Background: MeCP2 is an abundant methyl-CG-binding protein.
Results: Purified human MeCP2 is a potent, stoichiometric, HDAC-independent repressor that blocks preinitiation complex assembly. Methyl-CG binding is not sufficient for repression.
Conclusion: MeCP2 has the features of a global HDAC-independent repressor.
Significance: This HDAC-independent mechanism of MeCP2-mediated repression may be important in cells, such as mammalian neurons, that have high levels of MeCP2.
Keywords: DNA Methylation, Promoters, RNA Polymerase II, Transcription, Transcription Repressor, CG Methylation, CG-deficient Core Promoter, Core Promoter, Gel Mobility Shift, MeCP2
Abstract
MeCP2 is an abundant methyl-cytosine-guanine (CG)-binding protein and transcriptional repressor. We developed a biochemical system that exhibits CG methylation-specific transcriptional repression by purified human MeCP2. MeCP2 represses transcription by histone deacetylase (HDAC)-dependent and HDAC-independent mechanisms. Our system appears to recreate the HDAC-independent component of MeCP2-mediated repression and occurs via inhibition of the assembly of transcription preinitiation complexes. At a ratio of approximately one molecule of MeCP2 per two methyl-CG dinucleotides, as found in mammalian neurons, the magnitude of methylation-specific repression was greater than 10-fold. Notably, the HDAC inhibitor trichostatin A had no effect on MeCP2-mediated repression with either naked DNA or chromatin templates. We designed a CG-deficient core promoter that is resistant to MeCP2-mediated repression when placed in a plasmid lacking CG dinucleotides. By using this CG-deficient reporter as a reference, we found that eight CG dinucleotides in the core promoter region are sufficient for strong methylation-specific repression by MeCP2. In contrast, MeCP2 does not repress a construct with 13 CG dinucleotides located ∼1.7 kbp upstream of the promoter. Furthermore, by analysis of C-terminally truncated MeCP2 proteins, we found that binding of MeCP2 to methyl-CG dinucleotides is not sufficient for transcriptional repression. Hence, MeCP2-mediated repression is not due to the simple steric blockage of the transcriptional machinery. These experiments suggest that MeCP2 can function as a global methyl-CG-specific, HDAC-independent repressor. This HDAC-independent mechanism of MeCP2-mediated repression may be important in cells, such as mammalian neurons, that have high levels of CG methylation and MeCP2.
Introduction
Methyl-CpG-binding protein 2 (MeCP2) is a member of the methyl-CG-binding domain (MBD)5 family of proteins (1, 2). Mutations in the MECP2 gene cause Rett syndrome (3), an X-linked dominant neurodevelopmental disorder that is characterized by apparently normal development until 6–18 months of age, at which point the disorder is manifested (4). Rett syndrome patients exhibit developmental regression, seizures, and autistic behaviors. To understand the underlying molecular basis of Rett syndrome, it is important to elucidate the biochemical activities of MeCP2.
MeCP2 is a 53-kDa monomeric protein that binds to DNA containing methyl-CG dinucleotides, particularly in the vicinity of A/T-rich sequences (1, 5). MeCP2 is generally considered to be a transcriptional repressor, although genetic studies have also suggested that it may activate transcription (6). In addition to the MBD (7), MeCP2 has a transcriptional repression domain, termed the TRD (8). There are both histone deacetylase (HDAC)-dependent and HDAC-independent modes of transcriptional repression by MeCP2. The HDAC-dependent process involves the binding of MeCP2 to Sin3A-HDAC complexes (9, 10). However, MeCP2-mediated repression of transcription in cells is only partially relieved by treatment with HDAC inhibitors such as trichostatin A (TSA). Hence, a significant component of MeCP2-mediated repression occurs via an HDAC-independent mechanism that has been largely unexplored.
In humans, most CG dinucleotides are methylated, particularly in the brain (11), where the effects of mutation of MeCP2 on Rett syndrome patients are most evident. In the mouse brain, the genome-wide association of MeCP2 correlates with CG methylation (12). Moreover, in mouse neurons, there is nearly one molecule of MeCP2 per nucleosome (12). Hence, in the brain, MeCP2 may act as a global repressor of transcription. To investigate this possibility, we established a biochemical transcription assay with purified full-length, wild-type human MeCP2. With this system, we observed potent CG-methylation-specific, HDAC-independent repression of transcription by MeCP2. The biochemical properties of MeCP2 suggest that HDAC-independent repression may be an important component of MeCP2 function in the brain.
EXPERIMENTAL PROCEDURES
Synthesis and Purification of Recombinant Human MeCP2 Proteins
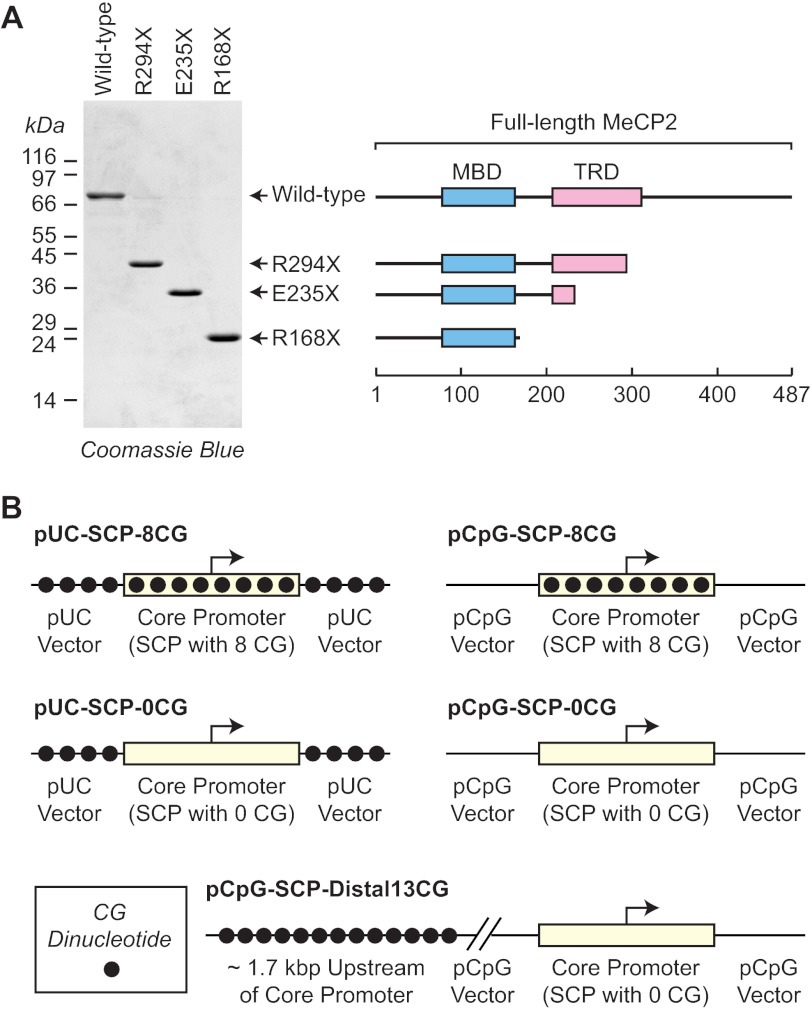
Recombinant wild-type and C-terminally truncated MeCP2 proteins (isoform 1, also known as MeCP2A, MeCP2e2, and MeCP2β) were synthesized and purified as described previously (13). Briefly, chitin-binding domain fusion proteins were synthesized in bacteria by overnight incubation at 16 °C. The bacteria were lysed by sonication, and the soluble fusion proteins were applied to chitin beads (New England Biolabs), which were subsequently washed to remove contaminating proteins. The MeCP2 proteins were cleaved from the beads by overnight incubation in buffer containing 50 mm DTT. The resulting proteins were dialyzed twice for 1 h against potassium storage buffer (25 mm HEPES-K+ (pH 7.6), 400 mm potassium acetate, 6 mm MgCl2, 20% (v/v) glycerol, 1 mm DTT, 1 mm benzamidine-HCl, 0.2 mm PMSF), dispensed into aliquots, frozen in liquid nitrogen, and stored at −80 °C. The purity of the proteins was assessed by polyacrylamide-SDS gel electrophoresis (Fig. 1A), and protein concentrations were measured by using the Bio-Rad protein assay with bovine serum albumin as the reference protein.
FIGURE 1.
Wild-type and mutant MeCP2 proteins and reporter constructs. A, purification of wild-type and mutant MeCP2 proteins. Shown is the polyacrylamide-SDS gel electrophoresis of full-length MeCP2 isoform 1 (also known as MeCP2A, MeCP2e2, and MeCP2β) as well as the indicated C-terminally truncated mutant proteins. The locations of the MBD and HDAC-dependent TRD are shown. B, reporter constructs. Two versions of the super core promoter (with sequences from −50 to +50 relative to the +1 transcription start site) were designed to contain either eight CG dinucleotides or 0 CG dinucleotides. These core promoters were inserted into pUC119 (which has 209 CG dinucleotides) or pCpG-Txn (which has 0 CG dinucleotides) to give the reporter constructs. In addition, the pCpG-SCP-Distal13CG construct has a stretch of 13 CG dinucleotides ∼1.7 kbp upstream of the SCP lacking CG dinucleotides.
Preparation of HeLa Nuclear Extract
All operations were carried out at 4 °C. HeLa cell pellets from 12 L of culture (∼50 ml of packed HeLa cells, National Cell Culture Center) were washed with 150 ml PBS containing 5 mm MgCl2. The cells were then pelleted by centrifugation at 3000 rpm for 10 min. The pellet was suspended in 150 ml PBS containing 5 mm MgCl2, and the cells were pelleted again by centrifugation at 3000 rpm for 10 min. The pellet was suspended in 60 ml of buffer H (10 mm Tris-HCl (pH 7.9), 10 mm KCl, 750 μm spermidine, 150 μm spermine, 0.1 mm EDTA, 0.1 mm EGTA). The cells were incubated on ice for 20 min and then lysed by 20 strokes with a 40-ml Wheaton Dounce homogenizer with the B (loose) pestle. The resulting lysate was subjected to centrifugation in a Sorvall SS-34 rotor at 8500 rpm for 10 min. The nuclei were suspended in 60 ml of buffer H and pelleted by centrifugation in an SS-34 rotor at 8500 rpm for 10 min. The nuclei were suspended in 80 ml of buffer AB (15 mm HEPES-K+ (pH 7.6), 110 mm KCl, 5 mm MgCl2, 0.1 mm EDTA, 2 mm DTT, 1 mm benzamidine-HCl, 0.2 mm PMSF, 1 mm NaHSO3). The nuclei were dispersed by three strokes with a 40-ml Wheaton Dounce homogenizer using the B (loose) pestle. Another 60 ml of buffer AB was added to the dispersed nuclei to give a total volume of ∼140 ml. The nuclei were then lysed by the addition of 1/10th volume of 4 m ammonium sulfate and incubated for 20 min on a rotating wheel. The nuclear debris was pelleted by ultracentrifugation in a Beckman 45 Ti rotor at 35,000 rpm for 60 min. The supernatant was transferred to a 250-ml beaker, and 0.3 g of pulverized ammonium sulfate was added per 1 ml of supernatant. The solution was stirred for 15 min, and the resulting suspension was subjected to centrifugation in an SS-34 rotor at 15,000 rpm for 20 min. The precipitate was suspended in 5 ml of HEG buffer (25 mm HEPES-K+ (pH 7.6), 0.1 mm EDTA, 10% (v/v) glycerol) containing 0.1 m KCl, 1 mm DTT, 1 mm benzamidine-HCl, 0.2 mm PMSF, and 1 mm NaHSO3. The mixture was dispersed by 10 strokes with a 15-ml Dounce homogenizer using the A (tight) pestle. Insoluble debris was pelleted by centrifugation in an SS-34 rotor at 10,000 rpm for 10 min. The supernatant was dialyzed three times for 50 min against 2 L of HEG buffer containing 0.1 m KCl, 1 mm DTT, 1 mm benzamidine-HCl, 0.2 mm PMSF, and 1 mm NaHSO3. Insoluble debris was pelleted by centrifugation in an SS-34 rotor at 10,000 rpm for 10 min. The supernatant was then dispensed into aliquots, frozen in liquid nitrogen, and stored at −80 °C.
pCpG-Txn Plasmid
The pCpG-Txn plasmid was derived from the pCpG-Luc plasmid (Invivogen) as follows. pCpG-Luc was digested with PstI to remove the mCMV enhancer and hEF1 promoter. This construct was digested with PciI, filled in with Escherichia coli RNA polymerase I (Klenow), and ligated to destroy the PciI restriction site. The resulting plasmid was then modified by the insertion of the sequence TACATGTACTTGTAGATCTTCTAGATGGGAACTGCAGGGATCCATGACATCATGAA into the PstI and XbaI sites. This procedure destroyed the existing multiple cloning site and generated a new proximal multiple cloning site. The resulting plasmid was modified by the insertion of the sequence TAAGCTTTGACTATGTACAACTAGTTGACTAATGCATGGGCCCTGACTACTTAAGACATG into the SphI site. This step created a distal multiple cloning site and yielded the pCpG-Txn plasmid.
Reporter DNA Templates
The reporter DNA templates were constructed with either pUC119 vector, which contains 209 CG dinucleotides (in the region used for subcloning), or the pCpG-Txn vector, which lacks CG dinucleotides. pUC-SCP-8CG and pCpG-SCP-8CG were generated by ligation of DNA containing the SCP-8CG promoter GCGTGTACGGTGGGAGGTCTATATAAGCAGAGCTCGTTTAGTGAACCGTCAGATCGCCTGGAGACGTCGAGCCGAGTGGTTGTGCCTCCATAGAAGACAC into the PstI and XbaI sites of pUC119 and pCpG-Txn, respectively. We also designed a CG-deficient version of the super core promoter and termed this promoter SCP-0CG. pUC-SCP-0CG and pCpG-SCP-0CG were constructed by ligation of DNA containing the SCP-0CG promoter GCTTGTACTATGGGAGGTCTATATAAGCAGAGCTGGTTTAGTGAACCCTCAGATTGCCTGGAGACCTCAAGCCAAGTGGTTGTGCCTCCATAGAAGACAC into the PstI and XbaI sites of pUC119 and pCpG-Txn, respectively. pCpG-SCP-Distal13CG was created by inserting the sequence AGCTTCGAACGACTAGTCGAACGTACGAACGTTCGAACGATCGTACGATCGAACGTACGT into the HindIII and SpeI sites of pCpG-SCP-0CG. The 13CG insert is located ∼1.7 kbp upstream (and 2.4 kbp downstream) of the core promoter. Schematic diagrams of these five reporter plasmids are shown in Fig. 1B. Methylation at C5 of CG dinucleotides in the reporter constructs was performed with the methyltransferase M.SssI (New England Biolabs) and S-adenosylmethionine. The methylation state of the M.SssI-treated DNAs, except for pCpG-SCP-Distal13CG, was verified by digestion of the constructs with the methylation-sensitive restriction enzymes BstUI and BsaHI (New England Biolabs). The methylation of the pCpG-SCP-Distal13CG construct was confirmed by resistance to the HpyCH4IV CG methylation-sensitive restriction enzyme (New England Biolabs).
In Vitro Transcription Analysis
For reactions with pUC119-based templates, 250 ng of template and 750 ng of unmethylated pUC119 competitor were included in each reaction. For reactions with pCpG-Txn-based templates, 250 ng of template, 750 ng of unmethylated pUC119 competitor, and 250 ng of methylated pUC119 competitor were added to the reaction. In the reactions with pCpG-Txn-based plasmids, the methylated pUC119 was added to provide approximately the same concentration of methylated CG dinucleotides as that in the reactions with pUC119-based plasmids. In a standard reaction, MeCP2 was incubated at 30 °C with template and competitor DNA for 20 min in a total volume of 36 μl in a buffer containing 20 mm HEPES-K+ (pH 8.0), 69 mm KOAc, 8 mm MgCl2, 3.5% (w/v) polyvinyl alcohol, 4.2 mm ATP, and 3.5% (v/v) glycerol. Next, HeLa nuclear extract (5.5 μl) and HEG containing 0.1 m KCl (4.5 μl) were added. Preinitiation complex (PIC) assembly was allowed to occur at 30 °C for 90 min in a total volume of 46 μl in buffer containing 21 mm HEPES-K+ (pH 8.0), 22 mm KCl, 54 mm KOAc, 6.5 mm MgCl2, 2.7% (w/v) polyvinyl alcohol, 3.3 mm ATP, and 5% (v/v) glycerol. Following PIC assembly, a solution (4 μl) containing 5 mm in each of the four ribonucleoside triphosphates was added, and the transcription reaction was performed at 30 °C for 20 min in a final volume of 50 μl in buffer containing a final concentration of 20 mm HEPES-K+ (pH 8.0); 20 mm KCl; 50 mm KOAc; 6 mm MgCl2; 2.5% (w/v) polyvinyl alcohol; 3.4 mm ATP; 0.4 mm CTP, GTP, and UTP; and 4.5% (v/v) glycerol. In reactions with Sarkosyl, buffer volumes were adjusted to allow the addition of 5% (w/v) Sarkosyl (2 μl) to give a final concentration of 0.2% (w/v) Sarkosyl. In reactions with TSA, initial buffer volumes were adjusted to allow the addition of TSA solution (2 μl) to give a final volume of 50 μl. Reactions were terminated by the addition of 100 μl of stop buffer (20 mm EDTA, 200 mm NaCl, 1% (w/v) SDS, 0.3 mg/ml glycogen) and 5 μl of 2.5 mg/ml proteinase K. The mixture was incubated for 20 min at 37 °C, and the nucleic acids were extracted with phenol-CHCl3 and precipitated with EtOH. The resulting transcripts were subjected to primer extension analysis as described previously (14) using 5′-32P-labeled M13 reverse sequencing primer AGCGGATAACAATTTCACACAGGA for reporters on the basis of pUC119 or 5′-32P-labeled pCpG-PE1 primer GGAAAGAGAAGAAGGTTAGTACAATTGT for pCpG-Txn-based reporters. Chromatin was assembled with purified ACF, NAP1, and histones and then characterized by DNA supercoiling and partial micrococcal nuclease digestion analyses, essentially as described by Fyodorov and Kadonaga (15). Transcription of the chromatin templates was carried out by the method of Jiang et al. (16). Quantification of reverse transcription products was performed with a Typhoon imager (GE Health Sciences). All transcription experiments were carried out a minimum of three independent times to ensure reproducibility of the results.
Gel Mobility Shift Assay
Electrophoretic mobility shift assays were performed by using conditions similar to those described previously (17) with the following modifications. First, the probe DNA has sequences from the promoter region of the human BDNF gene and has been designed as a single-stranded DNA with a double-stranded-like hairpin (dslHairpin) sequence from bacteriophage N4 promoters (18). The dslHairpin sequence facilitates the proper hybridization of the BDNF promoter sequence containing a single CG dinucleotide. The sequence of the BDNF-dslHairpin probe is ATCTTTTATTAGAAGAATTCCGTTCCAGGGCATTGCATGCTTGAAGCGGAGCTTCAAGCATGCAATGCCCTGGAACGGAATTCTTCTAATAAA. Unmethylated (normal) and methyl-CG versions of the probe were synthesized and used. The binding reactions contained 0–20 nm of wild-type or R168X truncation mutant MeCP2 protein, 3.13 nm of 5′-32P-labeled BDNF-dslHairpin probe, and 2.5 μm of unmethylated unlabeled competitor probe. Competitor DNA was not included in these experiments. The protein-DNA complexes were resolved on a 5% native polyacrylamide gel. Quantification of the protein-DNA complexes was performed with a Typhoon imager (GE Health Sciences). These experiments were carried out more than three independent times to ensure reproducibility of the data.
RESULTS
Stoichiometric, HDAC-independent, CG Methylation-specific Transcriptional Repression by Human MeCP2
To analyze the mechanism of transcriptional repression by MeCP2, we synthesized and purified full-length human MeCP2 protein (Fig. 1A). To test the activity of the purified MeCP2, we used pUC-SCP-8CG as the reporter construct (Fig. 1B). This plasmid, which is on the basis of the pUC119 vector, contains a version of the super core promoter (SCP) (19) with eight CG dinucleotides in the region from −50 to +50 relative to the +1 transcription start site. pUC-SCP-8CG has a total of 217 CG dinucleotides, 209 from pUC119 and eight from the SCP-8CG core promoter. When necessary, C5 methylation of the CG dinucleotides was performed with the M.SssI CG methyltransferase and S-adenosylmethionine. The methylation reactions were carried out essentially to completion, as the resulting M.SssI-treated templates were resistant to digestion by methylation-sensitive enzymes.
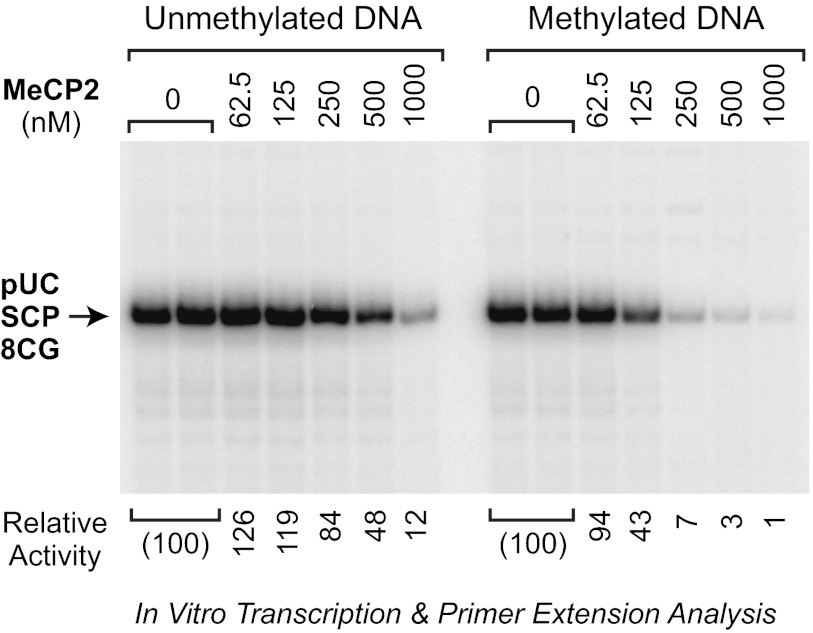
To assess the intrinsic transcriptional activity of the purified MeCP2, we subjected methylated or unmethylated pUC-SCP-8CG template DNAs to in vitro transcription analysis with a HeLa nuclear extract in the absence of MeCP2 or the presence of varying concentrations of MeCP2 (Fig. 2). In the absence of MeCP2, the level of transcription from the methylated template was nearly identical to that from the unmethylated template. Hence, the HeLa extract did not contain a significant level of methylation-specific repressors. In the presence of MeCP2, we observed potent, concentration-dependent transcriptional repression. MeCP2 represses transcription from both the unmethylated and methylated templates, but the magnitude of repression from the methylated template is substantially greater than that from the unmethylated template. For instance, at a MeCP2-to-CG dinucleotide ratio of 1:2 (Fig. 2, 250 nm), which is approximately the same as that seen in mouse neurons (12), the magnitude of methyl-CG-specific repression was over 10-fold. This preference for repression of methylated templates relative to unmethylated templates is similar to the higher affinity of MeCP2 for binding to DNA containing methylated CG dinucleotides relative to unmethylated CG dinucleotides (see, for example, Refs. 1, 7, 20). Thus, full-length, wild-type MeCP2 has the intrinsic ability to repress transcription in a CG methylation-specific manner. These findings are generally consistent with previous reports of transcriptional repression by MeCP2 in vitro (see, for example, Refs. 8, 21).
FIGURE 2.
Purified MeCP2 is a potent CG-methylation-specific repressor of transcription. Shown is transcriptional repression by purified full-length MeCP2. CG-methylated or unmethylated pUC-SCP-8CG template DNAs were transcribed with HeLa nuclear extract either in the absence of MeCP2 or in the presence of the indicated concentrations of wild-type MeCP2. At 500 nm MeCP2, there is approximately one molecule of MeCP2 per CG dinucleotide in the pUC-SCP-8CG template. The resulting transcripts were subjected to primer extension analysis. The image is a continuous single exposure of a series of reactions that were performed in parallel with equivalent amounts of methylated or unmethylated template DNA. The relative activity values are normalized to the amount of transcription observed in the absence of MeCP2.
The Histone Deacetylase Inhibitor TSA Does Not Relieve MeCP2-mediated Transcriptional Repression with Either Naked DNA or Chromatin Templates
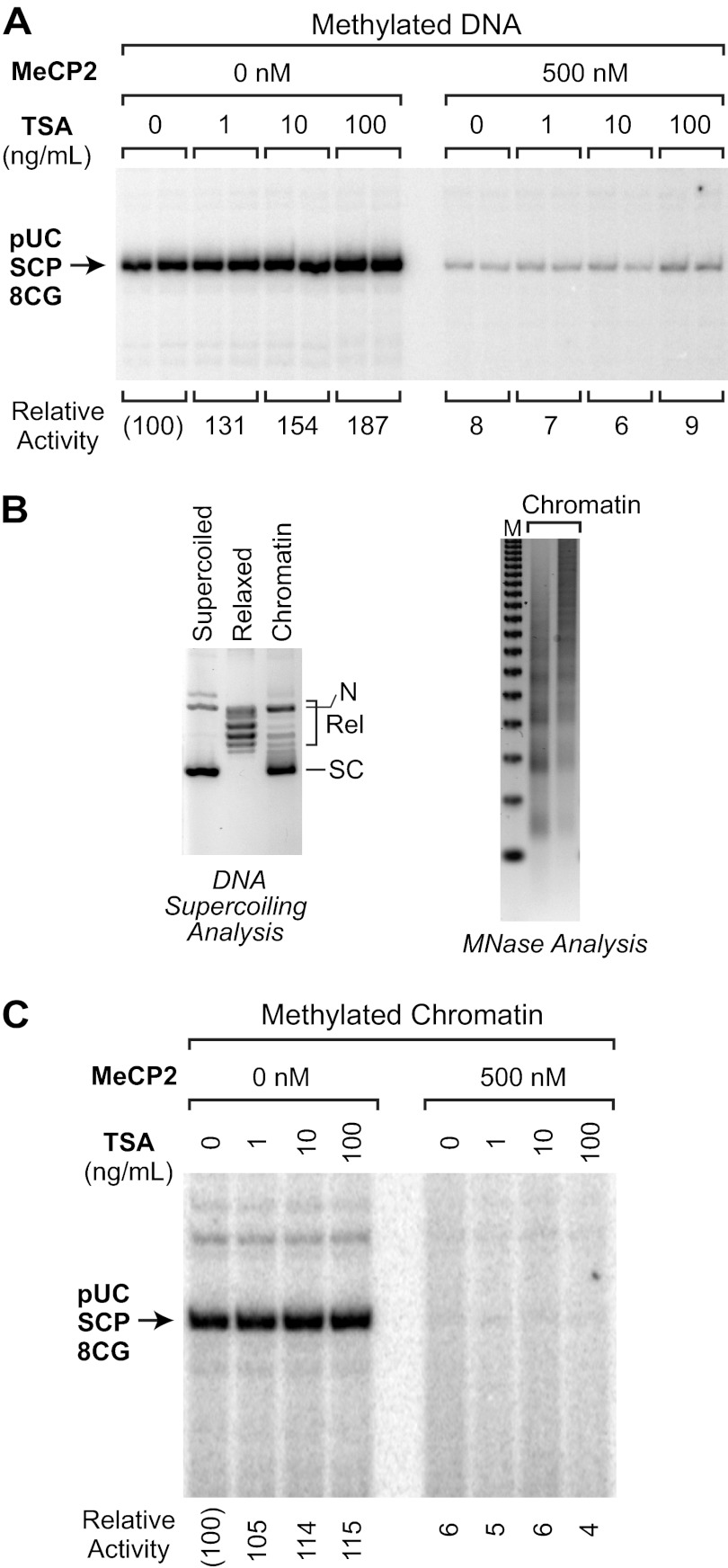
The study of MeCP2 as a transcriptional repressor has mainly focused on its HDAC-dependent mechanism of repression, but it is also known that a substantial fraction of MeCP2-mediated transcriptional repression is resistant to treatment with the HDAC inhibitor TSA (9, 10, 22). We therefore sought to determine whether the transcriptional repression that we observed in vitro occurs via HDAC-dependent and/or HDAC-independent mechanisms. To test for HDAC dependence of repression, we performed transcription reactions in the presence or absence of TSA. We first examined the effect of TSA on MeCP2-mediated repression of methylated pUC-SCP-8CG as naked DNA. Under these conditions, we found that the addition of TSA (at concentrations that are comparable with those used in vitro and in cells to inhibit HDAC activity (9, 10, 23)) did not affect the magnitude of MeCP2-mediated repression (Fig. 3A).
FIGURE 3.
MeCP2-mediated transcriptional repression is not affected by the histone deacetylase inhibitor TSA with either naked DNA or chromatin templates. A, TSA does not affect MeCP2-mediated transcriptional inhibition with naked DNA templates. Transcription reactions were performed, as in Fig. 2, with methylated pUC-SCP-8CG template in the absence or presence of the specified concentrations of MeCP2 and TSA. B, assembly of chromatin with purified ACF, NAP1, and histones. Methylated pUC-SCP-8CG was assembled into chromatin with purified ACF, NAP1, and histones by the method of Fyodorov and Kadonaga (15). The chromatin was analyzed by the DNA supercoiling and partial micrococcal nuclease digestion assays. In the DNA supercoiling gel, the positions of the supercoiled (SC), relaxed (Rel), and nicked open circular DNA (N) are indicated. In the micrococcal digestion gel, the size markers (M) are the 123 bp ladder (Invitrogen). C, TSA does not relieve MeCP2-mediated transcriptional repression with chromatin templates. Transcription reactions were performed as in A, with the same sample of chromatin shown in B.
We then tested the effect of the addition of TSA upon transcription of methylated pUC-SCP-8CG that is packaged into chromatin. In these experiments, chromatin was assembled with purified ACF, NAP1, and histones and then transcribed by methods established previously (15, 16). The efficiency of chromatin assembly was monitored by a DNA supercoiling assay, in which the change in the linking number of DNA that occurs upon formation of nucleosomes in the presence of topoisomerase I is observed (Fig. 3B). In addition, the periodicity of the nucleosomes was assessed by using a partial micrococcal nuclease digestion assay (Fig. 3B). These assays indicated that the purified ACF and NAP1 mediated the efficient assembly of methylated pUC-SCP-8CG into periodic arrays of nucleosomes. The resulting chromatin samples were transcribed, as in Fig. 3A, in the absence or presence of MeCP2 with varying concentrations of TSA. These experiments revealed that TSA does not relieve MeCP2-mediated transcriptional repression with chromatin templates (Fig. 3C). Hence, these findings suggest that the MeCP2-mediated repression that occurs in our system corresponds to the HDAC-independent component of MeCP2 function. Because we observed HDAC-independent repression by MeCP2 with both naked DNA and chromatin templates, we employed naked DNA templates throughout the remainder of this study.
Single-round Transcription Analysis Indicates That MeCP2 Inhibits the Assembly of the Transcription Preinitiation Complex
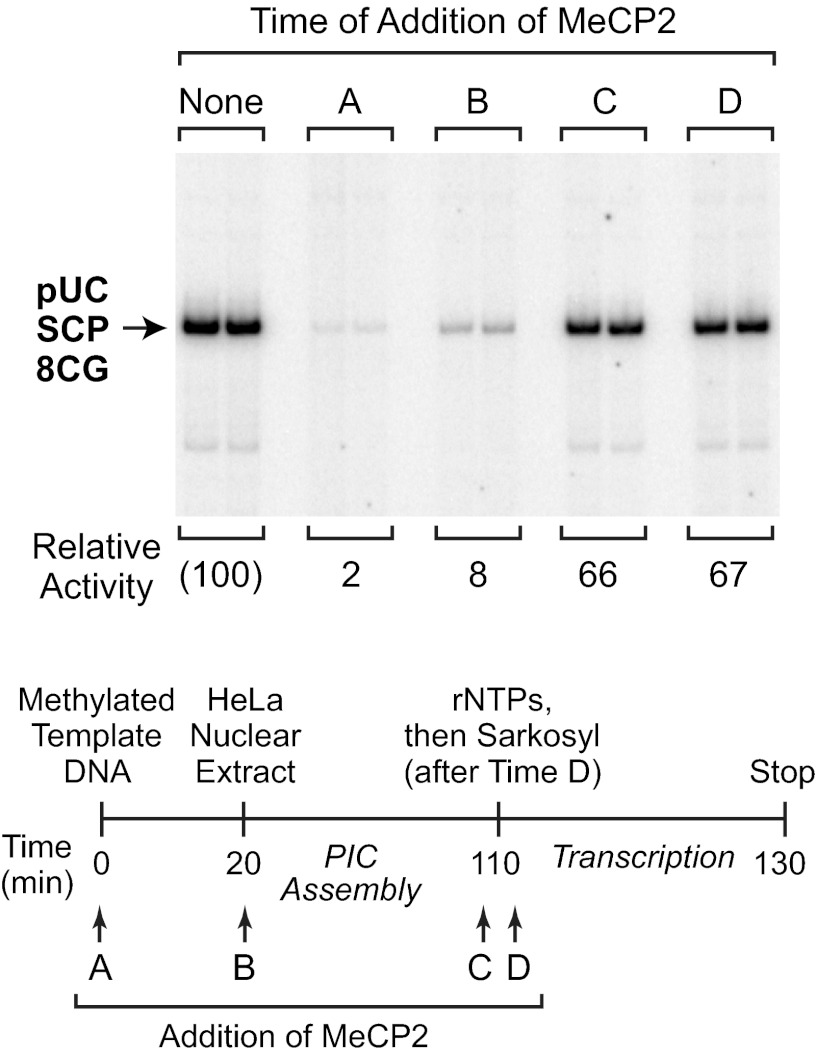
To clarify the mechanism of HDAC-independent repression, we performed single-round transcription analyses to identify the step in the transcription process that is affected by MeCP2. In these experiments, we used the detergent Sarkosyl, which inhibits PIC assembly but does not affect elongation of transcriptionally engaged RNA polymerase II (see, for example, Refs. 14, 24). As depicted in Fig. 4, MeCP2 was added at different time points before, during, or after PIC assembly. To limit transcription to a single round, Sarkosyl was added after the initiation of transcription by the addition of ribonucleoside triphosphates. These experiments revealed that MeCP2 inhibits transcription when it is added to the methylated DNA template prior to PIC assembly (time point A), but does not block transcription when added after PIC assembly (time point C) or immediately after transcription initiation (time point D) (Fig. 4). Hence, HDAC-independent repression of transcription by MeCP2 appears to occur via the inhibition of PIC assembly. It should be noted that the effect of MeCP2 upon PIC assembly was addressed in a separate study (19), but the results could not be interpreted unambiguously because the reactions were not performed as single-round transcriptions (for examples of single-round transcription analyses of PIC assembly, see Refs. 14, 24).
FIGURE 4.
Single-round transcription analysis indicates that MeCP2 inhibits PIC assembly. Single-round transcription reactions were performed with methylated pUC-SCP-8CG template DNA, as outlined in the schematic. MeCP2 (500 nm final concentration) was added at time points A, B, C, or D. Time point C is 10 s prior to the addition of the four ribonucleoside 5′-triphosphates (rNTPs), whereas time point D is 10 s after the addition of rNTPs. To limit transcription to a single round, Sarkosyl (0.2%, v/v, final concentration) was added 10 s after time point D. The image is a continuous single exposure of a set of reactions that were performed as indicated in the diagram. The relative activity values are normalized to the amount of transcription observed in the absence of MeCP2.
Eight CG Dinucleotides in the Core Promoter Region Are Sufficient for Methylation-specific Repression by MeCP2
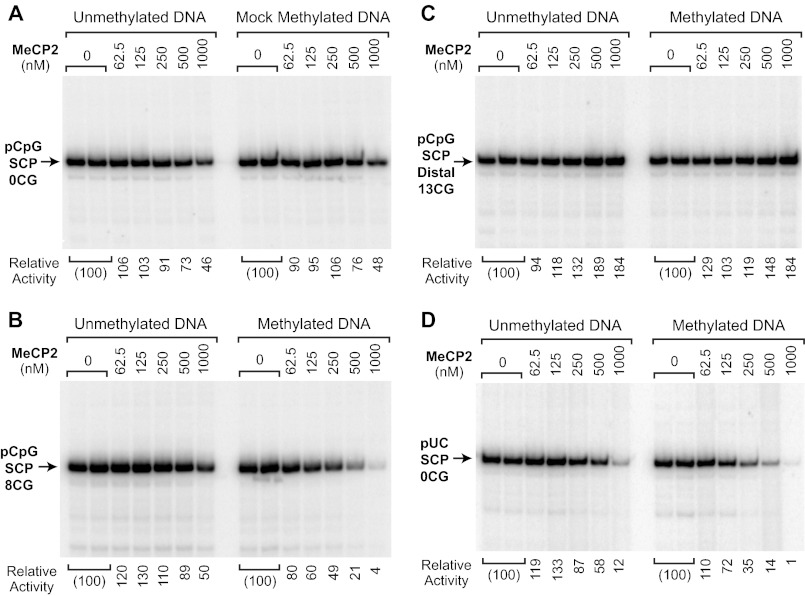
We next sought to determine the effect of the location of the CG dinucleotides upon MeCP2-mediated transcriptional repression. To this end, we constructed a reporter plasmid, termed pCpG-SCP-0CG (Fig. 1B), that is completely deficient in CG dinucleotides. To create pCpG-SCP-0CG, we designed SCP-0CG, which is a version of the super core promoter (17) that lacks CG dinucleotides, and inserted this core promoter into a CG-free vector. As might be anticipated (but, to our knowledge, never demonstrated experimentally), we observed that MeCP2 does not repress transcription from a completely CG-deficient reporter construct that is either unmethylated or mock-methylated (Fig. 5A). MeCP2 was unable to repress transcription from the mock-methylated template except at the highest concentration, in which a 2-fold repression was seen with both the unmethylated and mock-methylated template. Thus, these results provide further confirmation that MeCP2 is a CG dinucleotide-dependent transcriptional repressor. In addition, the pCpG-SCP-0CG construct served as a negative control or reference for the analysis of the CG dinucleotide requirements for MeCP2-mediated repression.
FIGURE 5.
CG methylation-specific repression by MeCP2 requires CG dinucleotides in the core promoter or the surrounding sequences. Transcription reactions were carried out with methylated and unmethylated template DNAs and the indicated concentrations of purified wild-type MeCP2. The DNA templates are depicted in Fig. 1B. Each image is a continuous single exposure of a series of reactions that were performed in parallel with equivalent amounts of methylated or unmethylated template DNA. At 500 nm MeCP2, there is approximately one molecule of MeCP2 per methyl-CG dinucleotide in the reaction mixture (see “Experimental Procedures”). The relative activity values are normalized to the amount of transcription observed in the absence of MeCP2. A, MeCP2 does not substantially repress transcription in the absence of CG dinucleotides. The pCpG-SCP-0CG plasmid lacks CG dinucleotides and, hence, cannot be methylated by M.SssI methyltransferase. In the mock methylation, the plasmid DNA was treated with M.SssI and S-adenosylmethionine under the standard methylation conditions. B, CG dinucleotides in the core promoter region are sufficient for transcriptional repression by MeCP2. The pCpG-SCP-8CG core promoter has eight CG dinucleotides in the core promoter region from −50 to +50 relative to the +1 transcription start site. C, MeCP2 does not repress transcription from a construct containing a stretch of 13 CG dinucleotides about 1.7 kbp upstream of a CG-deficient core promoter. D, a core promoter lacking CG dinucleotides can be repressed by MeCP2 via binding to the surrounding sequences. pUC-SCP-0CG lacks CG dinucleotides in the core promoter region (−50 to +50) but contains CG dinucleotides in the surrounding pUC vector sequences.
We then tested whether CG dinucleotides in the core promoter region would be sufficient for MeCP2-mediated repression. For these experiments we created pCpG-SCP-8CG, which has the SCP-8CG promoter in the CG-deficient vector. We performed transcriptional analyses with unmethylated or methylated versions of pCpG-SCP-8CG in the absence of MeCP2 or in the presence of varying concentrations of MeCP2 (Fig. 5B). These experiments revealed that eight CG dinucleotides in the core promoter region are sufficient for methylation-specific repression by MeCP2 (Fig. 2). To assess whether any particular CG dinucleotides in the promoter region were predominantly responsible for transcriptional repression of the pCpG-SCP-8CG construct, we constructed and analyzed a series of core promoters that contained only one or two CG dinucleotides. Each of these promoters exhibited weak repression by MeCP2 (data not shown), indicating that the observed repression was the combined effect of multiple CG dinucleotides rather than a single strategically located CG dinucleotide.
It is possible, however, that the repression observed with the pCpG-SCP-8CG construct (Fig. 5B) was due to a general inactivation of the DNA template, such as by nonspecific aggregation. To address this point, we analyzed the effect of the same concentrations of MeCP2 upon transcription from the pCpG-SCP-Distal13CG construct (see Fig. 1B), which contains 13 CG dinucleotides ∼1.7 kbp upstream of the SCP-0CG promoter. As shown in Fig. 5C, MeCP2 does not repress transcription from the pCpG-SCP-Distal13CG template. It therefore appears that MeCP2 does not indiscriminately inactivate the DNA template and that the repression observed with the pCpG-SCP-8CG construct is specific for the promoter.
We also investigated whether transcriptional repression would be observed with a CG-deficient core promoter that is surrounded by CG-containing sequences. To this end, we constructed pUC-SCP-0CG with the CG-deficient core promoter (from −50 to +50 relative to the +1 transcription start site) in pUC119, which has 209 CG dinucleotides. Analysis of this promoter indicated that MeCP2 can repress transcription in a methylation-specific manner via CG dinucleotides that are outside of the core promoter region (Fig. 5D). It should be noted, however, that the 209 CG dinucleotides flanking the core promoter in the pUC-SCP-0CG construct results in about the same extent of repression as that seen with the eight CG dinucleotides in the pCpG-SCP-8CG construct (compare Fig. 5, B and D). Hence, the CG dinucleotides in the core promoter region appear to be more important for MeCP2-mediated repression than those in the flanking DNA. Nevertheless, the ability of MeCP2 to repress the pUC-SCP-0CG reporter suggests that MeCP2-mediated repression can occur from outside of the core promoter region.
Binding of MeCP2 to DNA Is Not Sufficient for HDAC-independent Transcriptional Repression
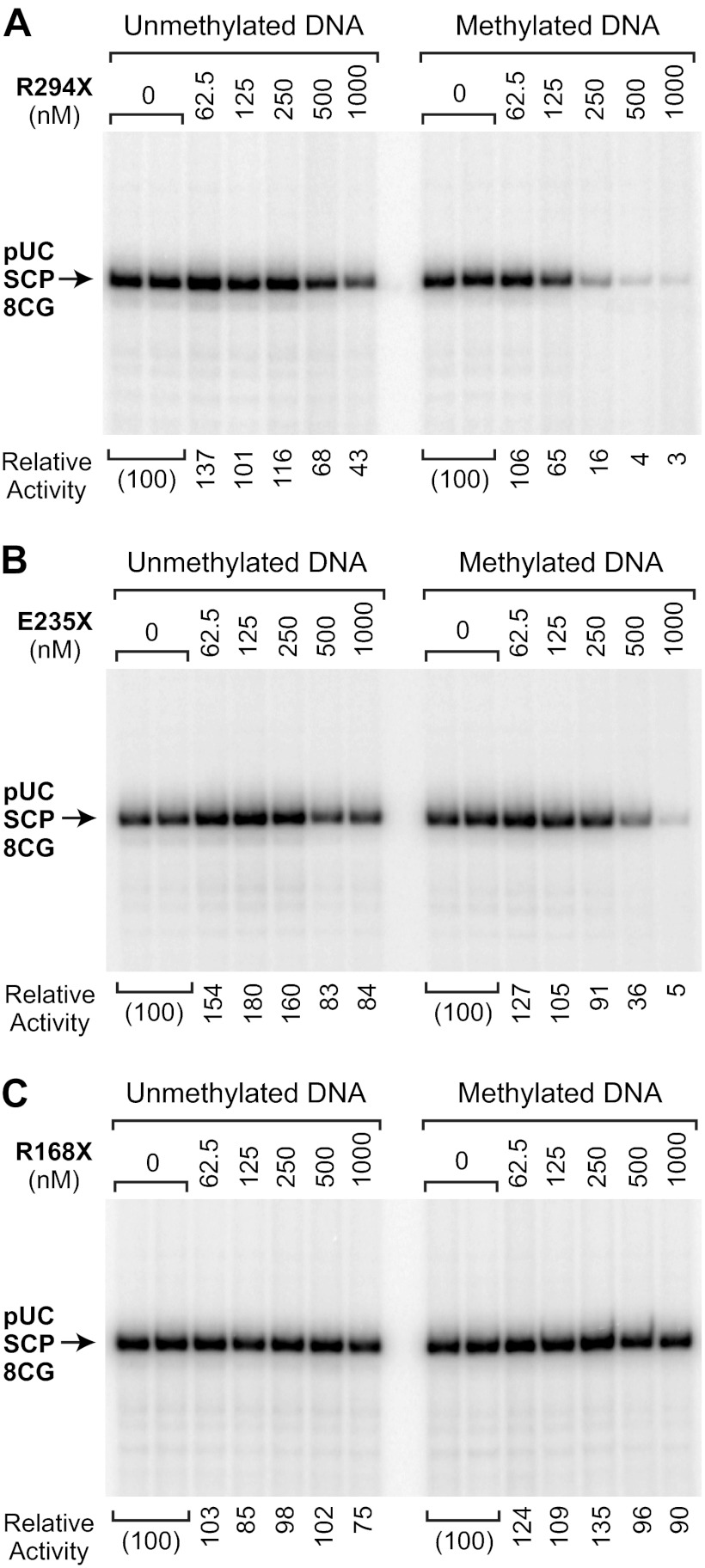
To investigate the process by which MeCP2 mediates HDAC-independent repression, we purified and analyzed three Rett syndrome-associated mutant versions of MeCP2, termed R294X, E235X, and R168X (Fig. 1A). These C-terminally truncated proteins lack part or all of the previously defined TRD (8), which appears to contain a composite of both HDAC-dependent and HDAC-independent repression activities. To identify the region of MeCP2 that is important for HDAC-independent repression, we carried out transcription reactions with unmethylated or methylated pUC-SCP-8CG template DNAs in the absence of MeCP2 or in the presence of varying concentrations of each mutant protein (Fig. 6). The R294X protein, which retains the majority of the TRD, represses transcription with an activity that is comparable with that of the wild-type protein (compare Figs. 2 and 6A). The E235X protein contains only a small portion of the TRD but is nevertheless able to repress transcription, albeit to a lesser extent than wild-type MeCP2-in a CG methylation-dependent manner (Fig. 6B). In contrast, the R168X protein, which lacks the TRD but retains the MBD, does not exhibit transcriptional repression activity (Fig. 6C).
FIGURE 6.
Analysis of transcriptional repression by C-terminally truncated MeCP2 proteins. Transcription reactions were performed with methylated and unmethylated pUC-SCP-8CG template DNA in the absence or presence of three Rett syndrome-associated mutant versions of MeCP2, termed R294X, E235X, and R168X. These C-terminally truncated MeCP2 proteins are shown in Fig. 1A. Each image is a continuous single exposure of a series of reactions that were performed in parallel with equivalent amounts of methylated or unmethylated template DNA. At 500 nm MeCP2, there is approximately one molecule of MeCP2 per CG dinucleotide in the pUC-SCP-8CG template. The relative activity values are normalized to the amount of transcription observed in the absence of MeCP2. A, MeCP2-R294X, which lacks the C-terminal region and a portion of the TRD, has transcriptional repression activity that is similar to that of the wild-type protein. B, MeCP2-E235X, which lacks the majority of the TRD, has a weak but distinct CG methylation-specific repression activity. C, MeCP2-R168X, which contains the MBD, does not repress transcription.
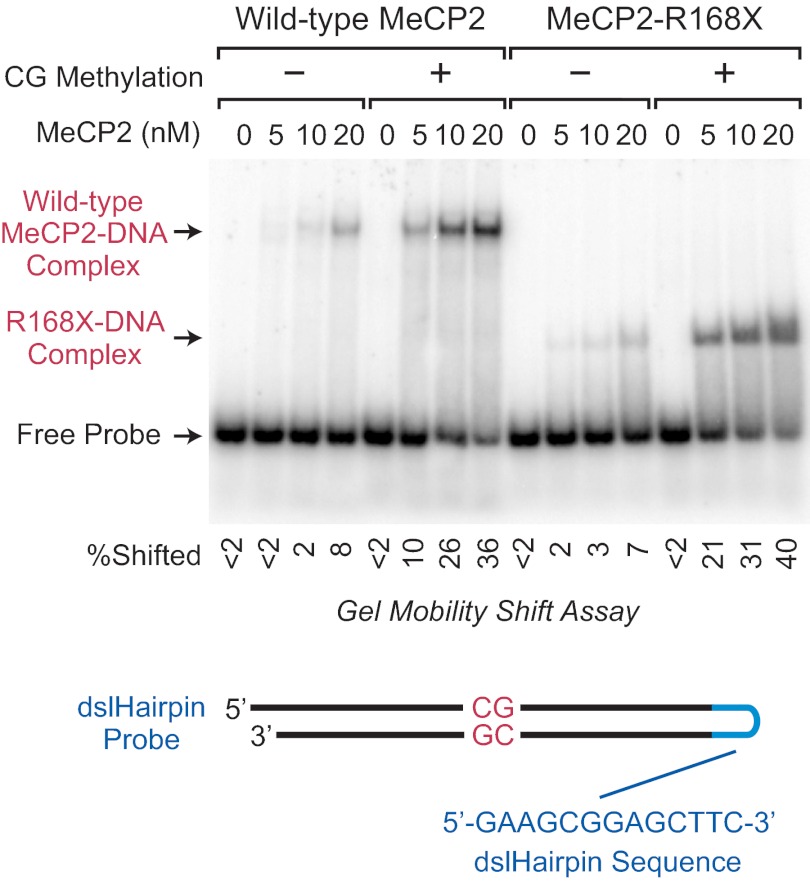
Because it was possible that the lack of repression activity with the R168X protein was due to a reduction or loss of DNA-binding affinity, we compared the DNA-binding affinities of wild-type and R168X proteins by gel mobility shift analysis with unmethylated or CG-methylated DNA probes (Fig. 7). To ensure that we were analyzing the direct binding of the proteins to DNA, we performed these experiments in the absence of competitor DNA. Under these conditions, both the wild-type and R168X proteins exhibit a severalfold preference for binding to CG-methylated DNA relative to unmethylated DNA, as seen previously with wild-type MeCP2 (see, for example, Refs. 18, 25). Importantly, we found that the wild-type and R168X proteins have comparable affinities for binding to CG-methylated DNA. Hence, the binding of MeCP2 to DNA via the MBD is not sufficient for HDAC-independent transcriptional repression. These findings suggest that HDAC-independent repression by MeCP2 occurs by a mechanism that is more complex than steric blockage of the RNA polymerase machinery.
FIGURE 7.
Both wild-type and truncated R168X MeCP2 proteins bind preferentially to CG-methylated DNA relative to unmethylated DNA. Gel mobility shift experiments were performed with a radiolabeled, single-stranded DNA probe with a dslHairpin DNA sequence derived from bacteriophage N4 promoters (18). The inclusion of the dslHairpin sequence enabled the proper hybridization of the BDNF promoter sequence containing a single CG dinucleotide. The protein concentrations and the absence or presence of CG methylation are shown. The image is a continuous single exposure of a series of binding reactions that were performed in parallel with equivalent amounts of methylated or unmethylated probe DNA. Competitor DNA was not included in these experiments. The percent of the probe that is shifted in each lane is indicated beneath the autoradiogram.
It is useful to note that we employed a novel method in the gel shift experiments, in which we designed the probe as a single-stranded DNA with an unusual hairpin that has double-stranded character (18) (Fig. 7). We term this structure a double-stranded-like hairpin, or dslHairpin. Our use of the dslHairpin probe was prompted by difficulties that we experienced in the proper hybridization of the complementary single DNA strands that we used for MeCP2 gel shift probes. The dslHairpin should be generally useful for gel mobility shift analyses.
DISCUSSION
In this study, we explored the mechanisms of transcriptional repression by MeCP2. To examine the properties of MeCP2, we characterized the wild-type protein via its binding to methyl-CG dinucleotides rather than a fusion protein via the binding of the DNA-binding domain of the hybrid protein to its recognition sites. We developed a biochemical system that exhibits CG methylation-specific, CG dinucleotide-dependent, and HDAC-independent transcriptional repression by purified full-length human MeCP2 (Figs. 1–3 and 5). Notably, MeCP2-mediated transcriptional repression was resistant to TSA with either naked DNA or chromatin templates (Fig. 3). How might HDAC-independent, MeCP2-mediated repression occur? Single-round transcription analyses revealed that MeCP2 inhibits the assembly of a productive transcription preinitiation complex (Fig. 4). Examination of the sequence requirements indicated that strong repression is achieved with eight CG dinucleotides in the core promoter region (with the pCpG-SCP-8CG construct), whereas little or no repression was observed with combinations of one or two CG dinucleotides throughout the same region (Fig. 5 and data not shown). We also did not observe MeCP2-mediated repression with a methylated reporter construct (pCpG-SCP-Distal13CG) in which a cluster of 13 CG dinucleotides is located ∼1.7 kbp upstream of the core promoter region (Fig. 5C). Hence, MeCP2 does not indiscriminately inactivate the DNA template. In addition, the methyl-CG-specific binding of a truncated MeCP2 protein to DNA is not sufficient for transcriptional repression (Figs. 6 and 7). These findings suggest a progressive mechanism of repression, which is a function of CG density and position relative to the core promoter, that is not strictly due to steric blockage of the transcriptional machinery by MeCP2.
It is relevant to consider our observation that MeCP2-mediated repression is not relieved by the HDAC inhibitor TSA with either naked DNA or chromatin templates (Fig. 3). Specifically, these results may appear to be in contrast to the findings that MeCP2-mediated repression can be partially relieved by TSA in cells and oocytes (9, 10). First, it should be noted that the earlier studies (9, 10) were performed with recombinant Gal4-MeCP2 fusion proteins in conjunction with reporter constructs containing five Gal4 binding sites. Hence, the behavior of the Gal4-MeCP2 fusion proteins that are recruited to the DNA template via the Gal4 DNA binding domain may not necessarily be identical to that of full-length wild-type MeCP2 that is recruited to the DNA template in a CG methylation-specific manner. Second, it is possible that MeCP2 may exhibit different activities in different biological contexts.
From a practical standpoint, it is useful to note that we developed a strong CG-deficient core promoter that we termed SCP-0CG. In a CG-deficient context, SCP-0CG is resistant to MeCP2-mediated transcriptional repression (Fig. 5A) and should provide stable, long-term gene expression in vertebrates. SCP-0CG could be easily incorporated into any existing expression system by replacement of the existing core promoter sequences, such as from −40 to +40 or −50 to +50 relative to the transcription start site, with the corresponding (i.e. −40 to +40 or −50 to +50) SCP-0CG sequences. For maximal resistance to methylation, CG dinucleotides should also be removed from the surrounding promoter region.
In addition, we employed a novel approach for gel mobility shift analysis in which we used a single-stranded DNA probe containing a dslHairpin structure (Fig. 7). The dslHairpin is advantageous for use in gel mobility shift probes for the following reasons. First, the single strand of DNA ensures the correct stoichiometry of each of the complementary single strands that comprise the DNA-binding region of the probe. Second, the dslHairpin ensures the proper hybridization of the complementary strands. This property is particularly useful with repetitive DNA sequences. Third, the resulting probe with the dslHairpin behaves as double-stranded DNA fragment, as it does not contain a single-stranded DNA loop that could be degraded by nucleases or bound by single-stranded DNA-binding proteins.
In conclusion, these studies highlight the potential importance of the HDAC-independent mechanism of MeCP2-mediated transcriptional repression. MeCP2 is of high biomedical importance because mutations in the gene encoding MeCP2 are responsible for most cases of Rett syndrome. It is therefore imperative to examine every possible aspect of the function of this factor. In this light and in the context of this work, the high levels of MeCP2 in neurons (12), the near complete methylation (98%) of CG dinucleotides in the brain (11) and the function of MeCP2 as a potent, stoichiometric, and HDAC-independent transcriptional repressor (this study) suggest that the HDAC-independent mechanism of transcription may be an important component of MeCP2 function in the brain.
Acknowledgments
We thank George Kassavetis, Yuan-Liang Wang, Jia Fei, Sharon Torigoe, and Sascha Duttke for critical reading of the manuscript. We also thank Lucia Rothman-Denes for sharing knowledge and expertise on the bacteriophage N4 promoter hairpin.
This work was supported, in whole or in part, by National Institutes of Health grant GM041249 (to J. T. K.). This project was initiated by an International Rett Syndrome Foundation (Rett Syndrome Research Foundation) grant.
- MBD
- methyl-CG binding domain
- CG
- cytosine-guanine
- TRD
- transcription repression domain
- HDAC
- histone deacetylase
- TSA
- trichostatin A
- PIC
- preinitiation complex
- ACF
- ATP-utilizing chromatin assembly and remodeling factor
- dslHairpin
- double-stranded-like hairpin DNA
- SCP
- super core promoter.
REFERENCES
- 1. Lewis J. D., Meehan R. R., Henzel W. J., Maurer-Fogy I., Jeppesen P., Klein F., Bird A. (1992) Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69, 905–914 [DOI] [PubMed] [Google Scholar]
- 2. Klose R. J., Bird A. P. (2006) Genomic DNA methylation. The mark and its mediators. Trends Biochem. Sci. 31, 89–97 [DOI] [PubMed] [Google Scholar]
- 3. Amir R. E., Van den Veyver I. B., Wan M., Tran C. Q., Francke U., Zoghbi H. Y. (1999) Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23, 185–188 [DOI] [PubMed] [Google Scholar]
- 4. Hagberg B., Aicardi J., Dias K., Ramos O. (1983) A progressive syndrome of autism, dementia, ataxia, and loss of purposeful hand use in girls. Rett's syndrome. Report of 35 cases. Ann. Neurol. 14, 471–479 [DOI] [PubMed] [Google Scholar]
- 5. Klose R. J., Sarraf S. A., Schmiedeberg L., McDermott S. M., Stancheva I., Bird A. P. (2005) DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol. Cell 19, 667–678 [DOI] [PubMed] [Google Scholar]
- 6. Chahrour M., Jung S. Y., Shaw C., Zhou X., Wong S. T., Qin J., Zoghbi H. Y. (2008) MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320, 1224–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nan X., Meehan R. R., Bird A. (1993) Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 21, 4886–4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nan X., Campoy F. J., Bird A. (1997) MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 88, 471–481 [DOI] [PubMed] [Google Scholar]
- 9. Nan X., Ng H.-H., Johnson C. A., Laherty C. D., Turner B. M., Eisenman R. N., Bird A. (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393, 386–389 [DOI] [PubMed] [Google Scholar]
- 10. Jones P. L., Veenstra G. J., Wade P. A., Vermaak D., Kass S. U., Landsberger N., Strouboulis J., Wolffe A. P. (1998) Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19, 187–191 [DOI] [PubMed] [Google Scholar]
- 11. Ehrlich M., Gama-Sosa M. A., Huang L.-H., Midgett R. M., Kuo K. C., McCune R. A., Gehrke C. (1982) Amount and distribution of 5-methylcytosine in human DNA from different types of tissues or cells. Nucleic Acids Res. 10, 2709–2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Skene P. J., Illingworth R. S., Webb S., Kerr A. R., James K. D., Turner D. J., Andrews R., Bird A. P. (2010) Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol. Cell 37, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yusufzai T. M., Wolffe A. P. (2000) Functional consequences of Rett syndrome mutations on human MeCP2. Nucleic Acids Res. 28, 4172–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kadonaga J. T. (1990) Assembly and disassembly of the Drosophila RNA polymerase II complex during transcription. J. Biol. Chem. 265, 2624–2631 [PubMed] [Google Scholar]
- 15. Fyodorov D. V., Kadonaga J. T. (2003) Chromatin assembly in vitro with purified recombinant ACF and NAP-1. Methods Enzymol. 371, 499–515 [DOI] [PubMed] [Google Scholar]
- 16. Jiang W., Nordeen S. K., Kadonaga J. T. (2000) Transcriptional analysis of chromatin assembled with purified ACF and dNAP1 reveals that acetyl-CoA is required for preinitiation complex assembly. J. Biol. Chem. 275, 39819–39822 [DOI] [PubMed] [Google Scholar]
- 17. Ballestar E., Yusufzai T. M., Wolffe A. P. (2000) Effects of Rett syndrome mutations of the methyl-CpG binding domain of the transcriptional repressor MeCP2 on selectivity for association with methylated DNA. Biochemistry 39, 7100–7106 [DOI] [PubMed] [Google Scholar]
- 18. Gleghorn M. L., Davydova E. K., Rothman-Denes L. B., Murakami K. S. (2008) Structural basis for DNA-hairpin promoter recognition by the bacteriophage N4 virion RNA polymerase. Mol. Cell 32, 707–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Juven-Gershon T., Cheng S., Kadonaga J. T. (2006) Rational design of a super core promoter that enhances gene expression. Nat. Methods 3, 917–922 [DOI] [PubMed] [Google Scholar]
- 20. Fraga M. F., Ballestar E., Montoya G., Taysavang P., Wade P. A., Esteller M. (2003) The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 31, 1765–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaludov N. K., Wolffe A. P. (2000) MeCP2 driven transcriptional repression in vitro. Selectivity for methylated DNA, action at a distance and contacts with the basal transcription machinery. Nucleic Acids Res. 28, 1921–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu F., Thiesen J., Strätling W. H. (2000) Histone deacetylase-independent transcriptional repression by methyl-CpG-binding protein 2. Nucleic Acids Res. 28, 2201–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang X., Kadonaga J. T. (2001) Biochemical analysis of transcriptional repression by Drosophila histone deacetylase 1. J. Biol. Chem. 276, 12497–12500 [DOI] [PubMed] [Google Scholar]
- 24. Hawley D. K., Roeder R. G. (1987) Functional steps in transcription initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J. Biol. Chem. 262, 3452–3461 [PubMed] [Google Scholar]
- 25. Ishibashi T., Thambirajah A. A., Ausió J. (2008) MeCP2 preferentially binds to methylated linker DNA in the absence of the terminal tail of histone H3 and independently of histone acetylation. FEBS Lett. 582, 1157–1162 [DOI] [PubMed] [Google Scholar]