Abstract
Exosomes are naturally occurring biological nanomembranous vesicles (∼40 to 100 nm) of endocytic origin that are released from diverse cell types into the extracellular space. They have pleiotropic functions such as antigen presentation and intercellular transfer of protein cargo, mRNA, microRNA, lipids, and oncogenic potential. Here we describe the isolation, via sequential immunocapture using anti-A33- and anti-EpCAM-coupled magnetic beads, of two distinct populations of exosomes released from organoids derived from human colon carcinoma cell line LIM1863. The exosome populations (A33-Exos and EpCAM-Exos) could not be distinguished via electron microscopy and contained stereotypical exosome markers such as TSG101, Alix, and HSP70. The salient finding of this study, revealed via gel-based LC-MS/MS, was the exclusive identification in EpCAM-Exos of the classical apical trafficking molecules CD63 (LAMP3), mucin 13 and the apical intestinal enzyme sucrase isomaltase and increased expression of dipeptidyl peptidase IV and the apically restricted pentaspan membrane glycoprotein prominin 1. In contrast, the A33-Exos preparation was enriched with basolateral trafficking molecules such as early endosome antigen 1, the Golgi membrane protein ADP-ribosylation factor, and clathrin. Our observations are consistent with EpCAM- and A33-Exos being released from the apical and basolateral surfaces, respectively, and the EpCAM-Exos proteome profile with widely published stereotypical exosomes. A proteome analysis of LIM1863-derived shed microvesicles (sMVs) was also performed in order to clearly distinguish A33- and EpCAM-Exos from sMVs. Intriguingly, several members of the MHC class I family of antigen presentation molecules were exclusively observed in A33-Exos, whereas neither MHC class I nor MHC class II molecules were observed via MS in EpCAM-Exos. Additionally, we report for the first time in any extracellular vesicle study the colocalization of EpCAM, claudin-7, and CD44 in EpCAM-Exos. Given that these molecules are known to complex together to promote tumor progression, further characterization of exosome subpopulations will enable a deeper understanding of their possible role in regulation of the tumor microenvironment.
The microenvironment in which a tumor originates plays a critical role in its initiation, progression, and metastasis (1–3). Recent advances have indicated that although the microenvironment provides crucial signaling to maintain tissue architecture, inhibit cell growth, and constrain the malignant phenotype, it can also promote and induce cancer (4). In addition to cancer cells, the tumor microenvironment comprises normal cells, blood cells, secreted proteins and peptides, and constituents of the extracellular matrix that actively influence cell behavior. Secreted proteins, peptides, and physiological small molecules such as soluble cytokines and chemokines are currently recognized as the main exocrine and juxtacrine factors underlying cell-to-cell communication within the tumor microenvironment and providing the metastatic niche in distant organs (5). In addition to soluble secreted proteins and peptides, most cell types also release extracellular membrane vesicles (eMVs)1 that transfer information between cells in the microenvironment; it is now recognized that eMVs can also influence cell-to-cell communication during tumor progression (6, 7). Another emerging means by which cells relay information to other cells is long, thin, interconnecting membranous bridges that connect neighboring cells through adhesion mechanisms (e.g. actin-based cytonemes or filopodial bridges) or tunneling nanotubes, which can establish direct tubular conduits between the cytoplasms of adjacent cells (for a review, see Ref. 8).
Over the past decade, eMVs have been shown to exhibit important pleiotropic roles in many biological processes. For example, eMVs are enriched in various bioactive molecules such as growth factors, lipids, membrane receptors (adhesion molecules, oncogenic receptors), mRNA, microRNA, transcriptional factors, splicing factors, and infectious particles (HIV, prions) (9; see reviews in Refs. 6 and 10–12). These bioactive molecules have been reported to (i) directly stimulate target cells via bioactive lipids or by acting like soluble cell-surface signaling complexes; (ii) transfer oncogenic cargo and cancer cell properties to nearby indolent or normal cells; (iii) epigenetically reprogram recipient cells via the transfer of mRNA, microRNA, and transcription factors; and (iv) serve as a delivery vehicle, in a “Trojan horse” manner, to transfer pathological cargo such as plant toxins, prions, or HIV particles. Although many of these properties have been ascribed to exosomes, it should be noted that functional studies were commonly performed on eMV preparations, which in many cases are heterogeneous mixtures of shed microvesicles (sMVs), exosomes, exosome-like particles, and apoptotic blebs (13).
Exosomes, along with sMVs that bud off from the plasma membrane (PM) and apotopic bodies, represent specific subtypes of eMVs (reviewed in Ref. 13). Of these, exosomes have been the most widely studied at both biochemical and functional levels. Exosomes are a small homogeneous population of intraluminal vesicles (ILVs) (40 to 100 nm in diameter) that derive from the inward budding of the luminal membranes of late endosomes and form within multivesicular bodies (MVBs). ILVs are constitutively exocytosed from the cell when the MVBs fuse with the PM; upon their release into the microenvironment, ILVs are referred to as exosomes. Exosomes are quite distinct from sMVs (heterogeneous 500 to 1000 nm diameter vesicles) being shed from the PM into the extracellular space upon cellular activation by various stimuli (14). Whereas exosomes typically float at a buoyant density of 1.08 to 1.22 g/cm3 (15) and their proteome profiles are defined from a variety of cell types and body fluids (10, 16), the biophysical properties of sMVs are less well understood.
Several strategies have been used for exosome isolation, including ultracentrifugation, density gradient separation, and immunoaffinity capture. Our group recently performed a proteomic analysis evaluating the ability of each of these techniques to enrich for exosome markers and proteins involved in exosome biogenesis, trafficking, and release from LIM1863 cells (17). Although exosomes prepared using all three isolation strategies contained 40 to 100 nm vesicles positive for exosome markers Alix, TSG101, and HSP70, gel-based liquid chromatography coupled with tandem mass spectrometry (GeLC-MS/MS) in combination with label-free spectral counting revealed that immunoaffinity capture enriched for exosome and exosome-associated proteins by at least 2-fold more than the other two methods studied. In that study, EpCAM, a ubiquitously expressed epithelial cancer marker (18), was the immunoaffinity capture target.
In our studies aimed at understanding the physiological role of exosomes in colorectal cancer (CRC) biology, we previously described a robust procedure for isolating and characterizing exosomes secreted from LIM1215 CRC cells (19). This study, using the colon epithelial cell-specific A33 antibody (20) for immunoaffinity capture, afforded the isolation of homogeneous A33-containing exosomes for biophysical characterization. A comparative proteome profiling of A33-positive LIM1215 exosomes with previously published murine mast cell (21) and human-urine-derived exosomes (22) revealed a subset of proteins common to the three exosome types and, for the first time, a human colon cancer exosomal proteome “signature”. As this signature might reflect the CRC exosomal profile of a restricted CRC subtype—LIM1215 cells were originally derived from a patient with inherited nonpolyposis colorectal cancer (23)—we have extended these studies to another CRC cell subtype and report here a robust proteome study of exosomes isolated from the CRC cell line LIM1863, which grows as organoids with spontaneous differentiation into crypt-like structures in vitro (24).
EXPERIMENTAL PROCEDURES
Cell Culture and Preparation of Concentrated Culture Medium
Human colon carcinoma LIM1863 cells grow as free-floating multicellular spheres (organoids) in which highly polarized cells localize around a central lumen. These organoids resemble colonic crypts in that they contain morphologically differentiated columnar and goblet cells (24). LIM1863 cells were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) containing 5% FCS, α-thioglycerol (10 μm), insulin (25 units/l), and hydrocortisone (1 mg/l), with 10% CO2 at 37 °C. LIM1863 cells (6 × 108 cells) were washed four times with 30 ml of RPMI 1640 medium and cultured for 24 h in 150 ml serum-free RPMI medium supplemented with 0.6% insulin-transferrin-selenium solution (Invitrogen). Culture medium (CM) was collected and centrifuged at 4 °C (480 × g for 5 min followed by 2,000 × g for 10 min) to remove intact cells and cell debris. CM was centrifuged at 10,000 × g for 30 min to isolate sMVs. CM was filtered using a VacuCap® 60 filter unit fitted with a 0.1 μm Supor® Membrane (Pall Life Sciences, Port Washington, NY) and then concentrated to 500 μl using an Amicon® Ultra-15 Ultracel centrifugal filter device with a 5K nominal molecular weight limit (Millipore, MA).
Preparation of hA33 Immunoaffinity Capture Dynabeads
Protein G Dynabeads™ (500 μl, 5 × 108 beads) (Invitrogen) in citrate-phosphate buffer (pH 5.0) were mixed with hA33 capture antibody (100 μl, 300 μg) (a kind gift from A. Scott, Ludwig Institute for Cancer Research Ltd. - Austin Campus) and incubated for 40 min at room temperature with gentle rotation, according to the manufacturer's instructions, (19). Briefly, hA33-Dynabeads were placed on a solid support magnet (SSM), separated for 2 min, and washed twice with 1 ml citrate-phosphate buffer (pH 5.0) followed by 1 ml 0.2 m triethanolamine (pH 8.2). Washed hA33-Dynabeads were suspended in 1 ml of freshly prepared 20 mm dimethyl pimelimidate in 0.2 m triethanolamine (pH 8.2) for 30 min with gentle agitation. hA33-Dynabeads were placed on the SSM for 2 min, the supernatant was discarded, and the beads were mixed with 1 ml of 50 mm Tris (pH 7.5) for 15 min with gentle agitation. The cross-linked hA33-Dynabeads were again magnetically bound using the SSM and washed three times with PBS containing 0.05% Tween-20.
Isolation of Exosomes Using hA33 Immunoaffinity Capture Dynabeads
LIM1863 concentrated culture medium (CCM) (500 μl, from 6 × 108 cells) was pre-incubated with Dynabeads (5 × 108 beads) for 2 h at 4 °C with gentle rotation to reduce nonspecific binding. The beads were harvested using the SSM, and the supernatant was retained and incubated with prepared hA33 immunoaffinity capture Dynabeads for 2 h at 4 °C with gentle rotation. The exosome-hA33-Dynabead complexes were magnetically held using the SSM and washed three times for 5 min in 1 ml PBS. Bound exosomes were eluted from the hA33-Dynabead complex with 0.2 m glycine (pH 2.8) for electron microscopy (EM) analysis or 100 μl 2 × SDS sample loading buffer for GeLC-MS/MS analysis (25).
Sequential A33/EpCAM Immunoaffinity Capture of Exosomes
After the removal of A33-Exos from the CCM, A33-depleted CCM (unbound material) was subjected to EpCAM immunoaffinity capture (EpCAM (CD326) magnetic microbeads from Miltenyi Biotec, Auburn, CA) according to the manufacturer's instructions. Briefly, 500 μl of A33-depleted CCM was incubated with EpCAM-microbeads (100 μl) for 4 h at 4 °C. An empty 3 ml LS Microcolumn was placed on a SSM and rinsed three times with Rinsing Solution (MACS® BSA Stock Solution diluted 1:20 with autoMACS® Rinsing Solution; Miltenyi Biotec). Exosome-bound microbeads were pipetted into the column and washed three times with 1 ml Rinsing Solution. The column was removed from the SSM, and exosome-bound microbeads were recovered by rinsing the column at room temperature with 3 × 1 ml Rinsing Solution. Exosome-bound microbeads were washed twice with 1 ml PBS and centrifuged at 100,000 × g for 1 h at 4 °C. The supernatant was removed, and EpCAM-Exos were eluted from the microbeads with 100 μl 0.2 m glycine, Tris-HCl, pH 2.8, for EM imaging or lysed with 100 μl of SDS sample buffer for GeLC-MS/MS analysis.
Protein Quantitation
The protein content of the sMVs, A33-Exos, and EpCAM-Exos was estimated via one-dimensional SDS-PAGE/SYPRO® Ruby protein staining densitometry (26). Briefly, 5 μl sample aliquots were solubilized in SDS sample buffer (2% (w/v) SDS, 125 mm Tris-HCl, pH 6.8, 12.5% (v/v) glycerol, 0.02% (w/v) bromphenol blue) and loaded into 1-mm 10-well NuPAGE™ 4–12% (w/v) Bis-Tris Precast gels (Invitrogen). Electrophoresis was performed at 150 V for 1 h in NuPAGE™ 1 × MES running buffer (Invitrogen) using an Xcell Surelock™ gel tank (Invitrogen). After electrophoresis, gels were removed from the tank and fixed in 50 ml fixing solution (40% (v/v) methanol, 10% (v/v) acetic acid in water) for 30 min on an orbital shaker and stained with 30 ml SYPRO® Ruby (Invitrogen) for 30 min. This was followed by destaining twice in 50 ml of 10% (v/v) methanol with 6% (v/v) acetic acid in water for 1 h. Gels were imaged on a Typhoon 9410 variable mode imager (Molecular Dynamics, Sunnyvale, CA) using a green (532 nm) excitation laser and a 610BP30 emission filter at 100-μm resolution. Densitometry quantitation was performed using ImageQuant software (Molecular Dynamics) to determine the protein concentration relative to a BenchMark™ Protein Ladder standard of known protein concentration (Invitrogen).
Western Blot Analysis
Exosome samples (∼10 μg protein) were lysed in SDS sample buffer, reduced with 50 mm DTT (when required), heated for 5 min at 95 °C, and subjected to electrophoresis using precast NuPAGE™ 4–12% (w/v) Bis-Tris Precast gels (Invitrogen) in MES running buffer at a constant 150 V for 1 h. Proteins were electrotransferred onto nitrocellulose membranes using the iBlot™ Dry Blotting System (Invitrogen), and the membranes were blocked with 5% (w/v) skim milk powder in Tris-buffered saline with 0.05% (v/v) Tween-20 (TTBS) for 1 h. Membranes were probed with primary mouse anti-TSG101 (1:500; BD Biosciences), mouse anti-Alix (1:1,000; Cell Signaling Technology Danvers, MA), rabbit anti-EpCAM (1:1,000; Abcam), and mouse anti-A33 (1 μg/ml) (a kind gift from A. Scott, Ludwig Institute for Cancer Research Ltd. - Austin Campus Cambridge, UK) for 1 h in TTBS, followed by incubation with the secondary antibody, IRDye 800 goat anti-mouse IgG or IRDye 700 goat anti-rabbit IgG (1:15,000; LI-COR Biosciences Lincoln, NE), for 1 h in darkness. All antibody incubations were carried out using gentle orbital shaking at room temperature. Western blots were washed three times in TTBS for 10 min after each incubation step and visualized using the Odyssey Infrared Imaging System, version 3.0 (LI-COR Biosciences).
EM
EM imaging of exosome preparations was performed as described elsewhere (19), with modifications. Briefly, exosome preparations (∼2 μg protein) were fixed in 1% (v/v) glutaraldehyde, layered onto Formvar coated 200 mesh copper grids (ProSciTech, Queensland, Australia), and allowed to dry. Grids were then washed twice with water for 5 min and stained with 1% (w/v) aqueous uranyl acetate (ProSciTech, Queensland, Australia) for 10 min. Imaging was performed using a Gatan UltraScan 1000 (2k × 2k) charge-coupled device camera coupled to a Tecnai F30 (FEI, Eindhoven, The Netherlands) electron microscope with an acceleration voltage of 200 kV.
GeLC-MS/MS
A33-Exos, EpCAM-Exos, and sMV samples (20 μg) were electrophoresed using SDS-PAGE (25), and proteins were visualized using Imperial™ Protein Stain (Thermo Fisher Scientific) according to the manufacturer's instructions. Gel lanes were cut into 20 × 2 mm bands using a GridCutter (The Gel Company, San Francisco, CA), and individual bands were subjected to in-gel reduction, alkylation, and trypsinization, as described elsewhere (25). Briefly, gel bands were reduced with 10 mm DTT (Calbiochem, San Diego, CA) for 30 min, alkylated for 20 min with 25 mm iodoacetic acid (Fluka, St. Louis, MO), and digested with 150 ng trypsin (Worthington Biochemical Corp, Freehold, NJ) for 4.5 h at 37 °C. Tryptic peptides were extracted with 50 μl 50% (v/v) acetonitrile and 50 mm ammonium bicarbonate concentrated to ∼10 μl via centrifugal lyophilisation and analyzed via LC-MS/MS. Reversed-phase HPLC was performed on a nanoAcquity® (C18) 150 × 0.15-mm internal diameter reversed phase ultra-performance liquid chromatography column (Waters, Milford, CT) using an Agilent 1200 HPLC coupled online to an LTQ-Orbitrap mass spectrometer equipped with a nanoelectrospray ion source (Thermo Fisher Scientific) (27). The column was developed with a linear 60-min gradient with a flow rate of 0.8 μl/min at 45 °C from 0%–100% solvent B, where solvent A was 0.1% (v/v) aqueous formic acid and solvent B was 0.1% (v/v) aqueous formic acid/60% acetonitrile. Survey MS scans were acquired with the resolution set to a value of 30,000. Real time recalibration was performed using a background ion from ambient air in the C-trap (28). Up to five selected target ions were dynamically excluded from further analysis for 3 min.
Database Searching and Protein Identification
Parameters used to generate peak lists, using Extract-MSn as part of Bioworks 3.3.1 (Thermo Fisher Scientific), were as follows: minimum mass 700; maximum mass, 5,000; grouping tolerance, 0 Da; intermediate scans, 200; minimum group count, 1; 10 peaks minimum; and total ion current of 100. Peak lists for each LC-MS/MS run were merged into a single MASCOT generic file. Automatic charge state recognition was used because of the high resolution survey scan (30,000). LC-MS/MS spectra were searched against the human RefSeq (29) protein database (38,791 sequences) using MASCOT (v2.2.01, Matrix Science, London, UK). Searching parameters included the following: fixed modification (carboxymethylation of cysteine; +58 Da), variable modifications (oxidation of methionine; +16 Da), up to three missed tryptic cleavages, 20 ppm peptide mass tolerance, and 0.8 Da fragment ion mass tolerance. Peptide identifications were deemed significant if the ion score was greater than the identity score. Significant protein identifications contained at least two unique peptide identifications. The false discovery rate (derived from a corresponding decoy database search) was less than 0.3% for each sample preparation. The UniProt database was used to classify identified proteins based on their annotated function and subcellular localization and to specify genes reportedly involved in colon cancer (30). Transmembrane-spanning alpha helices were predicted using the web-based prediction program Transmembrane Hidden Markov Model (TMHMM) v2.0 (31). The Human Protein Atlas was used as an annotated resource to assess the tissue expression of proteins identified in this study (32).
Label-free Spectral Counting
To compare the relative protein abundances of A33- and EpCAM-Exos, the ratio of normalized spectral counts (Rsc) was estimated (Eq. 1) as described elsewhere (33).
where n is the significant protein spectral count, t is the total number of significant MS/MS spectra in the sample, f is a correction factor set to 1.25, A is A33-Exos, and B is EpCAM-Exos.
LIM1863 Cell Imaging Using Confocal Microscopy
Approximately 1 × 107 LIM1863 cells were washed twice with 20 ml PBS before being fixed in 4% (v/v) formaldehyde in PBS for 10 min at room temperature. Cells were then permeabilized in 0.2% (v/v) Triton X-100 in PBS, washed twice with washing buffer (0.1% (w/v) BSA and 0.1% (v/v) Tween 20 in PBS), and blocked with Serum Blocking Buffer (5% (v/v) goat serum, 5% (w/v) BSA, 0.1% (w/v) cold fish gelatin, 0.05% (v/v) Tween 20, 0.05% (w/v) sodium azide, 0.01 m PBS pH 7.2) for 2 h at room temperature. Cells were incubated with primary antibodies (mouse anti-A33, 1 μg/ml; rabbit anti-EpCAM, 1 μg/ml) (Abcam, Cambridge, UK) in washing buffer for 1 h at room temperature, washed twice with washing buffer, and incubated with secondary antibodies Alexa Fluor® 488-conjugated goat anti-mouse IgG and Alexa Fluor® 546-conjugated goat anti-rabbit IgG (Invitrogen) diluted 1:200 in washing buffer for 30 min at room temperature in the dark. Cells were finally washed three times with washing buffer and imaged using a Nikon ECLIPSE TE2000-E confocal microscope equipped with a Nikon plan APO VC 60x/1.20 WI water-immersion lens.
RESULTS AND DISCUSSION
Immunoaffinity Capture Isolation of Two Populations of Exosomes from LIM1863 Cells
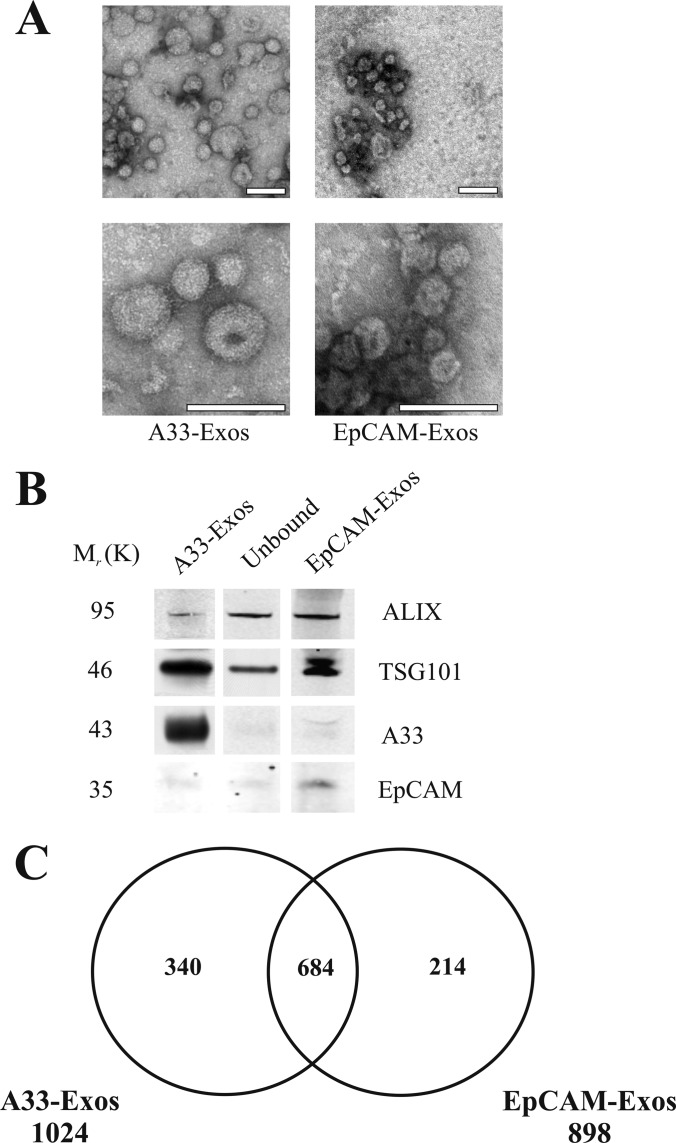
Immunoaffinity capture using magnetic bead technology is an effective method for isolating homogeneous exosomes for MS-based proteomics (17, 19). Here we used an hA33-immunoaffinity magnetic bead strategy that we previously applied to purify exosomes derived from LIM1215 CRC cells (19). LIM1863 cells were first grown to 80% confluence, and then cell culture was continued in serum-free medium for 24 h to minimize FCS contaminants in the CM. Under these culture conditions, cell viability was ∼95%, as previously reported (17). Intact cells, cell detritus, and large membranous particles were removed from the CM by means of low-speed centrifugation (480 × g for 5 min followed by 2,000 × g for 10 min). The CM was centrifuged at 10,000 × g for 30 min to isolate sMVs. The CM (600 ml) was then filtered through a 0.1-μm membrane using centrifugal ultrafiltration to remove large membranous vesicles. The filtered CM was concentrated to 500 μl via centrifugal filtration using a 5K nominal molecular weight limit membrane filter. The CCM was pretreated with control Dynabeads (i.e. beads not treated with antibodies) and then mixed with hA33mAb-coated beads for 2 h at 4 °C. The A33-coated beads were recovered magnetically. An outline of the purification strategy is shown in Fig. 1. EM revealed that the A33-Exos were essentially homogeneous and 40 to 60 nm in diameter (Fig. 2A); Western blot analysis revealed the presence of common exosome molecular markers Alix/PDCD6IP and TSG101 (Fig. 2B). Using a large excess of anti-A33-coated beads, ∼90% of A33-containing exosomes (A33-Exos) were captured from the CCM (Fig. 2B), with an overall yield of ∼50 μg protein from 6 × 108 LIM1863 cells. Because a significant amount of A33-negative exosomes that were positive for Alix, TSG101, and EpCAM remained in the CCM (Fig. 2B), we decided to further isolate and characterize these exosomes from the A33-Exos-depleted CCM using EpCAM immunocapture (Fig. 1). This second population of exosomes (EpCAM-Exos) were homogeneous in size (40 to 60 nm in diameter), as revealed by EM (Fig. 2B), and yielded ∼110 μg protein from 6 × 108 LIM1863 cells.
Fig. 1.
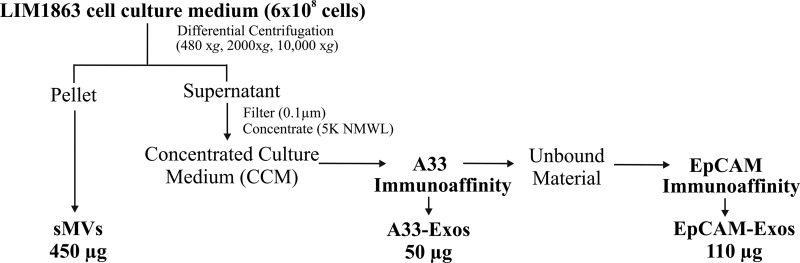
Isolation of exosomes and shed microvesicles from the colon carcinoma cell line LIM1863. LIM1863 cells were grown in serum-free medium supplemented with insulin-transferrin-selenium for 24 h, and then the CM was collected. Shed microvesicles (sMVs) were first isolated from the CM by means of differential centrifugation. The supernatant was then filtered (0.1 μm) and concentrated via centrifugal ultrafiltration through a 5K nominal molecular weight limit membrane. A33-positive exosomes (A33-Exos) were isolated from the CCM via anti-A33 antibody immunocapture. EpCAM-Exos were isolated from A33-Exos-depleted CCM by means of immunocapture using EpCAM-loaded magnetic beads.
Fig. 2.
Morphological characterization and proteome analysis of LIM1863 cell-derived A33- and EpCAM-Exos. A, electron micrographs of A33- and EpCAM-Exos negatively stained with uranyl acetate and examined at 200 kV; scale bar = 100 nm. B, Western blot analysis of A33-Exos, unbound material (flow-through of anti-A33 antibody capture beads), and EpCAM-Exos (10 μg per lane) for Alix (PDCD6IP), TSG101, A33, and EpCAM. C, two-way Venn diagram depicting the overlap of exosomal proteins derived from A33- and EpCAM-Exos. 684 proteins were common to both exosomal datasets, and 340 and 214 proteins were unique to A33- and EpCAM-Exos, respectively.
Proteomic Profiling of A33-Exos and EpCAM-Exos
We further compared the proteome profiles of the two populations of immune-captured LIM1863 exosomes using GeLC-MS/MS, as previously described (17, 25). This resulted in 1,024 and 898 proteins identified in A33-Exos and EpCAM-Exos, respectively. Of the 684 proteins in common, there was clear evidence at the MS level of typical exosome marker proteins such as ESCRT-I component TSG101, the ESCRT accessory protein Alix, HSP70, and CD9 (Fig. 2B and supplemental Table S1). The number of unique protein identifications in each of the A33- and EpCAM-exosome samples—340 and 214 proteins, respectively—is quite striking (Fig. 2C).
LIM1863 organoids comprise polarized epithelial cells (columnar and goblet cells) with well-formed brush borders containing microvilli located at the luminal membrane (24). Although accumulating evidence points to two general protein-sorting processes in MVB/ILV biogenesis in non-polarized cells—namely, partitioning into cholesterol/sphingolipid-rich microdomains (“lipid rafts”) (34) and higher-order oligomerization at the plasma membrane (35)—there is a paucity of information about exosome biogenesis in polarized cells (for a review, see Ref. 36). Because A33 and the tight junction glycoprotein EpCAM (37) localize to the basolateral and apical surfaces of LIM1863 cells, respectively (Fig. 3), we reasoned that A33-Exos and EpCAM-Exos might exocytose from these corresponding basolateral and apical regions. If indeed this is the case, A33-Exos and EpCAM-Exos are likely to harbor proteins involved in the organization of vesicular trafficking and tissue polarity (38). To test this hypothesis, we examined our A33-Exos and EpCAM-Exos datasets in the context of recycling pathways for apical and basolateral cell surface proteins and their associated sorting proteins. We first focused on the 214 proteins uniquely expressed in EpCAM-Exos (Fig. 2C) for the presence of apically sorted proteins.
Fig. 3.
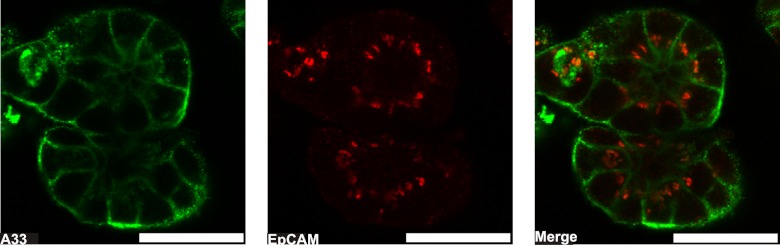
A confocal optical section through a preparation of LIM1863 organoids. LIM1863 cells were incubated with mouse anti-A33 antigen IgG and rabbit anti-EpCAM antigen IgG (1 μg/ml) followed by Alexa Fluor® 546-conjugated goat anti-rabbit IgG and Alexa Fluor® 488-conjugated goat anti-mouse IgG secondary antibodies (1:200). The A33 antigen (green) distributes to the basolateral cell periphery, and the EpCAM antigen to the apical ring (red). Scale bar, 30 μm.
EpCAM-Exos Are Enriched with Tetraspanins and Proteins with Apical Localization
Table I shows a selected list of proteins enriched in EpCAM-Exos (for a complete list of all proteins identified in this exosome preparation, see supplemental Table S1). Of note is the unique expression of the tetraspanin protein CD63 (LAMP3), a marker of the late endosome compartment (39) typically found in ILVs and secreted exosomes following the fusion of endosomes with the plasma membrane (40). In addition to CD63, the tetraspanins TSPAN3 and TSPAN6 were uniquely identified in EpCAM-Exos, and TSPAN14 (Rsc, −3.6), TSPAN15 (Rsc, −2.1), and CD81 (Rsc, −4.2), while also present in A33-Exos, were enriched in EpCAM-Exos (Table I). There is accumulating evidence that points to an important role for N-glycosylation in promoting trafficking from the trans-Golgi network and endosomes to the apical membrane of polarized cells (41–43). Although the tetraspanins TSPAN3, TSPAN6, and CD63 all contain N-glycosylation sequons, we did not identify any tryptic peptides containing these putative post-translational modification sites. The tetraspanins are a family of proteins that cross the plasma membrane four times and form complexes (referred to as a “tetraspanin web”) by interacting with other tetraspanins and a variety of transmembrane (principally the α3β1, α4β1, and α6β1 integrins) and cytosolic proteins required for their function (for a review, see Ref. 44). In our study there is MS evidence of the presence of α6β1 in EpCAM-Exos and α6β1/α3β1 in A33-Exos. Interestingly, the tetraspanin-like protein claudin-7 (CLDN7) (Rsc, −21.1) is uniquely expressed in the EpCAM-Exos (Table I). The adhesion protein CLDN7 regulates EpCAM-mediated functions in tumor progression (45), presumably through its complex with EpCAM and a CD44 isoform (46). This EpCAM-CLDN7 complex co-localizes with a tetraspanin-enriched membrane microdomain (47), and the complex, rather than EpCAM and/or CLDN7 alone, is reported to promote cell motility, proliferation, survival, tumorigenicity, and metastasis (45). To our knowledge, this is the first report of a co-localization of EpCAM/CLDN7/CD44 in exosomes (or extracellular vesicles). Interestingly, whereas both EpCAM (Rsc, −2.1) and CD44 (Rsc, −3.5) were present in the A33-Exos preparation, CLDN7 was not; CLDN3, CLDN4, and CLDN15 were present in both exosome preparations, but they were enriched in EpCAM-Exos (Rsc, −1.8 to −2.3).
Table I. Representative list of proteins either uniquely localized or significantly enriched in EpCAM-Exos.
| Gene symbol | Gene ID | Protein description | A33-Exos SpCa | EpCAM-Exos SpCb | RSC (A33-Exos/ EpCAM-Exos)c |
|---|---|---|---|---|---|
| CD44 | 960 | CD44 molecule | 20 | 47 | −3.5 |
| CD63 | 967 | CD63 molecule | 12 | −16.2 | |
| CD81 | 975 | CD81 molecule | 9 | 27 | −4.2 |
| CLDN3 | 1365 | Claudin 3 | 30 | 35 | −1.8 |
| CLDN4 | 1364 | Claudin 4 | 11 | 17 | −2.3 |
| CLDN7 | 1366 | Claudin 7 | 16 | −21.1 | |
| CLDN15 | 24146 | Claudin 15 | 8 | 11 | −2.0 |
| TSPAN3 | 10099 | Tetraspanin 3 | 3 | −5.2 | |
| TSPAN6 | 7105 | Tetraspanin 6 | 9 | −12.5 | |
| TSPAN14 | 81619 | Tetraspanin 14 | 6 | 16 | −3.6 |
| TSPAN15 | 23555 | Tetraspanin 15 | 7 | 10 | −2.1 |
| SI | 6476 | Sucrase isomaltase (alpha-glucosidase) | 9 | −12.5 | |
| DPEP1 | 1800 | Dipeptidase 1 (renal) | 29 | 49 | −2.5 |
| DPP4 | 1803 | Dipeptidyl-peptidase 4 (CD26) | 32 | 54 | −2.5 |
| MUC13 | 56667 | Mucin 13, cell-surface associated | 14 | −18.6 | |
| PROM1 | 8842 | Prominin 1 | 9 | 31 | −4.8 |
| PROM2 | 150696 | Prominin 2 | 3 | −5.2 | |
| EPCAM | 4072 | Epithelial cell adhesion molecule | 63 | 85 | −2.1 |
| CD46 | 4179 | CD46 molecule, complement regulatory protein | 7 | −10.1 | |
| CD59 | 966 | CD59 molecule, complement regulatory protein | 6 | −8.8 | |
| HMGB1 | 3146 | High mobility group box 1 | 11 | 190 | −24.0 |
| HMGB1L1 | 10357 | High mobility group box 1–like 1 | 3 | 105 | −38.3 |
| HMGB1L10 | 100130561 | High-mobility group box 1–like 10 | 3 | 136 | −49.5 |
| HMGB2 | 3148 | High mobility group box 2 | 123 | −152.3 | |
| HMGB3 | 3149 | High mobility group box 3 | 20 | −25.9 | |
| TOLLIP | 54472 | Toll interacting protein | 6 | 28 | −6.2 |
a Significant MS/MS spectral counts identified in A33-Exos.
b Significant MS/MS spectral counts identified in EpCAM-Exos.
c Relative spectral count ratio (Rsc) for proteins identified in A33-Exos in comparison with EpCAM-Exos (Eq. 1).
In the original LIM1863 cell line description, Whitehead and colleagues report that the intestinal enzymes sucrase isomaltase (α-glucosidase) and dipeptidyl peptidase IV localized to the luminal membrane and apical cytoplasm. In our present study, we observed unique expression of sucrase isomaltase in the EpCAM-Exos and a pronounced enrichment of dipeptidyl peptidase IV (Rsc, −2.5). Although there are at least five cell-surface mucins expressed in the gastrointestinal tract (48), only mucin 13 (MUC13) was observed in this study, uniquely expressed in the apical EpCAM-Exos (Table I). MUC13 has been shown via immunohistochemistry to be expressed exclusively on the apical membrane surface of normal columnar and goblet cells in the gastrointestinal tract, deep in crypts; it is aberrantly expressed in a variety of epithelial carcinomas, including gastric, colorectal, and ovarian cancers, where, in addition to apical surfaces, it also localizes to lateral and basolateral surfaces (49). There was no evidence of MUC13 expression in the A33-Exos preparation.
Prominin 1 (PROM1; CD133), an apically restricted pentaspan membrane glycoprotein, is enriched (Rsc, −4.8) in EpCAM-Exos (Table I). PROM1 localizes to cholesterol-based membrane microdomains and has been implicated in apical plasma membrane protrusion formation during (neuro) epithelial differentiation (50). Previously PROM1 has been observed in two types of microvesicles (∼600 nm P2 particles and 50 to 80 nm P4 particles) released from neuronal progenitor cells, as well as other epithelial cells such as the colon cancer Caco-2 cells (51). In contrast to the findings of Marzeco and colleagues (51) for the Caco-2 cell-derived 50 to 80 nm P4 microvesicles, the EpCAM-Exos contain both PROM1 and the exosomal marker CD63 (Table I). Interestingly, in our study, PROM2, which is structurally related to PROM1 and also released in extracellular vesicles (52), appears to be distributed in a polarized fashion, being restricted to EpCAM-Exos (but not A33-Exos).
We next focused on protein classes uniquely expressed/enriched in EpCAM-Exos. Intriguingly, the complement-mediated lysis inhibitors CD59 and CD46 (membrane cofactor protein) are uniquely expressed on EpCAM-Exos (Table I). In previous studies it has been shown that antigen-presenting exosomes can be protected from complement-mediated lysis by CD59 and CD55 (53), and human embryonic mesenchymal stem-cell-derived exosomes by CD59 (54). Another interesting observation was the significant enrichment in EpCAM-Exos of the high mobility group box class of proteins, HMGB1, -1L1, -1L10, -2, and -3 (Rsc, −24.0 to −152.3). Although the EpCAM-Exos are devoid of the adaptive immunity-related MHC class I and II molecules, the high mobility group box proteins have been reported to function as universal sentinels for nucleic-acid-mediated innate immune responses (55). Intriguingly, the EpCAM-Exos also contain the Toll-interacting protein (Rsc, −6.2), a component of the signaling pathway of Toll-like receptors, which mediate innate immune responses via nucleic acids.
A33-Exos Are Enriched for Basolateral Sorting Proteins
We next examined A33-Exos in the context of basolateral sorting signals (A33 localizes to the basolateral surface of LIM1863 cells (Fig. 3)). A33 has previously been reported to localize to the basolateral surface of polarized intestinal epithelial cells (56). Several basolateral trafficking/sorting proteins were observed to be uniquely expressed or significantly enriched in A33-Exos. For example, the early endosome antigen 1 (EEA1), a hydrophilic peripheral membrane protein that localizes to early endosomes (55) and a subset of basolateral-type endosomal compartments (56), is present only in A33-Exos (Table II). The Golgi membrane protein ADP-ribosylation factor 1, which recruits the cytosolic coatomer complex I to facilitate early-to-late endosome trafficking (57) of basolateral cargo proteins (58, 59), is uniquely identified in A33-Exos, along with enriched populations of several cytosolic coatomer complex I subunits (Rsc, 1.1 to 5.4). A striking observation was the significant enrichment of clathrin heavy and light chains (Rsc, 2.2 to 6.5) in A33-Exos and the clathrin adapter proteins (AP1/3) (Rsc, 1.7 to 13.3) (Table II). Clathrin, a key regulator of basolateral polarity (60) and a mediator of transport between the trans-Golgi network and endosomes (62), has been shown to associate directly with EEA1 on early endosomes (61). As mentioned earlier, EEA1 is uniquely expressed in A33-Exos. Rab13, also implicated in trans-Golgi network/basolateral endosome transport (63), uniquely localizes to A33-Exos (Table II).
Table II. Representative list of proteins either uniquely localized or significantly enriched in A33-Exos.
| Gene symbol | Gene ID | Protein description | A33-Exos SpCa | EpCAM-Exos SpCb | RSC (A33-Exos/EpCAM-Exos)c |
|---|---|---|---|---|---|
| AP1G1 | 164 | Adaptor-related protein complex 1, gamma 1 subunit | 24 | 13.3 | |
| AP1M1 | 8907 | Adaptor-related protein complex 1, mu 1 subunit | 8 | 4.9 | |
| AP1M2 | 10053 | Adaptor-related protein complex 1, mu 2 subunit | 20 | 11.2 | |
| AP1S1 | 1174 | Adaptor-related protein complex 1, sigma 1 subunit | 17 | 6 | 1.7 |
| AP3B1 | 8546 | Adaptor-related protein complex 3, beta 1 subunit | 5 | 3.3 | |
| ARF1 | 375 | ADP-ribosylation factor 1 | 44 | 23.8 | |
| CLSTN1 | 22883 | Calsyntenin 1 | 10 | 5.9 | |
| CLTC | 1213 | Clathrin, heavy chain (Hc) | 1,322 | 144 | 6.2 |
| CLTCL1 | 8218 | Clathrin, heavy chain-like 1 | 308 | 30 | 6.5 |
| CLTA | 1211 | Clathrin, light chain (Lca) | 10 | 5.9 | |
| CLTB | 1212 | Clathrin, light chain (Lcb) | 3 | 2.2 | |
| COPA | 1314 | Coatomer protein complex, subunit alpha | 57 | 14 | 2.5 |
| COPB1 | 1315 | Coatomer protein complex, subunit beta 1 | 33 | 7 | 2.7 |
| COPB2 | 9276 | Coatomer protein complex, subunit beta 2 (beta prime) | 9 | 5.4 | |
| COPG | 22820 | Coatomer protein complex, subunit gamma | 30 | 17 | 1.1 |
| EEA1 | 8411 | Early endosome antigen 1 | 8 | 4.9 | |
| GPA33 | 10223 | Glycoprotein A33 (transmembrane) | 47 | 25.3 | |
| HLA-A29.1 | 649853 | Major histocompatibility complex class I HLA-A29.1 | 13 | 7.5 | |
| HLA-A | 3105 | Major histocompatibility complex class I, A | 15 | 8.5 | |
| HLA-B | 3106 | Major histocompatibility complex class I, B | 5 | 3.3 | |
| HLA-C | 3107 | Major histocompatibility complex class I, C | 6 | 3.8 | |
| HLA-E | 3133 | Major histocompatibility complex class I, E | 4 | 2.8 | |
| RAB13 | 5872 | RAB13, member RAS oncogene family | 8 | 4.9 | |
| REEP6 | 92840 | Receptor accessory protein 6 | 24 | 13.3 |
a Significant MS/MS spectral counts identified in A33-Exos.
b Significant MS/MS spectral counts identified in EpCAM-Exos.
c Relative spectral count ratio (Rsc) for proteins identified in A33-Exos in comparison with EpCAM-Exos (Eq. 1).
In addition to well-recognized basolateral sorting proteins, several other protein classes are uniquely expressed/enriched in A33-Exos. Foremost among these are the major histocompatability complex I subunits A, B, C, E, and A29.1; these MHC class I molecules are uniquely expressed in A33-Exos (Table II). There was no evidence of MHC class II molecules in either A33- or EpCAM-Exos. These finding are in contrast to an earlier report that found MHC class I and class II molecules in both apical- and basolateral-derived exosomes from intestinal epithelial cells (56). It is tempting to speculate that exogenous protein antigens in the crypt lumen may be presented by MHC class I molecules, a process involving “cross presentation” (57), to CD8+ T cells present in the lamina propria. One means of conveying antigenic information from intestinal epithelial cells to the lamina propria or systemic immune cells might involve the transmission of 40 to 80 nm-diameter antigen-loaded MHC class I-positive basolateral exosomes though the 3 μm basement membrane pores; further studies will be required in order to clarify the function of MHC class I A33-Exos.
Colon-cancer-related Transmembrane Proteins Feature Highly in EpCAM- and A33-Exos
In order to determine the membrane topology of A33- and EpCAM-Exos, we performed a transmembrane domain analysis using the TMHMM algorithm (31). This analysis revealed a total of 119 transmembrane-containing proteins, of which 13 have been previously implicated in CRC (Table III). In addition to the integrins ITGA2, ITGAV, ITGA6, and ITGB1, the presence of the tumor-associated calcium signal transducer 2 protein and the sheddase ADAM metallopeptidase domain 10 involved in cell-surface substrate cleavage and the regulation of Notch (58) and HER2 signaling (59) were identified. Additionally, syndecan binding protein, E-cadherin, and basigin, identified in malignant ascites-derived exosomes and implicated in tumor progression (60), were observed.
Table III. List of colon-cancer-related and normal-colon-tissue-specific membrane proteins in LIM1863-derived exosomes.
| Gene symbol | Gene ID | Protein description | TMHMMa | A33-Exos SpCb | EpCAM-Exos SpCc | RSC (A33-Exos/ EpCAM-Exos)d |
|---|---|---|---|---|---|---|
| Colon cancer-related transmembrane proteins | ||||||
| ADAM10 | 102 | ADAM metallopeptidase domain 10 | 1 | 20 | 16 | −1.2 |
| BSG | 682 | Basigin (Ok blood group) | 2 | 25 | 40 | −2.4 |
| CD44 | 960 | CD44 molecule (Indian blood group) | 1 | 20 | 47 | −3.5 |
| CDH1 | 999 | Cadherin 1, type 1, E-cadherin (epithelial) | 1 | 20 | 23 | −1.7 |
| DPP4 | 1803 | Dipeptidyl-peptidase 4 | 1 | 32 | 54 | −2.5 |
| ITGA2 | 3673 | Integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) | 1 | 48 | 55 | −1.7 |
| ITGA6 | 3655 | Integrin, alpha 6 | 1 | 94 | 110 | −1.8 |
| ITGAV | 3685 | Integrin, alpha V (vitronectin receptor, antigen CD51) | 1 | 9 | 8 | −1.4 |
| ITGB1 | 3688 | Integrin, beta 1 (fibronectin receptor, antigen CD29) | 1 | 63 | 64 | −1.5 |
| MDK | 4192 | Midkine (neurite growth-promoting factor 2) | 1 | 6 | 48 | −10.4 |
| PROM1 | 8842 | Prominin 1 | 5 | 9 | 31 | −4.8 |
| SDCBP | 6386 | Syndecan binding protein (syntenin) | 1 | 21 | 63 | −4.4 |
| TACSTD2 | 4070 | Tumor-associated calcium signal transducer 2 | 1 | 17 | 13 | −1.2 |
| Normal-colon-tissue-specific transmembrane proteins | ||||||
| GPA33 | 10223 | Glycoprotein A33 (transmembrane) | 1 | 47 | 25.3 | |
| MUC13 | 56667 | Mucin 13, cell-surface associated | 1 | 14 | −18.6 | |
| TSPAN8 | 7103 | Tetraspanin 8 | 4 | 44 | 22 | 1.3 |
a Predicted number of transmembrane spanning domains, derived from TMHMM.
b Significant MS/MS spectral counts identified in A33-Exos.
c Significant MS/MS spectral counts identified in EpCAM-Exos.
d Relative spectral count ratio (Rsc) for proteins identified in A33-Exos in comparison with EpCAM-Exos (Eq. 1).
To morphologically distinguish colon-derived exosomes in blood and other body fluids from exosomes that derive from other tissue/cell types, we next interrogated our data for the presence of colon-tissue-specific membrane proteins. An examination of the Human Protein Atlas expression database (32) revealed three cell surface proteins specifically expressed on normal colon epithelium: MUC13, tetraspanin 8 (TSPAN8), and A33 (GPA33) (Table III). Although the tissue expression of these transmembrane proteins is colon specific in normal tissue, elevated expression is seen in many cancer tissues. For example, MUC13 is frequently expressed in gastric, colorectal, and ovarian cancers (48); TSPAN8 is overexpressed in colorectal, liver, lung, ovarian, prostate, and stomach cancers (61); and A33 has been observed in colorectal, stomach, liver, and ovarian cancers (32).
Protein Expression Profile of sMVs Is Distinct from That of A33- and EpCAM-Exos
Although the protein profile of EpCAM-Exos is consistent with that of stereotypical exosomes reported thus far (9, 13), to determine whether A33-Exos were sMVs, we analyzed LIM1863 sMVs (also referred to as microvesicles). For this undertaking, we used the 10,000 × g centrifugation method described for ARF6-regulated shedding of plasma membrane-derived microvesicles from LOX cell lines (62); for a more general purification protocol, see Ref. 63. A total of 1,392 proteins were seen in the sMV preparation (supplemental Table S2). Of these, 462 proteins were found to be unique to sMVs when compared with the A33- and EpCAM-Exos datasets. A majority of the apical and basolateral sorting proteins uniquely observed in the EpCAM- and A33-Exos preparations mentioned above were not identified via MS in the sMV preparation (supplemental Table S2). Intriguingly, the sMV preparation was enriched for several members of the ATP-binding cassette superfamily (e.g. ABC transport proteins such as ABCB1, ABCB4, ABCC1, ABCC2, ABCE1, and ABCG2) that are typically found in microsomal and plasma membrane preparations. The lack of cytochrome P450 proteins in our LIM1863 sMV preparation, however, would argue against significant microsomal contamination (64). It is interesting to note that human urinary exosomes extensively characterized by Knepper and colleagues (65, 66) are also enriched for ABC transporter proteins, as well as angiotensin converting enzyme isoforms and aldehyde dehydrogenase isoforms, all of which were observed in our sMV preparation but not the A33-Exos (or EpCAM-Exos) preparations. Taken together, our studies assert that the A33-Exos are not sMVs but, most likely, basolateral exosomes.
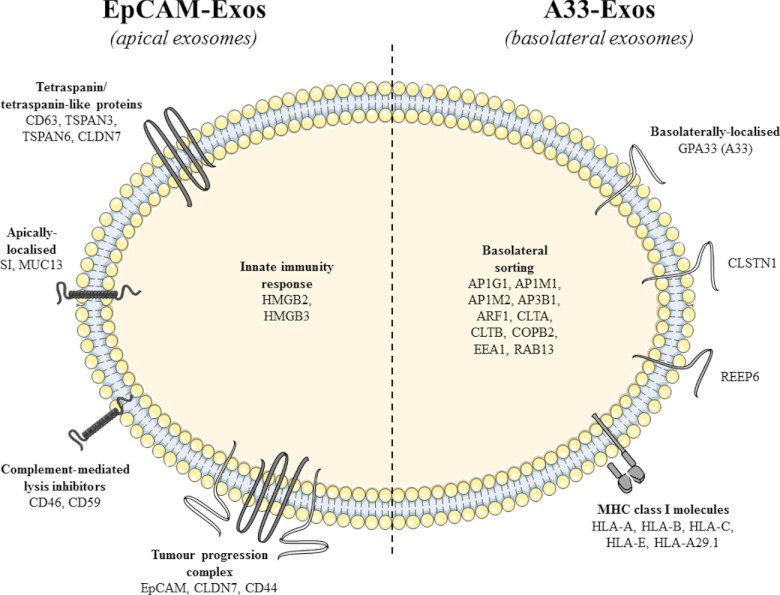
In summary, we have demonstrated that there are two distinct populations of exosomes released from LIM1863 organoids, one that can be purified to homogeneity using anti-EpCAM antibody loaded magnetic beads (EpCAM-Exos), and another that can be purified using anti-A33 antibody loaded magnetic beads (A33-Exos). We find that the proteome profile of EpCAM-Exos is consistent with stereotypical exosomes analyzed from diverse cell line/tissue sources. We have shown that EpCAM-Exos are enriched for apical sorting proteins, and A33-Exos for basolateral sorting proteins. Our observations are consistent with the notion that LIM1863 organoids release two distinct exosome populations: apical (EpCAM-Exos) and basolateral exosomes (A33-Exos) (Fig. 4). Because the LIM1863 organoids contain two morphologically differentiated cell types, it is not possible to establish whether EpCAM- and A33-Exos originate from a single or both cell types. (Extensive efforts by Hayward and Whitehead (67), as well as our own efforts, to isolate single cell populations (columnar and goblet cells) from LIM1863 cells thus far have been unsuccessful.) Protein profiling of LIM1863 sMVs revealed that A33- and EpCAM-Exos are distinct from sMVs. We show that only one of these exosome subtypes (A33-Exos) contains MHC class I molecules; there was no evidence of MHC class II molecules. Our study provides the first example of the colocalization of EpCAM, CLDN7, and CD44 in exosomes (EpCAM-Exos). Given that these molecules are known to complex together to promote tumor progression, further proteome characterization of tumor-derived exosome subpopulations will enable a deeper understanding of the emerging role of exosomes as mediators of tumorigenesis (68).
Fig. 4.
A model depicting the molecular structure of two discrete populations of LIM1863-derived exosomes. Selected proteins expressed in both apical exosomes (EpCAM-Exos) and basolateral exosomes (A33-Exos) include exosomal marker proteins (Alix, TSG101, HSP70, and CD9), integrins (ITGA2, ITGAV, ITGA6, and ITGB6), ephrin receptors (EPHB1, -B2, -B3, -B4, -A2, -A5, -A6, and -A7), tetraspanin proteins (TSPAN8, TSAN14, TSPAN15, and CD81), and the tetraspanin-like claudin proteins (CLDN3, CLDN4, and CLDN15). For a detailed list of ubiquitously expressed proteins found in both A33- and EpCAM-Exos, see supplemental Table S1. Apical exosomes also contained (i) tetraspanin proteins (CD63, TSPAN3, and TSPAN6) and tetraspanin-like claudin 7 (CLDN7), (ii) apically localized sucrase isomaltase (SI) and mucin 13 (MUC13), (iii) complement-mediated lysis inhibitors (CD46, CD59), and (iv) innate immunity response high-mobility group box proteins (HMGB2, HMGB3). Apical exosomes contained EpCAM, CLDN7, and CD44, which are reported to complex together to mediate tumor progression. Basolateral exosomes contained early endosome antigen 1 (EEA1), RAB13, and basolateral sorting proteins (clathrins CLTA and CLTB; clathrin adaptor proteins AP1G1, AP1M1, AP1M2, and AP3B1; coatomer subunit COPB2; and ADP-ribosylation factor 1). Basolateral exosomes also contained the cell-membrane-spanning proteins colon-specific antigen GPA33, calsyntenin 1 (CLSTN1), receptor accessory protein 6 (REEP6), and MHC class I molecules (HLA-A, -B, -C, -E, and -A29.1).
Footnotes
* This work was supported by the National Health & Medical Research Council of Australia program through Grant No. 487922 (R.J.S.), Fellowship No. 1016599 (S.M.), and Early Career CJ Martin Fellowship No. APP1037043 (R.A.M.). B.J.T. is supported by The University of Melbourne Research Scholarship (MRS). Analysis of proteomic data described in this work was supported using the Australian Proteomics Computational Facility funded by the National Health & Medical Research Council of Australia through Grant No. 381413. This work was supported by funds from the Operational Infrastructure Support Program provided by the Victorian Government Australia. We acknowledge the Australian Cancer Research Foundation for providing funds to purchase the Orbitrap™ mass spectrometer.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- A33-Exos
- exosomes isolated using anti-A33 immunoaffinity beads
- CCM
- concentrated culture medium
- CLN7
- claudin-7
- CM
- culture medium
- CRC
- colorectal cancer
- EEA1
- early endosome antigen 1
- EM
- electron microscopy
- eMV
- extracellular membrane vesicle
- EpCAM
- epithelial cell adhesion molecule
- EpCAM-Exos
- exosomes isolated using anti-EpCAM immunoaffinity beads
- ESCRT
- endosomal sorting complex required for transport
- ILV
- intraluminal vesicle
- MUC13
- mucin 13
- MVB
- multivesicular body
- PDCD6IP/Alix
- programmed cell death 6 interacting protein
- PM
- plasma membrane
- Rsc
- ratio of normalized spectral counts
- sMV
- shed microvesicle
- SSM
- solid support magnet
- TSG101
- tumor susceptibility gene 101.
REFERENCES
- 1. Mueller M. M., Fusenig N. E. (2004) Friends or foes—bipolar effects of the tumour stroma in cancer. Nat. Rev. Cancer 4, 839–849 [DOI] [PubMed] [Google Scholar]
- 2. Bhowmick N. A., Moses H. L. (2005) Tumor-stroma interactions. Curr. Opin. Genet. Dev. 15, 97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mbeunkui F., Johann D. J., Jr. (2009) Cancer and the tumor microenvironment: a review of an essential relationship. Cancer Chemother. Pharmacol. 63(4), 571–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bissell M. J., Hines W. C. (2011) Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat. Med. 17, 320–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Psaila B., Lyden D. (2009) The metastatic niche: adapting the foreign soil. Nat. Rev. Cancer 9, 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Al-Nedawi K., Meehan B., Rak J. (2009) Microvesicles: messengers and mediators of tumor progression. Cell Cycle 8, 2014–2018 [DOI] [PubMed] [Google Scholar]
- 7. Peinado H., Lavotshkin S., Lyden D. (2011) The secreted factors responsible for pre-metastatic niche formation: old sayings and new thoughts. Semin. Cancer Biol. 21, 139–146 [DOI] [PubMed] [Google Scholar]
- 8. Sherer N. M., Mothes W. (2008) Cytonemes and tunneling nanotubules in cell-cell communication and viral pathogenesis. Trends Cell Biol. 18, 414–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mathivanan S., Fahner C. J., Reid G. E., Simpson R. J. (2012) ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 40(1), D1241–D1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Simpson R. J., Lim J. W., Moritz R. L., Mathivanan S. (2009) Exosomes: proteomic insights and diagnostic potential. Expert Rev. Proteomics 6, 267–283 [DOI] [PubMed] [Google Scholar]
- 11. Thery C. (2011) Exosomes: secreted vesicles and intercellular communications. F1000 Biol. Rep. 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bobrie A., Colombo M., Raposo G., Thery C. (2011) Exosome secretion: molecular mechanisms and roles in immune responses. Traffic 12, 1659–1668 [DOI] [PubMed] [Google Scholar]
- 13. Mathivanan S., Ji H., Simpson R. J. (2010) Exosomes: extracellular organelles important in intercellular communication. J. Proteomics 73, 1907–1920 [DOI] [PubMed] [Google Scholar]
- 14. Cocucci E., Racchetti G., Meldolesi J. (2009) Shedding microvesicles: artefacts no more. Trends Cell Biol. 19(2), 43–51 [DOI] [PubMed] [Google Scholar]
- 15. Raposo G., Nijman H. W., Stoorvogel W., Liejendekker R., Harding C. V., Melief C. J., Geuze H. J. (1996) B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 183(3), 1161–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mathivanan S., Simpson R. J. (2009) ExoCarta: a compendium of exosomal proteins and RNA. Proteomics 9, 4997–5000 [DOI] [PubMed] [Google Scholar]
- 17. Tauro B. J., Greening D. W., Mathias R. A., Ji H., Mathivanan S., Scott A. M., Simpson R. J. (2012) Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 56, 293–304 [DOI] [PubMed] [Google Scholar]
- 18. Went P., Vasei M., Bubendorf L., Terracciano L., Tornillo L., Riede U., Kononen J., Simon R., Sauter G., Baeuerle P. A. (2006) Frequent high-level expression of the immunotherapeutic target Ep-CAM in colon, stomach, prostate and lung cancers. Br. J. Cancer 94, 128–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mathivanan S., Lim J. W., Tauro B. J., Ji H., Moritz R. L., Simpson R. J. (2010) Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol. Cell. Proteomics 9, 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Catimel B., Ritter G., Welt S., Old L. J., Cohen L., Nerrie M. A., White S. J., Heath J. K., Demediuk B., Domagala T., Lee F. T., Scott A. M., Tu G. F., Ji H., Moritz R. L., Simpson R. J., Burgess A. W., Nice E. C. (1996) Purification and characterization of a novel restricted antigen expressed by normal and transformed human colonic epithelium. J. Biol. Chem. 271(41), 25664–25670 [DOI] [PubMed] [Google Scholar]
- 21. Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J. J., Lotvall J. O. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 [DOI] [PubMed] [Google Scholar]
- 22. Pisitkun T., Shen R. F., Knepper M. A. (2004) Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. U.S.A. 101, 13368–13373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Whitehead R. H., Macrae F. A., St John D. J., Ma J. (1985) A colon cancer cell line (LIM1215) derived from a patient with inherited nonpolyposis colorectal cancer. J. Natl. Cancer Inst. 74, 759–765 [PubMed] [Google Scholar]
- 24. Whitehead R. H., Jones J. K., Gabriel A., Lukies R. E. (1987) A new colon carcinoma cell line (LIM1863) that grows as organoids with spontaneous differentiation into crypt-like structures in vitro. Cancer Res. 47, 2683–2689 [PubMed] [Google Scholar]
- 25. Simpson R. J., Connolly L. M., Eddes J. S., Pereira J. J., Moritz R. L., Reid G. E. (2000) Proteomic analysis of the human colon carcinoma cell line (LIM 1215): development of a membrane protein database. Electrophoresis 21, 1707–1732 [DOI] [PubMed] [Google Scholar]
- 26. Steinberg T. H., Lauber W. M., Berggren K., Kemper C., Yue S., Patton W. F. (2000) Fluorescence detection of proteins in sodium dodecyl sulfate-polyacrylamide gels using environmentally benign, nonfixative, saline solution. Electrophoresis 21, 497–508 [DOI] [PubMed] [Google Scholar]
- 27. Greening D. W., Simpson R. J. (2010) A centrifugal ultrafiltration strategy for isolating the low-molecular weight (<or=25K) component of human plasma proteome. J. Proteomics 73, 637–648 [DOI] [PubMed] [Google Scholar]
- 28. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 29. Pruitt K. D., Tatusova T., Maglott D. R. (2007) NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35, D61–D65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Apweiler R., Bairoch A., Wu C. H., Barker W. C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., Martin M. J., Natale D. A., O'Donovan C., Redaschi N., Yeh L. S. (2004) UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 32, D115–D119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305(3), 567–580 [DOI] [PubMed] [Google Scholar]
- 32. Uhlen M., Bjorling E., Agaton C., Szigyarto C. A., Amini B., Andersen E., Andersson A. C., Angelidou P., Asplund A., Asplund C., Berglund L., Bergstrom K., Brumer H., Cerjan D., Ekstrom M., Elobeid A., Eriksson C., Fagerberg L., Falk R., Fall J., Forsberg M., Bjorklund M. G., Gumbel K., Halimi A., Hallin I., Hamsten C., Hansson M., Hedhammar M., Hercules G., Kampf C., Larsson K., Lindskog M., Lodewyckx W., Lund J., Lundeberg J., Magnusson K., Malm E., Nilsson P., Odling J., Oksvold P., Olsson I., Oster E., Ottosson J., Paavilainen L., Persson A., Rimini R., Rockberg J., Runeson M., Sivertsson A., Skollermo A., Steen J., Stenvall M., Sterky F., Stromberg S., Sundberg M., Tegel H., Tourle S., Wahlund E., Walden A., Wan J., Wernerus H., Westberg J., Wester K., Wrethagen U., Xu L. L., Hober S., Ponten F. (2005) A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteomics 4, 1920–1932 [DOI] [PubMed] [Google Scholar]
- 33. Chen Y. S., Mathias R. A., Mathivanan S., Kapp E. A., Moritz R. L., Zhu H. J., Simpson R. J. (2011) Proteomics profiling of Madin-Darby canine kidney plasma membranes reveals Wnt-5a involvement during oncogenic H-Ras/TGF-beta-mediated epithelial-mesenchymal transition. Mol. Cell. Proteomics 10(2), M110.001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Gassart A., Geminard C., Fevrier B., Raposo G., Vidal M. (2003) Lipid raft-associated protein sorting in exosomes. Blood 102, 4336–4344 [DOI] [PubMed] [Google Scholar]
- 35. Fang Y., Wu N., Gan X., Yan W., Morrell J. C., Gould S. J. (2007) Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol. 5(6), e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lakkaraju A., Rodriguez-Boulan E. (2008) Itinerant exosomes: emerging roles in cell and tissue polarity. Trends Cell Biol. 18, 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Le Naour F., Zoller M. (2008) The tumor antigen EpCAM: tetraspanins and the tight junction protein claudin-7, new partners, new functions. Front. Biosci. 13, 5847–5865 [DOI] [PubMed] [Google Scholar]
- 38. Perret E., Lakkaraju A., Deborde S., Schreiner R., Rodriguez-Boulan E. (2005) Evolving endosomes: how many varieties and why? Curr. Opin. Cell Biol. 17, 423–434 [DOI] [PubMed] [Google Scholar]
- 39. Metzelaar M. J., Wijngaard P. L., Peters P. J., Sixma J. J., Nieuwenhuis H. K., Clevers H. C. (1991) CD63 antigen. A novel lysosomal membrane glycoprotein, cloned by a screening procedure for intracellular antigens in eukaryotic cells. J. Biol. Chem. 266(5), 3239–3245 [PubMed] [Google Scholar]
- 40. Escola J. M., Kleijmeer M. J., Stoorvogel W., Griffith J. M., Yoshie O., Geuze H. J. (1998) Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 273(32), 20121–20127 [DOI] [PubMed] [Google Scholar]
- 41. Vagin O., Kraut J. A., Sachs G. (2009) Role of N-glycosylation in trafficking of apical membrane proteins in epithelia. Am. J. Physiol. Renal Physiol. 296, F459–F469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Delacour D., Greb C., Koch A., Salomonsson E., Leffler H., Le Bivic A., Jacob R. (2007) Apical sorting by galectin-3-dependent glycoprotein clustering. Traffic 8, 379–388 [DOI] [PubMed] [Google Scholar]
- 43. Hara-Kuge S., Ohkura T., Ideo H., Shimada O., Atsumi S., Yamashita K. (2002) Involvement of VIP36 in intracellular transport and secretion of glycoproteins in polarized Madin-Darby canine kidney (MDCK) cells. J. Biol. Chem. 277(18), 16332–16339 [DOI] [PubMed] [Google Scholar]
- 44. Zoller M. (2009) Tetraspanins: push and pull in suppressing and promoting metastasis. Nat. Rev. Cancer 9, 40–55 [DOI] [PubMed] [Google Scholar]
- 45. Nubel T., Preobraschenski J., Tuncay H., Weiss T., Kuhn S., Ladwein M., Langbein L., Zoller M. (2009) Claudin-7 regulates EpCAM-mediated functions in tumor progression. Mol. Cancer Res. 7, 285–299 [DOI] [PubMed] [Google Scholar]
- 46. Kuhn S., Koch M., Nubel T., Ladwein M., Antolovic D., Klingbeil P., Hildebrand D., Moldenhauer G., Langbein L., Franke W. W., Weitz J., Zoller M. (2007) A complex of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol. Cancer Res. 5(6), 553–567 [DOI] [PubMed] [Google Scholar]
- 47. Ladwein M., Pape U. F., Schmidt D. S., Schnolzer M., Fiedler S., Langbein L., Franke W. W., Moldenhauer G., Zoller M. (2005) The cell-cell adhesion molecule EpCAM interacts directly with the tight junction protein claudin-7. Exp. Cell Res. 309, 345–357 [DOI] [PubMed] [Google Scholar]
- 48. Williams S. J., Wreschner D. H., Tran M., Eyre H. J., Sutherland G. R., McGuckin M. A. (2001) Muc13, a novel human cell surface mucin expressed by epithelial and hemopoietic cells. J. Biol. Chem. 276(21), 18327–18336 [DOI] [PubMed] [Google Scholar]
- 49. Maher D. M., Gupta B. K., Nagata S., Jaggi M., Chauhan S. C. (2011) Mucin 13: structure, function, and potential roles in cancer pathogenesis. Mol. Cancer Res. 9, 531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Corbeil D., Marzesco A. M., Wilsch-Brauninger M., Huttner W. B. (2010) The intriguing links between prominin-1 (CD133), cholesterol-based membrane microdomains, remodeling of apical plasma membrane protrusions, extracellular membrane particles, and (neuro)epithelial cell differentiation. FEBS Lett. 584, 1659–1664 [DOI] [PubMed] [Google Scholar]
- 51. Marzesco A. M., Janich P., Wilsch-Brauninger M., Dubreuil V., Langenfeld K., Corbeil D., Huttner W. B. (2005) Release of extracellular membrane particles carrying the stem cell marker prominin-1 (CD133) from neural progenitors and other epithelial cells. J. Cell Sci. 118(Pt 13), 2849–2858 [DOI] [PubMed] [Google Scholar]
- 52. Florek M., Bauer N., Janich P., Wilsch-Braeuninger M., Fargeas C. A., Marzesco A. M., Ehninger G., Thiele C., Huttner W. B., Corbeil D. (2007) Prominin-2 is a cholesterol-binding protein associated with apical and basolateral plasmalemmal protrusions in polarized epithelial cells and released into urine. Cell Tissue Res. 328, 31–47 [DOI] [PubMed] [Google Scholar]
- 53. Clayton A., Harris C. L., Court J., Mason M. D., Morgan B. P. (2003) Antigen-presenting cell exosomes are protected from complement-mediated lysis by expression of CD55 and CD59. Eur. J. Immunol. 33(2), 522–531 [DOI] [PubMed] [Google Scholar]
- 54. Lai R. C., Arslan F., Lee M. M., Sze N. S., Choo A., Chen T. S., Salto-Tellez M., Timmers L., Lee C. N., El Oakley R. M., Pasterkamp G., de Kleijn D. P., Lim S. K. (2010) Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 4, 214–222 [DOI] [PubMed] [Google Scholar]
- 55. Yanai H., Ban T., Wang Z., Choi M. K., Kawamura T., Negishi H., Nakasato M., Lu Y., Hangai S., Koshiba R., Savitsky D., Ronfani L., Akira S., Bianchi M. E., Honda K., Tamura T., Kodama T., Taniguchi T. (2009) HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature 462, 99–103 [DOI] [PubMed] [Google Scholar]
- 56. van Niel G., Raposo G., Candalh C., Boussac M., Hershberg R., Cerf-Bensussan N., Heyman M. (2001) Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology 121, 337–349 [DOI] [PubMed] [Google Scholar]
- 57. Cresswell P., Ackerman A. L., Giodini A., Peaper D. R., Wearsch P. A. (2005) Mechanisms of MHC class I-restricted antigen processing and cross-presentation. Immunol. Rev. 207, 145–157 [DOI] [PubMed] [Google Scholar]
- 58. Bozkulak E. C., Weinmaster G. (2009) Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol. Cell. Biol. 29, 5679–5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu P. C., Liu X., Li Y., Covington M., Wynn R., Huber R., Hillman M., Yang G., Ellis D., Marando C., Katiyar K., Bradley J., Abremski K., Stow M., Rupar M., Zhuo J., Li Y. L., Lin Q., Burns D., Xu M., Zhang C., Qian D. Q., He C., Sharief V., Weng L., Agrios C., Shi E., Metcalf B., Newton R., Friedman S., Yao W., Scherle P., Hollis G., Burn T. C. (2006) Identification of ADAM10 as a major source of HER2 ectodomain sheddase activity in HER2 overexpressing breast cancer cells. Cancer Biol. Ther. 5, 657–664 [DOI] [PubMed] [Google Scholar]
- 60. Keller S., Konig A. K., Marme F., Runz S., Wolterink S., Koensgen D., Mustea A., Sehouli J., Altevogt P. (2009) Systemic presence and tumor-growth promoting effect of ovarian carcinoma released exosomes. Cancer Lett. 278, 73–81 [DOI] [PubMed] [Google Scholar]
- 61. Boucheix C., Duc G. H., Jasmin C., Rubinstein E. (2001) Tetraspanins and malignancy. Expert Rev. Mol. Med. 2001, 1–17 [DOI] [PubMed] [Google Scholar]
- 62. Muralidharan-Chari V., Clancy J., Plou C., Romao M., Chavrier P., Raposo G., D'Souza-Schorey C. (2009) ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr. Biol. 19, 1875–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Thery C., Amigorena S., Raposo G., Clayton A. (2006) Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. Chapter 3, Unit 3.22 [DOI] [PubMed] [Google Scholar]
- 64. Peng L., Kapp E. A., Fenyo D., Kwon M. S., Jiang P., Wu S., Jiang Y., Aguilar M. I., Ahmed N., Baker M. S., Cai Z., Chen Y. J., Van Chi P., Chung M. C., He F., Len A. C., Liao P. C., Nakamura K., Ngai S. M., Paik Y. K., Pan T. L., Poon T. C., Salekdeh G. H., Simpson R. J., Sirdeshmukh R., Srisomsap C., Svasti J., Tyan Y. C., Dreyer F. S., McLauchlan D., Rawson P., Jordan T. W. (2010) The Asia Oceania Human Proteome Organisation Membrane Proteomics Initiative. Preparation and characterisation of the carbonate-washed membrane standard. Proteomics 10(22), 4142–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pisitkun T., Johnstone R., Knepper M. A. (2006) Discovery of urinary biomarkers. Mol. Cell. Proteomics 5, 1760–1771 [DOI] [PubMed] [Google Scholar]
- 66. Gonzales P. A., Pisitkun T., Hoffert J. D., Tchapyjnikov D., Star R. A., Kleta R., Wang N. S., Knepper M. A. (2009) Large-scale proteomics and phosphoproteomics of urinary exosomes. J. Am. Soc. Nephrol. 20, 363–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hayward I. P., Whitehead R. H. (1992) Patterns of growth and differentiation in the colon carcinoma cell line LIM 1863. Int. J. Cancer 50, 752–759 [DOI] [PubMed] [Google Scholar]
- 68. Peinado H., Aleckovic M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., Hergueta-Redondo M., Williams C., Garcia-Santos G., Ghajar C. M., Nitadori-Hoshino A., Hoffman C., Badal K., Garcia B. A., Callahan M. K., Yuan J., Martins V. R., Skog J., Kaplan R. N., Brady M. S., Wolchok J. D., Chapman P. B., Kang Y., Bromberg J., Lyden D. (2012) Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 18, 883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]