Background: Neuroglobins are expressed in vertebrate neurons.
Results: Neuroglobins are located in neural systems of two basal animals (acoels and jellyfish) and are ubiquitous in metazoan transcriptomes.
Conclusion: Neuroglobin was recruited in neural cell prototypes and later co-opted in hemoglobin-based blood systems.
Significance: The universality of neuroglobins sheds new light on the origin and evolution of globins.
Keywords: Animal Models, Confocal Microscopy, Crystal Structure, Evolution, Hemoglobin, Immunohistochemistry, Phylogenetics, Metazoans, Neuroglobin
Abstract
Neuroglobins, previously thought to be restricted to vertebrate neurons, were detected in the brain of a photosymbiotic acoel, Symsagittifera roscoffensis, and in neurosensory cells of the jellyfish Clytia hemisphaerica. For the neuroglobin of S. roscoffensis, a member of a lineage that originated either at the base of the bilateria or of the deuterostome clade, we report the ligand binding properties, crystal structure at 2.3 Å, and brain immunocytochemical pattern. We also describe in situ hybridizations of two neuroglobins specifically expressed in differentiating nematocytes (neurosensory cells) and in statocytes (ciliated mechanosensory cells) of C. hemisphaerica, a member of the early branching animal phylum cnidaria. In silico searches using these neuroglobins as queries revealed the presence of previously unidentified neuroglobin-like sequences in most metazoan lineages. Because neural systems are almost ubiquitous in metazoa, the constitutive expression of neuroglobin-like proteins strongly supports the notion of an intimate association of neuroglobins with the evolution of animal neural systems and hints at the preservation of a vitally important function. Neuroglobins were probably recruited in the first protoneurons in early metazoans from globin precursors. Neuroglobins were identified in choanoflagellates, sponges, and placozoans and were conserved during nervous system evolution. Because the origin of neuroglobins predates the other metazoan globins, it is likely that neuroglobin gene duplication followed by co-option and subfunctionalization led to the emergence of globin families in protostomes and deuterostomes (i.e. convergent evolution).
Introduction
Interest in the structure, function, and evolutionary relationships of circulating hemoglobins (Hbs)2 and intracellular myoglobins (Mbs) of animals dates back to the first three-dimensional structural determination of these proteins in the 1960s (1–3). The large range of animal globins and the extensive occurrence of globins in prokaryotes is now recognized (4). Prominent among the recently described metazoan globins is vertebrate neuroglobin (Ngb), which is expressed in neurons of the central and peripheral nervous systems (5). The in vivo function of Ngb remains undefined despite a major effort over the last decade. Suggested functions include the oxygen (O2) supply in hypoxia and ischemia (6), scavenging of reactive oxygen free radicals (7), protection from apoptosis (8), redox-regulated nitrite reductase activity (9), and involvement in respiratory chain function (10). In murine models of human neuropathology, Ngb is also expressed in reactive astrocytes, a subtype of glia cells in the nervous system (11).
In protostomes, globins have been observed in the nerve tissue of certain annelids, molluscs, and a nematode (12), but these have not been phylogenetically linked to vertebrate Ngbs or other deuterostome globins. Their O2 binding affinities resemble those of vertebrate Mbs, and their function is considered to be O2 storage and thus protection against hypoxia (13, 14).
Recent phylogenomic analyses of vertebrate globins have demonstrated that they can be separated into two groups, one derived from vertebrate-specific duplications (cytoglobins (Cygbs), globin E, globin Y, the Hb chains, and Mb), and another resulting from duplications preceding the emergence of chordates (Ngb and HbX) (15–17). The most recent molecular phylogenetic analysis of globin sequences from the five major groups comprising the deuterostomes, i.e. cephalochordates, echinoderms, hemichordates, urochordates, and vertebrates, suggests that all deuterostome globins occur in four clades (18).
Recently, Blank et al. (19) demonstrated that the functional hexacoordinated Globin X (GbX) protein of the cypriniform adult zebrafish is located in the nervous central system and retina, suggesting a neural-based function but contradicting a previous result obtained from the other cypriniform Carassius auratus GbX showing that mRNA GbX was not detected in brain and eye but in other tissues (muscle, heart, gut, and liver) (20). Thought to be restricted to vertebrate, GbX-like sequences have been recently identified in silico in other deuterostomes and in protostomes, supporting an early emergence of this gene family in metazoan evolution (21).
Despite the fact that a molecular analysis of metazoan globins (including echinoderm and cnidarian globins) suggested an ancestral connection to the nervous system (22), Ngbs have not been reported in deep branching metazoan lineages, and evolutionary patterns of emergence of metazoan globin lineages are still unresolved.
Symsagittifera roscoffensis is a photosymbiotic acoel (Fig. 1A), thus occupying a phylogenetic position either preceding the deuterostome-protostome split or branching at the base of deuterostomes (23, 24). This hermaphroditic marine flatworm has a simple body plan with a digestive syncytium (no epithelially lined gut), a ventral mouth, a muscle system, a nervous system with a simple central brain, but no excretory or blood circulatory systems (25).
FIGURE 1.
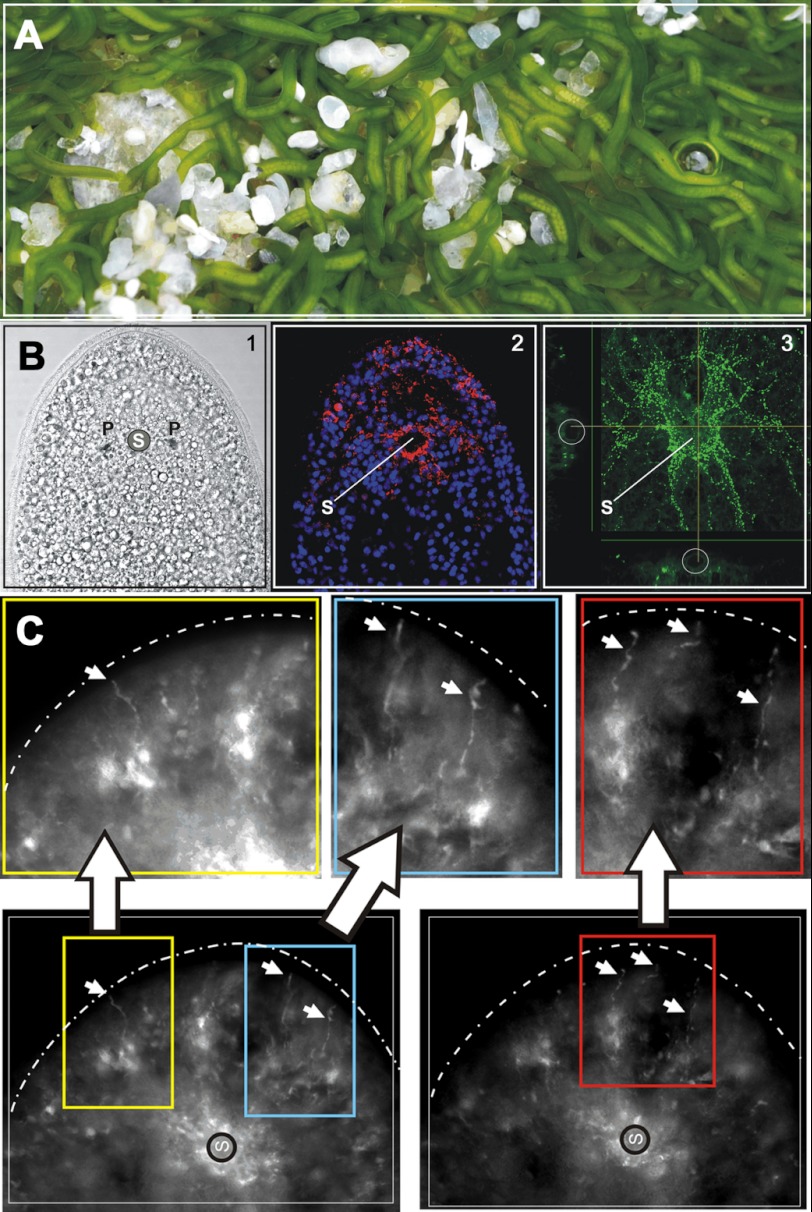
A, photograph of a colony of the symbiotic acoel S. roscoffensis (4–5 mm long) at low tide. The green color is due to the presence of approximately 50,000 photosymbionts (the unicellular green algae Tetraselmis convolutae) harbored within each adult acoel. B1, light micrograph of the anterior tip of a juvenile with the statocyst (S) flanked by two photoreceptors (P). Cilia are visible on the periphery of the head. B2, composite confocal image showing the red SrNgb1 antibody signal surrounding the statocyst with peripheral extensions. DAPI-stained nuclei appear in blue. B3, confocal image illustrating RF-amide stained S. roscoffensis nervous system. C, magnification of the extremity of the anterior tip (the head). Arrows indicate fiber-like structures labeled with SrNgb1 antibody occurring at the same place of the frontal glands (frontal sensors). These fiber-like structures are superimposable with serotonergic nervous system and especially neurites.
We report the discovery of Ngb-like sequences in EST libraries from S. roscoffensis and subsequent characterization and immunocytochemical localization of these globins. We also examined the sites of expression of putative Ngbs in the jellyfish Clytia hemisphaerica (Cnidaria, Hydrozoa), which, like the “higher” animals (the Bilateria), exhibits a complex body organization, including striated musculature, reproductive organs, and a specialized nervous system (26). Finally, to investigate the origin of globins in metazoan lineages, we conducted a broad in silico transcriptome survey to search for Ngb-like proteins across the diversity of metazoans and their unicellular ancestors.
EXPERIMENTAL PROCEDURES
Expression, Purification and Characterization of S. roscoffensis Ngb
The coding sequence of S. roscoffensis Ngb. (SrNgb1) (European Nucleotide Archive ID number HE972520) was amplified by PCR and subsequently cloned into a pET-3a cloning vector (Invitrogen). The construct was transformed into Escherichia coli BL21DE3 for protein expression in autoinducible medium (27). The protein was purified with an Akta purifier system (GE Healthcare). Due to the low pI of the S. roscoffensis globin, samples were loaded on a 5-ml HiTrap DEAE FF column (GE Healthcare) equilibrated with 50 mm Tris-HCl (pH 8.5) and eluted at a concentration of 25 mm NaCl. Resulting samples were loaded on a desalting Sephadex G-25 column (GE Healthcare) suspended in PBS (pH 7.4) and purified on a Superose 12 HR 16/50 (Amersham Biosciences) column equilibrated with PBS (pH 7.4). Finally, ferric and ferrous UV/visible spectra (O2 and CO) were measured with a Cary model 400 spectrophotometer.
Autoxidation Kinetics and Ligand Rebinding of SrNgb1
Full spectra were measured versus time on an HP 8453 diode-array spectrophotometer. The sample was first thoroughly deoxygenated in a sealed optical cuvette under a stream of N2. Then a slight excess of sodium dithionite was added to reduce the globin heme moiety. Finally the cuvette was equilibrated under air to obtain the oxy-reduced species and to allow depletion of residual unreacted dithionite. Ligand recombination kinetics were measured at a single wavelength after photodissociation by 10-ns pulses at 532 nm, as described previously (28). Samples in sealed cuvettes were equilibrated under various fractions of CO or O2. A mixed atmosphere of CO and O2 was used to study the O2 to CO replacement reaction after photolysis of CO.
Immunocyto-localization with SrNgb1 and RF-amide Antibodies
Acoel flatworms collected in Roscoff (Brittany, France) were anesthetized with 7% MgCl2 and fixed for 45 min in 4% paraformaldehyde at 4 °C. Animals were then washed with phosphate buffer (pH 7.4) and permeabilized with 0.1% Triton X-100 in PBS three times for 15 min at room temperature. They were then incubated with 5% BSA, 0.1% Triton X-100, and 0.05% Tween 20 in PBS for 2–3 h at room temperature and incubated overnight at 4 °C alternatively with 1/700 polyclonal S. roscoffensis anti-Ngb, produced against whole recombinant protein by Eurogentec (Speedy 28-day polyclonal packages), or with anti-RF-amide (courtesy of Thomas Leitz, Kaiserslautern). The following day, acoels were washed three times for 15 min in PBS and incubated with the appropriate secondary antibodies. They were then incubated for 10 min in a DAPI solution (2 μg/ml in PBS), washed three times in PBS, and mounted on a glass slide for microscope observation. Image acquisition of fluorescent-labeled specimens was undertaken with a confocal microscope (Leica sp5) equipped with a 20× objective and using Leica LAS-AF software.
Animal Collection and in Situ Hybridization
C. hemisphaerica colonies established from polyps provided by Evelyn Houliston (Observatoire Océanologique Villefranche-sur-Mer, France) were cultured in artificial seawater (Reef Crystals) as described previously (29). Animals were left unfed for 1 day before fixation. They were fixed for 40 min at 4 °C in 3.7% formaldehyde, 0.2% glutaraldehyde, PBT 1× (10 mm Na2HPO4, 150 mm NaCl, pH 7.5, 0.1% Tween 20). Digoxigenin-labeled antisense RNA probe synthesis and whole mount in situ hybridizations were carried out as described previously (30) The only modification to the in situ protocol was an acetic anhydride treatment before hybridization. Alkaline phosphatase activity was revealed using NBT/BCIP (blue staining) or fast red TR-naphthol reagent (Sigma, red staining). After postfixation and DAPI staining (31), samples were mounted in Citifluor. Double in situ hybridizations were performed as described in Ref. 32. Differential interference contrast images were obtained with an Olympus BX61 microscope using a Q-imaging camera with Image Pro plus software (Media Cybernetics).
Protein Crystallization
All crystallization experiments were carried out at 292 K. Initial crystallization trials were performed with the PACT, JCSG+, PEG I, and PEG II suites (Qiagen), i.e. a total of 384 conditions in four 96-well plates. The trials were set up using a Cartesian crystallization robot, and the sitting drops were made by mixing 300 nl of protein (13 mg/ml in 30 mm PBS buffer (pH 7.5), 100 mm NaCl) with 150 nl of reservoir solution. A single hit was identified in the PEG II screen, containing 1 m LiCl, 0.1 m sodium acetate, and 30% (w/v) PEG 6000. Subsequently, this crystallization condition was optimized in 24-well Linbro plates by the hanging-drop vapor-diffusion method, screening ranges from 0.6 to 1.0 m LiCl and 30 to 39% PEG 6000. These drops were prepared on siliconized coverslips by mixing 2 ml of protein with 1 ml of well solution. The drops were equilibrated against reservoir solutions of 0.75-ml volume. Best crystals were obtained for 32% PEG 6000, 1.0 m LiCl, and 0.1 m sodium acetate. For cryoprotection, 5% glycerol was added to the crystal drop solution before flash-freezing the crystals in the gaseous N2 stream at 100 K.
Data Collection and X-ray Diffraction Analysis
X-ray diffraction data were first collected from globin crystals at 100 K on beamline ID23-I at the ESRF (Grenoble, France) using an ADSC Quantum 4R CCD detector. All crystals were flash-cooled in a liquid nitrogen stream. The crystals were rotated through 120 °C with a 0.5 °C oscillation range per frame at a wavelength of 0.933 Å. All raw data were processed using the program XDS, and the resultant data were merged and scaled using the program XSCALE (33). Models for structure solution by molecular replacement were selected by a sequence search using BLAST against the Protein Data Bank. However, all attempts to solve the structure of this globin by molecular replacement performed with the program AMORE (34) using various Ngb or Mb models were unsuccessful. A second data set was therefore collected at the Fe absorption edge at a wavelength of 1.7387 Å on beamline BM30A, covering an angular section of 90° with an oscillation range of 1.0°. Data treatment was performed with XDS in the same way as for the native data set. All further data collection statistics are given in Table 1.
TABLE 1.
Data collection, phasing, and refinement statistics on globin crystals, space group P6222
| Beamline at ESRF | ID23-I | BM30A |
|---|---|---|
| Crystal parameters | ||
| Wavelength (Å) | 0.93 | 1.7389 |
| Unit cell parameters in Å and degrees (°) | a = b = 97.07, c = 140.10, α = β = 90, γ = 120 | a = b = 97.07, c = 140.10, α = β = 90, γ = 120 |
| Data collection | ||
| Resolution range (Å) | 30.06–2.3 (2.36–2.30)a | 50.06–3.2 (3.29–3.20)a |
| No. of observations | 57,993 (3605) | 60,276 (3599) |
| No. of unique reflections | 12,137 (688) | 11,480 (675) |
| Completeness (%) | 91.7 (77.4) | 93.3 (77.4) |
| <I/σ (I)> | 16.6 (2.2) | 14.9 (2.0) |
| Redundancy | 4.8 (5.2) | 5.2 (5.3) |
| Rsym (%)b | 4.8 (43.3) | 7.6 (56.7) |
| Phasing statistics | ||
| Anomalous difference (CC in %, given by SHELXE) | 35.21 | |
| Figure of merit | 0.335 | |
| Refinement statistics | ||
| Rcryst (%) | 21.4 | |
| Rfree (%)c | 25.1 | |
| Esu based on free R value | 0.17 | |
| Overall B-factor (Å2) | 38.1 | |
| Protein | 38.0 | |
| Heme | 33.4 | |
| Solvent | 41.2 | |
| Root mean square deviation in bond lengths (Å) | 0.029 | |
| Root means square deviation in bond angles (°) | 2.46 | |
a Values for the highest resolution shell are given in parentheses.
b Rsym = Σ|I − Iav|/ΣI|, where the summation is overall symmetry-equivalent reflections.
c Rfree values were calculated on 5% of the data (904 reflections) that were set aside in the minimization steps.
Crystal Structure Determination and Refinement
The iron atom substructure solution was calculated with SHELXD (35) followed by phasing and density modification performed with SHELXE, using the graphical interface HKL2MAP (36), and the resulting electron density map was displayed with Coot (37). Both possible enantiomorph space groups were tried, and the phasing procedure allowed a selection of a clear and contrasted structure solution in P6222. These starting phases were used to build the initial model using ARP/wARP and REFMAC as part of the CCP4 suite (38), and switching to the higher resolution data at 2.3 Å. Roughly 70% of the helices were constructed by the automatic procedure. The subsequent manual adjustment and model building were carried out with Coot and alternated with refinement cycles using REFMAC. Water molecules were added automatically with the REFMAC-ARP/wARP option and visually verified, one by one, using Coot. The final model contained residues ranging from 6 to 154, the prosthetic heme group, 98 water molecules, and an oxygen ligand bound to the iron atom. The asymmetric unit contains one globin molecule leading to a Matthews coefficient of 4.9 and a solvent content of 74.9%. The phasing and final refinement statistics are given in Table 1 (S. roscoffensis Ngb PDB ID code 4B4Y).
Phylogenomics and Molecular Phylogeny
The identification of Ngb-like/putative neural globin sequences was performed using S. roscoffensis Ngb1 and vertebrate Ngb sequence queries in blastp searches of the nonredundant nucleotide database maintained by NCBI and of nonannotated expressed sequence tag databases from various metazoans, deposited and archived at the National Institutes of Health Trace database.
A multiple alignment of a representative subset of Ngb-like sequences was automatically generated with HMMER v3.0 package (39) using the hmmalign program and the Globin (PF0042) raw HMM as a guide. Molecular phylogenetic analysis was carried out using the Maximum Likelihood approach with PhyMl software (40) with the LG option as the model of amino acid substitution, NNI moves option for tree topology search operation and SH-like support option for default branch support. Tree topology (Newick format) was edited with MEGA5.1 (41).
In addition to the results presented here, molecular phylogenic analyses were performed with the BioSide software and deposited at its website. For the molecular phylogeny procedure to be easily traceable and reproducible, a file including the original multiple alignment of sequences and PhyMl setups is available at BioSide website. Prediction of N-terminal myristoylation of Ngb-like sequences was performed with the program The MYR Predictor. This program calculates whether or not a protein is predicted as myristoylated with reliable/twilight zone confidence.
RESULTS AND DISCUSSION
The Ngb-like Protein 1 of S. roscoffensis Is a Functional Neuroglobin
SrNgb1 is expressed in the brain and nervous system of S. roscoffensis (Fig. 1, B and C). The acoel brain is formed by a layer of neuronal cell bodies surrounding a central neuropile, embedding the statocyst, a gravity sensor (25). The SrNgb1 signal mainly occurs in the anterior tip (“head”) where photoreceptors and frontal sensory organs collect environmental information. The signal surrounds the statocyst and the photoreceptors and is superimposable with the anti-RF-amide antibody pattern (Fig. 1B) and the serotonergic nervous system (42). Constitutive expression of SrNgb1 during embryogenesis and in juvenile and adult stages indicates its implication throughout nervous system development and in maintenance of brain activity.
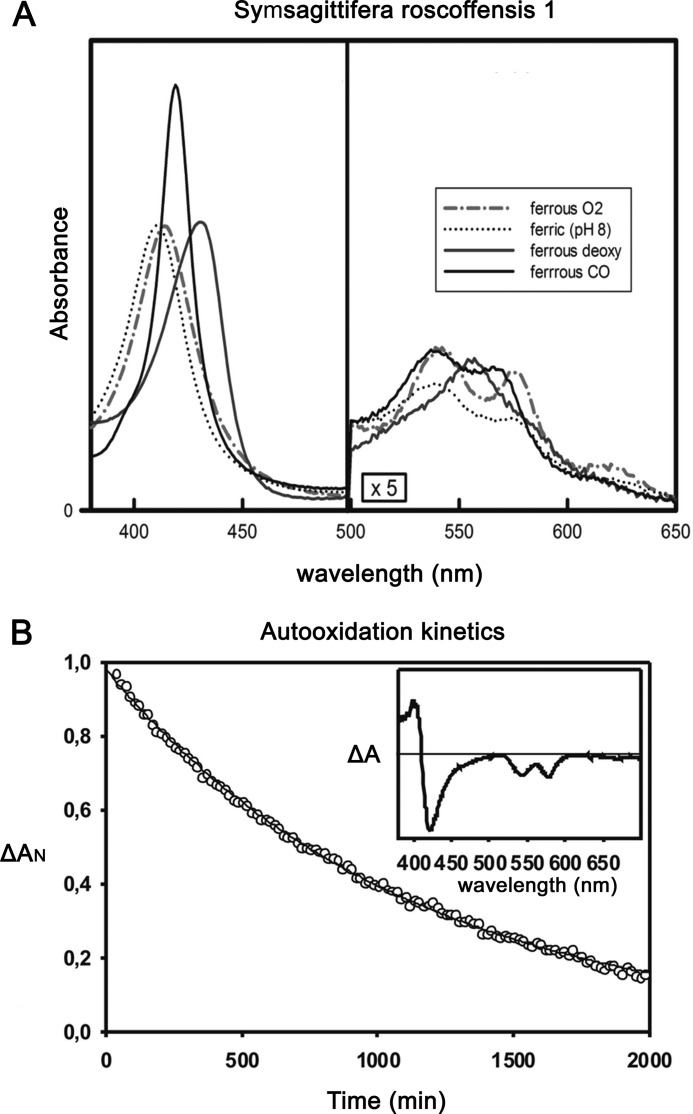
The spectroscopic properties of purified SrNgb1 (UV and visible absorption spectra of the ferrous and ferric forms) indicate that in the absence of external ligands it is pentacoordinated, in contrast to vertebrate Ngbs in which a sixth coordination bond is formed with a distal histidine (Fig. 2A). The rate constants of O2 and CO binding and of O2 dissociation are similar to those of vertebrate Mbs, and consequently so is its O2 binding affinity (Table 2). The rate of heme autoxidation under pure O2 at 25 °C is slow (first order rate 0.053 h−1; Fig. 2B), which is not surprising in view of the fact that there is a well established inverse relationship between O2 affinity and autoxidation rate for pentacoordinated globins. This reaction is much slower than those observed for vertebrate Ngbs, probably due to a higher capacity of the hexacoordinated form for transferring an electron to molecular O2 (43). Overall, these observations are consistent with an in vivo function involving reversible binding of the diatomic ligand rather than a redox reaction with O2 as a terminal electron acceptor.
FIGURE 2.
A, UV and visible spectra for SrNgb1. Inset, partially oxygenated spectrum measured under an oxygen tension of 10 Torr. The dashed lines refer to the maximum absorption for the full oxy and deoxy spectra. B, autoxidation of SrNgb1 in 50 mm Tris-HCl, 0.2 mm EDTA, 10 units superoxide dismutase, and catalase at pH 8.0 under 1 atmosphere of O2 at 25 °C.
TABLE 2.
O2 and CO binding data
Experimental conditions: 50 mm Tris-HCl, 100 mm NaCl, 5 mm DTT, pH 8.0. Human Ngb experimental conditions: 100 mm potassium phosphate, 2 mm DTT, pH 7.0. O2 solubility coefficient was 1.82 × 10−6 mol/liter at 25 °C, and for CO solubility the coefficient was 1.36 × 10−6 mol/liter at 25 °C. O2 affinity was estimated equal to 1.8 Torr.
| Binding data | SrNgb1 | Human Mb | Human Ngb (25 °C) |
|---|---|---|---|
| kon CO (μm/s) | 0.35 | 0.65 | 40 |
| kon O2 (μm/s) | 7 | 15 | 170 |
| koff O2 (/s) | 35 ± 5 | 27 | 0.7 |
| K O2 (μm) | 5.0 | 1.8 | 0.004 |
| P50 O2 (Torr) | 2.8 | 1 | 6.8 |
| kon His (s) | 1800 | ||
| koff His (s) | 0.6 | ||
| K His | 3000 |
The Structure of S. roscoffensis Neuroglobin
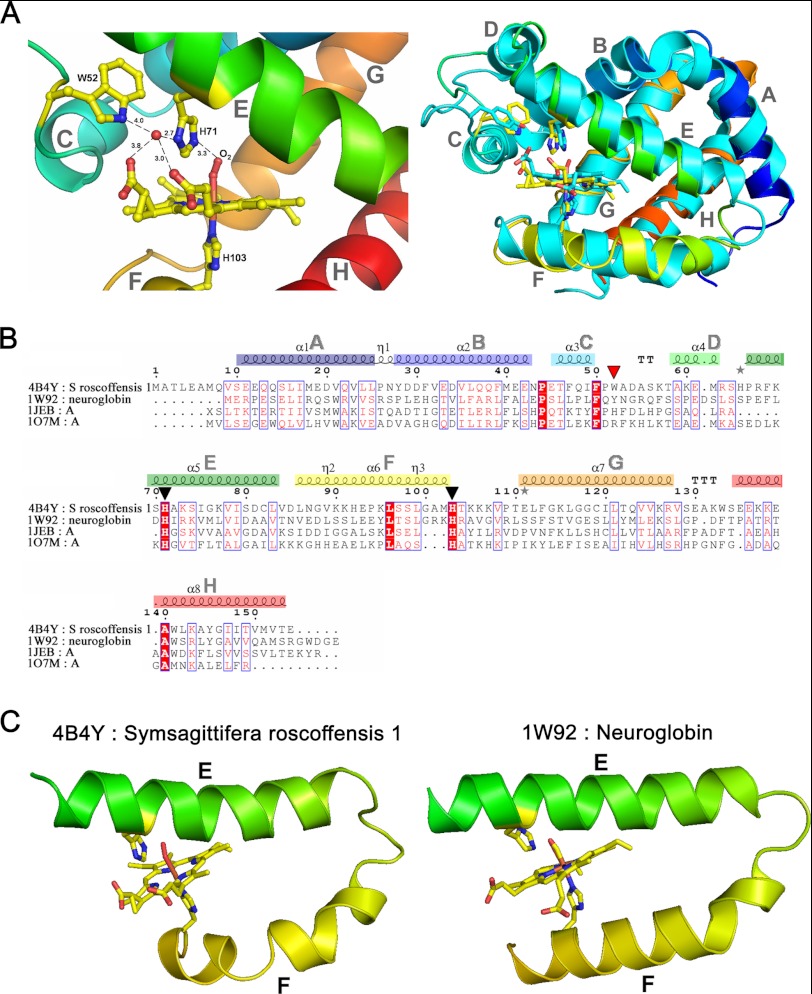
The structural model consists of 149 residues (including Ala-6 to Glu-154) that bind a heme b prosthetic group, with a bond between the heme iron and the proximal histidine (His-103), the distal ligand being an O2 molecule (Fig. 3A). The tertiary structure corresponds to the classical globin fold, consisting of eight helices (A–H, Fig. 3A), the heme binding cleft formed by helices E and F. Despite being deoxy-pentacoordinated, SrNgb1 shares certain structural features with vertebrate Ngbs that are distinct from classical Hb and Mb structures. Although the identity of SrNgb1 with mouse Ngb is only 19% (Fig. 3B), all of the conserved globin fold residues (44) are present, including the heme ligand residues E His-71 and F His-103. The C and D helix regions most closely resemble those described in murine Ngb (45). The Trp residue at position 52 in SrNgb1 (Fig. 3A) may present a ligand barrier and stabilize the heme pocket by forming a stable hydrogen bond to one of the heme propionates (distance 2.8 Å; Fig. 3A). In addition, a water molecule is located nearby (heme-propionate-O2D/HOH, distance 3.8 Å; HOH117), which is hydrogen-bonded to the distal histidine (ND1, 2.7 Å) and the second propionate group of the heme (HOH/heme-O2D, 3.0 Å). In murine Ngb, residues Lys-67 and Tyr-44 form a similar hydrogen-bonding network involving a water molecule also binding to the distal His (45). Structural equivalence is provided by superimposition of HOH117 with its murine counterpart and by superimposition of the Tyr-44 OH-group in murine Ngb with the Trp-52 NH-group in SrNgb1. Moreover, Tyr-44 in murine Ngb and Trp-52 in SrNgb1 are at equivalent positions in the sequence alignment (Fig. 3B). SrNgb1 also shares with murine Ngb the high flexibility of the connection between helices E and F (data not shown).
FIGURE 3.
A, ribbon representation of SrNgb1 crystal structure (4B4Y). Left, close-up view of the heme binding pocket of SrNgb1 highlighting the hydrogen bonding network involving the distal heme binding position and a tightly bound water molecule (HOH117). Right, three-dimensional structural super imposition with murine Ngb (1W92). B, multiple sequence alignment based on the structural superimposition with murine Ngb (1W92), bovine hemoglobin (1JEB), and sperm whale myoglobin (107M), as obtained with the program ESPRIPT. The conserved histidines (axial heme ligands) are marked by black triangles. The red triangle marks a Trp residue involved in the tight binding of a water molecule in the distal heme pocket. The eight helices that form the classical globin fold are labeled from A to H and color-coded from blue (N terminus) to red (C terminus) in the same manner as in the ribbon representation of SrNgb1 in Fig. 1B. C, extract of the SrNgb1 crystal structure highlighting the relative orientations of the heme-ligand containing helixes E and F. A proline at position 94 in helix F leads to a discontinuous and bent helix F in SrNgb1 (4B4Y). The same structural extract showing the relative orientations of helixes E and F in murine Ngb (1W92), where helix F is continuous and straight.
SrNgb1 displays a unique feature in that helix F is bent by the presence of a proline (Pro-94) (Fig. 3C). This could provide some flexibility for a conformational change, analogous to the transition of human Ngb structures triggered by a disulfide bond in the CD region (46). The closest match to SrNbg in the PDB database was ferrous CO-bound murine Ngb (1W92). Overall, the SrNgb1 structural sequence matches Ngbs and plant Hbs, with a slightly better Z-score (47) than to Mbs (data not shown).
In the Cnidarian C. hemisphaerica, Two Globins (CheNgb1 and CheNgb2) Are Expressed in Differentiating Neurosensory Cells
Nematocytes exhibit many characteristics of neurosensory cells, including mechanosensitive cilia, neurite-like outgrowths, and synapses. They contain a single-use dart specialized for killing prey. Nematogenesis (the generation of nematocytes) in Cnidaria is used as a model for non-bilaterian neurogenesis (26, 48), as these neural cells are continuously generated throughout larval and adult life.
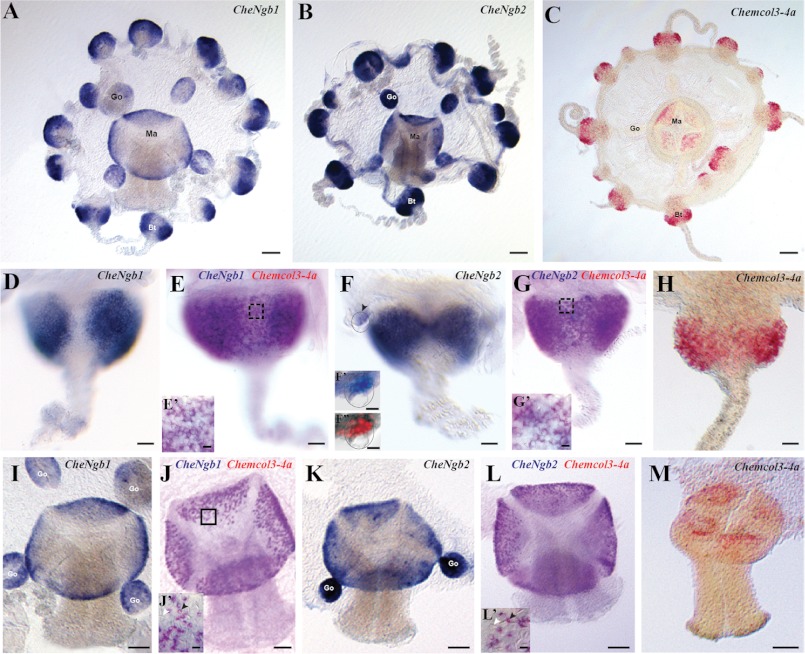
The CheNgb1 and CheNgb2 genes are mainly expressed in the nematogenic ectoderm of tentacle bulbs and manubrium (Fig. 4, A–C). In the tentacle bulbs, their expression patterns are crescent-shaped and interrupted on the external side of the bulb (blue staining in Fig. 4, D–F), thus exactly matching the expression of minicollagen 3-4a (red staining in Fig. 4H). The latter belongs to a family of small collagen-like proteins known in hydrozoans to be a major component of the nematocyst wall (32). Double in situ hybridizations revealed extensive co-expression of minicollagen 3-4a with both CheNgb1 (purple color in Fig. 4E) and CheNgb2 (purple color in Fig. 4G), indicating that both genes are expressed in differentiating nematoblasts over a large time window.
FIGURE 4.
Expression patterns of the two Clytia globin genes CheNgb1 and CheNgb2. A–C, whole mount in situ hybridizations for CheNg1, CheNg2 (blue = NBT/BCIP development) and Chemcol3–4a (red = fast red development) are shown. D–H, all bulbs have the same orientation, proximal area on the top and tentacle on the bottom. Crescent-shaped expression patterns are in the ectodermal layer of a tentacle bulb. All patterns are interrupted on the external side of the bulb, but sometimes the continuity of the staining on the inner face is visible (E and G). E and G, purple staining indicates expression of two genes. In each case, minicollagen 3-4a staining was revealed first with fast red, and then the other probe was revealed using NBT/BCIP. E′ and G′, details show staining in nematoblasts (black arrowhead). F′ and F″, higher magnification of a statocyst is delimited by the dotted circle. In situ hybridization (in black and white) and DAPI counterstaining (in red) merged after conversion of the in situ staining in gray scale. I–M, gene expression shown in the manubrium views (mouth on the bottom). The signal is concentrated in the ectoderm layer at the base of the manubrium. J′ and L′, detailed views show double-stained cells corresponding to nematoblasts (black arrowhead). Note that there is no signal in mature nematocytes (white arrowhead). go, gonad; ma, manubrium; tb, tentacle bulb. Scale bars: 100 μm (A–C); 25 μm (D–H), 50 μm (I–M), 5 μm (E′, G′, J′, L′), 10 μm (F′ and F″).
CheNgb2 mRNA was also detected in the statocysts (Fig. 4F, arrowhead, F′, and F″), the equilibration organs arranged regularly around the rim of the bell of the animal. CheNgb2-expressing cells are located in the basal epithelium of the statocyst, near the bell margin and interpreted as ciliated mechanosensory cells (Fig. 4, F′ and F″).
CheNgb1 and CheNgb2 transcripts were also abundant in the proximal part of the manubrium ectoderm and mimicked the expression pattern of minicollagen, with which they are co-expressed as demonstrated by double in situ hybridization. CheNgb1 and CheNgb2 were also localized in the female gonad in an unidentified cell type (not germ line cells) (Fig. 4, A and B).
Neuroglobins Are Ubiquitously Expressed in Metazoa
Using SrNgb1 as an in silico probe for blasting genomic resources, we identified 50 or so previously undescribed transcripts from different metazoan phyla (supplemental Table S1). These were mostly related to other Ngb/Ngb-like sequences according to classical blastp searches against the NCBI nonredundant nucleotide database. After systematic cross with the Panther predictive tool, all sequences were found to belong to the leghemoglobin (Lgb)-related family that encompasses 14 subfamilies including Ngb, GbX, nonsymbiotic Hb, and Lgb. None of the new sequences was related to the Hb family that includes vertebrate Hb, Cygb, and Mb.
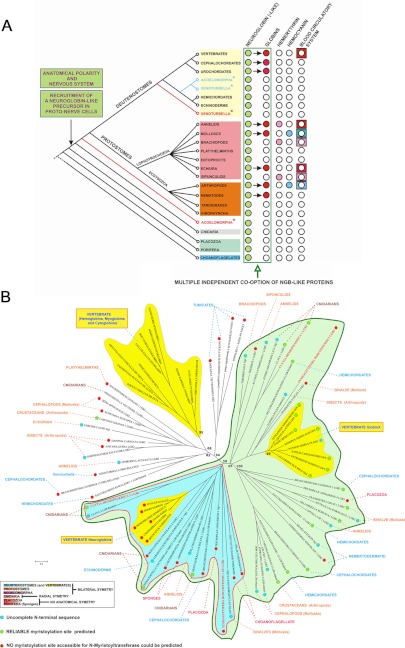
The taxonomic distribution of the Ngb-related sequences suggests broad conservation throughout metazoan evolution (Fig. 5A and supplemental Table S1). They were detected in nonsymmetrical body plan basal metazoans with neither nervous system nor circulatory system, i.e. in the metazoan lineages Porifera (the sponges Amphimedon queenslandica and Carterospongia foliascens) and Placozoa (Trichoplax adherens). In the radially symmetrical cnidarians which have a simple nervous system but no circulatory blood system, Ngb-like sequences were present in Anthozoa (the coral Montastraea faveolata and the sea anemones Anemonia viridis and Nematostella vectensis) and Hydrozoa (C. hemisphaerica and Hydra magnipapillata). No other types of globin (neither homologs of circulating Hbs nor Mb-like globins) were detected in these basal metazoans. In protostomes, expressed Ngb-like sequences were found in (i) cephalopod molluscs such as the cuttlefish Sepia officinalis and Euprimna scolopes and the squid Dorytuthis paeleii; (ii) many arthropods such as the hymenopter Apis mellifera (bee), the crustaceans Carcinus maenas (green shore crab) and Daphnia pulex (a common species of water flea) and the insect Harpegnathos saltator (ant); (iii) the sipunculid Themiste sp. (peanut worm); (iv) the brachiopod Terebratalia transversa (common lampshell); (v) various annelids such as the polychaetes Alvinella pompejana (Pompeii worm from deep-sea hydrothermal vents) or the hirudinea Hellobdella robusta (leech). Expressed Ngb-like sequences were also identified in so called “minor phyla” such as platyhelminthes, tardigrads, kinorhynchs, and nemertodermatids (a sister group of acoels) (supplemental Table S1). In deuterostomes, Ngb-like sequences were identified in all phyla preceding the emergence of vertebrates: in the echinoderms Strongilocentrotus purpuratus and Paracentrotus lividus (sea urchins), the hemichordates Saccoglossus kowalevskii (acorn worm) and Balanoglossus clavigerus, the cephalochordate Branchiostoma lanceolatum (amphioxus, also known as lancelet), and the urochordates Molgula tectiformis and Botrylus schlosseri (tunicates).
FIGURE 5.
A, schematic and consensual representation of metazoan phylogeny illustrating the presence/absence of Ngbs, other globins, the two other respiratory proteins (hemocyanin and hemerythin) and blood circulatory systems. The sporadic presence of globins in certain metazoan lineages can be explained by independent functional shifts from Ngb-like proteins (i.e. convergent evolution). Acoelomorphs and Xenoturbella are represented in two alternative phylogenetic positions, reflecting the ongoing debate as to their affiliations. B, unrooted molecular phylogeny based on multiple alignments of a subset of 84 sequences that comprise 138 amino acids of Ngbs, Lgb-related (Ngb-like), Hb, Mb, and Cygb sequences from diverse phyla. Dots indicate a possible myristoylated Ngb-like (green) or not (red). Vertebrate globins are in the yellow clusters. Hb, Mb, and Cygb appear to be an invention of vertebrates whereas vertebrate Ngbs and GbXs are embedded within the large green group where the functional Ngbs of S. roscoffensis and C. hemisphaerica occur (respective names are in bold red). The blue cluster that includes Ngb-like sequences from Choanoflagellates, Porifera (sponges), Placozoa, some Cnidaria, some protostome and deuterostomes including vertebrate Ngbs (yellow cluster) likely represents plesiomorphic Ngbs.
Vertebrate species have a single Ngb gene copy whereas many of the other metazoans have several copies, indicating gene duplication events correlated with subfunctionalization. The existence of a second Ngb sequence in both S. roscoffensis and C. hemisphaerica (supplemental Table S1) illustrates classical cases of diversification by gene duplication. The unrooted molecular phylogenetic tree (Fig. 5B) clearly shows that vertebrate Hbs, Mbs, and Cygbs form a distinct monophyletic group (Fig. 5B), in agreement with earlier results (18, 49). Vertebrate Ngbs and GbXs are included in a group of functional Ngbs and Ngb-related sequences that includes the Ngbs of S. roscoffensis and C. hemisphaerica characterized in this study. The presence of vertebrate GbX sequences in this group supports a connection of these proteins with neural systems. The cluster that contains the Lgb-related sequences of choanoflagellates (the closest living unicellular relative of metazoans (38)), as well as the poriferan and vertebrate Ngb sequences likely represents the ancestral Ngb lineage with plesiomorphic characteristics. In Blast results, choanoflagellate, poriferan, cnidarian, and S. roscoffensis Ngbs produced significant alignments with globins from protists, notably those of the unicellular green alga Micromonas and the diatom (unicellular brown alga) Thalassiosira that both exhibit a Lgb-related signature according to the Panther prediction system. These findings support the hypothesis that metazoan globins were likely inherited from a unicellular eukaryotic ancestor. The second cluster with SrNgb2, CheNgb1, and vertebrate GbX represents another cluster of Ngb-related sequences. The other sequences diagnosed as putative Ngb-related proteins (with a Lgb-related signature) that do not cluster specifically within the Ngb group reflect primary sequence divergence and likely species-specific diversification. Further exploratory approaches such as gene or protein expression localization will be required to formally establish the role of these proteins (including GbX) in the nervous system.
When the coding sequences we recovered were complete, we also noticed that some Ngb-related sequences exhibit a myristoylation site whereas others do not, with no clear pattern in the phylogenetic tree (Fig. 5B). Our molecular phylogeny is inevitably based on a heterogeneous subset of paralogous and orthologous Ngb-like sequences, but as transcriptomes do not reveal all transcripts (and especially those of cryptically expressed genes with a low number of corresponding transcripts), the number of Ngb-like proteins is likely to be significantly underestimated.
Neuroglobin Is Likely an Early Constitutive Actor in Nervous Systems and Brain Evolution
It is clear that Ngb-like proteins are ubiquitous in metazoans (Fig. 5A). The emergence of neural structures in metazoans leads to innovation in protein functions (50). Although the origin of nerve cells remains unknown, the Cnidaria, whose name derived from cnidocytes (i.e. nematocytes), occupy a key position with respect to early nervous system evolution in metazoans (51). Together with the ctenophores, the Cnidaria form the Coelenterata, the sister group of Bilateria (52). It is assumed that transduction of chemical and mechanical stimuli in nematocytes are hallmarks of primitive nerve cells and that nematocytes are thus representative of ancestral sensory cells that preceded the differentiation of neuronal cell types in animal evolution (53). The unequivocal expression of Ngbs in nematocytes of the jellyfish C. hemisphaerica appears to be a robust indication of the essential role of these proteins in early evolution of the nervous system. The fact that acoel and jellyfish statocysts (the sensory organs measuring pressure) are, respectively and specifically, targeted by Ngb antibody and Ngb probes illustrates the intimate connection of Ngbs with nerve nets and transmission of information. We assume that an original exaptation, i.e. the recruitment of a globin by proto-nervous cells and proto-nervous circuitry, laid the foundations for elaborate nervous systems and brains in the first metazoans displaying anatomical polarity (radial then bilateral symmetry) and differentiated nervous systems. Ngb precursors are likely homologous to those identified in unicellular eukaryotes (choanoflagellates) and simple metazoans (sponges and placozoans) devoid of neural cells, but possessing the basic genetic toolkit encoding proteins homologous to those involved in nervous system development in higher animals (54, 55) (Fig. 5A).
The deleterious effects on nerve cells of Ngb silencing (10, 56) and the conservation of this protein throughout metazoan evolution underline the pivotal function of Ngbs in development and physiology of neurons. Subcellular expression of Ngb in mitochondria of neuronal cells in regions of the brain with high metabolic activity (10, 57) is an indicator of the implication of Ngb in cellular homeostasis in extant organisms and, by extension, in early metazoan neuronal cells. The Ngb-like sequences of certain cnidarians, protostomes, and deuterostomes exhibit a predicted N-terminal myristoylation site, indicating a possible interaction with membranes, putatively including those of the mitochondria (Fig. 5B). The presence of such a site has already been described for the (Ngb-like) globin expressed in the gills of the crab C. maenas (58).
In the core of the globin fold, hexacoordination of the heme iron atom leads to a high autoxidation rate, suggesting that hexacoordinated vertebrate Ngbs are involved in redox metabolism connected to oxidative phosphorylation (59). Our results show that some Ngbs, such as SrNgb1, can be functionally pentacoordinated. SrNgb1, whose O2 binding affinity is similar to that of Mb, is likely to be involved in O2 storage. This proposal is in agreement with the most likely roles of nerve Hbs in the annelid (Aphrodite aculeata), the clams (Spisula solidissima and Tellina alternata) and the nemertean (Cerebratulus lacteus), which have been established to be the provision of O2 to the metabolically highly active neural cells and thus protection under hypoxic conditions (12, 13, 60, 61).
It remains to be determined which form of coordination (penta- or hexa-) of metazoan Ngbs was associated with neofunctionalization and which was the ancestral state. It is pertinent to note that human Ngb exists as an equilibrium between the two forms, with the hexacoordinated form being dominant (∼99:1) (9).
Neuroglobins Could Also Be Precursors of the Metazoan Globin Repertoire
The results of our survey highlight the presence of putative Ngb proteins in radial and bilateral animals irrespective of the presence or absence of a blood circulatory system and of the respiratory protein employed (hemocyanin in molluscs and arthropods, hemerythrin in sipunculids and brachiopods, hemoglobin in other metazoans). The presence of Ngb in ice fish, where circulating Hb has disappeared, is not paradoxical as claimed by Cheng et al. (62), but illustrates the separate evolutionary pathways of Ngbs and O2-binding Hbs, the mandatory constitutive expression of Ngb in the nervous system, and a clear case of disadaptation, i.e. loss of the circulating oxygen carrier.
Assuming that the ancestral bilaterian body plan had a simple nervous system but no blood circulatory system, it is obvious that the presence of Ngb predates the emergence of circulatory Hb. Given that Ngbs are ancestral and constitutively expressed in all metazoans (Fig. 5A), the sporadic presence of O2-binding Hb in individual metazoan lineages strongly suggests that they are polyphyletic (i.e. due to convergent evolution). The globin lineages other than Ngb found in many metazoan groups probably emerged as the result of functionalization (63) and co-option of a Ngb-like globin in early metazoans. Most of the metazoan transcriptomes analyzed in this study exhibit multiple Ngb-like paralogs, likely originating from gene duplication events.
CONCLUSION
We demonstrate the presence of a functional Ngb in neural cells of the acoel S. roscoffensis and expression of homologous Ngbs specific to neurosensory cells (differentiating nematoblasts) in the cnidarian jellyfish C. hemisphaerica. These results suggest that the first globins expressed in early bilaterians and symmetrically radial cnidarians were specifically linked to the metazoan nervous system. The pentacoordination of SrNgb1 vis à vis the hexacoordination of vertebrate Ngbs may be due to differences in function, with the acoel Ngb playing an O2 storage role providing neuroprotection during hypoxic periods. This interpretation is supported by reports of the functions of “nerve globin” in several protostomes.
Extensive in silico mining of genomic data using SrNgb1 as a probe revealed the occurrence of expressed Ngb-like sequences in most metazoan phyla, including sponges and placozoa, basal metazoans lacking neural and circulatory systems. Our results clearly demonstrate that the emergence of Ngb in metazoans chronologically preceded the emergence of other globin families. Consequently, we propose a novel scenario for metazoan globin evolution, based on two broad and complementary statements. On the one hand, our experimental and in silico results suggest that an ancestral globin-like gene was recruited in the emerging protoneural system in the ancestor of bilateria and diploblastic animals to become a functional Ngb. On the other hand, metazoan globins other than Ngbs, such as annelid, mollusc, arthropod, and vertebrate Hbs, likely originated independently from early Ngbs, via co-option of duplicated Ngb genes and functionalization during metazoan radiation, concomitant with increasing body plan complexity and the emergence of blood circulatory systems.
Access to multiple ontogenetic stages of emerging marine models, for which genomic resources and molecular tools are increasingly available (64), will be of a prime importance for functional genomic exploration using Ngbs as developmental markers in, for example, animal lineages exhibiting complex nervous tissues (cephalopods) or subject to anthropogenically induced stresses or diseases (corals, mussels, oysters).
Acknowledgments
Crystal structure determination was performed at the crystallography platform of the Station Biologique de Roscoff, supported by the Region Bretagne, the Centre National de la Recherche Scientifique, and Université Pierre et Marie Curie, Paris 06. We are indebted to the staff of the European Synchrotron Radiation Facilities (ESRF, Grenoble, France), beamline ID23-I and BM30A, for technical support during data collection and treatment. Confocal microscopy was performed in the imaging facility platform Merimage at the Station Biologique de Roscoff. Spectroscopic studies were supported by INSERM and the University Paris 11. We thank Ian Probert from the Marine Resource Center of the Station Biologique de Roscoff for discussions and critical reading of the manuscript.

This article contains supplemental Table S1.
The atomic coordinates and structure factors (code 4B4Y) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Hb
- hemoglobin
- BCIP
- bromochlorylindolophosphate
- Cyg
- cytoglobin
- GbX
- Globin X
- Lgb
- leghemoglobin
- Mb
- myoglobin
- NBT
- nitro blue tetrazolium
- Ngb
- neuroglobin
- PDB
- Protein Data Bank
- SrNgb1
- S. roscoffensis Ngb
- NBT
- nitro-blue tetrazolium chloride.
REFERENCES
- 1. Kendrew J. C., Bodo G., Dintzis H. M., Parrish R. G., Wyckoff H., Phillips D. C. (1958) A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature 181, 662–666 [DOI] [PubMed] [Google Scholar]
- 2. Perutz M. F., Rossmann M. G., Cullis A. F., Muirhead H., Will G., North A. C. (1960) Structure of haemoglobin: a three-dimensional Fourier synthesis at 5.5-Å resolution, obtained by x-ray analysis. Nature 185, 416–422 [DOI] [PubMed] [Google Scholar]
- 3. Zuckerkandl E., Pauling L. (1965) Molecules as documents of evolutionary history. J. Theor. Biol. 8, 357–366 [DOI] [PubMed] [Google Scholar]
- 4. Vinogradov S. N., Hoogewijs D., Bailly X., Arredondo-Peter R., Guertin M., Gough J., Dewilde S., Moens L., Vanfleteren J. R. (2005) Three globin lineages belonging to two structural classes in genomes from the three kingdoms of life. Proc. Natl. Acad. Sci. U.S.A. 102, 11385–11389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burmester T., Weich B., Reinhardt S., Hankeln T. (2000) A vertebrate globin expressed in the brain. Nature 407, 520–523 [DOI] [PubMed] [Google Scholar]
- 6. Burmester T., Gerlach F., Hankeln T. (2007) Regulation and role of neuroglobin and cytoglobin under hypoxia. Adv. Exp. Med. Biol. 618, 169–180 [DOI] [PubMed] [Google Scholar]
- 7. Li W., Wu Y., Ren C., Lu Y., Gao Y., Zheng X., Zhang C. (2011) The activity of recombinant human neuroglobin as an antioxidant and free radical scavenger. Proteins 79, 115–125 [DOI] [PubMed] [Google Scholar]
- 8. Brittain T., Skommer J., Henty K., Birch N., Raychaudhuri S. (2010) A role for human neuroglobin in apoptosis. IUBMB Life 62, 878–885 [DOI] [PubMed] [Google Scholar]
- 9. Tiso M., Tejero J., Basu S., Azarov I., Wang X., Simplaceanu V., Frizzell S., Jayaraman T., Geary L., Shapiro C., Ho C., Shiva S., Kim-Shapiro D. B., Gladwin M. T. (2011) Human neuroglobin functions as a redox-regulated nitrite reductase. J. Biol. Chem. 286, 18277–18289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lechauve C., Augustin S., Cwerman-Thibault H., Bouaita A., Forster V., Célier C., Rustin P., Marden M. C., Sahel J. A., Corral-Debrinski M. (2012) Neuroglobin involvement in respiratory chain function and retinal ganglion cell integrity. Biochim. Biophys. Acta 1823, 2261–2273 [DOI] [PubMed] [Google Scholar]
- 11. DellaValle B., Hempel C., Kurtzhals J. A., Penkowa M. (2010) In vivo expression of neuroglobin in reactive astrocytes during neuropathology in murine models of traumatic brain injury, cerebral malaria, and autoimmune encephalitis. Glia 58, 1220–1227 [DOI] [PubMed] [Google Scholar]
- 12. Wittenberg B. A., Briehl R. W., Wittenberg J. B. (1965) Haemoglobins of invertebrate tissues: nerve haemoglobins of Aphrodite, Aplysia and Halosydna. Biochem. J. 96, 363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Geuens E., Dewilde S., Hoogewijs D., Pesce A., Nienhaus K., Nienhaus G. U., Olson J., Vanfleteren J., Bolognesi M., Moens L. (2004) Nerve globins in invertebrates. IUBMB Life 56, 653–656 [DOI] [PubMed] [Google Scholar]
- 14. Hundahl C., Fago A., Dewilde S., Moens L., Hankeln T., Burmester T., Weber R. E. (2006) Oxygen binding properties of non-mammalian nerve globins. FEBS J. 273, 1323–1329 [DOI] [PubMed] [Google Scholar]
- 15. Hoffmann F. G., Opazo J. C., Storz J. F. (2010) Gene cooption and convergent evolution of oxygen transport hemoglobins in jawed and jawless vertebrates. Proc. Natl. Acad. Sci. U.S.A. 107, 14274–14279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Storz J. F., Opazo J. C., Hoffmann F. G. (2011) Phylogenetic diversification of the globin gene superfamily in chordates. IUBMB Life 63, 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoffmann F. G., Opazo J. C., Storz J. F. (2012) Whole-genome duplications spurred the functional diversification of the globin gene superfamily in vertebrates. Mol. Biol. Evol. 29, 303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoffmann F. G., Opazo J. C., Hoogewijs D., Hankeln T., Ebner B., Vinogradov S. N., Bailly X., Storz J. F. (2012) Evolution of the globin gene family in deuterostomes: lineage-specific patterns of diversification and attrition. Mol. Biol. Evol. 29, 1735–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blank M., Wollberg J., Gerlach F., Reimann K., Roesner A., Hankeln T., Fago A., Weber R. E., Burmester T. (2011) A membrane-bound vertebrate globin. PloS One 6, e25292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roesner A., Fuchs C., Hankeln T., Burmester T. (2005) A globin gene of ancient evolutionary origin in lower vertebrates: evidence for two distinct globin families in animals. Mol. Biol. Evol. 22, 12–20 [DOI] [PubMed] [Google Scholar]
- 21. Dröge J., Makałowski W. (2011) Phylogenetic analysis reveals wide distribution of globin X. Biol. Direct. 6, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bailly X., Vinogradov S. N. (2008) in Dioxygen Binding and Sensing Proteins: A Tribute to Beatrice and Jonathan Wittenberg (Bolognesi M., di Prisco G., Verde C., eds) pp. 191–201, Springer, New York [Google Scholar]
- 23. Bailly X. (2009) Focus on S. roscoffensis. Cahiers Biol. Marine 50, 106–107 [Google Scholar]
- 24. Lowe C. J., Pani A. M. (2011) Animal evolution: a soap opera of unremarkable worms. Curr. Biol. 21, R151–153 [DOI] [PubMed] [Google Scholar]
- 25. Bery A., Cardona A., Martinez P., Hartenstein V. (2010) Structure of the central nervous system of a juvenile acoel, Symsagittifera roscoffensis. Dev. Genes Evol. 220, 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Houliston E., Momose T., Manuel M. (2010) Clytia hemisphaerica: a jellyfish cousin joins the laboratory. Trends Genet. 26, 159–167 [DOI] [PubMed] [Google Scholar]
- 27. Studier F. W. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 28. Uzan J., Dewilde S., Burmester T., Hankeln T., Moens L., Hamdane D., Marden M. C., Kiger L. (2004) Neuroglobin and other hexacoordinated hemoglobins show a weak temperature dependence of oxygen binding. Biophys. J. 87, 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chevalier S., Martin A., Leclère L., Amiel A., Houliston E. (2006) Polarised expression of FoxB and FoxQ2 genes during development of the hydrozoan Clytia hemisphaerica. Dev. Genes Evol. 216, 709–720 [DOI] [PubMed] [Google Scholar]
- 30. Jager M., Quéinnec E., Le Guyader H., Manuel M. (2011) Multiple Sox genes are expressed in stem cells or in differentiating neuro-sensory cells in the hydrozoan Clytia hemisphaerica. Evodevo 2, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chiori R., Jager M., Denker E., Wincker P., Da Silva C., Le Guyader H., Manuel M., Quéinnec E. (2009) Are Hox genes ancestrally involved in axial patterning? Evidence from the hydrozoan Clytia hemisphaerica (Cnidaria). PloS One 4, e4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Denker E., Manuel M., Leclère L., Le Guyader H., Rabet N. (2008) Ordered progression of nematogenesis from stem cells through differentiation stages in the tentacle bulb of Clytia hemisphaerica (Hydrozoa, Cnidaria). Dev. Biol. 315, 99–113 [DOI] [PubMed] [Google Scholar]
- 33. Kabsch W. (1988) Evaluation of single-crystal X-ray diffraction data from a position-sensitive detector. J. Appl. Crystallogr. 21, 916–924 [Google Scholar]
- 34. Navaza J. (2001) Implementation of molecular replacement in AMoRe. Acta Crystallogr. D Biol. Crystallogr. 57, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 35. Schneider T. R., Sheldrick G. M. (2002) Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 36. Pape T., Schneider T. R. (2004) HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr. 37, 843–844 [Google Scholar]
- 37. Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 38. Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 39. Bateman A., Coin L., Durbin R., Finn R. D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E. L., Studholme D. J., Yeats C., Eddy S. R. (2004) The Pfam protein families database. Nucleic Acids Res. 32, D138–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyMl 3.0. Syst. Biol. 59, 307–321 [DOI] [PubMed] [Google Scholar]
- 41. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Semmler H., Chiodin M., Bailly X., Martinez P., Wanninger A. (2010) Steps towards a centralized nervous system in basal bilaterians: insights from neurogenesis of the acoel Symsagittifera roscoffensis. Dev. Growth Differ. 52, 701–713 [DOI] [PubMed] [Google Scholar]
- 43. Brunori M., Vallone B. (2007) Neuroglobin, seven years after. Cell Mol. Life Sci. 64, 1259–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bashford D., Chothia C., Lesk A. M. (1987) Determinants of a protein fold: unique features of the globin amino acid sequences. J. Mol. Biol. 196, 199–216 [DOI] [PubMed] [Google Scholar]
- 45. Vallone B., Nienhaus K., Brunori M., Nienhaus G. U. (2004) The structure of murine neuroglobin: novel pathways for ligand migration and binding. Proteins 56, 85–92 [DOI] [PubMed] [Google Scholar]
- 46. Hamdane D., Kiger L., Dewilde S., Green B. N., Pesce A., Uzan J., Burmester T., Hankeln T., Bolognesi M., Moens L., Marden M. C. (2003) The redox state of the cell regulates the ligand binding affinity of human neuroglobin and cytoglobin. J. Biol. Chem. 278, 51713–51721 [DOI] [PubMed] [Google Scholar]
- 47. Holm L., Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Galliot B., Quiquand M., Ghila L., de Rosa R., Miljkovic-Licina M., Chera S. (2009) Origins of neurogenesis, a cnidarian view. Dev. Biol. 332, 2–24 [DOI] [PubMed] [Google Scholar]
- 49. Burmester T., Hankeln T. (2009) What is the function of neuroglobin? J. Exp. Biol. 212, 1423–1428 [DOI] [PubMed] [Google Scholar]
- 50. Marlow H. Q., Srivastava M., Matus D. Q., Rokhsar D., Martindale M. Q. (2009) Anatomy and development of the nervous system of Nematostella vectensis, an anthozoan cnidarian. Dev. Neurobiol. 69, 235–254 [DOI] [PubMed] [Google Scholar]
- 51. Galliot B. (2010) in Key Transitions in Animal Evolution (Schierwater B., De Salle R., eds) pp. 127–161, Science Publishers & CRC Press, New York [Google Scholar]
- 52. Philippe H., Derelle R., Lopez P., Pick K., Borchiellini C., Boury-Esnault N., Vacelet J., Renard E., Houliston E., Quéinnec E., Da Silva C., Wincker P., Le Guyader H., Leys S., Jackson D. J., Schreiber F., Erpenbeck D., Morgenstern B., Wörheide G., Manuel M. (2009) Phylogenomics revives traditional views on deep animal relationships. Curr. Biol. 19, 706–712 [DOI] [PubMed] [Google Scholar]
- 53. Miljkovic-Licina M., Gauchat D., Galliot B. (2004) Neuronal evolution: analysis of regulatory genes in a first-evolved nervous system, the hydra nervous system. BioSystems 76, 75–87 [DOI] [PubMed] [Google Scholar]
- 54. Srivastava M., Simakov O., Chapman J., Fahey B., Gauthier M. E., Mitros T., Richards G. S., Conaco C., Dacre M., Hellsten U., Larroux C., Putnam N. H., Stanke M., Adamska M., Darling A., Degnan S. M., Oakley T. H., Plachetzki D. C., Zhai Y., Adamski M., Calcino A., Cummins S. F., Goodstein D. M., Harris C., Jackson D. J., Leys S. P., Shu S., Woodcroft B. J., Vervoort M., Kosik K. S., Manning G., Degnan B. M., Rokhsar D. S. (2010) The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Srivastava M., Begovic E., Chapman J., Putnam N. H., Hellsten U., Kawashima T., Kuo A., Mitros T., Salamov A., Carpenter M. L., Signorovitch A. Y., Moreno M. A., Kamm K., Grimwood J., Schmutz J., Shapiro H., Grigoriev I. V., Buss L. W., Schierwater B., Dellaporta S. L., Rokhsar D. S. (2008) The Trichoplax genome and the nature of placozoans. Nature 454, 955–960 [DOI] [PubMed] [Google Scholar]
- 56. Ye S. Q., Zhou X. Y., Lai X. J., Zheng L., Chen X. Q. (2009) Silencing neuroglobin enhances neuronal vulnerability to oxidative injury by down-regulating 14-3-3γ. Acta Pharmacol. Sin. 30, 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hundahl C. A., Allen G. C., Hannibal J., Kjaer K., Rehfeld J. F., Dewilde S., Nyengaard J. R., Kelsen J., Hay-Schmidt A. (2010) Anatomical characterization of cytoglobin and neuroglobin mRNA and protein expression in the mouse brain. Brain Res. 1331, 58–73 [DOI] [PubMed] [Google Scholar]
- 58. Ertas B., Kiger L., Blank M., Marden M. C., Burmester T. (2011) A membrane-bound hemoglobin from gills of the green shore crab Carcinus maenas. J. Biol. Chem. 286, 3185–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fago A., Mathews A. J., Moens L., Dewilde S., Brittain T. (2006) The reaction of neuroglobin with potential redox protein partners cytochrome b5 and cytochrome c. FEBS Lett. 580, 4884–4888 [DOI] [PubMed] [Google Scholar]
- 60. Kraus D. W., Colacino J. M. (1986) Extended oxygen delivery from the nerve hemoglobin of Tellina alternata (Bivalvia). Science 232, 90–92 [DOI] [PubMed] [Google Scholar]
- 61. Doeller J. E., Kraus D. W. (1988) A physiological comparison of bivalve mollusc cerebro-visceral connectives with and without neurohemoglobin. II. Neurohemoglobin characteristics. Biol. Bull. 174, 67–76 [DOI] [PubMed] [Google Scholar]
- 62. Cheng C. H., di Prisco G., Verde C. (2009) The “icefish paradox”: which is the task of neuroglobin in Antarctic hemoglobin-less icefish? IUBMB Life 61, 184–188 [DOI] [PubMed] [Google Scholar]
- 63. Ganfornina M. D., Sánchez D. (1999) Generation of evolutionary novelty by functional shift. Bioessays. 21, 432–439 [DOI] [PubMed] [Google Scholar]
- 64. Nosengo N. (2011) Marine biology network launches into choppy waters. Nature 470, 444–445 [DOI] [PubMed] [Google Scholar]