Background: Stereocilia tip-link protein cadherin 23 couples mechanical forces to sensory transduction in inner ear receptor hair cells.
Results: CNGA3 interacts with cadherin 23 and with cadherin 23 binding partner, myosin VIIa.
Conclusion: Cadherin 23 and myosin VIIa form alternate complexes with CNGA3.
Significance: CNGA3 interactions with cadherin 23 +68 and myosin VIIa predict a role for CNGA3 in hair-cell mechanotransduction.
Keywords: Cell Biology, Cyclic GMP (cGMP), Extracellular Matrix Proteins, Protein Structure, Protein-Protein Interactions, CNGA3 Channels, Cadherin 23, Mammalian Polarized Epithelial Cells, PCR, Surface Plasmon Resonance
Abstract
Previously, we obtained evidence for a photoreceptor/olfactory type of CNGA3 transcript in a purified teleost vestibular hair cell preparation with immunolocalization of CNGA3 protein to stereocilia of teleost vestibular and mammalian cochlear hair cells. The carboxyl terminus of highly Ca2+-permeable CNGA3 expressed in the mammalian organ of Corti and saccular hair cells was found to interact with an intracellular domain of microfibril interface-located protein 1 (EMILIN 1), a member of the elastin superfamily, also immunolocalizd to hair cell stereocilia (Selvakumar, D., Drescher, M. J., Dowdall, J. R., Khan, K. M., Hatfield, J. S., Ramakrishnan, N. A., and Drescher, D. G. (2012) Biochem. J. 443, 463–476). Here, we provide evidence for organ of Corti proteins, of Ca2+-dependent binding of the amino terminus of CNGA3 specifically to the carboxyl terminus of stereocilia tip-link protein CDH23 +68 (cadherin 23 with expressed exon 68) by yeast two-hybrid mating and co-transformation protocols, pulldown assays, and surface plasmon resonance analysis. Myosin VIIa, required for adaptation of hair cell mechanotransduction (MET) channel(s), competed with CDH23 +68, with direct Ca2+-dependent binding to the amino terminus of CNGA3. Based upon the premise that hair cell stereocilia tip-link proteins are closely coupled with MET, these results are consistent with the possibility that CNGA3 participates in hair-cell MET. Together with the demonstration of protein-protein interaction between HCN1 and tip-link protein protocadherin 15 CD3 (Ramakrishnan, N. A., Drescher, M. J., Barretto, R. L., Beisel, K. W., Hatfield, J. S., and Drescher, D. G. (2009) J. Biol. Chem. 284, 3227–3238; Ramakrishnan, N. A., Drescher, M. J., Khan, K. M., Hatfield, J. S., and Drescher, D. G. (2012) J. Biol. Chem. 287, 37628–37646), a protein-protein interaction for CNGA3 and a second tip-link protein, CDH23 +68, further suggests possible association of two different channels with a single stereocilia tip link.
Introduction
Cyclic nucleotide-gated (CNG)2 channels mediate sensory transduction in vision and olfaction (1, 2) and are highly permeable to Ca2+, representing a primary mechanism for Ca2+ entry. In previous work, we obtained three full-length sequences for a photoreceptor/olfactory type of CNGA3 transcript expressed by hair cells in a purified teleost vestibular hair cell preparation (3). CNGA3 subunit protein was immunolocalized to sites on the stereocilia and subcuticular plate region for both teleost saccular hair cells and mammalian cochlear cells. Protein-protein interaction protocols indicated for both models that the carboxyl terminus of CNGA3 binds to a cytoplasmic carboxyl terminus of elastin microfibril interface-located protein 1 (EMILIN1), an extracellular matrix protein and member of the elastin superfamily of proteins. EMILIN1 was also localized to hair cell stereocilia along with phosphodiesterase 6C, a phosphodiesterase regulating cGMP gating of CNGA3 in cone photoreceptors (3).
Here we present biochemical evidence that the cytoplasmic amino terminus of ion channel CNGA3 binds the cytoplasmic carboxyl terminus of cadherin 23 (CDH23), a transmembrane protein that forms the upper two-thirds of filaments constituting hair cell stereocilia tip links. The tip links are considered important in transducing the mechanical stimuli of inner ear hair cells without, however, the upper tip-link protein, cadherin 23, being previously identified as participating in protein-protein interactions with ion channels, although predicted (4). Mutations in the human CDH23 gene are responsible for Usher′s USH1D, a syndrome in which hearing loss is associated with retinitis pigmentosa (5). CDH23 is an atypical cadherin protein with up to 27 extracellular cadherin repeats, a transmembrane domain, and a short cytoplasmic domain. Alternative splicing of the CDH23 gene involves the expression of exon 68, which encodes part of the cytoplasmic domain (6) that is preferentially expressed in the inner ear but not in the retina (7). CDH23 with expressed exon 68 (CDH23 +68) was immunolocalized to hair cell stereocilia tips in the adult (4). The binding that we describe here for the first time between CNGA3 and CDH23 is specific to CDH23 that includes exon 68. CNGA3 did not interact with CDH23 minus exon 68 (CDH23 −68). Furthermore, myosin VIIa, a CDH23 binding partner (8), competes with CDH23 +68 for interaction sites on CNGA3.
EXPERIMENTAL PROCEDURES
Materials
Yeast two-hybrid plasmids pGBKT7 and pGADT7 were obtained from BD Biosciences Clontech (Palo Alto, CA). Glutathione S-transferase (GST) fusion plasmid pGEX-6P-1 and HBS-N, HBS-P, and HBS-EP buffers for surface plasmon resonance (SPR) were procured from GE Healthcare. Plasmids for hexahistidine-tagged fusion proteins (pRSET A, B, and C), anti-his-tag monoclonal antibody (recognizing polyhistidine sequences), anti-Xpress antibody (raised against the sequence DLYDDDDK), and MagicMark XP Western standards were obtained from Invitrogen. Anti-GST antibody (against Schistosoma japonicum GST sequence) was from GE. Phosphate-buffered saline (PBS), and PBS with Tween (PBST) buffer were from Sigma. Antibodies included: CDH23-specific goat polyclonal IgG, sc-26338; goat anti-mouse IgG-HRP, sc-2031; donkey anti-goat IgG-HRP, sc-2033 (Santa Cruz Biotechnology, Santa Cruz, CA).
Yeast Two-hybrid Analysis
Yeast two-hybrid screening for the interacting proteins of rat organ of Corti CNGA3 was performed (Matchmaker Gold Yeast Two-Hybrid System, Clontech, Mountain View, CA) using the cytoplasmic amino terminus of CNGA3 (aa 1–146) as bait (Fig. 1, A and C). PCR was carried out on a rat organ of Corti cDNA library with upstream primer 5′-GCCCCATGGATGGCCAAGGTCAATACCCAA-3′ and downstream primer 5′-TGCGAATTCTTAGTTGCTGGAGGGGTCCACCAC-3′ (restriction sites are underlined). The corresponding amplification product was purified, restriction-digested with NcoI and EcoRI enzymes, and inserted into a similarly digested pGBKT7 vector to produce a fusion construct with the GAL4 DNA binding domain. The fusion products were proven to be transcriptionally inactive in appropriate nutrient-deficient media.
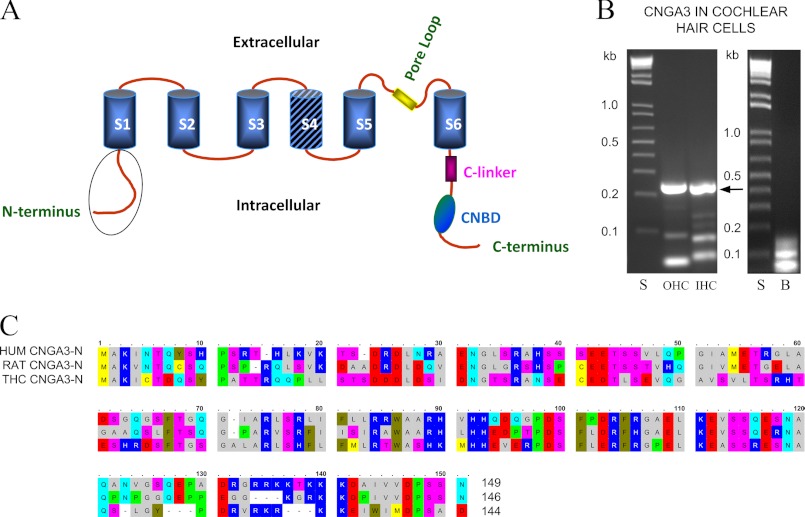
FIGURE 1.
CNGA3-predicted protein structure and mRNA expression in cochlear outer hair cells and inner hair cells. A, shown is a diagram illustrating the predicted structure of the CNGA3 protein (GenBankTM EDL99244, rat). The cytoplasmic amino terminus sequence is encircled in a schematic depiction of CNGA3 with S1-S6 transmembrane regions and S4 hatched representing the voltage sensor and CNBD referring to the cyclic nucleotide binding domain. B, shown is PCR amplification of a 242-bp cDNA product corresponding to the amino terminus of rat CNGA3, from the analysis of pooled, singly isolated rat cochlear outer hair cells (OHC) and inner hair cells (IHC). The primers were designed to target the CNGA3 sequence expressed in a subset of rat olfactory sensory neurons (16) (GenBankTM AJ272428) with an identity to human cone photoreceptor CNGA3 (NP_001073347) and trout saccular hair cell CNGA3–2 (HQ542178). The amplification product yielded rat CNGA3 sequence with 99% nucleotide identity for cochlear outer hair cell and 98% nucleotide identity for inner hair cells to rat AJ272428. Lanes S, 1-kb standards; lanes OHC and IHC, CNGA3 242-bp amino terminus products amplified from rat cochlear outer and inner hair cells, respectively, arrow; lane B, negative control in which cDNA was omitted. C, shown is alignment (with OMIGA 2.0) of the amino terminus amino acids of rat organ of Corti CNGA3 (EDL99244) used as bait (RAT CNGA3-N, second line) in yeast two-hybrid protocols aligned with human CNGA3 (HUM CNGA3-N, first line, NP_001073347) and trout saccular hair cell CNGA3–2 (THC CNGA3-N, third line, AER38241). CNGA3 amino termini display partial EF hand Ca2+ binding domains (17), DRDLDQ, aa 23–28, for rat organ of Corti sequence with comparable motifs appearing in the calcium-binding protein Cab45 (Q91ZS3), cadherin 23 (58), and EDH4 (36, NP_647540). In addition, there is a PDZ domain NGLG aa 31–34 (compare with piccolo, EDL99465). The amino terminus of trout saccular hair cell CNGA3–2 variant (third line) indicates identity to both human (first line) and rat CNGA3 (second line) sequences.
Prey Library Construction
A rat organ-of-Corti prey cDNA library was constructed from an organ of Corti subfraction from rat cochlea (1-month-old ACI Black Agouti rats, Harlan Sprague-Dawley), microdissected as previously described, and morphologically characterized (9). Equal proportions of oligo(dT) and random hexamer-derived cDNA obtained from 5 μg of total RNA were inserted into vector pGADT7-Rec by homologous recombination in the host strain Y187 (Matchmaker Library Construction and Screening Kit, Clontech) (3, 10). The transformants were plated on SD/−Leu medium, and after 5 days the colonies were collected and used for yeast mating experiments. Generally, 3 μg of pGADT7-Rec was mixed with 20 μl of the double-stranded cDNA for standard yeast transformation reactions.
Mating and Screening
To screen for interacting proteins, we mixed cells containing bait constructs in strain Y2H gold with the cells containing prey constructs in Y187. These cells were allowed to mate in 2× YPDA medium (Clontech) overnight at 30 °C and plated on a selection medium SD/−Ade/−His/−Leu/−Trp/aureobasidin A (QDO/A) (quadruple drop-out medium with aureobasidin A). After 5–7 days, the larger colonies that appeared on the plates were analyzed by PCR, amplification products were sequenced, and prey sequences were identified using BLAST (NCBI).
Yeast Two-hybrid Co-transformation Analysis
The rat CNGA3 amino-terminal cytoplasmic region (CNGA3-N) was prepared as a bait plasmid in pGBKT7 vector. The cytoplasmic carboxyl terminus of CDH23 expressed in rat organ of Corti was targeted in PCR using specific primers (upstream primer, 5′-GCCGAATTCTTCTGCCGGAACCTGGAGCTG-3′; downstream primer, 5′-TGCGGATCCTTACAGTTCCGTGATTTCCAGGG-3′; NM_053644, nucleotides 9747–10299). The PCR amplification revealed two fragments corresponding to CDH23 with and without exon 68. These fragments were cloned into the prey vector pGADT7. For the yeast two-hybrid assay, ∼500 ng of each vector was co-transformed into Y2H gold yeast strain using standard protocol (Clontech Yeast Two-hybrid Manual), and the transformants were plated on nutrient-deficient selection medium (QDO/A) for 3–5 days. Transformants were also plated on a less stringent deficient medium (SD/−Leu/−Trp) to select transformants. The colonies were picked and streaked on higher stringency medium (QDO/A). Negative controls contained either the bait or the prey along with the prey or bait empty vector, respectively. To further confirm the binding interaction of CNGA3-N with CDH23 +68, we performed the reversal system of bait and prey along with negative controls, CNGA3-N as prey and CDH23 +68 as bait in selection medium (QDO/A).
Western Blot Analysis
Purity of the fusion proteins was ascertained by SDS-PAGE (4–12% NuPAGE gel followed by overnight blocking with 5% nonfat milk at 4 °C) with Coomassie Blue staining. Blots were incubated with anti-Xpress monoclonal antibody (Invitrogen, 1:5,000) for 3 h at room temperature or overnight at 4 °C, washed with PBS containing 0.1% Tween 20, and incubated with HRP-conjugated anti-mouse IgG (1:10,000) for 1 h at room temperature, and the proteins were detected using Western Lightning chemiluminescence (Perkin-Elmer Life Sciences).
Cloning and Expression of GST Fusion Protein
CNGA3-N was amplified from the rat organ of Corti cDNA preparation using rat CNGA3 specific upstream (5′-GCCGAATTCATGGCCAAGGTCAATACCCAA-3′) and downstream (5′-TGCGCGGCCGCTTAGTTGCTGGAGGGGTCCACCAC-3′) primers (EcoRI and NotI restriction sites are underlined). The restriction-digested PCR products were ligated into similarly digested pGEX-6P-1 vector with a GST fusion tag. Carboxyl termini of rat organ of Corti CDH23 +68 and −68 were PCR-amplified with gene specific upstream (5′-GCCGGATCCTTCTGCCGGAACCTGGAGCTG-3′) and downstream (5′-TGCGAATTCTTACAGTTCCGTGATTTCCAGGG-3′) primers (BamH1 and EcoRI sites underlined). The restriction-digested PCR products were ligated into similarly digested pRSET-A vectors with polyhistidine fusion tags.
Pulldown Assay
Pulldown assays for rat organ of Corti proteins were performed as described previously (3) with GST fusion protein (GST-CNGA3-N) and hexahistidine-tagged fusion proteins (CDH23 +68 and CDH23 −68). GST conjugates were mixed with hexahistidine-tagged CDH23 +68 and −68 in binding buffer (PBST buffer, pH 7.4, 1× protease inhibitors) for 2 h at room temperature. Glutathione-Sepharose beads (30 μl of 50% slurry) were added to the reaction mix for 3 h at room temperature or overnight at 4 °C followed by centrifugation for 2 min at 1,000 × g. The beads were washed 5 times in the binding buffer, incubated at 70 °C for 5 min in gel loading buffer (30 μl), and centrifuged. The supernatant was then electrophoresed in a 4–12% NuPAGE gel (Invitrogen), electroblotted to a nylon membrane, and immunodetected using anti-Xpress antibody and CDH23-specific goat polyclonal antibody (sc-26338).
Protein Expression and Purification
cDNA was amplified from rat organ of Corti and inserted into pRSET vectors. PCR primers for protein expression in pRSET-B included CNGA3 amino terminus upstream (5′-GCCAGATCTATGGCCAAGGTCAATACCCAA-3′) and downstream (5′-TGCGAATTCTTAGTTGCTGGAGGGGTCCAC-3′) (AJ272428). For CDH23 +68 and −68 in pRSET-A, the primers were: upstream, 5′-GCCGGATCCTTCTGCCGGAACCTGGAGCTG-3′, and downstream, 5′-TGCGAATTCTTACAGTTCCGTGATTTCCAGGG-3′ (NM_053644). For the carboxyl terminus of myosin VIIa (amino acids 1608–2177, composed of SH3, MyTH4, and FERM domains) in pRSET-A, the upstream primer was 5′-GCCGGATCCGGCTGGGCCAATGGCATCAAC-3′, and downstream primer was 5′-TGCGAATTCTCACCTCCCGCTCCTGGAGTTCCT-3′ (NM_153473). The PCR products were purified and digested with restriction enzymes and cloned into similarly digested pRSET vectors in Escherichia coli DH5α cells (Invitrogen). Clones were selected and sequence-verified before protein expression studies. The pRSET expression vectors containing the desired sequences were used to transform E. coli BL21 (DE3) cells (Invitrogen). Selected clones were used for expression of fusion proteins induced by adding 1 mm isopropyl β-d-1-thiogalactopyranoside in LB culture medium. The cultures were incubated for 4 h at 37 °C, and the cells were harvested, lysed in 10 mm phosphate buffer, 8 m urea (pH 8.0), centrifuged, and analyzed via 4–12% SDS-PAGE with protein bands visualized by Coomassie Blue staining. Clones that showed robust expression were selected and stored for future use. For purification of fusion proteins, 100–500-ml cultures of each clone were grown for 4–6 h with isopropyl β-d-1-thiogalactopyranoside, lysed by sonication, and centrifuged at 20,818 × g. The clear supernatant was mixed with 0.5 ml Talon beads (Clontech), and the beads were sedimented and washed. The bound fusion proteins were eluted by the addition of elution buffer (His TALON buffer, Clontech) followed by mixing and centrifugation. The supernatant containing the fusion proteins was then exhaustively dialyzed against cold PBST buffer containing 1× protease inhibitors for 24 h at 4 °C with constant stirring and 5 or 6 buffer changes. Protein was determined with the Qubit fluorescent system (Invitrogen).
SPR Analysis
The binding interactions of CNGA3-N and CDH23 or myosin VIIa tail expressed in rat organ of Corti were measured using SPR with a Biacore 3000 instrument at 25 °C (Biacore, Piscataway, NJ) as previously described (3). Binding of the mobile molecule (analyte) to the immobilized molecule (ligand) produces changes in refractive index which can be detected. Affinity-purified hexahistidine-tagged fusion proteins were immobilized on a CM5 sensor chip by an amine-coupling reaction (cf. Ref. 11). Surfaces were prepared for each immobilized protein (ligand) with different concentrations of purified CDH23 +68 and −68 fusion proteins as analyte. The reference surface was blocked with ethanolamine and thus contained no ligand. The response was recorded as the ligand response units (RU) minus the reference RU.
SPR Kinetic Measurements of CNGA3-N Binding
The rate and equilibrium binding constants of the interaction of CNGA3-N with CDH23 +68 and myosin VIIa tail were determined in kinetic studies. The CNGA3-N fusion protein was immobilized as a ligand on a CM5 chip. Rat CDH23 +68 or myosin VIIa (analyte) was diluted in a series of concentrations in HBS-N buffer including 68 μm Ca2+. Kinetic values were determined using BIAevaluation 3.0 software (Biacore). A 1:1 Langmuir binding model was generally selected for calculations in the kinetic studies. Affinity-purified CDH23 +68 and myosin VIIa rat fusion proteins were analyzed as analytes, 0–320 nm and 0–100 nm, respectively. After each reaction, the chip was regenerated with HBS-N buffer.
Single Hair Cell RT-PCR
cDNA from a pool of 25–30 singly isolated outer hair cells or, separately, a similar number of inner hair cells was prepared according to “Make Your Own Mate & Plate Library System” (Clontech) with Moloney murine leukemia virus reverse transcriptase or, for selected experiments, SMARTScribe reverse transcriptase (12). The specific upstream and downstream primers for CNGA3 amplification were 5′-GCCAAGGTCAATACCCAATG-3′ and 5′-CGAATGGAGATGATGAAGCG-3′, respectively, designed to cross introns with the aid of Accelrys Gene 2.5 software (San Diego, CA) and a comparison with rat genomic sequence (NW_001084882.1). These PCR reactions were carried out using Advantage 2 GC-polymerase mix (Clontech) with denaturation at 95 °C for 20 s, annealing at 60 °C for 20 s, and extension at 72 °C for 30 s, with a total of 65 cycles. Negative controls were derived from the elimination of cDNA and substitution of sterilized water in the PCR reaction. The PCR products were analyzed with 1.5% agarose gel electrophoresis (NuSieve 3:1 agarose, Lonza, Rockland, ME). All amplified PCR products (253 bp) were sequenced.
RESULTS
The Amino Terminus of CNGA3 Binds to the Carboxyl Terminus of Cadherin 23 by Yeast Two-hybrid Analysis
Previously, a role for CNGA3 in vestibular hair cell function was implied from the finding of three forms of CNGA3 transcript expressed in a purified preparation of >106 hair cells (3) (GenBankTM accession numbers HQ542177, HQ542178, HQ542179). CNGA3 protein was immunolocalized to stereocilia, the site of mechanotransduction, in these saccular hair cells. A role for highly Ca2+-permeable CNGA3 in hair cells, across hair cell type from vestibular to cochlear hair cells, was suggested by immunolocalization of CNGA3 protein to stereocilia of cochlear outer hair cells (3) and cochlear inner hair cells. Evidence of transcript expression of CNGA3 in singly isolated cochlear inner and outer hair cells (Fig. 1B, arrow) further bolsters the conclusion that CNGA3 is expressed in cochlear as well as in vestibular hair cells.
Molecular function for ion channels can be deduced from protein-protein interactions of their intracellular domains. To identify CNGA3 interaction partners, we carried out yeast two-hybrid mating screens of a rat organ of Corti prey cDNA library using the intracellular amino terminus of CNGA3 (CNGA3-N) amplified from cDNA derived from rat organ of Corti, representing auditory sensory epithelium, as bait. The bait sequence corresponded to aa 1–146 of rat CNGA3 (GenBankTM accession number AJ272428) extending to the S1 transmembrane domain, with identity to both human and trout saccular hair cell CNGA3 (Fig. 1C, RAT CNGA3-N compared with HUM CNGA3-N and THC CNGA3-N sequence, respectively), indicating conservation of sequence across evolution. Transformation efficiency, based upon the number of colonies that appeared on the SD/−Leu plates after 5 days, was 8.4 × 105/3 μg of vector.
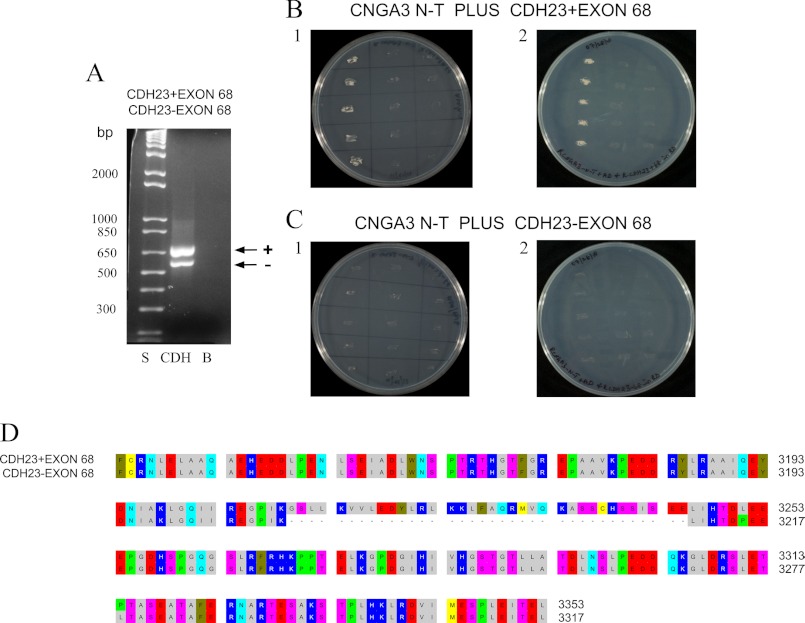
Overall, 92 prey clones were sequenced for the amino terminus bait (CNGA3-N). We noticed a strong interaction in quadruple drop-out media (i.e. multiple clones) for CNGA3-N and prey, which was identified as rat CDH23, a stereociliary tip-link protein. There are two splice variants for CDH23: ±exon 68, with and without an insert of 105 bp encoding 35 amino acids (aa 3210–3245, Fig. 2, A and D) into the putative internal PDZ binding domain of CDH23 (6). Both splice variants retain the same carboxyl terminus containing a second PDZ binding domain. And both splice variants are expressed in organ of Corti of the adult (Fig. 2A, arrow + and arrow − for PCR amplification of CDH23 +68 and CDH23 −68 transcripts, respectively). Transcript for CDH23 +68 is differentially found in sensory epithelia of the inner ear, specific for cochlear hair cells (4), not expressed in heart, kidney, spleen, brain, and retina, consistent with +68-specific function in the inner ear/hair cells (4, 7). Interestingly, it was CDH23 +68 that was identified as the binding partner of CNGA3-N, with 99, 97, and 95% aa identity, respectively, to rat (EDL93044), mouse (NP_001239564), and human (NP_071407.4) CDH23 +68, indicating again highly conserved sequence and theoretically highly conserved molecular function. These results point to hair cell-specific function that is modulated by CDH23 +68 protein-protein interaction with CNGA3, potentially placing CNGA3 in position to be affected by mechanical stimulus via the stereociliary tip-link protein CDH23 +68.
FIGURE 2.
Interaction of CNGA3 with CDH23 +68 and −68 in yeast two-hybrid mating protocols. A, CDH23 +68 and CDH23 −68 messages were expressed in rat organ of Corti. PCR amplification is shown of 660-bp product for CDH23 +68 (lane CDH, arrow +) and 552-bp for CDH23 −68 (lane CDH, arrow −) with lane S = 1 kb plus standards ladder (Invitrogen) and lane B = negative control for which cDNA was omitted. B1, yeast two-hybrid co-transformation assays with replicate streakings show CNGA3 binding to CDH23 +68. First lane, CNGA3 amino terminus (CNGA3 N-T) in bait vector and CDH23 +68 in prey vector; second lane, CNGA3 bait vector and empty prey vector as negative control; third lane, empty bait vector and CDH23 +68 in prey vector as another negative control. B2, shown is reversal of bait and prey sequence for binding of CNGA3 to CDH23 +68. First lane, CNGA3 in prey vector and CDH23 +68 in bait vector; second lane, CNGA3 in prey vector and empty bait vector as negative control; third lane, CDH23 +68 in bait vector and empty prey vector as another negative control. C1, CNGA3 does not bind to CDH23 −68 in yeast two-hybrid co-transformation assays. First lane, CNGA3 in bait vector and CDH23 −68 in prey vector; second lane, CNGA3 in bait vector and empty prey vector as negative control; third lane, empty bait vector and CDH23 −68 in prey vector as another negative control. C2, reversal of bait and prey sequence for CNGA3 and CDH23 −68 is shown. First lane, CNGA3 in prey vector and CDH23 −68 in bait vector; second lane, CNGA3 in prey vector and empty bait vector as negative control; third lane, CDH23 −68 in bait vector and empty prey vector as another negative control. D, shown is alignment (with OMIGA 2.0) of cytoplasmic carboxyl domain of rat CDH23 with amino acid numbering according to NP_446096.1. Alternative splicing generates CDH23 +68 (EDL93044) and CDH23 −68 (NP_446096.1) that differ in their cytoplasmic domain by inclusion/exclusion of exon 68. These sequences were used as prey/bait for binding to CNGA3.
Yeast two-hybrid co-transformation analysis confirmed specific interaction between CNGA3 and CDH23 +68 (Fig. 2B, lane 1 for B1 and B2) with no binding for CDH23 −68 (Fig. 2C, lane 1 for C1 and C2). There was no growth of negative controls on (QDO/A)-selective plates in combinations of bait with empty prey vector and the reverse when bait and prey transcripts were switched (Fig. 2, B1 and B2 and C1 and C2), emphasizing specificity. These results clearly indicate that the binding interaction between CNGA3-N and CDH23 requires exon 68.
Regulation of CNGA3-N and CDH23 Interactions by Alternative Splicing of CDH23 and Competition from Myosin VIIa
To further evaluate CNGA3-N and CDH23 interactions biochemically, we performed GST pulldown assays with standards, GST-CNGA3-N fusion protein at 42 kDa on Western blot (Fig. 3A, lane 2, arrow). Purified standard CNGA3-N in pRSET-B was observed on a Western blot at 20 kDa (Fig. 3B, lane 2, long arrow). CDH23 +68 and −68 in pRSET-A were observed at 30 kDa (Fig. 3B, lane 4, short arrow) and 25 kDa (Fig. 3B, lane 5, short arrow plus dot), respectively, on Western blot. All proteins were synthesized from transcripts expressed in rat organ of Corti. Standard His-tag fusion proteins of rat organ of Corti CDH23 +68 and −68 were detected with anti-Xpress (Fig. 3B) as well as with an anti-CDH23 antibody for further validation. Full-length CDH23 was detected in brain lysate at the predicted mass of 365 kDa with this antibody (Fig. 3C, arrow), and synthetic CDH23 +68 and −68 were also detected at 30 kDa (Fig. 3D, lane 2, short arrow) and 25 kDa (Fig. 3D, lane 3, short arrow plus dot), respectively, on Western blot.
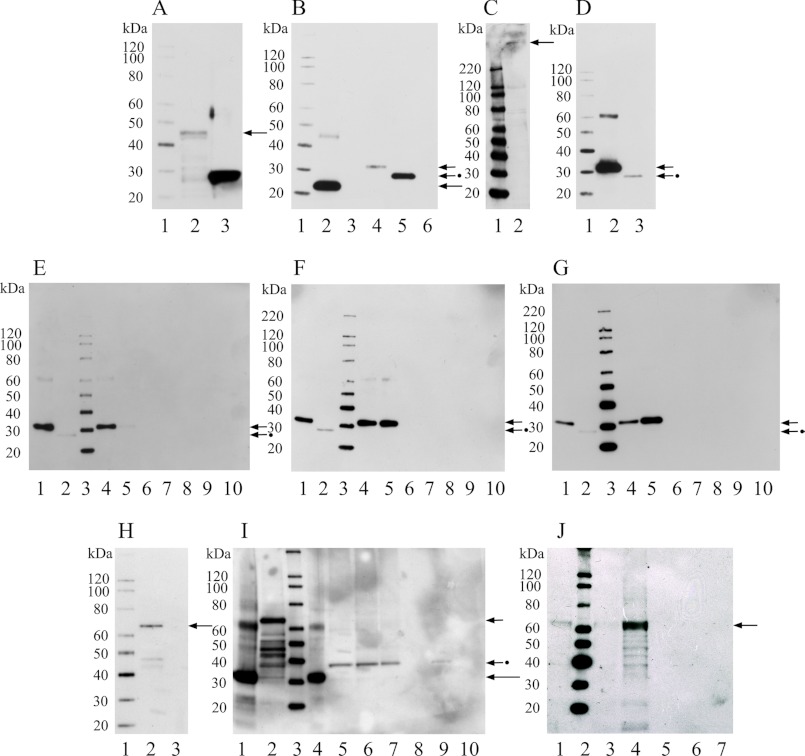
FIGURE 3.
Western blot and protein interactions of rat CNGA3-N with CDH23 +68 and CDH23 −68 by GST pulldown assays. A, shown is a Western blot of fusion protein CNGA3-N in GST vector. Lane 1, Magic XP Western protein standards; lane 2, CNGA3 amino terminus in GST vector detected with anti-GST antibody (GE) at 42 kDa (arrow); lane 3, GST vector bacterial lysate (26 kDa), as negative control detected with anti-GST antibody (GE). B, shown is a Western blot of fusion proteins CNGA3-N in pRSET-B and rat CDH23 +68/−68 in pRSET-A, detected with anti-Xpress monoclonal antibody (Invitrogen). Lane 1, Magic XP Western standards; lane 2, affinity purified CNGA3-N at ∼20 kDa, long arrow; lane 3, pRSET-B bacterial lysate as negative control; lane 4, affinity-purified CDH23 +68 (aa 3134–3353, Fig. 2D) at 30 kDa, short arrow; lane 5, CDH23 −68 (aa 3134–3317, Fig. 2D) at 25 kDa, short arrow plus dot; lane 6, pRSET-A bacterial lysate as negative control. C, shown is a Western blot for full-length CDH23 from rat brain lysate detected with an affinity-purified goat polyclonal antibody raised against a peptide in the carboxyl terminus of CDH23 of human origin, crossing to rat sequence (sc-26338, Santa Cruz; 1:500). Lane 1, Magic XP Western standards; lane 2, full-length CDH23 at 365 kDa (arrow). D, shown is a Western blot for CDH23 +68 and CDH23 −68 in pRSET-A detected with anti-CDH23 antibody (sc-26338, Santa Cruz; 1:500). Lane 1, Magic XP Western standards; lane 2, affinity-purified CDH23 +68 at 30 kDa, short arrow; lane 3, RCDH23 −68 at 25 kDa, short arrow plus dot. E, shown is a Western blot for GST pulldown assay for GST-CNGA3-N with either CDH23 +68 or CDH23 −68 pRSET fusion proteins (200 and 50 ng) detected with anti-Xpress monoclonal antibody. Lane 1, CDH23 +68 protein (30 kDa), short arrow; lane 2, CDH23 −68 protein (25 kDa) used in the pulldown assay, short arrow plus dot; lane 3, Magic XP Western standards; lanes 4 and 5 indicate pulldown for CDH23 +68 (∼200 ng and ∼50 ng, respectively) with GST-CNGA3-N, short arrow; lanes 6 and 7 indicate no pulldown for CDH23 −68 (∼200 and ∼50 ng, respectively) with GST-CNGA3-N; lane 8, a negative control, indicates no pulldown for CDH23 +68 with GST-CNGA3-N but no beads; lanes 9 and 10 are additional negative controls (lane 9, GST lysate without CNGA3-N plus CDH23 +68 plus glutathione beads; lane 10, CDH23 +68 and glutathione beads), all detected with anti-Xpress monoclonal antibody. F, shown is a Western blot for a GST pulldown assay for GST-CNGA3-N with CDH23 +68 and −68 pRSET fusion proteins, similar to E except for the CDH23 protein concentrations used (100 and 150 ng, respectively). G, shown is a Western blot for a GST pulldown assay: GST-CNGA3-N with CDH23 +68 and CDH23 −68 fusion peptides detected with anti-CDH23 antibody (Santa Cruz, 1: 500). Lane 1, CDH23 +68, short arrow; lane 2, CDH23 −68 used in the pulldown assay, short arrow plus dot; lane 3, Magic XP Western standards; lanes 4 and 5 indicate pulldown for CDH23 +68 (∼50 and ∼200 ng, respectively) with GST-CNGA3-N, short arrow, whereas lanes 6 and 7 indicate no pulldown for CDH23 −68 (∼50 and ∼200 ng, respectively) with GST-CNGA3-N; lane 8 indicates no pulldown for CDH23 +68 with GST-CNGA3-N (no beads); lanes 9 and 10 are negative controls (lane 9, GST lysate without CNGA3 and CDH23 +68 and glutathione beads; lane 10, CDH23 +68 and glutathione beads). H, shown is a Western blot for myosin VIIa fusion protein. Lane 1, Magic XP Western standards; lane 2, myosin-VIIa at 70 kDa detected with anti-Xpress monoclonal antibody (Invitrogen), arrow; lane 3, pRSET-A bacterial lysate as negative control, detected with anti-Xpress monoclonal antibody (Invitrogen). I, shown is a Western blot for competitive GST pulldown assay between CDH23 +68 and myosin VIIa fusion proteins in binding to GST-CNGA3-N. Lane 1, CDH23 +68 fusion protein at 30 kDa, long arrow; lane 2, myosin VIIa lysate used in the pulldown assay, myosin VIIa at 70 kDa (short arrow); lane 3, Magic XP Western standards; lane 4, pulldown of CDH23 +68 at 30 kDa (∼300 ng) by GST-CNGA3-N in the absence of myosin VII (∼100 ng, long arrow) lysate; lanes 5 and 6 (duplicates) indicate that in the presence of myosin VIIa (∼100 ng), CDH23 +68 (∼100 ng) at 30 kDa is not pulled down. However, a protein with a higher molecular mass of ∼38 kDa appears, putatively associated with myosin VIIa lysate (compare lane 2 with lanes 5 and 6, short arrow + dot); lane 7, myosin VIIa lysate protein in pRSET vector and GST-CNGA3-N. A protein with higher molecular mass potentially deriving from myosin VIIa (∼100 ng) lysate, compare lanes 2 and 7, interacts with GST-CNGA3-N fusion protein on glutathione beads. Lane 8, negative control consisting of GST-CNGA3-N with CDH23 +68 and myosin VIIa, where no beads were added; lanes 9 and 10 are negative controls: lane 9 indicates results for GST lysate (no CNGA3), glutathione beads, myosin VIIa and CDH23 +68; lane 10 illustrates results for glutathione beads, myosin VIIa, and CDH23 +68. J, shown is a Western blot for pulldown assay of GST-CNGA3-N with myosin VIIa pRSET fusion protein detected with anti-Xpress antibody. Lane 1, myosin VIIa standard (70 kDa), arrow; lane 2, Magic XP western standards; lanes 3 and 4 indicate pulldown for myosin VIIa (∼50 and ∼150 ng, respectively, arrow) with GST-CNGA3-N, arrow; lane 5 indicates no pulldown for myosin VIIa (∼150 ng) with GST-CNGA3-N (no beads); lanes 6 and 7 are negative controls. Lane 6 indicates results for GST lysate (no CNGA3) plus glutathione beads plus myosin VIIa at ∼150 ng; lane 7 shows results for glutathione beads plus myosin VIIa at ∼ 150 ng.
Pulldown assays showed that CNGA3 binds CDH23 +68 detected with anti-Xpress (Fig. 3E, lane 4, compared with standard CDH23 +68 in lane 1, short arrow; Fig. 3F, lanes 4 and 5 compared with standard in lane 1, short arrow). Similar results were obtained with the CDH23-specific antibody (Fig. 3G, lanes 4 and 5, compared with standard in lane 1, short arrow). There was no band in the pulldown assays corresponding to CDH23 −68 (Fig. 3, E--G, short arrow plus dot). In these experiments similar amounts of the CDH23 +68 and CDH23 −68 proteins were used to allow for a quantitative comparison of binding for +68 (Fig. 3E, lanes 4 and 5) versus no binding of −68 (Fig. 3E, lanes 6 and 7, respectively). Therefore, we conclude that GST-CNGA3-N interacted with CDH23 +68 and not CDH23 −68, which again suggests that amino acids encoded by exon 68 affect the interaction of CNGA3-N with CDH23. These results confirmed yeast two-hybrid mating and co-transformation findings. Negative controls included rat CDH23 +68 incubated with either GST bacterial lysate (not including CNGA3-N) or with Sepharose beads alone (Fig. 3, E and F, lanes 8–10) under conditions similar to those for the experimental samples.
The tip-link protein CDH23 is also known to bind myosin VIIa, which itself is required for the adaptation phase of hair cell mechanotransduction (8). This protein-protein interaction between CDH23 and myosin VIIa is also affected by the presence or absence of exon 68 in CDH23. Therefore, we wondered whether the carboxyl terminus of myosin VIIa, which binds CDH23, would disrupt the interaction between CDH23 +68 and CNGA3.
The carboxyl tail of myosin VIIa in pRSET-A, at 70 kDa on Western blots (Fig. 3H, lane 2, arrow), in competition with CDH23 +68 in GST pulldown assays, did prevent binding between CDH23 +68 and CNGA3. CDH23 +68 at 30 kDa was absent in pulldown assays with CNGA3 when myosin VIIa was included (Fig. 3I, lanes 5 and 6) with no band at the position of CDH23 +68 (demarked by the long arrow in Fig. 3I, lane 4, for CDH23 +68 interaction with CNGA3-N). Curiously, a band with a larger molecular mass (∼38 kDa) than that of CDH23 +68 (30 kDa) did appear in pulldown assays including both CDH23 +68 and myosin VIIa (Fig. 3I, lanes 5 and 6, short arrow with dot). This band was similar in mass to a protein in the myosin VIIa bacterial lysate (Fig. 3I, lane 2). Furthermore, a protein of very similar mass also appeared in negative controls with only myosin VIIa bacterial lysate and GST-CNGA3 attached to beads (Fig. 3I, lane 7), suggesting the possibility that a protein related to the carboxyl terminus of myosin VIIa may bind to the amino terminus of CNGA3. Confirmation of direct binding of myosin VIIa to the amino terminus of CNGA3 was obtained in multiple pulldown assays with GST-CNGA3-N as illustrated in Fig. 3J, lane 4 (myosin VIIa tail at 70 kDa, arrow). Overall, these results were consistent with the formation of alternate complexes of CNGA3 either with CDH23 +68, yielding a first protein complex, or with myosin VIIa, forming a second protein complex with both interactions for CNGA3 involving proteins with roles in hair cell mechanotransduction.
SPR Analysis of CNGA3-N and Cadherin 23 Interaction
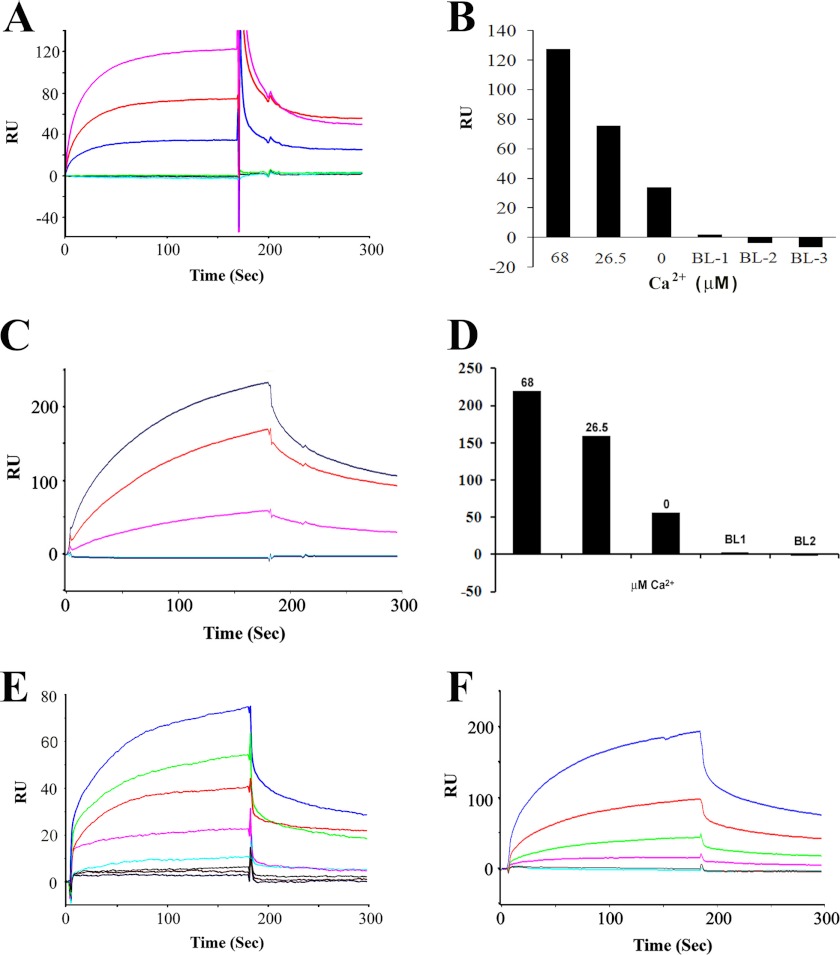
Interaction of the rat CNGA3-N with CDH23 +/−68 was also examined with SPR, a biochemical protocol capable of yielding kinetic constants for protein-protein interactions, and unequivocally, the dependence of the protein-protein interaction on Ca2+. Affinity-purified his-tag fusion proteins of the CDH23 +68 and −68 were utilized as analytes (aa 3076–3353 and 3076–3317, respectively, with amino acid numbering according to GenBankTM 446096.1), and affinity-purified CNGA3-N protein was immobilized on a CM5 sensor chip as ligand (11). Purified CDH23 +68 and −68 at 35 and 31 kDa on Western blot (Fig. 4A, lanes 2 and 3, respectively) were diluted to 100 nm in HBS-N SPR buffer (11). The CNGA3 amino terminus distinctly bound to CDH23 +68 in SPR analyses (Fig. 4, B, blue trace and C, magenta trace). There was no binding to CDH23 −68 (Fig. 4B, 26.5 μm Ca2+, magenta trace) or little binding (Fig. 4C, 68 μm Ca2+, red trace) consistent with results from yeast two-hybrid and pulldown assays. No binding was observed when HBS-N buffer alone was used as a negative control (Fig. 4B, red trace and C, purple trace).
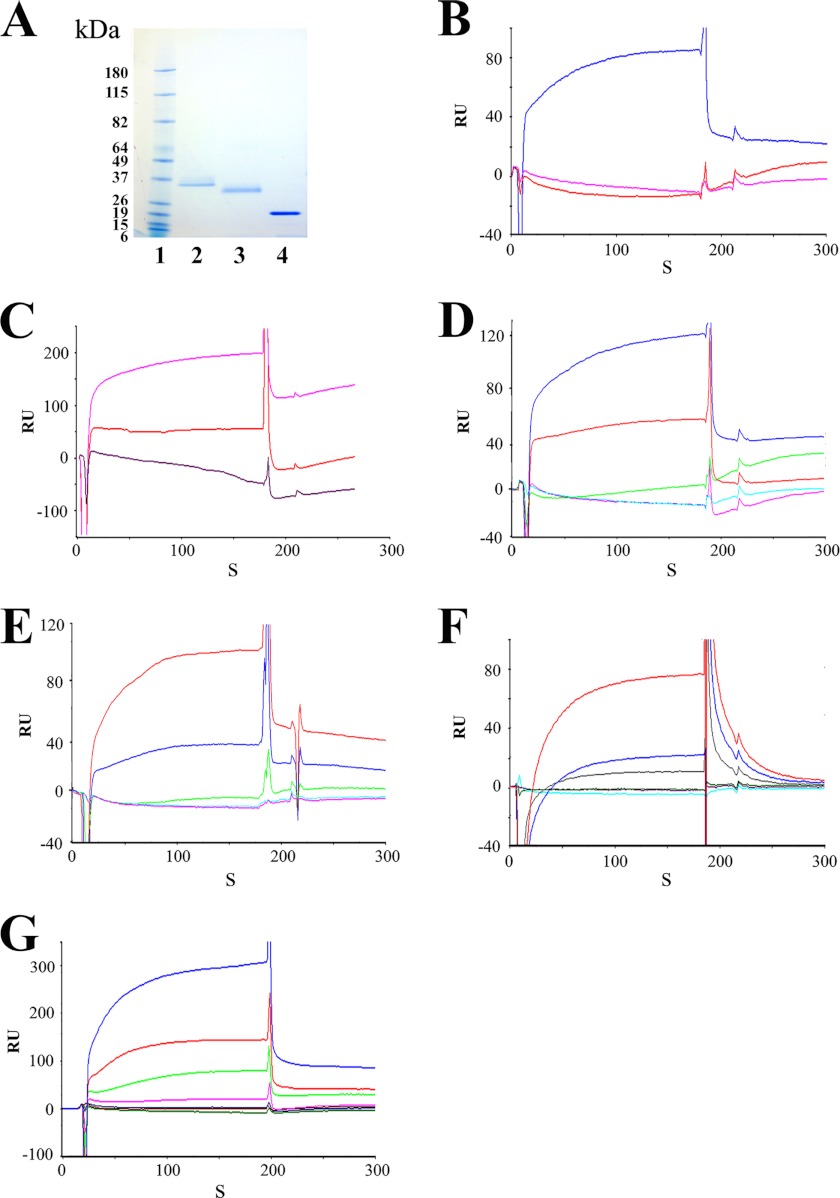
FIGURE 4.
CNGA3 channel and CDH23 interaction by SPR analysis. A, shown are affinity-purified fusion proteins used for SPR and detected with Coomassie Blue. Lane 1, prestained protein standards (Bench-Mark, Invitrogen); lane 2, CDH23 +68 (aa 3076–3353, NP_446096.1) at 35 kDa (30.9 kDa peptide + 4 kDa pRSET-A); lane 3, CDH23 −68 (aa 3076–3317) at 31 kDa (27.0 kDa peptide + 4 kDa pRSET-A, GenBankTM NP_446096.1 according to Boëda et al. (33); lane 4, CNGA3-N at 20 kDa. B, shown is SPR analysis of CNGA3 binding to CDH23 +68. Association and dissociation phases can be discerned. Affinity-purified CNGA3-N histidine-tagged fusion peptide, immobilized on the CM5 sensor chip, served as ligand, with CDH23 +68 or its splice variant CDH23 −68 as the analyte. The response was recorded as the RU for the ligand minus the RU for the reference. The SPR sensorgram shows maximum binding of ∼85 RU for 100 nm CDH23 +68 in 26.5 μm Ca2+ (blue). There was no binding when the 100 nm CDH23 −68 in 26.5 μm Ca2+ (magenta) was analyzed under the same conditions. HBS-N buffer served as the negative control (red). C, shown are CNGA3-N and CDH23 +68 interactions at 68 μm Ca2+. Binding was increased for CDH23 +68 as analyte (100 nm) with elevation of Ca2+ to 68 μm Ca2+ (magenta), whereas little/no binding was observed for CDH23 −68 (100 nm) as analyte (red). HBS-N buffer run under the same conditions served as the negative control (purple). D, Ca2+ dependence of CNGA3-N interaction with CDH23 +68 is shown. With CNGA3-N as ligand, CDH23 +68 (100 nm) dissolved in HBS-N buffer served as the analyte. Response units are indicated for binding with CDH23 +68 at 68 μm Ca2+ (blue), 26.5 μm Ca2+ (red), and 1 mm EGTA (green) compared with the response for HBS-N plus 1 mm EGTA (cyan) or HBS-N buffer alone (magenta). E, Ca2+ dependence of CDH23 +68 interactions with CNGA3-N and reversal of ligand and analyte is shown. With CDH23 +68 as ligand, 100 nm of CNGA3-N dissolved in HBS-N buffer served as the analyte. Response units are indicated for binding with CNGA3-N at 68 μm Ca2+ (red), 26.5 μm Ca2+ (blue), and 1 mm EGTA (green) compared with the response for HBS-N + 1 mm EGTA (cyan) or HBS-N buffer alone (magenta). F, Ca2+ dependence of CNGA3-N interaction with shorter fragment of CDH23 +68 (aa 3134–3353, with numbering according to GenBankTM NP_446096.1; see Fig. 2D). With CNGA3-N as ligand, CDH23 +68 (100 nm) dissolved in HBS-N buffer served as analyte. RU are indicated for binding of CNGA3 with CDH23 +68 at 68 μm Ca2+ (red), 26.5 μm Ca2+ (blue), and 1 mm EGTA (gray). Blank buffer with 10 μg/ml bovine serum albumin, run under the same conditions, served as a negative control (green) to be compared with the response for HBS-N + 1 mm EGTA (magenta) or HBS-N buffer alone (cyan). G, CNGA3-N and CDH23 +68 interaction kinetics are shown. Purified CNGA3-N was immobilized on a CM5 sensor chip, and CDH23 +68 (aa 3076–3353, NP_446096.1) was diluted in a series of concentrations with 68 μm Ca2+ in HBS-N buffer as analyte. Response units are indicated for 0 nm (green), 10 nm (red), 20 nm (blue), 40 nm (magenta), 80 nm (light green), 160 nm (light red), and 320 nm (light blue). See Table 1 for kinetic constants.
The binding of the purified CNGA3-N fusion protein as ligand to CDH23 +68 as analyte was Ca2+-dependent (Fig. 4D, 68 μm Ca2+, blue trace; 26.5 μm Ca2+, red trace). No binding was detected for 100 nm CDH23 +68 in HBS-N buffer with 1 mm EGTA (Fig. 4D, green trace). A dramatic increase in binding was observed when the CDH23 +68 was analyzed in HBS-N buffer containing 26.5 μm Ca2+ (Fig. 4D, red trace), with a further significant increase in binding at 68 μm Ca2+ (Fig. 4D, blue trace). The results were normalized by subtracting the SPR response (RU) for buffer alone and averaged from a single experiment performed in triplicate. All of the experiments were performed multiple times, and samples for different Ca2+ concentrations were analyzed randomly. There was no binding to the reference cell surface, which was blocked by ethanolamine. Thus, the present data indicate that CNGA3-N directly interacts with CDH23 +68 but only in the presence of a finite amount of Ca2+ (26.5 to 68 μm Ca2+). There was no evidence of interaction of CNGA3-N to CDH23 +68 in the absence of Ca2+ (1 mm EGTA). In a separate experiment we obtained similar results with SPR when ligand and analyte were reversed for further proof of protein-protein interaction (CDH23 +68 as ligand and CNGA3-N as analyte at 100 nm); binding again occurred between CNGA3-N and CDH23 +68 at 26.5 μm Ca2+ (Fig. 4E, blue trace), which was further enhanced at 68 μm Ca2+ (Fig. 4E, red trace). Response unit plots for binding of CNGA3-N to 0–320 nm CDH23 +68 as analyte at 68 μm Ca2+ (Fig. 4G) indicated a KD = 3.6 × 10−7 m (Table 1).
TABLE 1.
Kinetic constants for CNGA3 interaction
Rate constants for association and dissociation were obtained from original surface plasmon resonance plots by the use of BIAevaluation software. Equilibrium binding constants were calculated from the ratio of means of the rate constants. CNGA3-N, carboxyl terminus of CDH23 +68 and myosin VIIa tail his-tag fusion proteins were used in this study. Ca2+ concentration was 68 μm for each reaction. Kinetic constants were determined with CNGA3-N as ligand and CDH23 exon +68 as analyte, CNGA3-N as ligand and myosin VIIa as analyte, and CDH23 exon +68 as ligand and myosin VIIa as analyte.
| Binding partner | ka | kd | KD |
|---|---|---|---|
| m-1s−1 | s−1 | m | |
| CNGA3-N + CDH23 +68 | 2.6 × 104 | 9.2 × 10−3 | 3.6 × 10−7 |
| CNGA3-N + myosin VIIa | 3.2 × 105 | 7.3 × 10−3 | 2.3 × 10−8 |
| CDH23 +68 + myosin VIIa | 2.0 × 103 | 8.6 × 10−4 | 4.3 × 10−7 |
SPR conducted with shorter fragments of CDH23 +68 (amino acids 3134–3353, Fig. 2D with numbering according to GenBankTM 446096.1) yielded results for Ca2+-dependent binding of CNGA3-N to CDH23 +68 (Fig. 4F) similar to those obtained for the CDH23 +68 peptide which extended farther in the 5′ direction from the carboxyl terminus (i.e. Fig. 4E).
SPR Analysis of CNGA3-N and CDH23 +68 Interactions with the Myosin VIIa Carboxyl Tail
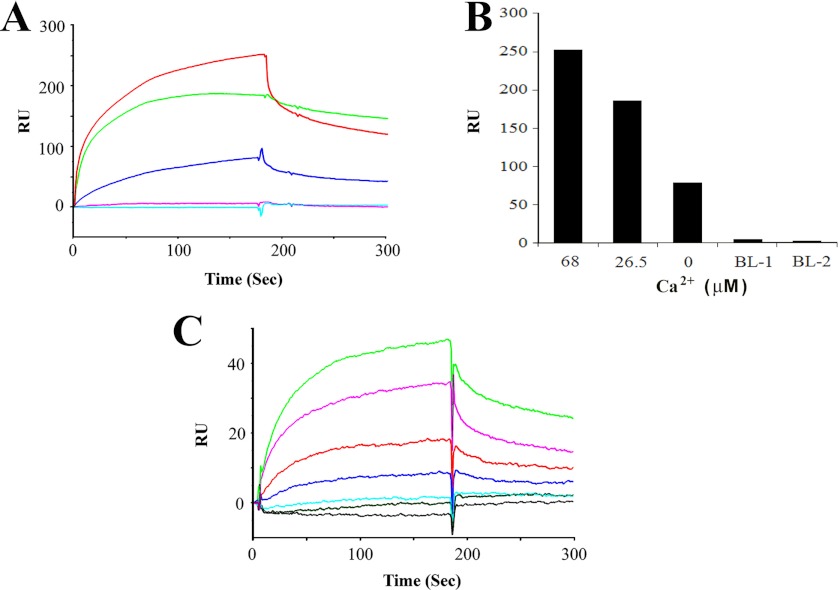
Confirming results of GST pulldown assays, the amino terminus of CNGA3 (CNGA3-N, ligand) was found by SPR to bind to the carboxyl terminus of myosin VIIa (100 nm, analyte). This interaction can occur in the absence of Ca2+ (Fig. 5A, blue trace) with increases in levels of binding observed at 26.5 μm Ca2+and further at 68 μm Ca2+ (Fig. 5A, red trace and magenta trace, respectively, indicating Ca2+ dependence Fig. 5B). Again, further proof of function was achieved by reversing ligand and analyte, demonstrating Ca2+-dependent interaction between CNGA3-N and myosin VIIa (Fig. 5, C and D).
FIGURE 5.
SPR analysis of interactions of CNGA3 with myosin VIIa. A, a SPR sensorgram shows Ca2+-dependent interaction between CNGA3-N (ligand) and myosin VIIa (100 nm, analyte) with maximum response of 130 RU at 68 μm Ca2+ (magenta) compared with 26.5 μm Ca2+ (red). A less but finite amount of binding was observed with 1 mm EGTA (blue). Negative controls included HBS-N + 1 mm EGTA (green), 10 μg/ml bovine serum albumin (gray), and HBS-N buffer alone (cyan). B, shown is a bar graph representation of results for A and in descending order. C, a SPR sensorgram shows a Ca2+-dependent interaction between myosin VIIa (ligand) and CNGA3-N (125 nm, analyte) with maximum response of 217 at 68 μm Ca2+ (blue) compared with that for 26.5 μm Ca2+ (red). Again, there was a smaller but finite response for 1 mm EGTA (magenta). Negative controls included HBS-N + 1 mm EGTA (cyan), HBS-N buffer alone (blue), overlapping in the bottom trace. D, shown is a bar graph representation of results for C in descending order. E, shown is quantitation of CNGA3-N (ligand) and myosin VIIa (analyte) interaction by SPR. Purified myosin VIIa was immobilized on a CM5 sensor chip, and CNGA3-N was diluted in a series of concentrations at 68 μm Ca2+: 100 nm (blue), 50 nm (green), 25 nm (red), 12.5 nm (magenta), 6.25 nm (cyan), 3.0 nm (gray), 1.5 nm (brown), and HBS-N buffer (black). Kinetic constants are summarized in Table 1. F, shown is quantitation of myosin VIIa (ligand) and CNGA3-N (analyte) interaction by SPR. Purified myosin VIIa was immobilized on a CM5 sensor chip, and CNGA3-N was diluted in a series of concentrations at 68 μm Ca2+: 320 nm (blue), 160 nm (red), 80 nm (green), 40 nm (magenta), 20 nm (red), 10 nm (cyan).
The interaction between CDH23 +68 (shorter fragment, amino acids 3134–3354, illustrated in Fig. 2D) and myosin VIIa, first demonstrated by Bahloul et al. (8), was confirmed and found also to be dependent on Ca2+ (Fig. 6, A and B). Similar to the interaction between CNGA3-N and myosin VIIa, there is finite binding for CDH23 +68 and myosin VIIa in the total absence of Ca2+ (1 mm EGTA) with increases at 26.5 μm Ca2+ (Fig. 6A, green trace) and further increases at 68 μm Ca2+ (Fig. 6A, red trace). No binding was detected in negative controls with HBS-N and 1 mm EGTA buffer negative controls (Fig. 6A, cyan and magenta traces, respectively). The results were normalized by subtracting the SPR response (RU) for buffer alone and averaged from a single experiment performed in triplicate.
FIGURE 6.
SPR analysis of interactions of CDH23 +68 with myosin VIIa. A, shown is CDH23 +68 interaction with myosin VIIa: Ca2+ dependence. CDH23 +68 (aa 3134–3353, Fig. 2D) was immobilized as ligand and myosin VIIa fusion peptide (300 nm) served as analyte. The sensorgram shows maximum binding of ∼255 RU at 68 μm Ca2+ (red) decreasing to 180 RU at 26.5 μm Ca2+ (green). Binding was reduced but finite with 1 mm EGTA (blue). HBS-N buffer and 1 mm EGTA (magenta) and HBS-N alone (cyan) served as negative controls. B, shown is a bar graph for results in A. C, shown is quantitation of CDH23 +68 and myosin VIIa interaction by SPR. Binding (68 μm Ca2+) was determined for CDH23 +68 (aa 3134–3353, Fig. 2D) as ligand to myosin VIIa as analyte at 100 nm (green), 50 nm (magenta), 25 nm (red), 12.5 nm (blue), 6.25 nm (cyan), 3.0 nm (gray-green), and HBS-N buffer (gray). See Table 1 for kinetic constants.
Kinetic analysis for CNGA3-N as the ligand interacting with myosin VIIa as analyte (1.5–100 nm, 68 μm Ca2+, Fig. 5E) yielded a KD = 2.3 × 10−8 (Table 1) or, for the reversal of ligand and analyte, a KD = 1.2 × 10−8 (calculated from results displayed in Fig. 5F) compared with the interaction of CDH23 +68 (shorter fragment, ligand) and myosin VIIa as analyte (3–100 nm, 68 μm Ca2+, Fig. 6C), with a KD of 4.3 × 10−7. It may be noted that with CNGA3-N as ligand, the KD for binding to myosin VIIa as analyte was a factor of 10× smaller (10−8), i.e. higher affinity, than for binding of CNGA3-N as ligand to CDH23 +68 as analyte (10−7) (Table 1), potentially influencing relative formation of alternate complexes with myosin VIIa replacing CDH23 +68 in interactions with CNGA3 dependent on Ca2+.
DISCUSSION
CNG channels mediate sensory transduction in olfaction, vision, and gustation, CNGA2 and CNGA4 in primary olfactory sensory neurons, CNGA1 and CNGA3 in rod and cone photoreceptors, respectively, and CNGA3 in chemosensory receptors. In addition, CNGA3 also mediates sensory transduction in a subpopulation of primary olfactory sensory neurons. The identity of the ion channel(s) responsible for sensory transduction in cochlear/vestibular receptor hair cells is unknown. However, transcript and protein for cAMP-gated olfactory forms of CNG, namely CNGA2 and CNGA4, have been detected in mammalian cochlear hair cells (13), and CNGA3 transcript was detected in an organ of Corti cochlear subdissected fraction (14).
Expression of CNGA3 transcript in teleost saccular hair cells has been documented (3). The molecular forms of three full-length amino acid sequences for CNGA3 expressed in teleost saccular hair cells (3), a model hair cell preparation free of supporting cells, show identity to CNGA3 expressed in human cone photoreceptors (15), trout pineal photoreceptors (AF393839), and human and rat primary olfactory sensory neurons (GenBankTM accession numbers NP_001073347 and EDL99244, respectively) (16), in contrast to the form of CNGA3 used in sensory transduction in rat gustation (NM_053495) (3). This assignment of the hair cell CNGA3 as an olfactory/cone photoreceptor CNGA3 subtype is primarily based upon amino terminus sequence obtained from a trout saccular hair cell preparation with its large numbers of hair cells as the only intact cell type. In the present investigation, it is the amino terminus of CNGA3, expressed in the rat organ of Corti and in rat cochlear outer and inner hair cells and corresponding to the mammalian olfactory/cone photoreceptor CNGA3 with high identity to trout saccular hair cell CNGA3, that was found as bait to bind to tip-link protein CDH23 +68 as prey in yeast two-hybrid mating protocols. Given that CDH23 +68 is specific to cochlear hair cells, the protein-protein interaction between CDH23 +68 and CNGA3 speaks to hair cell-specific function.
CNGA3 in Hair Cell Stereocilia and EMILIN1
Not only was there CNGA3 transcript in teleost saccular hair cells, but CNGA3 protein was immunolocalized to stereocilia and the subcuticular plate region in both teleost vestibular hair cells and mammalian auditory hair cells, predicting a role for this ion channel in stereocilia across hair cell types (3). The carboxyl terminus of CNGA3 was found to bind elastin microfibril interface-located protein (EMILIN1) in both teleost and mammalian hair cell models, pointing to conservation of molecular function across evolution. Although EMILIN1 is considered to be an extracellular matrix protein interacting with β1-integrin, we demonstrated that the carboxyl terminus of mammalian EMILIN1, preceded by a predicted transmembrane region, is likely to be intracellular. It was this carboxyl sequence that was specifically responsible for the binding of EMILIN1 to CNGA3, which points to intracellular binding between EMILIN1 and CNGA3 (3).
Transcript for phosphodiesterase 6C, the phosphodiesterase regulating intracellular levels of cGMP gating cone photoreceptor CNGA3, was identified in teleost saccular hair cells (3), consistent with a cone photoreceptor-like cGMP biochemical pathway in saccular hair cells, possibly correlating with the similarity in molecular form of CNGA3 that is expressed by these two sensory cells. EMILIN1 and phosphodiesterase 6C proteins were also immunolocalized to stereocilia of mammalian cochlear hair cells along with CNGA3, potentially reflecting a concentration of proteins required for CNGA3 action.
CNGA3 and Cadherin 23
In addition to interacting via its carboxyl terminus with EMILIN1, CNGA3 has now been found to bind via its amino terminus to CDH23 and in particular to the form of CDH23 containing the alternatively spliced exon 68 (CDH23 +68). The amino terminus of rat organ of Corti CNGA3 as bait in yeast two-hybrid mating protocols, applied to a rat organ of Corti prey cDNA library, interacted with prey protein CDH23 +68. These results were confirmed with yeast two-hybrid co-transformation protocols utilizing appropriate negative controls with empty vectors and the interchange of bait and prey. Pulldown assays and SPR analysis also confirmed the specificity of the interaction and its dependence on intracellular Ca2+. The amino terminus sequences of CNGA3 in human, trout hair cell, and rat organ of Corti contain calcium binding domains (e.g. DRDLDQ, aa 23–28 for rat organ of Corti sequence), representing partial EF-hand calcium binding motifs (17) present in calcium-binding proteins Cab45, EHD4, and CDH23 (Q91ZS3, NP_747540, EDL93044, respectively). The Ca2+ dependence of the CNGA3 interaction with CDH23 +68 may in part derive from this motif.
CDH23 is a transmembrane protein constituting the upper 2/3 of the hair cell stereociliary tip links, structures linking the stereocilia in the stereociliary array to the transducing mechanical stimulus. The tip links control gating of the mechanotransduction (MET) channels of receptor hair cells in the sensory epithelia of the inner ear (4, 18–23). The upper part of the filamentous tip link is formed by cadherin 23 homodimers and the lower part by protocadherin 15 homodimers. The two cadherins interact at their amino termini to form the tip link filament (24–26). CDH23 is thought to be inserted via its cytoplasmic carboxyl terminus at a lateral position on the taller stereocilia, whereas protocadherin 15 inserts, also via its carboxyl cytoplasmic terminus, into the top of the shorter stereocilia (24, 27) for adjacent stereocilia, which are arranged in ascending rows in the stereociliary array of each hair cell (see Fig. 7).
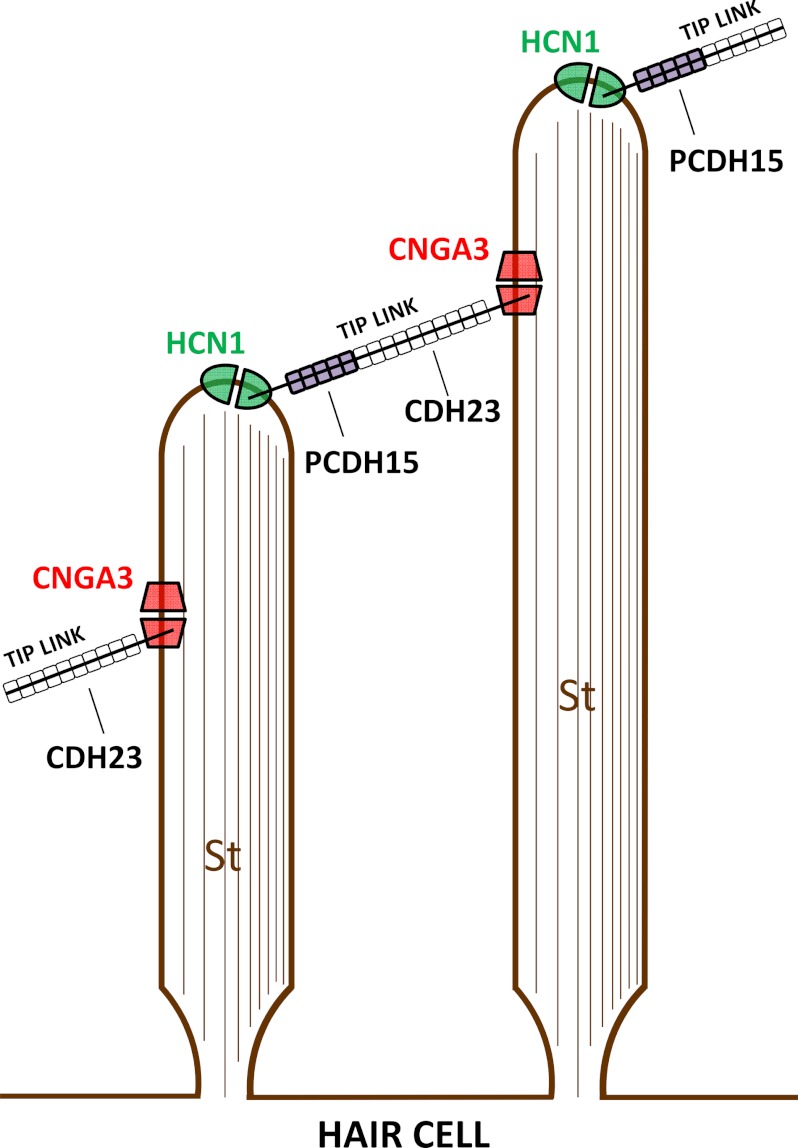
FIGURE 7.
Diagrammatic representation of one proposal for localization of protein-protein interactions of CNGA3 and HCN1 with tip-link proteins CDH23 and PCDH15, respectively. CDH23, constituting the upper 2/3 of the tip link, is inserted via its cytoplasmic carboxyl terminus at a lateral position on the taller stereocilium (St) in a stereociliary array (24–26). PCDH15, constituting the lower 1/3 of the tip link, is inserted via its cytoplasmic carboxyl terminus at the top of the shorter stereocilium (24). CNGA3 and HCN1, representing stereociliary proteins (3, 10, 12), interact with CDH23 +68 and PCDH15 CD3, respectively. In one configuration, as a consequence of specific Ca2+-dependent molecular interactions, HCN1 is localized to the top of the shorter stereocilium and CNGA3 to a lateral position on the next taller stereocilium.
CDH23 and protocadherin 15 are members of the cadherin superfamily, and both are protein targets in Usher syndrome Type I (USH1) loci (28, 29). Mutations in CDH23 are the specific cause of Usher syndrome deafness USH1D (4) and are also implicated in non-syndromic deafness DFNB12 (30–32). There are two alternatively spliced CDH23 isoforms differing with respect to the inclusion of exon 68, encoding part of the intracellular CDH23 domain (4, 6, 7). Messenger RNAs have been detected for both CDH23 +68 and CDH23 −68 in the inner ears of adult mice (4). CDH23 +68 is considered to be the hair cell-stereocilia-specific splice variant (4) and is not detected in retina (7).
Cadherin 23 Protein-Protein Interactions and Effect of the Presence or Absence of the +68 Exon
It has previously been shown that CDH23 interacts with the PDZ domain-containing proteins harmonin (7, 8, 33), MAGI-1 (34), PIST (35), and the non-PDZ domain-containing proteins protocadherin 15 (PCDH15) (24), myosin-VIIa (8), myosin-1c (4), and EHD4 (Eps15-homology domain-containing protein 4) (36). It was the EH calcium binding domain of EHD4 that interacted with CDH23. These CDH23 binding partners were identified in yeast two-hybrid protocols and verified in pulldown assays and/or SPR analysis using in vitro expression systems and brain as a source of protein. Immunofluorescence co-localizations of proteins were established in in vitro expression systems in some instances (24, 33, 35, 37). The individual proteins have been immunolocalized to positions on hair cell stereocilia relative to phalloidin-labeled actin, a stereocilia marker protein. Immunofluorescence co-localization of myosin VIIa, cadherin 23, and sans (a myosin VIIa-interacting protein) in hair cell stereocilia has been observed (38).
The interactions of the two PDZ domain-containing proteins harmonin and MAGI-1 with CDH23 are complex and speak to the issue of the +/−68 exon. Two PDZ domains in harmonin interact with two complementary binding surfaces in the CDH23 cytoplasmic domain, one internal to the cytoplasmic domain sequence and one at the carboxyl terminus. The internal site can be disrupted by sequences encoded by the alternatively spliced CDH23 +68 with the outcome of 10 times more binding for the −68 than for the +68 versions of CDH23 (7). The second and weaker binding site is at the carboxyl terminus. MAGI-1, a member of the membrane-associated guanylate kinase proteins, also utilizes the carboxyl-terminal PDZ binding domain of CDH23 +68 for its PDZ4 domain (34) and has been proposed to replace harmonin from CDH23 via its stronger binding. MAGI-1 is known to bind F-actin-binding proteins and has been suggested to play a role in actin cytoskeleton dynamics within polarized epithelial cells (39). The significance of this carboxyl-terminal PDZ binding domain was further pointed out by the action of PIST, a Golgi-associated PDZ protein that also binds this domain, thus retaining CDH23 in the Golgi. PIST is released from this carboxyl terminus site on CDH23 +68 by either MAGI-1 or harmonin (35), presumably reflecting relative binding affinities and possibly the existence of alternate protein complexes.
Curiously, the CNGA3 amino terminus appears to incorporate not only the possibility for Ca2+-dependent binding via a Ca2+-sensitive motif but also includes a PDZ domain NGLG (aa 31–34), corresponding to a polypeptide site on piccolo (EDL99465) recognized as having identity to the GLGF loop, a PDZ-carboxylate binding motif (CDD: 29049, see Ref. 40).
CNGA3, CDH23, and Myosin VIIa
CDH23 +68 also interacts with myosin VIIa, another targeted protein in Usher 1 syndrome, with mutations of myosin VIIa giving rise to USH1B. Myosin VIIa has been implicated as having a role in adaptation of hair cell (MET) (8, 41) and is localized to the upper tip-link density (38). Interestingly, a ternary complex can be formed between CDH23, harmonin, and myosin VIIa thought to be involved in “applying tension forces on hair bundle links” (8), affecting mechanotransduction. In this case the binding observed between CDH23 +68 and harmonin was high affinity, involving the harmonin amino terminus-PDZ1 supramodule and not requiring the CDH23 carboxyl terminus PDZ binding domain (8). The affinities between myosin VIIa and either CDH23 or harmonin A were a factor of 10 lower than that for CDH23 and harmonin A.
In our experiments the question that immediately arose was what effect alternative binding of myosin VIIa and CDH23 +68 (cf. Bahloul et al. (8)) would have on the interaction between CNGA3 and CDH23 +68. Pulldown assays with myosin VIIa and CDH23 +68 as competitors in binding to GST-CNGA3 indicated that the myosin VIIa tail (70 kDa) did in fact interfere with binding between CNGA3 and CDH23 +68, leading to the determination that myosin VIIa directly binds CNGA3 by both pulldown and SPR, and this interaction was found to be Ca2+-dependent. Thus, binding between the three proteins, CNGA3, CDH23 +68, and myosin VIIa, is modulated by separate interactions; elimination of binding of CNGA3 to CDH 23 + 68 by myosin VIIa in pulldown assays is not simply due to the competing interaction of CDH23 +68 with myosin VIIa, which we have confirmed, but is also the result of a direct interaction between CNGA3 and myosin VIIa. Curiously, in these experiments the KD for CNGA3 and CDH23 +68 was similar in magnitude to that for binding of myosin VIIa and CDH23 +68, whereas the KD for CNGA3 and myosin VIIa was smaller, i.e. affinity higher, by a factor of 10 (Table 1), consistent with the hypothesis that myosin VIIa could substitute for CDH23 +68 in binding to CNGA3 affecting formation of alternate protein complexes with CNGA3. Given that two of these proteins are involved in mechanotransduction, it may be pointed out that distinct alternate protein complexes are presumed to underlie cellular response to mechanical stimuli (discussed in Ref. 12).
Role of CNGA3 in Hair Cells; Ca2+ Permeability
The CNG channels admit Ca2+ in sensory and non-sensory cell types, representing a major source of Ca2+ influx. All CNG channels undergo Ca2+/calmodulin adaptation; Ca2+/calmodulin acts at the amino termini of CNGA1 (rod photoreceptors) and CNGA2 (olfactory sensory neurons), effecting adaptation. For heteromeric CNG3 channels comprising α and β subunits, adaptation occurs as a consequence of Ca2+/calmodulin modulation of CNGB3 involving an interaction between amino and carboxyl termini (42). Both high permeability to Ca2+ and the possibility for adaptation are known properties for the observed hair cell mechanosensory transduction channel. Also, the CNGA3 binding partner EMILIN1, theoretically, as a member of the elastin superfamily, could provide the elastic element, currently unidentified but predicted for hair cell mechanotransduction.
If CNGA3 were to be considered as necessary for hair cell function, there might be an expectation of deafness or vestibular deficits in individuals with CNGA3/CNGB3 mutations. Mutations of CNGA3 (on chromosome 2q11.2) (43) and CNGB3 (on chromosome 8q21.3) (44, 45) are known to give rise to the molecular heterogeneous photoreceptor retinal disease of achromatopsia (with degeneration of cone photoreceptors). CNGA3 and myosin VIIa are targeted genes in Leber congenital amaurosis (46). Both of these retinal diseases include congenital nystagmus.
Hair Cell MET Channels
The ion channel composition of the hair cell MET channels is unknown, but there is evidence for two channels per stereocilium (47), with the possibility of one channel at either end of the tip link (48). The two channels would derive from one positioned at the lower end of one tip link and the second associated with the upper end of the previous tip link (see tip link insertion points in Fig. 7). Although the initial rapid onset (<10 μs) and adaptation phases of MET are specific properties that need not invoke the participation of more than one type of channel (49), with CNG channels representing putative candidates (50), the possibility of more than one channel type contributing to what is measured as MET is not obviated. Van Netten et al. (51) have proposed that pairwise coupling of two hair cell transducer channels links auditory sensitivity and dynamic range. In this model two channels with different properties, one at either end of the tip link, could explain the electrophysiological characteristics of MET. The two channels would have different onset and adaptation characteristics, possibly representing different orientations of a single type of channel in shorter and taller stereocilia, respectively. The model did not preclude the possibility of two different channel types at the separate locations. Interestingly, in another study, reassembly of tip links after dissociation with BAPTA suggested a difference in timing, with CDH23 replacements (on the taller stereocilium) occurring at a later time than those for protocadherin 15, a phenomenon associated with alteration in MET adaptation (52), again possibly consistent with differential channel expression at either end of the tip link.
We previously obtained evidence that hyperpolarization-activated HCN1 binds tip-link protein protocadherin 15 CD3 (10, 12) and protocadherin 15 has been localized to the top of the shorter stereocilia (24), a putative site for mechanotransduction (53). HCN1 specifically interacts with F-actin-binding filamin A, and filamin A has recently been implicated as a mediator of cellular response to mechanical stimulation (54). Ehrlicher et al. (54) determined that mechanical strain in actin networks directly differentially regulates protein-protein interactions of filamin A. A correlate to this finding would be the existence of alternate protein complexes, a model that we have confirmed for proteins expressed in hair cells (10, 12). Filamin A, in immunoprecipitations of full-length proteins, binds HCN1 and tip link-protein protocadherin 15 CD3, but this protein complex does not include HCN2. HCN2 interacts with HCN1 via a conserved amino terminus region utilized in HCN channel formation, and this complex does not include protocadherin 15 CD3. HCN2 also interacts with fascin-2, and this complex includes HCN1 but not protocadherin 15 CD3.
In the study by Ehrlicher et al. (54), mechanical stimulation increased the binding of filamin A to the cytoplasmic tails of β-integrins. β1-integrin is a binding partner of EMILIN1 (55), that in turn undergoes protein-protein interactions with CNGA3 (3). Therefore, a pathway for HCN1 could extend directly to CNGA3 at one tip-link site, at the top of the shorter stereocilium, the presumed site of Ca2+ entry during mechanosensory transduction. We have obtained evidence that the cGMP pathway enzyme guanylate kinase is also a protein binding partner of protocadherin 15 CD3.3 Guanylate kinase is utilized to recycle GMP produced from cGMP, which is potentially required in conjunction with a cGMP-gated ion channel. The channels formed from HCN isoforms, while having the potential to contribute to the rapid component of MET (via Iinst (instantaneous current coded for by HCN isoforms)), admit Ca2+ but are not highly Ca2+-permeable as is MET. Therefore, a protein complex at the top of the shorter stereocilia incorporating highly Ca2+-permeable CNGA3 might support the property of MET having high Ca2+-permeability.
However, in addition to the above pathway considerations, the results from the present work are consistent with a second site for CNGA3 interaction, namely, at the upper tip-link CDH23 insert (Fig. 7). CNGA3 is not voltage-dependent; heterologously expressed human CNGA3 channel is activated by cGMP, and the cGMP-elicited current is enhanced by ATP and inhibited by phosphatidylinositol (3,4,5)-triphosphate (PIP3) (56). CNGA3/CNGB3 can undergo adaptation via a Ca2+-dependent mechanism, also observed for MET. One possible version of a two-channel model would include CNGA3 interacting with CDH23 at the upper tip-link density combined with HCN1 binding to protocadherin 15 CD3 at the lower tip-link density (Fig. 7). A further role for the described protein-protein interactions of CNGA3, CDH23, and myosin VIIa may exist in protein transport as modulated by Ca2+ (57), with all three proteins localized to hair cell subcuticular plate sites.
In conclusion, cGMP-gated CNGA3 binds tip-link protein CDH23 +68 and myosin VIIa, known components of the mechanotransduction apparatus (31). Based upon the premise that hair cell stereocilia tip-link proteins are closely coupled with the mechanotransduction channel(s), we suggest the results of this investigation are consistent with the possibility that CNGA3 participates in hair-cell MET.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DC004076 (to M. J. D.) and R01 DC000156 (to D. G. D.).
N. A. Ramakrishnan and M. J. Drescher, unpublished information.
- CNG
- cyclic nucleotide-gated channel
- CNGA3
- cyclic nucleotide-gated channel α-3
- CDH23 +/−68
- cadherin 23 +/− exon 68
- EMILIN1
- elastin microfibril interface-located protein 1
- HCN1
- hyperpolarization-activated, cyclic nucleotide-gated non-selective cation channel isoform 1
- USH1
- Usher syndrome Type I
- PCDH15
- protocadherin 15
- MET
- mechanoelectric transduction
- RU
- response units
- aa
- amino acids.
REFERENCES
- 1. Zagotta W. N., Siegelbaum S. A. (1996) Structure and function of cyclic nucleotide-gated channels. Annu. Rev. Neurosci. 19, 235–263 [DOI] [PubMed] [Google Scholar]
- 2. Biel M. (2009) Cyclic nucleotide-regulated cation channels. J. Biol. Chem. 284, 9017–9021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Selvakumar D., Drescher M. J., Dowdall J. R., Khan K. M., Hatfield J. S., Ramakrishnan N. A., Drescher D. G. (2012) CNGA3 is expressed in inner ear hair cells and binds to an intracellular C terminals domain of EMILIN1. Biochem. J. 443, 463–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Siemens J., Lillo C., Dumont R. A., Reynolds A., Williams D. S., Gillespie P. G., Müller U. (2004) Cadherin 23 is a component of the tip link in hair-cell stereocilia. Nature 428, 950–955 [DOI] [PubMed] [Google Scholar]
- 5. Bolz H., von Brederlow B., Ramírez A., Bryda E. C., Kutsche K., Nothwang H. G., Seeliger M., del C-Salcedó Cabrera M., Vila M. C., Molina O. P., Gal A., Kubisch C. (2001) Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat. Genet. 27, 108–112 [DOI] [PubMed] [Google Scholar]
- 6. Di Palma F., Pellegrino R., Noben-Trauth K. (2001) Genomic structure, alternative splice forms, and normal and mutant alleles of cadherin 23 (Cdh23). Gene 281, 31–41 [DOI] [PubMed] [Google Scholar]
- 7. Siemens J., Kazmierczak P., Reynolds A., Sticker M., Littlewood-Evans A., Müller U. (2002) The Usher syndrome proteins cadherin 23 and harmonin form a complex by means of PDZ-domain interactions. Proc. Natl. Acad. Sci. U.S.A. 99, 14946–14951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bahloul A., Michel V., Hardelin J. P., Nouaille S., Hoos S., Houdusse A., England P., Petit C. (2010) Cadherin-23, myosin VIIa and harmonin, encoded by Usher syndrome type I genes, form a ternary complex and interact with membrane phospholipids. Hum. Mol. Genet. 19, 3557–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oh C. K., Drescher M. J., Hatfield J. S., Drescher D. G. (1999) Selective expression of serotonin receptor transcripts in the mammalian cochlea and its subdivisions. Brain Res. Mol. Brain Res. 70, 135–140 [DOI] [PubMed] [Google Scholar]
- 10. Ramakrishnan N. A., Drescher M. J., Barretto R. L., Beisel K. W., Hatfield J. S., Drescher D. G. (2009) Calcium-dependent binding of HCN1 channel protein to hair cell stereociliary tip-link protein protocadherin 15 CD3. J. Biol. Chem. 284, 3227–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Drescher D. G., Ramakrishnan N. A., Drescher M. J. (2009) Surface plasmon resonance (SPR) analysis of binding interactions of proteins in inner-ear sensory epithelia. Methods Mol. Biol. 493, 323–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ramakrishnan N. A., Drescher M. J., Khan K. M., Hatfield J. S., Drescher D. G. (2012) HCN1 and HCN2 proteins are expressed in cochlear hair cells. HCN1 can form a ternary complex with protocadherin 15 CD3 and F-actin binding filamin A or can interact with HCN2. J. Biol. Chem. 287, 37628–37646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Drescher M. J., Barretto R. L., Chaturvedi D., Beisel K. W., Hatfield J. S., Khan K. M., Drescher D. G. (2002) Expression of subunits for the cAMP-sensitive “olfactory” cyclic nucleotide-gated ion channel in the cochlea. Implications for signal transduction. Brain Res. Mol. Brain Res. 98, 1–14 [DOI] [PubMed] [Google Scholar]
- 14. Drescher M. J., Chaturvedi D., Drescher D. G. (2001) Cyclic nucleotide gated ion channel subunit expression in the rat cochlea. J. Assoc. Res. Otolaryngol. Abstr. 24, 40–41 [Google Scholar]
- 15. Kohl S., Marx T., Giddings I., Jägle H., Jacobson S. G., Apfelstedt-Sylla E., Zrenner E., Sharpe L. T., Wissinger B. (1998) Total colour blindness is caused by mutations in the gene encoding the α-subunit of the cone photoreceptor cGMP-gated cation channel. Nat. Genet. 19, 257–259 [DOI] [PubMed] [Google Scholar]
- 16. Meyer M. R., Angele A., Kremmer E., Kaupp U. B., Muller F. (2000) A cGMP-signaling pathway in a subset of olfactory sensory neurons. Proc. Natl. Acad. Sci. U.S.A. 97, 10595–10600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou Y., Yang W., Kirberger M., Lee H. W., Ayalasomayajula G., Yang J. J. (2006) Prediction of EF-hand calcium-binding proteins and analysis of bacterial EF-hand protein. Proteins 65, 643–655 [DOI] [PubMed] [Google Scholar]
- 18. Zhao Y., Yamoah E. N., Gillespie P. G. (1996) Regeneration of broken tip links and restoration of mechanical transduction in hair cells. Proc. Natl. Acad. Sci. U.S.A. 93, 15469–15474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hudspeth A. J. (1997) How hearing happens. Neuron 19, 947–950 [DOI] [PubMed] [Google Scholar]
- 20. Gillespie P. G., Walker R. G. (2001) Molecular basis of mechanosensory transduction. Nature 413, 194–202 [DOI] [PubMed] [Google Scholar]
- 21. Corey D. (2003) Sensory transduction in the ear. J. Cell Sci. 116, 1–3 [DOI] [PubMed] [Google Scholar]
- 22. LeMasurier M., Gillespie P. G. (2005) Hair-cell mechanotransduction and cochlear amplification. Neuron 48, 403–415 [DOI] [PubMed] [Google Scholar]
- 23. Vollrath M. A., Kwan K. Y., Corey D. P. (2007) The micromachinery of mechanotransduction in hair cells. Annu. Rev. Neurosci. 30, 339–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kazmierczak P., Sakaguchi H., Tokita J., Wilson-Kubalek E. M., Milligan R. A., Müller U., Kachar B. (2007) Cadherin 23 and protocadherin 15 interact to form tip link filaments in sensory hair cells. Nature 449, 87–91 [DOI] [PubMed] [Google Scholar]
- 25. Sotomayor M., Weihofen W. A., Gaudet R., Corey D. P. (2010) Structural determinants of cadherin-23 function in hearing and deafness. Neuron 66, 85–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alagramam K. N., Goodyear R. J., Geng R., Furness D. N., van Aken A. F., Marcotti W., Kros C. J., Richardson G. P. (2011) Mutations in protocadherin 15 and cadherin 23 affect tip links and mechanotransduction in mammalian sensory hair cells. PLoS One 6, e19183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Senften M., Schwander M., Kazmierczak P., Lillo C., Shin J. B., Hasson T., Géléoc G. S., Gillespie P. G., Williams D., Holt J. R., Müller U. (2006) Physical and functional interaction between protocadherin 15 and myosin VIIa in mechanosensory hair cells. J. Neurosci. 26, 2060–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ahmed Z. M., Riazuddin S., Bernstein S. L., Ahmed Z., Khan S., Griffith A. J., Morell R. J., Friedman T. B., Riazuddin S., Wilcox E. R. (2001) Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am. J. Hum. Genet. 69, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alagramam K. N., Yuan H., Kuehn M. H., Murcia C. L., Wayne S., Srisailpathy C. R., Lowry R. B., Knaus R., Van Laer L., Bernier F. P., Schwartz S., Lee C., Morton C. C., Mullins R. F., Ramesh A., Van Camp G., Hageman G. S., Woychik R. P., Smith R. J. (2001) Mutations in the novel protocadherin PCDH15 cause Usher syndrome type 1F. Hum. Mol. Genet. 10, 1709–1718 [DOI] [PubMed] [Google Scholar]
- 30. Bork J. M., Peters L. M., Riazuddin S., Bernstein S. L., Ahmed Z. M., Ness S. L., Polomeno R., Ramesh A., Schloss M., Srisailpathy C. R., Wayne S., Bellman S., Desmukh D., Ahmed Z., Khan S. N., Kaloustian V. M., Li X. C., Lalwani A., Riazuddin S., Bitner-Glindzicz M., Nance W. E., Liu X. Z., Wistow G., Smith R. J., Griffith A. J., Wilcox E. R., Friedman T. B., Morell R. J. (2001) Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am. J. Hum. Genet. 68, 26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schwander M., Kachar B., Müller U. (2010) Review series. The cell biology of hearing. J. Cell Biol. 190, 9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schultz J. M., Bhatti R., Madeo A. C., Turriff A., Muskett J. A., Zalewski C. K., King K. A., Ahmed Z. M., Riazuddin S., Ahmad N., Hussain Z., Qasim M., Kahn S. N., Meltzer M. R., Liu X. Z., Munisamy M., Ghosh M., Rehm H. L., Tsilou E. T., Griffith A. J., Zein W. M., Brewer C. C., Riazuddin S., Friedman T. B. (2011) Allelic hierarchy of CDH23 mutations causing non-syndromic deafness DFNB12 or Usher syndrome USH1D in compound heterozygotes. J. Med. Genet. 48, 767–775 [DOI] [PubMed] [Google Scholar]
- 33. Boëda B., El-Amraoui A., Bahloul A., Goodyear R., Daviet L., Blanchard S., Perfettini I., Fath K. R., Shorte S., Reiners J., Houdusse A., Legrain P., Wolfrum U., Richardson G., Petit C. (2002) Myosin VIIa, harmonin and cadherin 23, three Usher 1 gene products that cooperate to shape the sensory hair cell bundle. EMBO J. 21, 6689–6699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu Z., Peng A. W., Oshima K., Heller S. (2008) MAGI-1, a candidate stereociliary scaffolding protein, associates with the tip link component cadherin 23. J. Neurosci. 28, 11269–11276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu Z., Oshima K., Heller S. (2010) PIST regulates the intracellular trafficking and plasma membrane expression of cadherin 23. BMC Cell Biol. 11, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sengupta S., George M., Miller K. K., Naik K., Chou J., Cheatham M. A., Dallos P., Naramura M., Band H., Zheng J. (2009) EHD4 and CDH23 are interacting partners in cochlear hair cells. J. Biol. Chem. 284, 20121–20129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zheng L., Zheng J., Whitlon D. S., García-Añoveros J., Bartles J. R. (2010) Targeting of the hair cell proteins cadherin 23, harmonin, myosin XVa, espin, and prestin in an epithelial cell model. J. Neurosci. 30, 7187–7201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Grati M., Kachar B. (2011) Myosin VIIa and sans localization at stereocilia upper tip-link density implicates these Usher syndrome proteins in mechanotransduction. Proc. Natl. Acad. Sci. U.S.A. 108, 11476–11481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Patrie K. M., Drescher A. J., Welihinda A., Mundel P., Margolis B. (2002) Interaction of two actin-binding proteins, synaptopodin and α-actinin-4, with the tight junction protein MAGI-1. J. Biol. Chem. 277, 30183–30190 [DOI] [PubMed] [Google Scholar]
- 40. Doyle D. A., Lee A., Lewis J., Kim E., Sheng M., MacKinnon R. (1996) Crystal structures of a complexed and peptide-free membrane protein-binding domain. Molecular basis of peptide recognition by PDZ. Cell 85, 1067–1076 [DOI] [PubMed] [Google Scholar]
- 41. Kros C. J., Marcotti W., van Netten S. M., Self T. J., Libby R. T., Brown S. D., Richardson G. P., Steel K. P. (2002) Reduced climbing and increased slipping adaptation in cochlear hair cells of mice with Myo7a mutations. Nat. Neurosci. 5, 41–47 [DOI] [PubMed] [Google Scholar]
- 42. Peng C., Rich E. D., Thor C. A., Varnum M. D. (2003) Functionally important calmodulin-binding sites in both NH2- and COOH-terminal regions of the cone photoreceptor cyclic nucleotide-gated channel CNGB3 subunit. J. Biol. Chem. 278, 24617–24623 [DOI] [PubMed] [Google Scholar]
- 43. Reuter P., Koeppen K., Ladewig T., Kohl S., Baumann B., Wissinger B., Achromatopsia Clinical Study Group (2008) Mutations in CNGA3 impair trafficking or function of cone cyclic nucleotide-gated channels, resulting in Achromatopsia. Hum. Mutat. 29, 1228–1236 [DOI] [PubMed] [Google Scholar]
- 44. Nishiguchi K. M., Sandberg M. A., Gorji N., Berson E. L., Dryja T. P. (2005) Cone cGMP-gated channel mutations and clinical findings in patients with achromatopsia, macular degeneration, and other hereditary cone diseases. Hum. Mutat. 25, 248–258 [DOI] [PubMed] [Google Scholar]
- 45. Thapa A., Morris L., Xu J., Ma H., Michalakis S., Biel M., Ding X. Q. (2012) Endoplasmic reticulum stress-associated cone photoreceptor degeneration in cyclic nucleotide-gated channel deficiency. J. Biol. Chem. 287, 18018–18029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang X., Wang H., Cao M., Li Z., Chen X., Patenia C., Gore A., Abboud E. B., Al-Rajhi A. A., Lewis R. A., Lupski J. R., Mardon G., Zhang K., Muzny D., Gibbs R. A., Chen R. (2011) Whole-exome sequencing identifies ALMS1, IQCB1, CNGA3, and MYO7A mutations in patients with Leber congenital amaurosis. Hum. Mutat. 32, 1450–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sul B., Iwasa K. H. (2010) Gating of two mechanoelectrical transducer channels associated with a single tip link. Biophys J. 99, 1027–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Denk W., Holt J. R., Shepherd G. M., Corey D. P. (1995) Calcium imaging of single stereocilia in hair cells. Localization of transduction channels at both ends of tip links. Neuron 15, 1311–1321 [DOI] [PubMed] [Google Scholar]
- 49. Fettiplace R. (2009) Defining features of the hair cell mechanoelectrical transducer channel. Pflugers Arch. 458, 1115–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ricci A. J., Kachar B., Gale J., Van Netten S. M. (2006) Mechano-electrical transduction. New insights into old ideas. J. Membr. Biol. 209, 71–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van Netten S. M., Meulenberg C. J., Lennan G. W., Kros C. J. (2009) Pairwise coupling of hair cell transducer channels links auditory sensitivity and dynamic range. Pflugers Arch. 458, 273–281 [DOI] [PubMed] [Google Scholar]
- 52. Indzhykulian A., Stepanyan R., Frolenkov G. (2012) Two-step regeneration of mechanotransduction and molecular composition of the tip links in mammalian cochlear inner hair cells. Assoc. Res. Otolaryngol. Abstr. 35, 204 [Google Scholar]
- 53. Beurg M., Fettiplace R., Nam J. H., Ricci A. J. (2009) Localization of inner hair cell mechanotransducer channels using high-speed calcium imaging. Nature Neurosci. 12, 553–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ehrlicher A. J., Nakamura F., Hartwig J. H., Weitz D. A., Stossel T. P. (2011) Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature 478, 260–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Spessotto P., Cervi M., Mucignat M. T., Mungiguerra G., Sartoretto I., Doliana R., Colombatti A. (2003) β1 integrin-dependent cell adhesion to EMILIN-1 is mediated by the gC1q domain. J. Biol. Chem. 278, 6160–6167 [DOI] [PubMed] [Google Scholar]
- 56. Bright S. R., Rich E. D., Varnum M. D. (2007) Regulation of human cone cyclic nucleotide-gated channels by endogenous phospholipids and exogenously applied phosphatidylinositol 3,4,5-trisphosphate. Mol. Pharmacol. 71, 176–183 [DOI] [PubMed] [Google Scholar]
- 57. Akhmanova A., Hammer J. A., 3rd (2010) Linking molecular motors to membrane cargo. Curr. Opin. Cell Biol. 22, 479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. de Brouwer A. P., Pennings R. J., Roeters M., Van Hauwe P., Astuto L. M., Hoefsloot L. H., Huygen P. L., van den Helm B., Deutman A. F., Bork J. M., Kimberling W. J., Cremers F. P., Cremers C. W., Kremer H. (2003) Mutations in the calcium binding motifs of CDH23 and the 35delG mutation in GJB2 cause hearing loss in one family. Hum. Genet. 112, 156–163 [DOI] [PubMed] [Google Scholar]