Abstract
Dietary restriction (DR) has many beneficial effects, but the detailed metabolic mechanism remains largely unresolved. As diet is essentially related to metabolism, we investigated the metabolite profiles of urines from control and DR animals using NMR and LC/MS metabolomic approaches. Multivariate analysis presented distinctive metabolic profiles and marker signals from glucuronide and glycine conjugation pathways in the DR group. Broad profiling of the urine phase II metabolites with neutral loss scanning showed that levels of glucuronide and glycine conjugation metabolites were generally higher in the DR group. The up-regulation of phase II detoxification in the DR group was confirmed by mRNA and protein expression levels of uridinediphospho-glucuronosyltransferase and glycine-N-acyltransferase in actual liver tissues. Histopathology and serum biochemistry showed that DR was correlated with the beneficial effects of low levels of serum alanine transaminase and glycogen granules in liver. In addition, the Nuclear factor (erythroid-derived 2)-like 2 signaling pathway was shown to be up-regulated, providing a mechanistic clue regarding the enhanced phase II detoxification in liver tissue. Taken together, our metabolomic and biochemical studies provide a possible metabolic perspective for understanding the complex mechanism underlying the beneficial effects of DR.
It has been known for more than 70 years that dietary restriction (DR)1 can extend the life span and delay the onset of age-related diseases, based on an early rodent study showing such effects (1). However, not until the 1980s was DR recognized as a good model for studying the mechanism of or inhibitory measures for aging (2). So far, extensive studies employing model organisms such as yeasts, nematodes, fruit flies, and rodents have shown that DR has beneficial effects in most of the species studied (for a review, see Ref. 3). Most notably, a recent 20-year-long study showed that monkeys, the species closest to humans, also benefit from DR similarly (4). Although there has not been (or could not have been) a systematic study on the effects of DR on the human life span, several longitudinal studies strongly suggest that changes in dietary intake can affect the life span and/or disease-associated marker values greatly (5–7).
This inverse correlation between dietary intake and long-term health strongly indicates that DR's effects should involve metabolism, and that DR elicits the reorganization of metabolic pathways. It also seems quite natural that something we eat should affect the body's metabolism. Despite this seemingly straightforward relationship between diet and metabolism, the mechanisms underlying the beneficial effects of DR are anything but simple. Intensive efforts, spanning decades, to understand the mechanisms of DR have identified several genes that might mediate the effects of DR, such as mTOR, IGF-1, AMPK, and SIRT1 (for a review, see Ref. 8). Still, most of them are involved in early nutrient-sensing steps, and specific metabolic pathways, especially those at the final steps actually responsible for the effects of DR, are largely unknown.
This might be at least partially due to the fact that previous studies have focused mostly on genomic or proteomic changes induced by DR, instead of looking at changes in metabolism or metabolites directly. Metabolomics, which has gained much interest in recent years (9–11), might be a good alternative for addressing the mechanistic uncertainty of DR's effects, with the direct profiling of metabolic changes elicited by environmental factors. In contrast to genomics or proteomics, which often employ DNA or proteins extracted from particular tissues, metabolomics studies mostly employ body fluids (i.e. urine or blood), which can reflect the metabolic status of multiple organs, enabling investigations at a more systemic level. In particular, urine has been used extensively to study the mechanism of external stimuli (i.e. drugs or toxic insults) at most major target organs, such as the lung, kidney, liver, or heart (12–18). Still, metabolomics studies of DR effects have been very limited. A few previous ones reported the changes in phenomenological urine metabolic markers with DR, without identification and/or validation of specific metabolic pathways reflected at the actual tissue or enzyme level (19, 20). Therefore, those studies fell short of providing a mechanistic perspective on DR's effects. In addition, they employed either NMR or LC/MS approaches without validation across the two analytical platforms.
Among the metabolic pathways that can directly affect the integrity of multiple organs, and hence long-term health, are phase II detoxification pathways (21). Typically, lipophilic endo/xenobiotics are metabolized first by a phase I system, such as cytochrome P450, which modifies the compounds so that they have hydrophilic functional groups for increased solubility. In many cases, though, these modifications might increase the reactivity of the compounds, leading to cellular damage. The phase II detoxification systems involve conjugation reactions that attach charged hydrophilic molecular moieties to reactive metabolites, thus facilitating the elimination of the harmful metabolites from body, ultimately reducing their toxicity (22). These systems are thus especially important in protecting cellular macromolecules, such as DNA and proteins, from reactive electrophilic or nucleophilic metabolites. The enzymes involved in these processes include glutathione-S-transferase (GST), sulfotransferase, glycine-N-acyltransferase (GLYAT), and uridinediphospho-glucuronosyltransferase (UGT), with the last enzyme being the most prevalent (23). The beneficial effects of phase II reactions have been particularly studied in relation to the mechanism of healthy dietary ingredients. It is well believed that many such foods can prevent cancers (hence the term “chemoprevention”) by inducing phase II detoxification systems (24–26). Although DR also substantially reduces the incidence of cancers, the exact mechanism remains elusive.
Here, we employed multi-platform metabolomics to obtain metabolic perspectives on the beneficial effects of DR on rats. Our results about urine metabolomics markers suggest that DR enhances the phase II detoxification pathway, which was confirmed by means of conjugation metabolite profiling and changes in mRNA/protein expression levels of phase II enzymes in actual liver tissues. A possible molecular mechanism was also addressed through the exploration of Nuclear factor (erythroid-derived 2)-like 2 (Nrf-2) pathway activation upon DR. We believe the current study provides new metabolic insights into DR's beneficial effects, as well as a workflow for studying DR's effects from a metabolic perspective.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
HPLC-grade acetonitrile and water were purchased from Burdick & Jackson (Morristown, NJ). Chemicals for NMR and LC/MS analysis were obtained from Sigma-Aldrich (St. Louis, MO). Vendors for biological reagents are indicated in the corresponding sections.
Animal and Diets
Male Sprague-Dawley rats (9 weeks of age) were purchased from Orient Bio (Sungnam, Seoul, Korea) and housed on a 12-h light/dark cycle prior to the experiment. Animal care and all experimental procedures were conducted in accordance with the guide for animal experiments edited by the Korea Academy of Medical Science. After 7 days of acclimatization, rats were randomly assigned to two groups: control (n = 8) and DR (n = 13). Rats in the control group had ad libitum access to standard rodent chow at all times. The DR practice was performed following the established protocols (27). Specifically, rats in the DR group received 60% of the food intake of the control group at specified feeding times. The food intake of the control group was calculated by subtracting the amount of the remaining food from the initial amount of food given. The amount of remaining food was carefully measured to include all the broken chow pieces in the cages. Food was given every Monday, Wednesday, and Friday, and a factor of 1.5 was considered in the calculation of the DR group's food allotment for Friday and Monday feeding.
Urine, Blood, and Tissue Collection
The pooling of urine samples was done over a 24-h period. The urine was collected in ice-cooled collection jars with sodium azide in them to prevent bacterial growth and sample deterioration. This collection was done once every 2 weeks. Fecal contamination was prevented by using commercially available metabolic cages specifically designed for that purpose. The pooled urine samples were frozen and stored at −80 °C for subsequent analysis. Blood was collected from the heart upon sacrifice at the end of the experiment. In order to obtain serum, the collected blood was incubated at room temperature for clotting and then centrifuged at 3000 rpm for 10 min at 4 °C. The supernatants (clear yellow fluid) were moved to centrifuge tubes, snap-frozen with liquid nitrogen, stored at −80 °C, and thawed just before analysis. For liver tissue collection, the same small parts of the liver tissue of each sacrificed rat were snap-frozen with liquid nitrogen and stored at −80 °C until use. The rest of the tissues were encased in paraffin blocks using routine procedures and were used for histopathological examination.
NMR Spectroscopic Analysis of Urine
For NMR analysis, urine samples were thawed at room temperature, and 500 μl of urine was mixed with 50 μl of potassium phosphate buffer (pH = 7.4). The insoluble parts were removed via centrifugation at 13,000 rpm for 10 min. A mixture of 500 μl of supernatant and 50 μl D2O containing sodium-3-trimethylsily-[2,2,3,3-2H4]-1-propionate (0.025%, w/v) as an internal standard was placed in a 5-mm NMR tube. All one-dimensional spectra of the urine samples were measured with an NMR spectrometer (Avance 500, Bruker Biospin, Rheinstetten, Germany) operating at a proton NMR frequency of 500.13 MHz. The NMR experiment was performed at the NMR facility at the Korea Basic Science Institute. We also used a 500 MHz machine (VNMRS500) at Varian Inc. Korea's facility for metabolite identification. The acquisition parameters were essentially the same as those previously reported (28–30). The metabolites were identified using Chenomx (spectral database; Edmonton, Alberta, Canada) by fitting the experimental spectra to those in the database and comparing them with spectra from standard compounds.
LC/MS Analysis of Urine
For LC/MS analysis, the thawed urine samples were centrifuged at 15,000 rpm for 10 min to remove the insoluble materials, and then the supernatants were injected with an injection volume of 5 μl. HPLC was performed on an Agilent 1100 Series liquid chromatography system equipped with a degasser, an auto-sampler, and a binary pump (Agilent, Santa Clara, CA). Chromatographic separation was performed on a Kinetex C18 analytical column (100 mm × 4.6 mm, 2.6 μm; Phenomenex, Torrance, CA) at 35 °C with the temperature of the auto-sampler set at 4 °C. For the solvent system, mobile phases A and B were 0.1% formic acid in de-ionized water and acetonitrile, respectively. The mobile phase was delivered at a flow rate of 0.35 ml/min, and the entire eluent was carried into a mass spectrometer. The linear gradient was as follows: 0% B at 0 min, 25% B at 14 min, 100% B at 23 min, 100% B at 28.50 min, 0% B at 29 min, and 0% B at 35 min.
An LTQ XL high performance linear ion trap mass spectrometer (Thermo Fisher Scientific Inc., Waltham, MA) equipped with an electrospray source was used in positive ion mode. The operating conditions of the mass spectrometer were as follows: 5 kV of ion spray voltage, heated capillary temperature of 275 °C, and sheath gas (nitrogen), auxiliary gas (nitrogen), and sweep gas (nitrogen) pressures of 35, 10, and 2 (arbitrary units), respectively. Full scanning analyses were performed in the range of m/z 75–1000, and a 35-V normalized collision energy was used for MS/MS. For neutral loss scanning, data-dependent scanning was started when the neutral loss of glucuronide or glycine was detected as a decrease in the m/z ratio of 176 (glucuronide) or 57 (glycine) Da. The chromatographic and mass spectral functions were controlled by Xcalibur software (Thermo Fisher Scientific Inc., Waltham, MA). The identification of metabolites was established using m/z values and MS/MS fragmentation patterns, which were compared with those in the Human Metabolome Project, METLIN, and MassBank databases. These identifications were further confirmed using standard samples for all but two metabolites (phenylalanylhydroxyproline and hydroxymethoxyindole glucuronide). For these two, ultra-high resolution MS spectra with 15T Fourier transform ion cyclotron resonance (with a resolution of 3,000,000 and an accuracy of 0.2 ppm) were obtained. The experiment gave the molecular formula for each measured m/z value (340.10269, C15H18NO8 for hydroxymethoxyindole glucuronide and 279.13393, C14H19N2O4 for phenylalanylhydroxyproline), and these matched nicely with the calculated monoisotopic masses of the metabolites (340.102693 and 279.133933, respectively).
Multivariate Data Analysis
All the obtained time domain NMR data were Fourier transformed, phase corrected, and baseline corrected manually. We used the region of 0.4–10.0 ppm, with the exclusion of water (4.6–5.0 ppm) and urea (5.6–6.0 ppm). 1H NMR chemical shifts were normalized against total integration values and 0.025% sodium-3-trimethylsily-[2,2,3,3-2H4]-1-propionate and then binned at a 0.044-ppm interval to reduce the complexity of the NMR data for pattern recognition. The MS raw data were processed using version 2.2 of the MZmine software (31). Peak detection was achieved by consecutively using the chromatogram builder and peak deconvolution functions. After the peak detection, peak lists of individual samples were aligned using the RANSAC aligner method. The retention time, m/z ratio, and peak height of the resulting peak list were then exported as a CSV file. The signals were then converted to an ASCII text file. The binning, normalization, and conversion were done using a Perl software written in-house. The resultant data sets were then imported into SIMCA-P, version 11.0 (Umetrics, Umeå, Sweden), and mean-centered with Pareto scaling for multivariate statistical analysis.
Reverse Transcription PCR and Western Blot Analysis
Total RNA was isolated from the liver tissues using the easy-spinTM Total RNA Extraction Kit (Intron, Seoul, Korea). The first-strand cDNA was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster, CA) according to manufacture instruction. The primer sequences of UGT1A, UGT2B, and GLYAT were as follows: UGT1A, sense 5′-ACACCGGAACTAGACCATCG-3′, antisense 3′-TTGGAACCCCATTGCATATT-5′; UGT2B, sense 5′-ATGCGCCACAAAGGGGC-3′, antisense 3′-GCAGGAATCCAATCACATCCAGAGAGTG-5′; GLYAT, sense 5′-CCATGGAAACCCATTCAATC-3′, antisense: 3′-GTGGGACTGGGAACTTTGAA-5′. The predicted sizes were 153 bp, 91 bp, and 223 bp, respectively. Beta-actin was used as a control, the sense and antisense primers of which were 5′-AGCCATGTACGTAGCCATCC-3′ and 3′-CTCTCAGCTGTGGTGGTGAA-5′, respectively. The predicted size was 228 bp. The PCR mixtures contained 1 μl of cDNA synthesized from 2 μg of total RNA, 3.2 μl of dNTP, 2 μl of 10 × ExTaq buffer, 0.1 μl of ExTaq DNA polymerase (Takara, Shiga, Japan), and 20 pmol each of sense and antisense primers from UGT1A, UGT2B, GLYAT, and beta-actin in a total volume of 20 μl. The PCR was performed with the following steps: initial denaturation at 95 °C for 5 min, followed by 33 cycles (denaturation at 95 °C for 1 min, annealing at 55 °C for 1 min, and extension at 72 °C for 1 min) and final extension at 72 °C for 10 min. The PCR products were separated on a 1% agarose gel.
For Western blots, 60 mg of liver tissue was measured and ground into powder using a mortar under liquid nitrogen. The powders of liver tissue were suspended in 1 ml of RIPA buffer containing protease inhibitor (2 μg/ml of Aprotinin, 1 μg/ml of Pepstatin, and 1 mm of PMSF) and put on ice for 30 min. After centrifugation at 13,000 rpm for 30 min, the supernatant containing the total protein released from the liver tissue was quantified using a BCATM Protein Assay Kit (Pierce, Appleton, WI). Twenty-five micrograms of total protein was subjected to 10% SDS-PAGE, and the resolved proteins were transferred to nitrocellulose membranes (Bio-Rad). The membrane was blotted with antibodies against UGT1A, UGT2B, multidrug resistance-associated protein 3 (MRP-3), NAD(P)H dehydrogenase 1 (NQO-1) (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA), Nrf-2 (1:100; Santa Cruz Biotechnology), and heme oxygenase-1 (HO-1) (1:1000; Enzo Life Sciences, Ann Arbor, MI), followed by treatment with anti-mouse, anti-goat, or anti-rabbit (1:10,000; Santa Cruz Biotechnology) secondary antibodies conjugated with HRP at room temperature for 1 h.
Liver Histopathology and Serum Biochemistry
After fixation for 48 h, liver tissues were embedded in paraffin according to routine procedures. Four-micrometer-thick sections were cut and stained with H&E and periodic acid-Schiff stains for histopathological evaluation. An expert pathologist at Inha University Hospital blindly analyzed the tissue slices. Serum alanine transaminase (ALT), aspartate transaminase (AST), alkaline phosphatase (ALP), triglyceride (TG), and low density lipoprotein (LDL) levels were measured using commercial kits at Inha University hospital (Incheon, Korea).
RESULTS
General Assessment of DR Effects
To ensure that our DR procedure led to a difference between the control and DR groups, we measured general parameters such as body weight, serum LDL, and serum TG (supplemental Fig. S1). Body weight showed time-dependent differences between the two groups. In addition, LDL and TG levels significantly decreased after 3 months of DR. Given that LDL and TG are important risk factors for age-associated diseases, our DR procedure established a clear difference between the control and DR groups.
NMR and LC/MS Metabolomics Analysis of DR Effects
Dietary intake is intimately related to metabolism, but the effects of DR on specific metabolic pathways, as related to the beneficial results, have been little explored. We measured the urine metabolomic profiles of the control and DR rat groups, because urine can reflect well the systemic effects of external stimuli and has been used extensively for organism-level metabolomics studies (13–15, 32, 33). We used both NMR and LC/MS to widen the metabolite coverage and cross-confirmation of detected metabolites.
Representative NMR spectra with identified metabolites (Figs. 1A and 1B) and an LC/MS chromatogram (Figs. 1C and 1D) are shown here. Although the overall signal profiles are quite similar for the two groups, there were seemingly specific signals in each group. To evaluate the statistical meaning of those signals and exclude possible confounding variables not related to the group difference, we applied the orthogonal projections to latent structure-discrimination analysis (OPLS-DA) multivariate analysis to all of the NMR and LC/MS data. The discrimination model for NMR and LC/MS could differentiate between the control and DR groups without any overlap (Figs. 2A and 2B) with the following statistical characteristics. For NMR, the model had one predictive and three orthogonal components with Q2(Y) = 0.637, R2(Y) = 0.936, and total R2(X) = 0.659, with 0.112 being predictive and 0.547 being orthogonal. For LC/MS, there were one predictive and two orthogonal components with Q2(Y) = 0.809, R2(Y) = 0.968, and total R2(X) = 0.508, with 0.205 being predictive and 0.303 orthogonal. These results indicate that the LC/MS approach performed slightly better in separating the class-specific signals from the confounding variables. Still, both models had quite high cross-validated predictability and goodness-of-fit values, meaning reliable differentiation between the groups.
Fig. 1.
Representative NMR and LC/MS spectra of urine collected from the control and DR groups. The NMR spectra were taken for urine samples containing 150 mm phosphate buffer (pH 7.4) and 0.025% sodium-3-trimethylsily-[2,2,3,3-2H4]-1-propionate as an internal standard for control (A) and DR (B) groups. The LC/MS experiments were performed with the injection of 5 μl urine from control (C) and DR (D) groups. The numbers on the spectra indicate assigned peaks corresponding to the metabolites listed in Table I. For NMR, the assignments were established using the spectra of the standard samples and the Chenomx database (Edmonton, Alberta, Canada).
Fig. 2.

Differentiation of control and DR groups using multivariate analysis. OPLS-DA score plot of control and DR groups from NMR (A) and LC/MS (B). Black circles: control group; open triangles: DR group. The models were established using one predictive and three orthogonal components for NMR, and one predictive and two orthogonal components for LC/MS.
Metabolites Related to DR
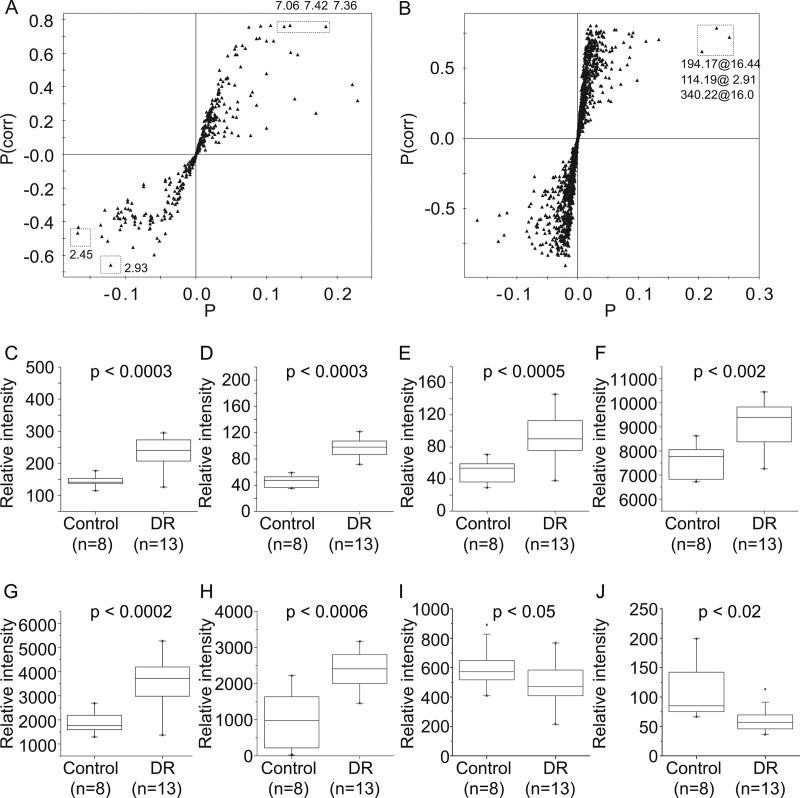
With the successful distinction between the control and DR groups, we tried to identify specific metabolites contributing to the difference. For this, we built the S-plot from the orthogonal projections to latent structure-discrimination analysis (OPLS-DA) model and picked the signals with high correlation and signal-to-noise ratio values. The distribution of the correlation and covariance values suggested that the DR group was represented by relatively conspicuous signals, whereas the control group was represented by the sum of weaker contributing signals (Figs. 3A and 3B). The analysis also showed that the NMR signals at 7.36, 7.42, 7.06, 2.45, and 2.93 ppm and LC/MS signals at m/z = 114.19, 194.17, and 340.22 were among the major contributors. The NMR spectral analysis and MS/MS analysis showed that these signals belonged to phenylacetylglycine, 1-methylhistidine, 2-oxoglutarate, N,N-dimethylglycine, creatinine, and hydroxymethoxyindole glucuronide (Table I). To confirm the significance of these metabolites found via the multivariate approach, we performed a Mann-Whitney U test on the levels of these metabolites (Figs. 3C–3J). The results reflected the statistical validity of these markers in the differentiation of the control and DR groups.
Fig. 3.
Marker signals contributing to the differentiation between the control and DR groups. S-plot analysis to identify the contributing signals for the control and DR groups from NMR (A) and LC/MS (B). P represents modeled covariation, and P(corr) represents modeled correlation. Potential marker signals that are significantly biased across the two groups are enclosed in boxes. The levels of the signals identified by the analysis were compared via Mann-Whitney U test, and the resulting p values are indicated. 7.36 ppm (C), 7.42 ppm (D) and 7.06 ppm (E) from NMR, m/z = 114.19 (F), m/z = 194.17 (G), and m/z = 340.22 (H) from LC/MS significantly increased in DR group; 2.45 ppm (I) and 2.93 ppm (J) from NMR decreased in DR group. The solid boxes represent the 25th and 75th percentile values with the median value inside. The whiskers represent outliers with a coefficient value of 1.5.
Table I. Metabolites identified via NMR and LC/MS analysis.
| Metabolites | Structural identifiers |
p value lower than | Fold change (%) | ||
|---|---|---|---|---|---|
| ppm (multiplicity) for NMR; m/z, Ret. time (min), daughter ions for LC/MS | |||||
| 1 | 1-methylhistidinea | 3.20(m), 3.25(m), 7.06(s) | 4.85 × 10−4 | 83.46 | ★ |
| 2 | 2-oxoglutarate | 2.45(t), 3.02(t) | 0.045 | −22.60 | ★ |
| 3 | 3-indoxylsulfate | 7.20(t), 7.28(t), 7.37(s), 7.51(d), 7.70(d) | 0.054 | 49.86 | |
| 4 | 3-methylglutarate | 0.93(d), 2.01(d), 2.26(m) | 0.943 | −0.52 | |
| 5 | Acetate | 1.93(s) | 0.750 | 7.01 | |
| 6 | Acetoacetate | 2.30(s), 3.52(s) | 0.012 | 31.23 | ★ |
| 7 | Adeninea | 8.32(s), 8.38(s) | 0.710 | 16.23 | |
| 8 | Alaninea | 1.49(d), 3.80(m) | 0.063 | −12.44 | |
| 9 | Allantoina | 5.38(s), 6.07(s) | 0.129 | 23.92 | |
| 10 | Argininea | 1.68(m), 1.93(m) | 0.036 | −9.42 | ★ |
| 11 | Citratea | 2.55(d), 2.69(d) | 0.078 | −47.57 | |
| 12 | Dimethylamine | 2.73(s) | 0.273 | −8.13 | |
| 13 | Ethanol | 1.17(t), 3.63(q) | 0.140 | 14.60 | |
| 14 | Formate | 8.47(s) | 0.292 | −19.42 | |
| 15 | Fucose | 1.21(d), 1.24(d) | 0.798 | −1.68 | |
| 16 | Glycine | 3.60(s) | 0.704 | −22.13 | |
| 17 | Guanidoacetatea | 3.80(s) | 0.228 | −10.42 | |
| 18 | Hippuratea | 3.97(d), 7.55(t), 7.64(t), 7.84(d), 8.54(s) | 0.856 | 2.62 | |
| 19 | Indole-3-acetatea | 7.13–7.30(m), 7.51(d) | 0.093 | 24.79 | |
| 20 | Lactatea | 1.35(d), 4.13(m) | 0.037 | −14.69 | ★ |
| 21 | N,N-dimethylglycinea | 2.93(s), 3.73(s) | 0.018 | −45.23 | ★ |
| 22 | N-acetylaspartate | 2.04(s), 2.53(m), 2.70(m), 4.40(m) | 0.251 | −9.28 | |
| 23 | Pyridoxinea | 2.41(s), 7.68(s) | 0.243 | −20.99 | |
| 24 | Pyruvate | 2.38(s) | 0.714 | 5.99 | |
| 25 | Succinatea | 2.41(s) | 0.243 | −20.99 | |
| 26 | Taurinea | 3.28(t), 3.43(t) | 0.064 | 49.03 | |
| 27 | Trigonellinea | 4.44(s), 8.84(t), 9.13(s) | 0.869 | 6.79 | |
| 28 | Tyrosinea | 6.87(d), 7.19(d) | 0.245 | −10.08 | |
| 29 | Uracila,b | 5.72(s) | |||
| 30 | Ureab | 5.81(s) | |||
| 31 | p-cresol | 2.25(s), 6.87(d) | 0.393 | −5.89 | |
| 32 | Trans-aconitate | 3.49(s), 6.53(s) | 0.531 | −6.79 | |
| 33 | Methylhippurate | 249.20 17.55 249, 119, 105 | 3.03 × 10−5 | 138.81 | ★ |
| 34 | Phenylacetylglycinea | 194.17 16.44 176, 76, 148, 91 | 1.32 × 10−4 | 86.78 | ★ |
| 3.66(s), 3.78(d), 7.36(t), 7.42(t) (ppm) | |||||
| 35 | Creatininea | 114.19 2.91 114, 86 | 1.44 × 10−3 | 18.90 | ★ |
| 3.05(s), 4.05(s) (ppm) | |||||
| 36 | hydroxymethoxyindole glucuronide | 340.22 16.0 164, 146, 122, 118 | 5.13 × 10−4 | 103.78 | ★ |
| 37 | Methoxytyrosine | 212.16 3.947 194, 166, 153, 109 | 8.03 × 10−4 | −11.15 | ★ |
The associated signal numbers (see Fig. 1), structural identifiers (NMR: ppm and multiplicity; MS: m/z, ret. time and daughter ions), and Mann-Whitney U-test (stars with p < 0.05) are presented. The fold-change values are from area-normalized peak intensities and represent percent changes in the DR group with respect to the control values, with negative values for decreases and positive for increases. All of the NMR-identified metabolites and LC/MS-identified metabolites with significant changes are included. Additional metabolites identified via LC/MS without significant changes are listed in supplementary Table S1. All the metabolites were confirmed with standard compounds and MS/MS analysis, except for two that were confirmed via 15T Fourier transform ion cyclotron resonance (see “Experimental Procedures” for details).
a Detected in both NMR and LC/MS studies.
b Not used in the normalization process.
Profiling of General Glucuronide and Glycine Conjugation via Neutral Loss Scanning
Two metabolites with high levels in the DR group, hydroxymethoxyindole glucuronide and phenylacetylglycine, turned out to be common products of phase II detoxification reactions, created through glucuronide and glycine conjugation mechanisms, respectively. As there could be many other conjugation products from these pathways, we decided to measure general profiles of compounds that have conjugated glucuronide or glycine. LC/MS enables the detection of these profiles through neutral loss scanning of 176 (glucuronide) and 57 (glycine). Fig. 4 shows that the peaks for the DR group were higher in number and intensity. Statistical analysis with Student's t test on the peak area showed that the general profiles of both glucuronide and glycine conjugated metabolites were enhanced in the DR group, with p values of 0.017 or 0.011, respectively, confirming that the DR group had higher activity in these pathways.
Fig. 4.
Neutral loss scanning for general assessment of glucuronide and glycine conjugation reactions. Glucuronide (A, B) and glycine (C, D) conjugation profiling based on neutral loss scanning of m/z 176 (glucuronide) and 57 (glycine) in control (A, C) and DR (B, D) groups. The MS/MS step was carried out using a 35-V normalized collision energy. The plots show the retention times and intensities of compounds that experience the loss of a specified common neutral (m/z 176 and 57) fragment. The numbers on the peaks indicate tentative assignments of conjugated metabolites (1: tyramine glucuronide; 2: indole acetylglycine).
Assessment of Phase II Detoxification Pathways in the Liver
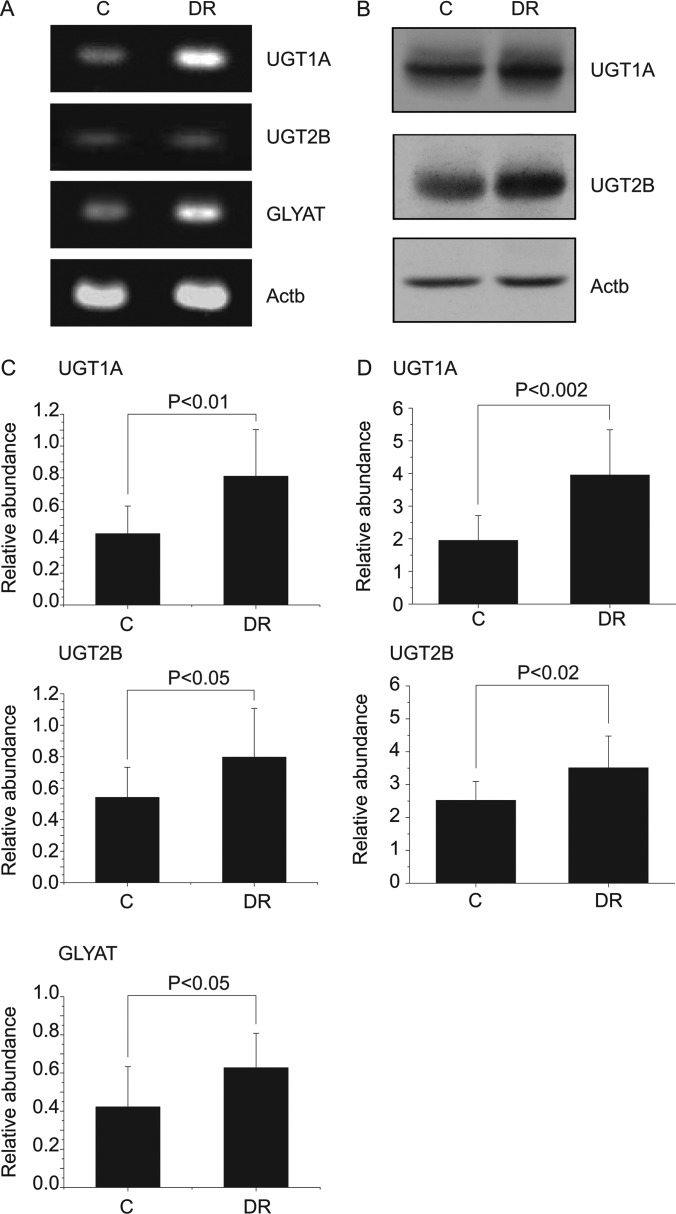
As the urine metabolite profile showed generally enhanced conjugation reactions, we tried to confirm that the differences also manifested in the actual tissues. For that, we first measured the levels of the two key markers, hydroxymethoxyindole glucuronide and phenylacetylglycine, in the liver tissue, as the liver is the main organ for the phase II detoxification pathways. The LC/MS measurement of the levels of these metabolites showed that they were elevated in the livers of the DR group (supplemental Fig. S2), indicating that the urine metabolomics results adequately reflected the metabolic changes in the liver tissues. For more detailed pathway analysis, we investigated the enhanced conjugation reactions at the metabolic enzyme level. We compared the mRNA levels of UGT1A, UGT2B, and GLYAT, primary enzymes for glucuronide and glycine conjugation reactions, respectively. Figs. 5A and 5C show that all the enzymes had significantly enhanced mRNA expression in the DR group. Then, we measured the protein levels of these enzymes via Western blot. Both UGT1A and UGT2B showed similarly enhanced protein levels in the DR group (Figs. 5B and 5D), consistent with the mRNA level. Although the GLYAT level could not be directly measured because of the failure of all available antibodies (data not shown), the increased mRNA level should reflect the protein level, because phase II enzymes are generally transcriptionally regulated (24), as shown for UGT1A and UGT2B. These data confirm that phase II detoxification pathways, particularly glucuronide and glycine conjugation, in the relevant organ (the liver) were up-regulated in the DR group.
Fig. 5.
Levels of the phase II detoxification enzymes for glucuronide and glycine conjugation in liver tissues. Reverse transcription PCR (A) and Western blot (B) represent the mRNA and the expressed protein levels of the phase II detoxification enzymes for glucuronide and glycine conjugation, such as uridinediphospho-glucuronosyltransferase 1A (UGT1A) and 2B (UGT2B) and glycine-N-acyltransferase (GLYAT), in liver tissues. Beta-actin (Actb) was used as a control. Bar charts represent the comparison of the mean band intensities for the levels of phase II enzymes in terms of mRNA (C) and protein (D) normalized to that of the Actb. Statistical analysis was performed using Student's t test, and the resulting p values are indicated. Error bars represent standard deviation. All the animals for which metabolomics data were obtained were tested (control, n = 8; DR, n = 13). A Western blot result for GLYAT could not be obtained, as none of the available antibodies reacted with rat GLYAT.
Biochemical and Histopathological Changes in the Liver
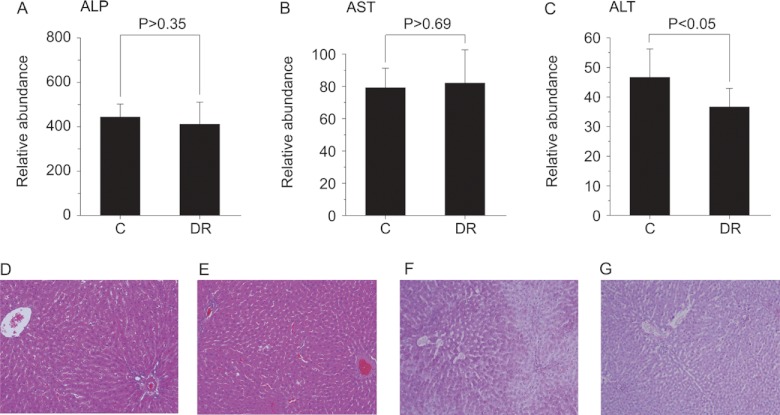
We next investigated the effects of DR on the integrity of the liver via blood biochemistry and direct tissue staining. We measured alkaline phosphatase (ALP), aspartate transaminase (AST), and alanine transaminase (ALT) levels, which are commonly used to assess the functional integrity of liver cells (Figs. 6A–6C). ALP and AST levels were similar in both groups, but there was small, yet significant, decrease in ALT level in the DR group. As ALT is more specific to liver than the others, this finding suggested that the liver cells in the DR group were somewhat healthier in a biochemical sense. We also looked at the changes in the actual tissue via histopathological staining. H&E staining showed no gross pathophysiological differences between groups (Figs. 6D and 6E), but the DR group exhibited noticeably denser cytoplasm, which might have been due to a decrease in glycogen granules. To test this, we performed periodic acid-Schiff staining, and the DR group showed glycogen depletion relative to the control group (Figs. 6F and 6G). This might well be due to a reduced insulin level, which is one of the well-documented beneficial effects of DR (34–36). These results indicate that DR induces beneficial biochemical and metabolic changes in the liver.
Fig. 6.
Blood biochemistry and histopathological staining. Serum alkaline phosphatase (ALP), aspartate transaminase (AST), and alanine transaminase (ALT) activities were measured using commercial kits employing spectrophotometric assays. Average values of the ALP (A), AST (B), and ALT (C) levels of each group are plotted, along with their standard deviations. Student's t test was also performed, and the resulting p values are indicated. Error bars represent standard deviation. All the animals for which metabolomics data were obtained were tested (control, n = 8; DR, n = 13). H&E and periodic acid-Schiff staining of liver tissues were performed on paraffin blocks of the samples: H&E staining control (D) and DR (E) groups (200×); periodic acid-Schiff staining control (F) and DR (G) groups (100×).
Up-regulation of Nrf-2 Signaling Pathway
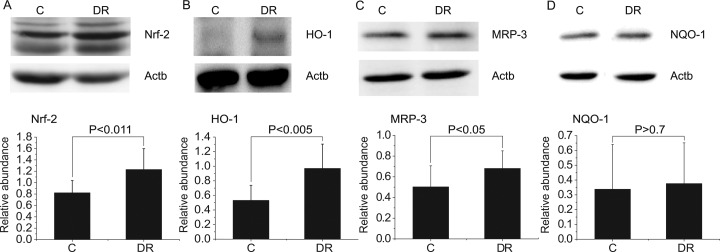
One of the important signaling pathways known to activate phase II detoxification metabolism is the Nrf-2 pathway (37). Nrf-2 is also important in protecting cells from oxidative stress (38). When activated, Nrf-2 translocates into the nucleus and regulates its downstream targets, such as HO-1, MRP-3, and NQO-1. To compare the activation status of the Nrf-2 pathway in the control and DR groups, we measured the expression levels of Nrf-2 and its downstream targets in the liver tissue. Fig. 7 shows that Nrf-2, along with its downstream targets (except NQO-1), was elevated in the DR group, showing that the Nrf-2 signaling pathway was up-regulated in the DR group. These results suggest that enhanced phase II detoxification in the liver tissue occur in the DR group, at least in part, through activation of the Nrf-2 pathway.
Fig. 7.
Up-regulation of the Nrf-2 signaling pathway. Western blots of Nrf-2 (A), HO-1 (B), MRP-3 (C), and NQO-1 (D) from the liver homogenates of the control and DR groups. Beta-actin (Actb) was used as a loading control. Bar charts below the blots represent the comparison of the mean band intensities for the levels of the proteins normalized to that of the Actb. Statistical analysis was performed using Student's t test, and the resulting p values are indicated. Error bars represent standard deviation. All the animals for which metabolomics data were obtained were tested (control, n = 8; DR, n = 13).
DISCUSSION
In identifying the marker metabolites for DR, we employed NMR and LC/MS approaches, most often used independently in metabolomics studies. NMR has merits for quantitation and reproducibility, and LC/MS in terms of sensitivity (39). Here, the use of both techniques expanded the coverage of the markers and gave added reliability to them. For example, hydroxymethoxyindole glucuronide, which gave an important clue regarding glucuronidation, was detected via LC/MS, and phenylacetylglycine, which led to the idea of enhanced glycine conjugation, was detected with both techniques. Creatinine was also detected by both methods, and 1-methylhistidine was observed in NMR studies.
Through a urine metabolomics approach, we showed that the creatinine level increased in the DR group, consistent with previous results (19, 20). The urine creatinine level has been used as a marker for kidney function, but the increase in our results seems to be due to an increase in muscle protein turnover in the DR condition rather than kidney impairment, as the amounts of collected urine, which often decrease in conditions with decreased urinary creatinine, were similar for both groups. Furthermore, we found that the level of 1-methylhistidine, which is produced from muscular actin and myosin, was higher in the DR group. Therefore, these two metabolic markers do not seem to contradict the idea of a beneficial effects of DR.
From a biological perspective, an important aspect of the current study is the finding of enhanced phase II metabolism and Nrf-2 pathway activation in the liver tissue of the DR group. The initial clue came from the metabolomic identification of increased hydroxymethoxyindole glucuronide and phenylacetylglycine in urine from the DR group. Although there have been isolated reports on the relationship between DR and UGT activities, those might be limited by their study of particular isotypes with the use of non-dietary chemical substrates (40, 41) or targeted investigation of DR's effects on those UGTs for a short period of time (42). Our non-targeted metabolomics study did not set any prior assumptions about DR's effects and generated a hypothesis of increased phase II reactions based on the identified markers in larger number of animals (n = 8 for control and 13 for DR). In addition, the hypothesis of a general increase in glucuronide and glycine conjugation was proven through global profiling of the conjugated metabolites with LC/MS-based neutral loss scanning, which is not limited to particular isotypes and products. Equally important, we addressed the mechanism of DR's effects by finding that the Nrf-2 pathway is up-regulated by DR, leading to phase II enhancement. Phase II enzymes, such as UGT and GLYAT, are essentially detoxification enzymes, and therefore it makes good sense that enhanced urinary elimination of toxic metabolites contributes to the beneficial effects of DR. In our experimental frame, the decreased serum ALT might well reflect protective effects on the liver cells by the increased phase II detoxification.
Specific life-beneficial roles of the phase II detoxification pathway can be also appreciated by its well-documented involvement in cancer prevention (24–26). It has been suggested that dietary flavonoids or unsaturated antioxidants can prevent a variety of cancers, which can be attributed to enhanced phase II metabolism. For example, limonene and sobrerol prevented the initiation of carcinogenesis in a rat breast cancer model by increasing phase II enzyme activities (43), and garden cress ingredients inhibited the formation of pre-neoplastic regions in chemically insulted rat colons through enhanced UGT activities (44). Therefore, the enhanced phase II pathway is likely to contribute to decreased cancer incidence in a future years-long experiment, although it was not directly measured in the current months-long experiment.
Very interesting species differences in phase II metabolism also support its importance, especially in terms of glucuronidation, in maintaining healthy life in living organisms that experience constant exposure to xenobiotics through their lifetime. Cats are very sensitive to drugs and toxins at levels not toxic to other species (45, 46). Notably, they are quite susceptible to the toxicity of acetaminophen, a widely used analgesic for humans. This species-unique toxicity of xenobiotics was found to be due to the unusually low activity of UGT in cats (47, 48). Although generally safe to humans, acetaminophen can be quite toxic to a minor population of humans, which also might be attributable to variable glucuronidation of the drug among individuals (49, 50). Considering the above and the well-known importance of the phase II pathways in detoxifying general xenobiotics, enhanced phase II detoxification could have a role in the mean life span of a population.
For the invertebrate C. elegans, enhanced phase II detoxification, to which UGT is a major contributor (23), was actually suggested to be responsible for anti-aging and life span extension (51), which are important beneficial effects of DR. It also has been suggested that an important cause of aging is molecular damage caused by toxic metabolites, and that enzymes involved in detoxification systems contribute to longevity assurance (52). The roles of enzymes in detoxification systems on longevity assurance also has been suggested by DNA microarray analysis of daf-2(-) worms (53). In addition, the Nrf-2 ortholog SKN-1, one of the longevity-assurance genes, regulates the expression of candidate phase II genes in C. elegans (54) and is required for life span extension by DR (55, 59).
For higher organisms, the roles of phase II detoxification in DR's beneficial effects have been much less studied. In addition, the relationships among the Nrf-2 pathway, its downstream target genes, and DR's effects are more complex. For example, one study suggested that the Nrf-2 pathway should be required for reducing carcinogen-induced tumor formation in the DR condition, but that it should not be required for life extension or increased insulin sensitivity (56). In addition, it was shown that Nrf-2 downstream effectors, such as NQO-1, HO-1, the glutamate-cysteine ligase catalytic subunit, and GST are not uniformly enhanced in the DR condition. Therefore, individual effector molecules might need to be evaluated in order to address a coherent pathway from DR and its beneficial effects through Nrf-2 signaling. Our observation of insignificant changes in NQO-1 may be understood in this sense. It seems that the increase in Nrf-2 and its targets HO-1 and MRP-3 might be the specific signature of DR-related Nrf-2 pathway activation. In particular, the elevation of the HO-1 level can be easily related to the beneficial effects of DR, as it protects cells from a variety of diseases caused by reactive oxygen species and inflammation by producing anti-oxidant and anti-inflammatory substances such as carbon monoxide and biliverdin.
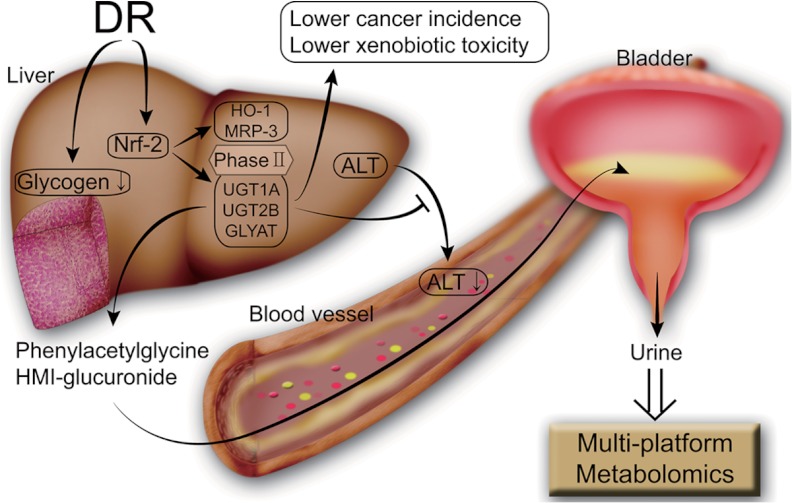
We observed many effects of DR related to liver functions (lower serum LDL and TG levels, lower glycogen, and reduced ALT level) that can ultimately contribute to long-term health. Our metabolomics and biochemical results show that the enhanced phase II detoxification in the liver of the DR group correlates well with the up-regulation of the Nrf-2 signaling pathway by DR. We showed the activation of Nrf-2 pathway by measuring the protein level of Nrf-2 itself and of its downstream target genes HO-1 and MRP-3. Specific correlation was also established by directly measuring the UGT and GLYAT levels of the liver tissue. Collectively, we suggest that DR up-regulates the Nrf-2 that turns up phase II detoxification genes in liver, which contributes to the beneficial effects of the DR (Fig. 8).
Fig. 8.
Overall pathways for DR's beneficial effects inferred from the metabolomics study. The DR procedure up-regulates phase II detoxification enzymes through the Nrf-2 pathway, and the products of this phase II metabolism were detected using a multi-platform metabolomics approach. The enhanced phase II metabolism could be responsible for various beneficial effects of DR such as lower ALT levels, reduced glycogen granules, reduced cancer incidence (58), and lower xenobiotic toxicity (37) as reported here and elsewhere.
Given the complexity of the mechanism of DR, which has hindered its detailed investigation for decades, we admit that the phase II detoxification pathway may not account for all of DR's beneficial effects, and that there should be other important metabolic pathways that could not be addressed by looking at urine metabolic profiles. For example, increased GST conjugation, suggested to be important in anti-aging because of its electrophile-scavenging role (57), was not detected in this study with significance. It might be that we did not use particular electrophile-generating compounds, or that UGT conjugation is a much more prevalent form of detoxification than others (i.e. GST conjugation) for the excretion of toxic materials (23). However, we note that GST A1 expression did not increase to a statistically significant degree in the DR group in a previous study (56), consistent with our results. In this sense, blood, multi-organ tissue extracts, or intact tissues themselves could be the next tier of targets for studies seeking a more systems-level understanding of the mechanism of DR. As DR essentially perturbs metabolism, studies of DR's effects with metabolomics, followed by the investigation of changes at protein and gene expression levels, as laid out in the current study, might be one efficient way to solve this complex puzzle.
Supplementary Material
Footnotes
* This study was supported by grants from the National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea (1120300); the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2011-0005826 and 2012-0009369); and the Korea Healthcare technology R&D Project, Ministry for Health & Welfare, Republic of Korea (A092006) to S.P. This research was also supported by a grant from the Basic Science Research Program through the NRF, funded by the Ministry of Education, Science and Technology (2011–0029572), to S.-w.P. The NMR facility at the Korea Basic Science Institute is supported by the Bio-MR Research Program of the Korean Ministry of Science and Technology (E29070).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ALP
- alkaline phosphatase
- ALT
- alanine transaminase
- AST
- aspartate transaminase
- DR
- dietary restriction
- GLYAT
- glycine-N-acyltransferase
- GST
- glutathione-S-transferase
- HO-1
- heme oxygenase-1
- LDL
- low density lipoprotein
- MRP-3
- multidrug resistance-associated protein 3
- Nrf-2
- Nuclear factor (erythroid-derived 2)-like 2
- NQO-1
- NAD(P)H dehydrogenase 1
- TG
- triglyceride
- UGT
- uridinediphospho-glucuronosyltransferase.
REFERENCES
- 1. McCay C. M., Crowell M. F., Maynard L. (1935) The effect of retarded growth upon the length of life span and upon the ultimate body size. J. Nutr. 10, 63–79 [PubMed] [Google Scholar]
- 2. Weindruch R., Walford R. L. (1988) The Retardation of Aging and Disease by Dietary Restriction, C.C. Thomas, Springfield, IL [Google Scholar]
- 3. Fontana L., Partridge L., Longo V. D. (2010) Extending healthy life span—from yeast to humans. Science 328, 321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Colman R. J., Anderson R. M., Johnson S. C., Kastman E. K., Kosmatka K. J., Beasley T. M., Allison D. B., Cruzen C., Simmons H. A., Kemnitz J. W., Weindruch R. (2009) Caloric restriction delays disease onset and mortality in rhesus monkeys. Science 325, 201–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Willcox B. J., Willcox D. C., Todoriki H., Fujiyoshi A., Yano K., He Q., Curb J. D., Suzuki M. (2007) Caloric restriction, the traditional Okinawan diet, and healthy aging: the diet of the world's longest-lived people and its potential impact on morbidity and life span. Ann. N.Y. Acad. Sci. 1114, 434–455 [DOI] [PubMed] [Google Scholar]
- 6. Holloszy J. O., Fontana L. (2007) Caloric restriction in humans. Exp. Gerontol. 42, 709–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heilbronn L. K., de Jonge L., Frisard M. I., DeLany J. P., Larson-Meyer D. E., Rood J., Nguyen T., Martin C. K., Volaufova J., Most M. M., Greenway F. L., Smith S. R., Deutsch W. A., Williamson D. A., Ravussin E. (2006) Effect of 6-month calorie restriction on biomarkers of longevity, metabolic adaptation, and oxidative stress in overweight individuals: a randomized controlled trial. JAMA 295, 1539–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mair W., Dillin A. (2008) Aging and survival: the genetics of life span extension by dietary restriction. Annu. Rev. Biochem. 77, 727–754 [DOI] [PubMed] [Google Scholar]
- 9. Patti G. J., Yanes O., Siuzdak G. (2012) Metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 13, 263–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nicholson J. K., Wilson I. D., Lindon J. C. (2011) Pharmacometabonomics as an effector for personalized medicine. Pharmacogenomics 12, 103–111 [DOI] [PubMed] [Google Scholar]
- 11. Nicholson J. K., Lindon J. C. (2008) Systems biology: metabonomics. Nature 455, 1054–1056 [DOI] [PubMed] [Google Scholar]
- 12. Yang H. J., Choi M. J., Wen H., Kwon H. N., Jung K. H., Hong S. W., Kim J. M., Hong S. S., Park S. (2011) An effective assessment of simvastatin-induced toxicity with NMR-based metabonomics approach. PloS One 6, e16641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kwon H. N., Kim M., Wen H., Kang S., Yang H. J., Choi M. J., Lee H. S., Choi D., Park I. S., Suh Y. J., Hong S. S., Park S. (2011) Predicting idiopathic toxicity of cisplatin by a pharmacometabonomic approach. Kidney Int. 79, 529–537 [DOI] [PubMed] [Google Scholar]
- 14. Clayton T. A., Baker D., Lindon J. C., Everett J. R., Nicholson J. K. (2009) Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. U.S.A. 106, 14728–14733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Manna S. K., Patterson A. D., Yang Q., Krausz K. W., Idle J. R., Fornace A. J., Gonzalez F. J. (2011) UPLC-MS-based urine metabolomics reveals indole-3-lactic acid and phenyllactic acid as conserved biomarkers for alcohol-induced liver disease in the Ppara-null mouse model. J. Proteome Res. 10, 4120–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liang X., Chen X., Liang Q., Zhang H., Hu P., Wang Y., Luo G. (2011) Metabonomic study of Chinese medicine Shuanglong formula as an effective treatment for myocardial infarction in rats. J. Proteome Res. 10, 790–799 [DOI] [PubMed] [Google Scholar]
- 17. Edmands W. M., Beckonert O. P., Stella C., Campbell A., Lake B. G., Lindon J. C., Holmes E., Gooderham N. J. (2011) Identification of human urinary biomarkers of cruciferous vegetable consumption by metabonomic profiling. J. Proteome Res. 10, 4513–4521 [DOI] [PubMed] [Google Scholar]
- 18. Nicholson J. K., Lindon J. C., Holmes E. (1999) “Metabonomics”: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 29, 1181–1189 [DOI] [PubMed] [Google Scholar]
- 19. Zhang Y., Yan S., Gao X., Xiong X., Dai W., Liu X., Li L., Zhang W., Mei C. (2012) Analysis of urinary metabolic profile in aging rats undergoing caloric restriction. Aging Clin. Exp. Res. 24, 79–84 [DOI] [PubMed] [Google Scholar]
- 20. Wang Y., Lawler D., Larson B., Ramadan Z., Kochhar S., Holmes E., Nicholson J. K. (2007) Metabonomic investigations of aging and caloric restriction in a life-long dog study. J. Proteome Res. 6, 1846–1854 [DOI] [PubMed] [Google Scholar]
- 21. Zimniak P. (2008) Detoxification reactions: relevance to aging. Ageing Res. Rev. 7, 281–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu C., Li C. Y., Kong A. N. (2005) Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch. Pharm. Res. 28, 249–268 [DOI] [PubMed] [Google Scholar]
- 23. Gibson G. G., Skett P. (2001) Introduction to Drug Metabolism, 3rd ed., Nelson Thornes, Bath, UK [Google Scholar]
- 24. Jana S., Mandlekar S. (2009) Role of phase II drug metabolizing enzymes in cancer chemoprevention. Curr. Drug Metab. 10, 595–616 [DOI] [PubMed] [Google Scholar]
- 25. Gullett N. P., Ruhul Amin A. R., Bayraktar S., Pezzuto J. M., Shin D. M., Khuri F. R., Aggarwal B. B., Surh Y. J., Kucuk O. (2010) Cancer prevention with natural compounds. Semin. Oncol. 37, 258–281 [DOI] [PubMed] [Google Scholar]
- 26. Sporn M. B., Suh N. (2002) Chemoprevention: an essential approach to controlling cancer. Nat. Rev. Cancer 2, 537–543 [DOI] [PubMed] [Google Scholar]
- 27. Pugh T. D., Klopp R. G., Weindruch R. (1999) Controlling caloric consumption: protocols for rodents and rhesus monkeys. Neurobiol. Aging 20, 157–165 [DOI] [PubMed] [Google Scholar]
- 28. Kang J., Choi M. Y., Kang S., Kwon H. N., Wen H., Lee C. H., Park M., Wiklund S., Kim H. J., Kwon S. W., Park S. (2008) Application of a 1H nuclear magnetic resonance (NMR) metabolomics approach combined with orthogonal projections to latent structure-discriminant analysis as an efficient tool for discriminating between Korean and Chinese herbal medicines. J. Agric. Food Chem. 56, 11589–11595 [DOI] [PubMed] [Google Scholar]
- 29. Kang J., Lee S., Kang S., Kwon H. N., Park J. H., Kwon S. W., Park S. (2008) NMR-based metabolomics approach for the differentiation of ginseng (Panax ginseng) roots from different origins. Arch. Pharm. Res. 31, 330–336 [DOI] [PubMed] [Google Scholar]
- 30. Wen H., Kang S., Song Y., Song Y., Sung S. H., Park S. (2010) Differentiation of cultivation sources of Ganoderma lucidum by NMR-based metabolomics approach. Phytochem. Anal. 21, 73–79 [DOI] [PubMed] [Google Scholar]
- 31. Pluskal T., Castillo S., Villar-Briones A., Oresic M. (2010) MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim K., Aronov P., Zakharkin S. O., Anderson D., Perroud B., Thompson I. M., Weiss R. H. (2009) Urine metabolomic analysis for kidney cancer detection and biomarker discovery. Mol. Cell. Proteomics 8, 558–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Legido-Quigley C., Stella C., Perez-Jimenez F., Lopez-Miranda J., Ordovas J., Powell J., van-der-Ouderaa F., Ware L., Lindon J. C., Nicholson J. K., Holmes E. (2009) Liquid chromatography-mass spectrometry methods for urinary biomarker detection in metabonomic studies with application to nutritional studies. Biomed. Chromatogr. 24, 737–743 [DOI] [PubMed] [Google Scholar]
- 34. Witte A. V., Fobker M., Gellner R., Knecht S., Flöel A. (2009) Caloric restriction improves memory in elderly humans. Proc. Natl. Acad. Sci. U.S.A. 106, 1255–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fontana L., Meyer T. E., Klein S., Holloszy J. O. (2004) Long-term calorie restriction is highly effective in reducing the risk for atherosclerosis in humans. Proc. Natl. Acad. Sci. U.S.A. 101, 6659–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ramsey J. J., Colman R. J., Binkley N. C., Christensen J. D., Gresl T. A., Kemnitz J. W., Weindruch R. (2000) Dietary restriction and aging in rhesus monkeys: the University of Wisconsin study. Exp. Gerontol. 35, 1131–1149 [DOI] [PubMed] [Google Scholar]
- 37. Shen G., Kong A.-N. (2009) Nrf2 plays an important role in coordinated regulation of Phase II drug metabolism enzymes and Phase III drug transporters. Biopharm. Drug Dispos. 30, 345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Taguchi K., Motohashi H., Yamamoto M. (2011) Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells 16, 123–140 [DOI] [PubMed] [Google Scholar]
- 39. Nicholson J. K., Connelly J., Lindon J. C., Holmes E. (2002) Metabonomics: a platform for studying drug toxicity and gene function. Nat. Rev. Drug Discov. 1, 153–161 [DOI] [PubMed] [Google Scholar]
- 40. Rajpurohit R., Krishnaswamy K. (1985) Effect of chronic undernutrition on glucuronide and glutathione conjugation in rat liver. Drug Nutr. Interact. 3, 121–128 [PubMed] [Google Scholar]
- 41. Wall K. L., Gao W., Qu W., Kwei G., Kauffman F. C., Thurman R. G. (1992) Food restriction increases detoxification of polycyclic aromatic hydrocarbons in the rat. Carcinogenesis 13, 519–523 [DOI] [PubMed] [Google Scholar]
- 42. Yu C., Ritter J. K., Krieg R. J., Rege B., Karnes H. T., Sarka M. A. (2004) Effect of food restriction on UDP-glucuronosyltransferase (UGT) enzymes in liver. Clin. Pharmacol. Ther. 75, P85 [Google Scholar]
- 43. Elegbede J. A., Maltzman T. H., Elson C. E., Gould M. N. (1993) Effects of anticarcinogenic monoterpenes on phase II hepatic metabolizing enzymes. Carcinogenesis 14, 1221–1223 [DOI] [PubMed] [Google Scholar]
- 44. Kassie F., Rabot S., Uhl M., Huber W., Qin H. M., Helma C., Schulte-Hermann R., Knasmuller S. (2002) Chemoprotective effects of garden cress (Lepidium sativum) and its constituents towards 2-amino-3-methyl-imidazo[4,5-f]quinoline (IQ)-induced genotoxic effects and colonic preneoplastic lesions. Carcinogenesis 23, 1155–1161 [DOI] [PubMed] [Google Scholar]
- 45. Watkins J. B., 3rd, Klaassen C. D. (1986) Xenobiotic biotransformation in livestock: comparison to other species commonly used in toxicity testing. J. Anim. Sci. 63, 933–942 [DOI] [PubMed] [Google Scholar]
- 46. Wilcke J. R. (1984) Idiosyncracies of drug metabolism in cats. Effects on pharmacotherapeutics in feline practice. Veterinary Clin. North Am. Small Anim. Pract. 14, 1345–1354 [DOI] [PubMed] [Google Scholar]
- 47. Court M. H., Greenblatt D. J. (1997) Biochemical basis for deficient paracetamol glucuronidation in cats: an interspecies comparison of enzyme constraint in liver microsomes. J. Pharm. Pharmacol. 49, 446–449 [DOI] [PubMed] [Google Scholar]
- 48. Court M. H., Greenblatt D. J. (2000) Molecular genetic basis for deficient acetaminophen glucuronidation by cats: UGT1A6 is a pseudogene, and evidence for reduced diversity of expressed hepatic UGT1A isoforms. Pharmacogenetics 10, 355–369 [DOI] [PubMed] [Google Scholar]
- 49. Court M. H., Duan S. X., von Moltke L. L., Greenblatt D. J., Patten C. J., Miners J. O., Mackenzie P. I. (2001) Interindividual variability in acetaminophen glucuronidation by human liver microsomes: identification of relevant acetaminophen UDP-glucuronosyltransferase isoforms. J. Pharmacol. Exp. Ther. 299, 998–1006 [PubMed] [Google Scholar]
- 50. Mutlib A. E., Goosen T. C., Bauman J. N., Williams J. A., Kulkarni S., Kostrubsky S. (2006) Kinetics of acetaminophen glucuronidation by UDP-glucuronosyltransferases 1A1, 1A6, 1A9 and 2B15. Potential implications in acetaminophen-induced hepatotoxicity. Chem. Res. Toxicol. 19, 701–709 [DOI] [PubMed] [Google Scholar]
- 51. McElwee J. J., Schuster E., Blanc E., Thomas J. H., Gems D. (2004) Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J. Biol. Chem. 279, 44533–44543 [DOI] [PubMed] [Google Scholar]
- 52. Gems D., McElwee J. J. (2005) Broad spectrum detoxification: the major longevity assurance process regulated by insulin/IGF-1 signaling? Mech. Ageing Dev. 126, 381–387 [DOI] [PubMed] [Google Scholar]
- 53. Murphy C. T., McCarroll S. A., Bargmann C. I., Fraser A., Kamath R. S., Ahringer J., Li H., Kenyon C. (2003) Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277–283 [DOI] [PubMed] [Google Scholar]
- 54. Oliveira R. P., Porter Abate J., Dilks K., Landis J., Ashraf J., Murphy C. T., Blackwell T. K. (2009) Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell 8, 524–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bishop N. A., Guarente L. (2007) Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature 447, 545–549 [DOI] [PubMed] [Google Scholar]
- 56. Pearson K. J., Lewis K. N., Price N. L., Chang J. W., Perez E., Cascajo M. V., Tamashiro K. L., Poosala S., Csiszar A., Ungvari Z., Kensler T. W., Yamamoto M., Egan J. M., Longo D. L., Ingram D. K., Navas P., de Cabo R. (2008) Nrf2 mediates cancer protection but not prolongevity induced by caloric restriction. Proc. Natl. Acad. Sci. U.S.A. 105, 2325–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McElwee J. J., Schuster E., Blanc E., Piper M. D., Thomas J. H., Patel D. S., Selman C., Withers D. J., Thornton J. M., Partridge L., Gems D. (2007) Evolutionary conservation of regulated longevity assurance mechanisms. Genome Biol. 8, R132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Martin-Montalvo A., Villalba J. M., Navas P., de Cabo R. (2011) NRF2, cancer and calorie restriction. Oncogene 30, 505–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mattison J. A., Roth G. S., Beasley T. M., Tilmont E. M., Handy A. M., Herbert R. L., Longo D. L., Allison D. B., Young J. E., Bryant M., Barnard D., Ward W. F., Qi W., Ingram D. K., de Cabo R. (2012) Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature 489, 318–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.