Abstract
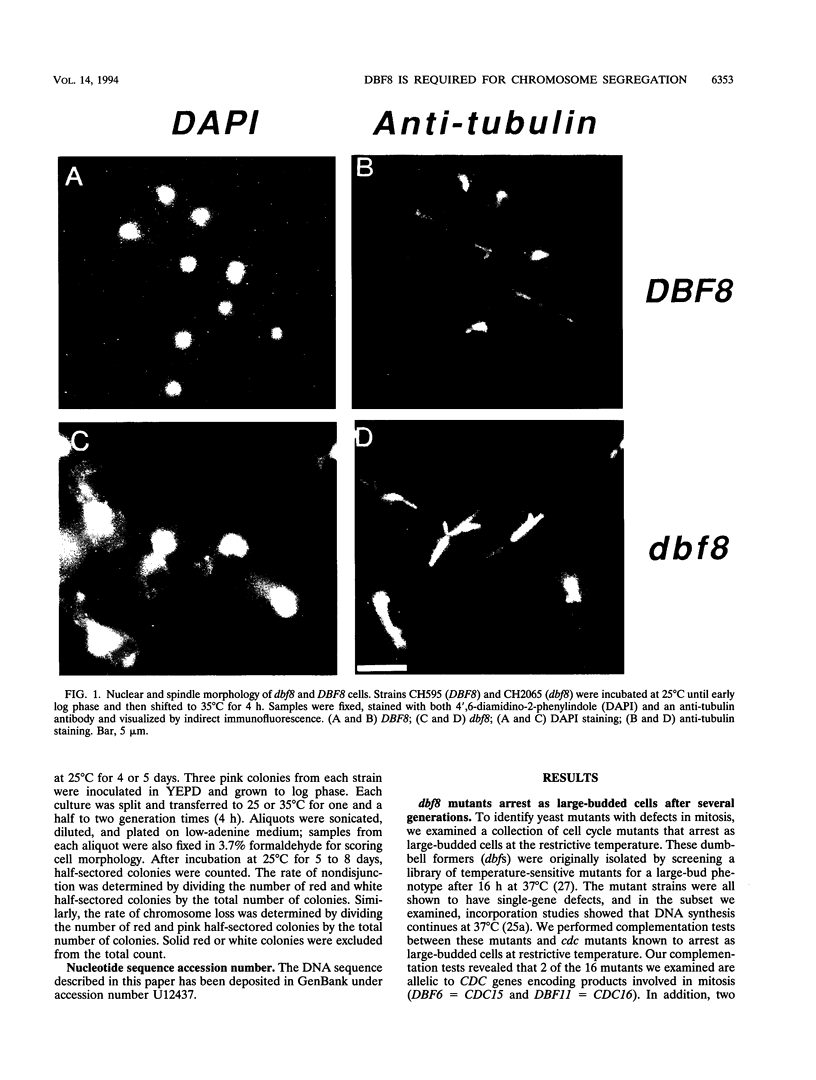
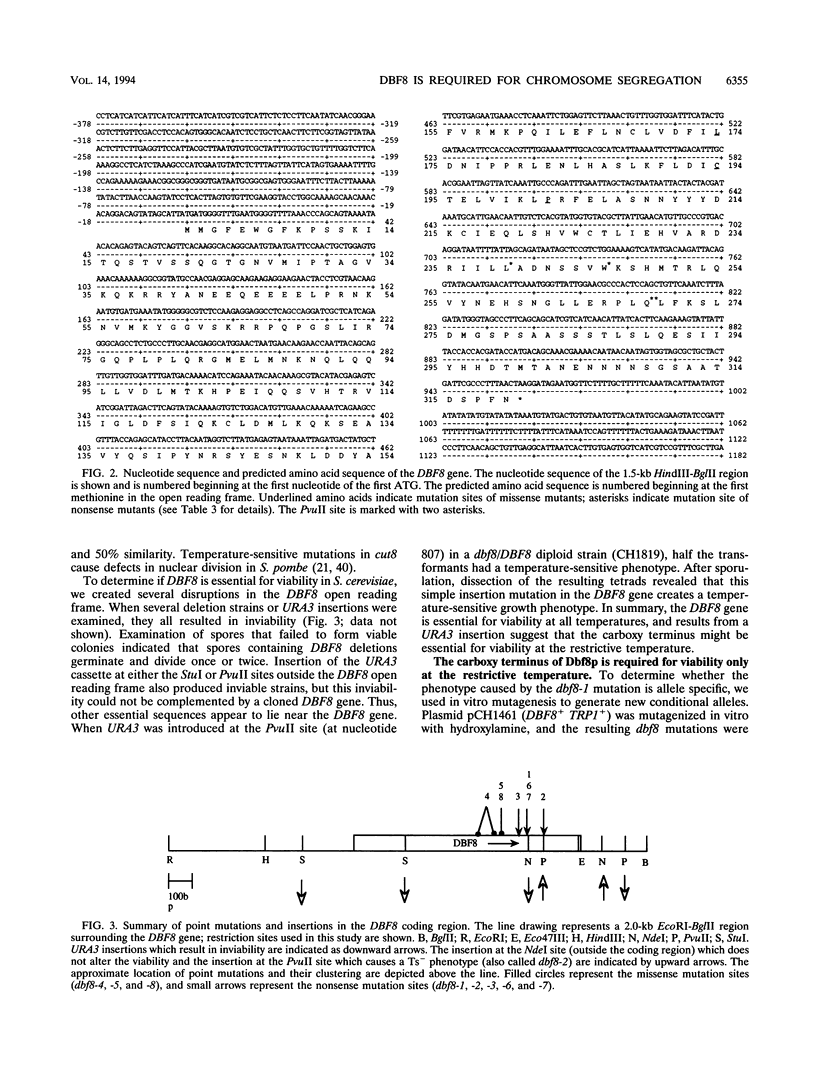
To investigate chromosome segregation in Saccharomyces cerevisiae, we examined a collection of temperature-sensitive mutants that arrest as large-budded cells at restrictive temperatures (L. H. Johnston and A. P. Thomas, Mol. Gen. Genet. 186:439-444, 1982). We characterized dbf8, a mutation that causes cells to arrest with a 2c DNA content and a short spindle. DBF8 maps to chromosome IX near the centromere, and it encodes a 36-kDa protein that is essential for viability at all temperatures. Mutational analysis reveals that three dbf8 alleles are nonsense mutations affecting the carboxy-terminal third of the encoded protein. Since all of these mutations confer temperature sensitivity, it appears that the carboxyl-terminal third of the protein is essential only at a restrictive temperature. In support of this conclusion, an insertion of URA3 at the same position also confers a temperature-sensitive phenotype. Although they show no evidence of DNA damage, dbf8 mutants exhibit increased rates of chromosome loss and nondisjunction even at a permissive temperature. Taken together, our data suggest that Dbf8p plays an essential role in chromosome segregation.
Full text
PDF



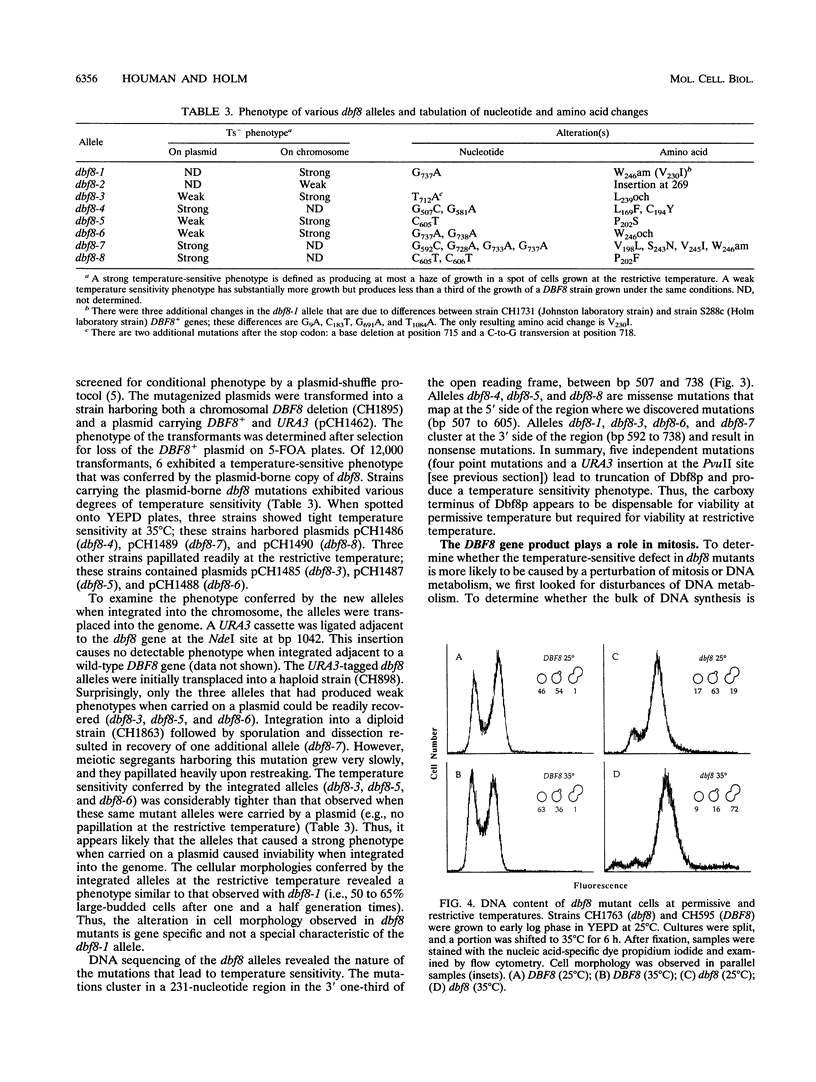

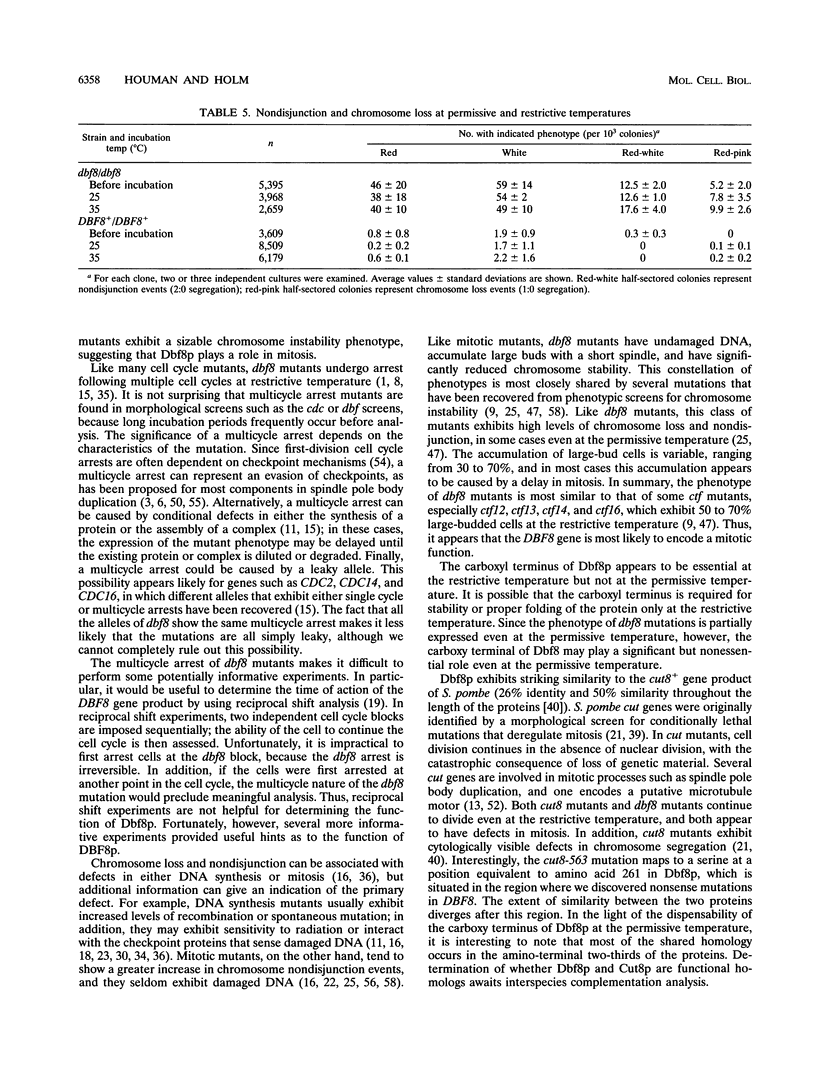


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Araki H., Awane K., Ogawa N., Oshima Y. The CDC26 gene of Saccharomyces cerevisiae is required for cell growth only at high temperature. Mol Gen Genet. 1992 Jan;231(2):329–331. doi: 10.1007/BF00279807. [DOI] [PubMed] [Google Scholar]
- Baum P., Yip C., Goetsch L., Byers B. A yeast gene essential for regulation of spindle pole duplication. Mol Cell Biol. 1988 Dec;8(12):5386–5397. doi: 10.1128/mcb.8.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Budd M., Campbell J. L. Temperature-sensitive mutations in the yeast DNA polymerase I gene. Proc Natl Acad Sci U S A. 1987 May;84(9):2838–2842. doi: 10.1073/pnas.84.9.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. S., Botstein D. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics. 1993 Nov;135(3):677–691. doi: 10.1093/genetics/135.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde J., Fink G. R. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson J. R. Biochemical and genetic studies on the function of, and relationship between, the PGI1- and CDC30-encoded phosphoglucose isomerases in Saccharomyces cerevisiae. J Gen Microbiol. 1991 Apr;137(4):765–770. doi: 10.1099/00221287-137-4-765. [DOI] [PubMed] [Google Scholar]
- Doheny K. F., Sorger P. K., Hyman A. A., Tugendreich S., Spencer F., Hieter P. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993 May 21;73(4):761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerring S. L., Spencer F., Hieter P. The CHL 1 (CTF 1) gene product of Saccharomyces cerevisiae is important for chromosome transmission and normal cell cycle progression in G2/M. EMBO J. 1990 Dec;9(13):4347–4358. doi: 10.1002/j.1460-2075.1990.tb07884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson S. I., Surosky R. T., Tye B. K. The phenotype of the minichromosome maintenance mutant mcm3 is characteristic of mutants defective in DNA replication. Mol Cell Biol. 1990 Nov;10(11):5707–5720. doi: 10.1128/mcb.10.11.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyllensten U. B., Erlich H. A. Generation of single-stranded DNA by the polymerase chain reaction and its application to direct sequencing of the HLA-DQA locus. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7652–7656. doi: 10.1073/pnas.85.20.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I., Yanagida M. Kinesin-related cut7 protein associates with mitotic and meiotic spindles in fission yeast. Nature. 1992 Mar 5;356(6364):74–76. doi: 10.1038/356074a0. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Mortimer R. K., Culotti J., Culotti M. Genetic Control of the Cell Division Cycle in Yeast: V. Genetic Analysis of cdc Mutants. Genetics. 1973 Jun;74(2):267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J Mol Biol. 1976 Jul 15;104(4):803–817. doi: 10.1016/0022-2836(76)90183-2. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics. 1985 Jul;110(3):381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Weinert T. A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989 Nov 3;246(4930):629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hennessy K. M., Lee A., Chen E., Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991 Jun;5(6):958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- Hereford L. M., Hartwell L. H. Sequential gene function in the initiation of Saccharomyces cerevisiae DNA synthesis. J Mol Biol. 1974 Apr 15;84(3):445–461. doi: 10.1016/0022-2836(74)90451-3. [DOI] [PubMed] [Google Scholar]
- Hieter P., Mann C., Snyder M., Davis R. W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985 Feb;40(2):381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- Hirano T., Funahashi S., Uemura T., Yanagida M. Isolation and characterization of Schizosaccharomyces pombe cutmutants that block nuclear division but not cytokinesis. EMBO J. 1986 Nov;5(11):2973–2979. doi: 10.1002/j.1460-2075.1986.tb04594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C., Stearns T., Botstein D. DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol Cell Biol. 1989 Jan;9(1):159–168. doi: 10.1128/mcb.9.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell E. A., McAlear M. A., Rose D., Holm C. CDC44: a putative nucleotide-binding protein required for cell cycle progression that has homology to subunits of replication factor C. Mol Cell Biol. 1994 Jan;14(1):255–267. doi: 10.1128/mcb.14.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M. A., He L., Loo K. K., Saunders W. S. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992 Jul;118(1):109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M. A., Stearns T., Botstein D. Chromosome instability mutants of Saccharomyces cerevisiae that are defective in microtubule-mediated processes. Mol Cell Biol. 1990 Jan;10(1):223–234. doi: 10.1128/mcb.10.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L. H., Eberly S. L., Chapman J. W., Araki H., Sugino A. The product of the Saccharomyces cerevisiae cell cycle gene DBF2 has homology with protein kinases and is periodically expressed in the cell cycle. Mol Cell Biol. 1990 Apr;10(4):1358–1366. doi: 10.1128/mcb.10.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L. H., Thomas A. P. The isolation of new DNA synthesis mutants in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1982;186(3):439–444. doi: 10.1007/BF00729466. [DOI] [PubMed] [Google Scholar]
- Jones J. S., Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990 Sep-Oct;6(5):363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- Kitada K., Johnston L. H., Sugino T., Sugino A. Temperature-sensitive cdc7 mutations of Saccharomyces cerevisiae are suppressed by the DBF4 gene, which is required for the G1/S cell cycle transition. Genetics. 1992 May;131(1):21–29. doi: 10.1093/genetics/131.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N., Kroll E., Bannikov V., Bliskovsky V., Gizatullin R., Kirillov A., Shestopalov B., Zakharyev V., Hieter P., Spencer F. CTF4 (CHL15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992 Dec;12(12):5736–5747. doi: 10.1128/mcb.12.12.5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N., Tsouladze A., Koryabin M., Hieter P., Spencer F., Larionov V. Identification and genetic mapping of CHL genes controlling mitotic chromosome transmission in yeast. Yeast. 1993 Jan;9(1):11–19. doi: 10.1002/yea.320090103. [DOI] [PubMed] [Google Scholar]
- Meluh P. B., Rose M. D. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990 Mar 23;60(6):1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Miles J., Formosa T. Evidence that POB1, a Saccharomyces cerevisiae protein that binds to DNA polymerase alpha, acts in DNA metabolism in vivo. Mol Cell Biol. 1992 Dec;12(12):5724–5735. doi: 10.1128/mcb.12.12.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabito P. M., Morris N. R. BIMA, a TPR-containing protein required for mitosis, localizes to the spindle pole body in Aspergillus nidulans. J Cell Biol. 1993 Feb;120(4):959–968. doi: 10.1083/jcb.120.4.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer R. E., Hogan E., Koshland D. Mitotic transmission of artificial chromosomes in cdc mutants of the yeast, Saccharomyces cerevisiae. Genetics. 1990 Aug;125(4):763–774. doi: 10.1093/genetics/125.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn M. D., Thireos G., Greer H. Temporal analysis of general control of amino acid biosynthesis in Saccharomyces cerevisiae: role of positive regulatory genes in initiation and maintenance of mRNA derepression. Mol Cell Biol. 1984 Mar;4(3):520–528. doi: 10.1128/mcb.4.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Fink G. R. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 1987 Mar 27;48(6):1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- Samejima I., Matsumoto T., Nakaseko Y., Beach D., Yanagida M. Identification of seven new cut genes involved in Schizosaccharomyces pombe mitosis. J Cell Sci. 1993 May;105(Pt 1):135–143. doi: 10.1242/jcs.105.1.135. [DOI] [PubMed] [Google Scholar]
- Samejima I., Yanagida M. Identification of cut8+ and cek1+, a novel protein kinase gene, which complement a fission yeast mutation that blocks anaphase. Mol Cell Biol. 1994 Sep;14(9):6361–6371. doi: 10.1128/mcb.14.9.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengstag C., Hinnen A. The sequence of the Saccharomyces cerevisiae gene PHO2 codes for a regulatory protein with unusual aminoacid composition. Nucleic Acids Res. 1987 Jan 12;15(1):233–246. doi: 10.1093/nar/15.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi N., Monteagudo M. C., Koshland D., Hogan E., Burke D. J. The CDC20 gene product of Saccharomyces cerevisiae, a beta-transducin homolog, is required for a subset of microtubule-dependent cellular processes. Mol Cell Biol. 1991 Nov;11(11):5592–5602. doi: 10.1128/mcb.11.11.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spencer F., Gerring S. L., Connelly C., Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990 Feb;124(2):237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Thomas J. H., Botstein D. A gene required for the separation of chromosomes on the spindle apparatus in yeast. Cell. 1986 Jan 17;44(1):65–76. doi: 10.1016/0092-8674(86)90485-x. [DOI] [PubMed] [Google Scholar]
- Toyn J. H., Araki H., Sugino A., Johnston L. H. The cell-cycle-regulated budding yeast gene DBF2, encoding a putative protein kinase, has a homologue that is not under cell-cycle control. Gene. 1991 Jul 31;104(1):63–70. doi: 10.1016/0378-1119(91)90465-n. [DOI] [PubMed] [Google Scholar]
- Uzawa S., Samejima I., Hirano T., Tanaka K., Yanagida M. The fission yeast cut1+ gene regulates spindle pole body duplication and has homology to the budding yeast ESP1 gene. Cell. 1990 Sep 7;62(5):913–925. doi: 10.1016/0092-8674(90)90266-h. [DOI] [PubMed] [Google Scholar]
- Wahle E., Keller W. The biochemistry of 3'-end cleavage and polyadenylation of messenger RNA precursors. Annu Rev Biochem. 1992;61:419–440. doi: 10.1146/annurev.bi.61.070192.002223. [DOI] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993 May;134(1):63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Goetsch L., Baum P., Byers B. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J Cell Biol. 1991 Aug;114(4):745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J. S., Hartwell L. H. A dependent pathway of gene functions leading to chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1982 Sep;94(3):718–726. doi: 10.1083/jcb.94.3.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z., McGrew J. T., Schroeder A. J., Fitzgerald-Hayes M. CSE1 and CSE2, two new genes required for accurate mitotic chromosome segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Aug;13(8):4691–4702. doi: 10.1128/mcb.13.8.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]