Abstract
Glycans, oligo- and polysaccharides secreted or attached to proteins and lipids, cover the surfaces of all cells and have a regulatory capacity and structural diversity beyond any other class of biological molecule. Glycans may have evolved these properties because they mediate cellular interactions and often face pressure to evolve new functions rapidly. We approach this idea two ways. First, we discuss evolutionary innovation. Glycan synthesis, regulation, and mode of chemical interaction influence the spectrum of new forms presented to evolution. Second, we describe the evolutionary conflicts that arise when alleles and individuals interact. Glycan regulation and diversity are integral to these biological negotiations. Glycans are tasked with such an amazing diversity of functions that no study of cellular interaction can begin without considering them. We propose that glycans predominate the cell surface because their physical and chemical properties allow the rapid innovation required of molecules on the frontlines of evolutionary conflict.
Keywords: Evolution, Glycoconjugate, Host Defense, Pathogen-associated Molecular Pattern (PAMP), Symbiosis
Glycan Diversity
All cellular organisms are coated by a glycocalyx, a “sugar shell” that defines the molecular frontier of cells, tissues, and whole organisms (1). Many biological interactions involve the molecules of this shell, including those between sperm and egg, hosts and pathogens, and the cells and extracellular matrices of developing organisms (2, 3). Glycans are oligo- or polysaccharide molecules that can be secreted or attached to proteins or lipids, forming glycoconjugates. Glycans form one of the four classes of biomolecules together with nucleic acids, lipids, and proteins (4). Glycoconjugates are common; >50% of all human proteins are glycosylated (5). The extracellular polysaccharides of plants, fungi, and arthropods (cellulose and chitin) are the most abundant biomolecules on earth. Glycan synthesis is not template-driven; genes do not encode them directly. Rather, glycans are assembled by synthetic enzymes in the endoplasmic reticulum Golgi or during direct secretion (6).
Glycans contain a variety of monosaccharide units and linkages joining them in structures that can be branched and further chemically modified. Sizes range from the single GalNAc monosaccharide of the Tn antigen to glycosaminoglycan polymers thousands of units long (7). Glycan functions include physical and structural roles (adding rigidity or charge), extracellular matrix formation and morphogenesis (glycosaminoglycans), protein folding, transcriptional regulation (by O-GlcNAcylation), and information exchange between cells (stage-specific embryonic antigens) (3). Some glycan types occur in most organisms (N-glycans), and others are restricted to certain lineages (hyaluronic acid in vertebrates and sialic acids in deuterostomes) (8). Glycan diversity could be a response to conflict: pathogens exploit host glycans for recognition, attachment, and entry, driving rapid evolution of both parties (8). Many important questions about glycan evolution remain, and new functions continue to be discovered. We review the roles of glycans in biological collaboration and conflict, emphasizing how their unique structural and regulatory properties influence constraints on evolution.
Evolutionary Constraints
The path of evolution is beset by constraints that limit the kinds of traits that can evolve. From below are constraints caused by mutation. The suite of variation presented by mutation is not random, and an abundance or absence of particular phenotypes can influence the direction of evolution or prevent organisms from reaching an optimal form (9). From above are constraints caused by selection itself. The optimal solution to one selective challenge is often a poor answer to another. This limits evolution to the set of phenotypes that optimally trade off opposing selective forces (10). Constraints hold organisms away from their fitness optima, so we expect selection to favor mechanisms of overcoming them. By delineating the limits that constraints impose, we may recognize familiar features of organisms as solutions to constraint (2). Many glycan features (their synthesis and degradation, regulation, molecular composition, and interactions) influence phenotypic variation in ways that overcome the limits that mutational and selective constraints place on evolution. We discuss each in turn.
Mutational Constraints on Evolution
Mutational constraints are limits on the spectrum of phenotypic changes offered to evolution. These include physical limits: many monosaccharide structures and glycosidic linkages are never observed in nature, though they can be generated artificially and incorporated into living organisms (11). Similarly, not all synthesis and breakdown reactions or ligand binding operations are chemically feasible. Vertebrate brain tissue is rich in the sialic acid Neu5Ac but contains only traces of Neu5Gc, likely because vertebrate sialidases cannot easily break down Neu5Gc polymers (12). Constraints also result from historical events. Loss of a synthetic enzyme can lock whole lineages into a subset of glycan structures (13–15). Glycans can also be constrained if all available changes impair obligate functions (1). Despite massive changes in sialic acid biology, all vertebrates make some form of sialic acid because it is needed for proper development (16).
Glycans face a different set of mutational constraints than proteins because of their different modes of synthesis and chemical interaction. This releases glycoconjugates from some constraints that typify the evolution of naked proteins and imposes other constraints more acutely. There are striking differences between proteins and glycans. Proteins are direct products of alleles encoded in DNA, whereas glycans take a variety of forms that reflect variation in their synthesis and attachment. Foreign proteins trigger adaptive immune reactions due to sampling and presentation by immune cells. Foreign glycans typically trigger less specific innate immune reactions. Modes of glycan and protein evolution can also differ. Viral proteins often evolve rapidly to escape detection by host antibodies (17, 18). Viral glycosylation can also change rapidly, but most viral glycans are built by the host synthesis machinery. Viruses can be selected to mimic the host or exploit host glycosylation of viral glycoproteins for immune evasion and tropism (19–21). Thus, the protein and glycan components of viruses can be simultaneously selected for opposite tasks, evasion and emulation, because of the different types of mutations available to each.
Evolution of New Glycoforms
Organisms evolve new functions by exploring the neighborhood of phenotypic variation created by mutation. Glycan synthesis involves many proteins, which can increase the number of phenotypes available by mutation and the rate that glycoproteins find new functions in two ways.
First, mutations affecting the regulation or activity of glycan-synthesizing proteins can alter patterns of glycosylation. About 2% of the mammalian genome encodes glycosyltransferases and glycosidases: enzymes for glycan assembly, remodeling, and degradation (22). Even in mammals, in which glycans are based on just 10 sugar types, the diversity of structures generated by sequential and combinatorial glycan synthesis is staggering (23). In glycolipids, both the lipid and glycan moieties are synthetic products that, when combined, yield an even larger potential diversity (24). Glycoforms can vary transiently within organisms, tracking diet, physiological state, or development (25–27), or they can vary among individuals. Parents and offspring (28, 29) or males and females (30, 31) can have different glycosylation patterns. Changes in the regulation of transferase enzymes, nucleotide sugar donors, or acceptor substrates can transform the glycoform of a cell and its secretions (32). Just as alternative splicing allows a gene to generate many protein structures, alternative glycosylation, both by different occupancy of glycosylation sites and with different glycans, can augment the phenotypic repertoire of an organism.
Second, glycan-synthesizing and glycan-binding protein families have remarkable patterns of evolution, involving every known mechanism of generating diversity. Alternative splicing, exon shuffling, retrotransposition, gene duplication, and gene loss typify glycosyltransferase (13, 33, 34) and glycosidase (35) gene families. Both glycosyltransferases (36) and glycosidases (37) sometimes evolve by positive selection, meaning that successful new mutations rapidly increase in frequency. Loss-of-function mutations can also be selected rapidly into populations (13, 14). Selection maintains balanced polymorphisms, where more than one allelic variant persists at intermediate frequency. Alleles encoding human blood group antigens may remain polymorphic because viral and bacterial pathogens target blood group-defining glycans for attachment (34, 38).
The proteins that bind glycans (lectins) can be similarly diverse. Oyster sperm contain bindin, an F-type lectin encoded by a single gene that produces >4500 protein variants in a single species by a combination of recombination, alternative splicing, and positive selection (39). Lectin gene families also change rapidly. In the human lineage, the family of sialic acid-binding Ig-like lectins (siglecs) has undergone changes to expression pattern and binding specificity and instances of gene duplication, gene conversion, and adaptive gene loss (14, 15, 40). Selection favoring diverse and labile patterns of glycosylation also favors genomic features that enable diversification of genes encoding glycan synthesis, degradation, and recognition.
Not All Glycan Changes Are Equally Likely
Glycans may experience some mutational constraints differently than naked proteins because they result from coordinated assembly pathways. Glycans are made by a network of enzymes, each building on or degrading the product of prior steps (41). Changing an enzyme's activity or regulation can alter product abundance and change rates of synthesis or degradation elsewhere in the pathway (42). Central steps tend to be conserved: enzymes at the base of the asparagine N-glycosylation pathway tolerate less variation than enzymes that catalyze terminal steps (43), perhaps because mutations that disrupt fewer glycan synthesis steps have milder phenotypes (44). Essential steps are not always encoded by redundant gene families. O-Glycan synthesis is initiated by 20 O-GalNAc glycosyltransferases (32), but a required step of N-glycan synthesis, transfer of dolichol-linked high mannose N-glycan, is catalyzed by a single protein (45). Single copy enzymes perform conserved functions inside the cell, such as O-GlcNAcylation and N-glycosylation for protein folding quality control. Extracellular glycans are often built by enzymes with partly redundant functions (32, 33).
The evolution of glycan synthesis pathways often involves loss-of-function mutations. However, some lineages evolve the ability to synthesize new glycans: the sialic acid variant Neu5Gc is found only in deuterostomes (46), and heparan sulfate is found only in metazoans (excluding sponges) (47). Changes sometimes occur repeatedly to the same genes. The ABO blood group glycosyltransferases were lost multiple times within humans (48), and the A and B antigens were independently gained by chimpanzees and gorillas (49). Neu5Gc was lost independently in humans, birds, and platypuses (46), and several bird groups have independently lost glycans terminating in Galα1–4Gal (50). Despite their capacity for structural diversity, glycans and the enzymes that process them do experience mutational constraint; some changes occur more readily and repeatedly than others.
Glycan Interactions Are Robust to Change
Robustness is the ability of a phenotype to tolerate evolutionary change and thus explore new functions by mutation (9). Several features cause glycan interactions to be robust: their expression can be finely regulated, the proteins that bind glycans are themselves robust, and glycan binding chemistry permits low affinity/high specificity receptor-ligand pairings. Spatial and temporal regulation increases robustness by separating conflicting tasks. Sperm glycans terminate with sialic acid to prevent their detection by female immunity. As sperm near the egg, these masks are removed, exposing the underlying glycans that function directly in fertilization (51). Masked glycans can change without immunological consequence because they are exposed only when they are functional (52). Glycans are subject to fine regulation in response to cellular location and metabolic state (25). This causes glycosylation to be robust because it allows variation to be masked in all but the appropriate contexts.
Lectins have stable folds, perhaps because they bind a diverse array of glycoforms. The C-type lectin fold can tolerate massive amino acid variation as shown by the major tropism determinant (Mtd) of the Bordetella bacteriophage. A retroelement-encoded reverse transcriptase generates ∼1013 different Mtd variants that bind receptors in different stages of the Bordetella life cycle (53). Although their folds are robust, the specificity of lectin-glycan interactions can be engineered by just a few mutations (54, 55). Lectins also rapidly evolve new functions in nature. Glycan-binding domains that evolve by positive selection include C-type lectins (56, 57), F-type lectins (39), and siglecs (15) among others. Lectins can tolerate mutational variation and thus explore the space of binding functions available by mutation, enabling their evolution and the co-evolution of their glycan targets.
The chemistry of glycan binding also creates robustness. Glycan binding often occurs with low affinity but high specificity (58). As a result, glycan binding is often multistep and cooperative. Glycans can be guided to their targets by interactions between nearby ligands, and spatial clustering can radically change glycan binding (59). This creates an extra control on the regulation of glycan binding: interaction intensity can be tuned by altering the distribution and density of glycans and lectins. Cooperative binding may increase robustness by placing molecules in position to physically interact even if their affinity for one another is low. Interactions between binding partners may therefore be more tolerant of variation in the affinity or selectivity of binding. The extraordinary specificity of some lectins could result from an increased ability to explore the space of binding functions because close physical interaction can be achieved even between molecules with low affinity.
Evolution of Uniqueness
Glycans are the frontline of one of the most interesting challenges facing organisms: distinguishing self from non-self. This requires generating unique molecular patterns and safeguarding their exclusivity against mimicry by pathogens. The ways that organisms create, recognize, and defend their own unique glycan patterns inform us about constraints on evolutionary innovation because markers of self are necessarily unique innovations. The space of possible unique molecular markers of self is huge (60, 61), so it is an interesting case to study how evolutionary constraints limit the variation available to evolution. Both mutational and selective constraints influence the evolution of unique glycans, and so we use this section as a bridge between these two categories of evolutionary constraint.
The most striking glycan difference in nature may be a consequence of mutational and selective constraints. Unicellular organisms make glycans of many different monosaccharide units, whereas glycan diversity in multicellular eukaryotes is usually a rearrangement of a handful of monosaccharides (62). Unicellular organisms can secrete novel monosaccharides outside their “bodies” and can thus use structures that cannot be degraded by existing cellular machinery (63). In multicellular organisms, new monosaccharides cannot simply be excreted and will be tolerated only if they are compatible with existing physiology. New glycoforms must not interfere with established patterns of self-recognition, innate immunity, or the synthesis and degradation of other metabolites (60). Many molecules may not be useful as markers of self because their accumulation causes disease (42). Thus, there are mutational constraints: many feasible glycoforms do not exist because no existing glycan-synthesizing enzyme can craft them. There are also selective constraints: glycans must be built, used, and degraded, and these different processes may conflict with one another.
The iconic example of self-recognition is a “green beard.” These genes act selfishly by helping other individuals only if they carry an identical allele (64). Green beard genes encode molecules that can (a) advertize their selfish allele, (b) recognize that allele in other individuals, and (c) send help specifically to these genetic relatives. Remarkably, these can all be properties of single molecules. In yeast, the Flo1 glycoprotein directs cooperation. Individuals with identical Flo1 molecules flocculate into groups that better survive environmental stress (65). Flocculation proteins are heavily N- and O- glycosylated, and glycan-glycan interaction plays a key role in the molecular handshake that identifies self (66). Self-recognition enables cooperation between cells, but this benefit carries a risk: signals of self are vulnerable to exploitation by pathogens and social partners. Self-recognition molecules are thus subject to interesting mutational and selective constraints. Rivals may engage in molecular jujutsu, evolving forms that can exploit others without themselves being exploited. The space of viable self-identity molecules is also limited to those that cannot be mimicked by pathogens. Some viruses have the ability to conceal themselves in host glycans (19–21). If disrupting cellular patterns of glycosylation harms the virus, hosts may be forced to balance the costs of disrupted glycosylation against the benefits of protection from pathogens (67). We elaborate on these and other selective constraints in the next section.
Selective Constraints on Evolution
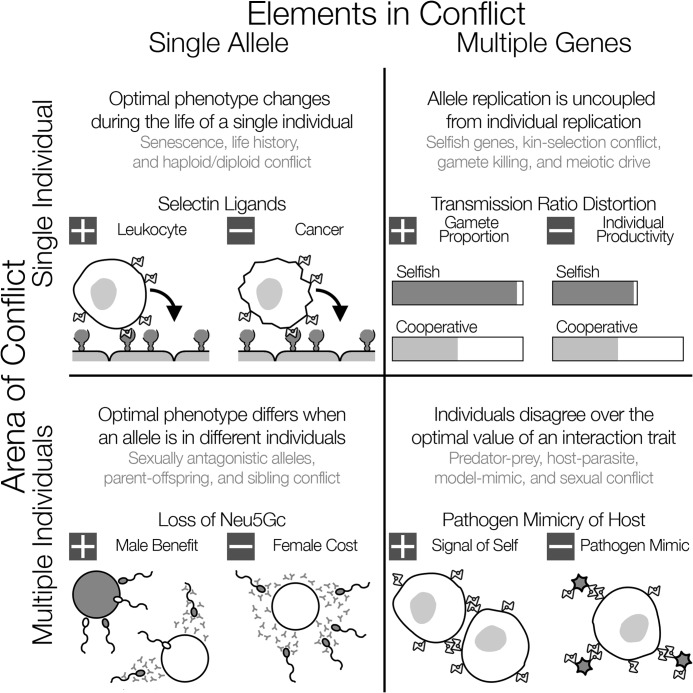
Selective constraint occurs when opposing selective forces influence the course of evolution (68–70). To recognize adaptations to selective constraint, we need a way to organize evolutionary conflicts so that we can identify responses to each type of conflict. We adopt a classification based on the elements that come into evolutionary conflict (the phenotypes of single alleles or multiple genes) and the arena in which conflict plays out (how these phenotypes interact in single or multiple individuals) (Fig. 1) (2).
FIGURE 1.
Evolutionary conflicts between alleles and individuals. For single allele-single individual, single alleles conflict with themselves when their positive effects in one context cause negative effects in another. Selectins on epithelial cells bind glycans on leukocytes and guide them to sites of inflammation; this can also be exploited by cancer cells (72). Regulatory or functional changes that separate conflicting tasks are expected to evolve in response. For single allele-multiple individuals, conflicts can extend across individuals that share an allele. Females that lack Neu5Gc raise antibodies against it. Males that lack Neu5Gc have higher rates of fertilization, and females have lower rates (14). Individual-specific regulation could resolve these conflicts. For multiple genes-single individual, selfish alleles can bias reproduction in their favor at the cost of individual reproduction, causing conflict with other genes in the genome. Mutant PMM2 alleles are passed to children more often than expected but increase the risk of congenital disorders of glycosylation (79). Other genes are selected to suppress the selfish allele, often by modification of chromosomal recombination and linkage. For multiple genes-multiple individuals, molecular markers of self cause cells to direct benefits toward identical genetic relatives, but they can be exploited by pathogen mimics. Co-evolution is a common outcome, as hosts develop more reliable markers of self, and pathogens develop more effective molecular mimics (8, 86).
For glycans, we must consider the term “allele” carefully. Glycan synthesis is not template-driven, so there is not always a one-to-one equivalence between an allele encoded in DNA and the glycan phenotype. The section on mutational constraints describes the effects that this mechanism of synthesis has on the evolution of glycan phenotypes. In the next section, we consider an allele to be a unique glycan variant, with more detail given if the mechanism of synthesis or regulation matters implicitly to the form or resolution of a selective constraint. We use glycan examples to explain evolutionary conflicts in each of four categories of selective constraint (Fig. 1). Our aim is to describe the properties of traits that could overcome each kind of selective constraint. We hope to prepare molecular biologists to recognize these properties in their study systems and to seek their evolutionary origins. We also hope to convince evolutionary biologists that glycans offer rich opportunities for examining mechanisms of phenotypic evolution.
Single Allele-Single Individual
Like most traits, glycans are pleiotropic, meaning that a single trait has effects on more than one part of an organism. Because of pleiotropy, an allele can conflict with itself if there is no version that functions perfectly in all of the contexts that it will encounter during an individual's lifetime. Alleles with positive effects on one task can cause negative effects on another. If so, we expect selection to favor mechanisms that separate the two selective contexts, allowing alleles to function differently where they have positive and negative effects on fitness. Changes to expression allow alleles to act only in specific contexts, and mechanisms of spatial or temporal regulation likely evolved in response to single allele-single individual conflicts (2, 71). For some of these conflicts, the positive and negative effects are inextricable; they simply cannot be resolved, and we recognize their accumulating consequences as symptoms of aging (70).
Glycan modifications can induce context-specific regulation or function in the products of single alleles. Glycosylation of the human chorionic gonadotropin protein results in five functionally unique forms that interact with at least two different receptors (26). The number and branching of N-glycans on mammalian growth receptors depend on hexose availability, allowing these glycoproteins to regulate cell growth in response to metabolic state (25). Stage-specific embryonic antigens are glycoproteins that guide development by tissue-specific expression (27). Glycans can themselves cause conflict when their function in one context negatively impacts another. Selectins are proteins that recognize sialylated and fucosylated glycans on leukocytes and arrest their movement through the bloodstream at sites of inflammation. This system of localization can be co-opted by cancers that express selectin-binding glycans that promote metastasis (Fig. 1) (72).
Single Allele-Multiple Individuals
Selective constraints also occur when the effect of an allele depends on which individual is carrying it (69). The allele that eliminated Neu5Gc in humans may have reached fixation because of its positive effect on males, despite its negative consequences for female fertility (14). Females that lack Neu5Gc raise antibodies against it and are partly incompatible with Neu5Gc-positive males. Males that lack Neu5Gc are compatible with all females in the population (Fig. 1). The pleiotropic effects of a single allele can thus extend across individuals. We expect selection to favor mechanisms that separate these conflicting roles. Glycans can allow proteins to take individual-specific functions. Glycodelin glycoforms function differently in male and female reproductive tracts. Glycodelin-A, -C, and -F are produced only by females and influence sperm-egg binding and the susceptibility of sperm to acrosome reaction. The glycodelin-S glycoform is made solely by males in seminal fluid and protects against premature sperm capacitation (73).
In mammals, placentation and lactation are prolonged intimate interactions between mother and child. Fetal resource uptake depends on the degree to which the mother accepts the trophoblast, and fetuses are capable of modifying the invasiveness and tolerance of trophoblasts to solicit more resources than the mother would optimally give (69). Trophoblast-specific glycosylation of membrane cofactor protein (CD46) reduces complement deposition, reducing the rate of maternal rejection (74). Trophoblast-specific N-glycosylation of α5β1 integrin promotes interaction between fetal extravillous trophoblast cells and the maternal extracellular matrix, increasing trophoblast invasiveness (75). Milk is a finely tuned secretion rich in glycoconjugates and free oligosaccharides (76). Milk glycans, especially those rich in sialic acid, contribute to infant brain growth (77). It is possible that mothers use milk glycans to manipulate infant behavior or development in ways that influence resource demand. Glycans allow fine-tuned individual-specific regulation, and so we expect that glycan composition and expression may be a major means by which parents and offspring negotiate conflicts over resource allocation.
Multiple Genes-Single Individual
Evolutionary conflicts also arise between different genes. Multicellular organisms are collaborations; their stability depends on fair allocation of reproductive effort to all cells. Selfish genes cheat and unfairly increase their own rate of replication at the cost of decreased individual replication (Fig. 1). Altruistic genes like green beards help other individuals with an identical allele at the cost of their own individual's survival (64). Transposons create additional copies of themselves in the genome, uncoupling their replication from that of other genes (78). Transmission ratio distorters over-represent themselves in the pool of gametes or offspring produced by an individual. Mutations of phosphomannomutase (PMM2) disrupt serum protein N-glycosylation. When homozygous, PMM2 mutations are lethal, but heterozygotes persist in populations at moderate frequency. Carriers transmit mutant PMM2 alleles to their offspring more often than is expected by chance, suggesting that the costs of this disease for individuals are offset by a transmission ratio that favors mutant alleles (79).
Masking the phenotypes of individual proteins with glycans may be an effective way to suppress selfish genes. The glycophenotype of a sperm is a “parliament of glycans,” the product of many proteins expressed in diploid cells. The sperm glycocalyx is not produced by the sperm itself; instead, sperm incorporate glycoproteins secreted by epididymal epithelial cells (80, 81). We expect that this kind of masking could counteract the effects of selfish alleles, whose expression in the haploid state could allow them to eliminate gametes that do not carry the selfish allele. Glycan masks thus function like the cytoplasmic bridges that connect developing sperm cells: they impart on all sperm an identical diploid phenotype (82).
Multiple Genes-Multiple Individuals
Many of the conflicts outlined above also occur when different genes act on behalf of different individuals. The battle of the sexes is waged on the molecular front: males and females manipulate fertilization using sperm- and egg-specific glycoproteins. The bindin protein of sea urchin sperm does not belong to any known family of lectins but does bind sulfated O-linked oligosaccharides on one of its egg receptors (83). Human sperm bind the oligosaccharide sialyl-LewisX on the egg zona pellucida (84). Parent-offspring conflicts are also fought between proteins expressed exclusively by mother or fetus. In humans, maternally expressed glycodelin-A suppresses the invasion of fetal cells by binding Siglec-6, which is expressed only on trophoblasts (85). These contests often result in co-evolution of proteins representing each party (86).
Interestingly, proteins that act on one side of a conflict can be co-opted into other conflicts that require similar features. CD46, a trophoblast glycoprotein that influences maternal tolerance, is also found on the exposed surface of sperm following acrosome reaction, although its function there is not yet known (87). Placental HLA-G glycoforms are key to avoiding rejection by the mother (88, 89), and HLA-G variants also help melanoma cells evade immune detection (90). Co-opted genes sometimes come from unlikely partners. Mammals with invasive implantation fuse the outer placental cells into a syncytium to increase contact with the mother and suppress her immune response. The glycoprotein responsible came from a retrovirus and has been domesticated independently by several mammal lineages (91).
The most recognizable evolutionary conflicts occur between hosts and parasites. Vertebrate innate immunity detects pathogen-associated molecular patterns, many of which are glycans. Innate immune cells target non-self glycans, such as high mannose glycans or β-glucans, using a suite of lectins (92). Some pathogen glycans are required for cellular function (62); others exist only to mimic or exploit the host (93, 94). Exploitation of host signaling may explain the existence of paired receptors, which are often lectins with similar sequences and specificity but opposite cellular responses on binding (95). Paired receptors often undergo rapid sequence evolution, duplication, pseudogenization, and homogenization by gene conversion (96).
When hosts lose a glycan, a self-identifying molecule becomes foreign. This can protect from viruses that camouflage themselves in host glycans (19–21). Loss of a glycan or receptor can also be adaptive if it eliminates a point of pathogen entry. Resurrected versions of human siglec pseudogenes bind sialylated bacterial pathogens (40). Host-parasite conflicts push the evolution of glycan signaling and recognition. These conflicts maintain glycan polymorphisms in populations (34, 38) and drive rapid differences between species (96). Conflict can also cause broad-scale diversification via the loss of glycoforms (13, 14, 40) and the evolution of new glycan innovations (1, 15).
Glycans as Evolved Mechanisms of Overcoming Evolutionary Constraint
Biological molecules are uniquely suited to their roles. DNA is an effective information storage device. Proteins have tremendous catalytic flexibility, and lipids self-assemble into membranes. Glycans carpet the outer surface of all cells and make up large parts of their extracellular matrices. Glycans have extraordinary structural diversity, biochemical specificity, and regulatory flexibility, properties that suit their role as negotiators of cellular interaction.
In this minireview, we have explored the idea that glycans evolved their biochemical flexibility to overcome the restrictions that mutation and selection place on the evolution of new glycoforms. We hope that this view will help biologists gain a deeper appreciation of glycobiology. For molecular biologists, it stresses the need to gather data that can be interpreted in light of evolution. We need information on glycan variation: polymorphisms within populations, differences between species, and broad patterns of diversity. We also need to integrate the selective contexts that shape glycan evolution: examining pleiotropic effects of glycans within and between individuals. For evolutionary biologists, glycans are an opportunity to test models of behavioral evolution on mechanistically tractable phenotypes (2). Behaviors caused by glycans result from simple molecular interactions, and there are many interesting examples that have received no evolutionary explanation. These are exciting prospects, and we watch the emerging synthesis of glycobiology and evolutionary biology with anticipation.
Acknowledgments
We thank Bernard Crespi and Ajit Varki for helpful comments on the manuscript.
Footnotes
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM095882. This work was also supported by a grant from the G. Harold and Leila Y. Mathers Charitable Foundation. This is the first article in the Thematic Minireview Series on Glycobiology and Extracellular Matrices: Glycan Functions Pervade Biology at All Levels.
REFERENCES
- 1. Gagneux P., Varki A. (1999) Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology 9, 747–755 [DOI] [PubMed] [Google Scholar]
- 2. Springer S. A., Crespi B. J., Swanson W. J. (2011) Beyond the phenotypic gambit: molecular behavioural ecology and the evolution of genetic architecture. Mol. Ecol. 20, 2240–2257 [DOI] [PubMed] [Google Scholar]
- 3. Varki A., Lowe J. B. (2009) in Essentials of Glycobiology (Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E., eds) 2nd Ed., pp. 75–88, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 4. Marth J. D. (2008) A unified vision of the building blocks of life. Nat. Cell Biol. 10, 1015–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Apweiler R., Hermjakob H., Sharon N. (1999) On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473, 4–8 [DOI] [PubMed] [Google Scholar]
- 6. Varki A.(2011) Evolutionary forces shaping the Golgi glycosylation machinery: why cell surface glycans are universal to living cells. Cold Spring Harb. Perspect. Biol. 3, a005462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Somerville C. (2006) Cellulose synthesis in higher plants. Annu. Rev. Cell Dev. Biol. 22, 53–78 [DOI] [PubMed] [Google Scholar]
- 8. Bishop J. R., Gagneux P. (2007) Evolution of carbohydrate antigens–microbial forces shaping host glycomes? Glycobiology 17, 23R–34R [DOI] [PubMed] [Google Scholar]
- 9. Wagner A. (2011) The molecular origins of evolutionary innovations. Trends Genet. 27, 397–410 [DOI] [PubMed] [Google Scholar]
- 10. Shoval O., Sheftel H., Shinar G., Hart Y., Ramote O., Mayo A., Dekel E., Kavanagh K., Alon U. (2012) Evolutionary trade-offs, Pareto optimality, and the geometry of phenotype space. Science 336, 1157–1160 [DOI] [PubMed] [Google Scholar]
- 11. Sletten E. M., Bertozzi C. R. (2009) Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew. Chem. Int. Ed. Engl. 48, 6974–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davies L. R. L., Pearce O. M. T., Tessier M. B., Assar S., Smutova V., Pajunen M., Sumida M., Sato C., Kitajima K., Finne J., Gagneux P., Pshezhetsky A., Woods R., Varki A. (2012) Metabolism of vertebrate amino sugars with N-glycolyl groups. Resistance of α2–8-linked N-glycolylneuraminic acid to enzymatic cleavage. J. Biol. Chem. 287, 28917–28931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koike C., Uddin M., Wildman D. E., Gray E. A., Trucco M., Starzl T. E., Goodman M. (2007) Functionally important glycosyltransferase gain and loss during catarrhine primate emergence. Proc. Natl. Acad. Sci. U.S.A. 104, 559–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ghaderi D., Springer S. A., Ma F., Cohen M., Secrest P., Taylor R. E., Varki A., Gagneux P. (2011) Sexual selection by female immunity against paternal antigens can fix loss of function alleles. Proc. Natl. Acad. Sci. U.S.A. 108, 17743–17748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Varki A. (2010) Colloquium paper: uniquely human evolution of sialic acid genetics and biology. Proc. Natl. Acad. Sci. U.S.A. 107, 8939–8946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schwarzkopf M., Knobeloch K.-P., Rohde E., Hinderlich S., Wiechens N., Lucka L., Horak I., Reutter W., Horstkorte R. (2002) Sialylation is essential for early development in mice. Proc. Natl. Acad. Sci. U.S.A. 99, 5267–5270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smith D. J., Lapedes A. S., de Jong J. C., Bestebroer T. M., Rimmelzwaan G. F., Osterhaus A. D. M. E., Fouchier R. A. M. (2004) Mapping the antigenic and genetic evolution of influenza virus. Science 305, 371–376 [DOI] [PubMed] [Google Scholar]
- 18. Price D. A., Goulder P. J., Klenerman P., Sewell A. K., Easterbrook P. J., Troop M., Bangham C. R., Phillips R. E. (1997) Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. U.S.A. 94, 1890–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vigerust D. J., Shepherd V. L. (2007) Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 15, 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Burton D. R., Poignard P., Stanfield R. L., Wilson I. A. (2012) Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science 337, 183–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rogers K. M., Heise M. (2009) Modulation of cellular tropism and innate antiviral response by viral glycans. J. Innate Immun. 1, 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stanley P., Schachter H., Taniguchi N. (2009) in Essentials of Glycobiology (Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E., eds) 2nd Ed., pp. 101–114, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 23. Cummings R. D. (2009) The repertoire of glycan determinants in the human glycome. Mol. BioSyst. 5, 1087–1104 [DOI] [PubMed] [Google Scholar]
- 24. Kolter T., Proia R. L., Sandhoff K. (2002) Combinatorial ganglioside biosynthesis. J. Biol. Chem. 277, 25859–25862 [DOI] [PubMed] [Google Scholar]
- 25. Lau K. S., Partridge E. A., Grigorian A., Silvescu C. I., Reinhold V. N., Demetriou M., Dennis J. W. (2007) Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell 129, 123–134 [DOI] [PubMed] [Google Scholar]
- 26. Cole L. A. (2012) hCG, the wonder of today's science. Reprod. Biol. Endocrinol. 10, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lanctot P. M., Gage F. H., Varki A. P. (2007) The glycans of stem cells. Curr. Opin. Chem. Biol. 11, 373–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ding N., Nie H., Sun X., Sun W., Qu Y., Liu X., Yao Y., Liang X., Chen C. C., Li Y. (2011) Human serum N-glycan profiles are age- and sex-dependent. Age Ageing 40, 568–575 [DOI] [PubMed] [Google Scholar]
- 29. Lityńska A., Przybyło M. (1998) Does glycosylation of lysosomal proteins show age-related changes in rat liver? Mech. Ageing Dev. 102, 33–43 [DOI] [PubMed] [Google Scholar]
- 30. Morris H. R., Dell A., Easton R. L., Panico M., Koistinen H., Koistinen R., Oehninger S., Patankar M. S., Seppala M., Clark G. F. (1996) Gender-specific glycosylation of human glycodelin affects its contraceptive activity. J. Biol. Chem. 271, 32159–32167 [DOI] [PubMed] [Google Scholar]
- 31. Wuhrer M., Koeleman C. A. M., Fitzpatrick J. M., Hoffmann K. F., Deelder A. M., Hokke C. H. (2006) Gender-specific expression of complex-type N-glycans in schistosomes. Glycobiology 16, 991–1006 [DOI] [PubMed] [Google Scholar]
- 32. Gill D. J., Clausen H., Bard F. (2011) Location, location, location: new insights into O-GalNAc protein glycosylation. Trends Cell Biol. 21, 149–158 [DOI] [PubMed] [Google Scholar]
- 33. Javaud C., Dupuy F., Maftah A., Julien R., Petit J.-M. (2003) The fucosyltransferase gene family: an amazing summary of the underlying mechanisms of gene evolution. Genetica 118, 157–170 [PubMed] [Google Scholar]
- 34. Marionneau S., Cailleau-Thomas A., Rocher J., Le Moullac-Vaidye B., Ruvoën N., Clément M., Le Pendu J. (2001) ABH and Lewis histo-blood group antigens, a model for the meaning of oligosaccharide diversity in the face of a changing world. Biochimie 83, 565–573 [DOI] [PubMed] [Google Scholar]
- 35. Bussink A. P., Speijer D., Aerts J. M. F. G., Boot R. G. (2007) Evolution of mammalian chitinase(-like) members of family 18 glycosyl hydrolases. Genetics 177, 959–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ivarsson Y., Mackey A. J., Edalat M., Pearson W. R., Mannervik B. (2003) Identification of residues in glutathione transferase capable of driving functional diversification in evolution. A novel approach to protein redesign. J. Biol. Chem. 278, 8733–8738 [DOI] [PubMed] [Google Scholar]
- 37. He B., Wang L., Wang J., Li G., Zhang S. (2009) Positive selection of three chitinase genes of the family 18 of glycoside hydrolases in mammals. Biologia 64, 819–825 [Google Scholar]
- 38. Fumagalli M., Cagliani R., Pozzoli U., Riva S., Comi G. P., Menozzi G., Bresolin N., Sironi M. (2009) Widespread balancing selection and pathogen-driven selection at blood group antigen genes. Genome Res. 19, 199–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Springer S. A., Moy G. W., Friend D. S., Swanson W. J., Vacquier V. D. (2008) Oyster sperm bindin is a combinatorial fucose lectin with remarkable intra-species diversity. Int. J. Dev. Biol. 52, 759–768 [DOI] [PubMed] [Google Scholar]
- 40. Wang X., Mitra N., Secundino I., Banda K., Cruz P., Padler-Karavani V., Verhagen A., Reid C., Lari M., Rizzi E., Balsamo C., Corti G., De Bellis G., Longo L., NISC Comparative Sequencing Program, Beggs W., Caramelli D., Tishkoff S. A., Hayakawa T., Green E. D., Mullikin J. C., Nizet V., Bui J., Varki A. (2012) Specific inactivation of two immunomodulatory SIGLEC genes during human evolution. Proc. Natl. Acad. Sci. U.S.A. 109, 9935–9940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rini J., Esko J., Varki A. (2009) in Essentials of Glycobiology (Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E., eds) 2nd Ed., pp. 63–74, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 42. Freeze H. H., Aebi M. (2005) Altered glycan structures: the molecular basis of congenital disorders of glycosylation. Curr. Opin. Struct. Biol. 15, 490–498 [DOI] [PubMed] [Google Scholar]
- 43. Dall'Olio G. M., Laayouni H., Luisi P., Sikora M., Montanucci L., Bertranpetit J. (2012) Distribution of events of positive selection and population differentiation in a metabolic pathway: the case of asparagine N-glycosylation. BMC Evol. Biol. 12, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dennis J. W., Granovsky M., Warren C. E. (1999) Protein glycosylation in development and disease. BioEssays 21, 412–421 [DOI] [PubMed] [Google Scholar]
- 45. Schwarz F., Aebi M. (2011) Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 21, 576–582 [DOI] [PubMed] [Google Scholar]
- 46. Schauer R., Srinivasan G. V., Coddeville B., Zanetta J.-P., Guérardel Y. (2009) Low incidence of N-glycolylneuraminic acid in birds and reptiles and its absence in the platypus. Carbohydr. Res. 344, 1494–1500 [DOI] [PubMed] [Google Scholar]
- 47. Yamada S., Sugahara K., Ozbek S. (2011) Evolution of glycosaminoglycans: comparative biochemical study. Commun. Integr. Biol. 4, 150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Calafell F., Roubinet F., Ramírez-Soriano A., Saitou N., Bertranpetit J., Blancher A. (2008) Evolutionary dynamics of the human ABO gene. Hum. Genet. 124, 123–135 [DOI] [PubMed] [Google Scholar]
- 49. O'hUigin C., Sato A., Klein J. (1997) Evidence for convergent evolution of A and B blood group antigens in primates. Hum. Genet. 101, 141–148 [DOI] [PubMed] [Google Scholar]
- 50. Suzuki N., Laskowski M., Jr., Lee Y. C. (2004) Phylogenetic expression of Galα1–4Gal on avian glycoproteins: glycan differentiation inscribed in the early history of modern birds. Proc. Natl. Acad. Sci. U.S.A. 101, 9023–9028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ma F., Wu D., Deng L., Secrest P., Zhao J., Varki N., Lindheim S., Gagneux P. (2012) Sialidases on mammalian sperm mediate deciduous sialylation during capacitation. J. Biol. Chem. 287, 38073–38079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kelm S., Schauer R. (1997) Sialic acids in molecular and cellular interactions. Int. Rev. Cytol. 175, 137–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McMahon S. A., Miller J. L., Lawton J. A., Kerkow D. E., Hodes A., Marti-Renom M. A., Doulatov S., Narayanan E., Sali A., Miller J. F., Ghosh P. (2005) The C-type lectin fold as an evolutionary solution for massive sequence variation. Nat. Struct. Mol. Biol. 12, 886–892 [DOI] [PubMed] [Google Scholar]
- 54. Iobst S. T., Drickamer K. (1994) Binding of sugar ligands to Ca2+-dependent animal lectins. II. Generation of high-affinity galactose binding by site-directed mutagenesis. J. Biol. Chem. 269, 15512–15519 [PubMed] [Google Scholar]
- 55. Yim M., Ono T., Irimura T. (2001) Mutated plant lectin library useful to identify different cells. Proc. Natl. Acad. Sci. U.S.A. 98, 2222–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ogawa T., Chijiwa T., Oda-Ueda N., Ohno M. (2005) Molecular diversity and accelerated evolution of C-type lectin-like proteins from snake venom. Toxicon 45, 1–14 [DOI] [PubMed] [Google Scholar]
- 57. Springer S. A., Crespi B. J. (2007) Adaptive gamete-recognition divergence in a hybridizing Mytilus population. Evolution 61, 772–783 [DOI] [PubMed] [Google Scholar]
- 58. Collins B. E., Paulson J. C. (2004) Cell surface biology mediated by low affinity multivalent protein-glycan interactions. Curr. Opin. Chem. Biol. 8, 617–625 [DOI] [PubMed] [Google Scholar]
- 59. Cohen M., Varki A. (2010) The sialome–far more than the sum of its parts. OMICS 14, 455–464 [DOI] [PubMed] [Google Scholar]
- 60. Freeze H. H. (2006) Genetic defects in the human glycome. Nat. Rev. Genet. 7, 537–551 [DOI] [PubMed] [Google Scholar]
- 61. Varki A. (2011) Since there are PAMPs and DAMPs, there must be SAMPs? Glycan “self-associated molecular patterns” dampen innate immunity, but pathogens can mimic them. Glycobiology 21, 1121–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Adibekian A., Stallforth P., Hecht M. L., Werz D. B., Gagneux P., Seeberger P. H. (2011) Comparative bioinformatics analysis of the mammalian and bacterial glycomes. Chem. Sci. 2, 337–344 [Google Scholar]
- 63. Patnaik S. K., Stanley P. (2006) Lectin-resistant CHO glycosylation mutants. Methods Enzymol. 416, 159–182 [DOI] [PubMed] [Google Scholar]
- 64. Hamilton W. D. (1964) The genetical evolution of social behaviour. J. Theor. Biol. 7, 1–16 [DOI] [PubMed] [Google Scholar]
- 65. Smukalla S., Caldara M., Pochet N., Beauvais A., Guadagnini S., Yan C., Vinces M. D., Jansen A., Prevost M. C., Latgé J.-P., Fink G. R., Foster K. R., Verstrepen K. J. (2008) FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell 135, 726–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Goossens K., Willaert R. (2010) Flocculation protein structure and cell-cell adhesion mechanism in Saccharomyces cerevisiae. Biotechnol. Lett. 32, 1571–1585 [DOI] [PubMed] [Google Scholar]
- 67. Freeze H. H., Westphal V. (2001) Balancing N-linked glycosylation to avoid disease. Biochimie 83, 791–799 [DOI] [PubMed] [Google Scholar]
- 68. Dawkins R., Krebs J. R. (1979) Arms races between and within species. Proc. R. Soc. Lond. B Biol. Sci. 205, 489–511 [DOI] [PubMed] [Google Scholar]
- 69. Trivers R. L. (1974) Parent-offspring conflict. Amer. Zool. 14, 249–264 [Google Scholar]
- 70. Williams G. C. (1957) Pleiotropy, natural selection, and the evolution of senescence. Evolution 11, 398–411 [Google Scholar]
- 71. Carroll S. B. (2005) Evolution at two levels: on genes and form. PLoS Biol. 3, e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Läubli H., Borsig L. (2010) Selectins promote tumor metastasis. Semin. Cancer Biol. 20, 169–177 [DOI] [PubMed] [Google Scholar]
- 73. Yeung W. S. B., Lee K.-F., Koistinen R., Koistinen H., Seppälä M., Chiu P. C. N. (2009) Effects of glycodelins on functional competence of spermatozoa. J. Reprod. Immunol. 83, 26–30 [DOI] [PubMed] [Google Scholar]
- 74. Vanderpuye O. A., Labarrere C. A., McIntyre J. A. (1992) Glycosylation of membrane cofactor protein (CD46) in human trophoblast, kidney and platelets. Biochim. Biophys. Acta 1121, 301–308 [DOI] [PubMed] [Google Scholar]
- 75. Yamamoto E., Ino K., Miyoshi E., Inamori K., Abe A., Sumigama S., Iwase A., Kajiyama H., Shibata K., Nawa A., Kikkawa F. (2009) N-Acetylglucosaminyltransferase V regulates extravillous trophoblast invasion through glycosylation of α5β1 integrin. Endocrinology 150, 990–999 [DOI] [PubMed] [Google Scholar]
- 76. Lefèvre C. M., Sharp J. A., Nicholas K. R. (2010) Evolution of lactation: ancient origin and extreme adaptations of the lactation system. Annu. Rev. Genomics Hum. Genet. 11, 219–238 [DOI] [PubMed] [Google Scholar]
- 77. Wang B. (2009) Sialic acid is an essential nutrient for brain development and cognition. Annu. Rev. Nutr. 29, 177–222 [DOI] [PubMed] [Google Scholar]
- 78. Feschotte C., Pritham E. J. (2007) DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41, 331–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Schollen E., Kjaergaard S., Martinsson T., Vuillaumier-Barrot S., Dunoe M., Keldermans L., Seta N., Matthijs G. (2004) Increased recurrence risk in congenital disorders of glycosylation type Ia (CDG-Ia) due to a transmission ratio distortion. J. Med. Genet. 41, 877–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Schröter S., Osterhoff C., McArdle W., Ivell R. (1999) The glycocalyx of the sperm surface. Hum. Reprod. Update 5, 302–313 [DOI] [PubMed] [Google Scholar]
- 81. Kirchhoff C., Schröter S. (2001) New insights into the origin, structure and role of CD52: a major component of the mammalian sperm glycocalyx. Cells Tissues Organs 168, 93–104 [DOI] [PubMed] [Google Scholar]
- 82. Braun R. E., Behringer R. R., Peschon J. J., Brinster R. L., Palmiter R. D. (1989) Genetically haploid spermatids are phenotypically diploid. Nature 337, 373–376 [DOI] [PubMed] [Google Scholar]
- 83. Vacquier V. D. (2012) The quest for the sea urchin egg receptor for sperm. Biochem. Biophys. Res. Commun. 425, 583–587 [DOI] [PubMed] [Google Scholar]
- 84. Pang P.-C., Chiu P. C. N., Lee C.-L., Chang L.-Y., Panico M., Morris H. R., Haslam S. M., Khoo K.-H., Clark G. F., Yeung W. S. B., Dell A. (2011) Human sperm binding is mediated by the sialyl-LewisX oligosaccharide on the zona pellucida. Science 333, 1761–1764 [DOI] [PubMed] [Google Scholar]
- 85. Lam K. K. W., Chiu P. C. N., Lee C.-L., Pang R. T. K., Leung C. O. N., Koistinen H., Seppälä M., Ho P.-C., Yeung W. S. B. (2011) Glycodelin-A protein interacts with Siglec-6 protein to suppress trophoblast invasiveness by down-regulating extracellular signal-regulated kinase (ERK)/c-Jun signaling pathway. J. Biol. Chem. 286, 37118–37127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Clark N. L., Gasper J., Sekino M., Springer S. A., Aquadro C. F., Swanson W. J. (2009) Coevolution of interacting fertilization proteins. PLoS Genet. 5, e1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Cummerson J. A., Flanagan B. F., Spiller D. G., Johnson P. M. (2006) The complement regulatory proteins CD55 (decay accelerating factor) and CD59 are expressed on the inner acrosomal membrane of human spermatozoa as well as CD46 (membrane cofactor protein). Immunology 118, 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. McMaster M., Zhou Y., Shorter S., Kapasi K., Geraghty D., Lim K. H., Fisher S. (1998) HLA-G isoforms produced by placental cytotrophoblasts and found in amniotic fluid are due to unusual glycosylation. J. Immunol. 160, 5922–5928 [PubMed] [Google Scholar]
- 89. Hunt J. S., Petroff M. G., McIntire R. H., Ober C. (2005) HLA-G and immune tolerance in pregnancy. FASEB J. 19, 681–693 [DOI] [PubMed] [Google Scholar]
- 90. Paul P., Rouas-Freiss N., Khalil-Daher I., Moreau P., Riteau B., Le Gal F. A., Avril M. F., Dausset J., Guillet J. G., Carosella E. D. (1998) HLA-G expression in melanoma: a way for tumor cells to escape from immunosurveillance. Proc. Natl. Acad. Sci. U.S.A. 95, 4510–4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Dupressoir A., Lavialle C., Heidmann T. (2012) From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta 33, 663–671 [DOI] [PubMed] [Google Scholar]
- 92. van Kooyk Y., Rabinovich G. A. (2008) Protein-glycan interactions in the control of innate and adaptive immune responses. Nat. Immunol. 9, 593–601 [DOI] [PubMed] [Google Scholar]
- 93. Lewis A. L., Desa N., Hansen E. E., Knirel Y. A., Gordon J. I., Gagneux P., Nizet V., Varki A. (2009) Innovations in host and microbial sialic acid biosynthesis revealed by phylogenomic prediction of nonulosonic acid structure. Proc. Natl. Acad. Sci. U.S.A. 106, 13552–13557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. van Die I., Cummings R. D. (2010) Glycan gimmickry by parasitic helminths: a strategy for modulating the host immune response? Glycobiology 20, 2–12 [DOI] [PubMed] [Google Scholar]
- 95. Fossum S., Saether P. C., Vaage J. T., Daws M. R., Dissen E. (2011) Paired opposing leukocyte receptors recognizing rapidly evolving ligands are subject to homogenization of their ligand binding domains. Immunogenetics 63, 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Angata T. (2006) Molecular diversity and evolution of the Siglec family of cell-surface lectins. Mol. Divers. 10, 555–566 [DOI] [PubMed] [Google Scholar]