibition on PLB-SERCA binding (Fig. 2D).
PLB and SLN Interact with Different Sets of Transmembrane Residues in SERCA
It has been shown that point mutations in the transmembrane helices M2, M4, and M6 of SERCA1 affect the interaction between SERCA1 and PLB (13, 41). To determine whether SLN binds to the same transmembrane residues as PLB, the transmembrane residues were mutated to alanine. The SERCA1 transmembrane mutants (V89C, L321A, V795A, L802A, T805A, and F809A) were co-expressed together with either E7C-SLN or N30C-PLB in HEK cells. Our cross-linking studies showed that SLN binding with SERCA1 is not significantly affected by any of these mutations except F809A (Fig. 3A). On the other hand, mutations L321A, V795A, L802A, T805A, and F809A in SERCA significantly decrease PLB binding with SERCA (Fig. 3, A and B) as has been reported previously (13, 41). These results suggest that although they bind to the same groove on SERCA, the binding sites for SLN and PLB are not identical.
FIGURE 3.
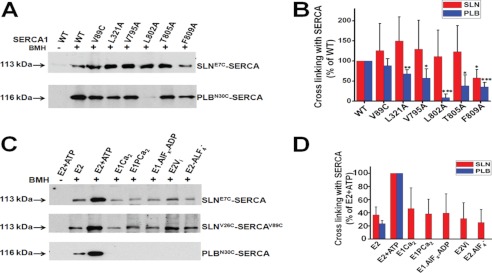
SLN and PLB do not bind to the same residues on SERCA TM domain, and SLN can bind to the different kinetic states of SERCA. A, representative immunoblotting of cross-linked E7C-SLN (upper panel) and N30C-PLB (lower panel) to SERCA1 transmembrane point mutations (V89C, L321A, V795A, L802A, T805A, and F809A). The 113-kDa band refers to SLN cross-linking to SERCA, and the 116-kDa band refers to PLB cross-linking to SERCA1. B, the bar graph shows the band intensity of E7C-SLN or N30C-PLB cross-linking to WT SERCA and TM mutants presented as the percentage of cross-linking to WT type SERCA (n = 3 for each SERCA mutant, data are mean ± S.D.) * (p < 0.05), ** (p < 0.01), and *** (p < 0.001) indicate statistical difference with WT. C, cross-linking of SLN to SERCA following induction of different kinetics states of SERCA as described under “Experimental Procedures.” Upper panel, E7C-SLN; middle panel, V26C-SLN; lower panel, N30C-PLB. The 113-kDa band refers to SLN cross-linking to SERCA, and the 116-kDa band refers to PLB cross-linking to SERCA1. D, the bar graph shows the band intensity of E7C-SLN or N30C-PLB cross-linking to SERCA presented as the percentage of E2+ATP state (n = 3 for each condition, data are mean ± S.D.).
Only SLN Can Bind to Various Kinetic States of SERCA Pump
Our finding that SLN decreases the Vmax of Ca2+ uptake and competitively binds to SERCA in the presence of high Ca2+ (Figs. 1 and 2) indicated that SLN may be affecting one or more kinetic steps during the SERCA reaction cycle. Recent studies also suggested that SLN binding to SERCA could promote uncoupling of Ca2+ transport from the ATP hydrolysis activity of the SERCA pump resulting in heat production (28, 30). To investigate whether SLN can bind to the various kinetic steps of SERCA during the catalytic cycle, we chemically induced various SERCA transition steps (kinetic isomers) by exposing SERCA to metal fluorides and vanadate and performed cross-linking (35, 36, 42, 43). Our results show that SLN can be cross-linked to SERCA in the Ca2+-free E2 state, and the addition of ATP significantly increases the interaction between SERCA and SLN (Fig. 3C). Binding of both ATP and Ca2+ transitions SERCA to E1·Ca2 and E1PCa2 as a result of ATP hydrolysis (43). We found that SLN is able to interact with SERCA at these kinetic states. As shown in Fig. 2C, SLN also interacts to the Ca2+-free E2P state induced by E2Vi or E2·AlF4−, suggesting that SLN occupies SERCA during the whole catalytic cycle. PLB interaction with SERCA was further investigated using the same chemical modifications; it was found that PLB also interacts with the Ca2+-free E2 form of SERCA and that binding of ATP further enhances the interaction as reported previously (12, 15, 18). The binding of Ca2+ to ATP-bound SERCA abolished the interaction between PLB and SERCA. PLB is also unable to bind the subsequent phospho-intermediates, E1PCa2 and E2P of the catalytic cycle (Fig. 3D). Surprisingly, even the Ca2+-free phospho-intermediate (E2P) did not interact with PLB. Importantly, these studies suggest that SLN functions very differently from PLB; it can interact with the SERCA intermediates tested here, and its ability to remain bound during the catalytic cycle may facilitate uncoupling of the pump from Ca2+ transport.
SLN Can Interact with SERCA Even after Both Ca2+ Binding Sites Are Occupied
SERCA has two Ca2+ binding sites, I and II, located adjacent to each other, surrounded by transmembrane helices M4, M5, M6, and M8 (44–46). These sites show cooperativity in Ca2+ binding, and site II can bind the second Ca2+ ion only when site I is filled by the first Ca2+ ion, to trigger the kinetic cycle of the SERCA pump in the presence of ATP. Therefore, the site I mutant (E771Q) precludes any Ca2+ binding to SERCA pump, whereas the site II mutant (E309Q) is able to bind one Ca2+ ion at site I (21, 42, 44). To determine how Ca2+ binding to each of these sites in SERCA affects the ability of SLN to interact with SERCA, we mutated site I (E771Q) and site II (E309Q) individually as reported earlier (21, 44). D351A-SERCA mutant, which binds Ca2+ at both sites as well as ATP but is unable to hydrolyze the ATP, was further utilized to test whether SLN binds to Ca2+-bound SERCA (47). The data shown here demonstrate that only SLN can bind to SERCA that is already bound to Ca2+ and ATP (Fig. 4). This is evident from the results showing that D351A-SERCA interacts with SLN, but not PLB, at all Ca2+ concentrations tested. Interestingly, PLB binding with D351A-SERCA is abolished at a low Ca2+ concentration of 0.1 μm. We also observed that SERCA with site I mutation (E771Q) binds with SLN as well as PLB under all the Ca2+ concentrations tested. Cross-linking of SLN to either of these SERCA mutants (E771Q or E309Q) showed that loss of Ca2+ binding sites increased the level of SLN interaction with SERCA even at high Ca2+, in contrast to SLN binding to WT SERCA (Fig. 4A). On the other hand, PLB was able to bind to the site I mutant (E771Q SERCA) at all Ca2+ concentrations, whereas when site II was mutated (E309Q SERCA), PLB interaction with SERCA could be detected even at 5 μm Ca2+ but was competed out at higher Ca2+ (Fig. 4B) (21). SLN, however, continues to bind E309Q SERCA even at high Ca2+ (100 μm). These findings provide direct evidence that SLN binding to SERCA is distinct from PLB and that its ability to interact with Ca2+-bound SERCA can promote uncoupling of the pump.
FIGURE 4.
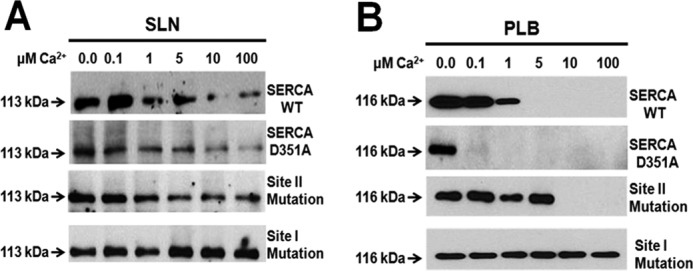
SLN, but not PLB, can interact with Ca2+-bound SERCA. A and B, cross-linking of SLN (A) or PLB (B) to WT SERCA, phosphorylation-defective mutant (D351A-SERCA), and Ca2+ binding site mutants, site II (E309Q-SERCA) and site I (E771Q-SERCA), under increasing Ca2+ concentrations. Mutation of D351A in SERCA allows binding of both Ca2+ and ATP to SERCA but blocks the catalytic cycle by preventing ATP hydrolysis. Mutation to site II allows Ca2+ binding to site I, abolishes PLB binding, but does not affect SLN binding to SERCA. Mutation to site I arrests SERCA in a Ca2+-free state and favors maximal binding of SLN or PLB to SERCA.
DISCUSSION
Using both gain of function and loss of function of SLN mouse models, we recently demonstrated that SLN plays a unique role in muscle physiology and that the SLN-SERCA interaction is an important contributor to muscle-based thermogenesis (31). Our goals in this study were two-fold: 1) to determine whether PLB plays a role in muscle thermogenesis and 2) to understand how the SLN interaction with SERCA differs from that of PLB and the basis for SLN-mediated SERCA uncoupling and muscle thermogenesis. Our studies showed that PLB is not essential for thermogenesis. Therefore, we focused our efforts on defining the uniqueness of SLN-SERCA interaction by comparing it with that of PLB-SERCA. We chose to employ the chemical cross-linking strategy over co-immunoprecipitation (13) because co-immunoprecipitation requires solubilization and disruption of the SR membrane architecture, which will destroy the native interaction between SERCA and SLN. Moreover cross-linking agents have been shown to be reliable reagents not only for deciphering accurate distances between key residues of interacting protein molecules, but also for monitoring the dynamic changes between protein molecules that affect the protein-protein interactions (15, 18, 19, 21).
Data from this study reveal that SLN interaction with SERCA differs significantly from PLB; SLN can bind to SERCA even at high Ca2+ (up to 100 μm), and it remains bound to SERCA during the SERCA kinetic cycle, whereas PLB does not bind to SERCA at high Ca2+ (above 1 μm) or bind to SERCA kinetic intermediate states. A notable finding of our study is that the presence of SLN significantly decreases the Vmax of Ca2+ uptake; however, ATP hydrolysis is unaffected. Our results are in agreement with recent studies showing that SLN has no effect on ATPase activity at saturating concentrations of Ca2+ in unsealed or sealed membrane preparations (28, 30). Collectively, our data showing that despite inhibition of Vmax, ATP hydrolysis is unchanged, suggest that in the presence of SLN, SERCA continues to hydrolyze ATP but less Ca2+ is transported to the lumen of the SR, thus implicating SLN as an uncoupler of SERCA (30). PLB, on the other hand, decreases SERCA Ca2+ transport and ATP hydrolysis only at lower Ca2+ concentrations, but has no effect at higher Ca2+ concentrations (47–49)
Previous modeling studies have shown that the PLB interaction site lies in a groove on the surface of SERCA formed by TMs M2, M4, M6, and M9 (13). Using mutagenesis in the SERCA TM regions and co-immunoprecipitation assays, Asahi et al. (13) showed that PLB and SLN bind to the same groove of SERCA and that the same set of amino acids interacts with SLN or PLB. Our studies showed that SLN can be cross-linked to SERCA at Cys-318 and Val-89 in SERCA as found for PLB, indicating that they bind to the same groove on SERCA. Moreover our studies employing SERCA TM mutants suggested that except for mutation F809A, other mutations including L321A and L802A had no effect on cross-linking between the two molecules (Fig. 3, A and B). Interestingly, although the L802A-SERCA1 mutation did not interfere with SLN binding to SERCA, it completely abolished the interaction of PLB with SERCA (Fig. 3, A and B). These and other published results suggest that SLN and PLB may bind to the same groove in SERCA (26, 50); however, our studies suggest that they may not interact with the same set of amino acids in the TM domain of SERCA. In addition, differences in the N terminus of the two molecules may determine whether it can remain bound to Ca2+-bound SERCA. On the other hand, the protruding C terminus of SLN composed of five unique residues (RSYQY) could provide additional points of interaction for stronger binding with SERCA (51). A detailed understanding of SLN contact points with SERCA requires additional structural studies.
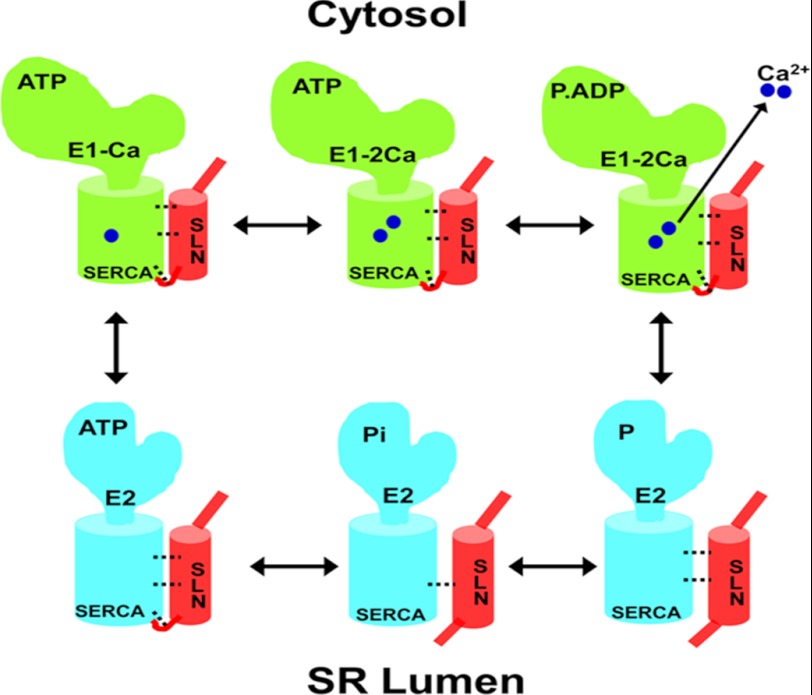
An important goal of this study was to determine how the SLN interaction with SERCA differs from PLB during the Ca2+ transport cycle. Following Ca2+ binding, SERCA hydrolyzes ATP and transitions into E1PCa2+, and subsequently to E2P, before releasing Ca2+ into the lumen (46, 52–54). We found that SLN is able to interact with different SERCA phospho-intermediates during the kinetic cycle (Fig. 3C), whereas PLB can only interact with the Ca2+-free E2 state. Further, our data showing that Ca2+ effectively competes out PLB, but not the SLN interaction with SERCA, suggest that SLN has a higher affinity to SERCA. These novel findings suggest that SLN alone can promote the uncoupling of SERCA ATPase activity from Ca2+ transport. Thus SLN binding to SERCA prevents release of Ca2+ into the lumen by promoting slippage of Ca2+ back to the cytosol (Fig. 5).
FIGURE 5.
Schematic diagram depicting SLN uncoupling of SERCA. Ca2+-bound SERCA is shown in green, and the Ca2+-free form is shown in blue. The functional interaction between SLN (shown in red) and SERCA is shown as dotted bridges. SLN binds strongly to ATP-bound SERCA in the E2 state and remains bound during Ca2+ binding to sites I and II. Ca2+ binding activates ATP hydrolysis and transition to the E1P Ca2+. The continued presence of SLN inhibits normal transition from E1P to E2P and promotes the premature release of Ca2+ to the cytosol. Following slippage of Ca2+, SLN loses its affinity for SERCA, and the pump transitions to the E2P and E2Pi states. The subsequent release of Pi returns SERCA to the E2 state. The model predicts that both SLN and Ca2+ bind simultaneously to SERCA but that the presence of SLN prevents luminal opening of SERCA.
We also studied how Ca2+ binding influences SLN interaction with SERCA using mutagenesis. During Ca2+ transport, sequential binding of Ca2+ to sites I and II promotes conformational changes resulting in occlusion of Ca2+ in the presence of ATP (42, 43). Our mutagenesis studies of Ca2+binding sites I and II indicate that Ca2+ binding to SERCA modifies the ability of PLB and SLN to bind to SERCA. When site I is mutated, SERCA remains in a Ca2+-free (E2) state, allowing maximal binding of SLN or PLB. However, mutation of site II allows Ca2+ binding to site I (requires higher Ca2+) and abolishes PLB interaction (21). The finding that SLN continues to bind to the site II mutant further confirms that SLN can bind to Ca2+-bound SERCA. These data, along with the data showing that SLN interacts with SERCA throughout the kinetic cycle, further support that SLN alone can promote uncoupling of SERCA.
This study reveals important insights on how SLN differs from PLB. Our data conclusively show that 1) SLN interacts with SERCA in the presence of high Ca2+, 2) only SLN remains bound to SERCA throughout the kinetic cycle, and 3) PLB cannot interact with Ca2+-bound SERCA. Based on our findings, we propose a model to illustrate how SLN binding leads to uncoupling of the SERCA pump in Fig. 5. The model shows that SLN and SERCA are co-localized in the SR and that their functional interaction is modulated by Ca2+. In the absence of Ca2+, SLN interacts maximally with the E2 state of SERCA. When the Ca2+ level is increased, the majority of the SLN is released from SERCA, but a fraction of the pumps remains bound to SLN. SLN binding to SERCA allows ATP hydrolysis to proceed but interferes with Ca2+ transport into SR, instead promoting release of Ca2+ back to the cytosol (54).
These studies additionally suggest how PLB and SLN regulation of SERCA can produce distinct physiological outcomes in muscle. PLB is predominant in the heart, and its ability to modulate Ca2+ affinity in a phosphorylation-dependent manner is well suited to regulate cardiac function during rest and exercise, requiring different rates of Ca2+ transport. SLN, on the other hand, does not affect pump affinity for Ca2+ but primarily affects Vmax by causing inefficiency and promoting uncoupling of SERCA contributing to enhanced heat production.
Acknowledgments
We thank Dr Litsa Kranias (University of Cincinnati) for providing PLB−/− mice, Jonathan Lytton (University of Calgary) for rat SERCA1a, and Anthony G. Lee (University of Southampton) for advice.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 HL080551 (to M. P.).

This article contains supplemental Figs. 1 and 2.
- SERCA
- sarco(endo)plasmic reticulum Ca2+ATPase
- SR
- sarcoplasmic reticulum
- PLB
- phospholamban
- SLN
- sarcolipin
- BMH
- 1,6-bismaleimidohexane
- TG
- thapsigargin
- BAT
- brown adipose tissue
- iBAT
- interscapular BAT
- aa
- amino acids
- TM
- transmembrane.
REFERENCES
- 1. Periasamy M., Bhupathy P., Babu G. J. (2008) Regulation of sarcoplasmic reticulum Ca2+ ATPase pump expression and its relevance to cardiac muscle physiology and pathology. Cardiovasc. Res. 77, 265–273 [DOI] [PubMed] [Google Scholar]
- 2. Bhupathy P., Babu G. J., Periasamy M. (2007) Sarcolipin and phospholamban as regulators of cardiac sarcoplasmic reticulum Ca2+ ATPase. J. Mol. Cell. Cardiol. 42, 903–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Babu G. J., Bhupathy P., Carnes C. A., Billman G. E., Periasamy M. (2007) Differential expression of sarcolipin protein during muscle development and cardiac pathophysiology. J. Mol. Cell. Cardiol. 43, 215–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vangheluwe P., Schuermans M., Zádor E., Waelkens E., Raeymaekers L., Wuytack F. (2005) Sarcolipin and phospholamban mRNA and protein expression in cardiac and skeletal muscle of different species. Biochem. J. 389, 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Butler J., Lee A. G., Wilson D. I., Spalluto C., Hanley N. A., East J. M. (2007) Phospholamban and sarcolipin are maintained in the endoplasmic reticulum by retrieval from the ER-Golgi intermediate compartment. Cardiovasc. Res. 74, 114–123 [DOI] [PubMed] [Google Scholar]
- 6. Gramolini A. O., Kislinger T., Asahi M., Li W., Emili A., MacLennan D. H. (2004) Sarcolipin retention in the endoplasmic reticulum depends on its C-terminal RSYQY sequence and its interaction with sarco (endo) plasmic Ca2+-ATPases. Proc. Natl. Acad. Sci. U.S.A. 101, 16807–16812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. MacLennan D. H., Asahi M., Tupling A. R. (2003) The regulation of SERCA-type pumps by phospholamban and sarcolipin. Ann. N.Y. Acad. Sci. 986, 472–480 [DOI] [PubMed] [Google Scholar]
- 8. MacLennan D. H., Kranias E. G. (2003) Phospholamban: a crucial regulator of cardiac contractility. Nat. Rev. Mol. Cell Biol. 4, 566–577 [DOI] [PubMed] [Google Scholar]
- 9. Traaseth N. J., Ha K. N., Verardi R., Shi L., Buffy J. J., Masterson L. R., Veglia G. (2008) Structural and dynamic basis of phospholamban and sarcolipin inhibition of Ca2+-ATPase. Biochemistry 47, 3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chu G., Lester J. W., Young K. B., Luo W., Zhai J., Kranias E. G. (2000) A single site (Ser16) phosphorylation in phospholamban is sufficient in mediating its maximal cardiac responses to β-agonists. J. Biol. Chem. 275, 38938–38943 [DOI] [PubMed] [Google Scholar]
- 11. Luo W., Grupp I. L., Harrer J., Ponniah S., Grupp G., Duffy J. J., Doetschman T., Kranias E. G. (1994) Targeted ablation of the phospholamban gene is associated with markedly enhanced myocardial contractility and loss of β-agonist stimulation. Circ. Res. 75, 401–409 [DOI] [PubMed] [Google Scholar]
- 12. Asahi M., McKenna E., Kurzydlowski K., Tada M., MacLennan D. H. (2000) Physical interactions between phospholamban and sarco (endo) plasmic reticulum Ca2+-ATPases are dissociated by elevated Ca2+, but not by phospholamban phosphorylation, vanadate, or thapsigargin, and are enhanced by ATP. J. Biol. Chem. 275, 15034–15038 [DOI] [PubMed] [Google Scholar]
- 13. Asahi M., Sugita Y., Kurzydlowski K., De Leon S., Tada M., Toyoshima C., MacLennan D. H. (2003) Sarcolipin regulates sarco (endo) plasmic reticulum Ca2+-ATPase (SERCA) by binding to transmembrane helices alone or in association with phospholamban. Proc. Natl. Acad. Sci. U.S.A. 100, 5040–5045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Autry J. M., Rubin J. E., Pietrini S. D., Winters D. L., Robia S. L., Thomas D. D. (2011) Oligomeric interactions of sarcolipin and the Ca-ATPase. J. Biol. Chem. 286, 31697–31706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen Z., Akin B. L., Stokes D. L., Jones L. R. (2006) Cross-linking of C-terminal residues of phospholamban to the Ca2+ pump of cardiac sarcoplasmic reticulum to probe spatial and functional interactions within the transmembrane domain. J. Biol. Chem. 281, 14163–14172 [DOI] [PubMed] [Google Scholar]
- 16. Chen Z., Stokes D. L., Rice W. J., Jones L. R. (2003) Spatial and dynamic interactions between phospholamban and the canine cardiac Ca2+ pump revealed with use of heterobifunctional cross-linking agents. J. Biol. Chem. 278, 48348–48356 [DOI] [PubMed] [Google Scholar]
- 17. James P., Inui M., Tada M., Chiesi M., Carafoli E. (1989) Nature and site of phospholamban regulation of the Ca2+ pump of sarcoplasmic reticulum. Nature 342, 90–92 [DOI] [PubMed] [Google Scholar]
- 18. Jones L. R., Cornea R. L., Chen Z. (2002) Close proximity between residue 30 of phospholamban and cysteine 318 of the cardiac Ca2+ pump revealed by intermolecular thiol cross-linking. J. Biol. Chem. 277, 28319–28329 [DOI] [PubMed] [Google Scholar]
- 19. Toyoshima C., Asahi M., Sugita Y., Khanna R., Tsuda T., MacLennan D. H. (2003) Modeling of the inhibitory interaction of phospholamban with the Ca2+ ATPase. Proc. Natl. Acad. Sci. U.S.A. 100, 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Buffy J. J., Buck-Koehntop B. A., Porcelli F., Traaseth N. J., Thomas D. D., Veglia G. (2006) Defining the intramembrane binding mechanism of sarcolipin to calcium ATPase using solution NMR spectroscopy. J. Mol. Biol. 358, 420–429 [DOI] [PubMed] [Google Scholar]
- 21. Chen Z., Akin B. L., Jones L. R. (2010) Ca2+ binding to site I of the cardiac Ca2+ pump is sufficient to dissociate phospholamban. J. Biol. Chem. 285, 3253–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Babu G. J., Bhupathy P., Timofeyev V., Petrashevskaya N. N., Reiser P. J., Chiamvimonvat N., Periasamy M. (2007) Ablation of sarcolipin enhances sarcoplasmic reticulum calcium transport and atrial contractility. Proc. Natl. Acad. Sci. U.S.A. 104, 17867–17872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Odermatt A., Becker S., Khanna V. K., Kurzydlowski K., Leisner E., Pette D., MacLennan D. H. (1998) Sarcolipin regulates the activity of SERCA1, the fast-twitch skeletal muscle sarcoplasmic reticulum Ca2+-ATPase. J. Biol. Chem. 273, 12360–12369 [DOI] [PubMed] [Google Scholar]
- 24. Tupling A. R., Asahi M., MacLennan D. H. (2002) Sarcolipin overexpression in rat slow twitch muscle inhibits sarcoplasmic reticulum Ca2+ uptake and impairs contractile function. J. Biol. Chem. 277, 44740–44746 [DOI] [PubMed] [Google Scholar]
- 25. Tupling A. R., Bombardier E., Gupta S. C., Hussain D., Vigna C., Bloemberg D., Quadrilatero J., Trivieri M. G., Babu G. J., Backx P. H., Periasamy M., MacLennan D. H., Gramolini A. O. (2011) Enhanced Ca2+ transport and muscle relaxation in skeletal muscle from sarcolipin-null mice. Am. J. Physiol. Cell Physiol. 301, C841–C849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Asahi M., Nakayama H., Tada M., Otsu K. (2003) Regulation of sarco (endo) plasmic reticulum Ca 2+ adenosine triphosphatase by phospholamban and sarcolipin: implication for cardiac hypertrophy and failure. Trends Cardiovasc. Med. 13, 152–157 [DOI] [PubMed] [Google Scholar]
- 27. Asahi M., Kurzydlowski K., Tada M., MacLennan D. H. (2002) Sarcolipin inhibits polymerization of phospholamban to induce superinhibition of sarco (endo) plasmic reticulum Ca2+-ATPases (SERCAs). J. Biol. Chem. 277, 26725–26728 [DOI] [PubMed] [Google Scholar]
- 28. Mall S., Broadbridge R., Harrison S. L., Gore M. G., Lee A. G., East J. M. (2006) The presence of sarcolipin results in increased heat production by Ca2+-ATPase. J. Biol. Chem. 281, 36597–36602 [DOI] [PubMed] [Google Scholar]
- 29. Reis M., Farage M., de Souza A. C. L., de Meis L. (2001) Correlation between uncoupled ATP hydrolysis and heat production by the sarcoplasmic reticulum Ca2+-ATPase. J. Biol. Chem. 276, 42793–42800 [DOI] [PubMed] [Google Scholar]
- 30. Smith W. S., Broadbridge R., East J. M., Lee A. G. (2002) Sarcolipin uncouples hydrolysis of ATP from accumulation of Ca2+ by the Ca2+-ATPase of skeletal-muscle sarcoplasmic reticulum. Biochem. J. 361, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bal N. C., Maurya S. K., Sopariwala D. H., Sahoo S. K., Gupta S. C., Shaikh S. A., Pant M., Rowland L. A., Bombardier E., Goonasekera S. A., Tupling A. R., Molkentin J. D., Periasamy M. (2012) Sarcolipin is a newly identified regulator of muscle-based thermogenesis in mammals. Nat. Med. 18, 1575–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maruyama K., MacLennan D. H. (1988) Mutation of aspartic acid-351, lysine-352, and lysine-515 alters the Ca2+ transport activity of the Ca2+-ATPase expressed in COS-1 cells. Proc. Natl. Acad. Sci. U.S.A. 85, 3314–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ji Y., Loukianov E., Periasamy M. (1999) Analysis of sarcoplasmic reticulum Ca2+ transport and Ca2+ ATPase enzymatic properties using mouse cardiac tissue homogenates. Anal. Biochem. 269, 236–244 [DOI] [PubMed] [Google Scholar]
- 34. Andersen J. P. (1995) Functional consequences of alterations to amino acids at the M5S5 boundary of the Ca-ATPase of sarcoplasmic reticulum. J. Biol. Chem. 270, 908–914 [DOI] [PubMed] [Google Scholar]
- 35. Daiho T., Yamasaki K., Danko S., Suzuki H. (2007) Critical role of Glu40-Ser48 loop linking actuator domain and first transmembrane helix of Ca2+-ATPase in Ca2+ deocclusion and release from ADP-insensitive phosphoenzyme. J. Biol. Chem. 282, 34429–34447 [DOI] [PubMed] [Google Scholar]
- 36. Inesi G., Lewis D., Toyoshima C., Hirata A., de Meis L. (2008) Conformational fluctuations of the Ca2+-ATPase in the native membrane environment. J. Biol. Chem. 283, 1189–1196 [DOI] [PubMed] [Google Scholar]
- 37. Asahi M., Otsu K., Nakayama H., Hikoso S., Takeda T., Gramolini A. O., Trivieri M. G., Oudit G. Y., Morita T., Kusakari Y. (2004) Cardiac-specific overexpression of sarcolipin inhibits sarco (endo) plasmic reticulum Ca2+ ATPase (SERCA2a) activity and impairs cardiac function in mice. Proc. Natl. Acad. Sci. U.S.A. 101, 9199–9204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scarpa A., Baldassare J., Inesi G. (1972) The effect of calcium ionophores on fragmented sarcoplasmic reticulum. J. Gen. Physiol. 60, 735–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sagara Y., Fernandez-Belda F., de Meis L., Inesi G. (1992) Characterization of the inhibition of intracellular Ca2+ transport ATPases by thapsigargin. J. Biol. Chem. 267, 12606–12613 [PubMed] [Google Scholar]
- 40. Xu C., Ma H., Inesi G., Al-Shawi M. K., Toyoshima C. (2004) Specific structural requirements for the inhibitory effect of thapsigargin on the Ca2+ ATPase SERCA. J. Biol. Chem. 279, 17973–17979 [DOI] [PubMed] [Google Scholar]
- 41. Asahi M., Kimura Y., Kurzydlowski K., Tada M., MacLennan D. H. (1999) Transmembrane helix M6 in sarco (endo) plasmic reticulum Ca2+-ATPase forms a functional interaction site with phospholamban. J. Biol. Chem. 274, 32855–32862 [DOI] [PubMed] [Google Scholar]
- 42. Inesi G., Kurzmack M., Coan C., Lewis D. (1980) Cooperative calcium binding and ATPase activation in sarcoplasmic reticulum vesicles. J. Biol. Chem. 255, 3025–3031 [PubMed] [Google Scholar]
- 43. Inesi G., Lewis D., Ma H., Prasad A., Toyoshima C. (2006) Concerted conformational effects of Ca2+ and ATP are required for activation of sequential reactions in the Ca2+ ATPase (SERCA) catalytic cycle. Biochemistry 45, 13769–13778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Andersen J. P., Vilsen B. (1992) Functional consequences of alterations to Glu309, Glu771, and Asp800 in the Ca2+-ATPase of sarcoplasmic reticulum. J. Biol. Chem. 267, 19383–19387 [PubMed] [Google Scholar]
- 45. Clarke D. M., Loo T. W., Inesi G., MacLennan D. H. (1989) Location of high affinity Ca2+-binding sites within the predicted transmembrane domain of the sarco-plasmic reticulum Ca2+-ATPase. Nature 339, 476–478 [DOI] [PubMed] [Google Scholar]
- 46. Toyoshima C., Inesi G. (2004) Structural basis of ion pumping by Ca2+-ATPase of the sarcoplasmic reticulum. Ann. Rev. Biochem. 73, 269–292 [DOI] [PubMed] [Google Scholar]
- 47. Akin B. L., Chen Z., Jones L. R. (2010) Superinhibitory phospholamban mutants compete with Ca2+ for binding to SERCA2a by stabilizing a unique nucleotide-dependent conformational state. J. Biol. Chem. 285, 28540–28552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hughes G., Starling A. P., Sharma R. P., East J. M., Lee A. G. (1996) An investigation of the mechanism of inhibition of the Ca2+-ATPase by phospholamban. Biochem. J. 318, 973–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reddy L. G., Autry J. M., Jones L. R., Thomas D. D. (1999) Co-reconstitution of phospholamban mutants with the Ca-ATPase reveals dependence of inhibitory function on phospholamban structure. J. Biol. Chem. 274, 7649–7655 [DOI] [PubMed] [Google Scholar]
- 50. Morita T., Hussain D., Asahi M., Tsuda T., Kurzydlowski K., Toyoshima C., Maclennan D. H. (2008) Interaction sites among phospholamban, sarcolipin, and the sarco (endo) plasmic reticulum Ca2+-ATPase. Biochem. Biophys. Res. Comm. 369, 188–194 [DOI] [PubMed] [Google Scholar]
- 51. Hughes E., Clayton J. C., Kitmitto A., Esmann M., Middleton D. A. (2007) Solid-state NMR and functional measurements indicate that the conserved tyrosine residues of sarcolipin are involved directly in the inhibition of SERCA1. J. Biol. Chem. 282, 26603–26613 [DOI] [PubMed] [Google Scholar]
- 52. Toyoshima C., Mizutani T. (2004) Crystal structure of the calcium pump with a bound ATP analogue. Nature 430, 529–535 [DOI] [PubMed] [Google Scholar]
- 53. Toyoshima C., Nakasako M., Nomura H., Ogawa H. (2000) Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Ä resolution. Nature 405, 647–655 [DOI] [PubMed] [Google Scholar]
- 54. Toyoshima C., Nomura H. (2002) Structural changes in the calcium pump accompanying the dissociation of calcium. Nature 418, 605–611 [DOI] [PubMed] [Google Scholar]
- 55. Lee A. G. (2002) A calcium pump made visible. Curr. Opin. Struct. Biol. 12, 547–554 [DOI] [PubMed] [Google Scholar]