Background: For sorting to late endosomes, caveolin-1 is ubiquitinated and requires the VCP ATPase.
Results: Mutation of the lysines in the N-terminal region prevents ubiquitination, binding to VCP, and sorting to late endosomes.
Conclusion: Endosomal trafficking of CAV1 depends on ubiquitination of the N-terminal region and the subsequent recruitment of VCP-UBXD1.
Significance: This study identifies the functional ubiquitination site in CAV1 and its significance.
Keywords: Caveolin, Endosomes, Lysosomes, Membrane Trafficking, Ubiquitin, Cdc48, Valosin-containing Protein, p97
Abstract
Caveolin-1 (CAV1) is the defining constituent of caveolae at the plasma membrane of many mammalian cells. For turnover, CAV1 is ubiquitinated and sorted to late endosomes and lysosomes. Sorting of CAV1 requires the AAA+-type ATPase VCP and its cofactor UBXD1. However, it is unclear in which region CAV1 is ubiquitinated and how ubiquitination is linked to sorting of CAV1 by VCP-UBXD1. Here, we show through site-directed mutagenesis that ubiquitination of CAV1 occurs at any of the six lysine residues, 5, 26, 30, 39, 47, and 57, that are clustered in the N-terminal region but not at lysines in the oligomerization, intramembrane, or C-terminal domains. Mutation of Lys-5–57 to arginines prevented binding of the VCP-UBXD1 complex and, importantly, strongly reduced recruitment of VCP-UBXD1 to endocytic compartments. Moreover, the Lys-5–57Arg mutation specifically interfered with trafficking of CAV1 from early to late endosomes. Conversely and consistently, depletion of VCP or UBXD1 led to accumulation of ubiquitinated CAV1, suggesting that VCP acts downstream of ubiquitination and is required for transport of the ubiquitinated form of CAV1 to late endosomes. These results define the N-terminal region of CAV1 as the critical ubiquitin conjugation site and, together with previous data, demonstrate the significance of this ubiquitination for binding to the VCP-UBXD1 complex and for sorting into lysosomes.
Introduction
Caveolae are invaginations of the plasma membrane that define cholesterol-rich microdomains that are implicated in many physiological processes, including endo- and transcytosis, pathogen entry, lipid regulation, signaling, and cancer (1–3). They can bud from the plasma membrane and dock onto endosomes for cargo delivery (4). Structurally, they are characterized by a proteinaceous coat that, in contrast to the clathrin coat, stays tightly associated with the membrane (5). The major constituents of this coat are caveolin proteins that in humans are represented by caveolin-1 (CAV1), caveolin-2, as well as the muscle-specific caveolin-3 (2, 6). Caveolins are integral membrane proteins with a central hydrophobic hairpin domain inserted inside the membrane and both N and C termini facing the cytosol (7) (Fig. 1A). The C-terminal cytosolic domain is palmitoylated. The N-terminal cytosolic moiety contains the conserved oligomerization and scaffolding domain (amino acids 61–101) that mediates lipid binding, protein-protein interactions with signaling molecules, and homo-oligomerization (7). This region alone oligomerizes when incubated as a fragment in vitro, suggesting that it mediates caveolar coat assembly (8). Preceding the oligomerization domain is the N-terminal region that comprises 60 amino acids in CAV1 and has varying length in the other isoforms (7). This N-terminal region is not required for formation of caveolae, suggesting that it has regulatory function (9, 10).
FIGURE 1.
Monoubiquitination of CAV1 on endosomes occurs in the N-terminal region. A, domain structure of the caveolin-1 (CAV1) protein with the positions of all 12 lysine residues. Brackets indicate lysine residues that are exchanged to arginines in CAV1 variants used in this study. CAV1(K5–57R) is lacking the N-terminal lysines, the variant CAV1(K65–176R) is harboring only the N-terminal lysines, and CAV1(K5–176R) is the lysine-less variant. B, U2OS cells were transfected with constructs coding for CAV1-HA wild-type or indicated variants and lysed after 24 h. Cell lysates were analyzed by Western blotting using a CAV1-specific antibody (N20). The slower-migrating double band representing monoubiquitinated CAV1 is indicated (CAV1-HA-Ub). Note that ubiquitination is lost if the lysines of the N-terminal region (K5–57R) or all lysines (K5–176R) are mutated to arginine but not lysines 65–176 (K65–176). α-Tubulin served as a loading control. The asterisk indicates an unspecific band. C, U2OS cells were transfected with constructs coding for CAV1-HA as in B, formaldehyde-fixed, and processed for immunofluorescence microscopy with anti-HA and anti-ubiquitin (FK2) antibodies. Images were taken with a spinning disk confocal microscope using a ×100 objective. Note that although localization to endosomal compartments was comparable for all CAV1-HA variants (supplemental Fig. S1A), only the CAV1-HA variants harboring lysines in the N-terminal region effectively colocalized with the ubiquitin signal. Scale bars = 10 μm. D, quantification of C. Automated identification of CAV1-HA vesicles and colocalization with the ubiquitin (FK2) signal was carried out as detailed under “Experimental Procedures.” Whiskers indicate the 10th/90th percentile, and (●) indicates outliers. ***, p < 0.001. n = 20 cells/condition.
CAV1 is inserted cotranslationally into the ER2 where it rapidly forms SDS-resistant oligomers. During transport through the Golgi apparatus, the oligomers associate with cholesterol and self-assemble to form larger caveolar domains (11–13) that subsequently travel between the plasma membrane and endosomes (5, 14). When maturation fails in the ER, caveolin is polyubiquitinated and degraded by the proteasome (15). In contrast, the plasma membrane and endosomal pools of CAV1 are turned over in the lysosome (9, 16).
During the process, CAV1 is modified mostly with monoubiquitin in addition to short ubiquitin chains (16), consistent with monoubiquitin and short lysine 63-linked chains being signals for endolysosomal sorting (17, 18). Consequently, a lysine-less variant of CAV1 that cannot be ubiquitinated fails to be efficiently degraded in lysosomes (16). Consistently, CAV1 transport to late endosomes and lysosomes depends on components of the endosomal sorting complex required for transport (ESCRT) pathway that binds and packages ubiquitinated cargo into intraluminal vesicles of multivesicular bodies (16, 19). In addition, however, endosomal sorting of CAV1 requires VCP (20).
VCP (also called valosin-containing protein, p97, or Cdc48) is a multifunctional hexameric AAA+-type ATPase that uses the energy of ATP hydrolysis to structurally remodel and segregate protein complexes (21, 22). Its function is best studied during ER-associated degradation where it helps deliver misfolded proteins to the proteasome for degradation in cooperation with its heterodimeric cofactor Ufd1-Npl4 (23). The VCP-Ufd1-Npl4 complex binds substrate proteins after they have been modified with lysine 48-linked polyubiquitin chains at the cytosolic side of the membrane (24). Upon ATP hydrolysis, VCP segregates the substrates from the membrane to make them available to the proteasome (25). In addition, VCP cooperates with alternative cofactors in many other cellular processes (26, 27). These include endosomal trafficking pathways where VCP has been reported to physically interact with clathrin and early endosomal antigen 1 (EEA1) (28, 29). Moreover, VCP is essential for efficient autophagy, a process intimately linked with endosomal trafficking (30–32).
We showed recently that VCP binds monoubiquitinated CAV1 and that this complex involves the UBXD1 cofactor that can also be detected at CAV1-containing endosomes (20). The fact that cellular depletion of VCP or UBXD1, overexpression of dominant-negative mutants of VCP, or pharmacological inhibition of VCP lead to accumulation of CAV1 at the limiting membrane of late endosomes demonstrates that the VCP-UBXD1 complex is required for proper trafficking of CAV1 to lysosomes (20). However, the functional relationship between CAV1 ubiquitination and its sorting by the VCP-UBXD1 ATPase complex is unknown. Moreover, it is unclear in which of the functional domains CAV1 is ubiquitinated. Here we show that CAV1 is ubiquitinated at lysines within the flexible N-terminal region but not in the other functional domains and that this constitutes the signal for targeting CAV1 from early to late endosomes. Moreover, we provide evidence that the VCP-UBXD1 complex is recruited to endosomes by this specific ubiquitination. Importantly, we show that in the absence of VCP-UBXD1 activity ubiquitinated CAV1 accumulates, thus providing evidence that VCP-UBXD1 binds ubiquitinated CAV1 to facilitate downstream turnover of specifically this form of CAV1.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
CAV1-myc, UBXD1-mCherry, GFP-RAB5, and pcDNA-VCP-myc-strep/FRT/TO were described before (16, 20). A construct coding for HA-tagged human CAV1 was generated in two steps. First, a primer pair coding for a single HA tag and flanking the NotI and XhoI restriction sites (5′-GGCCGCATACCCATACGACGTCCCAGACTACGCTAGCTAGC-3′ (sense) and 5′-TCGAGCTAGCTAGCGTAGTCTGGGACGTCGTATGGGTATGC-3′ (antisense)) was annealed and cloned into a pcDNA3.1 + (Invitrogen) vector to yield pcDNA3.1/HA. The human CAV1 cDNA was then subcloned from CAV1-myc into pcDNA3.1/HA using the EcoRI/NotI restriction sites. The pcDNA-CAV1-HA/FRT/TO construct for the generation of stable cell lines was produced by PCR cloning of the CAV1-HA cDNA flanked by the HindIII and BamHI sites (5′-ATTGAAGCTTGGCACGAGGGGGCCAG-3′ (sense) and 5′-TACGGATCCCGAGCGGCCGCTATTTC-3′ (antisense)) into the HindIII and BamHI restriction sites of pcDNA5/FRT/TO (Invitrogen).
Site-directed Mutagenesis
Single lysine-to-arginine (KR) mutations were introduced into CAV1-myc by site-directed mutagenesis using the QuikChange protocol (Stratagene) and confirmed by sequencing. The HA-tagged CAV1 mutant variants were subcloned from CAV1-myc into pcDNA-HA as described above. The CAV1-HA K5–176R plasmid was used as a template to reintroduce arginine-to-lysine (RK) mutations in the N-terminal domain. All mutagenesis primer sequences are available upon request.
Cell Culture and Transfections
U2OS cells with a single integrated FRT site (U2OS-FRT) were generated by transfection of the pFRT/LacZEO plasmid (Invitrogen) according to the instructions of the manufacturer. Single integration was confirmed by Southern blotting. U2OS-FRT cells were stably transfected with the pcDNA6/TR plasmid according to the instructions of the manufacturer. Stable inducible U2OS cell lines expressing either VCP-myc-strep (U2OS-VCP) or CAV1-HA (U2OS-CAV1-HA) were generated from the U2OS-FRT cells. The pcDNA-VCP-myc-strep-FRT/TO or pcDNA-CAV1-HA-FRT/TO plasmids were transfected according to the protocol of the manufacturer for Flip-In recombination, and single clones were selected. U2OS and HEK293 cells were maintained in DMEM supplemented with 10% FCS in the presence of penicillin/streptomycin. In addition, stable cell lines were supplemented with 4 μg/ml blasticidin S and 130 μg/ml hygromycin B (U2OS-VCP) or 4 μg/ml blasticidin S and 120 μg/ml hygromycin B (U2OS-CAV1-HA). Expression was induced with 1 μg/ml doxycycline. HEK293 and U2OS cells were transfected using the JetPrime (Polyplus) or the JetPEI reagents (Polyplus), respectively. For JetPEI transfection reactions, the medium was replaced with fresh culture medium 4 h post-transfection. Cells were analyzed after 24 h.
RNAi
The siRNA oligomers targeting UBXD1 (CCAGGUGAGAAAGGAACUUTT), VCP (AACAGCCAUUCUCAAACAGAATT), and a non-targeting control oligomer (UUCUCCGAACGUGUCACGUTT) were purchased from Microsynth and were characterized previously (20, 33), Transfection into U2OS-CAV1-HA cells was done at 10 nm final concentration using Lipofectamine RNAiMAX (Invitrogen) according to the recommendations of the manufacturer. Cells were analyzed after 48 h.
Antibodies and Other Reagents
Rabbit polyclonal anti-VCP (HME8) and anti-UBXD1 (E43) antibodies were described previously (20). Rabbit polyclonal anti-CAV1 (N20, catalog no. sc-894), anti-LAMP1 (catalog no. sc-5570), and mouse monoclonal anti-myc (9E2, catalog no. sc-40) antibodies were purchased from Santa Cruz Biotechnology, Inc. Rabbit polyclonal anti-HA (H6908) and mouse monoclonal anti-α-tubulin (T-5168) antibodies were from Sigma-Aldrich. Mouse monoclonal anti-ubiquitin antibody (FK2, catalog no. 04-263) was from Millipore. Mouse monoclonal anti-HA antibody (HA.11, catalog no. MMS-101P) was from Covance. HRP-coupled secondary antibodies were purchased from Bio-Rad and Alexa Fluor-conjugated secondary antibodies from Invitrogen. DBeQ was purchased from Interbioscreen, MG132 from Calbiochem, and bafilomycin A1 from Sigma. Ammonium chloride (NH4Cl) was purchased from Fluka.
Pharmacological Treatments
U2OS-CAV1-HA cells were induced to express CAV1-HA for 24 h. The cells were treated with 200 ng/ml bafilomycin A1 in DMSO or 20 mg/ml NH4Cl in PBS for 18 h prior to lysis or fixation. Treatment with 10 μg/ml MG132 in DMSO or 10 μg/ml DBeQ in DMSO was carried out for 4 h prior to lysis or fixation.
Immunoprecipitation and Western Blot Analysis
Transfected cells were harvested in extraction buffer (150 mm KCl, 50 mm Tris, 5 mm MgCl2, 5% glycerol, 1% Triton X-100, and 2 mm β-mercaptoethanol (pH 7.4) supplemented with Roche complete EDTA-free protease inhibitors) and incubated on ice for 20 min. To investigate the ubiquitination of caveolin-1, the extraction buffer was additionally supplemented with 10 mm N-ethylmaleimide (Sigma) to inhibit deubiquitinating enzymes. Lysates were cleared by centrifugation (14,000 × g, 15 min, 4 °C), and the protein concentration was determined in a BCA assay (Interchim). Cleared lysates were either directly resolved by SDS-PAGE (13% Tris/glycine or MiniProtean TGX 4–15% Tris/glycine gels, Bio-Rad) and analyzed by Western blotting or subjected to immunoprecipitation. For immunoprecipitation, the protein concentrations were adjusted to 1 mg/ml in extraction buffer, and lysates were supplemented with 1 μg/ml BSA. For the coimmunoprecipitation experiments of CAV1-myc, 200 μg of lysate protein was used, of which 5% was taken as an input sample. The lysates were precleared with 5 μl of Dynabeads protein G (Invitrogen) for 1 h and incubated overnight at 4 °C with 0.2 μg of anti-myc primary antibody (9E2). Antigen-antibody complexes were captured with 20 μl of Dynabeads protein G at 4 °C for 1 h. The beads were washed three times in extraction buffer and eluted by boiling in 2 × SDS sample buffer. Samples were resolved by SDS-PAGE and analyzed by Western blotting.
Immunofluorescence Imaging
U2OS cells were grown on glass coverslips. 24 h after transfection, the cells were fixed in 4% formaldehyde in PBS and permeabilized with 0.1% Triton X-100 in PBS. After blocking with 3% BSA/PBS, the coverslips were incubated with primary antibodies (diluted 1:500 except for mouse anti-myc (1:1000) in 3% BSA/PBS) for 1.5 h and Alexa Fluor-conjugated secondary antibodies (diluted 1:500 in 3% BSA/PBS) for 30 min. Coverslips were mounted in Mowiol (Calbiochem) containing 0.5 μg/ml DAPI. Imaging was performed on an inverted spinning disc confocal microscope (Nikon Eclipse Ti with a Yokogawa CSU X-1 spinning disc unit) using a ×100, 1.49 NA (numerical aperture) or ×40, 0.95 NA objective. Images were acquired with an Andor iXon X3 EMCCD (electron multiplying charge-coupled device) camera.
Automated Colocalization Analysis
The colocalization between CAV1-HA-positive endosomal structures (Alexa Fluor 568 or Alexa Fluor 488) and ubiquitin, LAMP1, VCP-myc (Alexa Fluor 488), or UBXD1-mCherry signal was analyzed using an ImageJ (1.45 k, National Institutes of Health) routine that is available upon request. Images were acquired as series with equal laser intensities and exposure time. In detail, the image of the CAV1-HA channel was thresholded (mean of HA signal + twice the standard deviation of the HA signal; in the case of the Lamp1 and FK2 colocalization experiment, mean of HA signal + three times the standard deviation of the HA signal) to remove the background signal. To discriminate vesicles in close proximity, the ImageJ Watershed algorithm was used. Vesicles were identified using the Analyze Particles function in ImageJ (Size 10–400 pixels, roundness 0.5–1.0). For each vesicle, the colocalizing signal of ubiquitin, LAMP1, VCP-myc, or UBXD1-mCherry was analyzed individually and considered positive if the signal intensity was above the threshold (mean intensity + three times the standard deviation of the signal over the whole image) and the variation of the signal was not more than three times (four times for colocalization with VCP-myc or UBXD1) greater than the signal variation over the whole picture. For each cell, the total number of vesicles and of vesicles with colocalization was given as a result.
Statistical Analysis
Statistical analyses were performed in SigmaPlot software (Systat) as Mann-Whitney U tests. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 were considered significant. Box plots show medians, lower and upper quartiles (line and box), 10th and 90th percentiles (whiskers), and outliers (●).
RESULTS
CAV1 Is Monoubiquitinated in the N-terminal Region
Human CAV1 is a 178-amino acid protein that contains 12 lysines in four distinct functional domains (Fig. 1A). To identify the functional acceptor site of ubiquitin in CAV1, we undertook a systematic mutagenesis analysis in which we exchanged individual or groups of lysines to arginines. Mutagenized HA-tagged CAV1 variants were overexpressed in U2OS osteosarcoma cells and lysates analyzed by Western blotting with CAV1-specific antibodies. An experiment summarizing the results is shown in Fig. 1B. Overexpressed wild-type CAV1 migrated slightly above endogenous CAV1 because of the HA-tag. Importantly, a prominent slower-migrating double band was apparent that we previously showed to correspond to monoubiquitin-modified CAV1 in addition to a minor species above the doublet, most likely representing oligoubiquitinated CAV1 (16, 20). As expected, these bands were absent in lysates from cells expressing a CAV1 variant lacking all 12 lysines (K5–176R). Importantly, mutation of the six lysines that cluster in the N-terminal region (K5–57R) was sufficient to abolish ubiquitination. Conversely, mutation of all lysine residues in the other regions of CAV1 did not affect ubiquitination. To confirm this result and assess subcellular distribution of CAV1 ubiquitination by light microscopy, we transfected the four constructs into U2OS cells and colocalized CAV1 with the endosomal marker GFP-RAB5 or ubiquitin-specific antibodies. In all cases, CAV1 localized to endosomal compartments (supplemental Fig. S1A), as shown previously (16, 20). In the case of CAV1 wild-type and the K65–176R mutant that are both proficient in ubiquitination, frequent colocalization of CAV1-containing vesicles with the ubiquitin signal was apparent (Fig. 1C). Automated image analysis revealed that this was the case for a median of about 20% of CAV1-positive vesicles/cell (Fig. 1D). In contrast, the ubiquitination-deficient CAV1(K5–176R) and, importantly, also the N-terminal CAV1(K5–57R) mutant displayed a significantly reduced ubiquitin signal on CAV1-positive vesicles, as apparent in micrographs (Fig. 1C) and automated image quantification (D). These results demonstrate that the N-terminal region is the exclusive acceptor site for ubiquitination in CAV1 on endosomes.
Ubiquitination within the N-terminal Region Is Promiscuous
To assess the relevance of the individual lysine residues within the N-terminal region more exactly, we mutated single residues to arginine and again expressed the constructs in U2OS cells. Western blot analysis revealed that none of the single mutations significantly affected ubiquitination (Fig. 2A). In a converse experiment, we took the lysine-less CAV1(K5-K176R) construct as a template and reintroduced individual lysine residues in the N-terminal region. It should be noted that CAV1(K5–176R) runs slightly faster in SDS gels than the wild-type because of the extent of mutations (16), and we observed a gradual shift with increasing number of mutations (data not shown). As expected, CAV1(K5–176R) was not ubiquitinated (Fig. 2B). Crucially, however, reintroduction of individual lysines at any of the six residues in the N-terminal region resulted in ubiquitination of CAV1, although the lysine at position 39 was ubiquitinated most strongly, whereas position 57 was only ubiquitinated poorly (Fig. 2B). These results, which were again confirmed by immunofluorescence microscopy (supplemental Fig. S2) and automated quantification (Fig. 2C), suggest that ubiquitination can occur at any residue of the N-terminal region, although to a lesser extent at its border. The experiment also provided an explanation for the occurrence of the double band for monoubiquitinated wild-type CAV1. We found that single lysine residues at positions 5, 26, or 30 resulted in the upper band of the doublet, whereas ubiquitination at position 39, 47, or 57 resulted in the lower band (Fig. 2B). This suggests that formation of an isopeptide bond with ubiquitin causes different migration behavior in CAV1 in SDS gels, depending in which of the two subregions conjugation occurs. Indeed, reintroduction of two lysines at position 30 and 39 in the CAV1(R30,39K) construct resulted in ubiquitination at both sites with similar probability and, consequently, in the occurrence of a double band (Fig. 2B).
FIGURE 2.
Ubiquitination in the N-terminal region of CAV1 is promiscuous. A, individual lysine residues in the N-terminal region of CAV1 were mutated as indicated and CAV1-HA variants expressed in U2OS cells for 24 h along with the CAV1 variants described in Fig. 1. Ubiquitination was determined by Western blot analysis of cell lysates using a CAV1-specific antibody. Note that single lysine mutations had little or no effect on the CAV1 ubiquitination compared with wild-type CAV1 or CAV1(K65–176R) when variations of CAV1 expression levels are taken into account (short exposure). α-Tubulin served as a loading control. B, individual lysine residues of the N-terminal region were reintroduced into the lysine-less variant CAV1(K5–176R) as indicated, and ubiquitination was analyzed by Western blotting using lysates of transfected U2OS cells as in A. Note the presence of only a single band of ubiquitinated CAV1 when just one lysine is available in contrast to a double band for the wild-type or when lysines at position 30 and 39 are reintroduced (R30,39K). C, U2OS cells were transfected with the constructs coding for CAV1 with single lysines in the N-terminal region as in B. Cells were processed for immunofluorescence microscopy with anti-HA and anti-ubiquitin (FK2) antibodies, and images were taken by spinning disk confocal microscopy (supplemental Fig. S2). Colocalization of the ubiquitin signal with CAV1-HA-containing vesicles was quantified by automated image analysis as in Fig. 1D. Whiskers indicate the 10th/90th percentile, and (●) indicates outliers. n = 15 cells/condition.
CAV1 Ubiquitination in the N-terminal Region Mediates Recruitment of VCP and UBXD1 to Endosomes
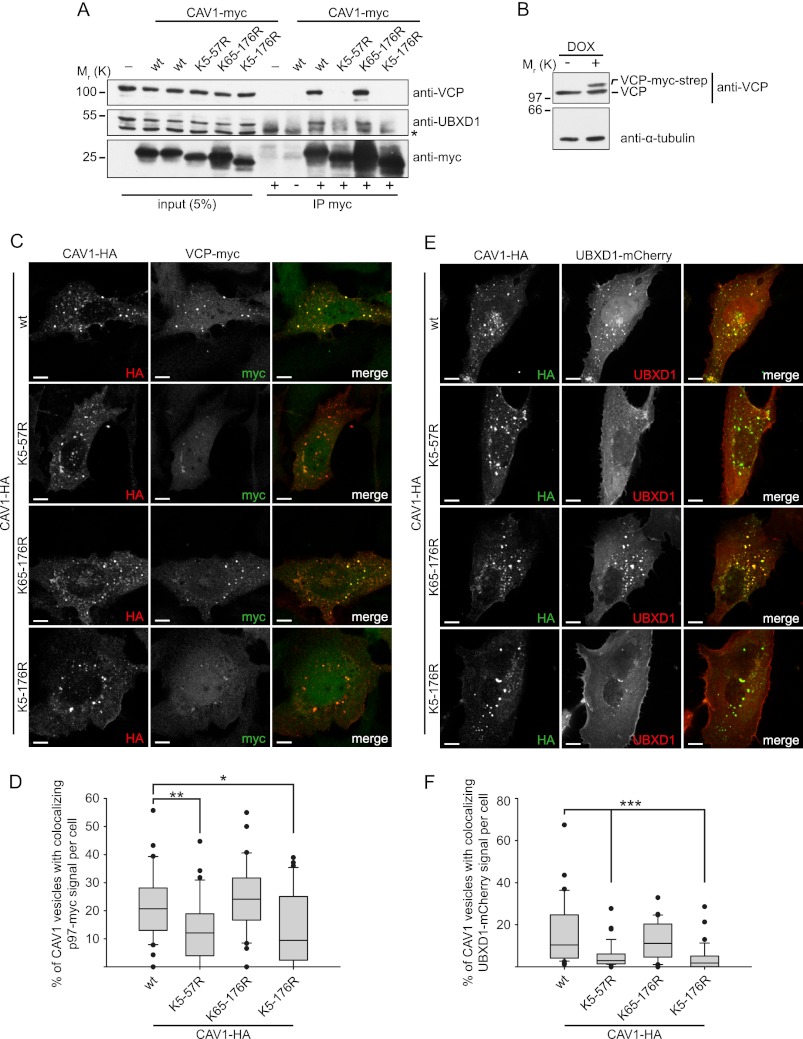
We next investigated the impact of CAV1 ubiquitination in the N-terminal region on interaction with VCP and UBXD1. To do so, we performed coimmunoprecipitation experiments with CAV1 variants from HEK293 cell lysates (Fig. 3A). HEK293 cells were used to increase protein yield. We confirmed previous results that both VCP and UBXD1 specifically cosedimented with the CAV1 wild type but not with the lysine-less CAV1(K5–176R) variant (20). Crucially, mutation of the N-terminal region in the K5–57R mutant was sufficient to largely reduce VCP and UBXD1 binding, whereas mutation of all other lysines in the construct CAV1(K65–176R) had no effect (Fig. 3A), consistent with CAV1(K65–176R) being ubiquitinated as well as the wild type (Fig. 1, B–D). Next, we analyzed recruitment of VCP to CAV1 containing endosomes. We were previously unable to detect endogenous VCP or transiently overexpressed VCP on endosomes for technical reasons. We therefore generated stable U2OS cells that overexpressed myc-tagged VCP at low levels upon doxycycline induction (Fig. 3B). These cells were transfected with the four different CAV1 variants and induced to express VCP-myc for 1 day. Colocalization of VCP-myc with CAV1-positive vesicles was then determined by immunofluorescence microscopy and automated image analysis. VCP-myc localization to CAV1-positive vesicles was apparent in cells expressing the CAV1 wild type and detected with a median of more than 20% (Fig. 3, C and D). In contrast, transfection of the lysine-less variant CAV1(K5–176K) or the N-terminal lysine mutant CAV1(K5–57R), but not CAV1(K65–176R), significantly reduced localization of VCP to endosomes, as apparent in micrographs and detected by automated quantification (Fig. 3, C and D). Interestingly, localization of VCP to endosomes was not completely abolished under these conditions, suggesting the presence of additional factors that target VCP to endosomes. In a similar manner, we investigated colocalization of transiently overexpressed mCherry-tagged UBXD1 with the CAV1-containing variant (Fig. 3, E and F). We confirmed our previous results that UBXD1-mCherry colocalized with CAV1 wild-type but not the lysine-less CAV1(K5–176R) variant (20). Crucially and consistently, however, we found that mutation of K5–57R but not K65–176R in CAV1 was sufficient to abolish recruitment of UBXD1-mCherry to CAV1-containing vesicles. These data suggest that ubiquitination of the N-terminal region of CAV1 mediates recruitment of VCP and UBXD1 to endosomes.
FIGURE 3.
CAV1 interaction with VCP-UBXD1 and recruitment to endosomes is regulated through ubiquitination of the N-terminal region. A, HEK293 cells were transfected with myc-tagged CAV1 wild-type or the K5–57R, K65–176R, or the K5–176R variants. Cells were detergent-extracted, and CAV1-myc was immunoprecipitated (IP) with an anti-myc antibody. Anti-HA antibody was used as a negative control (-). Efficiency of CAV1-myc isolation and coimmunoprecipitation of VCP and UBXD1 were probed with specific antibodies by Western blot analysis. Note the large reduction of VCP and UBXD1 coisolation with CAV1 variants lacking the N-terminal lysines. The asterisk indicates the antibody heavy chain. B, characterization of a stable U2OS cell line generated to express VCP-myc-strep under control of a doxycycline-inducible (DOX) promoter (U2OS-VCP). The cells were left untreated or induced with DOX for 24 h, and cell lysates were analyzed by Western blotting. C, U2OS-VCP cells were induced with doxycycline to express VCP-myc-strep and transiently transfected with CAV1-HA for 24 h. Cells were formaldehyde-fixed and stained with tag-specific antibodies as indicated. Images were taken with a spinning disk confocal microscope using a ×100 objective. Note the localization of VCP-myc to CAV1-HA-positive vesicles in cells expressing CAV1-HA wild-type or the K65–176R variant, whereas localization is reduced in cells expressing the non-ubiquitinatable CAV1-HA variants K5–57R or K5–176R. Scale bars = 10 μm. D, quantification of C. Colocalization between VCP-myc- and CAV1-HA-positive vesicles was quantified by automated image analysis as detailed under “Experimental Procedures.” Whiskers indicate the 10th/90th percentile and (●) indicates outliers. *, p < 0.05; **, p < 0.01. n = 35 cells/condition from three independent experiments. E, U2OS cells were transiently transfected with UBXD1-mCherry and CAV1-HA. Cells were fixed after 24 h and stained with HA-specific antibodies. Note reduced colocalization in cells expressing non-ubiquitinatable CAV1-HA variants. F, quantification of E as in D. Whiskers indicate the 10th/90th percentile, and (●) indicates outliers. ***, p < 0.001. n = 30 cells/condition from three independent experiments.
Ubiquitinated CAV1 Accumulates in the Absence of VCP and UBXD1 Activity
To address the role of VCP-UBXD1 after binding to ubiquitinated CAV1 in regulating its subsequent turnover, we monitored the ubiquitination status of CAV1 after inactivation of the VCP-UBXD1 complex in two ways. In the first approach, we used siRNA oligonucleotides to deplete VCP or UBXD1. To facilitate the approach, we generated stable U2OS cells that express CAV1-HA upon doxycycline induction (Fig. 4A). Cells were either left untreated or treated with control siRNA or siRNA targeting VCP or UBXD1 for a total of 2 days. Expression of CAV1-HA was induced in all cell populations 1 day prior to cell lysis. Western blot analysis of cell lysates confirmed specific and efficient depletion of VCP and UBXD1 in the respective samples (Fig. 4B). As before, CAV1-HA and the slower-migrating bands representing ubiquitinated CAV1-HA were detectable in control lysates. Crucially, however, the CAV1 ubiquitination levels were strongly increased in VCP- and UBXD1-depleted cells. Also, the total level of unmodified CAV1 was increased in these samples, consistent with the role of VCP and UBXD1 in CAV1 turnover. To confirm this result, we applied a pharmacological approach with the VCP inhibitor DBeQ (34). As DBeQ is toxic because of caspase activation and apoptosis (34), the treatment could only be extended to 4 h. Nevertheless, accumulation of ubiquitinated CAV1 was observed by Western blot analysis even during this short period (Fig. 4C). We compared the effect on the ubiquitination status of CAV1 of these treatments with that of lysosome inhibition. Indeed, bafilomycin A1 or NH4Cl treatment led, like VCP or UBXD1 inactivation, to accumulation of ubiquitinated CAV1 (Fig. 4D). In contrast and as expected, proteasome inhibition with MG132 resulted in a general increase of cellular ubiquitin conjugates but left CAV1 ubiquitination unaffected (Fig. 4D). We confirmed that CAV1 accumulated in LAMP1-positive compartments upon bafilomycin A1 or DBeQ inhibition but not upon MG132 treatment (Fig. 4E) and that these populations were ubiquitinated (data not shown). These data suggest that VCP-UBXD1 acts downstream of CAV1 ubiquitination and, subsequently, helps to clear the ubiquitinated CAV1 population in lysosomes.
FIGURE 4.
Cellular depletion of VCP or UBXD1 or pharmacological inhibition of VCP leads to accumulation of monoubiquitinated CAV1. A, a stable U2OS cell line was generated to express CAV1-HA under the control of a doxycycline-inducible (DOX) promoter (U2OS-CAV1-HA). The cells were left untreated or induced by DOX for 24 h, and cell lysates were analyzed by Western blotting with a CAV1-specific antibody (N20). α-Tubulin served as loading control. B, U2OS-CAV1-HA cells were either left untreated (no siRNA), treated with control (si-ctrl) siRNA oligonucleotides, or with siRNAs targeting VCP or UBXD1, as indicated, for a total of 48 h. 24 h prior to lysis, cells were induced to express CAV1-HA. Lysates were subjected to Western blot analysis with antibodies against CAV1, VCP, and UBXD1 as indicated. α-Tubulin served as loading control. Note the accumulation of ubiquitinated CAV1 upon VCP or UBXD1 depletion. C, U2OS-CAV1-HA cells were induced as in A. The cells were mock treated (m.t.) or treated with DMSO or DBeQ for 4 h prior to lysis in extraction buffer. CAV1 ubiquitination was analyzed by Western blotting as in A. After DBeQ treatment, an accumulation of ubiquitinated CAV1-HA was observed. D, U2OS-CAV1-HA cells were induced as in A and mock-treated or treated with DMSO, bafilomycin A1, or NH4Cl for 18 h or with MG132 for 4 h. Note the accumulation of ubiquitinated CAV1-HA specifically after treatment with the lysosome inhibitors bafilomycin A1 or NH4Cl, but not MG132, which caused a general increase of ubiquitin conjugates. E, U2OS-CAV1-HA cells were induced and treated as in C and D prior to fixation and staining with HA- and LAMP1-specific antibodies. Images were taken with a spinning disk confocal microscope using a ×100 objective. Note the increase of CAV1 in LAMP1-positive vesicles after bafilomycin A1 or DBeQ treatment. Scale bars = 10 μm.
Ubiquitination of the N-terminal Region Is Essential for CAV1 Transport to Late Endosomes
To test the role of CAV1 ubiquitination and the significance of the N-terminal lysine residues on subsequent transport to late endosomes, we devised automated image analysis to score how efficiently the different CAV1-HA variants reached late endosomes on the basis of colocalization with the late endosome and lysosome marker protein LAMP1. CAV1 wild-type or the three variants K5–57R, K65–176, or K5–176R were expressed in U2OS cells for 24 h. Cells were fixed and stained with HA- and LAMP1-specific antibodies (Fig. 5, A and B). CAV1 wild-type as well as the K65–176R variant that both contain the critical lysines in the N-terminal region colocalized frequently and to the same extent with LAMP1, indicating equally efficient transport to late endosomes. In contrast and expectedly, colocalization was reduced significantly for the lysine-less K5–176R variant. Crucially, however, mutation of the lysines in the N-terminal region (K5–57R) was sufficient to cause an equally significant reduction. These results demonstrate that ubiquitination of the N-terminal region of CAV1 is essential for efficient transport of CAV1 to late endosomal compartments.
FIGURE 5.
Ubiquitination of CAV1 in the N-terminal region is required for transport to late endosomes. A, U2OS cells were transiently transfected with the indicated CAV1-HA variants. After 24 h, cells were formaldehyde-fixed and stained with specific antibodies for HA or the late endosome and lysosome marker LAMP1. Images were taken with a spinning disk confocal microscope using a ×100 objective. The arrows indicate CAV1-HA-positive vesicles colocalizing with LAMP1. Note that colocalization is decreased in the case of the non-ubiquitinatable variants CAV1(K5–57R) and CAV1(K5–176R). Scale bars =10 μm. B, quantification of A. Identification of CAV1-HA vesicles and colocalization with LAMP1 was carried out by automated image analysis as detailed under “Experimental Procedures.” Whiskers indicate the 10th/90th percentile, and (●) indicates outliers. ***, p < 0.001. n = 60 cells/condition from two independent experiments.
DISCUSSION
Our results identify the acceptor site for monoubiquitination within the CAV1 protein at endosomes and thus allow detailed investigation of the functional role of this modification for interaction with the VCP-UBXD1 complex and sorting into late endosomes for subsequent turnover in lysosomes.
We find that CAV1 is exclusively ubiquitinated in the N-terminal region. Among the six lysines within this region, ubiquitination appears promiscuous. Reintroduction of lysines into the lysine-less CAV1 variant showed that, in principle, ubiquitination can occur at any of the six lysines. Ubiquitination of Lys-57, however, is less efficient, probably because of the fact that this residue is at the border to the oligomerization domain. Our findings are consistent with two large-scale mass spectrometry approaches published during the course of our study, which together identify di-glycine profiles indicative for ubiquitin modification for all six lysines in the N-terminal region (35, 36). In addition, one of the studies identified modification at residue Lys-176 (35). We did not find evidence for Lys-176 ubiquitination in Western blot analysis or immunofluorescence microscopy, as monoubiquitination was fully abolished already by the K5–57R mutations that leave Lys-176 intact. One explanation is, therefore, that Lys-176 is not used for monoubiquitination at the endosome but for modification with lysine 48-linked polyubiquitin chains during ER-associated degradation of misfolded species of CAV1 (15), which cannot be discriminated by the mass spectrometry approach in that study. Interestingly, isopeptide bond formation between ubiquitin and CAV1 resulted in slightly different migration behavior in SDS gels depending on whether ubiquitin is conjugated to Lys-5, Lys-26, or Lys-30 or whether it is conjugated to Lys-39, Lys-47, or Lys-57. The observed double band for monoubiquitinated wild-type CAV1, therefore, indicates an approximate equal probability of ubiquitin conjugation to one or the other sequence segment within the N-terminal region. Interestingly, structure predictions indicate that the second segment comprising Lys-39–57 assumes an amphipathic α helix that is tightly associated with the membrane, whereas the first segment is purely hydrophilic and points into the cytosol (7). Phosphorylation of tyrosine 14 in this region has been described (37). However, we observed no effect of the phosphorylation-deficient Y14F mutation on ubiquitination levels (data not shown), suggesting that ubiquitination is not regulated by tyrosine 14 phosphorylation. The lysines in the N-terminal region of CAV1 are conserved in zebrafish, chicken, and mouse (data not shown). Moreover, the muscle-specific CAV3 variant that lacks much of the N-terminal region also contains lysine residues equivalent to Lys-47 and Lys-57 in CAV1 plus an additional lysine upstream. This is at least consistent with ubiquitin-mediated regulation being a general feature for caveolin proteins. Conversely, fusion of the CAV1 N-terminal region alone to GFP did not results in detectable monoubiquitination (data not shown), suggesting that this region is not sufficient to trigger ubiquitination and targeting to the endosomal system.
The identification of the N-terminal region as the ubiquitin acceptor site has functional implications. Given that ubiquitination occurs outside of the oligomerization domain, it is unlikely that ubiquitination interferes with or even directly regulates formation of CAV1 oligomers or larger assemblies. Conversely, the fact that the N-terminal region is not directly involved in oligomerization and may remain accessible in CAV1 oligomers (7, 10) suggests that CAV1 can be ubiquitinated while engaged in oligomers. This raises the possibility that oligomerized CAV1 can be targeted for lysosomal turnover through ubiquitination and that ubiquitination may recruit factors such as VCP that assist CAV1 trafficking, possibly by disassembly of caveolar assemblies or CAV1 oligomers.
In fact, our data show that ubiquitination in the N-terminal region is essential for CAV1 interaction with VCP and UBXD1. More importantly, in this study we show for the first time that VCP, in addition to UBXD1, is recruited to CAV1-containing endosomes and that CAV1 ubiquitination in the N-terminal region constitutes the essential signal for this recruitment. Localization of VCP to endosomes was also reported elsewhere, and it was speculated that this is mediated by ubiquitinated early endosomal antigen 1 (EEA1) (29). This suggests that ubiquitination of other endosomal proteins can also mediate recruitment of VCP, which would explain the remaining VCP on endosomes in the absence of CAV1 ubiquitination. Besides binding monoubiquitinated CAV1, we provide further evidence that the VCP-UBXD1 complex, in fact, actively controls the turnover of the ubiquitinated form of CAV1. This notion is on the basis of the observation that inactivation of the VCP-UBXD1 complex by siRNA-mediated depletion or pharmacological inhibition of VCP leads to accumulation of ubiquitinated CAV1. This demonstrates that VCP-UBXD1 acts after CAV1 ubiquitination and then allows targeting to downstream steps, presumably by virtue of its protein segregation activity. This order of events is well established for polyubiquitinated substrates of the VCP-Ufd1-Npl4 complex, for example during ER-associated degradation (22, 23, 25). It therefore seems to be a general principle that VCP binds ubiquitinated proteins and directs them for degradation, albeit by different mechanisms and with distinct cofactors. In the case of polyubiquitinated proteins VCP directs them to the proteasome, while in the case of mono- or oligoubiquitinated endosomal proteins it directs them to the lysosome.
So far it is unclear what exactly the molecular function of VCP is that facilitates endosomal sorting of CAV1. One possibility is that VCP and UBXD1 are recruited to CAV1 to disassemble CAV1 oligomers or larger caveolar assemblies. Protein complex disassembly has been proposed in many cases for VCP/p97 and its homologue Cdc48, including topologically similar remodeling of SNARE (soluble N-ethylmaleimide-sensitive-factor attachment receptor) complexes or segregation of Spt23 dimers in the membrane (38, 39). Here we show that ubiquitination in the N-terminal region of CAV1 is at least compatible with ubiquitination of CAV1 oligomers and therefore consistent with the notion that CAV1 complexes are also disassembled. Given that CAV1 assemblies are believed to induce membrane curvature toward the cytosol (7), CAV1 oligomer disassembly may indeed be required for sorting of CAV1 into intraluminal vesicles whose formation involve curvature in the opposite direction (19). Another possibility is that VCP is recruited to CAV1 to modulate the endosomal sorting machinery to release CAV1 or other cargo from the extensive network of protein-protein interactions among these factors (40). Further work is required to address this question.
Acknowledgments
We thank Sabine Effenberger for technical assistance.
This work was funded by Deutsche Forschungsgemeinschaft (DFG) Priority Program SPP1365/2 Grant Me1626/2-1.

This article contains supplemental Figs. S1 and S2.
- ER
- endoplasmic reticulum
- VCP
- valosin-containing protein
- DMSO
- dimethyl sulfoxide
- DBeQ
- N2N4-dibezylquinazoline-2,4-diamine
- FRT
- FLP recombination target.
REFERENCES
- 1. Razani B., Woodman S. E., Lisanti M. P. (2002) Caveolae. From cell biology to animal physiology. Pharmacol. Rev. 54, 431–467 [DOI] [PubMed] [Google Scholar]
- 2. Parton R. G., Simons K. (2007) The multiple faces of caveolae. Nat. Rev. Mol. Cell Biol. 8, 185–194 [DOI] [PubMed] [Google Scholar]
- 3. Parat M. O. (2009) The biology of caveolae. Achievements and perspectives. Int. Rev. Cell Mol. Biol. 273, 117–162 [DOI] [PubMed] [Google Scholar]
- 4. Pelkmans L., Bürli T., Zerial M., Helenius A. (2004) Caveolin-stabilized membrane domains as multifunctional transport and sorting devices in endocytic membrane traffic. Cell 118, 767–780 [DOI] [PubMed] [Google Scholar]
- 5. Tagawa A., Mezzacasa A., Hayer A., Longatti A., Pelkmans L., Helenius A. (2005) Assembly and trafficking of caveolar domains in the cell. Caveolae as stable, cargo-triggered, vesicular transporters. J. Cell Biol. 170, 769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anderson R. G. (1998) The caveolae membrane system. Annu. Rev. Biochem. 67, 199–225 [DOI] [PubMed] [Google Scholar]
- 7. Parton R. G., Hanzal-Bayer M., Hancock J. F. (2006) Biogenesis of caveolae. A structural model for caveolin-induced domain formation. J. Cell Sci. 119, 787–796 [DOI] [PubMed] [Google Scholar]
- 8. Fernandez I., Ying Y., Albanesi J., Anderson R. G. (2002) Mechanism of caveolin filament assembly. Proc. Natl. Acad. Sci. U.S.A. 99, 11193–11198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hill M. M., Bastiani M., Luetterforst R., Kirkham M., Kirkham A., Nixon S. J., Walser P., Abankwa D., Oorschot V. M., Martin S., Hancock J. F., Parton R. G. (2008) PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell 132, 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kirkham M., Nixon S. J., Howes M. T., Abi-Rached L., Wakeham D. E., Hanzal-Bayer M., Ferguson C., Hill M. M., Fernandez-Rojo M., Brown D. A., Hancock J. F., Brodsky F. M., Parton R. G. (2008) Evolutionary analysis and molecular dissection of caveola biogenesis. J. Cell Sci. 121, 2075–2086 [DOI] [PubMed] [Google Scholar]
- 11. Hayer A., Stoeber M., Bissig C., Helenius A. (2010) Biogenesis of Caveolae: Stepwise Assembly of Large Caveolin and Cavin Complexes. Traffic 11, 361–382 [DOI] [PubMed] [Google Scholar]
- 12. Scheiffele P., Verkade P., Fra A. M., Virta H., Simons K., Ikonen E. (1998) Caveolin-1 and -2 in the exocytic pathway of MDCK cells. J. Cell Biol. 140, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Monier S., Parton R. G., Vogel F., Behlke J., Henske A., Kurzchalia T. V. (1995) VIP21-caveolin, a membrane protein constituent of the caveolar coat, oligomerizes in vivo and in vitro. Mol. Biol. Cell 6, 911–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mundy D. I., Machleidt T., Ying Y. S., Anderson R. G., Bloom G. S. (2002) Dual control of caveolar membrane traffic by microtubules and the actin cytoskeleton. J. Cell Sci. 115, 4327–4339 [DOI] [PubMed] [Google Scholar]
- 15. Galbiati F., Volonte D., Minetti C., Bregman D. B., Lisanti M. P. (2000) Limb-girdle muscular dystrophy (LGMD-1C) mutants of caveolin-3 undergo ubiquitination and proteasomal degradation. Treatment with proteasomal inhibitors blocks the dominant negative effect of LGMD-1C mutanta and rescues wild-type caveolin-3. J. Biol. Chem. 275, 37702–37711 [DOI] [PubMed] [Google Scholar]
- 16. Hayer A., Stoeber M., Ritz D., Engel S., Meyer H. H., Helenius A. (2010) Caveolin-1 is ubiquitinated and targeted to intraluminal vesicles in endolysosomes for degradation. J. Cell Biol. 191, 615–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Raiborg C., Stenmark H. (2009) The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458, 445–452 [DOI] [PubMed] [Google Scholar]
- 18. MacGurn J. A., Hsu P. C., Emr S. D. (2012) Ubiquitin and membrane protein turnover. From cradle to grave. Annu. Rev. Biochem. 81, 231–259 [DOI] [PubMed] [Google Scholar]
- 19. Hurley J. H., Emr S. D. (2006) The ESCRT complexes. Structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35, 277–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ritz D., Vuk M., Kirchner P., Bug M., Schütz S., Hayer A., Bremer S., Lusk C., Baloh R. H., Lee H., Glatter T., Gstaiger M., Aebersold R., Weihl C. C., Meyer H. (2011) Endolysosomal sorting of ubiquitinated caveolin-1 is regulated by VCP/p97 and UBXD1 and impaired by VCP disease mutations. Nat. Cell Biol. 13, 1116–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chapman E., Fry A. N., Kang M. (2011) The complexities of p97 function in health and disease. Mol. Biosyst. 7, 700–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meyer H., Bug M., Bremer S. (2012) Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 14, 117–123 [DOI] [PubMed] [Google Scholar]
- 23. Stolz A., Hilt W., Buchberger A., Wolf D. H. (2011) Cdc48: a power machine in protein degradation. Trends Biochem. Sci. 36, 515–523 [DOI] [PubMed] [Google Scholar]
- 24. Ye Y., Meyer H. H., Rapoport T. A. (2003) Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 162, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ye Y. (2006) Diverse functions with a common regulator. Ubiquitin takes command of an AAA ATPase. J. Struct. Biol. 56, 29–40 [DOI] [PubMed] [Google Scholar]
- 26. Schuberth C., Buchberger A. (2008) UBX domain proteins. Major regulators of the AAA ATPase Cdc48/p97. Cell Mol. Life Sci. 65, 2360–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yeung H. O., Kloppsteck P., Niwa H., Isaacson R. L., Matthews S., Zhang X., Freemont P. S. (2008) Insights into adaptor binding to the AAA protein p97. Biochem. Soc. Trans 36, 62–67 [DOI] [PubMed] [Google Scholar]
- 28. Pleasure I. T., Black M. M., Keen J. H. (1993) Valosin-containing protein, VCP, is a ubiquitous clathrin-binding protein. Nature 365, 459–462 [DOI] [PubMed] [Google Scholar]
- 29. Ramanathan H. N., Ye Y. (2012) The p97 ATPase associates with EEA1 to regulate the size of early endosomes. Cell Res. 22, 346–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ju J. S., Weihl C. C. (2010) p97/VCP at the intersection of the autophagy and the ubiquitin proteasome system. Autophagy 6, 283–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tresse E., Salomons F. A., Vesa J., Bott L. C., Kimonis V., Yao T. P., Dantuma N. P., Taylor J. P. (2010) VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy 6, 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rusten T. E., Stenmark H. (2009) How do ESCRT proteins control autophagy? J. Cell Sci. 122, 2179–2183 [DOI] [PubMed] [Google Scholar]
- 33. Meerang M., Ritz D., Paliwal S., Garajova Z., Bosshard M., Mailand N., Janscak P., Hübscher U., Meyer H., Ramadan K. (2011) The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double strand breaks. Nat. Cell Biol. 13, 1376–1382 [DOI] [PubMed] [Google Scholar]
- 34. Chou T. F., Brown S. J., Minond D., Nordin B. E., Li K., Jones A. C., Chase P., Porubsky P. R., Stoltz B. M., Schoenen F. J., Patricelli M. P., Hodder P., Rosen H., Deshaies R. J. (2011) Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways. Proc. Natl. Acad. Sci. U.S.A. 108, 4834–4839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., Choudhary C. (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, M111 013284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. del Pozo M. A., Balasubramanian N., Alderson N. B., Kiosses W. B., Grande-García A., Anderson R. G., Schwartz M. A. (2005) Phospho-caveolin-1 mediates integrin-regulated membrane domain internalization. Nat. Cell Biol. 7, 901–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rabouille C., Kondo H., Newman R., Hui N., Freemont P., Warren G. (1998) Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell 92, 603–610 [DOI] [PubMed] [Google Scholar]
- 39. Rape M., Hoppe T., Gorr I., Kalocay M., Richly H., Jentsch S. (2001) Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell 107, 667–677 [DOI] [PubMed] [Google Scholar]
- 40. Bug M., Meyer H. (2012) Expanding into new markets. VCP/p97 in endocytosis and autophagy. J. Struct. Biol. 179, 78–82 [DOI] [PubMed] [Google Scholar]