Background: The Ku70/80-DNA complex recruits DNA-PKcs to DSBs and results in activation of DNA-PKcs kinase activity.
Results: Truncation fragments of DNA-PKcs show that different regions of the protein are required for complete functionality of DNA-PKcs.
Conclusion: The N-terminal region of DNA-PKcs is required for its ability to interact with the Ku-DNA complex and its full activation.
Significance: We provide insights into the biochemical mechanism required for DNA-PKcs activation.
Keywords: DNA Damage, DNA Damage Response, DNA Repair, DNA-Protein Interaction, Molecular Biology, DNA Double-stranded Breaks, DNA-PKcs, Kinase Activity, Ku70/80
Abstract
DNA-dependent protein kinase (DNA-PK) plays an essential role in the repair of DNA double-stranded breaks (DSBs) mediated by the nonhomologous end-joining pathway. DNA-PK is a holoenzyme consisting of a DNA-binding (Ku70/Ku80) and catalytic (DNA-PKcs) subunit. DNA-PKcs is a serine/threonine protein kinase that is recruited to DSBs via Ku70/80 and is activated once the kinase is bound to the DSB ends. In this study, two large, distinct fragments of DNA-PKcs, consisting of the N terminus (amino acids 1–2713), termed N-PKcs, and the C terminus (amino acids 2714–4128), termed C-PKcs, were produced to determine the role of each terminal region in regulating the activity of DNA-PKcs. N-PKcs but not C-PKcs interacts with the Ku-DNA complex and is required for the ability of DNA-PKcs to localize to DSBs. C-PKcs has increased basal kinase activity compared with DNA-PKcs, suggesting that the N-terminal region of DNA-PKcs keeps basal activity low. The kinase activity of C-PKcs is not stimulated by Ku70/80 and DNA, further supporting that the N-terminal region is required for binding to the Ku-DNA complex and full activation of kinase activity. Collectively, the results show the N-terminal region mediates the interaction between DNA-PKcs and the Ku-DNA complex and is required for its DSB-induced enzymatic activity.
Introduction
DNA double-stranded breaks (DSBs)3 are the most lethal damage among the different kind of DNA damages because unrepaired DSBs can result in genomic instability, cell death, and tumorigenesis (1). In humans, nonhomologous end joining (NHEJ) plays an important role in repairing DSBs (2). The DNA-dependent protein kinase (DNA-PK) plays an essential role in NHEJ-mediated DSB repair (3). DNA-PK consists of two subunits, a scaffold subunit (Ku70/Ku80) and a catalytic subunit (DNA-PKcs). Upon creation of a DSB, Ku70/80 recognizes and quickly binds to the DNA double-stranded break ends (4). The DNA-bound Ku protein then recruits DNA-PKcs to the DNA ends where the catalytic activity of DNA-PKcs is increased (5). The large DNA-PKcs molecule forms a distinct structure at the DNA termini and likely plays an active role in the formation of a synaptic complex that holds the two ends of the broken DNA molecule together (6, 7). In low salt conditions, DNA-PKcs can bind to dsDNA ends, and its kinase activity is activated independently of the Ku heterodimer, but in physiological salt conditions, DNA-PKcs has limited affinity for DNA ends in the absence of Ku70/80 and has no to limited kinase activity (8, 9). Thus, the Ku heterodimer plays a significant role in stabilizing DNA-PKcs at DNA ends. Inactivation of DNA-PKcs kinase activity via point mutations or small molecule chemical inhibition results in radiosensitivity and a defect in DSB repair, indicating that the kinase activity is essential for NHEJ (10).
Once activated, DNA-PKcs can phosphorylate a number of substrates including XRCC4, H2AX, p53, and Ku70/80 (11). The best characterized DNA-PKcs substrate is DNA-PKcs itself. A large number of the phosphorylation sites are clustered in different regions of DNA-PKcs (12–15). Two prominent phosphorylation clusters that have been identified to be phosphorylated and autophosphorylated in response to IR are the Thr-2609 (12, 16) and Ser-2056 clusters (17–19). Phosphorylation of Ser-2056 is a bona fide autophosphorylation site (18), whereas phosphorylation of the Thr-2609 cluster can be phosphorylated by DNA-PKcs itself, ATM, and ATR (19, 20). Inhibiting phosphorylation at Ser-2056 together with the Thr-2609 cluster leads to severe radiosensitivity and alters the dynamics of DNA-PKcs at DSB sites, resulting in a rigid binding of DNA-PKcs to DNA ends in vivo, which interferes with the NHEJ process (21). A number of phosphorylation sites have been discovered in the C terminus, but most are uncharacterized. One that has been characterized is threonine 3950 (13). Ablation of this site by alanine (T3950A) substitution does not affect the kinase activity of DNA-PKcs, but the phospho-mimic aspartic acid (T3950D) substitution results in ablation of DNA-PKcs kinase activity. Finally, a solution structure of DNA-PKcs obtained by small angle x-ray scattering revealed that autophosphorylation of DNA-PKcs induces a conformational change, which likely releases DNA-PKcs from DNA ends (22).
DNA-PKcs is a member of PI-3K-like kinase (PIKK) family (23). Three PIKK family members, DNA-PKcs, ATM, and ATR, play important roles in the cellular response to DNA damage (24). The N-terminal region of the family members is not well conserved, but it has been postulated to be composed mostly of HEAT (Huntington, Elongation, A-subunit, TOR) repeats (25). The N-terminal region of DNA-PKcs contains a leucine zipper, which was shown to contribute to its innate DNA affinity (26). The C-terminal region of the family contains a FAT (FRAP, ATM, TRRAP) domain, a PI3K domain, and a FATC (FAT C-terminal) domain (27). The FAT domain is not highly conserved between the family members, but the PI3K and FATC domains are. The FAT and FATC domains likely play an important role in regulating kinase activity of the PIKK family members (28). A low resolution structure showed that binding to the Ku-DNA complex induces a conformational change in the FAT domain of DNA-PKcs and enhancement of DNA-PKcs kinase activity (29). Deletions in the FATC domain of ATM, ATR, and mammalian target of rapamycin result in a significant decrease of kinase activity, suggesting that this domain is important for the activity of the entire PIKK family (30–32). The importance of the FATC domain in regulating DNA-PKcs activity was first delineated when it was discovered that scid mice contain a premature termination mutation at amino acid 4045, which results in the deletion of the last 83 residues encompassing the FATC domain (33, 34). Predictions from a low resolution structure show that binding to the Ku-DNA complex results in a conformational change in the FATC domain of DNA-PKcs (35, 36). This conformation change is predicted to result in the alteration of the catalytic groups and/or the ATP-binding pocket of DNA-PKcs, resulting in full activation of its kinase activity. However, because of the large size DNA-PKcs (450 kDa), limited structure to function experiments have been performed, in particular experiments to determine the role the N-terminal region plays in regulating DNA-PKcs kinase activity.
Here, we show the purification and biochemistry of two large fragments of DNA-PKcs. The N-terminal region (amino acids 1–2713) of DNA-PKcs serves an important role in regulating DNA-PKcs because it is required for binding to the Ku-DNA complex, for the ability to localize to DSBs in vivo, and for full activation of the enzymatic activity of the protein. The C-terminal fragment (amino acids 2714–4128) of DNA-PKcs (C-PKcs) has kinase activity, but it is differentially regulated compared with full-length DNA-PKcs. First, C-PKcs has higher basal activity than DNA-PKcs, and unlike DNA-PKcs, C-PKcs is not activated by Ku and DNA. The data suggest that the N terminus is important for keeping the basal activity of DNA-PKcs low and is required for the Ku and DNA binding-induced conformational change required for activation of the protein. The ability of C-PKcs to phosphorylate known DNA-PKcs substrates is limited, suggesting that the N terminus may play a role in substrate recognition either directly or via an N terminus-mediated conformational change. Collectively, the data show that the N-terminal region of DNA-PKcs is required for full and complete activation of the protein.
EXPERIMENTAL PROCEDURES
Purification of Full-length DNA-PKcs, N-terminal fragment (N-PKcs), C-terminal fragment (C-PKcs), Ku70/Ku80, and XRCC4 C-terminal Fragment
Full-length DNA-PKcs was purified from HeLa cells as previously described with modifications (37). HeLa nuclear extracts were prepared and sequentially bound to phosphocellulose (PC11) resin. Proteins were eluted with a 100 mm step gradient from 0.1 to 1 m KCl. Peak DNA-PKcs containing fractions were pooled and equilibrated to column buffer (CB) (50 mm Tris-HCl, pH 7.9, 1 mm EDTA, 2% glycerol, 1 mm DTT, and 0.1 m KCl and protease inhibitors) before progressing to the next round of chromatography. The peak DNA-PKcs-containing fractions were next bound to a DEAE-Sepharose column and then a heparin-agarose column. As stated above, for each of these columns, the samples were eluted with a 100 mm step gradient from 0.1 to 1 m KCl, and peak DNA-PKcs-containing fractions were pooled and equilibrated to CB buffer. DNA-PKcs-containing fractions were further purified by two final rounds of chromatography: an ssDNA cellulose matrix followed by a Mono Q anion exchange column. Each column was eluted using a linear salt gradient from 0.1 to 1 m KCl, and peak fractions were collected and dialyzed in CB buffer before the next column. Purified DNA-PKcs was flash frozen and stored at −80 °C.
The N-PKcs and C-PKcs of DNA-PKcs were generated by PCR and subsequently subcloned into pFASTBac. Recombinant baculovirus expressing N-PKcs or C-PKcs were generated by using Bac-to-Bac baculovirus expression system (Invitrogen). Sf9 cells were infected with either N-PKcs or C-PKcs recombinant baculovirus and harvested 60 h after infection. Harvested Sf9 cells were lysed in 50 mm Tris-HCl, pH 7.9, 150 mm KCl, 1 mm EDTA, 10% glycerol, 20 μg/ml PMSF, 1 μg/ml pepstatin A, 1 μg/ml leupeptin, 1 μg/ml soybean trypsin inhibitor, and 1 mm DTT. Total cell lysate was centrifuged at 35,000 rpm for 1 h, and the supernatant was saved. NH2SO4 (33%) was added to the cleared supernatant with stirring for 1 h. The precipitate was pelleted via centrifugation at 14,000 rpm for 30 min. For N-PKcs, the pelleted precipitate was resuspended in CB buffer and bound to a DEAE column. N-PKcs was eluted via a step gradient of KCl (0.1, 0.3, and 0.5 m), and the samples were subsequently dialyzed in CB buffer. The samples were then applied to a heparin-agarose column, and the samples were eluted via a step gradient (0.1, 0.3, and 0.5 m), dialyzed, and then bound to a DEAE column and eluted as above. The dialyzed samples were then bound to a single-stranded DNA cellulose column and eluted via a linear gradient of KCl (0–1 m) and dialyzed. Finally, the samples were separated using a Mono Q column and eluted via a linear gradient of KCl (0–1 m). Fractions containing N-PKcs was confirmed by Western blotting with anti-DNA-PKcs (18-2) or anti-FLAG antibodies. N-PKcs-containing fractions were flash frozen and stored at −80 °C. For C-PKcs, the pelleted precipitate was resuspended in CB buffer and bound to a phosphocellulose (PC11) resin. C-PKcs was eluted via a linear gradient of KCl (0–1 m), and the sample was subsequently dialyzed in CB buffer overnight. The sample was applied to a Mono Q (GE Healthcare) column chromatography and eluted via a linear gradient of KCl (0–1 m) and then applied to Superdex 200 16/60 column. Finally, the samples were applied to a Mono Q column and eluted via a linear gradient of KCl (0–1 m). Fractions containing C-PKcs were confirmed by Western blotting with anti-DNA-PKcs (25-4) or anti-FLAG antibodies. C-PKcs containing fractions were flash frozen and stored at −80 °C. Ku70/Ku80 and GST fusion XRCC4 C-terminal fragments were prepared as previously described (37).
Cell Culture and Transfection
V3 cells and V3 cells stably expressing YFP-tagged DNA-PKcs were cultured in α-minimum Eagle's medium supplemented with 5% (v/v) fetal bovine serum, 5% (v/v) fetal calf serum, 100 units/ml penicillin, and 100 mg/ml streptomycin at humidified 5% CO2 incubation. V3 cells expressing YFP-DNA-PKcs were generated as previously described (21). For transient expression of YFP-N-PKcs or YFP-C-PKcs and DsRed-PCNA, V3 cells were transfected with YFP-N-PKcs or DsRed-PCNA and YFP-C-PKcs via electroporation using the reagents supplied in the Amaxa Nucleofector kit (Lonza) according to the manufacturer's instructions.
dsDNA Cellulose Pulldown Assay
dsDNA cellulose (GE Healthcare) was used for pulldown assays. Purified DNA-PKcs, N-PKcs, or C-PKcs were subjected to pulldown in the presence or absence of Ku70/80 protein. Pulldowns with or without 50 μg/ml ethidium bromide were also performed with DNA-PKcs and C-PKcs. Purified proteins were incubated with 40 μl of 50% slurry of DNA cellulose in binding buffer containing 50 mm Tris-HCl, pH 8.0, 0.5% EDTA, 150 mm KCl, 1% Nonidet P-40 at 4 °C for 1 h. DNA cellulose was pulled down by centrifugation at 2,000 rpm for 2 min and washed with binding buffer three times. Proteins pulled down via the dsDNA cellulose were resolved by a SDS-PAGE gel and transferred to a nitrocellulose membrane, and Western blot analysis was performed using the antibodies indicated in figure legends.
Live Cell Imaging and Laser Microirradiation
Live cell imaging combined with laser microirradiation was carried out as described previously (21, 38, 39).
In Vitro Kinase Assay
In vitro kinase assay was performed as previously described (37). Phosphorylation reactions contained 25 mm Tris-HCl, pH 7.9, 25 mm MgCl2, 1.5 mm DTT, 50 mm KCl, 10% glycerol, 100 ng of sonicated herring DNA, 0.16 μm [γ-32P]ATP (6000 Ci/mmol), 8 nm DNA-PKcs, N-PKcs, or C-PKcs, 20 nm Ku70/80, and GST-tagged XRCC4 C-terminal domain protein. The final volume was 10 μl. The reactions were incubated for 30 min at 30 °C and terminated by the addition of SDS-PAGE sample buffer. In vitro kinase assays with DNA-PKcs or C-PKcs in the presence concentrations of N-PKcs were performed as above, but N-PKcs at the following concentrations (8, 16, and 24 nm) was added. In vitro kinase assays with ssDNA oligonucleotides were performed as indicated above except in 10 mm KCl. The reactions were resolved via a 12% SDS-PAGE, and γ-32P incorporation was detected by PhosphorImager analysis (Amersham Biosciences).
RESULTS
Two Large, Distinct Fragments of DNA-PKcs Purified from Sf9 Cells
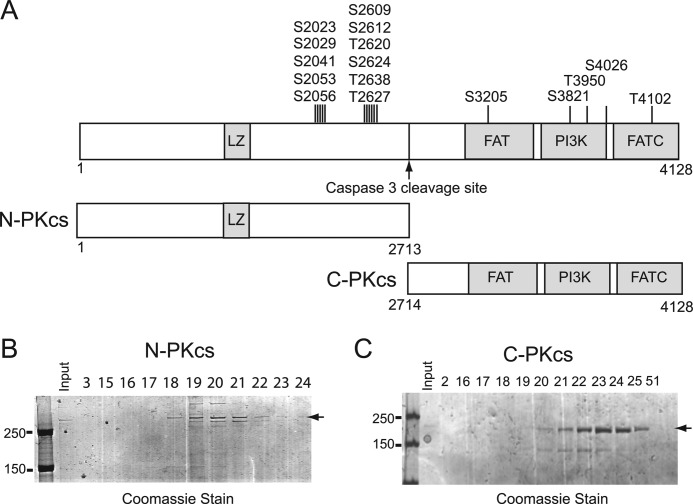
Deciphering the mechanisms regulating the activities of DNA-PKcs has been mostly limited because of the inability to express and purify recombinant DNA-PKcs protein fragments. DNA-PKcs is cleaved by caspase 3 following induction of apoptosis into two stable fragments (40, 41). Using the cleavage site as a guide, two large fragments of DNA-PKcs termed N-PKcs (amino acids 1–2713) and C-PKcs (amino acids 2714–4128) were expressed in and purified from Sf9 insect cells. N-PKcs contains the leucine zipper motif and the well characterized serine 2056 and threonine 2609 phosphorylation clusters, whereas C-PKcs contains the FAT domain, catalytic kinase domain (PI3K), and the FATC domain (Fig. 1A). N-PKcs and C-PKcs were purified by a series of column chromatography steps (supplemental Fig. S1, A and B, respectively) to near homogeneity (Fig. 1, B and C, respectively). Western blot analysis using monoclonal antibodies that specifically recognizes the C-terminal (25–4) and N-terminal (18-2) regions of DNA-PKcs was performed to confirm the identity of the purified fragments (supplemental Fig. S1, A and B, respectfully). The data show the successful purification of two large, distinct fragments of DNA-PKcs.
FIGURE 1.
Purification of N-PKcs and C-PKcs. A, schematic diagram of domains of DNA-PKcs, N-PKcs, and C-PKcs. LZ, leucine zipper. B, purification of N-PKcs. 5 μl of the indicated fractions were resolved via a 6% SDS-PAGE gel and stained with Coomassie Brilliant Blue stain. The full-length N-PKcs protein is indicated. C, purification of C-PKcs. 5 μl of the indicated fractions were resolved via a 6% SDS-PAGE gel and stained with Coomassie Brilliant Blue stain. The full-length C-PKcs protein is indicated.
The N-terminal Fragment of DNA-PKcs Interacts with the Ku-DNA Complex and Localizes to DSBs
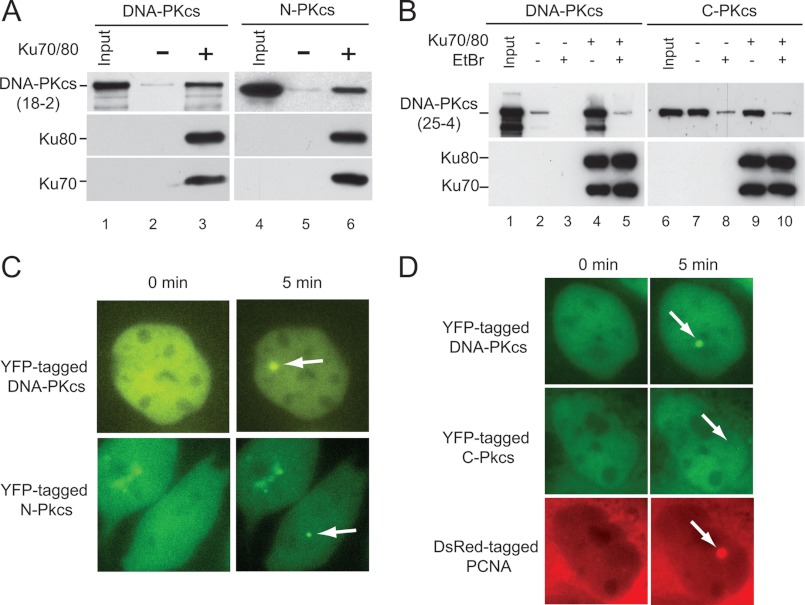
DNA-PKcs is recruited to DSBs by the Ku heterodimer where binding to Ku70/80 stabilizes DNA-PKcs at the DSB and results in activation of the kinase activity of DNA-PKcs (5, 21). Previous data showed that Ku70/80 interacts with amino acids 3002–3850 in the C-terminal region of DNA-PKcs, but it is not known whether this region of DNA-PKcs is required to interact with the Ku-DNA complex, which is the true DNA-PKcs binding partner in vivo (42). To test which portion of DNA-PKcs is required to interact with the Ku-DNA complex, pulldown assays were performed with dsDNA cellulose which was or was not prebound by the Ku heterodimer. Similar to previous studies, full-length DNA-PKcs has weak binding affinity for DNA in the absence of Ku70/80 (9), but the addition of the Ku heterodimer resulted in a marked increase in the amount of DNA-PKcs, which is bound to the dsDNA cellulose (Fig. 2A, lanes 2 and 3). Similar to full-length DNA-PKcs, N-PKcs has low affinity for dsDNA in vitro (Fig. 2A, compare lanes 2 and 5), and the addition of the Ku heterodimer on the dsDNA cellulose resulted in a marked increase in the amount of N-PKcs bound to the dsDNA cellulose (Fig. 2A, lane 6). dsDNA cellulose pulldown assays showed that C-PKcs interacts with dsDNA cellulose in the absence of Ku70/80, and this binding appears to be stronger than full-length DNA-PKcs (Fig. 2B, compare lanes 2 and 7). The addition of Ku70/80 did not result in an increase in the amount of C-PKcs, which bound to the dsDNA cellulose, suggesting that C-PKcs does not interact with the DNA-Ku complex (Fig. 2B, compare lanes 7 and 9). Ethidium bromide was also added in the dsDNA pulldown assays to disrupt DNA-protein interactions. The addition of ethidium bromide results in a marked decrease in the amount of DNA-PKcs, which is pulled down with the dsDNA cellulose even in the presence of Ku70/80, which is consistent with previous results showing that DNA is required for the DNA-PKcs-Ku70/80 interaction (Fig. 2B, lane 5) (5). The addition of ethidium bromide results in a marked decrease in the dsDNA-C-PKcs interaction (Fig. 2B, lanes 8 and 10), and C-PKcs could not interact with shorter dsDNA molecules (data not shown), which, taken together, suggests the DNA binding affinity of C-PKcs may not be specific. Together, the data show that the N-terminal region of DNA-PKcs interacts with the Ku-DNA complex and that the previously identified Ku-binding domain (amino acids 3002–3850), which is present in C-PKcs is not sufficient to interact with the Ku-DNA complex on its own in vitro.
FIGURE 2.
The N-terminal region of DNA-PKcs mediates the interaction between DNA-PKcs and the DNA-Ku complex. A and B, double-stranded DNA-cellulose was prebound with or without purified Ku70/80. Purified DNA-PKcs, N-PKcs, or C-PKcs were incubated with the DNA-cellulose with our without prebound Ku heterodimer for 1 h at room temperature. Additionally, DNA-PKcs or C-PKcs reactions were incubated with or without ethidium bromide. The samples were washed three times, resolved via a 6% SDS-PAGE gel, and transferred to nitrocellulose paper for Western blot analysis. Western blot analysis was performed using antibodies to DNA-PKcs (18-2 or 25-4), N-PKcs (18-2), and C-PKcs (25-4), Ku70, and Ku80. C and D, YFP-tagged full-length DNA-PKcs, N-PKcs, or C-PKcs with DsRed-PCNA were transiently expressed in the DNA-PKcs-deficient CHO cell line, V3. The nucleus was microirradiated using a 365-nm laser, and accumulation of the YFP-tagged and DsRed-tagged proteins to the DNA damage site was monitored. Localizations of DNA-PKcs, N-PKcs, or C-PKcs and DsRed-PCNA to laser-generated DSBs are marked by white arrows.
DNA-PKcs rapidly localizes to laser-generated DSBs, and this localization is dependent on the Ku heterodimer (21). To test whether N-PKcs or C-PKcs localizes to laser-induced DSBs, each fragment was tagged with a YFP. YFP-tagged N-PKcs or C-PKcs expressed in the DNA-PKcs-deficient cell line, V3, was strictly cytoplasmic (data not shown). The addition of a nuclear localization signal (NLS) resulted in diffuse localization of N-PKcs to the nucleus and the cytoplasm and C-PKcs to the nucleus in a relatively low number of V3 cells (Fig. 2, C and D, respectively). It was next determined whether the YFP-tagged N-PKcs or C-PKcs with a NLS could localize to laser-generated DSBs in the nucleus. YFP-tagged full-length DNA-PKcs and N-PKcs + NLS both rapidly localize to laser-generated DSBs (Fig. 2C), whereas YFP-tagged C-PKcs + NLS did not (Fig. 2D). Exogenously expressed DsRed-PCNA localized to the laser-induced DNA damage in the YFP-C-PKcs + NLS-expressing cells, which shows that DNA damage was produced in these cells (Fig. 2D). The data show that N-PKcs, but not C-PKcs, is able to localize to DSBs in vivo, which further supports that the N-terminal region of DNA-PKcs is required to interact with the Ku-DNA complex.
The C-terminal Fragment of DNA-PKcs Has Kinase Activity
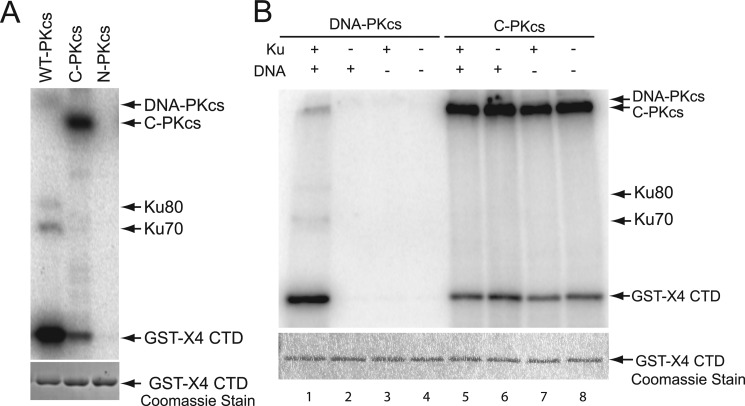
The C-terminal fragment of DNA-PKcs contains the catalytic domain of DNA-PKcs; therefore, it was assessed whether C-PKcs has kinase activity in vitro. Furthermore, assays with C-PKcs could reveal a possible role that the N-terminal region of the protein may play in regulating the kinase activity of DNA-PKcs. The kinase activity of C-PKcs was determined by performing standard DNA-PKcs in vitro kinase assays and compared with full-length DNA-PKcs and N-PKcs, which is not predicted to contain a kinase domain (43). C-PKcs has kinase activity in vitro, but N-PKcs does not (Fig. 3A). Unlike DNA-PKcs, C-PKcs prefers to autophosphorylate than phosphorylate the established DNA-PKcs substrate, the C-terminal domain (CTD) of XRCC4 (Fig. 3A). C-PKcs has ∼10% activity toward the XRCC4 CTD (supplemental Fig. S2A) and another established DNA-PKcs substrate, H2AX (data not shown), compared with full-length DNA-PKcs. Overall, the kinase activity of C-PKcs was reduced by ∼80% compared with full-length DNA-PKcs (supplemental Fig. S2B). Because C-PKcs has reduced activity compared with full-length DNA-PKcs in our assays, it was next tested whether this was due to inactivation via being autophosphorylated at threonine 3950. Threonine 3950 is located in the highly conserved PI3K kinase domain, and autophosphorylation at this site results in inactivation of DNA-PKcs kinase activity (13). Western blot analysis showed that the purified DNA-PKcs and C-PKcs were not phosphorylated at threonine 3950, illustrating that the lower activity of C-PKcs is not due to being autophosphorylated at this site in Sf9 cells or during purification of the protein (supplemental Fig. S3, lanes 1 and 3). Because C-PKcs autophosphorylates in vitro, it was assessed whether C-PKcs phosphorylated itself at threonine 3950. Surprisingly, in contrast to full-length DNA-PKcs, C-PKcs does not autophosphorylate at threonine 3950 (supplemental Fig. S3, lanes 2 and 4). C-PKcs has kinase activity in vitro, and to our knowledge C-PKcs is the first purified fragment of DNA-PKcs to retain kinase activity. The marked decrease in substrate phosphorylation suggests that the N terminus may play a role in the proper phosphorylation of DNA-PKcs substrates.
FIGURE 3.
C-PKcs has kinase activity in vitro and activity is not activated by Ku70/80and dsDNA. A, in vitro kinase assay using purified DNA-PKcs, N-PKcs, or C-PKcs. The kinase assays were performed using standard DNA-PKcs kinase assay conditions, which include adding purified Ku70/80 heterodimer, DNA, and a 1:5 32P-ATP:cold ATP mixture to the reaction. GST-tagged C-terminal domain of XRCC4 (GST-X4 CTD) was used as a substrate. 32P-Labeled proteins were observed using phosphorimaging, and Coomassie staining was performed to verify equal amounts of substrate were included. B, kinase activity of C-PKcs is Ku70/80- and DNA-independent. In vitro kinase assays were performed using purified DNA-PKcs or C-PKcs either in the presence or absence of Ku70/80, dsDNA, and Ku70/80 and dsDNA, and activity was assessed as described above.
C-PKcs Has Constitutive Kinase Activity and Is Not Activated by Ku70/80 and DNA
One characterized mechanism for regulating DNA-PKcs activity is its interaction with the Ku-DNA complex, which stimulates DNA-PKcs kinase activity (5). In the absence of the Ku heterodimer and DNA, full-length DNA-PKcs has limited kinase activity at physiological salt concentrations (5). It was next determined whether the Ku-DNA complex activated C-PKcs kinase activity. In vitro kinase assays were performed in the presence Ku70/80, dsDNA, or Ku70/80 and dsDNA. As shown in Fig. 3B, full-length DNA-PKcs has very little kinase activity in the absence of the Ku heterodimer (lane 2), DNA (lane 3), and both Ku70/80 and DNA (lane 4) but was activated in their presence (lane 1). C-PKcs had kinase activity in the absence of Ku70/80 and DNA, and the addition of the Ku-DNA complex did not further activate the kinase activity of C-PKcs (Fig. 3B, lanes 5–8). A previous study showed that in low salt concentrations, free ssDNA ends can activate DNA-PKcs in vitro in the absence of the Ku heterodimer (44). Furthermore, a structural study of DNA-PKcs suggested that the C-terminal crown structure of the protein may contain a ssDNA-binding pocket that was hypothesized to possibly be responsible for ssDNA-mediated activation of DNA-PKcs (45). Thus, kinase assays were performed in the presence of ssDNA in low salt conditions or dsDNA and Ku70/80 in normal salt conditions to determine whether ssDNA activates the kinase activity C-PKcs. The kinase assays showed that the addition of ssDNA did not further activate either full-length DNA-PKcs or C-PKcs (supplemental Fig. S4A, lanes 1–4), whereas dsDNA and Ku70/80 activated DNA-PKcs but not C-PKcs (supplemental Fig. S4A, lanes 5–8). Lastly, kinase assays were performed with C-PKcs in the presence of ssDNA of various lengths, but none were able to stimulate the enzymatic activity of C-PKcs (supplemental Fig. S4B). The data implicate that ssDNA is unlikely able to activate DNA-PKcs in vitro. Taken together, the data suggest that the N-terminal region of DNA-PKcs plays a role in regulating the kinase activity by keeping the basal activity of the protein low because removal of the N-terminal region of the protein results in spontaneous activation of the kinase. Furthermore, the ability of the N-terminal region of DNA-PKcs to interact with the Ku-DNA complex is required for full activation of DNA-PKcs. Overall, the data show that the kinase activity of C-PKcs is differentially regulated compared with DNA-PKcs.
Activity of C-PKcs Is Inhibited by the DNA-PKcs Specific Inhibitor NU7026 but C-PKcs Is Differentially Inhibited by PI3K Inhibitors Compared with DNA-PKcs
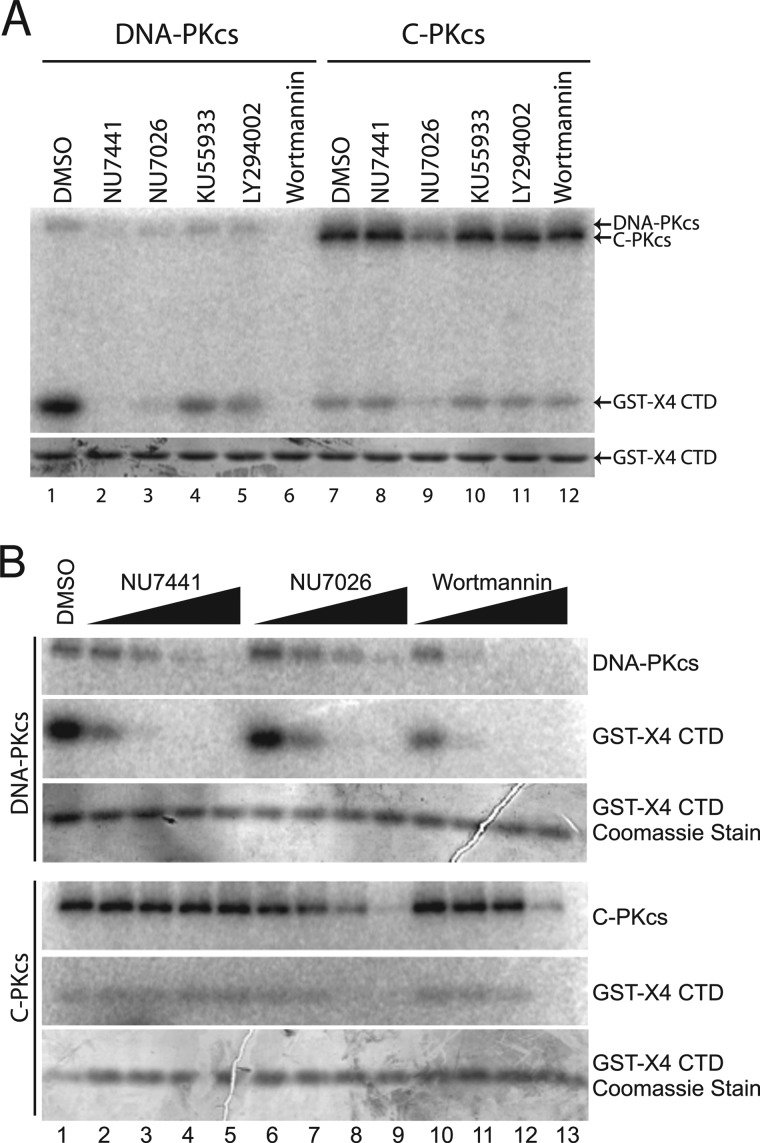
Because the kinase activity of C-PKcs is differentially regulated compared with DNA-PKcs, it was next assessed using inhibitors whether the activity which was observed was due to the C-PKcs protein or due to a contaminating kinase. Furthermore, experiments with the inhibitors could give clues to the integrity of the kinase domain, in particular the ATP-binding pocket of C-PKcs compared with DNA-PKcs. Both competitive inhibitors (LY294002 and those derived from LY294002, which include the DNA-PKcs specific inhibitors NU7441 and NU7026 and the ATM specific inhibitor KU55933) and a noncompetitive inhibitor (wortmannin) for the ATP-binding pocket of DNA-PKcs were used to determine whether the kinase activity of C-PKcs could be inhibited (46–49). In vitro kinase assays were performed with full-length DNA-PKcs or C-PKcs with a panel of kinase inhibitors which included Me2SO (mock), NU7441 (DNA-PKcs), NU7026 (DNA-PKcs), KU55933 (ATM), LY294002 (broad PI3K inhibitor), and wortmannin (broad PI3K inhibitor). Full-length DNA-PKcs was inhibited by NU7441, NU7026, and wortmannin (Fig. 4A, compare lanes 1 with lanes 2, 3, and 6). KU55933 and LY294002 slightly inhibited DNA-PKcs kinase activity at the concentration (10 μm) used in this assay (Fig. 4A, lanes 4 and 5) but were not able to inhibit DNA-PKcs at lower concentrations (data not shown). Surprisingly, C-PKcs was only effectively inhibited by NU7026 (Fig. 4A, lane 9). To further expand on this, full-length DNA-PKcs and C-PKcs were incubated with increasing concentrations (0.1–10 μm) of NU7441, NU7026, or wortmannin. Comparing inhibition of autophosphorylation of DNA-PKcs and C-PKcs shows that DNA-PKcs is inhibited by each inhibitor at low concentrations (Fig. 4B), whereas C-PKcs autophosphorylation is only comparably inhibited by NU7026 (Fig. 4B, compare lanes 6–9 for DNA-PKcs and C-PKcs). Wortmannin can inhibit C-PKcs kinase activity only at high concentrations of the drug, but the potent DNA-PKcs inhibitor NU7741 does not appear to inhibit C-PKcs. C-PKcs kinase activity is blocked by the DNA-PKcs specific inhibitor NU7026, suggesting that the observed activity in our assays is due to C-PKcs. Binding to the Ku-DNA complex produces a conformational change that opens the ATP-binding pocket and results in activation of the kinase activity of DNA-PKcs (5, 29). Enzymatic activity in the absence of the Ku-DNA complex and inhibition by NU7026 illustrates that deletion of the N-terminal region of the protein likely produces a conformational change in the catalytic core of the protein, which results in its activation. However, the inability of wortmannin and NU7441 to effectively inhibit C-PKcs implies that this conformational change at or near the ATP-binding pocket of the protein is not the same as the one induced by binding to the Ku-DNA complex in full-length DNA-PKcs.
FIGURE 4.
C-PKcs is differentially blocked by PI3K inhibitors compared with DNA-PKcs. A, in vitro kinase assays were performed and assessed as described for Fig. 3A except 10 μm of the indicated PI3K kinase inhibitor was preincubated with the reaction mixture before the addition of the ATP mix. B, in vitro kinase assays were performed as previously described except in the presence of the control dimethyl sulfoxide (DMSO), the broad PI3K inhibitor wortmannin, or specific DNA-PKcs inhibitor NU7441 or NU7026. Increasing concentrations (0.1, 1, 10, or 100 μm) of each inhibitor were used. Equal loading was confirmed by Coomassie staining of each SDS-PAGE gel.
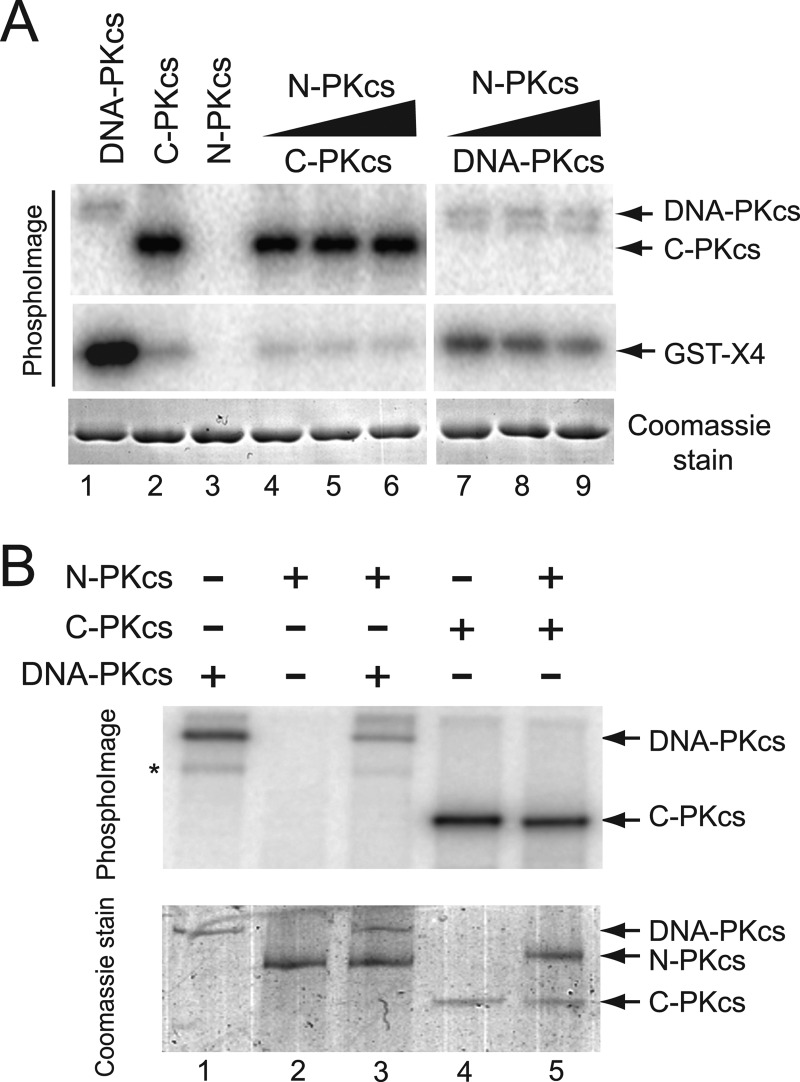
Addition of N-PKcs to Kinase Assays Does Not Correct Misregulation of C-PKcs and Inhibits Full-length DNA-PKcs Kinase Activity
Because N-PKcs is able to interact with the Ku-DNA complex, it was next tested whether addition of N-PKcs to the in vitro kinase assays could correct the apparent misregulation of the C-PKcs kinase activity. In vitro kinase assays were performed as outlined above, but with increasing concentrations of purified N-PKcs protein. The addition of N-PKcs did not affect the kinase activity of C-PKcs because C-PKcs still had little activity toward the substrate XRCC4 CTD (Fig. 5A, compare lanes 2 and lanes 4–6), and its autophosphorylation was unaffected (Fig. 5B, lanes 4 and 5). Surprisingly, the addition of N-PKcs to the kinase assays with full-length DNA-PKcs resulted in a decrease in phosphorylation of the XRCC4 CTD (Fig. 5A, compare lanes 1 and lanes 7–9) and its autophosphorylation (Fig. 5B, lanes 1 and 3). The decrease in DNA-PKcs-mediated phosphorylations in the presence of N-PKcs is likely because N-PKcs is completing with DNA-PKcs for the Ku-DNA complex, which is required for activation of the kinase activity of DNA-PKcs. The data further suggest that N-PKcs interacts with the DNA-Ku complex.
FIGURE 5.
A, in vitro kinase assays were performed and assessed as described in Fig. 3A except kinase assays with DNA-PKcs or C-PKcs were performed either in the absence or with increasing concentrations of N-PKcs. B, in vitro assays were performed as described above, but in the absence of the substrate XRCC4 CTD, in order to assess autophosphorylation of DNA-PKcs or C-PKcs in the absence or presence of N-PKcs.
DISCUSSION
To study the structure to function relationship of DNA-PKcs, we attempted to make truncation fragments of the protein. The first site chosen to cut was the caspase cleavage site at aspartic acid 2713. Previous work from our laboratory (data not shown) and others (40, 41) has shown that following apoptosis, the two apoptotic-induced fragments of DNA-PKcs are present in cells and appeared to be stable. Subautomic structure of DNA-PKcs modeled that the caspase cleavage site is located near a hinge region of the protein, which links the two main phosphorylation clusters, Ser-2056 and Thr-2609, to the C-terminal PI3K kinase, FAT, and FATC domains (50). Using this as a clue to make stable fragments of DNA-PKcs, two large and distinct protein fragments were expressed and purified from Sf9 insect cells. Testing the kinase activity first showed that the C-terminal, but not the N-terminal, fragment has kinase activity. To our knowledge, these are the first large recombinant fragments of DNA-PKcs to be purified and the first fragment of the protein, which still retains catalytic activity.
The kinase activity we observed in our assays is likely from the C-PKcs protein because it was inhibited by the DNA-PKcs specific inhibitor NU7026 and the general PI3K kinase inhibitor wortmannin. Furthermore, it was able to phosphorylate, albeit weakly compared with full-length DNA-PKcs, three known substrates of DNA-PKcs, the CTD of XRCC4, H2AX (data not shown), and the Ku heterodimer (data not shown). Taken together, the kinase activity purified from the Sf9 insect cells is in fact a PI3K-like kinase and is likely from the C-PKcs molecule. NU7026 is a competitive inhibitor for the ATP-binding pocket of DNA-PKcs; thus inhibition by NU7026 and kinase activity of C-PKcs suggests the C-PKcs ATP-binding pocket is intact (46). Surprisingly, the DNA-PKcs specific inhibitor NU7441 does not inhibit C-PKcs. NU7441 and NU7026 were both derived from the chromenone backbone of LY29004 (47). Because NU7026 is a smaller molecule than NU7441, the data suggest that the conformational change in the ATP-binding pocket produced by deletion of the N terminus does not result in a fully open ATP-binding pocket, resulting in the ability of the smaller molecules, ATP and NU7026, to bind but occlusion of the larger NU7441. At high concentrations, C-PKcs is inhibited by wortmannin. Wortmannin is a noncompetitive inhibitor of DNA-PKcs, which covalently attaches to DNA-PKcs at lysine 2751 via a nucleolyitc attack by a furan group (48). Inhibition by wortmannin further suggests that C-PKcs retains a structural integrity similar to full-length DNA-PKcs.
The major step in the recruitment of DNA-PKcs to DSBs is its ability to interact with the DNA-Ku70/80 complex (5). This stabilizes DNA-PKcs at the DSB and results in a conformational change that results in activation of its kinase activity (5). Experimental evidence in this study shows that the N-terminal region of DNA-PKcs mediates the interaction between DNA-PKcs and the Ku-DNA complex. This notion is supported by structural studies showing that the N-terminal region of DNA-PKcs forms a central cavity that has been implicated to produce a putative dsDNA-binding channel (45, 50). Eliminating the N-terminal portion of DNA-PKcs results in the loss of this central cavity and thus a loss in DNA threading through the channel, which is likely required to stabilize the DNA-PKcs-Ku-DNA complex. A previous study identified a Ku-binding motif in the C-terminal portion of DNA-PKcs (amino acids 3002–3850), but our data show that C-PKcs, which contains this motif, cannot interact with the Ku-DNA complex, suggesting that this motif is not sufficient for the interaction with Ku70/80 in vivo (42). Predictions from low resolution structure show that Ku has multiple contacts with DNA-PKcs including contacts with the N- and C-terminal regions of the protein (29, 36). We hypothesize that both the N- and C-terminal regions of the DNA-PKcs make contacts with Ku70/80, but that the N-terminal region is absolutely necessary for the ability of the protein to interact with and/or be stabilized by the Ku-DNA complex. Finally, the N-terminal region of the DNA damage-responsive PI3K-like protein kinase family members may play a general role in mediating the association of the proteins to DNA damage or the area near the damage site. A previous report showed that, similar to C-PKcs, a C-terminal domain fragment of ATM (amino acids 1303–3056) is strictly cytoplasmic, but addition of an exogenous NLS to the fragment results in its localization to the nucleus (51). Furthermore, this C-terminal fragment of ATM even after the addition of a NLS could not localize to DNA damage and had significantly decreased ability to phosphorylate known ATM substrates, similar to the C-terminal fragment of DNA-PKcs. Taken together, the data strongly suggest that the N-terminal regions of both DNA-PKcs and ATM play a significant role in mediating their association with DNA damage and play an important role in modulating the ability of each protein to phosphorylate substrates.
C-PKcs showed increased basal level of kinase activity compared with DNA-PKcs. Activation of kinase activity of DNA-PKcs is dependent on binding to the DNA-Ku complex as shown in Fig. 3A. However, deletion of the N-terminal region of DNA-PKcs not only abrogates binding to the Ku-DNA complex, but also results in spontaneous activation of its kinase activity. A recent study showed DNA and Ku-independent activation of a N-terminally constrained DNA-PKcs (52). The data suggest that a conformational change mediated by the N terminus of the protein results in enzyme activation. Our data suggest that deletion of the N-terminal region deletes a physical restraint, which keeps DNA-PKcs basal activity low. It is likely that the deletion of the N-terminal portion results in a slight conformational change, which opens the ATP-binding pocket of the protein, which results in an increase in basal kinase activity. Furthermore, similar to C-PKcs, DNA-PKcs tethered by its N terminus also autophosphorylates efficiently in the absence of DNA and Ku. C-PKcs preferred to autophosphorylate over phosphorylating substrates, suggesting that binding to the DNA-Ku complex results in a conformational change, which affects substrate binding and its DNA-PKcs-mediated phosphorylation. C-PKcs does not autophosphorylate at a known site (Thr-3950), which further supports the hypothesis that substrate recognition is affected by deletion of the N-terminal region of DNA-PKcs. Taken together, the two studies clearly show that the N terminus plays a role in modulating the enzymatic activity of DNA-PKcs.
The truncation of DNA-PKcs was designed using the known caspase-3 cleavage site (40). Previous studies have shown that apoptotic-mediated cleavage results in a near elimination in DNA-PKcs kinase activity at 24 h after treatment with etoposide (40). Although we do see a significant (∼90%) decrease in activity toward known substrates (XRCC4 and H2AX), autophosphorylation is still observed, and total activity is ∼20% compared with full-length DNA-PKcs. Our data implies that apoptosis-driven caspase cleavage of DNA-PKcs abrogates the ability of DNA-PKcs to bind to the DNA-Ku complex and thus likely completely inhibits the ability of NHEJ to be completed. Because apoptosis results in fragmentation of the chromosomal DNA and thus generates a large number of DSBs, blocking DSB repair and DNA damaging signaling would be required for apoptosis to be completed. Therefore, apoptosis-mediated cleavage of DNA-PKcs may be one method by which an apoptotic cell blocks DSB repair because cleaved DNA-PKcs cannot interact with the Ku-DNA complex and assist in mediating NHEJ. The fact that C-PKcs retains kinase activity but its regulation is different from DNA-PKcs suggests that there is a possibility that this truncated DNA-PKcs may play some other significant role in apoptosis, because DNA-PKcs has been implicated in preventing and also being required for apoptosis (53, 54). Recently, a study showed that a large number of proteins are phosphorylated in response to apoptosis in a DNA-PKcs-dependent manner (55). It was found that DNA-PKcs was cleaved early following the induction of apoptosis, and the C-terminal fragment produced by caspase cleavage relocated from the nucleus to the cytoplasm, where, it is assumed, it phosphorylates a number of proteins. Taken together with our study, C-PKcs likely has its own unidentified substrates, and its total activity is completely different from that of DNA-PKcs and may play a role in the communication between DSB repair pathways and apoptosis.
Taken together, our study shows that the N-terminal region (amino acids 1–2712) of DNA-PKcs serves an important role in regulating DNA-PKcs. The N-terminal portion of DNA-PKcs is required for binding to the DNA-Ku complex and activation of the enzymatic activity of the protein. The C-terminal fragment of DNA-PKcs lacking the N-terminal region of the protein has kinase activity in vitro, but its activity is differentially regulated. C-PKcs has higher basal activity than DNA-PKcs, and unlike DNA-PKcs, C-PKcs is not activated by Ku and DNA. The data suggest that the N terminus is important for keeping the basal activity of DNA-PKcs low and is required for the Ku and DNA binding conformational change required for activation of the protein. Lastly, we propose that the apoptotic cleaved fragment of DNA-PKcs may also be involved in other cellular functions during apoptosis through the differentially regulated kinase activity of C-PKcs.
Acknowledgments
We thank Miaw-Sheue Tsai and the Expression and Molecular Biology Core (Lawrence Berkeley National Laboratory, Berkeley, CA) for creation of the N-PKcs and C-PKcs baculovirus and expression of the protein. We are also grateful to Zhengping Shao for assistance with the experiment testing the real time recruitment kinetics of DNA-PKcs, N-PKcs, and C-PKcs to laser-generated DNA damage. We also thank Benjamin B. P. Chen and Kazi R. Fattah for offering suggestions on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants CA50519 and CA13499. This work was also supported by Cancer Prevention Research Institute of Texas Grant RP110465.

This article contains supplemental Figs. S1–S4.
- DSB
- double-stranded break
- DNA-PK
- DNA-dependent protein kinase
- DNA-PKcs
- DNA-PK catalytic subunit
- N-PKcs
- N-terminal fragment of DNA-PKcs
- C-PKcs
- C-terminal fragment of DNA-PKcs
- NHEJ
- nonhomologous end-joining
- PIKK
- PI3K-like kinase
- FAT
- FRAP, ATM, TRRAP domain
- FATC
- FAT-C-terminal domain
- ssDNA
- single-stranded DNA
- YFP
- yellow fluorescence protein
- NLS
- nuclear localization signal
- CTD
- C-terminal domain
- CB
- column buffer
- ATM
- Ataxia telangiectasia mutated
- ATR
- Ataxia telangiectasia and Rad3 related.
REFERENCES
- 1. Khanna K. K., Jackson S. P. (2001) DNA double-strand breaks. Signaling, repair and the cancer connection. Nat. Genet. 27, 247–254 [DOI] [PubMed] [Google Scholar]
- 2. Burma S., Chen B. P., Chen D. J. (2006) Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair 5, 1042–1048 [DOI] [PubMed] [Google Scholar]
- 3. Lieber M. R. (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walker J. R., Corpina R. A., Goldberg J. (2001) Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412, 607–614 [DOI] [PubMed] [Google Scholar]
- 5. Gottlieb T. M., Jackson S. P. (1993) The DNA-dependent protein kinase. Requirement for DNA ends and association with Ku antigen. Cell 72, 131–142 [DOI] [PubMed] [Google Scholar]
- 6. Cary R. B., Peterson S. R., Wang J., Bear D. G., Bradbury E. M., Chen D. J. (1997) DNA looping by Ku and the DNA-dependent protein kinase. Proc. Natl. Acad. Sci. U.S.A. 94, 4267–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weterings E., van Gent D. C. (2004) The mechanism of non-homologous end-joining. A synopsis of synapsis. DNA Repair 3, 1425–1435 [DOI] [PubMed] [Google Scholar]
- 8. Hammarsten O., Chu G. (1998) DNA-dependent protein kinase. DNA binding and activation in the absence of Ku. Proc. Natl. Acad. Sci. U.S.A. 95, 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. West R. B., Yaneva M., Lieber M. R. (1998) Productive and nonproductive complexes of Ku and DNA-dependent protein kinase at DNA termini. Mol. Cell. Biol. 18, 5908–5920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kurimasa A., Kumano S., Boubnov N. V., Story M. D., Tung C. S., Peterson S. R., Chen D. J. (1999) Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol. Cell. Biol. 19, 3877–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mahaney B. L., Meek K., Lees-Miller S. P. (2009) Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 417, 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Merkle D., Douglas P., Moorhead G. B., Leonenko Z., Yu Y., Cramb D., Bazett-Jones D. P., Lees-Miller S. P. (2002) The DNA-dependent protein kinase interacts with DNA to form a protein-DNA complex that is disrupted by phosphorylation. Biochemistry 41, 12706–12714 [DOI] [PubMed] [Google Scholar]
- 13. Douglas P., Cui X., Block W. D., Yu Y., Gupta S., Ding Q., Ye R., Morrice N., Lees-Miller S. P., Meek K. (2007) The DNA-dependent protein kinase catalytic subunit is phosphorylated in vivo on threonine 3950, a highly conserved amino acid in the protein kinase domain. Mol. Cell. Biol. 27, 1581–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3. [DOI] [PubMed] [Google Scholar]
- 15. Ma Y., Pannicke U., Lu H., Niewolik D., Schwarz K., Lieber M. R. (2005) The DNA-dependent protein kinase catalytic subunit phosphorylation sites in human Artemis. J. Biol. Chem. 280, 33839–33846 [DOI] [PubMed] [Google Scholar]
- 16. Chan D. W., Chen B. P., Prithivirajsingh S., Kurimasa A., Story M. D., Qin J., Chen D. J. (2002) Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 16, 2333–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cui X., Yu Y., Gupta S., Cho Y. M., Lees-Miller S. P., Meek K. (2005) Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol. Cell. Biol. 25, 10842–10852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen B. P., Chan D. W., Kobayashi J., Burma S., Asaithamby A., Morotomi-Yano K., Botvinick E., Qin J., Chen D. J. (2005) Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J. Biol. Chem. 280, 14709–14715 [DOI] [PubMed] [Google Scholar]
- 19. Chen B. P., Uematsu N., Kobayashi J., Lerenthal Y., Krempler A., Yajima H., Löbrich M., Shiloh Y., Chen D. J. (2007) Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J. Biol. Chem. 282, 6582–6587 [DOI] [PubMed] [Google Scholar]
- 20. Yajima H., Lee K. J., Chen B. P. (2006) ATR-dependent phosphorylation of DNA-dependent protein kinase catalytic subunit in response to UV-induced replication stress. Mol. Cell. Biol. 26, 7520–7528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Uematsu N., Weterings E., Yano K., Morotomi-Yano K., Jakob B., Taucher-Scholz G., Mari P. O., van Gent D. C., Chen B. P., Chen D. J. (2007) Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J. Cell Biol. 177, 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hammel M., Yu Y., Mahaney B. L., Cai B., Ye R., Phipps B. M., Rambo R. P., Hura G. L., Pelikan M., So S., Abolfath R. M., Chen D. J., Lees-Miller S. P., Tainer J. A. (2010) Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J. Biol. Chem. 285, 1414–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abraham R. T. (2004) PI 3-kinase related kinases. “Big” players in stress-induced signaling pathways. DNA Repair 3, 883–887 [DOI] [PubMed] [Google Scholar]
- 24. Bakkenist C. J., Kastan M. B. (2004) Initiating cellular stress responses. Cell 118, 9–17 [DOI] [PubMed] [Google Scholar]
- 25. Perry J., Kleckner N. (2003) The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell 112, 151–155 [DOI] [PubMed] [Google Scholar]
- 26. Gupta S., Meek K. (2005) The leucine rich region of DNA-PKcs contributes to its innate DNA affinity. Nucleic Acids Res. 33, 6972–6981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hartley K. O., Gell D., Smith G. C., Zhang H., Divecha N., Connelly M. A., Admon A., Lees-Miller S. P., Anderson C. W., Jackson S. P. (1995) DNA-dependent protein kinase catalytic subunit. A relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell 82, 849–856 [DOI] [PubMed] [Google Scholar]
- 28. Lempiäinen H., Halazonetis T. D. (2009) Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J. 28, 3067–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Spagnolo L., Rivera-Calzada A., Pearl L. H., Llorca O. (2006) Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol. Cell 22, 511–519 [DOI] [PubMed] [Google Scholar]
- 30. Banin S., Moyal L., Shieh S., Taya Y., Anderson C. W., Chessa L., Smorodinsky N. I., Prives C., Reiss Y., Shiloh Y., Ziv Y. (1998) Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281, 1674–1677 [DOI] [PubMed] [Google Scholar]
- 31. Peterson R. T., Beal P. A., Comb M. J., Schreiber S. L. (2000) FKBP12-rapamycin-associated protein (FRAP) autophosphorylates at serine 2481 under translationally repressive conditions. J. Biol. Chem. 275, 7416–7423 [DOI] [PubMed] [Google Scholar]
- 32. Sun Y., Jiang X., Chen S., Fernandes N., Price B. D. (2005) A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. U.S.A. 102, 13182–13187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Beamish H. J., Jessberger R., Riballo E., Priestley A., Blunt T., Kysela B., Jeggo P. A. (2000) The C-terminal conserved domain of DNA-PKcs, missing in the SCID mouse, is required for kinase activity. Nucleic Acids Res. 28, 1506–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Blunt T., Gell D., Fox M., Taccioli G. E., Lehmann A. R., Jackson S. P., Jeggo P. A. (1996) Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proc. Natl. Acad. Sci. U.S.A. 93, 10285–10290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rivera-Calzada A., Maman J. D., Spagnolo L., Pearl L. H., Llorca O. (2005) Three-dimensional structure and regulation of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs). Structure 13, 243–255 [DOI] [PubMed] [Google Scholar]
- 36. Rivera-Calzada A., Spagnolo L., Pearl L. H., Llorca O. (2007) Structural model of full-length human Ku70-Ku80 heterodimer and its recognition of DNA and DNA-PKcs. EMBO Rep. 8, 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee K. J., Jovanovic M., Udayakumar D., Bladen C. L., Dynan W. S. (2004) Identification of DNA-PKcs phosphorylation sites in XRCC4 and effects of mutations at these sites on DNA end joining in a cell-free system. DNA Repair 3, 267–276 [DOI] [PubMed] [Google Scholar]
- 38. So S., Davis A. J., Chen D. J. (2009) Autophosphorylation at serine 1981 stabilizes ATM at DNA damage sites. J. Cell Biol. 187, 977–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shao Z., Davis A. J., Fattah K. R., So S., Sun J., Lee K. J., Harrison L., Yang J., Chen D. J. (2012) Persistently bound Ku at DNA ends attenuates DNA end resection and homologous recombination. DNA Repair 11, 310–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Song Q., Lees-Miller S. P., Kumar S., Zhang Z., Chan D. W., Smith G. C., Jackson S. P., Alnemri E. S., Litwack G., Khanna K. K., Lavin M. F. (1996) DNA-dependent protein kinase catalytic subunit. A target for an ICE-like protease in apoptosis. EMBO J. 15, 3238–3246 [PMC free article] [PubMed] [Google Scholar]
- 41. Mukherjee B., Kessinger C., Kobayashi J., Chen B. P., Chen D. J., Chatterjee A., Burma S. (2006) DNA-PK phosphorylates histone H2AX during apoptotic DNA fragmentation in mammalian cells. DNA Repair 5, 575–590 [DOI] [PubMed] [Google Scholar]
- 42. Jin S., Kharbanda S., Mayer B., Kufe D., Weaver D. T. (1997) Binding of Ku and c-Abl at the kinase homology region of DNA-dependent protein kinase catalytic subunit. J. Biol. Chem. 272, 24763–24766 [DOI] [PubMed] [Google Scholar]
- 43. Lee K. J., Dong X., Wang J., Takeda Y., Dynan W. S. (2002) Identification of human autoantibodies to the DNA ligase IV/XRCC4 complex and mapping of an autoimmune epitope to a potential regulatory region. J. Immunol. 169, 3413–3421 [DOI] [PubMed] [Google Scholar]
- 44. Hammarsten O., DeFazio L. G., Chu G. (2000) Activation of DNA-dependent protein kinase by single-stranded DNA ends. J. Biol. Chem. 275, 1541–1550 [DOI] [PubMed] [Google Scholar]
- 45. Williams D. R., Lee K. J., Shi J., Chen D. J., Stewart P. L. (2008) Cryo-EM structure of the DNA-dependent protein kinase catalytic subunit at subnanometer resolution reveals alpha helices and insight into DNA binding. Structure 16, 468–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hollick J. J., Golding B. T., Hardcastle I. R., Martin N., Richardson C., Rigoreau L. J., Smith G. C., Griffin R. J. (2003) 2,6-disubstituted pyran-4-one and thiopyran-4-one inhibitors of DNA-Dependent protein kinase (DNA-PK). Bioorg. Med. Chem. Lett. 13, 3083–3086 [DOI] [PubMed] [Google Scholar]
- 47. Leahy J. J., Golding B. T., Griffin R. J., Hardcastle I. R., Richardson C., Rigoreau L., Smith G. C. (2004) Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg. Med. Chem. Lett. 14, 6083–6087 [DOI] [PubMed] [Google Scholar]
- 48. Izzard R. A., Jackson S. P., Smith G. C. (1999) Competitive and noncompetitive inhibition of the DNA-dependent protein kinase. Cancer Res. 59, 2581–2586 [PubMed] [Google Scholar]
- 49. Hickson I., Zhao Y., Richardson C. J., Green S. J., Martin N. M., Orr A. I., Reaper P. M., Jackson S. P., Curtin N. J., Smith G. C. (2004) Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 64, 9152–9159 [DOI] [PubMed] [Google Scholar]
- 50. Sibanda B. L., Chirgadze D. Y., Blundell T. L. (2010) Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature 463, 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Young D. B., Jonnalagadda J., Gatei M., Jans D. A., Meyn S., Khanna K. K. (2005) Identification of domains of ataxia-telangiectasia mutated required for nuclear localization and chromatin association. J. Biol. Chem. 280, 27587–27594 [DOI] [PubMed] [Google Scholar]
- 52. Meek K., Lees-Miller S. P., Modesti M. (2012) N-terminal constraint activates the catalytic subunit of the DNA-dependent protein kinase in the absence of DNA or Ku. Nucleic Acids Res. 40, 2964–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gurley K. E., Moser R., Gu Y., Hasty P., Kemp C. J. (2009) DNA-PK suppresses a p53-independent apoptotic response to DNA damage. EMBO Rep. 10, 87–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hill R., Leidal A. M., Madureira P. A., Gillis L. D., Waisman D. M., Chiu A., Lee P. W. (2008) Chromium-mediated apoptosis. Involvement of DNA-dependent protein kinase (DNA-PK) and differential induction of p53 target genes. DNA Repair 7, 1484–1499 [DOI] [PubMed] [Google Scholar]
- 55. Dix M. M., Simon G. M., Wang C., Okerberg E., Patricelli M. P., Cravatt B. F. (2012) Functional interplay between caspase cleavage and phosphorylation sculpts the apoptotic proteome. Cell 150, 426–440 [DOI] [PMC free article] [PubMed] [Google Scholar]