Background: The GreenCut is a group of ∼600 green-lineage-specific proteins hypothetically involved in photosynthesis.
Results: A Chlamydomonas reinhardtii mutant disrupted for GreenCut gene CPLD38 has a marked reduction in cytochrome b6f and an increase in chlororespiration.
Conclusion: CPLD38 is essential for accumulating cytochrome b6f and balancing chlororespiration and photosynthesis.
Significance: This analysis demonstrates the importance of a GreenCut protein in photosynthesis.
Keywords: Algae, Chlamydomonas, Chloroplast, Electron Transport, Genomics, Photosynthesis, Plant Physiology, Comparative Genomics, Cytochrome b6f Complex, Chlororespiration
Abstract
Based on previous comparative genomic analyses, a set of nearly 600 polypeptides was identified that is present in green algae and flowering and nonflowering plants but is not present (or is highly diverged) in nonphotosynthetic organisms. The gene encoding one of these “GreenCut” proteins, CPLD38, is in the same operon as ndhL in most cyanobacteria; the NdhL protein is part of a complex essential for cyanobacterial respiration. A cpld38 mutant of Chlamydomonas reinhardtii does not grow on minimal medium, is high light-sensitive under photoheterotrophic conditions, has lower accumulation of photosynthetic complexes, reduced photosynthetic electron flow to P700+, and reduced photochemical efficiency of photosystem II (ΦPSII); these phenotypes are rescued by a wild-type copy of CPLD38. Single turnover flash experiments and biochemical analyses demonstrated that cytochrome b6f function was severely compromised, and the levels of transcripts and polypeptide subunits of the cytochrome b6f complex were also significantly lower in the cpld38 mutant. Furthermore, subunits of the cytochrome b6f complex in mutant cells turned over much more rapidly than in wild-type cells. Interestingly, PTOX2 and NDA2, two major proteins involved in chlororespiration, were more than 5-fold higher in mutants relative to wild-type cells, suggesting a shift in the cpld38 mutant from photosynthesis toward chlororespiratory metabolism, which is supported by experiments that quantify the reduction state of the plastoquinone pool. Together, these findings support the hypothesis that CPLD38 impacts the stability of the cytochrome b6f complex and possibly plays a role in balancing redox inputs to the quinone pool from photosynthesis and chlororespiration.
Introduction
Over 50 years of in-depth biochemical, genetic, and structural analyses have yielded a significant body of information on polypeptides and cofactors involved in photosynthesis (1, 2). Crystal structures now exist for the four major protein complexes involved in light-driven electron transport and ATP generation; these include photosystem II (PSII)3 (3–5), the cytochrome b6f complex (6, 7), photosystem I (PSI) (8), and the ATP synthase (9–11). However, there are still major gaps in our understanding of photosynthesis, especially with respect to the biogenesis of the photosynthetic complexes, regulatory processes that control individual activities critical for photosynthesis and the ways in which these regulatory processes are impacted by environmental conditions.
Currently, there are ongoing attempts to identify novel genes involved in the regulation of photosynthesis. These efforts have most frequently relied on forward genetic screens (12) and expression studies (13). However, the use of informatic resources may also facilitate identification of key activities and novel components that impact photosynthetic processes. Clues concerning the physiological roles of specific proteins have been gleaned from the presence of targeting sequences that route these proteins to specific subcellular sites, tissue- and development-specific patterns of gene expression, and the ways in which environmental conditions impact expression patterns. Many proteins associated with photosynthetic functions are expressed at high levels in plant leaves, are targeted to chloroplasts, and may be up-regulated under conditions that favor increased photosynthetic activity. However, there are many exceptions to some or all of these criteria. For example, proteins essential for regulating expression of genes encoding key photosynthetic components and integrating photosynthetic activity with other cellular processes may not necessarily be targeted to chloroplasts. Furthermore, some photosynthetic constituents may be required at near constant levels under most conditions, and thus, their expression patterns may not be informative.
A novel approach to identify proteins important for photosynthetic function exploits the wealth of available sequence information and comparative genomic analyses. Proteins potentially involved in plastid-specific processes can be identified based on their presence in a diverse group of photosynthetic organisms and their absence in heterotrophic organisms. The inventory of proteins present in green-lineage eukaryotic organisms, but not present in nonphotosynthetic organisms, was only first determined in 2007, at which time it was designated the GreenCut (14), and then subsequently refined (15); prior to this time, the number of plant genomes available had been limited. The most recent comparative genomic analysis (15) identified 597 proteins that were present in plants and green algae (based on mutual best BLAST hits and similarity criteria) but were not identified in bacteria, archaea, fungi, and mammals. Of these 597 proteins, 311 were not assigned a biological function, although multiple bioinformatic techniques were used to probe their importance in plant-specific processes. Sequence motifs, identified by BLAST analyses, had been associated with 231 of these GreenCut “unknowns.” This information along with assignments of subcellular location, co-expression analysis, and transcript abundance information from both Arabidopsis thaliana and Chlamydomonas reinhardtii (Arabidopsis and Chlamydomonas throughout) have resulted in hypotheses concerning potential functions of some of these proteins (15); testing such hypotheses requires extensive experimentation.
Many of the GreenCut proteins may not be directly involved in catalytic aspects of photosynthetic function, and some may not participate in photosynthetic processes at all, but they may be critical for plant-specific regulatory or signaling activities and/or maintenance of chloroplast structure and function. To discover those GreenCut proteins that were most likely involved in photosynthesis, we identified proteins present in the GreenCut that were also present in red algae, diatoms, and at least 36 of 37 cyanobacterial species of diverse morphologies and different niche specificities (15). This assemblage, represented by 68 polypeptides, is hypothesized to harbor those proteins that were present in the cyanobacterial progenitor of the modern-day plastid (15). In addition, it is likely that these proteins function in processes shared by all oxygenic photoautotrophs (e.g. photosynthesis). This group of 68 proteins contains 27 with unknown functions, but many have conserved domains with predicted activities. One protein in this group with no predicted function is CPLD38. Because this protein is present in the GreenCut (14, 15), is predicted to be targeted to the chloroplast (16)), is shown to be retained in thylakoid extracts (17), and is encoded by a gene adjacent to ndhL (18) in cyanobacteria, we hypothesized that it is important for photosynthetic function. Furthermore, a strain previously identified with an insertion in the CPLD38 gene (12) allowed us to characterize the phenotype of a cpld38 mutant; we expected the analysis of the mutant to provide information about novel photosynthetic processes and to help assess the validity of the use of bioinformatic approaches. This study describes the characterization of the cpld38 mutant. Based on our analyses, this thylakoid protein appears to be essential for proper photosynthetic function. Although the mutant cells have a reduction in chlorophyll and the major photosynthetic complexes (∼50% of wild-type (WT) levels of PSI and PSII), the cytochrome b6f complex is reduced well below the levels of the other complexes. The absence of the CPLD38 protein appears to impact the specific activity of the cytochrome b6f complex, the stability of the individual subunits of the complex, and the levels of transcripts for the nucleus-encoded subunits of the complex. In addition, PTOX2 and NDA2 proteins, which are involved in chlororespiration, are up-regulated ∼5-fold in the cpld38 mutant. Experimental analyses also indicate an accelerated rate of chlororespiration in the mutant strain.
EXPERIMENTAL PROCEDURES
Mutant Generation and Complementation
The cpld38 mutant (strain CAL026.02.10) was generated by insertional mutagenesis as part of a screen to discover novel genes involved in photosynthesis (12). This mutant proved to require a fixed carbon source (acetate-requiring or ac-) for growth and to be sensitive to moderate/high light (>100 μmol photon m−2 s−1). To complement the cpld38 mutant, the WT CPLD38 gene was amplified from Chlamydomonas genomic DNA (strain D66+; CC4425), and the amplification product was purified (Qiagen catalog no. 28704). The primers used for amplification of the genomic sequence annealed to regions ∼1,000 bp upstream of the CPLD38 transcription start site (5′-GGGCCTGCGCGTTTTGACAG-3′) and 400 bp downstream of the 3′-UTR (5′-CCGTTGCACACGACACTTCACTG-3′). The amplification product contains the native promoter, the complete gene sequence with introns, and the native 3′-UTR. For transformation of the cpld38 strain, mutant cells were grown in TAP medium to early exponential phase (1 × 106 cells ml−1) and collected by centrifugation at room temperature at 3,500 × g for 10 min, and the cell pellet was resuspended in TAP medium containing 40 mm sucrose (TAP + sucrose) to a final cell concentration of 1 × 108 cells ml−1. The CPLD38 PCR product (200 ng) plus 2 μg of salmon sperm DNA and 150 ng of the AphVIII gene under the control of the RBCSII promoter (19) was incubated with cpld38 cells for 20 min. The cells were then electroporated at 0.9 kV/25 microfarad capacitance and immediately diluted in 5 ml of TAP + sucrose medium. The electroporated cultures were incubated overnight in the dark with gentle agitation, then spread on solid minimal medium containing 5 μg ml−1 paromomycin, and placed in the light (40 μmol photons m−2 s−1). After a week of growth at low light, the plates were moved to relatively high light (300 μmol photons m−2 s−1) and incubated for an additional 7 days; only transformants harboring the introduced CPLD38 gene were able to grow under the high light conditions.
Cell Counts, Chlorophyll, P700, and Cytochrome f Determinations
To reduce cell aggregation in the various algal strains for cell counting, cultures were pelleted by centrifugation (3,500 × g) and incubated for 30 min at room temperature with autolysin (kindly provided by M. Aksoy) before analysis. Chlorophyll concentrations were determined following extraction of the pigments in methanol as described previously (20). In brief, cells were pelleted at 10,000 × g for 2 min, and the resulting pellet was resuspended in 1 ml of 100% methanol (vortexing was used to resuspend the pellet) and then placed in the dark (tubes were wrapped in aluminum foil). After incubating the sample in the dark at room temperature for 5 min, the cellular debris was pelleted by centrifugation at full speed in a microcentrifuge (21,000 × g) for 5 min. To determine total chlorophyll in the sample, the absorbance of the extract was quantified at 652 and 665 nm, and the absorbance values were used in Equation 1.
P700 and cytochrome f concentrations were determined in whole cells following a 20-min incubation with 1 mm methyl viologen and 5 μm 2,5-dibromo-3-methyl-6-isopropylbenzoquinone using a JTS-10 spectrophotometer (BioLogic, France). For quantification of P700, optical changes were monitored at 705 nm during a brief period of illumination. The molar extinction coefficient used in this analysis for P700+ (50,000 m−1 cm−1) was estimated from a previous report (21). Cytochrome f was determined from the optical absorption change at 554 nm with respect to a base line drawn from optical absorption changes at 546 and 573 nm. The molar extinction coefficient used in this analysis for cytochrome f, 18,000 m−1 cm−1, was taken from a previous report (22).
In Vivo Measurement of PSII Activity
Chlorophyll fluorescence was analyzed using a Walz Dual-PAM-100 fluorometer on the “light curve” setting. Various parameters were determined during a stepped illumination regime; samples were illuminated for 30 s at each step followed by a saturating light pulse. Individual samples were analyzed in triplicate (technical replicates), with each sample being tested on at least three separate occasions (biological replicates). The ΦPSII value, which reflects the yield of PSII at different light intensities, was derived from Equation 2 (23, 24).
In Vivo Measurements of P700 Redox State
The oxidation rate of P700 was determined using a JTS-10 spectrophotometer. To assess the pool of electron donors to PSI, cells were incubated with 3-(3,4-dichlorophenyl)-1–1-dimethylurea (10 μm) and hydroxylamine (1 mm) to completely block PSII activity, and optical changes at 705 nm were monitored following saturating flashes of light from an Nd:YAG laser.
Cytochrome b6f Complex and ATP Synthase Activities
Spectroscopic measurements using the JTS-10 spectrophotometer allowed us to monitor the activities of the cytochrome b6f complex and ATP synthase. To quantify charge separation that occurs as a consequence of cytochrome b6f complex activity, whole cells were treated with 3-(3,4-dichlorophenyl)-1–1-dimethylurea (DCMU) and hydroxylamine, which block PSII activity, left in the dark for 30 min (moderately reducing conditions), and then excited with a single flash from an Nd:YAG laser. The extent of charge separation in the cytochrome b6f complex can be inferred indirectly through the electrogenicity change, which is monitored as a change in absorbance at 520 nm. To quantify cytochrome b6f complex activity, cells were pelleted by centrifugation (3,200 × g) at room temperature and resuspended in 20 mm HEPES (pH 7.5). Cells were broken by bead beating and analyzed for cytochrome b6f activity using a previously published protocol (25).
To quantify proton movement through the ATP synthase complex, cells were shaken vigorously to ensure aerobic conditions and then excited with a single flash from an Nd:YAG laser. The decay of the signal that appears at 520 nm following this excitation is indicative of the translocation of protons associated with ATP synthase function.
Measurements of Chlororespiratory Electron Flow
Chlororespiration was measured according to previously published protocols (26, 27). In brief, samples were incubated in 0.9 m sucrose and then illuminated for 3 s at 165 μmol photons m−2 s−1 in a JTS-10 spectrophotometer. Fluorescence induction curves were then recorded after the cells were maintained in the dark for various times. The size of the oxidized plastoquinone pool was estimated by determining the area over the fluorescence rise curves. The area corresponding to 1 electron was determined by following a fluorescence rise curve in the presence of DCMU for 200 ms.
SDS-PAGE, Immunoblot Analysis, and Heme Staining
For immunoblot analysis, Chlamydomonas cells were collected in exponential phase by centrifugation (3,500 × g, 5 min) and resuspended in protein extraction buffer (100 mm Na2CO3, 100 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 1 mm α-amino-n-caproic acid, and 1 mm benzamidine HCl). Samples were aliquoted, flash-frozen, and stored at −80 °C until use. Prior to electrophoresis, samples were thawed, and the SDS and sucrose concentrations were increased to 2 and 12%, respectively. After boiling for 1 min, cellular debris was removed by centrifugation at 21,000 × g for 2 min at 4 °C. Solubilized polypeptides were resolved by SDS-PAGE on a 15% polyacrylamide gel (1 h, 125 V constant voltage) using the Laemmli buffer system (28) and then transferred from the gel to polyvinylidene difluoride membranes and probed with specific antibodies as described previously (29). Briefly, the blots were blocked in 5% milk in Tris-buffered saline solution (TBS) with 0.1% Tween 20 prior to a 1-h incubation with the primary antibodies (in TBS and 0.1% Tween 20). All primary antibodies were used at a dilution of 1:10,000. All primary antibodies were purchased from Agrisera except α-tubulin, which was purchased from Sigma (T5168) Horseradish peroxidase-conjugated anti-rabbit IgG (Promega, Madison, WI), at a 1:10,000 dilution, was used as the secondary antibody. Peroxidase activity was detected by an enhanced chemiluminescence assay (GE Healthcare). Relative protein levels were quantified using ImageJ.
Heme staining was performed according to a previously published protocol with some modifications (30). In brief, cultures were grown to exponential phase and pelleted by centrifugation at 3,500 × g for 10 min. Samples were prepared as described above and resolved by SDS-PAGE, and the polyacrylamide gels with the resolved proteins were incubated in the dark at room temperature in a staining solution (30% methanol, 2.6 mm 3,3′,5,5′-tetramethylbenzidine, and 175 mm NaOAc (pH 5.0)) for 50 min. To develop the heme signal, 750 μl of 30% H2O2 was added to the sample, and after 15 min at room temperature, the gel was incubated overnight in the dark at 4 °C.
RNA Isolation and Quantification
Cells were harvested in exponential phase and pelleted by centrifugation at 4 °C at 3,500 × g for 10 min; the resulting pellet was flash-frozen in liquid nitrogen and stored at −80 °C. Pellets were dissolved in RNA-free lysis buffer (50 mm Tris (pH 7.5), 150 mm NaCl, 15 mm EDTA, and 2% SDS (w/v)) and extracted with RNase-free phenol/chloroform (phenol was equilibrated in sodium citrate (pH 4.0)). The phases were separated by centrifugation (10,000 × g for 10 min) in baked glass centrifuge tubes (Corex tubes, Corning Inc., Corning, NY). Three additional phenol/chloroform extractions were performed, followed by two chloroform extractions. After the final chloroform extraction, the RNA solution was precipitated overnight at −20 °C following the addition of 2 volumes of ethanol. The precipitate was collected by centrifugation (10,000 × g at 4 °C) for 30 min, and the pelleted RNA was washed with 70% ethanol and then resuspended in sterile H2O. LiCl was added to the RNA solution to a final concentration of 4 m, and the RNA was allowed to precipitate overnight at 4 °C. The precipitate was collected by centrifugation (10,000 × g for 30 min at 4 °C) and then washed with 70% ethanol. Once again, the pelleted RNA was resuspended in sterile water, and all contaminating DNA was degraded with RNase-free DNase (Qiagen catalog no. 79254). The purity of the final RNA was evaluated on a formaldehyde-agarose gel (RNeasy mini-handbook).
Quantification of Specific Transcripts
First-strand cDNA was synthesized from 0.5 μg of total RNA using oligo(dT)12–18 for priming the SuperScript III reverse transcriptase reaction, as described in the manual (Invitrogen). RT-quantitative PCR was performed with a LightCycler 480 (Roche Applied Science). PCRs, in a final volume of 20 μl, contained 10 μl of LightCycler 480 SYBR Green Master Mix (Qiagen, Frederick, MD), 5 μl of a 1:20 cDNA dilution, 400 nm of each primer, and distilled water to make up the remainder of the 20-μl volume. Conditions used for amplification in the thermocycler were as follows: preincubation at 95 °C for 5 min, followed by 50 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 20 s, elongation at 72 °C for 20 s, and measurement of fluorescence after 80 °C for 5 s (the last step was incorporated into the protocol to avoid background signals resulting from the formation of primer dimers). A melt-curve analysis program (60–99 °C, heating rate of 2.2 °C/s, with florescence measurements every 0.2 s) was used to evaluate the specificity of the amplification reactions. All reactions were performed in triplicate (technical replicates) with at least two biological replicates. Two genetically different complemented strains (cpld38-CPLD38_1 and cpld38-CPLD38_2) were used as biological replicates to demonstrate transcript accumulation in the rescued strains. The CBLP gene was used as a housekeeping gene control (31). Transcript abundances were evaluated using the ΔΔCt method, with all reactions assumed to be operating at 100% efficiency (32). The primer pairs used for RT-quantitative PCR analysis were as follows: 5′-CTTCTCGCCCATGACCAC-3′ and 5′-CCCACCAGGTTGTTCTTCAG-3′ for CBLP; 5′-GCTACGGTGCCTTCTTCGTGCC-3′ and 5′-TTCTCGGCCGCCACCCAAG-3′ for PETC; 5′-ACCGCCGGCCAGAAGGCTG-3′ and 5′-CAACCAGAGACTCCACGCGCA-3′ for PETM; 5-ACCCACGTGAGGCTAATGGTCG-3′ and 5′-GGAACGCGATCTGGTGGTGCTA-3′for petA; 5′-CGTTCAATTCACCGTTGGTCAGC-3′ and 5′-ACCAACACCTGGGATTGCATCAG-3′ for petB; 5′-GCGCCAGGCGGTGAAGGTC-3′ and 5′-AGCGGCCCCAGACCACCAG-3′ for PETN; and 5′-ACTACCAGCGCCACAGCCC-3′ and 5′-GCTAGGCAGTAAAATGGCGACCG-3′for CPLD38.
RESULTS
Bioinformatic Analyses
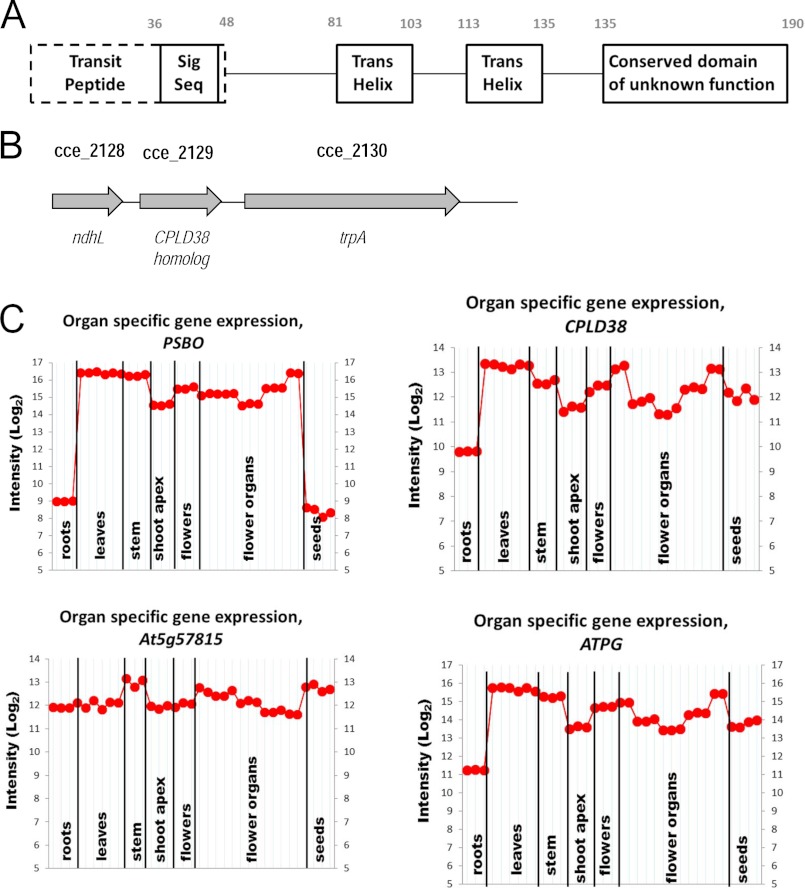
The CPLD38 protein was originally identified as a member of the PlastidCut, which contains orthologs in green lineage organisms, red algae, and diatoms (14). This protein has a predicted N-terminal transit peptide (16, 33) followed by a signal sequence, two predicted transmembrane domains, and a conserved domain of unknown function (Fig. 1A). Based on previous proteomic analyses (17, 34), CPLD38 was determined to be localized to thylakoid membranes in Arabidopsis. The N-terminal transit peptide and signal sequence are predicted to be removed as the protein becomes localized to its site of function, although this will require experimental validation (53). The conserved coiled-coil domain at the C terminus of CPLD38 is rich in acidic amino acids; it contains nine glutamate and two aspartate residues. A complete sequence of the CPLD38 protein, with the C-terminal glutamate and aspartate residues indicated with an asterisk, is given in supplemental Fig. S1.
FIGURE 1.
Features of the CPLD38 protein and CPLD38 gene and expression of CPLD38 transcript. A, domains of CPLD38 showing the putative transit peptide and signal sequence (Sig Seq), two transmembrane helices (Trans Helix), and a conserved domain of unknown function. The numbering of the first and last amino acids of each domain is indicated above the domains. B, structure of the cyanobacterial operon containing the gene encoding the CPLD38 ortholog (cce_2129 in Cyanothece sp. ATCC 51142) (28). A similar operon structure was identified in 29 of the 30 cyanobacterial genomes analyzed. C, expression pattern of the Arabidopsis CPLD38 ortholog (At3g17930) in the various tissue types of Arabidopsis compared with the expression of genes encoding a protein involved in PSII (PSBO) and respiratory (At5g57815, cytochrome c oxidase subunit VIb) functions.
We observed that the CPLD38 protein is encoded by at least 29 fully sequenced cyanobacterial genomes (35); genes encoding the CPLD38 ortholog in some of these cyanobacteria and the immediately adjacent genes are presented in supplemental Table S1. In 97% of the cyanobacterial genomes analyzed, the cyanobacterial orthologs are in close proximity to ndhL (in some genomes this gene is designated ictA or hypothetical protein). In 80% of the cyanobacterial genomes analyzed, it is also in close proximity to trpA (Fig. 1B and supplemental Table S1). The protein product of ndhL is an essential component of the NDH complex. In cyanobacteria, this complex is involved in respiration, cyclic electron flow around PSI, and the transport of inorganic carbon (36). Chlamydomonas does not have this complex in the chloroplast, although it does have type II NDH proteins that contribute to cyclic electron transport (37, 38). The trpA gene encodes the α subunit of the enzyme tryptophan synthase, which catalyzes the second to last step of tryptophan biosynthesis (39). The CPLD38 orthologous gene in cyanobacteria likely forms an operon with both ndhL and trpA because these genes cluster together, are closely spaced in numerous cyanobacterial genomes, and their respective locations in those clusters are conserved. In addition, there is no obvious promoter region immediately preceding either trpA or the CPLD38 ortholog. Therefore, it is reasonable to hypothesize that the putative cyanobacterial CPLD38 genes are co-expressed with ndhL and trpA.
The pattern of expression of the CPLD38 ortholog in Arabidopsis is generally similar to that of genes encoding proteins that function in chloroplasts except, compared with some of the other transcripts for proteins involved in photosynthesis, the level of the CPLD38 transcript is high in seeds relative to roots. This is shown in the tissue-specific transcript profile of CPLD38 compared with that of PSBO (Fig. 1C), a component of the PSII-associated O2-evolving complex, in Arabidopsis. The expression patterns of five other genes encoding proteins associated with photosynthetic activities are presented in supplemental Fig. S2; the ATPG transcript has a pattern most similar to that of CPLD38. Additionally, this pattern of gene expression is very different from that observed for most other genes, including At5g57815, which encodes subunit VIb of cytochrome oxidase (Fig. 1C). The finding that CPLD38 has a tissue-specific transcript profile similar to other genes known to play crucial roles in photosynthesis suggests that CPLD38 has an undetermined activity associated with the function/biogenesis of photosynthetic complexes or chloroplasts (or potentially other plant-specific processes that predominantly occur in photosynthetic tissue).
Growth/Pigmentation Phenotype and Complementation
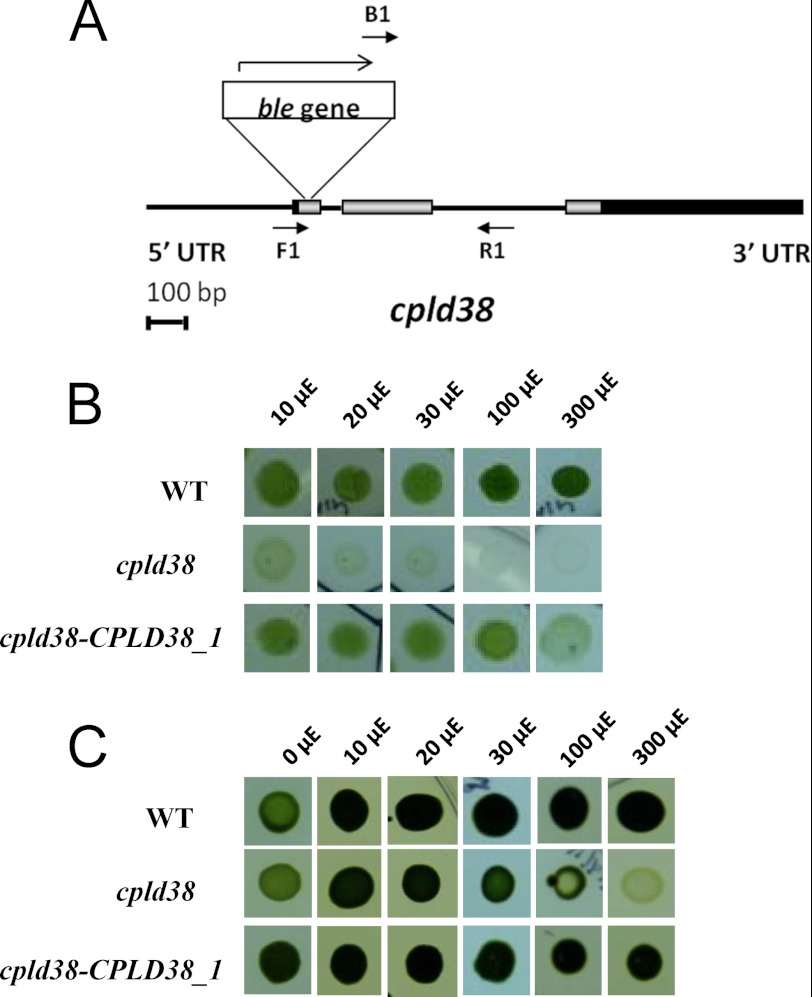
The cpld38 mutant was identified in a forward screen for mutants defective in photosynthesis, pigment accumulation, and/or light sensitivity (12). The mutants were generated by random insertion of the ble gene (for Zeocin or bleomycin resistance) into the Chlamydomonas genome. Because of the large number of mutants acquired in this screen and the possibility of multiple insertions and secondary lesions, we filtered the collection for mutants with disruptions in genes encoding GreenCut proteins (15) and targeted those mutants for additional analyses. Based on TAIL PCR (6, 40), the cpld38 mutant was found to have an insertion in the first exon of the gene (Fig. 2A), which is very likely to result in a null phenotype. A study of the growth properties of this mutant was performed in the presence and absence of a fixed carbon source (TAP medium and minimal medium, respectively). As shown in Fig. 2B, the cpld38 mutant exhibited very little growth on minimal medium; some growth may occur at low light intensities (e.g. 10 μmol photons m−2 s−1; this is the same as 10 microeinsteins in Fig. 2B). When the strain is grown on solid TAP medium at low light intensities, its growth is comparable with that of WT cells (Fig. 2C, 10–20 μE). However, it shows signs of chlorosis and death with increased levels of irradiation. At 30 μmol photons m−2 s−1, the colony size of the mutant was slightly decreased relative to that of WT cells, whereas at 100 μmol photons m−2 s−1, the mutant displayed marked chlorosis, with clearing in the central part of the colony. These results suggest that the cpld38 mutant has some deficiency in photosynthetic electron flow that markedly slows the rate of carbon fixation, making the growth of the cells acetate-dependent. In addition, this deficiency causes cell death even under mixotrophic conditions at high light intensities, indicating a potential block in photosynthetic electron flow that may result in the accumulation of reactive oxidative species. Reactive oxidative species can cause severe damage to many cellular constituents, including the components of photosynthetic electron transport. This potential block also appears to affect accumulation of chlorophyll as the cpld38 mutant has a 57% decrease in total chlorophyll/cell content (Table 1). This observation suggests that the mutant may have an overall reduction in components of the photosynthetic apparatus.
FIGURE 2.
cpld38 mutant and its responses to light under photoautotrophic and mixotrophic conditions. A, map showing insertion of the ble gene in the cpld38 mutant. Exons are shown as gray boxes, introns and intergenic regions as black lines, and the 5′- and 3′-UTRs as black boxes. The ble gene is inserted into the first exon, as indicated. Primers used for the analysis of progeny from genetic crosses are represented by short arrows (see supplemental Fig. S3). B and C, WT cells, cpld38, and the cpld38-CPLD38_1 rescued mutant were grown to exponential phase in TAP medium and then diluted to 1 × 105 cells ml−1. Five μl of culture was spotted onto solid (1.5% agar), minimal (B), or TAP (C) medium. Cultures were grown at the light intensities indicated for either 10 days (minimal medium) or 7 days (TAP medium). The designation μE is equivalent to μmol photons m−2 s−1.
TABLE 1.
In vitro and in vivo analysis of chlorophyll/cell ratios, chlorophyll/pigment ratios, maximum photosynthetic efficiency, and cytochrome b6f activity
Samples were taken for measurement during exponential growth in low light (40 μmol photons m−2 s−1) on TAP medium with moderate shaking (150 rpm).
| Chl/106 cellsa | PSI/PSII ratio | P700/Chlb | PSII/cellc | PSI/celld | Cyt f/Chle | FV/FM | PSI act/Chlf | Cyt b6f act/Chlg | |
|---|---|---|---|---|---|---|---|---|---|
| WT | 2.98 ± 0.41 | 0.8 | 0.88 ± 0.08 | 2.09 | 2.62 | 0.36 ± 0.06 | 0.66 ± 0.01 | 636 ± 44 | 156 ± 14 |
| cpld38 | 1.28 ± 0.27 | 0.93 | 1.12 ± 0.07 | 1.33 | 1.43 | 0.16 ± 0.06 | 0.68 ± 0.01 | 545 ± 61 | 17 ± 18 |
| cpld38-CPLD38_3 | 3.11 ± 0.3 | 1.06 | 0.96 ± 0.05 | 3.16 | 2.98 | 0.51 ± 0.08 | 0.66 ± 0.01 | 722 ± 27 | 94 ± 17 |
a Data were expressed in micrograms of total chlorophyll/106 cell; values are the average of 8–11 replicates ± S.D.
b Data were expressed in nanomoles/mg total chlorophyll; values are the average of 10 replicates ± S.D.
c The estimated value was determined from the chlorophyll/106 cells, PSI/PSII ratio, and P700/ chlorophyll (expressed in nanomoles of PSII/109 cell).
d The estimated value was determined from the chlorophyll/106 cells and P700/chlorophyll (expressed in nanomoles of PSI/109 cell).
e Data were expressed in nanomoles/mg of total chlorophyll; values are the average of 10 replicates ± S.D.
f Data were expressed in micromoles of O2 consumed per mg of chlorophyll/h; values are the average of three replicates ± S.D.
g Data were expressed in micromoles of O2 consumed per mg of chlorophyll/h; values are the average of 5–9 replicates ± S.D.
To determine the genetic basis for its phenotypes, the cpld38 mutant was crossed to WT cells. The acetate-requiring and high light-sensitive phenotypes co-segregated as a 2:2 pattern in each of 16 tetrads that were analyzed from the third backcross of the mutant to the parental WT strain. These results demonstrated that the observed phenotypes are the consequence of a single nuclear mutation. PCR analysis of each of these 16 tetrads demonstrated that the insertion (illustrated for tetrads 1 and 2 in supplemental Fig. S3) co-segregated with an aberrant fluorescence phenotype (2:2 segregation); the mutant has much lower ΦPSII (described below). These results indicate that the phenotype is closely linked to the ble insertion. To further show that the growth phenotype is a direct consequence of the ble insertion, we co-transformed cpld38 with a WT copy of the CPLD38 gene along with the paromomycin resistance cassette (AphVIII gene). The presence of the CPLD38 transcript in transformants (three different transformants with a rescued growth phenotype) was verified by RT-PCR (supplemental Fig. S4). The cpld38 mutant harboring a second normal copy of CPLD38 was able to grow on minimal medium at light intensities above 30 μmol photons m−2 s−1, although at higher intensities the rescued strain grew somewhat more slowly than WT cells (Fig. 2B). In addition, on TAP medium the rescued strain showed no chlorosis at any of the light intensities tested (Fig. 2C) and had a chlorophyll/cell ratio that was statistically similar to that of WT cells (Table 1). These data suggest that the mutant has been rescued for its inability to grow in the absence of fixed carbon and partially rescued for light sensitivity; although the rescued strain appears to show some light sensitivity under photoautotrophic conditions, it does not display light sensitivity in the presence of acetate (at light intensities of up to 300 μmol photons m−2 s−1).
Analysis of the Photosynthetic Reaction Centers
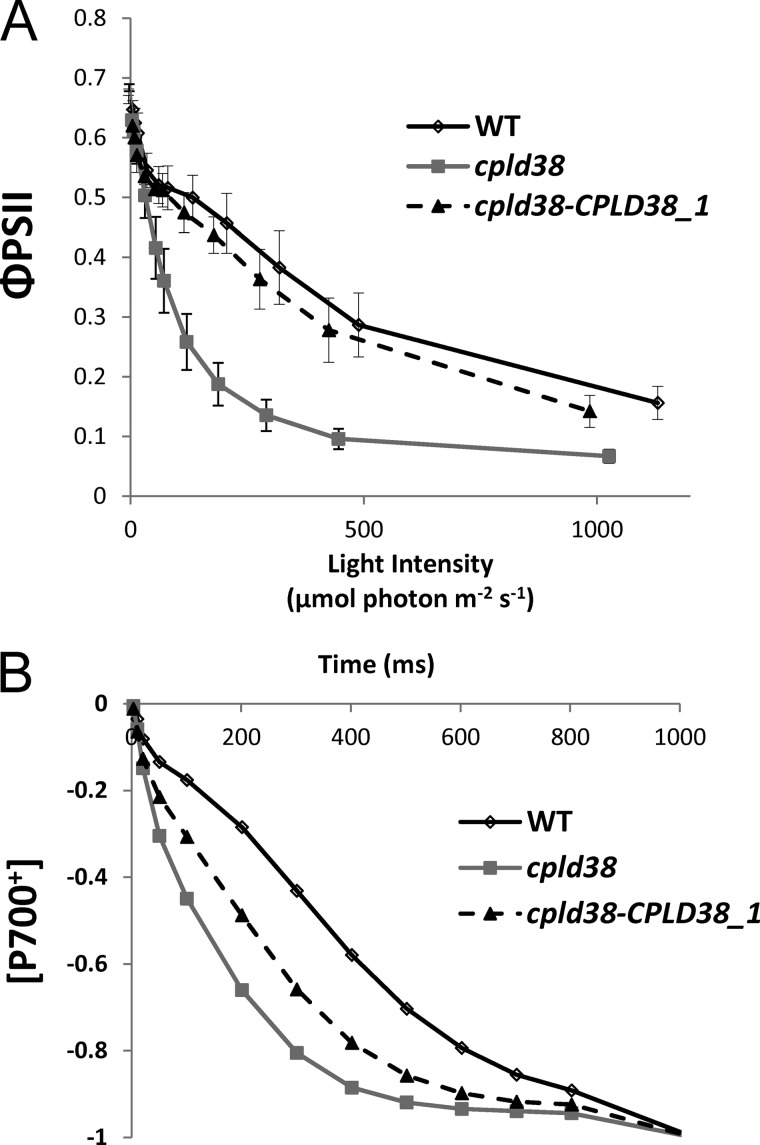
To further characterize the phenotype of the mutant, photosynthetic electron flow was assayed by spectroscopic and fluorescent assays. The efficiency of PSII (ΦPSII) as measured by modulated chlorophyll fluorescence indicated that the plastoquinone pool was significantly more reduced in cpld38 than in either WT cells or the rescued strains at irradiances exceeding 50 μmol photons m−2 s−1 (Fig. 3A). However, the quantum yield of PSII after a dark incubation, or FV/FM (Fig. 3A, and Table 1), was approximately equal for the WT and cpld38 mutant, suggesting that PSII is not impaired in the mutant and that the decrease in electron transport is a consequence of an aberration downstream of PSII. In addition, as shown in Fig. 3B, P700 oxidized at a far more rapid rate in the mutant than in the WT or complemented strains in the presence of DCMU and hydroxylamine; this likely reflects diminished availability of electrons at the donor side of PSI. Direct examination of PSI activity showed that the complex present in the membranes had activity similar to that of WT cells (Table 1), indicating the decrease in electron flow through the complex in the mutant is a consequence of an aberration upstream of PSI. Together, the data indicate that a dominant feature of the mutant strain is a marked reduction in the flow of electrons between PSII and PSI.
FIGURE 3.
Measurements of photosynthetic electron flow in WT, the cpld38 mutant, and the rescued mutant cpld38-CPLD38_1. A, efficiency of light-driven electron transport through PSII measured as the fluorescence parameter ΦPSII (FM′ − Ft′)/FM′). Cells were illuminated with actinic light at intensities indicated on the x axis for 1 min. B, rate of P700+ oxidation. Lower values on the y axis represent a greater proportion of oxidized P700. The inhibitors of PSII, DCMU, and hydroxylamine were added at concentrations of 10 μm and 1 mm, respectively. For all experiments, values on the y axis are the average of three measurements (technical replicates). Experiments were repeated with a biological replicate (data not shown), which gave nearly identical results. Error bars in A represent the standard deviation. Errors bars are not shown in B, but each data point in the displayed figure does not deviate from any of the individual replicates by more than 5% of the maximum value.
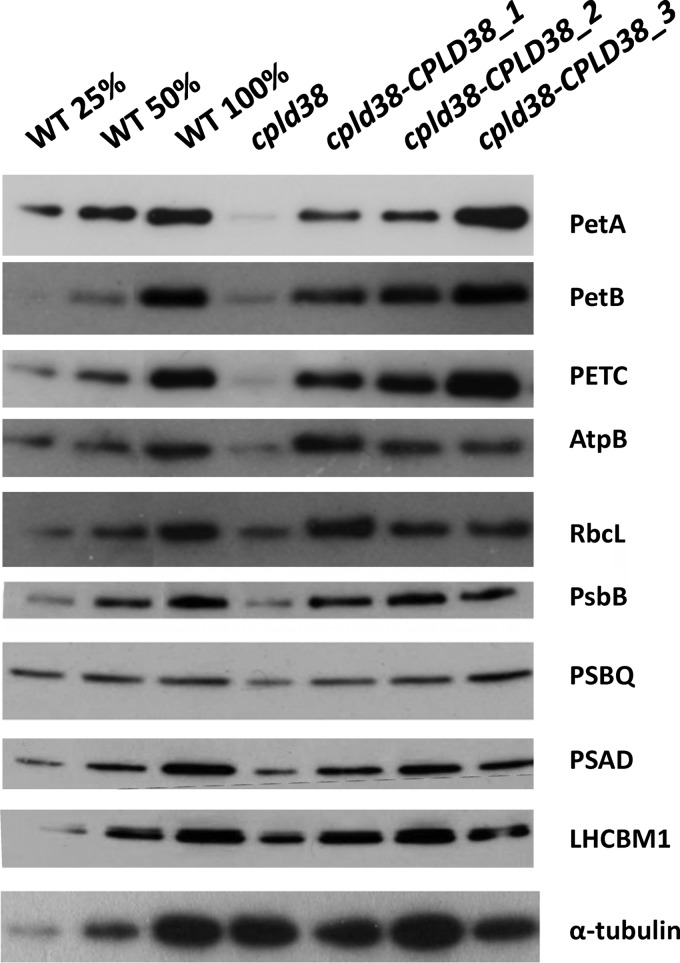
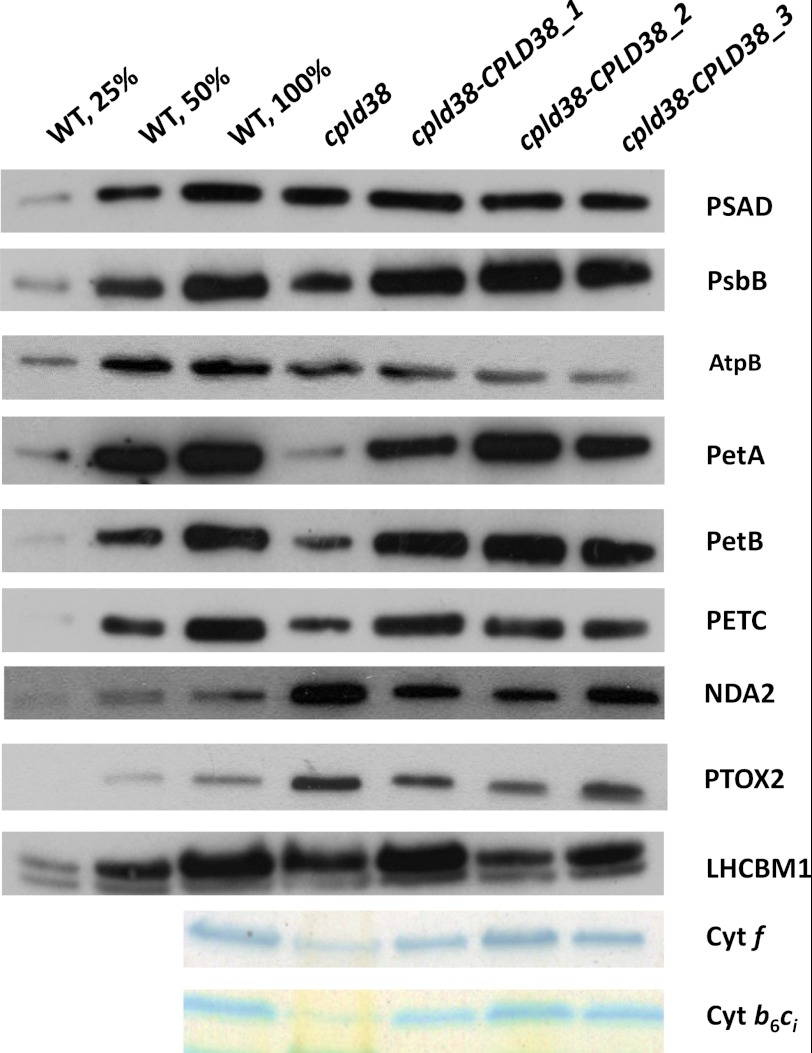
PSI/PSII ratios were also measured in the mutant and WT strains to ensure that differences in the electron transport rates determined by fluorescence and P700 oxidation kinetics were not a consequence of aberrant stoichiometries of the photosynthetic reaction centers (Table 1). Although this measurement indicated that the cpld38 mutant had a slightly increased level of PSI relative to PSII (16%) compared with WT cells, it is unlikely that this increase would account for the dramatic changes observed in Fig. 3, A and B. When normalized to chlorophyll, the mutant had a similar [P700+] compared with the WT strain, further indicating that the in vivo differences in P700 oxidation kinetics are likely due to a difference in electron flow; electrons from PSII were not efficiently transferred to PSI. In accord with the chlorophyll/cell ratio for the cpld38 mutant and WT cells, the total amount of the PSI subunit PSAD, determined by Western blot analyses, appeared to be significantly decreased in the cpld38 mutant (Fig. 4). However, the ratio of PSAD to the chlorophyll concentration was similar in mutant and WT cells (Fig. 5), which is in agreement with what was determined for [P700+] in the cpld38 mutant. A similar situation was observed with respect to the level of PSII proteins. For example, Western blots showed that there was a significant reduction in the levels of CP47 (PsbB) and PSBQ compared with the WT when normalized by cell number (Fig. 4), but when samples were normalized to chlorophyll, the mutant showed a less dramatic change in the PsbB polypeptide content (Fig. 5). All of the findings presented above strongly indicate that the levels of PSII and PSI are reduced in the cpld38 mutant relative to WT cells, although based on total chlorophyll content the differences in activities, stoichiometries, and amounts of the photosynthetic proteins between mutant and WT cells are small.
FIGURE 4.
Western blot analyses of proteins critical for photosynthetic function, including polypeptides of PSI, PSII, the light-harvesting complex (LHCBM1), and ATP synthase polypeptides (AtpB). Chlamydomonas proteins from the WT, cpld38, and the various cpld38-CPLD38 rescued strains were resolved by SDS-PAGE on a 15% polyacrylamide gel, and various proteins were detected immunologically. Samples were taken for analysis when the strains were in their exponential growth phase in low light (40 μmol photons m−2 s−1) on TAP media with moderate shaking (150 rpm). The antibodies used for this analysis are to the polypeptides indicated on the right side of the figure. Samples were normalized by cell number (∼2 × 105 cells per lane). α-Tubulin was used as a loading control.
FIGURE 5.
Western blot and heme staining analyses to identify photosynthetic and chlororespiratory components. Chlamydomonas proteins from the WT, cpld38, and the various cpld38-CPLD38 rescued strains were resolved by SDS-PAGE on a 15% polyacrylamide gel and detected immunologically or by heme staining. Samples were taken for analysis when the strains were in their exponential growth phase in low light (40 μmol photons m−2 s−1) on TAP media with moderate shaking (150 rpm). The antibodies used for this analysis are against polypeptides indicated on the right side of the figure. Approximately 1 μg of chlorophyll was loaded for each sample analyzed by Western blotting, whereas∼30 μg of chlorophyll was loaded for each sample analyzed for heme-containing polypeptides. The LHCBM1 protein was used as a loading control.
Analysis of the Cytochrome b6f Complex
Because the ratios of reaction center to chlorophyll appeared similar in WT and the cpld38 mutant, it seemed unlikely that a defect in these complexes caused the observed mutant phenotype. Therefore, the decrease in electron flow to PSI in the mutant is likely a consequence of a deficiency in plastocyanin or the cytochrome b6f complex; such a defect reduces the rate at which electrons reach P700 from PSII. To measure the activity of the cytochrome b6f complex, samples were incubated under moderately reducing conditions with DCMU and hydroxylamine to block PSII activity and were excited with a single turnover saturating light pulse. Following this pulse, optical changes at 520 nm were monitored to detect the light-induced electrochromic shift of carotenoids. This optical signal reflects the electrochemical potential across the thylakoid membrane that is generated by the activity of the cytochrome b6f complex and PSI. As shown in Fig. 6, there was a large increase in absorbance at 520 nm in WT cells that occurs 0–20 ms following the saturating pulse. The initial and very rapid rise in absorbance (occurring within 1 ms) was a consequence of PSI activity (because DCMU and hydroxylamine were blocking any contribution from PSII). The slower rise, observed between 1 and 20 ms, was a consequence of proton and electron transport within the cytochrome b6f complex (41). The signal was normalized to the initial and very rapid PSI-dependent rise in all samples analyzed (Fig. 6). The electrogenicity attributable to cytochrome b6f turnover was extremely low in the cpld38 mutant relative to WT cells, suggesting that the activity of the cytochrome b6f complex is severely compromised in the mutant strain, either because it is not functioning properly and/or it does not readily acquire electrons when there is no contribution from PSII (electrons would have to come from cyclic electron flow and reductant in the stroma). This effect was mostly mitigated in the rescued strain, where the slower component of the 520 nm signal appeared to be mostly re-established.
FIGURE 6.
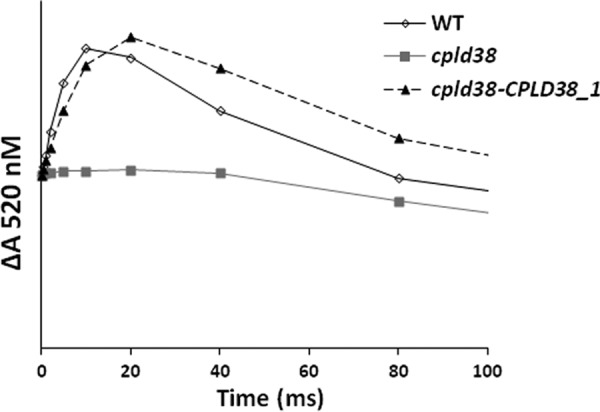
Measurements of cytochrome b6f complex activity by monitoring electrochromic absorption changes at 520 nm. Cells were incubated under moderately reducing conditions (unperturbed for 30 min in dark) to reduce cytochrome bh. Following this incubation, samples were excited with a saturating light pulse from an Nd:YAG laser to induce a single electron turnover. The PSII inhibitors DCMU and hydroxylamine were added at concentrations of 10 μm and 1 mm, respectively. The increase in absorption observed in the first 20 ms (excluding the value at time 0) corresponds to a charge separation within the cytochrome b6f complex. The traces are the average of three replicates with each data point in the displayed figure not deviating from any of the individual replicates by more than 5% of the maximum value. Samples were taken for measurement when the strains were in their exponential growth phase in low light (40 μmol photons m−2 s−1) on TAP media with moderate shaking (150 rpm).
To determine whether the cytochrome b6f complex in the mutant was also impaired under steady-state conditions, the complex was measured in continuous illumination in the presence of duroquinone (25, 42). In this assay (performed with broken cells in the presence of DCMU), duroquinone reduces the cytochrome b6f complex directly (25). When illuminated, electrons are transported to PSI through plastocyanin and ultimately to the artificial electron acceptor methyl viologen. Methyl viologen reduces O2 to O2− (which can then form H2O2), a reaction that can be monitored as O2 uptake by a Clark-type oxygen electrode (43). These measurements show that there is an 89% decrease in cytochrome b6f complex activity in the mutant strain (Table 1).
It is possible (although not likely) that the defect in cytochrome b6f function is simply a secondary effect of lowered ATP synthase activity or altered plastocyanin/cytochrome c6 levels. Experiments measuring the proton translocation through the ATP synthase complex and electron transport from luminal donors to P700+ both indicated that these aspects of the photosynthetic apparatus are unimpaired (supplemental Figs. S5 and S6).
Immunoblots were performed to evaluate the levels of three subunits of the cytochrome b6f complex (PetA, PetB, and PetC) in WT cells and the cpld38 mutant. Fig. 4 shows that the levels of PetA (cytochrome f), PetB (cytochrome b6), and PETC (Rieske iron-sulfur protein) were significantly reduced in cpld38 relative to WT cells. There was also a decrease in the levels of these subunits in the mutant compared with the WT strain when the samples were normalized to chlorophyll concentrations (Fig. 5). However, when normalized to chlorophyll, the amount of PetA accumulates to a somewhat lower level (less than 25%) than the amount of PetB and PetC (∼50%). Considering that these three subunits accumulate to stoichiometric levels in the complex (44, 45), this observation is unusual. A quantification of light-oxidizable cytochrome f in the cpld38 mutant showed a decrease that is more in accord with the amounts of PetB and PetC, decreasing to 42% based on chlorophyll levels (Table 1). Heme staining also showed an amount of cytochrome f and cytochrome b6 that was more similar to the levels of PetB and PetC than PetA (Fig. 5). One possible explanation for this effect could include the turnover of the subunits of the complex at slightly different rates due to oxidative stress caused by singlet oxygen formed by PSII. This hypothesis seems to be supported by the observation that the level of PetA is closer to PetB when cpld38 is grown in very low light (2 microeinsteins, supplemental Fig. S7).
Turnover and Transcript Analysis of Subunits of the Cytochrome b6f Complex
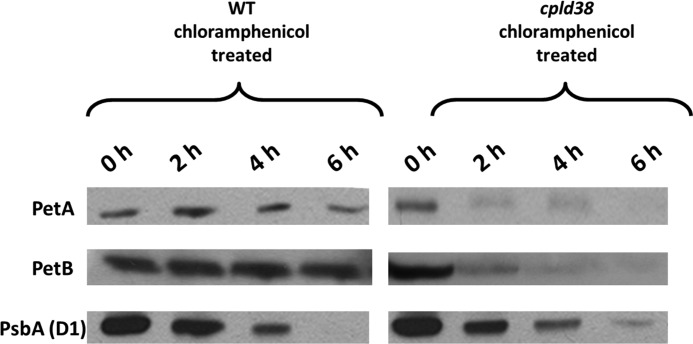
A possible explanation for the decreased activity and lowered accumulation of cytochrome b6f complex could be improper assembly of the subunits of the complex. A defect in the native structure may be sensed and the complex degraded. To examine the stability of the polypeptide components of the complex, the rate of turnover of individual subunits of the cytochrome b6f complex was measured in Chlamydomonas cells following treatment of the cultures with chloramphenicol, an inhibitor of translation on chloroplast 70 S ribosomes (46). The decay of PsbA, which has a well characterized degradation pattern, was monitored in this experiment to ensure that the chloramphenicol was inhibiting chloroplast protein synthesis; its rate of degradation was the same in WT and cpld38 mutant cells, as shown in Fig. 7. Furthermore, although there was little degradation of PetA (cytochrome f) and PetB (cytochrome b6) subunits in WT cells over the course of the 6-h incubation in the presence of chloramphenicol, there was a marked reduction in the level of these polypeptides in the mutant strain. As shown in Fig. 7, there was a rapid decline in the level of PetA and PetB polypeptides even just 2 h after adding the translational inhibitor, with a nearly complete loss of the polypeptides after ∼6 h. These results demonstrate that the steady-state level of the cytochrome b6f complex is lower in mutant cells most likely because the polypeptides of the complex are destabilized.
FIGURE 7.
Turnover of cytochrome f, cytochrome b6, and PsbA in thylakoid membranes. Following exposure of cells to chloramphenicol for the times indicated in the figure, total Chlamydomonas protein from WT and cpld38 mutant cells was fractionated by SDS-PAGE on a 15% polyacrylamide gel and detected immunologically. The antibodies used for this analysis are to polypeptides indicated on the left side of the figure. The 0-h time points were normalized to the cytochrome f intensities.
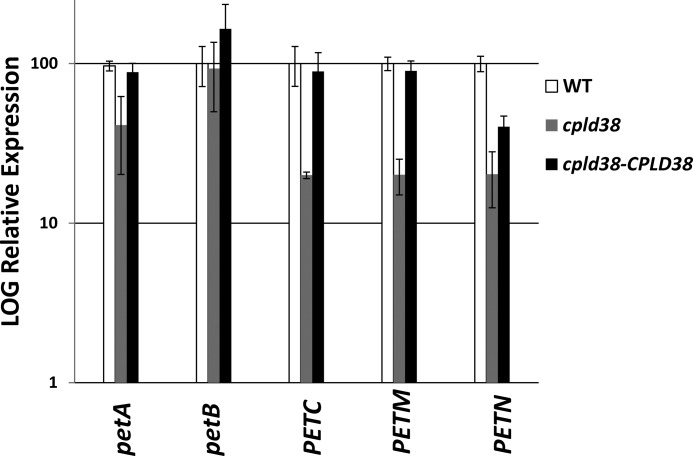
It is possible that CPLD38 plays a role in accumulation of transcripts encoding subunits of the cytochrome b6f complex. To test this possibility, petA, petB, PETC, PETM, and PETN expressions were compared in the WT, cpld38, and rescued cpld38-CPLD38 strains. As shown in Fig. 8, the levels of the three nucleus-encoded transcripts (PETC, PETM, and PETN) were lower in the cpld38 mutant by ∼5-fold. This reduction in transcript abundance could at least in part be responsible for decreased accumulation of cytochrome b6f subunits in the mutant, along with the more rapid turnover of the individual subunits of the complex. Therefore, the loss of CPLD38 results in both altered biogenesis of the cytochrome b6f complex as well as altered regulation of the nuclear genes encoding the subunits of the complex; this control of gene expression can be either a direct or indirect consequence of the primary lesion.
FIGURE 8.
RT-quantitative PCR analysis of the accumulation of transcript for subunits of the cytochrome b6f complex in WT, cpld38, and cpld38-CPLD38. The transcript levels for cpld38-CPLD38_1 and cpld38-CPLD38_2 were similar and were averaged to give the level of transcript in the complemented strain. The accumulation of petA, petB, PETC, PETM, and PETN transcripts was measured when the strains were in their exponential growth phase in low light (40 μmol photons m−2 s−1). Levels of individual transcripts are evaluated with respect to the transcript abundance of the housekeeping control gene (CBLP). The WT transcript level has been set to 100% with each strain adjusted accordingly.
One condition that causes degradation of the cytochrome b6f complex and a decrease in accumulation of PETC, PETM, and PETN polypeptides in Chlamydomonas is nitrogen starvation (47, 48). To ensure that the mutant was not limited for nitrogen, transcripts encoding nitrate and nitrite reductase were analyzed and found to be unchanged in the cpld38 mutant (data not shown); these transcripts increase in abundance during nitrogen starvation.
Analysis of Chlororespiratory Proteins and Activity
Although many photosynthetic proteins were decreased in the cpld38 mutant, some proteins involved in chlororespiration were increased. As can be seen in Fig. 5, both the levels of NDA2 and PTOX2 were elevated in the cpld38 mutant. This increase in protein levels is not likely due to decreases in cytochrome b6f complex as mutants lacking this complex have no change in the levels of these polypeptides (26).
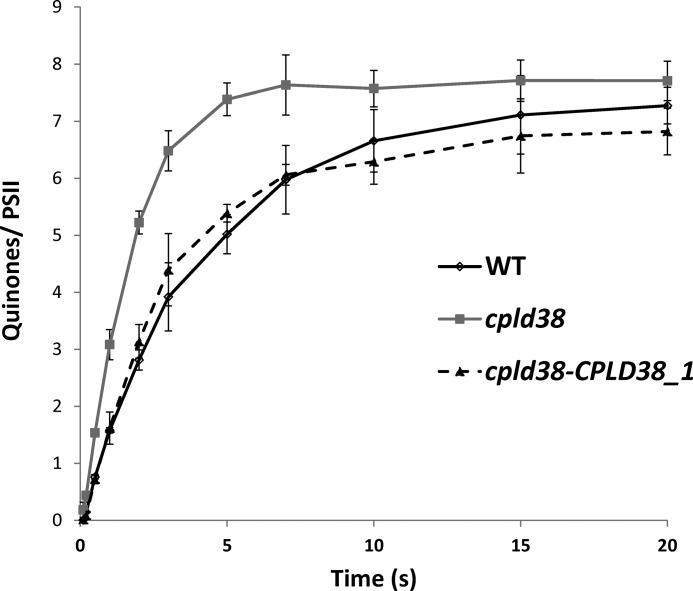
To determine whether PTOX2 and NDA2 activities are accelerated in the mutant strain, chlororespiration was measured after exposing the cells to osmotic shock (26). In these experiments, Chlamydomonas cells were treated with 0.9 m sucrose. The resulting osmotic shock inhibits electron flow from the cytochrome b6f complex to PSI (49). Once this inhibition was achieved, cells were illuminated to reduce the entire plastoquinone pool. Following this illumination, reoxidation of the pool (via the enzyme designated the Plastoquinol Terminal Oxidase, PTOX) was evaluated for the periods of time indicated on the x axis. The rate at which the plastoquinol pool is oxidized was determined by integrating the area above a subsequent fluorescence induction curve (the area corresponding to 1 electron was determined using an induction curve generated in the presence of DCMU). This analysis demonstrated that the plastoquinol pool is oxidized at a significantly faster rate in the cpld38 mutant relative to WT cells (Fig. 9). The rate of PTOX-catalyzed plastoquinol oxidation determined from this experiment was 4.80 ± 0.30 e− s−1 PSII−1 for the mutant, ∼2.5 times faster than the rate determined for the WT (1.85 ± 0.32 e− s−1 PSII−1). This increased rate of re-oxidation, coupled with the increase in PTOX2 and NDA2 proteins, suggests a metabolic shift in the chloroplast that favors electron flow through chlororespiration in the mutant strain.
FIGURE 9.
In vivo measurements of chlororespiratory electron flow. Kinetics of the reoxidation of the plastoquinol pool after a pre-illumination. For all experiments, values on the y axis are the average of three measurements (technical replicates). Error bars represent the standard deviation. The data were fit to a single exponential decay to give fluxes of 1.85 ± 0.32, 4.80 ± 0.30, and 1.86 ± 0.35 e− s−1 PSII−1 for the WT, cpld38 mutant, and cpld38-CPLD38-1 strain, respectively. Samples were taken for measurement when the strains were in their exponential growth phase in low light (40 μmol photons m−2 s−1) on TAP media with moderate shaking (150 rpm).
DISCUSSION
Many tools are available that provide novel insights into the biological role of genes that encode proteins of unknown function. Two techniques that have been used in photosynthetic organisms to identify genes involved in photosynthesis are forward genetics and expression analysis. Both of these approaches have noteworthy drawbacks. In some cases, mutations generated in a forward genetic screen are not linked to the insertion of the antibiotic resistance cartridge (selectable marker), making the mapping of these alterations difficult, and subtle phenotypic effects may go unnoticed. In addition, differential expression analyses will not provide clues to the functions of constitutively expressed genes. The use of comparative genomics approaches offers a distinct approach to identify novel genes that are essential for photosynthesis, and in combination with the aforementioned techniques, it can provide new insights into the activity, assembly, and regulation of the photosynthetic apparatus.
A comparative genomic analysis led to the identification of proteins, designated GreenCut proteins, potentially associated with chloroplast function, biogenesis, and regulation. Combined with previous -omics studies and physiological and biochemical analyses, we are gaining insights into potential functions of many of the GreenCut proteins, including CPLD38. CPLD38 was previously determined to be localized to thylakoid membranes, and it is predicted to contain two putative transmembrane domains and a C-terminal conserved domain of unknown function. In addition, the CPLD38 orthologous gene was found across a diverse group of cyanobacterial species to be in a putative operon that contains one gene encoding a protein that has been associated with photosynthetic/respiratory electron flow and a second gene whose protein product is likely involved in amino acid biosynthesis; these genes are designated ndhL and trpA, respectively. Additionally, tissue-specific expression data indicate that the CPLD38 transcript is present in photosynthetic tissue as well as in some nonphotosynthetic tissue (seeds). Despite these interesting findings, both genetic and biochemical approaches are required to determine the function of CPLD38.
A forward genetic screen identified a cpld38 mutant of Chlamydomonas. This mutant was impaired in growth under photoautotrophic conditions (12), but the linkage between the interruption of this gene and the aberrant phenotype had not been established. We focused our attention on characterization of the cpld38 mutant. Spectroscopic, genetic, and biochemical data support the idea that CPLD38 is critical to sustain photosynthetic function. The cpld38 mutant of Chlamydomonas is light-sensitive and acetate-requiring, even at relatively low light intensities, and it has lower chlorophyll content per cell than the WT strain. The lower chlorophyll reflects reduced levels of PSI and PSII (by about 50%), but the photosystems that are present appear to be functioning properly as determined by biochemical, fluorescent, and spectroscopic measurements. However, both fluorescence and spectroscopic analyses indicate that the mutant has a severe block in linear electron flow. Based on analysis of the functions of the photosystems, the block was hypothesized to be in the intersystem electron transport chain. There are two electron transport complexes that could have been responsible for the electron transport phenotype of cpld38; these are plastocyanin and the cytochrome b6f complex. Detailed spectroscopic analysis has indicated that the mutant is not defective in the donation of electrons by plastocyanin to PSI. Secondary effects caused by an aberrant ATP synthase complex were also deemed unlikely due to the unaltered flow of protons through that complex. Hence, it became most likely that the absence of CPLD38 caused an alteration in cytochrome b6f function. Immunological and spectroscopic data indicated that the cpld38 mutant has reduced levels of PetA, PetB, and PETC and photooxidizable cytochrome f when normalized to the WT levels on a chlorophyll basis. Based on the PetB, PetC, and photooxidizable cytochrome f, the level of cytochrome b6f complex/chlorophyll is 40–50% that of the WT. However PetA, as detected by Western blot analysis, accumulated to a lower level (as low as 10–20%). As shown above, these subunits of the cytochrome b6f complex were degraded by the cell at an enhanced rate, and this degradation or a separate chemical modification could lead to a less immunodetectable PetA polypeptide. It seems unlikely that this protein accumulates to a level significantly less than PetB and PetC as these three protein subunits have been previously found to accumulate to stoichiometric levels (44, 45). Furthermore, the mutant has a marked defect in the electrochromic shift following a single turnover flash, which is dependent on a charge separation within the cytochrome b6f complex. Steady-state measurements of cytochrome b6f complex activity also showed a marked decrease in enzymatic function. These results suggest the cytochrome b6f complex in the mutant strain is synthesized and integrates into the thylakoid membranes, but it is not functioning properly and is rapidly degraded.
A recent publication on a cpld38 mutant in Arabidopsis (called DAC) showed some of the same characteristics that we have presented (50). In both Arabidopsis and Chlamydomonas mutants, the amounts of PSI and PSII are decreased by ∼50%, and the amount of cytochrome b6f complex is decreased by ∼85%. In addition, both fluorescence and P700 oxidation data indicate a block in electron flow downstream of PSII and upstream of PSI. However, there are also some differences in the results obtained for the Chlamydomonas and Arabidopsis mutants. In Arabidopsis, the cytochrome b6f complex does not appear to be degraded at an accelerated rate in the DAC mutant in the presence of lincomycin. Based on this finding, the lowered accumulation of radiolabeled PetA and PetD was attributed to decreased biosynthetic capabilities (50). In contrast, the cytochrome b6f complex is turned over very rapidly in the cpld38 mutant in Chlamydomonas. Degradation of these subunits would likely be attributed to protein instability. Our spectroscopic and biochemical results support the concept that the cytochrome b6f complex is unstable in the mutant strain. Additionally, in the DAC mutant there appears to be no change in the levels of nuclear transcripts that encode cytochrome b6f complex subunits, whereas a 5-fold decrease in nuclear transcripts encoding cytochrome b6f subunits (PETC, PETM, and PETN) was observed in the corresponding mutant in Chlamydomonas. It is possible that these significant discrepancies are accounted for by differences in the physiology of Chlamydomonas and Arabidopsis. Interestingly, nitrogen limitation also causes accelerated degradation of the cytochrome b6f complex (47), reduction in the levels of the PETC, PETM, and PETN transcripts (48), and up-regulation in PTOX and NDA24 in Chlamydomonas. Therefore, a conserved mechanism activated during nitrogen deprivation may become active in the cpld38 mutant; this hypothesis could be validated by further experimentation.
Xiao et al. (50) also demonstrated that the DAC protein was present in chloroplasts and interacts with PetD, based on yeast two-hybrid screens and pulldown analyses. These observations support the hypothesis that the DAC protein is involved in the biosynthesis of the cytochrome b6f complex; however, our additional findings make it unclear whether “assembly” of the complex is a primary role of CPLD38. Alternatively, the interaction of CPLD38 with the cytochrome b6f complex could modulate the role of the complex in linear and cyclic electron flow with the concomitant regulation of chlororespiratory processes. PTOX2 and NDA2 are markedly elevated in the cpld38 mutant. These increases, coupled with an ∼2.5-fold increase in the chlororespiration rate, demonstrate that the lack of CPLD38 has elicited a strong metabolic shift in chloroplast metabolism. This shift causes an increase in chlororespiration at the expense of photosynthesis. Based on this shift, we suggest that CPLD38 is potentially a regulatory factor that is associated with the cytochrome b6f complex that modulates the ratio of electron flow between photosynthesis and chlororespiration. This shift is unlikely to be caused by the absence of the cytochrome b6f complex, as previous analyses have shown that Chlamydomonas strains lacking this complex do not have more chlororespiration (26). However, this shift likely causes the light sensitivity and photosynthetic deficiencies observed in the mutant phenotype as it appears to decrease the content of the cytochrome b6f complex. Mutants in both Arabidopsis and Chlamydomonas having reduced amount of this complex share this phenotype (51, 52). It is less clear if the increase in chlororespiration contributes to the growth phenotypes, although it would seem unlikely considering the aforementioned mutants. It is possible that these proteins act as a switch that down-regulates multiple photosynthetic processes in the cell; however, this hypothesis requires rigorous experimentation.
A role of CPLD38 in the regulation of photosynthesis/chlororespiration could help explain the coupling of the CPLD38 gene with ndhL in an operon that appears to have been conserved through evolutionary time (occurring in nearly all cyanobacterial genomes analyzed). Co-transcription of ndhL and CPLD38 could be important for maintaining the ratios between respiratory, linear, and cyclic photosynthetic electron flow. In both respiratory and cyclic electron flow, the NDH complex reduces the quinone pool with reduced nicotine adenine dinucleotides generated through the oxidation of various metabolites. CPLD38 would serve as a lever that controls whether the reducing equivalents entering the chloroplast through the NDH complex (or NDA2 in Chlamydomonas) are used to reduce PSI or oxygen (through PTOX). However, based on the mutant phenotype, the loss of CPLD38 would have to also impact the stability of the cytochrome b6f complex, which can be linked to its association with the complex, as determined by Xiao et al. (50). The finding that the trpA gene is also part of the cpld38-ndhL operon in many cyanobacteria is perplexing given our current knowledge of the functions of the encoded proteins. However, it is not unreasonable to conjecture that these affiliations could represent a completely unexplained aspect of photosynthetic/respiratory processes and potentially the integration of the biogenesis/degradation of various metabolic complexes (a major nitrogen sink in the cells) with nitrogen metabolism.
At this point, it is clear that the cpld38 lesion causes a significant reduction in the level of the cytochrome b6f complex in the thylakoid membranes, a decline in the stability of the complex, a loss of functionality of the complex that is present, and unexpectedly, also impacts expression of nuclear genes encoding the subunits of the complex. The lesion also appears to result in an overall decline in the level of all complexes associated with photosynthetic electron transport. Furthermore, there is a corresponding increase in the rate of chlororespiration and the proteins that participate in that process. Elucidation of the precise biochemical function of CPLD38 will surely expand our understanding of factors that regulate both the level and activity of complexes of the photosynthetic apparatus, but it will also provide new insights into the role of chlororespiration in chloroplast metabolism.
Acknowledgments
We acknowledge Sabeeha Merchant and Dudley Page for kindly providing antibodies for cytochrome f. We also thank Mary Beth Mudgett for kindly providing antibodies for α-tubulin.
This work was supported in part by National Science Foundation Grants MCB0824469 and MCB0235878 (to A. R. G.) and by private funds donated by Brigitte Berthelemot.

This article contains supplemental Figs. S1–S7 and Table S1.
F. A. Wollman, personal communication.
- PSII
- photosystem II
- PSI
- photosystem I
- DCMU
- 3-(3,4-dichlorophenyl)-1–1-dimethylurea
- NDH
- NADH dehydrogenase.
REFERENCES
- 1. Eberhard S., Finazzi G., Wollman F. A. (2008) The dynamics of photosynthesis. Annu. Rev. Genet. 42, 463–515 [DOI] [PubMed] [Google Scholar]
- 2. Nelson N., Ben-Shem A. (2004) The complex architecture of oxygenic photosynthesis. Nat. Rev. Mol. Cell Biol. 5, 971–982 [DOI] [PubMed] [Google Scholar]
- 3. Umena Y., Kawakami K., Shen J. R., Kamiya N. (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55–60 [DOI] [PubMed] [Google Scholar]
- 4. Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. (2009) Cyanobacterial photosystem II at 2.9-Å resolution and the role of quinones, lipids, channels, and chloride. Nat. Struct. Mol. Biol. 16, 334–342 [DOI] [PubMed] [Google Scholar]
- 5. Ferreira K. N., Iverson T. M., Maghlaoui K., Barber J., Iwata S. (2004) Architecture of the photosynthetic oxygen-evolving center. Science 303, 1831–1838 [DOI] [PubMed] [Google Scholar]
- 6. Kurisu G., Zhang H., Smith J. L., Cramer W. A. (2003) Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science 302, 1009–1014 [DOI] [PubMed] [Google Scholar]
- 7. Stroebel D., Choquet Y., Popot J. L., Picot D. (2003) An atypical haem in the cytochrome b6f complex. Nature 426, 413–418 [DOI] [PubMed] [Google Scholar]
- 8. Jordan P., Fromme P., Witt H. T., Klukas O., Saenger W., Krauss N. (2001) Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 411, 909–917 [DOI] [PubMed] [Google Scholar]
- 9. Abrahams J. P., Leslie A. G., Lutter R., Walker J. E. (1994) Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 [DOI] [PubMed] [Google Scholar]
- 10. Gibbons C., Montgomery M. G., Leslie A. G., Walker J. E. (2000) The structure of the central stalk in bovine F1-ATPase at 2.4 Å resolution. Nat. Struct. Biol. 7, 1055–1061 [DOI] [PubMed] [Google Scholar]
- 11. Stock D., Leslie A. G., Walker J. E. (1999) Molecular architecture of the rotary motor in ATP synthase. Science 286, 1700–1705 [DOI] [PubMed] [Google Scholar]
- 12. Dent R. M., Haglund C. M., Chin B. L., Kobayashi M. C., Niyogi K. K. (2005) Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol. 137, 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ledford H. K., Baroli I., Shin J. W., Fischer B. B., Eggen R. I., Niyogi K. K. (2004) Comparative profiling of lipid-soluble antioxidants and transcripts reveals two phases of photo-oxidative stress in a xanthophyll-deficient mutant of Chlamydomonas reinhardtii. Mol. Genet. Genomics 272, 470–479 [DOI] [PubMed] [Google Scholar]
- 14. Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., Witman G. B., Terry A., Salamov A., Fritz-Laylin L. K., Maréchal-Drouard L., Marshall W. F., Qu L. H., Nelson D. R., Sanderfoot A. A., Spalding M. H., Kapitonov V. V., Ren Q., Ferris P., Lindquist E., Shapiro H., Lucas S. M., Grimwood J., Schmutz J., Cardol P., Cerutti H., Chanfreau G., Chen C. L., Cognat V., Croft M. T., Dent R., Dutcher S., Fernández E., Fukuzawa H., González-Ballester D., González-Halphen D., Hallmann A., Hanikenne M., Hippler M., Inwood W., Jabbari K., Kalanon M., Kuras R., Lefebvre P. A., Lemaire S. D., Lobanov A. V., Lohr M., Manuell A., Meier I., Mets L., Mittag M., Mittelmeier T., Moroney J. V., Moseley J., Napoli C., Nedelcu A. M., Niyogi K., Novoselov S. V., Paulsen I. T., Pazour G., Purton S., Ral J. P., Riano-Pachón D. M., Riekhof W., Rymarquis L., Schroda M., Stern D., Umen J., Willows R., Wilson N., Zimmer S. L., Allmer J., Balk J., Bisova K., Chen C. J., Elias M., Gendler K., Hauser C., Lamb M. R., Ledford H., Long J. C., Minagawa J., Page M. D., Pan J., Pootakham W., Roje S., Rose A., Stahlberg E., Terauchi A. M., Yang P., Ball S., Bowler C., Dieckmann C. L., Gladyshev V. N., Green P., Jorgensen R., Mayfield S., Mueller-Roeber B., Rajamani S., Sayre R. T., Brokstein P., Dubchak I., Goodstein D., Hornick L., Huang Y. W., Jhaveri J., Luo Y., Martínez D., Ngau W. C., Otillar B., Poliakov A., Porter A., Szajkowski L., Werner G., Zhou K., Grigoriev I. V., Rokhsar D. S., Grossman A. R. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Karpowicz S. J., Prochnik S. E., Grossman A. R., Merchant S. S. (2011) The GreenCut2 resource, a phylogenomically derived inventory of proteins specific to the plant lineage. J. Biol. Chem. 286, 21427–21439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 17. Friso G., Giacomelli L., Ytterberg A. J., Peltier J. B., Rudella A., Sun Q., Wijk K. J. (2004) In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16, 478–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grossman A. R., Karpowicz S. J., Heinnickel M., Dewez D., Hamel B., Dent R., Niyogi K. K., Johnson X., Alric J., Wollman F. A., Li H., Merchant S. S. (2010) Phylogenomic analysis of the Chlamydomonas genome unmasks proteins potentially involved in photosynthetic function and regulation. Photosyn. Res. 106, 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gonzalez-Ballester D., Pootakham W., Mus F., Yang W., Catalanotti C., Magneschi L., de Montaigu A., Higuera J. J., Prior M., Galván A., Fernandez E., Grossman A. R. (2011) Reverse genetics in Chlamydomonas: a platform for isolating insertional mutants. Plant Methods 7, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Porra R. J., Thompson W. A., Kriedemann P. E. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394 [Google Scholar]
- 21. Hiyama T., Ke B. (1972) Difference spectra and extinction coefficients of P700. Biochim. Biophys. Acta 267, 160–171 [DOI] [PubMed] [Google Scholar]
- 22. Pierre Y., Breyton C., Kramer D., Popot J. L. (1995) Purification and characterization of the cytochrome b6f complex from Chlamydomonas reinhardtii. J. Biol. Chem. 270, 29342–29349 [DOI] [PubMed] [Google Scholar]
- 23. Maxwell K., Johnson G. N. (2000) Chlorophyll fluorescence–a practical guide. J. Exp. Bot. 51, 659–668 [DOI] [PubMed] [Google Scholar]
- 24. Genty B., Briantais J., Baker N. R. (1989) The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta 990, 87–92 [Google Scholar]
- 25. White C. C., Chain R. K., Malkin R. (1978) Duroquinol as an electron donor for chloroplast electron transfer reactions. Biochim. Biophys. Acta 502, 127–137 [DOI] [PubMed] [Google Scholar]
- 26. Houille-Vernes L., Rappaport F., Wollman F. A., Alric J., Johnson X. (2011) Plastid terminal oxidase 2 (PTOX2) is the major oxidase involved in chlororespiration in Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 108, 20820–20825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bennoun P. (2001) Chlororespiration and the process of carotenoid biosynthesis. Biochim. Biophys. Acta 1506, 133–142 [DOI] [PubMed] [Google Scholar]
- 28. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 29. Catalanotti C., Dubini A., Subramanian V., Yang W., Magneschi L., Mus F., Seibert M., Posewitz M. C., Grossman A. R. (2012) Altered fermentative metabolism in Chlamydomonas reinhardtii mutants lacking pyruvate formate lyase and both pyruvate formate lyase and alcohol dehydrogenase. Plant Cell, 24, 692–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thomas P. E., Ryan D., Levin W. (1976) An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75, 168–176 [DOI] [PubMed] [Google Scholar]
- 31. Pootakham W., Gonzalez-Ballester D., Grossman A. R. (2010) Identification and regulation of plasma membrane sulfate transporters in Chlamydomonas. Plant Physiol. 153, 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dussault A. A., Pouliot M. (2006) Rapid and simple comparison of messenger RNA levels using real-time PCR. Biol. Proced. Online 8, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lamesch P., Berardini T. Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D. L., Garcia-Hernandez M., Karthikeyan A. S., Lee C. H., Nelson W. D., Ploetz L., Singh S., Wensel A., Huala E. (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Peltier J. B., Ytterberg A. J., Sun Q., van Wijk K. J. (2004) New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J. Biol. Chem. 279, 49367–49383 [DOI] [PubMed] [Google Scholar]
- 35. Nakao M., Okamoto S., Kohara M., Fujishiro T., Fujisawa T., Sato S., Tabata S., Kaneko T., Nakamura Y. (2010) CyanoBase: the cyanobacteria genome database update 2010. Nucleic Acids Res. 38, D379–D381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Battchikova N., Eisenhut M., Aro E. M. (2011) Cyanobacterial NDH-1 complexes: novel insights and remaining puzzles. Biochim. Biophys. Acta 1807, 935–944 [DOI] [PubMed] [Google Scholar]
- 37. Desplats C., Mus F., Cuiné S., Billon E., Cournac L., Peltier G. (2009) Characterization of Nda2, a plastoquinone-reducing type II NAD(P)H dehydrogenase in Chlamydomonas chloroplasts. J. Biol. Chem. 284, 4148–4157 [DOI] [PubMed] [Google Scholar]
- 38. Jans F., Mignolet E., Houyoux P. A., Cardol P., Ghysels B., Cuiné S., Cournac L., Peltier G., Remacle C., Franck F. (2008) A type II NAD(P)H dehydrogenase mediates light-independent plastoquinone reduction in the chloroplast of Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 105, 20546–20551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yanofsky C., Platt T., Crawford I. P., Nichols B. P., Christie G. E., Horowitz H., VanCleemput M., Wu A. M. (1981) The complete nucleotide sequence of the tryptophan operon of Escherichia coli. Nucleic Acids Res. 9, 6647–6668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu Y. G., Mitsukawa N., Oosumi T., Whittier R. F. (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463 [DOI] [PubMed] [Google Scholar]
- 41. Joliot P., Delosme R. (1974) Flash-induced 519 nm absorption change in green algae. Biochim. Biophys. Acta 357, 267–284 [DOI] [PubMed] [Google Scholar]
- 42. Wykoff D. D., Davies J. P., Melis A., Grossman A. R. (1998) The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol. 117, 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chua N. H. (1971) The methyl viologen-catalyzed Mehler reaction and catalase activity in blue-green algae and Chlamydomonas reinhardtii. Biochim. Biophys. Acta 245, 277–287 [DOI] [PubMed] [Google Scholar]
- 44. Choquet Y., Stern D. B., Wostrikoff K., Kuras R., Girard-Bascou J., Wollman F. A. (1998) Translation of cytochrome f is autoregulated through the 5′-untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 95, 4380–4385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kuras R., Wollman F. A. (1994) The assembly of cytochrome b6/f complexes: an approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J. 13, 1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yildiz F. H., Davies J. P., Grossman A. R. (1994) Characterization of sulfate transport in Chlamydomonas reinhardtii during sulfur-limited and sulfur-sufficient growth. Plant Physiol. 104, 981–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bulté L., Wollman F. A. (1992) Evidence for a selective destabilization of an integral membrane protein, the cytochrome b6f complex, during gametogenesis in Chlamydomonas reinhardtii. Eur. J. Biochem. 204, 327–336 [DOI] [PubMed] [Google Scholar]
- 48. Miller R., Wu G., Deshpande R. R., Vieler A., Gärtner K., Li X., Moellering E. R., Zäuner S., Cornish A. J., Liu B., Bullard B., Sears B. B., Kuo M. H., Hegg E. L., Shachar-Hill Y., Shiu S. H., Benning C. (2010) Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 154, 1737–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cruz J. A., Salbilla B. A., Kanazawa A., Kramer D. M. (2001) Inhibition of plastocyanin to P700+ electron transfer in Chlamydomonas reinhardtii by hyperosmotic stress. Plant Physiol. 127, 1167–1179 [PMC free article] [PubMed] [Google Scholar]
- 50. Xiao J., Li J., Ouyang M., Yun T., He B., Ji D., Ma J., Chi W., Lu C., Zhang L. (2012) DAC is involved in the accumulation of the cytochrome b6/f complex in Arabidopsis thaliana. Plant Physiol., 160, 1911–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Malnoë A., Wollman F. A., de Vitry C., Rappaport F. (2011) Photosynthetic growth despite a broken Q-cycle. Nat. Commun. 2, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lennartz K., Bossmann S., Westhoff P., Bechtold N., Meierhoff K. (2006) HCF153, a novel nuclear-encoded factor necessary during a post-translational step in biogenesis of the cytochrome bf complex. Plant J. 45, 101–112 [DOI] [PubMed] [Google Scholar]
- 53. Sun Q., Zybailov B., Majeran W., Friso G., Olinares P. D., van Wijk K. J. (2009) PPDB, the Plant Database at Cornell. Nucleic Acid Res. 37, D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]