Abstract
Protein glycosylation is widespread throughout all three domains of life. Bacterial protein N-glycosylation and its application to engineering recombinant glycoproteins continue to be actively studied. Here, we focus on advances made in the last 2 years, including the characterization of novel bacterial N-glycosylation pathways, examination of pathway enzymes and evolution, biological roles of protein modification in the native host, and exploitation of the N-glycosylation pathways to create novel vaccines and diagnostics.
Keywords: Campylobacter, Evolution, Glycoconjugate, Glycosyltransferases, Vaccines, Glycoengineering, HMW1C, N-Linked Protein Glycosylation, PglB, Di-N-acetylbacillosamine
Introduction
Protein modification with carbohydrates is common in all domains of life and provides mechanisms for the control of various cellular processes, including signal transduction, protein folding, targeting, stability, cell-cell and virus-cell interactions, and host immune responses (1–4). First thought to be exclusively of eukaryotic origin, it is now well established that protein glycosylation also occurs in Archaea (for review, see Ref. 5) and in Bacteria (for review, see Ref. 6). Despite the fact that an increasing number of bacterial protein modification systems are being discovered (6), the best studied pathway remains the one in Campylobacter jejuni, where a general N-linked protein glycosylation (pgl) system was first described more than a decade ago (7). We now know that >60 periplasmic and membrane-bound proteins are N-glycosylated with a conserved heptasaccharide in this gastrointestinal pathogen (8).
The heptasaccharide α-d-GalNAc(1–4)α-d-GalNAc(1-4)α-d-GalNAc(β-d-Glc(1–3))(1–4)α-d-GalNAc(1–4)α-d-GalNAc(1–3)α-d-di-N-acetylbacillosamine (diNAcBac)3-β1 is assembled from nucleotide-activated sugars onto an undecaprenyl phosphate (Und-P) lipid carrier. The reducing end sugar, diNAcBac or 2,4-diamino-2,4,6-trideoxy-d-glucose (also called N-acetyl-quinovosamine), is a derivative of the amino sugar bacillosamine, which was discovered >50 years ago by Nathan Sharon in Bacillus licheniformis (9). diNAcBac is synthesized by the consecutive action of a dehydratase (PglF), an aminotransferase (PglE), and an acetyltransferase (PglD) on UDP-GlcNAc (10, 11) and transferred to Und-P by PglC (12). After stepwise extension mediated by four glycosyltransferases (GTases), PglA, PglJ, PglH, and PglI (13, 14), the full-length Und-PP-linked heptasaccharide (lipid-linked oligosaccharide (LLO)) is then flipped by the ATP-dependent flippase PglK from the cytosolic side of the bacterial inner membrane toward the periplasmic space (14, 15). The oligosaccharyltransferase (OTase) PglB is the key enzyme of the pathway and is homologous to the STT3 subunit from the eukaryotic OTase complex (16). PglB catalyzes glycan transfer from the lipid carrier to specific asparagines within flexible loops of folded acceptor proteins (17) that carry the bacterial glycosylation sequon (D/E)XNZ(S/T) (where X and Z can be any amino acid except proline) (18). PglB also releases the heptasaccharide as free oligosaccharides into the periplasmic space, and this hydrolase activity is influenced by the osmotic environment of the cell (19).
In eukaryotes, the OTase complex transfers a tetradecasaccharide unit (Glc3Man9GlcNAc2) to proteins, and these sugars are further modified by additional glycosyl hydrolases and GTases to yield a heterologous mixture of N-glycoproteins (1, 20, 21). Although the C. jejuni N-glycans are homogeneous in structure, the OTase is capable of recognizing various substrates (22–24) and is limited only by the requirement for an acetyl group at C-2 of the reducing end sugar (24, 25). The promiscuity of the C. jejuni PglB OTase coupled with the ability to function in Escherichia coli (16) in combination with enzymes from lipopolysaccharide O-antigen biosynthetic pathways and various GTases of bacterial and eukaryotic origin provides opportunities to produce defined sugar structures on select acceptor proteins (see below for details).
Bacterial N-Linked Protein Glycosylation: An Update
There have been several publications and reviews describing the classical N-glycosylation pathway in the ϵ-subdivision of Proteobacteria over the last decade (for literature selection, see Refs. 6, 26, and 27). It has been demonstrated that nearly all bacteria within this class, which includes Campylobacter, Helicobacter, and Wolinella species, possess at least one ortholog of the bacterial OTase (for review, see Ref. 6). In addition, a subset of Deltaproteobacteria, including Desulfovibrio species, also possess the N-glycan pathway (28, 29).
Periplasmic N-Glycosylation Pathways in Epsilonproteobacteria: Variations of a Common Theme
In contrast to C. jejuni, in which a single pglB gene is located within a gene cluster encoding all enzymes required for biosynthesis of the N-linked glycan, in other Campylobacter and Helicobacter species, the pgl genes are more dispersed across the genome (6, 30–32) and may contain two copies of pglB (6, 31). The composition of the N-glycans is also more variable than originally thought (Fig. 1) (6, 33, 34). Helicobacter pullorum produces a linear pentasaccharide with unknown sugar residues with masses of 216 and 217 Da (31). Wolinella succinogenes produces a hexasaccharide containing three 216-Da monosaccharides and an unusual 232-Da residue at the nonreducing end and is the only species so far that has a sugar branching off the second sugar from the reducing end (30).
FIGURE 1.
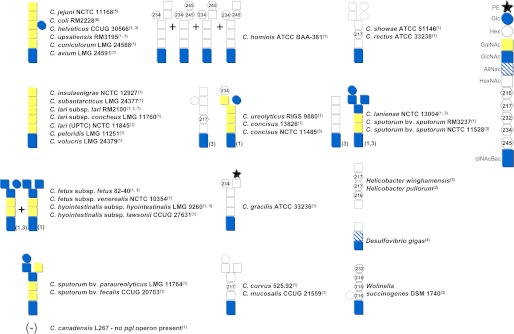
Known bacterial N-glycan structures from strains possessing orthologs of the pgl gene cluster. The numbers in parentheses following each strain designation refer to the publication in which the structure was first described: 1, Ref. 32; 2, Ref. 31; 3, Ref. 30; 4, Ref. 29; 5, Ref. 92; 6, Ref. 34; and 7, Ref. 81. The numbers next to certain structures indicate that differences in the composition of the oligosaccharides were reported. Symbols representing individual monosaccharides are explained on the right. PE, pEtN. AllNAc, N-acetylallosamine.
At least 16 different structures were identified in the Campylobacter genus (Fig. 1) (30, 32). In all species examined, the first two to three reducing end sugars (which include diNAcBac) were conserved, and all of the variation occurred at the nonreducing end. On that basis, Campylobacter species were divided into two groups (32). Group I (Cj group) includes thermotolerant strains that (except for the Campylobacter lari-like species, which lacks the glucose branch) produce the C. jejuni heptasaccharide. The non-thermotolerant Campylobacter species (Group II) express different N-glycan chain lengths from hexasaccharides to octasaccharides and, in some cases, produce from two to four structural variants. The presence of monosaccharides with unusual masses of 217 Da (as observed in H. pullorum (31), Hilicobacter winghamensis (31), Campylobacter rectus, Campylobacter showae, Campylobacter curvus, Campylobacter mucosalis (32), and Campylobacter concisus (30) and predicted to be 2-acetamido-2-deoxy-d-galacturonic acid (32)), 234 Da (identified as glucolactilic acid in C. concisus but also present in other species (32)), and 245 Da (present in Campylobacter hominis (32) and predicted to be an O-acetylated HexNAc (32)) suggests that some Campylobacter species have acquired these unique sugars independently (32).
Interestingly, the overlap in the N-glycoproteome was shown to be marginal, with the exception of the SecG protein, which is part of the general protein secretory machinery and has been demonstrated to be a target for N-glycosylation in every Campylobacter species examined (32). Although a variety of phenotypes are associated with loss of protein glycosylation (see “Biological Significance” below), it appears that this machinery has specific targets requiring N-glycosylation, and modification of other proteins may occur simply because they possess a sequon for N-glycosylation.
In addition to the variety in N-glycan structures, non-carbohydrate modifications can be found. Scott et al. (35) identified a phosphoethanolamine (pEtN) residue attached to the terminal GalNAc of the N-glycan of C. jejuni on multiple proteins. Although pEtN was not an immunodominant epitope, deletion of the gene in C. jejuni JHH1 resulted in reduced motility and increased sensitivity toward polymyxin B (35). Interestingly, the glycans of Campylobacter gracilis were found to be nearly 100% pEtN-modified on the free oligosaccharides and ∼50% modified on the glycoprotein, with non-modified forms being present on the same acceptor peptide (32). pEtN modifications are frequently present on lipid A and lipopolysaccharide core structures and also have been recently identified to modify the C. jejuni flagellar hook protein FlgG (36).
However, the question of why this modification is added to the free oligosaccharides and N-glycans of only two species when all Campylobacter species possess a pEtN transferase homolog remains to be answered. Interestingly, Neisseria species that have a general pathway for O-glycosylation further modify their O-glycosylated pili and at least two other glycoproteins with pEtN, phosphocholine, and phosphoglycerol, but these modifications are added to unoccupied Ser/Thr residues on the protein and not directly to the sugars (37, 38).
Biological Significance
Disruption of the C. jejuni N-glycosylation pathway results in pleiotropic phenotypes, including loss of protein recognition by both human (39) and rabbit (7, 40) immune sera (Fig. 2). The observation that multiple proteins lose antibody reactivity when the pgl genes are disrupted led to the initial discovery of this general glycosylation pathway (7). Subsequently, it was demonstrated by van Sorge et al. (41) that the C. jejuni N-glycan can also be recognized by the innate immune system through the C-type lectin known as MGL (macrophage galactose lectin receptor with specificity for N-acetylgalactosamine) on dendritic cells. Mutation of the pgl genes also influences bacterial adherence and invasion of cultured intestinal cells in vitro as well as mouse and chicken colonization in vivo (40, 42, 43). Of the >60 proteins that have been annotated and demonstrated to be modified with N-glycans in C. jejuni (8), the majority are part of protein complexes. Despite the fact that several studies have demonstrated the importance of N-glycosylation in C. jejuni pathogenesis, the general role of the N-glycans is still unknown. Site-directed mutagenesis within the N-glycosylation sequon has been used to study proteins such as Cj1496c, ZnuA, and VirB10 (44–46). Removal of the N-linked glycans from Cj1496c, a protein involved in epithelial cell invasion and chicken colonization, and ZnuA, which is important for survival in low-zinc environments, showed no significant influence on the function of these proteins (42, 45). Similarly, the N-glycan on the surface-associated lipoprotein JlpA, an adhesin required for chicken colonization, was not required for protein antigenicity (47, 48), and N-glycosylation of two mechanosensitive channel proteins had no major effect on their function and stability (49). However, N-glycosylation at one site of VirB10, a periplasmic component of the type IV secretion system, was shown to be important for DNA uptake and stability of the complex (44). This was the first study to suggest that N-glycans are potentially involved in the proper assembly and function of protein complexes in C. jejuni.
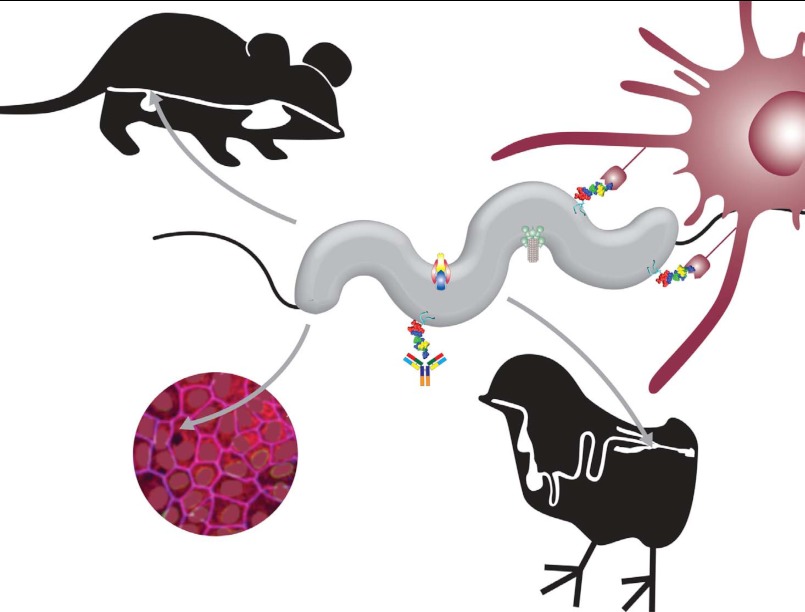
FIGURE 2.
Importance of N-glycosylation for C. jejuni. Disruption of the bacterial N-glycosylation pathway leads to reduced intestinal epithelial cell adherence and invasion in vitro and reduced colonization of mice and chickens in vivo. The N-glycans at the nonreducing end are also recognized by both adaptive (i.e. antibodies) and innate (i.e. MGL receptors on dendritic cells) immune mechanisms. Although removal of the N-glycan does not influence the biological activity of several proteins, the N-glycans are important for the function of the type IV secretion system and for the CmeABC multidrug efflux pump. Graphics were done by Thuan Nguyen.
CmeABC is the major efflux pump in C. jejuni responsible for conferring resistance to a broad range of antibiotics and bile salts and is also required for colonization of chickens in vivo (50). The three components of the CmeABC multidrug efflux pump in C. jejuni are also N-glycosylated. Mutagenesis of the Asn123 and Asn273 sites of the periplasmic component of this efflux pump, CmeA, demonstrated similar patterns of antimicrobial sensitivity and reduced chicken colonization levels as a cmeA mutant.4 Because CmeA protein levels were not altered in the N-glycosylation mutant, studies are under way to determine whether N-glycosylation influences pump assembly.
Evolutionary Perspective
Because all Campylobacter species but one possess a pgl gene cluster with counterparts showing high levels of similarity and synteny (32), this suggests that the N-glycan pathway was likely present in the common ancestor of the Campylobacter genus prior to the division into species. Thus, it is not surprising that the first two to three reducing end sugars are conserved in all species capable of protein glycosylation (Fig. 1). This is in contrast to the diverse N-glycan structures at the nonreducing end, which are subjected to pressures to evolve due to immune recognition. This is emphasized by the observation that C. jejuni N-glycan-specific antibodies recognize only the full-length heptasaccharide and do not react against pathway mutants devoid of the nonreducing end sugars (19, 51). We have also shown that C. jejuni N-glycan antibodies do not react against N-glycans produced by non-thermotolerant Campylobacter species, and conversely, antibodies raised against the N-glycan of Campylobacter fetus fetus react only with N-glycans of species with the same terminal glycans (Fig. 1), although the three reducing end sugars are identical between these species (32).
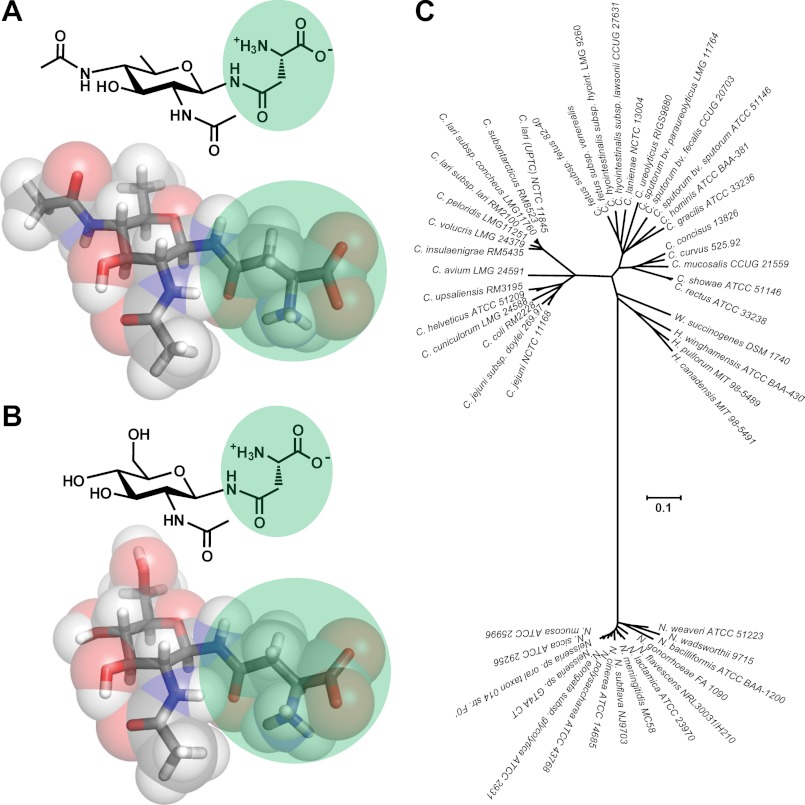
Of the 29 Campylobacter species capable of N-glycosylating their proteins, all form the N-linkage to asparagine through the unusual diNAcBac sugar (Figs. 1 and 3) (32). In contrast, all eukaryotes form N-linkages through GlcNAc. To synthesize diNAcBac, bacteria start with UDP-GlcNAc and make the conversion using three enzymes, PglD, PglE, and PglF (as outlined above). All Neisseria species examined to date O-glycosylate their proteins. Synthesis of the oligosaccharide occurs on Und-P, followed by flipping across the membrane and addition to Ser/Thr residues of multiple target proteins by the promiscuous PglL OTase (52–54). Although this genus belongs to the Betaproteobacteria, they also predominantly use diNAcBac at the reducing end (referred to as DATDH (diacetamidotrideoxyhexose) in the literature because the absolute conformation of the hexose has not been determined), although other variants such as GATDH (glyceramidoacetamidotrideoxyhexose (55) and DADDGlc (diacetamidodideoxyglucopyranose (56)) have been detected. Thus, Neisseria species also possess homologs of the PglFED enzymes. Phylogenetic analyses of these three enzymes, which convert UDP-GlcNAc to UDP-diNAcBac from all strains known to produce N-glycans and Neisseria species containing the pgl locus, showed the formation of two distinctive clades (Fig. 3) (data not shown). These results suggest that the Campylobacter and Neisseria protein glycosylation pathways have convergently evolved to use diNAcBac. Why this unusual sugar is produced by two different bacterial protein glycosylation systems remains a mystery. It is also surprising to see that the enzymes from W. succinogenes (which uses diNAcBac) (30) cluster with those from Helicobacter species possessing pgl gene loci, including pglFED, but use HexNAc as the reducing end sugar (31). Desulfovibrio species were excluded from these analyses because they do not possess obvious PglFED orthologs. This is supported by x-ray crystallography data demonstrating that the reducing end sugar of Desulfovibrio gigas is GlcNAc (29).
FIGURE 3.
Use of diNAcBac for bacterial protein glycosylation. Shown is a structural comparison (space-filling models provided by Michelle Richards) of the bacterial diNAcBac residue (A) and the eukaryotic GlcNAc residue (B) attached to asparagine for N-glycosylation (highlighted in green). C, dendrogram of proteobacterial PglE (the aminotransferase involved in diNAcBac synthesis) amino acid sequences. Protein sequences were extracted from the draft or completed genome sequences for the taxa illustrated in the tree. The dendrogram was constructed using a neighbor-joining algorithm and the Poisson distance estimation method. The scale bar represents substitutions per site. Similar dendrograms were obtained for PglD (acetyltransferase) and PglE (dehydratase) (data not shown).
Gammaproteobacteria Do It, Too! The Cytoplasmic N-Glycosylation Pathway
In 2003, St Geme and co-workers (57) demonstrated that the extracellular high-molecular weight non-pilus adhesin HMW1 of the Gram-negative bacterium Haemophilus influenzae was modified with mono- or dihexoses on asparagine residues. HMW1 mediates attachment to human epithelial cells, an essential step in the establishment of H. influenzae disease (58–60). HMW1C is a unique GTase that is required for glycosylation of HMW1 in the cytoplasm, and glycosylation was shown to be essential for HMW1 stability and translocation to the bacterial surface (57). Thirty-one modification sites were identified in HMW1, and all but one of the asparagines is within the conventional consensus sequence for eukaryotic N-glycosylation, NX(S/T) (61). In a subsequent study, HMW1C was demonstrated to have substrate specificity for UDP-α-d-glucose and UDP-α-d-galactose, but with the prerequisite that glucose must be the first hexose linked to asparagine in the glycopeptides containing dihexose modifications prior to further modification by the same enzyme (62). Because HMW1C orthologs can be found in several other Gram-negative pathogens, including enterotoxigenic E. coli, Yersinia pseudotuberculosis, Yersinia enterocolitica, Yersinia pestis, Haemophilus ducreyi, Actinobacillus pleuropneumoniae, Mannheimia spp., Xanthomonas spp., and Burkholderia spp., among others (62), these proteins were suggested to constitute a new family of bacterial enzymes capable of creating N-linked carbohydrate-protein and carbohydrate-carbohydrate bonds in the cytoplasm that do not require the formation of the monosaccharide units on a lipid-linked intermediate (61).
Recently, the structure of the HMW1C ortholog of A. pleuropneumoniae that is capable of glycosylating the H. influenzae HMW1 adhesin in vitro and in vivo (63) was solved (64). Structure-guided modeling in combination with site-directed mutagenesis and glycosylation assays demonstrated that the H. influenzae HMW1C protein adopts the same structure as A. pleuropneumoniae HMW1C, with critical residues for UDP-hexose binding and residues for acceptor protein binding being conserved (64). Schwarz et al. (65) confirmed that the HMW1C ortholog from A. pleuropneumoniae is an inverting GTase that transfers a glucose or galactose moiety (with specificity for UDP-glucose) to asparagine within the eukaryotic NX(S/T) consensus sequence, but the enzyme did not further elongate the N-linked monosaccharide. An additional, glucose-specific, polymerizing glucosyltransferase elongates the N-linked glucose, adding two glucose residues (and up to six residues in excess of donor) to generate Glcα(1–6)Glcα(1–6)Glc (65). Despite lacking the need for Asp/Glu at the −2 position of the sequon, HMW1C proteins could not glucosylate short peptides that work for C. jejuni PglB (22), indicating that binding of the substrate to the enzyme might require an extended contact surface (65). A proline in the +1 position of the acceptor peptide impaired glycosylation (like C. jejuni PglB). HMW1C does not require metal ions (unlike C. jejuni PglB) but operates on folded proteins (like C. jejuni PglB). Because HMW1C does not share any sequence identity with PglB and because of the fact that the two enzymes belong to two structurally unrelated GTase families, these bacterial N-glycosylation systems seem to have evolved independently in two genera within two different bacterial classes and in different compartments of the cell.
Bacterial Glycoengineering: A New Discipline Is on the Rise
Conjugate vaccines, which are composed of polysaccharide antigens covalently linked to carrier proteins, have been demonstrated to be the most effective and safest vaccines against bacterial pathogens. In contrast to isolated bacterial polysaccharides, conjugate vaccines induce a long-lasting T-lymphocyte-dependent immunological memory (66, 67) and thus have played an enormous role in preventing infectious diseases (67, 68). However, the majority of these vaccines are based on chemical conjugation of a protein to either purified polysaccharide from a natural source or carbohydrates produced by multiple chemical synthesis steps, an expensive and time-consuming process yielding variable products (69). Bacterial glycoengineering may provide a solution to facilitate the production of such compounds in a cost-effective and homogeneous manner.
The functional transfer of the C. jejuni glycosylation machinery into E. coli started the era of bacterial glycoengineering (16). The C. jejuni-derived glycoprotein CmeA, a periplasmic component of the multidrug efflux pump, was the preferred N-glycan acceptor protein to be modified with various bacterial oligosaccharides in a PglB-dependent manner (23, 24). Now, researchers can take advantage of this system for the creation of novel glycoconjugates (70).
Vaccines, Therapeutic Proteins, and Novel Glycans
A novel vaccine against Brucella abortus, a major cause of brucellosis in humans and livestock (71, 72), was described recently (73). The purified glycoconjugate, produced by PglB-dependent coupling of an N-formylperosamine homopolymer to C. jejuni CmeA, induced a pathogen sugar-specific IgG response in mice but was not protective in challenge experiments. However, when the recombinant glycoprotein was coated onto magnetic beads, it was efficient in differentiating between naïve and infected bovine sera, thus acting a useful diagnostic tool (73). Ihssen et al. (74) produced two glycoconjugates as potential vaccine candidates against Shigella dysenteriae type 1 by PglB-dependent coupling of the Shigella O1-antigen to either C. jejuni CmeA or a toxoid form of Pseudomonas aeruginosa exotoxin A with engineered N-glycosylation sites. Similarly, Hug et al. (75) engineered E. coli to generate a Lewis X (Galβ(1–4)(Fucα1–3)GlcNAc-) glycan-containing glycoprotein (C. jejuni CmeA) in a combined in vivo and in vitro approach.
These examples, together with previous results (16, 23, 24), demonstrate that it is now feasible to produce a variety of bacterial glycans attached to protein acceptors with relative ease. However, the synthesis of eukaryote-like oligosaccharides to generate humanized glycoproteins is still a challenge within the field of bacterial glycoengineering. Recently, a combination of in vivo and chemoenzymatic approaches produced homogeneous eukaryotic N-glycoproteins by in vitro trimming and remodeling of a bacterially generated N-linked glycan by transglycosylation (76). In this proof-of-concept study, the authors were able to generate a natural high mannose-type glycan (Man9GlcNAc2) and a biantennary complex-type N-glycan attached to CmeA as well as two eukaryotic proteins, the CH2 domain of human IgG-Fc and the single-chain antibody F8 (76). A big step forward was the complete in vivo synthesis of a eukaryotic trimannosylchitobiose glycan by expressing four eukaryotic GTases and the subsequent transfer (via C. jejuni PglB) to acceptor proteins in E. coli (77). Despite the fact that only a very small fraction of each acceptor protein was glycosylated under the tested conditions, this study sets the stage for further engineering of eukaryotic glycoproteins using bacterial systems (77).
N-Linked protein glycosylation can also improve the biophysical and pharmacokinetic properties of existing therapeutic proteins such as antibodies when expressed and N-glycosylated in eukaryotic cell lines (78, 79). Using the bacterial system, Lizak et al. (80) demonstrated that introduction of two N-glycan acceptor sites and subsequent N-glycosylation of a single-chain antibody (scFv 3D5) with the C. jejuni N-glycan in E. coli significantly increased the proteolytic stability and solubility without influencing the affinity and serum stability of the single-chain antibody. However, N-glycosylation did not affect the overall clearance rate in the murine model system, which was probably due to the properties of the attached C. jejuni N-glycan (80).
Understanding and Improving N-Glycosylation
The x-ray crystal structure of C. lari PglB, which is 56% identical to C. jejuni PglB and complements C. jejuni PglB function in E. coli (80, 81), was solved in complex with the DQNATF acceptor peptide. This structure greatly advanced our understanding of the detailed mechanism for N-linked protein glycosylation at a molecular level (82). The structure not only identified the acceptor peptide-binding and catalytic sites in the transmembrane domain but provided insights into the mechanism of sugar transfer from LLOs to acceptor proteins. The structural requirements of the acceptor peptide (flexible loop formation), as well as important amino acid residues within C. lari PglB that explain the peptide sequon requirements and the catalytic mechanism (including divalent metal ion complexing), were identified.
Topology predictions further demonstrated the commonality of three motifs in bacterial, archaeal, and eukaryotic OTases that were the target for mutagenesis and functional analysis within C. jejuni PglB (83, 84). C. jejuni PglB activity was either low, medium, or completely abolished in point mutants with substitutions of amino acid residues that are involved in substrate binding and catalysis, confirming and supporting the observations made by Lizak et al. (82) for C. lari PglB. Genetic and functional analysis suggested that a 568MXXI571 motif has an impact on C. jejuni PglB function (84); in addition, single point mutations introduced into the conserved 475DXXK478 motif, earlier identified within the OTase of Pyrococcus furiosus (85), implied that Asp475 is involved but does not play an essential role in PglB activity (83). However, double valine or alanine substitutions of the Asp475 and Lys478 sites resulted in the complete loss of C. jejuni CmeA glycosylation activity in E. coli, an effect more drastic than a single Asp475 substitution.5 The fact that D475V and K478V single and double point mutants displayed a significant reduction in PglB-mediated free oligosaccharide release and N-glycosylation levels when analyzed in the native C. jejuni host suggests that these residues are indeed important for PglB activity.5
The PglB structure in combination with in vitro evolution systems such as cell-surface and phage display (86, 87) will lead to the creation of OTases that efficiently glycosylate eukaryotic acceptor sites. The identification and characterization of novel OTases (88), especially those with more relaxed substrate specificities (28, 81), will assist in engineering novel antigens. The use of codon-optimized GTase- and OTase-encoding genes, better glycoconjugate production strains, and improved culture conditions and glycoconjugate purification methods (74, 80, 89–91) will further aid to optimize novel and already existing glycoconjugate expression systems.
Conclusions
Over the last decade, an incredible amount of progress has been made in our understanding of bacterial N-glycosylation systems. However, several key questions still remain. For instance, what are the main targets for N-glycosylation? Although tremendous progress has been made in the field of bacterial glycoengineering, there is still room for improvement and it needs to be determined whether it will be possible to produce humanized therapeutic proteins in the bacterial system in large enough quantities to make it feasible to replace existing technologies. Analogous to eukaryotes, which require flipping of a LLO from the cytoplasm to the lumen of the endoplasmic reticulum, protein N-glycosylation has been detected only in Gram-negative bacteria, which contain two membranes. Why do only some Gram-negative bacteria modify their proteins with sugars, whereas others use the same proteins without modification? Why was diNAcBac selected twice during bacterial evolution to act as the sugar that links oligosaccharides to proteins? Are there additional protein targets for the cytoplasmic HMW1C-dependent glycosylation pathway, or do these organisms use this pathway solely for the purpose of glycosylating the adhesion needed for human cell attachment? What are the exact roles of the sugars on the adhesin? The answers to the many remaining questions will provide more insight into the process of asparagine-linked protein glycosylation in bacteria.
Acknowledgments
We thank William Miller for providing the protein sequences for the Campylobacter PglD, PglE, and PglF homologs and for helpful discussions. We also thank Michelle Richards and Thuan Nguyen for help with figures.
This is the second article in the Thematic Minireview Series on Glycobiology and Extracellular Matrices: Glycan Functions Pervade Biology at All Levels.
R. Dubb, H. Nothaft, J. Wong, T. L. Ravio, and C. M. Szymanski, manuscript in preparation.
Y. Barre, H. Nothaft, X. Liu, J. Li, K. Ng, and C. M. Szymanski, manuscript in preparation.
- diNAcBac
- di-N-acetylbacillosamine
- Und-P
- undecaprenyl phosphate
- GTase
- glycosyltransferase
- LLO
- lipid-linked oligosaccharide
- OTase
- oligosaccharyltransferase
- pEtN
- phosphoethanolamine.
REFERENCES
- 1. Helenius A., Aebi M. (2004) Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049 [DOI] [PubMed] [Google Scholar]
- 2. Vigerust D. J., Shepherd V. L. (2007) Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 15, 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rudd P. M., Elliott T., Cresswell P., Wilson I. A., Dwek R. A. (2001) Glycosylation and the immune system. Science 291, 2370–2376 [DOI] [PubMed] [Google Scholar]
- 4. Mitra N., Sinha S., Ramya T. N., Surolia A. (2006) N-Linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends Biochem. Sci. 31, 156–163 [DOI] [PubMed] [Google Scholar]
- 5. Calo D., Kaminski L., Eichler J. (2010) Protein glycosylation in Archaea: sweet and extreme. Glycobiology 20, 1065–1076 [DOI] [PubMed] [Google Scholar]
- 6. Nothaft H., Szymanski C. M. (2010) Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 8, 765–778 [DOI] [PubMed] [Google Scholar]
- 7. Szymanski C. M., Yao R., Ewing C. P., Trust T. J., Guerry P. (1999) Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32, 1022–1030 [DOI] [PubMed] [Google Scholar]
- 8. Scott N. E., Parker B. L., Connolly A. M., Paulech J., Edwards A. V., Crossett B., Falconer L., Kolarich D., Djordjevic S. P., Højrup P., Packer N. H., Larsen M. R., Cordwell S. J. (2011) Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics 10, M000031–MCP201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sharon N. (2007) Celebrating the golden anniversary of the discovery of bacillosamine, the diamino sugar of a Bacillus. Glycobiology 17, 1150–1155 [DOI] [PubMed] [Google Scholar]
- 10. Schoenhofen I. C., McNally D. J., Vinogradov E., Whitfield D., Young N. M., Dick S., Wakarchuk W. W., Brisson J. R., Logan S. M. (2006) Functional characterization of dehydratase/aminotransferase pairs from Helicobacter and Campylobacter: enzymes distinguishing the pseudaminic acid and bacillosamine biosynthetic pathways. J. Biol. Chem. 281, 723–732 [DOI] [PubMed] [Google Scholar]
- 11. Olivier N. B., Chen M. M., Behr J. R., Imperiali B. (2006) In vitro biosynthesis of UDP-N,N′-diacetylbacillosamine by enzymes of the Campylobacter jejuni general protein glycosylation system. Biochemistry 45, 13659–13669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Glover K. J., Weerapana E., Chen M. M., Imperiali B. (2006) Direct biochemical evidence for the utilization of UDP-bacillosamine by PglC, an essential glycosyl-1-phosphate transferase in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry 45, 5343–5350 [DOI] [PubMed] [Google Scholar]
- 13. Glover K. J., Weerapana E., Imperiali B. (2005) In vitro assembly of the undecaprenylpyrophosphate-linked heptasaccharide for prokaryotic N-linked glycosylation. Proc. Natl. Acad. Sci. U.S.A. 102, 14255–14259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kelly J., Jarrell H., Millar L., Tessier L., Fiori L. M., Lau P. C., Allan B., Szymanski C. M. (2006) Biosynthesis of the N-linked glycan in Campylobacter jejuni and addition onto protein through block transfer. J. Bacteriol. 188, 2427–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alaimo C., Catrein I., Morf L., Marolda C. L., Callewaert N., Valvano M. A., Feldman M. F., Aebi M. (2006) Two distinct but interchangeable mechanisms for flipping of lipid-linked oligosaccharides. EMBO J. 25, 967–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wacker M., Linton D., Hitchen P. G., Nita-Lazar M., Haslam S. M., North S. J., Panico M., Morris H. R., Dell A., Wren B. W., Aebi M. (2002) N-Linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298, 1790–1793 [DOI] [PubMed] [Google Scholar]
- 17. Kowarik M., Numao S., Feldman M. F., Schulz B. L., Callewaert N., Kiermaier E., Catrein I., Aebi M. (2006) N-Linked glycosylation of folded proteins by the bacterial oligosaccharyltransferase. Science 314, 1148–1150 [DOI] [PubMed] [Google Scholar]
- 18. Kowarik M., Young N. M., Numao S., Schulz B. L., Hug I., Callewaert N., Mills D. C., Watson D. C., Hernandez M., Kelly J. F., Wacker M., Aebi M. (2006) Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 25, 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nothaft H., Liu X., McNally D. J., Li J., Szymanski C. M. (2009) Study of free oligosaccharides derived from the bacterial N-glycosylation pathway. Proc. Natl. Acad. Sci. U.S.A. 106, 15019–15024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Burda P., Aebi M. (1999) The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426, 239–257 [DOI] [PubMed] [Google Scholar]
- 21. Trombetta E. S. (2003) The contribution of N-glycans and their processing in the endoplasmic reticulum to glycoprotein biosynthesis. Glycobiology 13, 77R–91R [DOI] [PubMed] [Google Scholar]
- 22. Chen M. M., Glover K. J., Imperiali B. (2007) From peptide to protein: comparative analysis of the substrate specificity of N-linked glycosylation in C. jejuni. Biochemistry 46, 5579–5585 [DOI] [PubMed] [Google Scholar]
- 23. Feldman M. F., Wacker M., Hernandez M., Hitchen P. G., Marolda C. L., Kowarik M., Morris H. R., Dell A., Valvano M. A., Aebi M. (2005) Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 102, 3016–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wacker M., Feldman M. F., Callewaert N., Kowarik M., Clarke B. R., Pohl N. L., Hernandez M., Vines E. D., Valvano M. A., Whitfield C., Aebi M. (2006) Substrate specificity of bacterial oligosaccharyltransferase suggests a common transfer mechanism for the bacterial and eukaryotic systems. Proc. Natl. Acad. Sci. U.S.A. 103, 7088–7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen M. M., Weerapana E., Ciepichal E., Stupak J., Reid C. W., Swiezewska E., Imperiali B. (2007) Polyisoprenol specificity in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry 46, 14342–14348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Larkin A., Imperiali B. (2011) The expanding horizons of asparagine-linked glycosylation. Biochemistry 50, 4411–4426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schwarz F., Aebi M. (2011) Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 21, 576–582 [DOI] [PubMed] [Google Scholar]
- 28. Ielmini M. V., Feldman M. F. (2011) Desulfovibrio desulfuricans PglB homolog possesses oligosaccharyltransferase activity with relaxed glycan specificity and distinct protein acceptor sequence requirements. Glycobiology 21, 734–742 [DOI] [PubMed] [Google Scholar]
- 29. Santos-Silva T., Dias J. M., Dolla A., Durand M. C., Gonçalves L. L., Lampreia J., Moura I., Romão M. J. (2007) Crystal structure of the 16-heme cytochrome from Desulfovibrio gigas: a glycosylated protein in a sulphate-reducing bacterium. J. Mol. Biol. 370, 659–673 [DOI] [PubMed] [Google Scholar]
- 30. Jervis A. J., Butler J. A., Lawson A. J., Langdon R., Wren B. W., Linton D. (2012) Characterization of the structurally diverse N-linked glycans of Campylobacter species. J. Bacteriol. 194, 2355–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jervis A. J., Langdon R., Hitchen P., Lawson A. J., Wood A., Fothergill J. L., Morris H. R., Dell A., Wren B., Linton D. (2010) Characterization of N-linked protein glycosylation in Helicobacter pullorum. J. Bacteriol. 192, 5228–5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nothaft H., Scott N. E., Vinogradov E., Liu X., Hu R., Beadle B., Fodor C., Miller W. G., Li J., Cordwell S. J., Szymanski C. M. (2012) Diversity in the protein N-glycosylation pathways within the Campylobacter genus. Mol. Cell. Proteomics 11, 1203–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fouts D. E., Mongodin E. F., Mandrell R. E., Miller W. G., Rasko D. A., Ravel J., Brinkac L. M., DeBoy R. T., Parker C. T., Daugherty S. C., Dodson R. J., Durkin A. S., Madupu R., Sullivan S. A., Shetty J. U., Ayodeji M. A., Shvartsbeyn A., Schatz M. C., Badger J. H., Fraser C. M., Nelson K. E. (2005) Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 3, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Szymanski C. M., Michael F. S., Jarrell H. C., Li J., Gilbert M., Larocque S., Vinogradov E., Brisson J. R. (2003) Detection of conserved N-linked glycans and phase-variable lipooligosaccharides and capsules from Campylobacter cells by mass spectrometry and high resolution magic angle spinning NMR spectroscopy. J. Biol. Chem. 278, 24509–24520 [DOI] [PubMed] [Google Scholar]
- 35. Scott N. E., Nothaft H., Edwards A. V., Labbate M., Djordjevic S. P., Larsen M. R., Szymanski C. M., Cordwell S. J. (2012) Modification of the Campylobacter jejuni N-linked glycan by EptC protein-mediated addition of phosphoethanolamine. J. Biol. Chem. 287, 29384–29396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cullen T. W., Trent M. S. (2010) A link between the assembly of flagella and lipooligosaccharide of the Gram-negative bacterium Campylobacter jejuni. Proc. Natl. Acad. Sci. U.S.A. 107, 5160–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aas F. E., Egge-Jacobsen W., Winther-Larsen H. C., Lovold C., Hitchen P. G., Dell A., Koomey M. (2006) Neisseria gonorrhoeae type IV pili undergo multisite, hierarchical modifications with phosphoethanolamine and phosphocholine requiring an enzyme structurally related to lipopolysaccharide phosphoethanolamine transferases. J. Biol. Chem. 281, 27712–27723 [DOI] [PubMed] [Google Scholar]
- 38. Anonsen J. H., Egge-Jacobsen W., Aas F. E., Børud B., Koomey M., Vik A. (2012) Novel protein substrates of the phospho-form modification system in Neisseria gonorrhoeae and their connection to O-linked protein glycosylation. Infect. Immun. 80, 22–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Szymanski C. M., Goon S., Allan B., Guerry P. (2005) in Campylobacter: Molecular and Cellular Biology (Ketley J. M., Konkel M. E., eds) pp. 259–273, Horizon Bioscience, Norwich, United Kingdom [Google Scholar]
- 40. Szymanski C. M., Burr D. H., Guerry P. (2002) Campylobacter protein glycosylation affects host cell interactions. Infect. Immun. 70, 2242–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van Sorge N. M., Bleumink N. M., van Vliet S. J., Saeland E., van der Pol W. L., van Kooyk Y., van Putten J. P. (2009) N-Glycosylated proteins and distinct lipooligosaccharide glycoforms of Campylobacter jejuni target the human C-type lectin receptor MGL. Cell. Microbiol. 11, 1768–1781 [DOI] [PubMed] [Google Scholar]
- 42. Hendrixson D. R., DiRita V. J. (2004) Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52, 471–484 [DOI] [PubMed] [Google Scholar]
- 43. Szymanski C. M., Wren B. W. (2005) Protein glycosylation in bacterial mucosal pathogens. Nat. Rev. Microbiol. 3, 225–237 [DOI] [PubMed] [Google Scholar]
- 44. Larsen J. C., Szymanski C., Guerry P. (2004) N-Linked protein glycosylation is required for full competence in Campylobacter jejuni 81-176. J. Bacteriol. 186, 6508–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Davis L. M., Kakuda T., DiRita V. J. (2009) A Campylobacter jejuni znuA orthologue is essential for growth in low-zinc environments and chick colonization. J. Bacteriol. 191, 1631–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kakuda T., DiRita V. J. (2006) Cj1496c encodes a Campylobacter jejuni glycoprotein that influences invasion of human epithelial cells and colonization of the chick gastrointestinal tract. Infect. Immun. 74, 4715–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Flanagan R. C., Neal-McKinney J. M., Dhillon A. S., Miller W. G., Konkel M. E. (2009) Examination of Campylobacter jejuni putative adhesins leads to the identification of a new protein, designated FlpA, required for chicken colonization. Infect. Immun. 77, 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Scott N. E., Bogema D. R., Connolly A. M., Falconer L., Djordjevic S. P., Cordwell S. J. (2009) Mass spectrometric characterization of the surface-associated 42 kDa lipoprotein JlpA as a glycosylated antigen in strains of Campylobacter jejuni. J. Proteome Res. 8, 4654–4664 [DOI] [PubMed] [Google Scholar]
- 49. Kakuda T., Koide Y., Sakamoto A., Takai S. (2012) Characterization of two putative mechanosensitive channel proteins of Campylobacter jejuni involved in protection against osmotic downshock. Vet. Microbiol. 160, 53–60 [DOI] [PubMed] [Google Scholar]
- 50. Lin J., Sahin O., Michel L. O., Zhang Q. (2003) Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect. Immun. 71, 4250–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nothaft H., Liu X., Li J., Szymanski C. M. (2010) Campylobacter jejuni free oligosaccharides: function and fate. Virulence 1, 546–550 [DOI] [PubMed] [Google Scholar]
- 52. Vik A., Aas F. E., Anonsen J. H., Bilsborough S., Schneider A., Egge-Jacobsen W., Koomey M. (2009) Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U.S.A. 106, 4447–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ku S. C., Schulz B. L., Power P. M., Jennings M. P. (2009) The pilin O-glycosylation pathway of pathogenic Neisseria is a general system that glycosylates AniA, an outer membrane nitrite reductase. Biochem. Biophys. Res. Commun. 378, 84–89 [DOI] [PubMed] [Google Scholar]
- 54. Faridmoayer A., Fentabil M. A., Mills D. C., Klassen J. S., Feldman M. F. (2007) Functional characterization of bacterial oligosaccharyltransferases involved in O-linked protein glycosylation. J. Bacteriol. 189, 8088–8098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chamot-Rooke J., Rousseau B., Lanternier F., Mikaty G., Mairey E., Malosse C., Bouchoux G., Pelicic V., Camoin L., Nassif X., Duménil G. (2007) Alternative Neisseria spp. type IV pilin glycosylation with a glyceramido acetamido trideoxyhexose residue. Proc. Natl. Acad. Sci. U.S.A. 104, 14783–14788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Craig L., Volkmann N., Arvai A. S., Pique M. E., Yeager M., Egelman E. H., Tainer J. A. (2006) Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell 23, 651–662 [DOI] [PubMed] [Google Scholar]
- 57. Grass S., Buscher A. Z., Swords W. E., Apicella M. A., Barenkamp S. J., Ozchlewski N., St Geme J. W., 3rd (2003) The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis. Mol. Microbiol. 48, 737–751 [DOI] [PubMed] [Google Scholar]
- 58. Barenkamp S. J., Leininger E. (1992) Cloning, expression, and DNA sequence analysis of genes encoding nontypeable Haemophilus influenzae high-molecular-weight surface-exposed proteins related to filamentous hemagglutinin of Bordetella pertussis. Infect. Immun. 60, 1302–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. St Geme J. W., 3rd, Falkow S., Barenkamp S. J. (1993) High-molecular-weight proteins of nontypable Haemophilus influenzae mediate attachment to human epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 90, 2875–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. St Geme J. W., 3rd, Kumar V. V., Cutter D., Barenkamp S. J. (1998) Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect. Immun. 66, 364–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gross J., Grass S., Davis A. E., Gilmore-Erdmann P., Townsend R. R., St Geme J. W., 3rd (2008) The Haemophilus influenzae HMW1 adhesin is a glycoprotein with an unusual N-linked carbohydrate modification. J. Biol. Chem. 283, 26010–26015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Grass S., Lichti C. F., Townsend R. R., Gross J., St Geme J. W., 3rd (2010) The Haemophilus influenzae HMW1C protein is a glycosyltransferase that transfers hexose residues to asparagine sites in the HMW1 adhesin. PLoS Pathog. 6, e1000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Choi K. J., Grass S., Paek S., St Geme J. W., 3rd, Yeo H. J. (2010) The Actinobacillus pleuropneumoniae HMW1C-like glycosyltransferase mediates N-linked glycosylation of the Haemophilus influenzae HMW1 adhesin. PLoS ONE 5, e15888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kawai F., Grass S., Kim Y., Choi K. J., St Geme J. W., 3rd, Yeo H. J. (2011) Structural insights into the glycosyltransferase activity of the Actinobacillus pleuropneumoniae HMW1C-like protein. J. Biol. Chem. 286, 38546–38557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schwarz F., Fan Y. Y., Schubert M., Aebi M. (2011) Cytoplasmic N-glycosyltransferase of Actinobacillus pleuropneumoniae is an inverting enzyme and recognizes the NX(S/T) consensus sequence. J. Biol. Chem. 286, 35267–35274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lockhart S. (2003) Conjugate vaccines. Expert Rev. Vaccines 2, 633–648 [DOI] [PubMed] [Google Scholar]
- 67. Weintraub A. (2003) Immunology of bacterial polysaccharide antigens. Carbohydr. Res. 338, 2539–2547 [DOI] [PubMed] [Google Scholar]
- 68. Trotter C. L., McVernon J., Ramsay M. E., Whitney C. G., Mulholland E. K., Goldblatt D., Hombach J., Kieny M. P. (2008) Optimising the use of conjugate vaccines to prevent disease caused by Haemophilus influenzae type b, Neisseria meningitidis and Streptococcus pneumoniae. Vaccine 26, 4434–4445 [DOI] [PubMed] [Google Scholar]
- 69. Frasch C. E. (2009) Preparation of bacterial polysaccharide-protein conjugates: analytical and manufacturing challenges. Vaccine 27, 6468–6470 [DOI] [PubMed] [Google Scholar]
- 70. Feldman M. F. (2009) in Microbial Glycobiology, Structures, Relevance and Applications (Moran A. P., Holst O., Brennan P. J., von Itzstein M., eds) pp. 903–914, Elsevier, Burlington, MA [Google Scholar]
- 71. Corbel M. J. (1997) Brucellosis: an overview. Emerg. Infect. Dis. 3, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Young E. J., Tarry A., Genta R. M., Ayden N., Gotuzzo E. (2000) Thrombocytopenic purpura associated with brucellosis: report of 2 cases and literature review. Clin. Infect. Dis. 31, 904–909 [DOI] [PubMed] [Google Scholar]
- 73. Iwashkiw J. A., Fentabil M. A., Faridmoayer A., Mills D. C., Peppler M., Czibener C., Ciocchini A. E., Comerci D. J., Ugalde J. E., Feldman M. F. (2012) Exploiting the Campylobacter jejuni protein glycosylation system for glycoengineering vaccines and diagnostic tools directed against brucellosis. Microb. Cell Fact. 11, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ihssen J., Kowarik M., Dilettoso S., Tanner C., Wacker M., Thöny-Meyer L. (2010) Production of glycoprotein vaccines in Escherichia coli. Microb. Cell Fact. 9, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hug I., Zheng B., Reiz B., Whittal R. M., Fentabil M. A., Klassen J. S., Feldman M. F. (2011) Exploiting bacterial glycosylation machineries for the synthesis of a Lewis antigen-containing glycoprotein. J. Biol. Chem. 286, 37887–37894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Schwarz F., Huang W., Li C., Schulz B. L., Lizak C., Palumbo A., Numao S., Neri D., Aebi M., Wang L. X. (2010) A combined method for producing homogeneous glycoproteins with eukaryotic N-glycosylation. Nat. Chem. Biol. 6, 264–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Valderrama-Rincon J. D., Fisher A. C., Merritt J. H., Fan Y. Y., Reading C. A., Chhiba K., Heiss C., Azadi P., Aebi M., DeLisa M. P. (2012) An engineered eukaryotic protein glycosylation pathway in Escherichia coli. Nat. Chem. Biol. 8, 434–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Elliott S., Lorenzini T., Asher S., Aoki K., Brankow D., Buck L., Busse L., Chang D., Fuller J., Grant J., Hernday N., Hokum M., Hu S., Knudten A., Levin N., Komorowski R., Martin F., Navarro R., Osslund T., Rogers G., Rogers N., Trail G., Egrie J. (2003) Enhancement of therapeutic protein in vivo activities through glycoengineering. Nat. Biotechnol. 21, 414–421 [DOI] [PubMed] [Google Scholar]
- 79. Stork R., Zettlitz K. A., Müller D., Rether M., Hanisch F. G., Kontermann R. E. (2008) N-Glycosylation as novel strategy to improve pharmacokinetic properties of bispecific single-chain diabodies. J. Biol. Chem. 283, 7804–7812 [DOI] [PubMed] [Google Scholar]
- 80. Lizak C., Fan Y. Y., Weber T. C., Aebi M. (2011) N-Linked glycosylation of antibody fragments in Escherichia coli. Bioconjug. Chem. 22, 488–496 [DOI] [PubMed] [Google Scholar]
- 81. Schwarz F., Lizak C., Fan Y. Y., Fleurkens S., Kowarik M., Aebi M. (2011) Relaxed acceptor site specificity of bacterial oligosaccharyltransferase in vivo. Glycobiology 21, 45–54 [DOI] [PubMed] [Google Scholar]
- 82. Lizak C., Gerber S., Numao S., Aebi M., Locher K. P. (2011) X-ray structure of a bacterial oligosaccharyltransferase. Nature 474, 350–355 [DOI] [PubMed] [Google Scholar]
- 83. Jaffee M. B., Imperiali B. (2011) Exploiting topological constraints to reveal buried sequence motifs in the membrane-bound N-linked oligosaccharyl transferases. Biochemistry 50, 7557–7567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Maita N., Nyirenda J., Igura M., Kamishikiryo J., Kohda D. (2010) Comparative structural biology of eubacterial and archaeal oligosaccharyltransferases. J. Biol. Chem. 285, 4941–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Igura M., Maita N., Kamishikiryo J., Yamada M., Obita T., Maenaka K., Kohda D. (2008) Structure-guided identification of a new catalytic motif of oligosaccharyltransferase. EMBO J. 27, 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Celik E., Fisher A. C., Guarino C., Mansell T. J., DeLisa M. P. (2010) A filamentous phage display system for N-linked glycoproteins. Protein Sci. 19, 2006–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Dürr C., Nothaft H., Lizak C., Glockshuber R., Aebi M. (2010) The Escherichia coli glycophage display system. Glycobiology 20, 1366–1372 [DOI] [PubMed] [Google Scholar]
- 88. Nasab F. P., Schulz B. L., Gamarro F., Parodi A. J., Aebi M. (2008) All in one: Leishmania major STT3 proteins substitute for the whole oligosaccharyltransferase complex in Saccharomyces cerevisiae. Mol. Biol. Cell 19, 3758–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pandhal J., Desai P., Walpole C., Doroudi L., Malyshev D., Wright P. C. (2012) Systematic metabolic engineering for improvement of glycosylation efficiency in Escherichia coli. Biochem. Biophys. Res. Commun. 419, 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Pandhal J., Wright P. C. (2010) N-Linked glycoengineering for human therapeutic proteins in bacteria. Biotechnol. Lett. 32, 1189–1198 [DOI] [PubMed] [Google Scholar]
- 91. Pandhal J., Ow S. Y., Noirel J., Wright P. C. (2011) Improving N-glycosylation efficiency in Escherichia coli using shotgun proteomics, metabolic network analysis, and selective reaction monitoring. Biotechnol. Bioeng. 108, 902–912 [DOI] [PubMed] [Google Scholar]
- 92. Young N. M., Brisson J. R., Kelly J., Watson D. C., Tessier L., Lanthier P. H., Jarrell H. C., Cadotte N., St Michael F., Aberg E., Szymanski C. M. (2002) Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 277, 42530–42539 [DOI] [PubMed] [Google Scholar]