Background: Pathways involved in neuron-dependent GLT1 regulation in astrocytes remain to be characterized.
Results: Neuronal microRNA 124a can be transferred into astrocytes through neuronal exosomes and significantly increases GLT1 protein expression in an indirect manner.
Conclusion: Neuronal exosomal miRNA 124a is able to regulate astroglial GLT1 expression.
Significance: We characterize a novel pathway in neuron-to-astrocyte communication and identify a microRNA that modulates GLT1 protein expression.
Keywords: Amyotrophic Lateral Sclerosis (Lou Gehrig's Disease), Astrocytes, Exosomes, MicroRNA, Transporters, GLT1, Glutamate Transporter
Abstract
Perisynaptic astrocytes express important glutamate transporters, especially excitatory amino acid transporter 2 (EAAT2, rodent analog GLT1) to regulate extracellular glutamate levels and modulate synaptic activation. In this study, we investigated an exciting new pathway, the exosome-mediated transfer of microRNA (in particular, miR-124a), in neuron-to-astrocyte signaling. Exosomes isolated from neuron-conditioned medium contain abundant microRNAs and small RNAs. These exosomes can be directly internalized into astrocytes and increase astrocyte miR-124a and GLT1 protein levels. Direct miR-124a transfection also significantly and selectively increases protein (but not mRNA) expression levels of GLT1 in cultured astrocytes. Consistent with our in vitro findings, intrastriatal injection of specific antisense against miR-124a into adult mice dramatically reduces GLT1 protein expression and glutamate uptake levels in striatum without reducing GLT1 mRNA levels. MiR-124a-mediated regulation of GLT1 expression appears to be indirect and is not mediated by its suppression of the putative GLT1 inhibitory ligand ephrinA3. Moreover, miR-124a is selectively reduced in the spinal cord tissue of end-stage SOD1 G93A mice, the mouse model of ALS. Subsequent exogenous delivery of miR-124a in vivo through stereotaxic injection significantly prevents further pathological loss of GLT1 proteins, as determined by GLT1 immunoreactivity in SOD1 G93A mice. Together, our study characterized a new neuron-to-astrocyte communication pathway and identified miRNAs that modulate GLT1 protein expression in astrocytes in vitro and in vivo.
Introduction
Despite the important modulatory roles of astrocytes in synaptic functions, the molecular regulation of the neuron-astrocyte functional unit is largely undetermined. As one of the most important astroglial functional synaptic proteins, excitatory amino acid transporter 2 (EAAT2, rodent analog GLT1) expression regulation has become an excellent model to understand how neuronal signals coordinate astrocyte function at synapses. EAAT2/GLT1 protein is expressed predominantly and abundantly in astrocytes across the adult CNS (1, 2), although GLT1 mRNA has also been detected in embryonic neurons and a small subset of adult neurons (3, 4). Physiological induction of GLT1 in astrocytes is strongly dependent upon neuronal signals (5–7). Primary astrocytes maintained in culture express very low levels of GLT1 mRNA or protein. However, astrocytes cocultured with neurons or treated with neuron-conditioned medium (NCM)3 show a dramatic induction in both mRNA and protein expression levels of GLT1 (7, 8). By using a novel microfluidic culture platform, we have demonstrated previously that presynaptic axonal signals are necessary and sufficient for neuron-dependent transcriptional activation of the EAAT2/GLT1 promoter (6).
In parallel with transcriptional regulation of EAAT2/GLT1 expression, translational regulation has also been implicated in EAAT2/GLT1 protein expression. Multiple EAAT2/GLT1 mRNA transcripts have been identified in cultured astrocytes and CNS tissue (9–11). These transcripts give rise to differential GLT1 protein isoforms that can form homomeric and heteromeric complexes (10). Differential translation efficiency among EAAT2/GLT1 mRNA transcripts has also been found in an immortalized astroglial cell line following treatment with corticosterone or retinol (12). Interestingly, all these EAAT2/GLT1 mRNA transcripts have a uniquely long 3′ untranslated region (UTR) (9.7kb) (3, 13), implicating potential 3′UTR-dependent regulation, especially through microRNA-mediated translational inhibition or mRNA degradation. In pathological conditions, abnormal EAAT2 splicing has been linked to the pathogenesis of ALS (14), although it is inconclusive whether aberrant EAAT2 transcripts contribute to the reduced EAAT2 protein expression in ALS patients (15, 16). The exact mechanisms of EAAT2/GLT1 translational regulation remain to be investigated. A recent study has demonstrated that genetic ablation of the neuronal EphA4 receptor results in a dramatic increase in GLT1 protein (but not mRNA) levels in the hippocampus (17), implicating a neuronal role in translational regulation of GLT1 expression.
MicroRNAs (miRNAs) are a class of non-coding RNAs with a length of 20–25 nucleotides that anneal their seed sequences (nucleotides 2–8 at its 5′ end) to the complementary seed match sequences located in the 3′UTR region of target mRNAs (18, 19). Following a multistep maturation process, mature miRNAs are incorporated into the RNA-induced silencing complex and actively modulate gene expression by either inhibiting mRNA translation or inducing mRNA degradation in the cell (20). Although miRNAs act mainly in a cell-autonomous manner, accumulating evidence has recently suggested that miRNAs can also be packed into vesicular structures known as exosomes for intercellular signaling (21–24). Exosomes are 40- to 100-nm membrane vesicles of endocytic origin secreted by most cell types and contain various active biomolecules, including mRNAs, proteins, lipids, and miRNAs (25–27). Exosomes can be secreted into the extracellular space and then fused to neighboring cells to release their contents, thus serving as a novel intercellular genetic regulatory mechanism (23, 28, 29). Exosome-mediated secretion of signals from neurons and their regulatory effect on EAAT2/GLT1 expression is unknown. Although CNS-specific miRNAs have been identified through miRNA expression arrays (30), potential regulatory roles of these miRNAs in the translational regulation of EAAT2/GLT1 expression have not yet been explored. In this study, we investigated the transfer of neuronal exosomal miRNA (especially miR-124a) into astrocytes and miRNA-mediated translational regulation of GLT1 protein expression.
EXPERIMENTAL PROCEDURES
Animals
FVB WT and SOD1G93A transgenic mice were obtained from The Jackson Laboratory. The ephrinA3−/− (knockout) mice were a kind gift of Dr. Elena B. Pasquale (Burnham Institute for Medical Research, La Jolla, CA). The care and treatment of animals in all procedures strictly followed the National Institutes of Health Guide for the Care and Use of Laboratory Animals and the Guidelines for the Use of Animals in Neuroscience Research and the Tufts University IACUC.
Generation of GLT1 and EphrinA3 mRNA 3′UTR Luciferase Reporter Constructs and Luciferase Assay
GLT1 mRNA 3′UTR luciferase reporter constructs were generated via PCR using mouse genomic DNA as a template. PCR products were cloned into the pmirGLO vector (Promega). As the GLT1 mRNA 3′UTR (NM_001077514.3) is 9.7 kb, five partially overlapping regions (∼2 kb each, named 3′UTR-1 to 5) were separately amplified and cloned to ensure that the entire 3′UTR was assessed. Primer sequences used for amplification of GLT1 3′UTR were as follows: 3′UTR-1-F, 5′ CCGGAGCTCGTAGTCGCCTATTCACTTTCA 3′; 3′UTR-1-R, 5′ CCGGTCGACGTAAGTAGATTTCTAGAGGGC 3′; 3′UTR-2-F, 5′ GCCGCTAGCCCTGGATCATAGGTGAGAATC 3′; 3′UTR-2-R, 5′ CCGGTCGACGTTTATGAATAGATGAGAATG 3′; 3′UTR-3-F, 5′ GCCGAGCTCACTGTTGCCTGCTTTAGGGAG 3′; 3′UTR-3-R, 5′ CCCGTCGACCTATACAGTAATGCCTGGGTG 3′; 3′UTR-4-F, 5′ CGGGTCGACGGAGATGGAATATCTCAAGGC 3′; 3′UTR-4-R, 5′ CCCGTCGACCTGCTGAGCAGGGCTGCTGCC 3′; 3′UTR-5-F, 5′ GGGGAGCTCGCAGAGTTCAAGTCAGATGGG 3′; and 3′UTR-5-R, 5′ CCCGTCGACCAGAGATAATGGGATGTAGAG 3′. For the ephrinA3 luciferase reporter construct, the ephrinA3 mRNA 3′UTR (NM_010108.1, 500 bp) was amplified, cloned into the pmirGLO vector, and named ephrinA3 3′UTR-luci. Primer sequences used for the amplification of ephrinA3 3′UTR were as follows: EphrinA3–3′UTR-F, 5′ ATATATATATGCTAGCTCTGCCCCTTCCTATGACA 3′ and EphrinA3–3′UTR-R, 5′ ATATATATATCTCGAGTGTGGGAGTGGAGGGAGA 3′. Reporter constructs were confirmed by sequencing. Luciferase activity was measured 24 h after transfection using the Dual-Glo luciferase kit (Promega). Coexpressed Renilla luciferase on the pmirGLO vector was used as an internal control to normalize the firefly luciferase activity.
Cell Culture, Treatment, and Transfection
For cortical astrocyte cultures, P0-P3 mouse pups were decapitated, and cerebral cortices were removed and transferred into cold Hanks' balanced salt solution for dissection. Meninges were stripped, and cortices were minced and placed into 0.05% trypsin solution for 10 min in a 37 °C water bath. The enzymatic reaction was stopped by addition of astrocyte culture medium (DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin). The tissue was washed twice with astrocyte medium and then dissociated gently by trituration with a fire-polished Pasteur pipette. Dissociated cells were filtered through a 70-μm strainer to collect a clear astrocyte cell suspension. For neuronal primary culture, cortical neuronal cells were isolated from embryonic 14- to 16-day-old mouse brains. The neuron culture medium was composed of neurobasal medium, 2% B27 neurobasal supplement, 2 mm glutamine, 1% of 100× Glutamax, and 1% penicillin-streptomycin. The brain dissociation procedure was similar to the astrocyte isolation procedure described above. Freshly prepared neurons were then plated on cell culture dishes (1–5 × 107 cells). Tetrodotoxin (final concentration 2 μm) was added daily into the neuronal cultures for 48 h to block neuronal activity. KCL (15 mm) was added into neuronal cultures for 24 h to depolarize neurons. For mixed cultures, brains of E20-P0 pups were dissected and dissociated as described above. HEK293 cells were maintained in DMEM supplemented with 10% FBS.
Primary astrocyte transfection in a 6-well plate with miR-124a, miR-124a-A/S (antisense), or a mixture of miR-124a and miR-124a-A/S (final concentration for all 25 nm) was performed with Dharma FECT reagent (Thermo Fisher) following the instructions of the manufacturer. Neuronal exosomes were added into the miR-124a-A/S pretransfected astrocytes 72 h following transfection. For HEK293 cell transfection, HEK293 cells that were seeded 1 × 104 cells/well in 96-well plates the day prior to transfection were incubated with 0.1 μg of luciferase reporter construct alone, luciferase reporter construct with 25 nm miR-124a mimics, or luciferase reporter construct with miR-124a/miR-124a-A/S mimics (Thermo Scientific) using Dharma FECT reagent. Mock-transfected cells were transfected with the 3′UTR-free pmirGLO vector alone. Transfections were replicated five times in each of three independent experiments.
Exosome Purification and EM Analysis
Exosomes were prepared from NCM or astrocyte-conditioned medium from neuronal or astrocyte primary culture as described previously (21). Briefly, NCM or astrocyte-conditioned medium was collected from cultures (1–2 × 107 cells) that were maintained for 5–8 days in vitro. NCM or astrocyte-conditioned medium was first spun at 500 × g for 10 min to remove cell debris and then underwent a 16,500 × g centrifugation step for 20 min. Finally, exosomes were pelleted by ultracentrifugation at 100,000 × g for 70 min. For in vitro astrocyte treatment experiments, exosomes were resuspended with astrocyte culture medium and added into normal confluent or miR-124a-A/S pretransfected astrocytes (72 h following transfection). Astrocytes were collected 48 h following exosome treatment for immunoblot analysis. EM analysis of exosomes was performed in the Structural Electron Microscopy Facility at the Boston University School of Medicine.
Time-lapse Imaging
Freshly prepared neuronal exosomes were labeled with PKH67 green fluorescent dye (Sigma) according to the instructions of the manufacturer. Astrocytes grown on a glass-bottomed dish were visualized under a Nikon A1-R confocal scanning microscope. Fluorescence images were obtained using an argon laser that excites PKH67 with 488nm and the emission light passes through a 482/35nm band filter. Images were acquired every 1 min for 60 min. After a 1-min baseline acquisition, 50 μl of dye-labeled exosomes was applied, and images were acquired for the following 59 min.
MiRNA Isolation, Detection, Quantification, and Synthesis
MicroRNA was extracted from the exosome pellet following a chloroform extraction procedure. Briefly, exosome pellets were resuspended in 600 μl of RNase-free PBS buffer, and then 200 μl of chloroform was added. After vortexing for 20 s, the emulsion was kept at room temperature for 5 min before centrifugation at 12,000 rpm for 15 min at 4 °C. The aqueous phase was collected carefully, mixed with an equal volume of isopropanol (550 μl) and 3 μl of linear acrylamide (5 μg/μl), and then placed at −20 °C overnight. To recover precipitated RNA, the preparation was centrifuged for 20 min at 12, 000 rpm at 4 °C. The RNA pellet was washed with 70% ethanol and resuspended with 10 μl of RNase-free water. The quantity and quality of isolated miRNA was determined using the Agilent BioAnalyzer with the pico RNA kit according to the instructions of the manufacturer. Total RNA (including miRNA) was isolated from primary astrocyte or neuronal cultures with a similar chloroform approach after lysing the cells with lysis buffer. The miRNA was converted to cDNA using the TaqMan MicroRNA reverse transcription kit (Applied Biosystems) with specific primers for miR-124a and control snoRNA202. The quantity of miR-124a was measured by QRT-PCR using SYBR Green after a standard curve was established via QRT-PCR on a synthesized miR-124a oligo (5′ UAAGGCACGCGGUGAAUGCC 3′). MiR-124a double-stranded mimics were synthesized by Thermo Scientific. Cy3-labeled miR-124a and scrambled RNA were synthesized by IDT DNA Technologies. Antisense miR-124a (5′ TGGCATTCACCGCGTGCCTTAA 3′) was synthesized by Isis Pharmaceuticals, Inc. (Carlsbad, CA).
Immunoblot Analysis
The following antibodies were used in immunoblot analysis: anti-GLT1 (1:2000, homemade), anti-GLAST (1:100, homemade), anti-EAAC1 (1:100, homemade), anti-Alix (1:100, BioLegend), anti-Tsg101 (1:2000, Abcam), anti-ephrinA3 (1:250, Invitrogen), anti-β-Actin (1:1000, Sigma), anti-ALDH1L1 (1:1, NeuroMab), and anti-GFAP (1:1000, Dako). Glutamate transporter immunoblots have often shown monomer (62-kDa), dimer (120-kDa), and sometimes multimer (250-kDa) bands (1, 2). All transporter bands were included for quantification. Freshly dissected striatum tissue or in vitro neuronal or astrocyte cultures were homogenized in lysis buffer (Tris-HCl 20 mm, NaCl 140 mm, EDTA 1 mm, SDS 0.1%, Triton 1% and glycerol 10%). Exosomes collected from individual confluent 10-cm dishes (1–2 × 107 cells) were used as individual samples for the immunoblot analysis. The total protein amount was determined by Bradford protein assay using the Gen 5 data analysis program. Two (striatum) or 50 (astrocyte cultures) μg of total cell lysate was loaded onto a 4–15% gradient SDS-PAGE gel. Separated proteins were transferred to a PVDF membrane for 1 h. The membrane was blocked with 3% BSA in 1× TBST (Tris buffer saline with 0.1% Tween 20) and then incubated with the appropriate primary antibody overnight at 4 °C. The following day, the membrane was exposed to HRP-conjugated goat anti-rabbit or anti-mouse secondary antibody (1:2000) dissolved in 1× TBST. Signals were visualized on CL-XPosureTM film by ECL Plus chemiluminescent substrate.
Preparation of Crude Synaptosomes and Glutamate Uptake Assay
Glutamate uptake was performed with crude synaptosome preparations from mouse striatum using the 0.32 m sucrose centrifugation method (31). After total protein determination by Bradford assay (Bio-Rad), 1μCi l-3H glutamate and 100 μm non-labeled glutamate were mixed with Na+ uptake buffer (total volume 275 μl) and then added into 25 μl of each synaptosome sample in 96-well multiscreen HTS filter plates (Millipore). After a 6-min incubation, uptake was terminated by 5 min rapid cooling at 4 °C. Samples were then filtered by the Steriflip vacuum filtration system (Millipore) and washed 6X with ice-cold PBS while continuing to filter the samples. Each filtered 96-well membrane was excised and transferred for scintillation counting. Dihydrokainate (500 μm) or dl-threo-β-benzyloxyaspartic acid (500 μm) was added into appropriate wells in the uptake assay. The disintegration/minute value was normalized by total protein concentration and converted to a fmol/μg/min unit.
Stereotaxic Delivery of Oligos in Vivo
For delivery of miR-124a into the spinal cord of SOD1G93A mice, SOD1G93A mice (105 days old) were anesthetized with a mixture of ketamine/xylazine (ketamine 95 mg/kg and xylazine 10 mg/kg). The spinal cord was exposed by laminectomy of C5 and C6 as described previously (32). Cy3-labeled miR-124a or scrambled miRNA (5 μg/μl) was stereotaxically injected bilaterally at the C5–6 level (2 μl/injection). Each animal received a total of 4 μl of the miRNA. Postoperative care included injections of buprenorphine according to the IACUC requirements. Animals were perfused 7 days after the stereotaxic injection of oligos. For delivery of miR-124a-A/S into the striatum, a unilateral intrastriatal injection of 2 μl containing vehicle, 100 μg (50 μg/μl) of miR-124a-A/S, or control oligo was performed on adult FVB mice 6–8 weeks old. Mice were sacrificed, and brain was freshly sliced (1 mm) using a brain slicer 24 days following injection. Striatum tissue was then carefully dissected out for immunoblot analysis.
Tissue Preparation and Immunostaining
Animals were deeply anesthetized with a ketamine/xylazine mix and perfused intracardially with 4% paraformaldehyde in PBS. The spinal cords were dissected, postfixed in 4% paraformaldehyde overnight at 4 °C, and then cryoprotected in PBS containing 30% sucrose for 48 h. Cervical and lumbar segments of spinal cords were embedded and frozen in the mounting medium. Coronal sections (20 μm) were taken with a cryostat at −20 °C and mounted on glass slides. Prior to immunostaining, slides were rinsed three times in 1× PBS. Sections were then treated with blocking buffer (1% BSA, 5% goat serum, and 0.2% Triton X-100 in 1× PBS) for 20 min at room temperature. The primary antibodies anti-GLT1 (rabbit 1:2000), GFAP (mouse, Calbiochem IF03L, 1:1000), and NeuN (mouse Millipore MAB377, 1:500) were incubated overnight at 4 °C in blocking buffer. After washing three times in 1× PBS, a corresponding secondary antibody (anti-mouse IgG or anti-rabbit IgG, Alexa Fluor 488- or 633-conjugated antibody, 1:2,000) was added for 90 min at room temperature. The sections were rinsed three times in 1× PBS before proceeding to the next antibody, repeating the same sequence of incubation and rinsing steps for each one individually. Images were obtained with a confocal laser scanning microscopy (A1R Nikon).
Data Analysis
For quantification of GLT1 immunoreactivity in vivo, individual astrocytes were circled using the GFAP staining pattern to define the GLT1 staining within each individual astrocyte. The average intensity of GLT1 immunoreactivity in a selected area (individual astrocyte) was then measured using ImageJ software. The intensity of GLT1 immunoreactivity in GFAP and miR-124a-Cy3-positive cells in the cervical region was normalized using the intensity of GLT1 immunoreactivity in GFAP-positive cells in the lumbar cord in each mouse. All the resulting raw data were graphed and plotted in GraphPad 5 (GraphPad Software, La Jolla, CA). Unless stated otherwise, all values are reported as mean ± S.E., with n indicating the number of replicates.
RESULTS
Cultured Primary Neurons Secrete Exosomes That Are Enriched with Small RNA
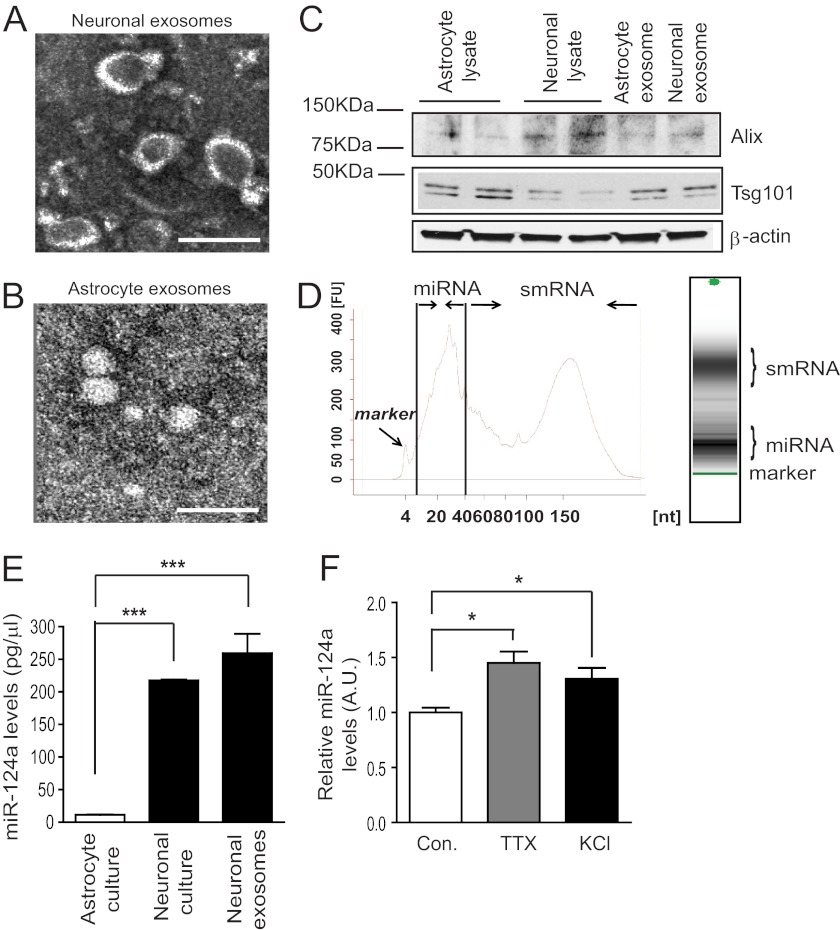
Neurons are known to communicate with neighboring cells through exocytosis-mediated release of signals such as the activity-dependent release of neurotransmitters. Secretion of exosomes has been observed previously from cultured primary neurons (33). We first isolated exosomes through serial steps of centrifugation from NCM. Isolated exosomes were negatively stained, and their representative EM image is shown in Fig. 1A. Several heavily stained vesicle structures were found in the representative EM image, suggesting the presence of abundant exosomes in NCM. Interestingly, only smaller vesicles were found in astrocyte-conditioned medium (Fig. 1B). Subsequent immunoblot analysis of the exosome homogenates detected the expression of Alix and Tsg101 (Fig. 1C), selective exosome markers (21, 34), in neuronal and astroglial exosomes, respectively, which confirms the EM analysis.
FIGURE 1.
Secreted neuronal exosomes contain miR-124a. Representative electron microscopy images of neuronal (A) and astroglial (B) exosome preparation. Scale bar = 200 nm in neuronal exosomes and 50 nm in astroglial exosomes. C, detection of exosome markers Alix and Tsg101 in neuronal and astroglial exosomes. D, bioanalyzer pico analysis of neuronal exosome RNA. smRNA, small RNA. E, relative miR-124a levels in astrocytes, neurons, and neuronal exosomes. n = 3–4. ***, p < 0.001 from one-way ANOVA with Bonferroni post-test. F, MiR-124a levels in neuronal exosomes following tetrodotoxin or KCL treatment. A.U., arbitrary unit. n = 4. *, p < 0.05 from Student's t test with control separately.
To determine whether neuronal exosomes contain microRNA, total RNA was isolated from neuronal exosomes and subjected to bioanalyzer pico analysis. As shown in Fig. 1D, abundant miRNA and small RNA (smRNA) were found in neuronal exosome preparations, indicated by the absorbance peaks. As miR-124a (miRBase code mmu-miR-124) is one of the most abundant miRNAs in the CNS and is highly expressed in neurons (35, 36), we compared the expression levels of miR-124a in neurons, neuronal exosomes, and astrocytes. Total RNA was isolated from cultured primary astrocytes, primary neurons, and neuronal exosome preparations. Although small nucleolar RNA 202 (snoRNA 202) is often used as the endogenous control to normalize input RNA concentration in QRT-PCR, it is unclear whether snoRNA 202 is packed into exosomes, so we instead generated a miR-124a standard curve using serially diluted synthesized miR-124a oligos with the miR-124a-specific TaqMan QRT-PCR kit (Applied Biosystems, Carlsbad, CA). The absolute quantity of miR-124a from these samples was then determined by comparing the threshold cycle (Ct) value from the miR-124a QRT-PCR with the standard curve. Cultured neurons and neuronal exosomes express comparably high levels of miR-124a, whereas only minimal miR-124a levels were detected in cultured astrocytes (Fig. 1E). As neuronal exocytosis is often associated with synaptic activity, we next investigated whether neuronal activity is involved in exosomal miR-124a secretion in neuronal cultures. Interestingly, effective blockade of neuronal activity by tetrodotoxin (2 μm) or increasing neuronal activity by KCL (15 mm) treatment both significantly increased exosomal miR-124a levels (Fig. 1F), suggesting that exosomal miRNA secretion is not closely associated with neuronal activity changes in vitro.
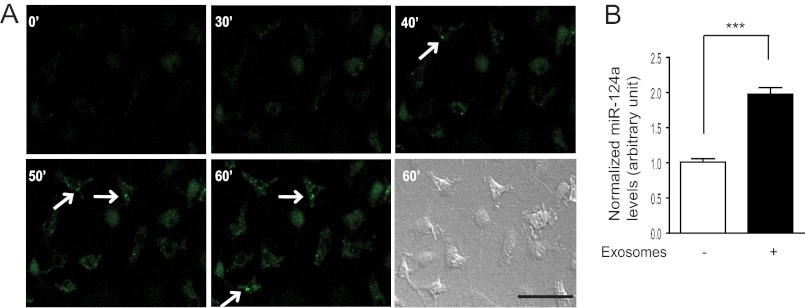
To further demonstrate whether neuronal exosomes can be directly internalized into astrocytes, membranes of neuronal exosomes were labeled with the lipophilic green fluorescent reagent PKH67 (Sigma). PKH67-labeled exosomes were added to primary astrocyte cultures grown on glass-bottomed dishes. Time-lapse fluorescent microscopy images (1 scan/min for a total of 60 min) were collected to monitor PKH67 fluorescence in cultured astrocytes. As shown in Fig. 2A, a gradual increase in PKH67 fluorescence was found within the cultured astrocytes starting 30–40 min after addition of exosomes. The Dynamic view (supplemental movie) of the time-lapse images also revealed active movement of PKH-labeled puncta inside astrocytes (starting at 20 min) as a result of the internalized exosome vesicles. Internalization of neuronal exosomes also significantly increases miR-124a levels in cultured astrocytes (Fig. 2B). Taken together, these results suggest that neuronal microRNA, such as miR-124a, is packed into neuronal exosomes, which are actively secreted and potentially serve as an intercellular signal.
FIGURE 2.
Internalization of neuronal exosomes into astrocytes. A, time-lapse images of cultured astrocytes treated with labeled neuronal exosomes. Scale bar = 50 μm. B, neuronal exosome treatment increases miR-124a levels in cultured astrocytes. n = 3. ***, p < 0.001 from Student's t test.
Neuronal miR-124a Regulates GLT1 Protein Expression in Vitro
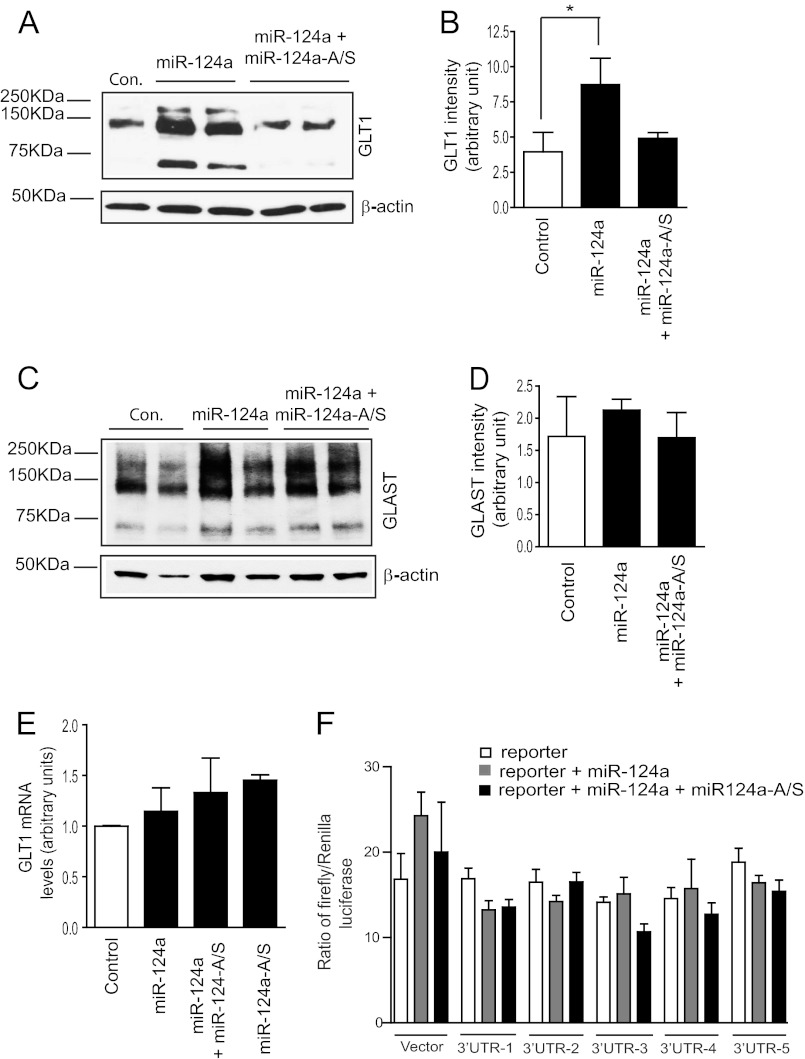
Previous sequencing and Northern blot analysis of EAAT2/GLT1 transcripts found that the EAAT2/GLT1 mRNA transcripts have a uniquely long 3′UTR (9.7 kb) (3, 13), making them potential targets for miRNA-mediated inhibition. As miR-124a can be transferred from neurons to astrocytes through exosomes, we investigated whether miR-124a regulates GLT1 protein expression in astrocytes. Synthesized miR-124a mimics (Thermo Scientific) were transfected directly into cultured primary astrocytes. Surprisingly, direct transfection of miR-124a significantly and consistently increases GLT1 protein expression levels (Fig. 3, A and B). The miR-124a-induced increase in GLT1 protein was completely blocked by cotransfection of specific miR-124a antisense (miR-124a-A/S), confirming the specific up-regulation effect of miR-124a on GLT1 protein expression in cultured astrocytes. Although protein expression of another astroglial glutamate transporter, GLAST, was also slightly increased in astrocytes following miR-124a transfection (Fig. 3C), this increase was not specifically attenuated by the co-transfected miR-124a-A/S (D), suggesting that the slight GLAST increase is unlikely to be miR-124a-dependent. These results indicate that miR-124a selectively affects GLT1 protein expression in astrocytes. Interestingly, although GLT1 protein is significantly increased, there is no significant change in GLT1 mRNA levels following transfection of miR-124a, miR-124a-A/S, or a mixture of miR-124a and miR-124a-A/S (Fig. 3E).
FIGURE 3.
Neuronal miR-124a up-regulates astroglial GLT1 protein expression levels. A, expression of GLT1 protein levels in astrocyte cultures following transfection of miR-124a. B, quantification of GLT1 expression levels following miR-124a transfection. n = 4–6. *, p < 0.05 from one-way ANOVA with Bonferroni post-test. C, expression of GLAST protein levels in astrocyte cultures following transfection of miR-124a. D, quantification of GLAST expression levels following miR-124a transfection. n = 5–6. The three band sizes represent trimers, dimers, and monomers of GLT1 or GLAST. E, GLT1 mRNA expression levels in cultured astrocytes following miR-124a transfection. n = 3–5. F, activity of the GLT1 mRNA 3′UTR luciferase reporter following miR-124a or mixture of miR-124a/miR-124a-A/S transfection. n = 5/experiment for three independent experiments
MiRNA-mediated regulation of protein expression is mostly through direct binding of miRNA to target mRNA transcripts to either inhibit the mRNA translation or induce mRNA degradation (20). To determine whether miR-124a acts through direct interaction with the GLT1 mRNA 3′UTR, a luciferase reporter-based analysis was performed in HEK293 cells. The 3′UTR sequence (9.7 kb) of GLT1 mRNA was amplified in five ∼2 kb fragments that were cloned individually downstream of the luciferase reporter to serve as an artificial 3′UTR sequence for the luciferase reporter. Potential modulation of miR-124a on the GLT1 mRNA 3′UTR was assessed by the luciferase activity changes. We observed no significant and specific changes (Fig. 3F) in the luciferase activity of any GLT1 mRNA 3′UTR luciferase reporter following transfection of miR-124a (or mixture of miR-124a/miR-124a-A/S), suggesting that miR-124a is unlikely to directly bind to the GLT1 mRNA 3′UTR and modulate its translation/degradation. We also analyzed the seed sequence of miR-124a using the TargetScan program (37) and found no direct match with GLT1 mRNA transcripts. These results indicate that the up-regulation effect of miR-124a on GLT1 protein expression is likely to be indirect.
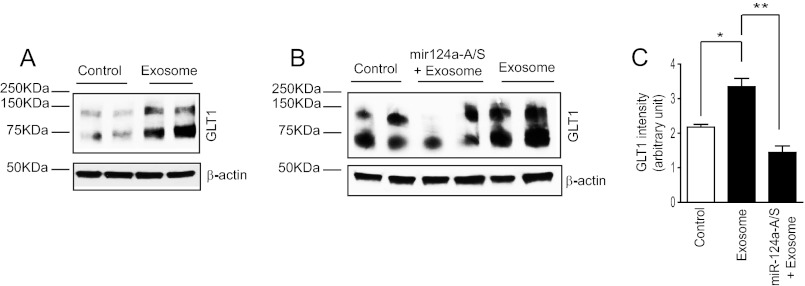
We next determined whether neuronal exosomes that contain abundant miR-124a are sufficient to increase GLT1 expression. As shown in Fig. 4A, direct application of neuronal exosomes to astrocyte cultures significantly increases GLT1 protein expression levels. Moreover, pretransfection of miR-124a-A/S in astrocyte cultures attenuated neuronal exosome-induced up-regulation of GLT1 (Fig. 4, B and C), suggesting that miR-124a directly mediates the effect of neuronal exosomes in up-regulating GLT1 expression in astrocytes. Taken together, these in vitro results show that neuronal exosome-mediated transfer of miR-124a from neurons to astrocytes represents a new pathway in neuron-dependent GLT1 regulation.
FIGURE 4.
MiR-124a mediates neuronal exosome-dependent up-regulation of GLT1 expression in astrocytes. A, neuronal exosome treatment increases GLT1 protein levels in cultured astrocytes. B, GLT1 protein expression levels in miR-124a-A/S-transfected astrocytes following treatment with neuronal exosomes. Neuronal exosomes collected from one 10-cm dish were added into four wells of astrocyte cultures in a 6-well plate. C, quantification of the GLT1 protein levels in astrocytes following treatment with neuronal exosomes/pre-transfection of miR-124a-A/S. n = 4–6. *, p < 0.05 from one-way ANOVA with Bonferroni post-test
MiRNA-dependent Translational Regulation of GLT1 Expression in Vivo
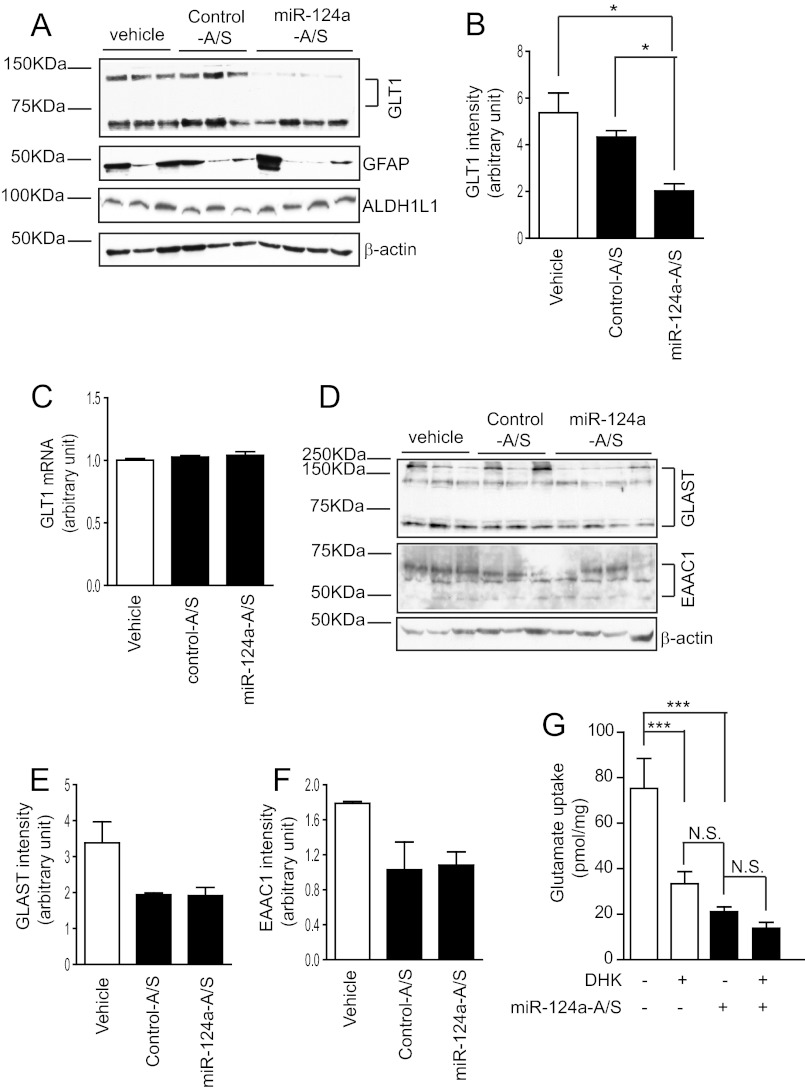
On the basis of these in vitro findings, we examined the effect of miR-124a on GLT1 protein expression in vivo. Because miRNA unwinds to expose the active strand for its specific recognition and binds to the target sequence once incorporated into the RNA-induced silencing complex (20), specific miRNA antisense can inhibit the function of miRNA by binding to its active strand. Loss-of-function analysis of miR-124a was performed using its specific antisense oligo. Specific miR-124a antisense (miR-124a-A/S) oligos (2 μl, 50 μg/μl) were injected intrastriatally into the brain of adult mice (P45–60, n = 10/group). Mice were sacrificed 24 days following oligo injection, and striata were sliced into 1-mm sections using a brain matrix and blade. GLT1 protein expression levels were examined by immunoblot analysis following antisense oligo injection. Injection of scrambled oligos served as a control for miR-124a-A/S. As shown in Fig. 5A, in vivo injection of miR-124a-A/S significantly reduced GLT1 protein expression levels, consistent with the in vitro observation that miR-124a transfection increases GLT1 protein expression levels. Quantitative analysis of the GLT1 immunoblot analysis further shows that GLT1 protein levels decreased 55% following miR-124a-A/S injection (Fig. 5B). Interestingly, there were no significant changes in GLT1 mRNA levels following miR-124a-A/S injection (Fig. 5C), suggesting that the regulatory effect of miR-124a on GLT1 expression is at the translational level. These in vivo and in vitro results provide strong and consistent evidence that miR-124a has a significant role in regulating GLT1 protein expression.
FIGURE 5.
Intrastriatal injection of miR-124a antisense (miR-124a-A/S) significantly reduces GLT1 protein expression levels in striatum. A, GLT1 expression levels in striatum following intrastriatal injection of miR-124a-A/S. B, quantification of the GLT1 expression levels from immunoblot analysis. n = 5–7. *, p < 0.05 from one-way ANOVA with Bonferroni test. C, GLT1 mRNA levels following miR-124a-A/S injection. D, expression changes of other glutamate transporters following miR-124a-A/S injection. Quantification of the GLAST (E) and EAAC1 (F) expression levels from immunoblot analysis. n = 3–5. G, measurement of functional glutamate uptake in striatum tissue following miR-124a-A/S injection. n = 3–4. ***, p < 0.001, one-way ANOVA with Bonferroni post-test. N.S., not significant.
We also examined the effect of miR-124a-A/S on other glutamate transporters in the CNS, including the astroglial GLAST (human analog, excitatory amino acid transporter 1, EAAT1) and neuronal EAAC1 (human analog, excitatory amino acid transporter 3, EAAT3), neither of which has 3′UTR binding sites for miR-124a according to the TargetScan program. Immunoblot analyses of GLAST and EAAC1 showed that neither GLAST nor EAAC1 protein levels were altered following miR-124a-A/S injection when compared with the control antisense injection (Fig. 5, D–F). Expression of other astroglial specific proteins, including glial fibrillary acidic protein (GFAP) and aldehyde dehydrogenase 1 member L1 (ALDH1L1) (38), was not altered (Fig. 5A) following miR-124a-A/S injection. These results suggest that the effect of miR-124a on GLT1 expression is likely to be specific.
We next determined whether GLT1-mediated functional glutamate uptake is altered following antisense injection. Crude synaptosomes were prepared from mouse striatal brain slices following miR-124a-A/S injection, and l-[3H] glutamate uptake was measured by a scintillation counter. As shown in Fig. 5G, miR-124a-A/S reduced functional glutamate uptake by 72% (from 75.3 to 21.1 pmol/mg) compared with control antisense. Similarly, functional glutamate uptake was reduced significantly (from 75.3 to 33.4 pmol/mg) in samples from the control antisense injection in the presence of the GLT1-specific inhibitor dihydrokainate. As GLT1 is the dominant glutamate transporter in the mature CNS and is responsible for more than half of the total glutamate uptake in control tissue, dramatic decrease of glutamate uptake following miR-124a-A/S injection suggested that miR-124a-A/S significantly reduced GLT1-dependent glutamate uptake, consistent with the GLT1 immunoblot analysis results above. In addition, similarly reduced glutamate uptake levels in miR-124a-A/S injection alone or dihydrokainate treatment confirmed that miR-124a-A/S-induced decrease of glutamate uptake is indeed a consequence of the reduction of GLT1 protein levels. Functional glutamate uptake was almost completely abolished by adding the pan-glutamate transporter inhibitor TBOA to the reaction or by removing Na+ in the reaction buffer (data not shown), confirming that the glutamate uptake measured by our assay was mediated specifically by Na+-dependent glutamate transporters.
EphrinA3 Is Not the Downstream Target of miR-124a in Astrocytes
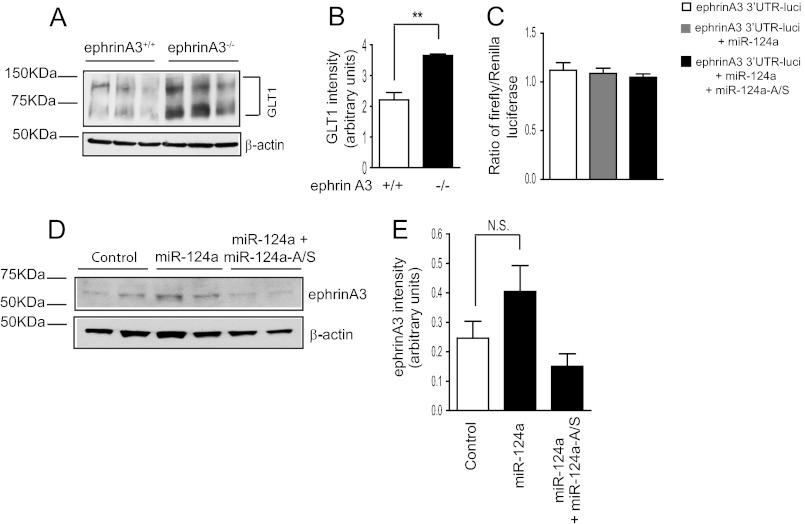
Although miR-124a consistently up-regulates GLT1 protein expression in vitro and in vivo, miR-124a is selectively expressed in neurons, and its expression in astrocytes is minimal (35, 36) (Fig. 1E). Our sequence and luciferase reporter analysis results (Fig. 3F) also suggested that miR-124a is unlikely to directly bind to the GLT1 mRNA 3′UTR. As introduction of exogenous miR-124a into cultured astrocytes is sufficient to increase GLT1 protein (but not mRNA) levels, it is likely that miR-124a acts on astroglial factors that are involved in the translational regulation of GLT1 protein. Interestingly, a recent study (17) found that deletion of the astroglial ephrinA3 ligand increases GLT1 protein (but not mRNA) expression levels, implicating its inhibitory role in GLT1 translational regulation. We first examined GLT1 expression levels in ephrinA3−/− astrocytes. As shown in Fig. 6, A and B, GLT1 protein (but not mRNA) expression levels were significantly increased in the absence of ephrinA3 in astrocytes, confirming that astroglial ephrinA3 negatively regulates GLT1 protein expression.
FIGURE 6.
Astroglial ephrinA3 is not the downstream target of miR-124a in astrocytes in GLT1 expression regulation. A, increased GLT1 protein levels in cultured ephrinA3−/− astrocytes. B, quantitative analysis of the GLT1 expression from the ephrinA3−/− astrocytes. n = 6. **, p < 0.01, Student's t test. C, luciferase activity of the ephrinA3 mRNA 3′UTR luciferase reporter in astrocytes following miR-124a transfection. n = 10 from two independent experiments. D, representative immunoblot analysis of ephrinA3 expression in astrocytes following miR-124a transfection. E, quantification of ephrinA3 expression in astrocytes following miR-124a transfection. n = 6. N.S., not significant.
We then tested whether miR-124a can reduce ephrinA3 expression, therefore suppressing its inhibitory effect on GLT1 protein expression. We performed the luciferase reporter assay using the mouse ephrinA3 mRNA 3′UTR sequence. As shown in Fig. 6C, direct transfection of miR-124a or a mixture of miR-124a/miR-124a-A/S has no effect on the luciferase reporter activity, suggesting that miR-124a does not modulate the ephrinA3 3′UTR to suppress ephrinA3 mRNA translation. In addition, a modest increase, but not decrease, of ephrinA3 protein expression was found in astrocytes following miR-124a transfection (Fig. 6, D and E), confirming that miR-124a does not suppress ephrinA3 expression in astrocytes. Therefore, it is unlikely that ephrinA3 is a downstream target of miR-124a in astrocytes. Although ephrinA3 negatively regulates GLT1 protein expression, our results suggest that the up-regulation of miR-124a of GLT1 protein expression is not mediated by suppression of ephrinA3.
Exogenously Delivered miR-124a Increases GLT1 Protein Levels in the Spinal Cord of SOD1G93A Mice
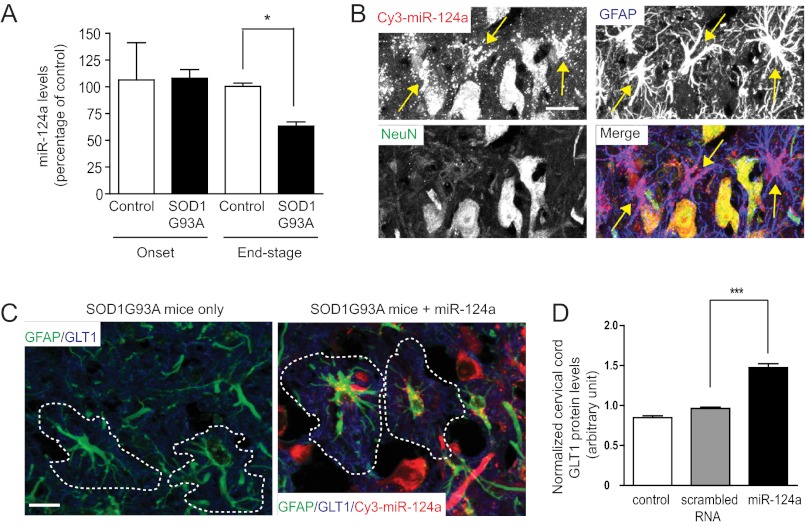
GLT1 expression is highly dynamic under pathological conditions in vivo. In particular, its expression is decreased significantly (40% to 100%) in the end stage of ALS patients or rodent models (39, 40). As miR-124a has a significant role in the translational regulation of GLT1 expression, we examined expression changes of miR-124a in the spinal cord of SOD1G93A mice. Small RNAs were prepared from spinal cord tissue from wild-type or SOD1G93A mice at different stages (onset and end stages). Expression levels of miR-124a were determined using TaqMan miRNA QRT-PCR. SnoRNA202 was used as the endogenous control for normalizing RNA concentration. As shown in Fig. 7A, miR-124a expression is decreased significantly (40%) in the end stage of SOD1G93A mice compared with that in age-matched control mice, whereas the miR-124a expression levels are essentially unchanged at disease onset.
FIGURE 7.
Stereotaxically injected miR-124a increases GLT1 protein expression in SOD1G93A mice. A, down-regulation of miR-124a in the spinal cord of end-stage SOD1G93A mice. n = 5–6. *, p < 0.05, Student's t test. B, high abundance of injected miR-124a in astrocytes of cervical spinal cord of SOD1G93A mice. Scale bar = 50 μm. Astrocytes with abundant miR-124a are highlighted with yellow arrows. C, GLT1 immunostaining in cervical cord following miR-124a injection. Scale bar = 50 μm. GLT1 immunoreactivity from individual astrocyte (circled in a dashed line from the GFAP staining) was quantified. D, quantitative analysis of GLT1 protein levels following miR-124a injection. GLT1 immunoreactivity was measured in miR-124a+GFAP+ cells in the cervical cord and normalized with the GLT1 immunoreactivity in the lumbar cord of the same SOD1 G93A mouse. n = 78–114 cells from three SOD1 G93A mice. **, p < 0.01 from Student's t test.
We also investigated the effect of miR-124a on GLT1 protein expression in vivo by direct stereotaxic injection of miR-124a oligo into the ventral gray matter of the spinal cord. Previous studies have shown that synthesized small RNAs can be stable in vivo for several weeks (41). We performed stereotaxic injections of Cy3-labeled miR-124a (bilateral, 10 μg/2 μl/injection) into the ventral gray matter of cervical cord (C5-C6) of SOD1G93A mice at P105 to examine the effect of miR-124a on increasing GLT1 protein expression in vivo. We also injected scrambled RNA as the oligo control. In SOD1G93A mice, loss of GLT1 protein in lumbar or cervical cords generally follows the disease progression pattern, which is first reduced in lumbar cord and then in cervical cord. On the basis of the lumbar and cervical pathology, we decided to inject miR-124a into the cervical cord at P105, the point at which the cervical pathology is just beginning in SOD1G93A mice. This would allow up to a 2-week window to test the effect of injected miR-124a on GLT1 protein expression levels. During this 2-week window, GLT1 expression continues to decrease in deteriorating cervical cord, which might be prevented or delayed by the injected miR-124a. In addition, stereotaxic injection into the cervical cord is surgically less challenging than lumbar injection, making it more feasible for efficient delivery of miR-124a into the right gray matter area. Mice were sacrificed 7 days following Cy3-labeled miR-124a and scrambled RNA injection. Abundant Cy3 fluorescence signals were observed in the ventral gray matter of the cervical cord (Fig. 7B). Cy3 fluorescence signals were enriched in both neurons and astrocytes, indicated by the NeuN and GFAP staining (yellow arrows, Fig. 7B). No apparent oligo-induced toxicity was observed in the spinal cord sections, indicated by NeuN staining. Double immunostaining of GLT1 and GFAP was performed to quantify the GLT1 immunoreactivity changes in astrocytes following miR-124a injection. High magnification (×60) confocal images were acquired, and the mean GLT1 immunoreactivity was measured from individual astrocyte surface areas on the basis of the GFAP immunostaining signals (Fig. 7C) in ImageJ software. Progressive and severe loss of GLT1 protein levels during the disease progression in SOD1G93A spinal cord makes it more feasible to quantify changes of GLT1 protein expression levels by its immunoreactivity. Because the rate of disease progression and loss of GLT1 protein vary in individual SOD193A mouse, GLT1 immunoreactivity levels in the lumbar cord of each SOD1G93A mouse were used as endogenous controls to normalize the base-line differences in GLT1 protein levels in different SOD1G93A mice. Using this approach, we found that miR-124a injection resulted in a 30–40% increase in GLT1 protein levels in the cervical cord compared with that in the age-matched control (uninjected or scrambled RNA injected) SOD1G93A mice (Fig. 7D). These results directly demonstrate that exogenously delivered miR-124a is sufficient to increase GLT1 protein expression in vivo.
DISCUSSION
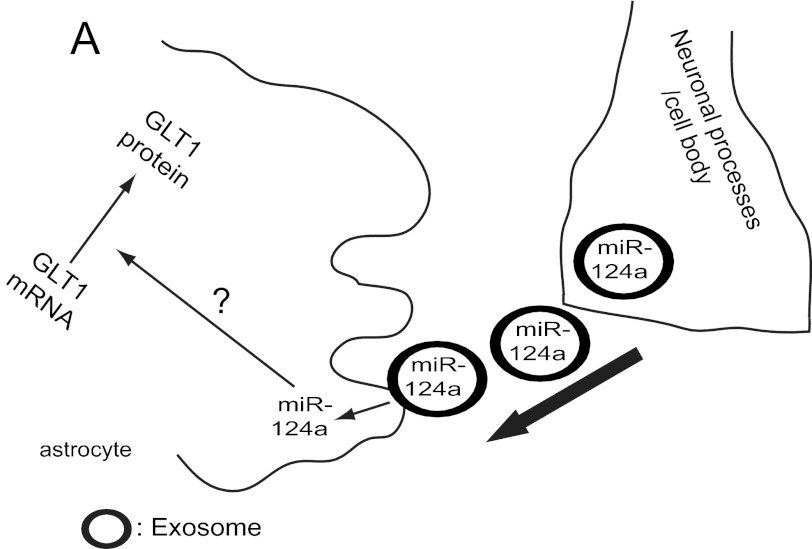
Despite the importance of EAAT2/GLT1 in both physiological and pathological conditions, relatively little is known about the regulation of EAAT2/GLT1 expression. Expression of EAAT2/GLT1 is highly dynamic during development and in pathophysiological conditions, which is likely to be delicately regulated through differential pathways at multiple levels. Although global disruption of astroglial miRNA biogenesis through selective deletion of Dicer in cerebellar astrocytes significantly reduces GLT1 expression (42), specific miRNA-mediated regulation of GLT1 protein expression has not been explored previously. Our results in this study unveiled a new pathway in EAAT2/GLT1 expression regulation: through the exosomal transfer of neuronal miR-124a into astrocytes (Fig. 8). This study also provides the first direct evidence for the miRNA-mediated translational regulation of EAAT2/GLT1 expression, opening a new frontier to understanding EAAT2/GLT1 expression regulation.
FIGURE 8.
Exosome-mediated miR-124a transfer from neurons to astrocytes and its regulation of GLT1 protein expression.
Exosomes have been recently considered as a new mechanism for intercellular communication (43). Various signal molecules, including thousands of proteins, lipids, and RNAs, have been found in exosomes isolated from many mammalian cell types and model organisms Caenorhabditis elegans (44) and Drosophila (45). These exosomes are actively involved in many (patho)physiological processes, including immune signaling (46), development (44), cancer (47), and viral infections (48). Exosome-mediated communication and physiological significance in the CNS is largely unknown, although exosomes have been found in neuronal and glial cultures (33, 34). This study demonstrates that this exosome-mediated transfer of microRNA and subsequent regulation of astroglial GLT1 expression represents an alternative communication pathway from neurons to astrocytes in regulating astrocyte functions (Fig. 8), apart from the synaptic release of glutamate from excited neurons. The mechanisms for exosome release from neurons are unclear at this point. We found no evidence that neuronal activity is directly involved in regulating the release of exosomes or miRNAs. In addition, we have also found that exosome-like vesicles are secreted from cultured astrocytes. Because astrocytes are known to release various signals to modulate synaptic/neuronal functions (49, 50) and survival (51, 52), it is likely that astroglial exosomes may also play a modulatory role in synaptic functions. The exact functions of astroglial exosomes remain to be investigated in the future. Given the potentially diverse composition of signals in exosomes that have been profiled previously, it is conceivable that neuronal or astroglial exosomes also contain multiple signals and that exosome-mediated transfer of signals may play significant roles in mediating neuron-to-astrocyte communication.
MicroRNA-dependent translational regulation is largely cell-autonomous and is mainly mediated by its direct binding to the target mRNA (20). Interestingly, although miR-124a consistently up-regulates GLT1 protein expression in vitro and in vivo, its effect on GLT1 protein expression is likely to be indirect and is mediated through other astroglial factors. Although a recent study has implicated an inhibitory role of astroglial ephrinA3 in the translational regulation of GLT1 (17), we found that miR-124a has no significant effect on ephrinA3 protein expression levels or on ephrinA3 mRNA 3′UTR function (indicated by a 3′UTR luciferase reporter assay, Fig. 6). Astroglial factors and pathways, especially miRNAs that directly mediate the action of miR-124a in GLT1 up-regulation in astrocytes, remain to be characterized. In addition, although miR-124 appears to specifically and significantly up-regulate GLT1 protein expression in astrocytes, as a single miRNA usually modulates translation expression of multiple target mRNAs, it is conceivable that miR-124a can broadly modulate astroglial gene expression. Characterization of the downstream targets of miR-124a in astrocytes will then help identify astroglial genes that are regulated by neuronal signals, like miR-124a. Ultimately, this knowledge will increase our understanding how neuronal signals coordinate astroglial functions at developing and mature synapses.
Severe and selective reduction of EAAT2/GLT1 protein has been observed in ALS patients and rodent models of ALS, promoting excitotoxicity to motor neuron degeneration (40). Our observation that exogenously delivered miR-124a is sufficient to increase GLT1 protein expression in the spinal cord of SOD1G93A mice provides encouraging results that miR-124a may hold potential for developing GLT1-based neuroprotective strategies for ALS. In addition, the selective reduction of miR-124a, presumably as a result of the loss of neurons, may provide a new biomarker for detecting early motor neuron loss in ALS.
Overall, this study explored a unique and novel regulatory mechanism in neuron-to-astrocyte communication. It also provides new insight into miRNA-mediated translational regulation of the key astroglial glutamate transporter EAAT2/GLT1 in physiological and pathological conditions. Ultimately, these results will increase our understanding of EAAT2/GLT1 expression regulation, which could facilitate the development of drugs that up-regulate GLT1 expression in ALS.
Acknowledgments
We thank the Tufts Center for Neuroscience Research (NIH P30 NS047243; PI, Rob Jackson) for providing valuable core facilities and a new faculty recruitment grant (NIH P30 5P30NS069254-02; PI, Phil Haydon) in the Tufts Neuroscience Department.
This work was supported, in whole or in part, by National Institutes of Health Grants MH099554 (to Y. Y.) and NS233958 and NS36465 (to J. D. R.).

This article contains a supplemental movie.
- NCM
- neuron-conditioned medium
- UTR
- untranslated region
- miRNA
- microRNA
- P0
- postnatal day 0
- E20
- embryonic day 20
- A/S
- antisense
- QRT-PCR
- quantitative RT-PCR
- GFAP
- glial fibrillary acidic protein
- ANOVA
- analysis of variance.
REFERENCES
- 1. Danbolt N. C. (2001) Glutamate uptake. Prog. Neurobiol. 65, 1–105 [DOI] [PubMed] [Google Scholar]
- 2. Rothstein J. D., Martin L., Levey A. I., Dykes-Hoberg M., Jin L., Wu D., Nash N., Kuncl R. W. (1994) Localization of neuronal and glial glutamate transporters. Neuron 13, 713–725 [DOI] [PubMed] [Google Scholar]
- 3. Chen W., Aoki C., Mahadomrongkul V., Gruber C. E., Wang G. J., Blitzblau R., Irwin N., Rosenberg P. A. (2002) Expression of a variant form of the glutamate transporter GLT1 in neuronal cultures and in neurons and astrocytes in the rat brain. J. Neurosci. 22, 2142–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen W., Mahadomrongkul V., Berger U. V., Bassan M., DeSilva T., Tanaka K., Irwin N., Aoki C., Rosenberg P. A. (2004) The glutamate transporter GLT1a is expressed in excitatory axon terminals of mature hippocampal neurons. J. Neurosci. 24, 1136–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Swanson R. A., Liu J., Miller J. W., Rothstein J. D., Farrell K., Stein B. A., Longuemare M. C. (1997) Neuronal regulation of glutamate transporter subtype expression in astrocytes. J. Neurosci. 17, 932–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang Y., Gozen O., Watkins A., Lorenzini I., Lepore A., Gao Y., Vidensky S., Brennan J., Poulsen D., Won Park J., Li Jeon N., Robinson M. B., Rothstein J. D. (2009) Presynaptic regulation of astroglial excitatory neurotransmitter transporter GLT1. Neuron 61, 880–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schlag B. D., Vondrasek J. R., Munir M., Kalandadze A., Zelenaia O. A., Rothstein J. D., Robinson M. B. (1998) Regulation of the glial Na+-dependent glutamate transporters by cyclic AMP analogs and neurons. Mol. Pharmacol. 53, 355–369 [DOI] [PubMed] [Google Scholar]
- 8. Gegelashvili G., Danbolt N. C., Schousboe A. (1997) Neuronal soluble factors differentially regulate the expression of the GLT1 and GLAST glutamate transporters in cultured astroglia. J. Neurochem. 69, 2612–2615 [DOI] [PubMed] [Google Scholar]
- 9. Berger U. V., DeSilva T. M., Chen W., Rosenberg P. A. (2005) Cellular and subcellular mRNA localization of glutamate transporter isoforms GLT1a and GLT1b in rat brain by in situ hybridization. J. Comp. Neurol. 492, 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peacey E., Miller C. C., Dunlop J., Rattray M. (2009) The four major N- and C-terminal splice variants of the excitatory amino acid transporter GLT-1 form cell surface homomeric and heteromeric assemblies. Mol. Pharmacol. 75, 1062–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Utsunomiya-Tate N., Endou H., Kanai Y. (1997) Tissue specific variants of glutamate transporter GLT-1. FEBS Lett. 416, 312–316 [DOI] [PubMed] [Google Scholar]
- 12. Tian G., Lai L., Guo H., Lin Y., Butchbach M. E., Chang Y., Lin C. L. (2007) Translational control of glial glutamate transporter EAAT2 expression. J. Biol. Chem. 282, 1727–1737 [DOI] [PubMed] [Google Scholar]
- 13. Kim S. Y., Chao W., Choi S. Y., Volsky D. J. (2003) Cloning and characterization of the 3′-untranslated region of the human excitatory amino acid transporter 2 transcript. J. Neurochem. 86, 1458–1467 [DOI] [PubMed] [Google Scholar]
- 14. Lin C. L., Bristol L. A., Jin L., Dykes-Hoberg M., Crawford T., Clawson L., Rothstein J. D. (1998) Aberrant RNA processing in a neurodegenerative disease. The cause for absent EAAT2, a glutamate transporter, in amyotrophic lateral sclerosis. Neuron 20, 589–602 [DOI] [PubMed] [Google Scholar]
- 15. Bristol L. A., Rothstein J. D. (1996) Glutamate transporter gene expression in amyotrophic lateral sclerosis motor cortex. Ann. Neurol. 39, 676–679 [DOI] [PubMed] [Google Scholar]
- 16. Flowers J. M., Powell J. F., Leigh P. N., Andersen P., Shaw C. E. (2001) Intron 7 retention and exon 9 skipping EAAT2 mRNA variants are not associated with amyotrophic lateral sclerosis. Ann. Neurol. 49, 643–649 [PubMed] [Google Scholar]
- 17. Carmona M. A., Murai K. K., Wang L., Roberts A. J., Pasquale E. B. (2009) Glial ephrin-A3 regulates hippocampal dendritic spine morphology and glutamate transport. Proc. Natl. Acad. Sci. U.S.A. 106, 12524–12529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ambros V. (2004) The functions of animal microRNAs. Nature 431, 350–355 [DOI] [PubMed] [Google Scholar]
- 19. Rogelj B., Giese K. P. (2004) Expression and function of brain specific small RNAs. Rev. Neurosci. 15, 185–198 [DOI] [PubMed] [Google Scholar]
- 20. Inui M., Martello G., Piccolo S. (2010) MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 11, 252–263 [DOI] [PubMed] [Google Scholar]
- 21. Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., Lötvall J. O. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 [DOI] [PubMed] [Google Scholar]
- 22. Vlassov A. V., Magdaleno S., Setterquist R., Conrad R. (2012) Exosomes. Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta 1820, 940–948 [DOI] [PubMed] [Google Scholar]
- 23. Record M., Subra C., Silvente-Poirot S., Poirot M. (2011) Exosomes as intercellular signalosomes and pharmacological effectors. Biochem. Pharmacol. 81, 1171–1182 [DOI] [PubMed] [Google Scholar]
- 24. Lotvall J., Valadi H. (2007) Cell to cell signalling via exosomes through esRNA. Cell Adh. Migr. 1, 156–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schaeffer D., Clark A., Klauer A. A., Tsanova B., van Hoof A. (2011) Functions of the cytoplasmic exosome. Adv. Exp. Med. Biol. 702, 79–90 [DOI] [PubMed] [Google Scholar]
- 26. O'Loughlin A. J., Woffindale C. A., Wood M. J. (2012) Exosomes and the emerging field of exosome-based gene therapy. Curr. Gene Ther. 12, 262–274 [DOI] [PubMed] [Google Scholar]
- 27. Pant S., Hilton H., Burczynski M. E. (2012) The multifaceted exosome. Biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem. Pharmacol. 83, 1484–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keller S., Sanderson M. P., Stoeck A., Altevogt P. (2006) Exosomes. From biogenesis and secretion to biological function. Immunol. Lett. 107, 102–108 [DOI] [PubMed] [Google Scholar]
- 29. Bobrie A., Colombo M., Raposo G., Théry C. (2011) Exosome secretion. Molecular mechanisms and roles in immune responses. Traffic 12, 1659–1668 [DOI] [PubMed] [Google Scholar]
- 30. Bak M., Silahtaroglu A., Møller M., Christensen M., Rath M. F., Skryabin B., Tommerup N., Kauppinen S. (2008) MicroRNA expression in the adult mouse central nervous system. RNA 14, 432–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Robinson M. B., Hunter-Ensor M., Sinor J. (1991) Pharmacologically distinct sodium-dependent L-[3H]glutamate transport processes in rat brain. Brain Res. 544, 196–202 [DOI] [PubMed] [Google Scholar]
- 32. Lepore A. C., Rauck B., Dejea C., Pardo A. C., Rao M. S., Rothstein J. D., Maragakis N. J. (2008) Focal transplantation-based astrocyte replacement is neuroprotective in a model of motor neuron disease. Nat. Neurosci. 11, 1294–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fauré J., Lachenal G., Court M., Hirrlinger J., Chatellard-Causse C., Blot B., Grange J., Schoehn G., Goldberg Y., Boyer V., Kirchhoff F., Raposo G., Garin J., Sadoul R. (2006) Exosomes are released by cultured cortical neurones. Mol. Cell Neurosci. 31, 642–648 [DOI] [PubMed] [Google Scholar]
- 34. Wang G., Dinkins M., He Q., Zhu G., Poirier C., Campbell A., Mayer-Proschel M., Bieberich E. (2012) Astrocytes secrete exosomes enriched with proapoptotic ceramide and prostate apoptosis response 4 (PAR-4). Potential mechanism of apoptosis induction in Alzheimer disease (AD). J. Biol. Chem. 287, 21384–21395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Conaco C., Otto S., Han J. J., Mandel G. (2006) Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl. Acad. Sci. U.S.A. 103, 2422–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Deo M., Yu J. Y., Chung K. H., Tippens M., Turner D. L. (2006) Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev. Dyn. 235, 2538–2548 [DOI] [PubMed] [Google Scholar]
- 37. Lewis B. P., Burge C. B., Bartel D. P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 38. Cahoy J. D., Emery B., Kaushal A., Foo L. C., Zamanian J. L., Christopherson K. S., Xing Y., Lubischer J. L., Krieg P. A., Krupenko S. A., Thompson W. J., Barres B. A. (2008) A transcriptome database for astrocytes, neurons, and oligodendrocytes. A new resource for understanding brain development and function. J. Neurosci. 28, 264–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Howland D. S., Liu J., She Y., Goad B., Maragakis N. J., Kim B., Erickson J., Kulik J., DeVito L., Psaltis G., DeGennaro L. J., Cleveland D. W., Rothstein J. D. (2002) Focal loss of the glutamate transporter EAAT2 in a transgenic rat model of SOD1 mutant-mediated amyotrophic lateral sclerosis (ALS). Proc. Natl. Acad. Sci. U.S.A. 99, 1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rothstein J. D., Van Kammen M., Levey A. I., Martin L. J., Kuncl R. W. (1995) Selective loss of glial glutamate transporter GLT-1 in amyotrophic lateral sclerosis. Ann. Neurol. 38, 73–84 [DOI] [PubMed] [Google Scholar]
- 41. Xia X., Zhou H., Huang Y., Xu Z. (2006) Allele-specific RNAi selectively silences mutant SOD1 and achieves significant therapeutic benefit in vivo. Neurobiol. Dis. 23, 578–586 [DOI] [PubMed] [Google Scholar]
- 42. Tao J., Wu H., Lin Q., Wei W., Lu X. H., Cantle J. P., Ao Y., Olsen R. W., Yang X. W., Mody I., Sofroniew M. V., Sun Y. E. (2011) Deletion of astroglial Dicer causes non-cell-autonomous neuronal dysfunction and degeneration. J. Neurosci. 31, 8306–8319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Belting M., Wittrup A. (2008) Nanotubes, exosomes, and nucleic acid-binding peptides provide novel mechanisms of intercellular communication in eukaryotic cells. Implications in health and disease. J. Cell Biol. 183, 1187–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kolotuev I., Apaydin A., Labouesse M. (2009) Secretion of Hedgehog-related peptides and WNT during Caenorhabditis elegans development. Traffic 10, 803–810 [DOI] [PubMed] [Google Scholar]
- 45. Korkut C., Ataman B., Ramachandran P., Ashley J., Barria R., Gherbesi N., Budnik V. (2009) Trans-synaptic transmission of vesicular Wnt signals through Evi/Wntless. Cell 139, 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Théry C., Ostrowski M., Segura E. (2009) Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593 [DOI] [PubMed] [Google Scholar]
- 47. Yu X., Riley T., Levine A. J. (2009) The regulation of the endosomal compartment by p53 the tumor suppressor gene. FEBS J. 276, 2201–2212 [DOI] [PubMed] [Google Scholar]
- 48. Logozzi M., De Milito A., Lugini L., Borghi M., Calabrò L., Spada M., Perdicchio M., Marino M. L., Federici C., Iessi E., Brambilla D., Venturi G., Lozupone F., Santinami M., Huber V., Maio M., Rivoltini L., Fais S. (2009) High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS ONE 4, e5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Halassa M. M., Fellin T., Haydon P. G. (2007) The tripartite synapse. Roles for gliotransmission in health and disease. Trends Mol. Med. 13, 54–63 [DOI] [PubMed] [Google Scholar]
- 50. Eroglu C., Barres B. A. (2010) Regulation of synaptic connectivity by glia. Nature 468, 223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nagai M., Re D. B., Nagata T., Chalazonitis A., Jessell T. M., Wichterle H., Przedborski S. (2007) Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat. Neurosci. 10, 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Di Giorgio F. P., Carrasco M. A., Siao M. C., Maniatis T., Eggan K. (2007) Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat. Neurosci. 10, 608–614 [DOI] [PMC free article] [PubMed] [Google Scholar]