Abstract
Numerous studies highlight the fact that concerted proteolysis is essential for skin morphology and function. The cysteine protease cathepsin L (Ctsl) has been implicated in epidermal proliferation and desquamation, as well as in hair cycle regulation. In stark contrast, mice deficient in cathepsin B (Ctsb) do not display an overt skin phenotype. To understand the systematic consequences of deleting Ctsb or Ctsl, we determined the protein abundances of >1300 proteins and proteolytic cleavage events in skin samples of wild-type, Ctsb−/−, and Ctsl−/− mice via mass-spectrometry-based proteomics. Both protease deficiencies revealed distinct quantitative changes in proteome composition. Ctsl−/− skin revealed increased levels of the cysteine protease inhibitors cystatin B and cystatin M/E, increased cathepsin D, and an accumulation of the extracellular glycoprotein periostin. Immunohistochemistry located periostin predominantly in the hypodermal connective tissue of Ctsl−/− skin. The proteomic identification of proteolytic cleavage sites within skin proteins revealed numerous processing sites that are underrepresented in Ctsl−/− or Ctsb−/− samples. Notably, few of the affected cleavage sites shared the canonical Ctsl or Ctsb specificity, providing further evidence of a complex proteolytic network in the skin. Novel processing sites in proteins such as dermokine and Notch-1 were detected. Simultaneous analysis of acetylated protein N termini showed prototypical mammalian N-alpha acetylation. These results illustrate an influence of both Ctsb and Ctsl on the murine skin proteome and degradome, with the phenotypic consequences of the absence of either protease differing considerably.
Cathepsins B and L are ubiquitously expressed papain-like cysteine proteases belonging to the C1a papain family (clan CA), with 11 members in humans (1) and 18 members in mice (2). Most cysteine cathepsins like cathepsin L are endopeptidases, whereas cathepsin B shows both endopeptidase and carboxydipeptidase activity (3). Mainly localized in the endosomal/lysosomal compartment, cathepsins have traditionally been thought to play important roles in lysosomal protein turnover. Additional specific functions have been postulated that link cathepsins to different physiological and pathological processes.
Studies using cathepsin L (Ctsl)-gene-deficient mice1 revealed an important role of Ctsl in cardiac homeostasis (4–6) and a contribution of Ctsl to MHC II-mediated antigen presentation (7, 8) and prohormone processing (9, 10). In a mouse model of pancreatic neuroendocrine cancer, Ctsl promoted tumor growth and invasiveness (11, 12). In stark contrast, Ctsl was found to attenuate tumor progression in mouse models of skin cancer, highlighting the context-specific function of this protease (13, 14).
The most prominent phenotype of Ctsl-deficient mice is periodic hair loss together with epidermal hyperplasia, acanthosis, and hyperkeratosis (15). These alterations in skin morphology are assumed to be keratinocyte specific, as controlled re-expression of Ctsl under a keratin 14 promoter results in inconspicuous epidermal proliferation (16). The hair loss phenotype is caused by increased apoptosis and proliferation of hair follicle keratinocytes during the regression phase (catagen) of the hair follicle (17).
Cathepsin B (Ctsb)-gene-deficient mice do not display a spontaneous phenotype (18, 19), but if pathologically challenged these mice are less susceptible to disease in pancreatitis (20) and are less affected by TNFα-induced hepatocyte apoptosis (21). In tumor models of metastasizing breast cancer and pancreatic neuroendocrine neoplasias, mice deficient in Ctsb showed delayed cancer progression and reduced invasion (11, 22, 23). As good corroboration, the overexpression of Ctsb in the mouse mammary tumor virus–polyoma middle T breast cancer model promotes a more severe tumor phenotype (24).
In contrast to single-gene-deficient mice, mice with a double deficiency in both Ctsb and Ctsl die 4 weeks after birth as a result of pronounced lysosomal storage disease leading to neuron death in the cerebral cortex and the degeneration of cerebellar Purkinje cells (25). Because single-gene-deficient mice do not show autophagolysosomal and lysosomal accumulations in neurons, mutual compensation between Ctsb and Ctsl in vivo has been suggested (26).
The present proteomic study focuses on the molecular roles of Ctsb and Ctsl in skin homeostasis. We applied a 2-fold strategy consisting firstly of a gel-free quantitative proteomic approach (27, 28) to investigate protein alterations. Secondly, we performed terminal amine isotopic labeling of substrates (TAILS) (29) to determine changes in the skin proteome cleavage pattern and to identify Ctsb- and Ctsl-dependent cleavage events. Selected proteomic data were corroborated by means of immunodetection and immunohistochemistry. Selected mRNA levels were determined via qPCR in order to discriminate expression changes from posttranslational alterations. We identified specific proteomic and degradomic effects stemming from the deletion of either Ctsb or Ctsl. Our findings highlight the pivotal function of these proteases in maintaining proteome homeostasis and in balancing the proteolytic network. This is one of the first studies investigating how the deletion of individual proteases affects proteolytic processing in vivo.
EXPERIMENTAL PROCEDURES
Mice
The generation of Ctsb- and Ctsl-gene-deficient mice has been described previously (15, 19). All mice were backcrossed to the FVB/N background for at least 10 generations. All animal experiments were performed in compliance with institutional guidelines.
Preparation of Skin Samples
Dorsal skin was dissected after intracardiac perfusion with 0.9% NaCl and 0.4 units/ml heparin. Fresh tissue samples (200 mg) were lysed in 1 ml of homogenization buffer (100 mm Na-acetate, 5 mm EDTA, 1 mm dithiothreitol, 0.01 mm trans-epoxysuccinyl-l-leucylamido(4-guanidino)butane (E64), 1 mm phenylmethanesulfonyl fluoride, 0,05% Brij, pH 5.5) using an Ultra-Turrax and centrifuged at 1000g for 15 min at 4 °C. Protein concentrations were determined via Bradford assay (Bio-Rad, Munich, Germany).
Quantitative Proteome Comparison
For proteome comparison, samples from two Ctsb-deficient mice, two Ctsl-deficient mice, and four wild-type mice were prepared (two wild-type mice were used for comparison to two Ctsb-deficient mice, and two different wild-type mice were compared with two Ctsl-deficient mice). The preparation of mass spectrometry samples was performed as described previously, including stable isotope labeling with either d213C-formaldehyde (“heavy,” employed for wild-type skin samples) or d012C formaldehyde (“light,” employed for cathepsin-deficient skin samples) for quantitative comparison and pre-fractionation via strong cation exchange chromatography (30). LC-MS/MS analysis is described in the corresponding section. Data were converted to mzXML format (31) using ProteoWizard (32) with centroiding of MS1 and MS2 data. Peptide sequences were identified using X!Tandem (version 2010.12.01) (33), including cyclic permutation, in conjunction with PeptideProphet (part of version 4.3.1 of the Trans Proteomic Pipeline) (34) and a decoy search strategy: the complete mouse proteome file was downloaded from UniProt (35) on October 16, 2011, comprising 44,819 protein sequences. It was appended with an equal number of randomized sequences derived from the original mouse proteome entries. The decoy database was generated with DBToolkit (36). Tryptic cleavage specificity with no missed cleavage sites was applied. The mass tolerance was 10 ppm for parent ions and 0.5 for fragment ions. Static modifications were cysteine carboxyamidomethylation (+57.02 Da), lysine, and N-terminal dimethylation (light formaldehyde, 28.03 Da; heavy formaldehyde, 34.06 Da). X!Tandem results were further validated using PeptideProphet at a confidence level of >95%. Corresponding protein identifications are based on the ProteinProphet algorithm with a protein false discovery rate of <1.0%. Only protein identifications stemming from at least two peptide identifications were used. The relative quantitation for each protein was calculated from the relative areas of the extracted ion chromatograms of the precursor ions and their isotopically distinct equivalents using the ASAPratio algorithm (37). In our experience (shared by others (38)), ASAPratio occasionally displays inaccuracies with regard to background removal and separation of neighboring peaks along a given mass trace. To prevent inaccurate protein quantization, protein ratios were also analyzed using the XPRESS algorithm (39). Proteins were considered if XPRESS and ASAPratio yielded convergent results. Reported fold change (Fc) values are based on normalized ASAPratio.
Generation and Culturing of Cell Lines
Mouse embryonic fibroblast (MEF) cell lines have been described previously (30). A retroviral, bicistronic expression system with an internal ribosomal entry site was used for polyclonal, dosable expression of Ctsl. Mouse Ctsl cDNA was cloned into a pMIG expression vector for retroviral transfection (40). The resulting construct encodes for both Ctsl and green fluorescent protein (GFP). They are controlled by the same promoter, yielding a combined mRNA. Because of an internal ribosomal entry site between both coding sequences, Ctsl is translated as an untagged protein. However, Ctsl and GFP expression levels correlate. GFP expression was used to fractionate the cell population by means of preparative fluorescence-assisted cell sorting. Individual fluorescence-assisted cell sorting fractions were then probed for Ctsl expression via Western blot.
Western Blot
Cell conditioned media was harvested and concentrated as described elsewhere (30). 5 μg proteins from cell conditioned media or 80 μg proteins from skin lysates were loaded on to 12% SDS-polyacrylamide gels. Actin served as an internal loading control. After electrophoretic separation, proteins were transferred on polyvinylidene fluoride membranes using a semidry blot system (Bio-Rad, Munich, Germany). After blocking, the membranes were exposed to the primary antibodies (actin, 1:10,000; Ctsb, 1:500; Ctsd, 1:1000; Ctsl, 1:500; Ctsz, 1:500; dermokine, 1:1000; tubulin, 1:1000; notch-1, 1:500; periostin, 1:500) overnight at 4 °C. After washing, the membranes were incubated for 2 h with the secondary antibody. The membranes were washed and developed with the West Pico Chemiluminescent substrate (Pierce). Peroxidase activity was detected with a LumiImager device (Roche Applied Science, Mannheim, Germany). The primary antibodies were purchased from MP Biomedicals (Illkirch, France) (actin: Catalog No. 691001), Sigma-Aldrich (St. Louis, MO) (tubulin: Catalog No. T6199), Abcam (Cambridge, MA) (notch-1: Catalog No. ab27526), Proteintech (Chicago, IL) (dermokine: Catalog No. 16252–1-AP), and R&D Systems (Minneapolis, MN) (Ctsb: Catalog No. AF965; Ctsd: Catalog No. AF1014; Ctsl: Catalog No. AF1515; Ctsz: Catalog No. AF1033; periostin: Catalog No. AF2955).
RNA Isolation and Quantitative Real-time PCR Measurement
RNA was isolated using the SV Total RNA Isolation System (Promega, Madison, WI) and reversely transcribed to cDNA using the iScript cDNA Synthesis system (Bio-Rad). The primer sequences for quantitative real-time PCR (by MyiQ, Bio-Rad) were as follows: actin forward → 5′-ACC CAG GCA TTG CTG ACA GG-3′, actin reverse → 5′-GGA CAG TGA GGC CAG GAT GG-3′, Ctsd forward → 5′-GTG CAC ATG GAC CAG TTG GA-3′, Ctsd reverse → 5′-CAA TAG CCT CAC AGC CTC CCT-3′, cystatin B forward → 5′-CAC GGC CGA GAC GCA GGA G-3′, cystatin B reverse → 5′-CAG CCA CTA TCT GTC TCT TGA AG -3′, cystatin C forward → 5′-TCG CTG TGA GCG AGT ACA AC-3′, cystatin C reverse → 5′-ATC TGG AAG GAG CAG AGT GC-3′, periostin forward → 5′-AAC CAA GGA CCT GAA ACA CG-3′, periostin reverse → 5′-GTG TCA GGA CAC GGT CAA TG-3′. Cycling conditions were as follows: 1 cycle of 72 °C for 1 min; 50 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s; and 1 cycle of 72 °C for 5 min.
Histology and Immunohistochemistry
Paraffin sections of back skin were deparaffinized in xylene, hydrated in graded ethanol solutions, and stained with hematoxylin and eosin. Visualization of the primary antibody (periostin, Catalog No. AF2955, R&D Systems, Minneapolis, MN) was via peroxidase reaction using the Vectastain Elite ABC Kit (Vector Laboratories, Burlingame, CA). Microscopy was performed using an Axioplan microscope (Zeiss, Stuttgart, Germany), and digital images were acquired with an Axiocam camera (Zeiss, Stuttgart, Germany). Adobe Photoshop software was used for slight contrast enhancement.
Preparative Fluorescence-assisted Cell Sorting
MEFs were sorted according to their GFP expression level using a MoFlo high speed cell sorter (Beckman Instruments).
Cleavage Site Analysis with TAILS
TAILS was performed using formaldehyde labeling according to the original publications (29, 41). After tryptic digest samples were desalted using a reversed phase C18 column, prefractionated via strong cation exchange chromatography as described, and desalted using self-packed C18 STAGE tips (Empore 3M, Minneapolis, MN) (42). LC-MS/MS analysis is described in the corresponding section. Data were converted to mzXML format (31) using mzWiff (version 4.3.1) with centroiding of MS1 and MS2 data, precursor charge determination, and deisotoping of MS2 data. Peptide sequences were identified by X!Tandem (version 2010.12.01) (33), including cyclic permutation, in conjunction with PeptideProphet (part of version 4.3.1 of the Trans Proteomic Pipeline) (25). The same mouse proteome database as described in the section “Quantitative Proteome Comparison” was used. Semi Arg-C specificity with up to three missed cleavage sites was applied. Static modifications are +57.02 Da, lysine, and N-terminal dimethylation (light formaldehyde, +28.03 Da; heavy formaldehyde, +34.06 Da). For acetylated N termini (+42.01 Da), modifications were cysteine carboxyamidomethylation, N-terminal acetylation, and lysine dimethylation. Asymmetric mass tolerance was 0–120 ppm for precursor ions and 0.05 Da for fragment ions. X!Tandem results were further validated by PeptideProphet at a confidence level of >95%. The relative quantification for each peptide was calculated using the ASAPratio algorithm (37) as described in the section “Quantitative Proteome Comparison.” Fc values were based on ASAPratio normalized for all peptide ratios. Two biological replicates with independent sample preparation and mass spectrometric measurement were analyzed. Samples from two Ctsb-deficient mice, two Ctsl-deficient mice, and four wild-type mice were prepared. Ratios were divided into the following quantiles: 0–15, 15–25, 25–75, 75–85, and 85–100 (43). N termini were considered as decreased if found in the quantile 0–15 in one biological replicate and in the quantile 0–25 in the second replicate. Similiarly, N termini were considered as increased if found in the quantile 75–100 in one biological replicate and in the quantile 85–100 in the second replicate. Analysis and graphs were done in R (v2.15.0) with Rstudio as a front-end integrated development environment.
LC-MS/MS Analysis
For mass spectrometric measurements, two nanoflow-LC-MS/MS systems were used: system A, an Orbitrap XL (Thermo Scientific GmbH, Bremen, Germany), for quantitative proteome comparison, and system B, a Qstar Elite (AB Sciex, Darmstadt, Germany), for cleavage site analysis with TAILS. Each was coupled to an Ultimate3000 micro pump (Dionex, Idstein, Germany). As mobile phases, 0.5% acidic acid and 0.5% acetic acid (ACS reagent; Sigma, Steinheim, Germany) in 80% acetonitrile were used (water and acetonitrile were of HPLC grade or better). HPLC column tips (fused silica) with a 75 μm inner diameter (New Objective, Woburn, MA) were self-packed (44) with Reprosil-Pur 120 ODS-3 (Dr. Maisch, Ammerbuch, Germany). The mass spectrometers were operated in the data-dependent mode for MS and MS/MS. For the Orbitrap XL, each MS scan was followed by a maximum of five MS/MS scans, and in the case of the Qstar Elite by four MS/MS scans. For the Qstar Elite, the smart mode of the Analyst 2.0 software for MS/MS accumulation was used.
Statistical Analysis of Protein Abundance Data
Ratios of each dataset were calculated as cathepsin depleted over control condition. For data analysis and transformation, ASAPratio data were log2-transformed. Mean Fc values, including the standard deviation and 90% confidence interval (based on a Student's t test distribution), were calculated for each protein identified in both replicates. ASAPratio p values were combined using Fisher's method. Protein abundance was considered as significantly altered if the following three criteria were met: (i) 90% confidence did not cross zero, (ii) Fisher's method yielded a combined p value < 0.1, and (iii) manual extracted ion chromatogram inspection corroborated the calculated heavy/light value.
TAILS Ranking Scheme
The scoring scheme was based on a recent proteomic characterization of Ctsb and Ctsl specificity (45). In the Ctsb ranking scheme, one point was awarded for P, F, Y, W, I, V, or A in P2; R, K, G, A, or T in P1; G, F, L, or V in P1′; and I, V, G, A, S, or T in P2′ position. Two points were given for a G residue at position P3′. One point was subtracted (penalty) if an E, D, or G residue was located at P2, P1′, or P2′ and if a V residue was located at position P1. Two points were subtracted (penalty) if a P residue was located at position P1 or P1′. In the Ctsl ranking scheme, two points were given for P, F, Y, W, I, V, or L in P2; G, A, S, T, or Q in P1; G, A, S, T, or H in P1′; and G, A, S, T, or P in P2′. Two points were subtracted if G, A, S, T, E, K, or R residues were located in P2 and if a P residue was located at position P1 or P1′.
In Vitro Cleavage Assays
Recombinant human Ctsl (0.5 ng/μl) and murine periostin (50 ng/μl) were incubated overnight at 37 °C in sodium acetate buffer (200 mm sodium acetate, 1 mm EDTA, 0.05% Brij 35, pH 5.5) or sodium phosphate buffer (100 mm sodium phosphate, 0.4 m sodium chloride, 10 mm EDTA, pH 7). Dextrane sulfate was added at a concentration of 10 ng/μl. Samples were loaded on to 12% SDS-polyacrylamide gels, and bands were visualized by means of silver staining.
Aortic Ring Assay
Rat aortic explant cultures were performed as described in Ref. 46. Briefly, rat aortas were prepared, cleared from periaortic fibroadipose tissue, and sectioned into 1-mm-thick aortic rings. The aortic rings were embedded in a collagen I matrix (1.5 mg/ml) (collagen R from rat tail tendons, Serva, Heidelberg, Germany) and cultured in serum-free DMEM (GIBCO, Carlsbad, CA). Murine recombinant VEGF (20 ng/ml) (Peprotech, Rocky Hill, NJ) was supplemented to induce vessel growth. The cysteine cathepsin inhibitor E64d (Bachem, Bubendorf, Switzerland) was administered at different concentrations (5 μm, 20 μm). Media were replaced every third day. Microscopic images were taken at day 9, and the vessel density was computationally quantified as described elsewhere (47).
Microarray Processing and Data Analysis
RNA from mouse embryonic fibroblasts was isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany). RNA integrity and quality were estimated on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA), and the RNA integrity number index was calculated for each sample using the Agilent 2100 Expert software. All samples reached an integrity number > 9.0 and were further processed. For transcriptomic analysis, the MouseWG-6 v2.0 Expression BeadChip Kit (Illumina, San Diego, CA) was used according to the manufacturer's instructions.
Microarray scanning was done using a Beadstation array scanner, with the setting adjusted to a scaling factor of 1 and photomultiplier tubes, settings at 430. Data extraction was done for all beads individually, and outliers were removed at >2.5 median absolute deviation. All remaining data points were used for the calculation of the mean average signal for a given probe, and the standard deviation for each probe was calculated. Data analysis was done via normalization of the signals using the quantile normalization algorithm without background subtraction. Probes with a bad quality score according to the manufacturer or those having a low detection significance (p < 0.05) were discarded. For further analysis, only probes having an Entrez ID were retained, always choosing probes with the largest interquartile range across experiments if multiple probes matched the same Entrez ID.
To compare the fold differences of two wild-type cell lines and the two respective Ctsb-deficient or Ctsl-deficient cell lines, we calculated the gene-wise fold expression difference for each wild-type-knockout pair. The resulting fold difference distributions were fitted with a skewed t-distribution to account for the asymmetry in differentially up- and down-regulated genes (48). Overall p values for differential regulation were calculated from the one-tailed p values of the individual skew-t distributions using Fisher's method. Overall fold expression differences denote the mean of the experimental fold differences. Genes were considered significantly up- or down-regulated at a combined p value < 0.01.
RESULTS AND DISCUSSION
Impact of Ctsb or Ctsl Ablation on Skin Proteome Composition
To elucidate the contribution of Ctsb and Ctsl to the skin proteome, we quantitatively compared the proteomes of skin lysates of wild-type and Ctsb or Ctsl knockout mice, respectively. Each quantitative proteome comparison was performed in two biological replicates comparing skin from two different wild-type and two different knockout mice. Skin samples comprised epidermis, dermis, and hypodermis. Stable isotope labeling with either d213C-formaldehyde (heavy) or d012C formaldehyde (light) in combination with LC-MS/MS was applied. Comparison of wild-type and Ctsb-deficient skin samples identified 1472 proteins in the first biological replicate (supplemental Table S1) and 1546 proteins in the second biological replicate (supplemental Table S2). 1191 proteins were identified in both experiments (Fig. 1A). Comparison of wild-type and Ctsl−/− skin samples identified 1635 proteins in the first biological replicate (supplemental Table S3) and 1658 proteins in the second biological replicate (supplemental Table S4). 1317 proteins were detected in both replicates (Fig. 1A). Alterations of protein abundance in all quantitative proteome comparisons are presented as fold change values (Fc; log2 of heavy/light ratio). Average Fc values, together with their distribution for all identified proteins, are shown in supplemental Figs. S1A and S1B and supplemental Tables S5 and S6. All replicates showed a near-normal distribution (distribution profile in supplemental Figs. S1A and S1B) and an average Fc close to 0 (Fig. 1B). Both Ctsb and Ctsl had a pronounced impact on the skin proteome composition. 1191 proteins were identified in both Ctsb replicates, and 1317 proteins were identified in both Ctsl replicates. 356 proteins were changed more than 2-fold (Fc < −0.58 or Fc > 0.58) in both Ctsb replicates, and 517 proteins were changed more than 2-fold in both Ctsl replicates.
Fig. 1.
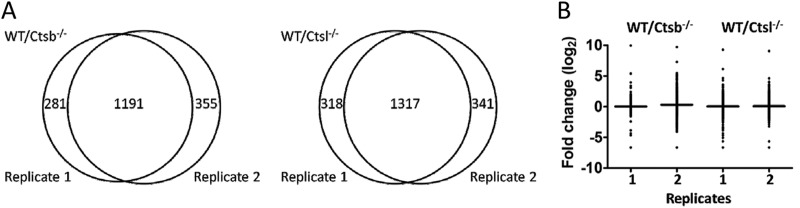
Protein identification and quantification for each biological replicate. A, 1191 proteins were identified in both biological replicates for the comparison of wild-type and Ctsb−/− skin samples. 1317 proteins were identified in both biological replicates for the comparison of wild-type and Ctsl−/− skin samples. B, distribution and geometric mean (horizontal bar) of fold change values (log2) of proteins from each replicate of the Ctsb and Ctsl experiments, respectively.
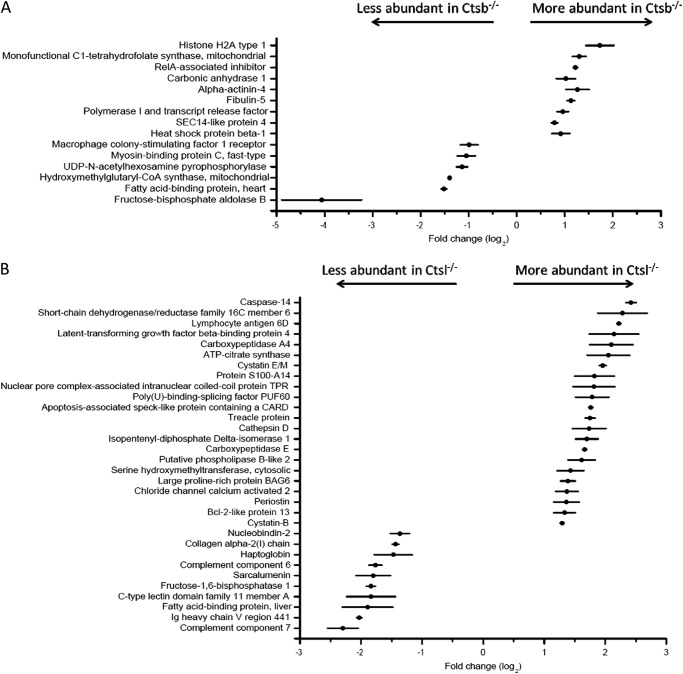
In order to consider individual protein abundances as significantly altered, we combined the three following criteria: (a) The 90% confidence interval of the averaged Fc value does not span zero. (b) Combination of the ASAPratio p values (37) using Fisher's method yields a merged p value < 0.1. Noteworthy, ASAPratio p values smaller than 0.1 to 0.2 are typically considered as indicating altered protein quantity (37, 49). (c) Manual inspection of the extracted ion chromatograms corroborates the heavy/light ratio calculated by ASAPratio. With these strict criteria employed, 15 proteins displayed significantly altered abundance in Ctsb-depleted skin (Fig. 2A), and 32 proteins displayed altered abundance in Ctsl-depleted skin (Fig. 2B).
Fig. 2.
Proteins with significantly altered abundance in the Ctsb (A) and Ctsl experiments (B). Horizontal bars indicate the 90% confidence interval; black circles indicate average fold change values (log2). For each protein shown in this figure, Fisher's test yielded a merged p value < 0.1. See “Experimental Procedures” for details.
Ctsl ablation affects the abundances of several proteins that are associated with overt skin phenotypes in the respective gene knockout mice. This is the case for cathepsin D, cystatin M/E, periostin, and caspase 14 (50–54). In contrast, only one protein affected by Ctsb is associated with a murine skin phenotype (RelA-associated inhibitor (55)). The more pronounced impact of Ctsl ablation on important components in mouse skin biology corresponds nicely to the overt skin phenotype in Ctsl−/− mice and the lack thereof in Ctsb−/− mice.
Ctsl Deletion Results in Altered Abundances of Several Proteases and Protease Inhibitors
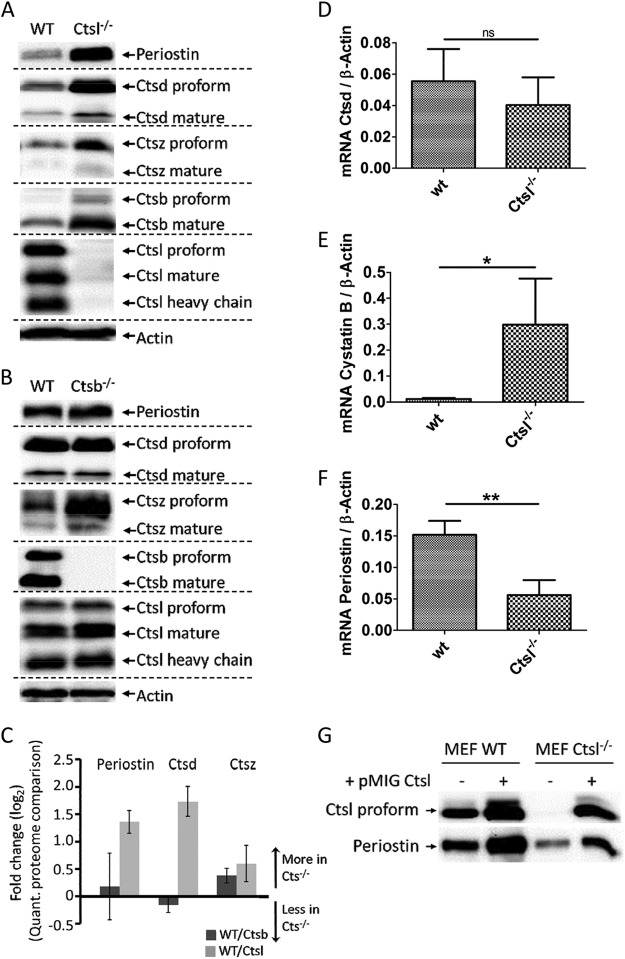
Ctsl deficiency significantly affected the abundances of several proteases and protease inhibitors. This includes increased abundances of cathepsin D (Ctsd), carboxypeptidase A4, carboxypeptidase E, caspase 14, and cystatins E/M and B. Immunoblotting corroborated increased Ctsd levels in Ctsl−/− skin (Fig. 3A), whereas qPCR analysis indicated unaffected Ctsd transcript levels, suggesting that Ctsd accumulation is a posttranslational effect (Fig. 3D). Ctsd is known as a positive regulator of epidermal differentiation and barrier function (51, 52, 56). In skin, its abundance is specifically affected by Ctsl deficiency and not by the absence of Ctsb (Fig. 3B). This gives further evidence of the long-standing hypothesis that Ctsl contributes significantly to the degradation, and thereby inactivation, of Ctsd (57). The ability of Ctsl to degrade Ctsd has already been shown in vitro (57, 58). Increased levels of active Ctsd have been found in Ctsl−/− MEFs (59) and, most notably, in the brains of Ctsb/Ctsl double deficient mice (25, 26). However, it is unresolved whether the Ctsd increase is a compensatory safeguard or promotes pathological tissue damage.
Fig. 3.
Immunoblot analysis of periostin, Ctsb, Ctsd, Ctsl, and Ctsz comparing wild-type (WT) with (A) Ctsl−/− and (B) Ctsb−/− skin samples. Actin was used as a loading control. C, averaged MS fold changes (log2) of periostin, Ctsd, and Ctsz for the quantitative proteome comparison of WT/Ctsb−/− (dark gray bars) and WT/Ctsl−/− (light gray bars). mRNA expression for Ctsd (D), cystatin B (E), and periostin (F) in WT and Ctsl-depleted skin as measured by qPCR. Gene expression data were normalized to the expression of β-actin and are presented as mean ± S.E. (n = 3). ns, not significant (p > 0.05; unpaired t test); * significant (p < 0.05; unpaired t test); ** significant (p < 0.01; unpaired t test). G, re-expression of Ctsl in Ctsl−/− MEFs and over-expression in WT MEFs. Re-expression of Ctsl to WT-like expression levels using the retroviral expression vector pMIG restores WT-like periostin expression in cell conditioned media. Mild overexpression of Ctsl further increases periostin abundance.
Cystatins are tight-binding inhibitors of cysteine proteases like Ctsb and Ctsl (60). Cystatin E/M is important for epidermal differentiation. Mice lacking cystatin E/M display an ichthyosis-like phenotype due to impaired epidermal differentiation and die within 2 weeks after birth (53, 61). The known target proteases of cystatin E/M in the epidermis are legumain and cathepsins L and V (62). The deletion of cathepsin L but not of legumain prevents the lethality of cystatin E/M deficiency in mice (63). While cystatin E/M is secreted, cystatin B is predominantly located in the cytosol. It inhibits a wider range of proteases including cathepsins B, H, L, and S (64). qPCR analysis showed an increase of cystatin B mRNA levels in the absence of Ctsl (Fig. 3E), suggesting that its accumulation is due to increased gene expression.
As outlined above, we chose strict criteria to identify proteins with altered abundance. The strictness of our criteria is illustrated by proteins that fail to meet these conditions but for which immunodetection still corroborates altered abundance. This is the case for cathepsin Z, which is more abundant in both Ctsb- and Ctsl-deficient skin (Figs. 3A, 3B).
The examples above suggest that the removal of a single protease can result in altered abundance of further proteases and protease inhibitors. To evaluate this observation, we performed genome-wide transcriptomic analysis of MEFs lacking either Ctsb or Ctsl in comparison to wild-type fibroblasts. As shown in supplemental Figs. S2A and S2B, the removal of either Ctsl or Ctsb significantly affects the transcript levels of a large number of proteases and protease inhibitors.
Periostin Accumulates in Ctsl−/− Skin, Superseding Transcriptional Down-regulation
Periostin is an extracellular matrix protein with important involvement in skin physiology and cancer biology. Periostin-deficient mice display impaired collagen fibrillogenesis leading to increased skin stiffness (50). Periostin mediates cancer stem cell maintenance and metastatic dissemination by facilitating increased Wnt signaling (65). Our data reveal that periostin protein is significantly enriched in Ctsl−/− skin. Its accumulation was corroborated by immunoblotting (Fig. 3A). This effect was specific for Ctsl and was not observed in Ctsb−/− skin (Fig. 3B).
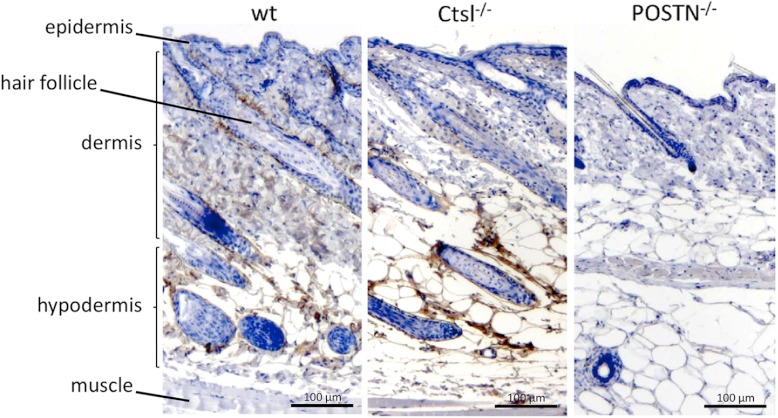
qPCR analysis showed a significant decrease of periostin mRNA levels in Ctsl deficiency (Fig. 3F). We previously described a similar effect of Ctsl on periostin transcript levels in MEFs (30). In MEFs, the decrease in periostin mRNA is accompanied by a corresponding decrease in periostin protein (30). In murine skin, however, the decrease in periostin mRNA is superseded by an accumulation of the corresponding protein, predominantly in the hypodermis (Fig. 4), and to a lesser extent around hair follicles.
Fig. 4.
Immunohistochemical analysis for periostin in wild-type and Ctsl-depleted skin. Periostin (POSTN)-deficient skin served as the control. One representative staining out of four independent experiments is shown.
These results indicate that in vivo, Ctsl plays an important role in regulating periostin abundance on both the transcriptional and the post-transcriptional level. Whereas the transcriptional impact likely represents a secondary or downstream effect of Ctsl deletion, the accumulation of periostin protein in skin indicates impaired degradation, either by Ctsl or by other proteases with impeded activity upon Ctsl ablation. The latter possibility includes decreased protease abundance or increased abundance of protease inhibitors. We are not aware of another case in which transcriptional down-regulation is superseded in such a prominent manner by post-translational accumulation of the corresponding protein.
To test whether Ctsl itself has the ability to degrade periostin, we performed in vitro cleavage assays. Ctsl degraded periostin at pH 5.5. Only minor processing was observed at pH 7.0 (supplemental Fig. S3A), even in the presence of dextrane sulfate (supplemental Fig. S3B), which reportedly stabilizes cathepsin activity at neutral and basic pH levels (66). Periostin predominantly accumulates in the hypodermis with a neutral to basic extracellular pH. Possibly, Ctsl degrades endocytosed, internalized periostin. In fact, periostin is known to bind to cell surface integrins (67).
To further support our observation that Ctsl has a strong impact on periostin transcript levels, we re-expressed Ctsl in Ctsl−/− MEFs and mildly overexpressed Ctsl in wild-type MEFs. As outlined above, Ctsl deletion in MEFs led to decreased periostin mRNA (similar to the situation in skin) and a corresponding decrease of periostin protein. We chose a bi-cistronic expression system with retroviral transfection to yield polyclonal, stable, and dosable Ctsl expression. In Ctsl−/− MEFs, we achieved wild-type-like re-expression of Ctsl (Fig. 3G). In wild-type MEFs, additional expression of Ctsl increased its abundance more than 2-fold (Fig. 3G). Re-expression of Ctsl rescued the periostin phenotype, whereas Ctsl overexpression in MEFs led to increased periostin abundance (Fig. 3G). These data underline the strong effect of Ctsl on periostin expression.
Pathways and Protein Classes Affected by Cathepsin Depletion
To obtain a global picture of pathways and protein classes affected by the depletion of either Ctsb or Ctsl, we analyzed all averaged protein ratios with PANTHER gene expression tools (68) (supplemental Tables S5 and S6). This approach focused on the combination and classification of individual proteins and their ratios, with the aim of identifying common biological themes. Importantly, the individual proteins that are found for a biological theme might each be only moderately affected by cathepsin ablation.
Generally, pathway analysis for Ctsb and Ctsl achieved similar results (supplemental Figs. S4A, S4B). For example, the ablation of either Ctsb or Ctsl led to elevated levels of proteins involved in the ubiquitin proteasome pathway, possibly indicating a compensatory mechanism for protein turnover. A specific effect of Ctsb, but not of Ctsl, was observed in the VEGF signaling pathway and in angiogenesis. This effect involved elevated levels of several MAPKs, GSK3α, and Stat1. A specific role of Ctsb, but not of Ctsl, in angiogenesis was found previously in a study investigating the roles of different cathepsins in a mouse model of pancreatic islet cell carcinogenesis (11). Matrigel-embedded human umbilical vein endothelial cells showed a significant reduction in the length of capillary-like tubes upon application of the cathepsin B specific inhibitor CA074Me (69). In line with this observation, VEGF-induced angiogenesis is reduced upon the application of CA074Me in cathepsin B–null mice (70). By performing an aortic ring assay, we observed reduced vessel sprouting from rat aortic rings upon VEGF induction using the cysteine cathepsin inhibitor E64d (supplemental Fig. S5).
PANTHER protein class analysis revealed that the removal of either Ctsb or Ctsl had a strong effect on the abundance of numerous proteins associated with proteolysis (supplemental Figs. S4C, S4D). In addition to the aforementioned cystatins (affected predominantly by Ctsl), this includes the decreased abundance of several serine protease inhibitors upon the deletion of either cathepsin. An interconnection between cathepsin and serpine abundance is further supported by the aforementioned study investigating the contribution of cathepsin L to the secretome composition of MEFs. Here, the removal of cathepsin L repeatedly reduced the abundance of serpine E2 by more than 50% (30). These observations exemplify cross-talk between mechanistically different classes of proteolysis. In fact, cross-class inhibition between some serpins and several cysteine cathepsins has been reported (71, 72). Decreased serpine abundance coincides with increased proteolytic processing of serpins in cathepsin-deficient skin (see below), presenting a putative explanation for the altered abundance of this protein class.
Systemic Effect of Ctsb and Ctsl on Skin Proteolysis: N-terminal Cleavage Site Analysis
Because both cathepsins are active proteases and are important for regulating the abundance of further proteases and protease inhibitors, we applied a quantitative N-terminomic technique in order to better understand how Ctsb and Ctsl contribute to proteolytic processes in the skin. TAILS (29, 73) has been applied to assess the influence of Ctsl depletion on extracellular proteolysis in MEFs (30). The TAILS approach allows for the quantitative comparison of protein N termini from different biological samples based on a negative selection strategy. Both naturally unmodified and naturally modified (e.g. acetylated) N termini can be identified. “Unmodified” protein N termini possess an amino group (–NH2) and might include native as well as proteolytically generated protein N termini. These naturally unmodified protein N termini are chemically dimethylated using isotopic labeled formaldehyde. Thus, natively unmodified protein termini are identified as being dimethylated. In most cases, natively unmodified protein termini correspond to sites of proteolytic processing.
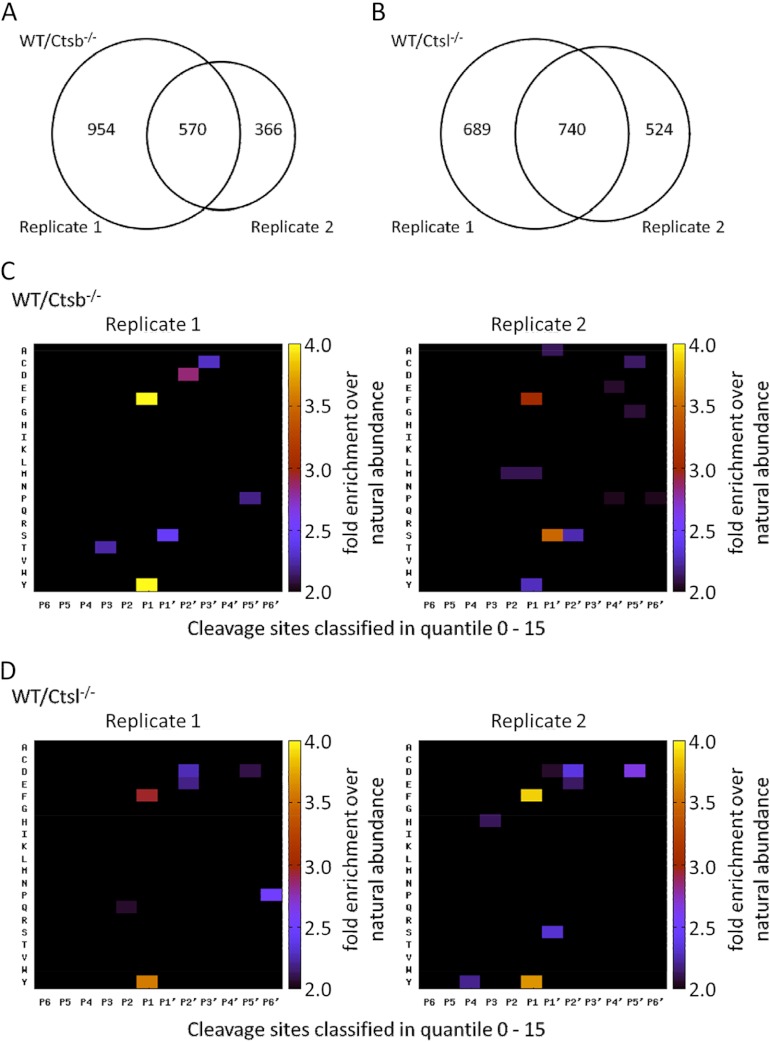
TAILS was performed in two biological replicates comparing skin from two different wild-type mice and two different mice deficient in either Ctsb or Ctsl. All replicates showed a near-normal distribution (distribution profile in supplemental Figs. S6A and S6B). For the Ctsb experiment, TAILS identified 1524 naturally unmodified (chemically dimethylated) N termini in the first replicate and 936 naturally unmodified (chemically dimethylated) N termini in the corresponding second replicate (supplemental Tables S7 and S8; annotated spectra in supplemental files 1 and 2). Of these, 570 N termini were present in both replicates (Fig. 5A, supplemental Table S9). In the Ctsl experiment, we identified 1429 dimethylated N termini in the first biological replicate and 1264 N termini in the second biological replicate (supplemental Tables S10 and S11; annotated spectra in supplemental files 3 and 4). 740 dimethylated N termini were detected in both replicates (Fig. 5B and supplemental Table S12).
Fig. 5.
N-terminal peptides identified and quantified for each biological replicate in the TAILS experiment. A, 570 peptides were identified in both biological replicates of the Ctsb experiment. B, 740 peptides were identified in both biological replicates of the Ctsl experiment. Numbers refer to naturally unmodified, chemically dimethylated N termini. C, D, global specificity pattern upon Ctsb deficiency (C) and Ctsl deficiency (D). Graphical presentation of the secretome specificity profile of all N termini found in the 0–15 quantile and therefore decreased upon Ctsb ablation (C) or Ctsl ablation (D). Positional occurrences are shown as enrichment over natural abundance of murine amino acid abundances as derived from the International Protein Index (97). TAILS identifies prime-site sequences of proteineous cleavage sites. The corresponding non-prime sequences were derived bioinformatically through database searches similar to the proteomic identification of protease cleavage sites for protease specificity characterization (74, 75).
To discern natively unmodified cleavage sites with reduced abundance upon cathepsin removal (cathepsin-dependent cleavage events), we chose a quantile-based approach as described under “Experimental Procedures.” In the Ctsb experiment, 37 N termini were decreased and 61 N termini were increased in both biological replicates. In the Ctsl experiment, 80 N termini were decreased and 116 N termini were increased upon Ctsl deficiency (supplemental Tables S7, S8, S10, and S11). This result is in good agreement with the proteome data, where we observed stronger effects upon Ctsl depletion than upon Ctsb depletion. Nevertheless, the removal of either Ctsb or Ctsl has a systemic and substantial effect on global skin proteolysis.
In some cases, the altered abundance of a proteolytically processed N terminus reflects altered abundance of the corresponding protein rather than impaired proteolytic processing. For example, several cleavage sites in collagen α-2 (type I) are less abundant in Ctsl−/− skin. This is best explained, however, by a reduced overall abundance of collagen α-2 (Fig. 2B), rather than assuming altered proteolysis of this protein. Similarly, altered abundances of processing sites in procollagen C-endopeptidase enhancer 1 in Ctsb−/− skin are best explained by the decreased overall abundance of this protein.
To test whether decreased or increased N termini predominantly represent altered protein abundance (as in the above examples), we selected those proteins for which the TAILS procedure identified an N terminal peptide (analysis restricted to proteins and N termini identified in both biological replicates). We found only a weak correlation (Ctsb: r = 0.106; Ctsl: r = 0.399) between Fc values of N termini and the corresponding proteins (supplemental Figs. S7A and S7B). This shows that quantitatively altered N termini typically do not represent altered protein abundance and rather reflect altered proteolysis on a global scale.
Cathepsin Deficiency Results in Dominant “Secondary” or “Downstream” Effects on Skin Proteolysis
Although quantitatively decreased cleavage events in Ctsb−/− or Ctsl−/− skin evidently depend on the presence of either Ctsb or Ctsl, they do not necessarily represent actual Ctsb or Ctsl cleavage sites. As shown in this study, both Ctsb and Ctsl represent important nodes in the protease web that regulate the abundance of several other proteases and protease inhibitors. Thus, the aforementioned decreased cleavage events might represent the altered activity of further proteases rather than primary cathepsin cleavage sites. In fact, we have previously shown for MEFs that Ctsl strongly contributes to extracellular proteolysis by altering the activity of other proteolytic enzymes (30).
TAILS identifies the C terminal (prime site) part of a cleavage site. For a more comprehensive analysis, we derived the corresponding N terminal (non-prime site) sequences using a bioinformatic strategy that has been established for the proteomic identification of protease cleavage sites (74–76).
For those cleavage sites that depend on the presence of either of Ctsb or Ctsl, we show the global cleavage pattern in Figs. 5C and 5D. N-terminal to the scissile peptide bond, the removal of either Ctsb or Ctsl predominantly affected cleavage sites with aromatic residues in P1 (Schechter and Berger nomenclature, Ref. 77). Noteworthy, the “specificity patterns” are reproducible for the biological replicates of the Ctsb or Ctsl experiment. Similarly, the TAILS specificity pattern for the Ctsl−/− secretome has shown robust reproducibility (30). Together, these data underline that cathepsins B and L systematically and robustly contribute to skin proteolysis.
Importantly, the global TAILS profiles of cathepsin-dependent cleavage sites do not correspond to the well-characterized specificity of Ctsb and Ctsl (74). Both cathepsins prefer aromatic residues in P2. In addition, Ctsb strongly prefers P3′ glycine. Thus, global analysis of cathepsin-dependent cleavage events suggests that the majority of these cleavage events do not represent actual Ctsb or Ctsl cleavage sites but rather stem from the altered activity of other proteases. This finding is in good agreement with our results obtained via quantitative proteomic analysis of Ctsb- or Ctsl-deficient skin; the outcome of these experiments points to an important role of both cathepsins in controlling the abundance of a multitude of proteases and protease inhibitors. The loss of either Ctsb or Ctsl causes substantial perturbations of protease activity, which in turn outshine actual Ctsb or Ctsl cleavage sites.
At the same time, the lack of a pronounced cathepsin specificity fingerprint also might indicate that Ctsb and Ctsl have predominantly degradative functions and yield only a few stable cleavage products.
Processing Sites in Dermokine and NOTCH1
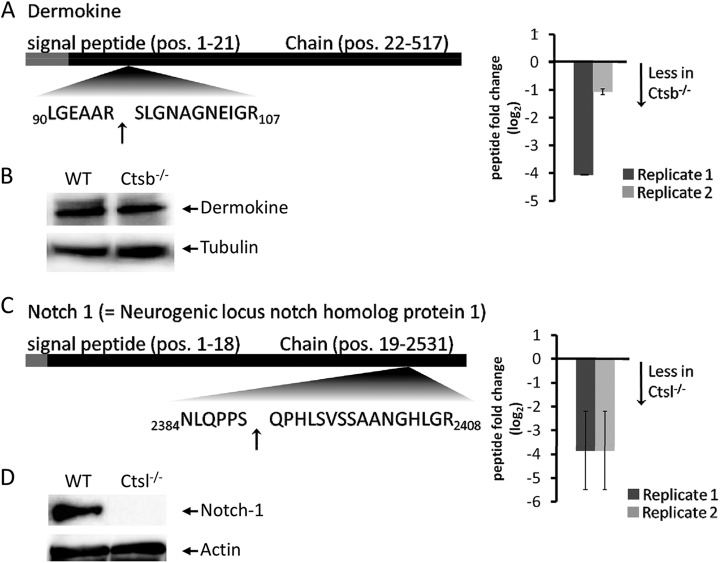
Cathepsins are broad-specificity proteases. In contrast to highly specific proteases such as caspases, they display positional preferences rather than strict selectivity for individual amino acids. To account for this broad specificity and to identify depleted cleavage sites that correspond to Ctsb or Ctsl specificity, we implemented a scoring scheme for P2-P3′ based on the active site specificity of Ctsb and Ctsl (74). The score is augmented for residues in P2-P3′ that match Ctsb or Ctsl specificity, whereas residues that contradict cathepsin specificity lead to a penalty (details provided under “Experimental Procedures”). We applied this ranking scheme to Ctsb- or Ctsl-dependent N termini that were decreased in abundance according to our quantile analysis in both biological replicates. (Ctsb experiment: supplemental Table S13; Ctsl experiment: supplemental Table S14). In agreement with the global pattern of Ctsb- or Ctsl-dependent cleavage sites, few cleavage sites achieved a score > 0, with maximum scores of 6 for Ctsb and 5 for Ctsl. A Ctsb-dependent cleavage site in dermokine corresponds closely to Ctsb specificity (e.g. through a P3′ glycine) (Fig. 6A). Dermokine was not identified in the quantitative proteome comparison, but immunoblotting revealed equal dermokine protein levels in Ctsl-depleted and wild-type skin (Fig. 6B). Cleavage products reflecting cleavage events in dermokine were not detected. Possibly, cleavage products are present in substantially lower amounts than in intact dermokine and are therefore not detected by immunoblotting. Nevertheless, the processing of dermokine by Ctsb needs to be investigated further. Dermokine is secreted by keratinocytes and is discussed as a biomarker for early-stage colorectal cancer (78, 79). Proteolytic processing of dermokine has not yet been reported (80).
Fig. 6.
A, novel processing site in dermokine that is affected by deletion of Ctsb. N termini were identified and quantified via TAILS in both biological replicates, including their fold changes (log2) (black bars = replicate a; gray bars = replicate b; arrows indicate the identified cleavage site). B, immunoblot analysis of dermokine comparing wild-type and Ctsb−/− skin samples. Tubulin was used as a loading control. C, novel processing site in notch-1 that is affected by the deletion of Ctsl. N termini were identified and quantified via TAILS in both biological replicates, including their fold changes (log2) (black bars = replicate a; gray bars = replicate b; arrows indicate the identified cleavage site). D, immunoblot analysis revealed a 15-kDa notch-1 cleavage product comparing wild-type and Ctsl−/− skin samples using an C-terminal antibody. Actin was used as a loading control.
We also detected a novel cleavage site in the Notch 1 intracellular domain (Fig. 6C). In both biological replicates, this cleavage event strongly depended on the presence of Ctsl. However, the cleavage sequence lacked the prototypical Ctsl specificity determinant of hydrophobic residues in P2. Notch signaling is critical for epidermal and hair follicle differentiation (81–83). The identified neo-N terminus reflects a cleavage event at residue 2390 in the Notch intracellular domain, leading to a C-terminal fragment of 141 amino acids. This fragment represents the conserved domain DUF3454 with unknown functionality. The presence of this fragment was corroborated via immunoblotting using an antibody detecting the C terminus of notch-1 (supplemental Fig. S6D). Notch-1 was not identified in the quantitative proteome comparison. However, immunoblotting uncovered globally depleted notch-1 levels in Ctsl-deficient skin (data not shown). This has been observed using the C-terminal antibody and an antibody detecting cleaved notch-1 (Val1744). Thus the C-terminal fragment of 141 amino acids represents altered notch-1 levels in Ctsl-deficient skin, rather than Ctsl-dependent processing. With respect to the hair and skin phenotype of Ctsl-deficient mice, altered Notch signaling is of great interest. However, it is currently not known whether this novel Notch intracellular domain processing site affects Notch signaling. Granzyme B has been reported to process the Notch intracellular domain at several sites (84); however, cleavage at residue 2390 does not correspond to the strict granzyme specificity for P1 aspartate. Interestingly, periostin has been found to interact with notch-1, thereby influencing its extracellular proteolytic processing (85).
Increased Proteolysis of Serpines Coincides with Their Decreased Protein Abundance
Terminomic approaches such as TAILS are designed to detect N termini of stable cleavage products. Although limited proteolysis differs from degradation processes, the lack of a specific cleavage event might result in an accumulation of the corresponding protein. We examined whether underrepresented cleavage sites in cathepsin-deficient skin are observed for proteins with increased abundance (cleavage site detected in both biological replicates in quantile 0–25; averaged protein Fc > 1). For Ctsl, this is the case for epidermal fatty-acid-binding protein and cadherin-1. For Ctsb, decreased proteolytic processing, together with increased protein abundance, is observed in calsequestrin-1, hemoglobin subunits β-1 and -2, and apolipoprotein C-III. Analogous to the above reasoning, an increase in proteolytic processing can result in decreased protein abundance. In Ctsl−/− skin, this is the case for a surprisingly large number of protease inhibitors, albeit with a milder effect on protein abundance (cleavage site detected in both replicates in quantile 0–25; averaged protein Fc < −0.5). The affected protease inhibitors include serpine H1, α-1-antitrypsin, α-2-macroglobulin, antithrombin-III, serpine C1, and peptidase inhibitor 16. The identification of the N termini mentioned above provides evidence of processing by Ctsb or Ctsl but needs further investigation.
N-terminal Protein Acetylation Corresponds to Prototypical Mammalian N-α Acetylation
TAILS is a negative selection technique and is suitable for the analysis of N-terminal protein acetylation (86). Independently of our work on cathepsin biology, we examined the N-α acetylation pattern in order to contribute to recent studies focusing on N-terminal protein acetylation (87–93). In this context, we present the first proteomic overview of N-α acetylation in mice. In the Ctsb experiments, TAILS identified between 411 and 637 acetylated protein N termini (supplemental Tables S15 and S16; annotated spectra in supplemental files 5 and 6). In the Ctsl experiment, TAILS identified 284 to 574 acetylated protein N termini (supplemental Tables S17 and S18; annotated spectra in supplemental files 7 and 8). In all four experiments, >75% of all N-α acetylations occurred after the removal of a methionine residue. More than 95% of all N-α acetylated proteins were intracellular. These results correspond to several of the aforementioned studies.
To characterize the specificity of N-α acetylation in mice, we generated sequence logos of those N-terminal acetylation events with a P1 methionine (Fig. 7). In all four TAILS experiments, there was a striking preference for small (alanine, serine) or acidic (aspartate, glutamate) residues in P1′ and, to a lesser extent, P2′. This profile bears close resemblance to the recently presented N-α acetylation pattern from human A2780 cells (94). Thus we have provided experimental evidence that mice share a prototypical N-α acetylation pattern.
Fig. 7.
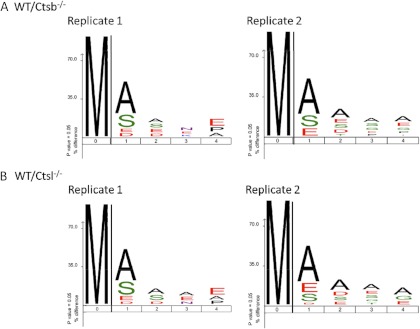
N-α acetylation pattern upon (A) Ctsb deficiency and (B) Ctsl deficiency. Sequence logos were generated with iceLogo (98).
CONCLUSION
A wealth of studies have demonstrated that balanced proteolysis is essential for skin development and function. The deficiency of numerous proteolytic enzymes, as well as of endogenous protease inhibitors, leads to severe skin phenotypes in mice (15, 51, 53, 95, 96). The present study constitutes the first comprehensive investigation of how the ablation of cathepsin B or cathepsin L affects the murine skin proteome and degradome. Generally, it is one of the first studies investigating how the deletion of individual proteases affects proteolytic processing in vivo. Both cathepsins are deeply involved in maintaining skin proteome composition. In some cases (e.g. cystatin B in Ctsl−/− skin), altered protein abundance stems from affected gene expression rather than impaired degradation. The accumulation of periostin in Ctsl−/− skin is of particular note: here a transcriptional down-regulation is superseded by impaired periostin degradation. The ablation of Ctsb or Ctsl has a pronounced effect on further proteases and protease inhibitors. This characterizes Ctsb and, more prominently, Ctsl as important regulatory modules of the proteolytic network in skin. Consequently, their genetic deficiency affects a multitude of proteolytic processing sites, albeit predominantly through dominant downstream effects.
The present study highlights the necessity of combining cleavage site analysis with global quantitative proteome analysis. In some cases, altered protein abundance might be caused by adversely affected proteolytic processing. In other cases, quantitatively affected cleavage sites represent altered protein abundance rather than impaired proteolysis.
Supplementary Material
Acknowledgments
We thank Florian Sigloch and Tom Michoel for critical discussion. Bettina Mayer, Heidi Bräuner, Franz Jehle, Silke Kowar, and Guy Roland are acknowledged for excellent technical assistance, and we thank Silvia Blacher for quantification of the aortic ring assay. We thank Dr. Simon Conway, Indiana, for the kind gift of periostin−/− paraffin sections and Dr. Ulrich Maurer for assistance with the pMIG system. The core facility of the Universitätsklinikum Freiburg (Dr. Marie Follo and Klaus Geiger) is acknowledged for sorting cell lines, and the DKFZ Microarray Core Facility in Heidelberg is acknowledged for performing the transcriptomic analysis.
Footnotes
* O.S. is supported by an Emmy-Noether Grant from the Deutsche Forschungsgemeinschaft (DFG) (SCHI 871/2) and a starting grant from the European Research Council (Programme “Ideas”; call identifier: ERC-2011-StG 282111-ProteaSys). A.G.A. and F.W. are supported in part by the Excellence Initiative of the German Federal and State Governments (GSC-4, Spemann Graduate School). J.N.K. acknowledges the Michael Smith Foundation for Health Research (MSFHR) Career Investigator Scholar Award. T.R. and A.N. are supported by European Commission FP7 Grant No. 201279 (Microenvimet). T.R. is further supported by Deutsche Forschungsgemeinschaft SFB 850 Project B7 and by the Excellence Initiative of the German Federal and State Governments (EXC 294 and GSC-4, Spemann Graduate School). M.B. and H.B. are supported by the Excellence Initiative of the German Federal and State Governments (FRIAS - LifeNet) and the German Federal Ministry of Education and Research (Gerontosys - “Stromal Aging,” 0315576D).
The data associated with this manuscript may be downloaded from the Proteome Commons Tranche using the following hash: S8JVSxYQ+JuqUvu/uafqO4MEmjcckxjO2egV+RvhQM9kLLDXux2yxCP0lZ0kbOJnCLjMgbKMUynImxS5v5vad7/0BF8AAAAAAAAC8Q==. The hash may be used to prove exactly what files were published as part of this manuscript's data set, and it may also be used to check that the data have not changed since publication.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- Ctsb
- cathepsin B
- Ctsd
- cathepsin D
- Ctsl
- cathepsin L
- E64
- trans-epoxysuccinyl-L-leucylamido(4-guanidino)butane
- Fc
- fold change
- GFP
- green fluorescent protein
- LC-MS/MS
- liquid chromatography–tandem mass spectrometry
- MEF
- mouse embryonic fibroblast
- TAILS
- terminal amine isotopic labeling of substrates.
REFERENCES
- 1. Turk V., Turk B., Turk D. (2001) Lysosomal cysteine proteases: facts and opportunities. EMBO J. 20, 4629–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rawlings N. D., Barrett A. J., Bateman A. (2010) MEROPS: the peptidase database. Nucleic Acids Res. 38, D227–D233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aronson N. N., Jr., Barrett A. J. (1978) The specificity of cathepsin B. Hydrolysis of glucagon at the C-terminus by a peptidyldipeptidase mechanism. Biochem. J. 171, 759–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Petermann I., Mayer C., Stypmann J., Biniossek M. L., Tobin D. J., Engelen M. A., Dandekar T., Grune T., Schild L., Peters C., Reinheckel T. (2006) Lysosomal, cytoskeletal, and metabolic alterations in cardiomyopathy of cathepsin L knockout mice. FASEB J. 20, 1266–1268 [DOI] [PubMed] [Google Scholar]
- 5. Spira D., Stypmann J., Tobin D. J., Petermann I., Mayer C., Hagemann S., Vasiljeva O., Gunther T., Schule R., Peters C., Reinheckel T. (2007) Cell type-specific functions of the lysosomal protease cathepsin L in the heart. J. Biol. Chem. 282, 37045–37052 [DOI] [PubMed] [Google Scholar]
- 6. Tang Q., Cai J., Shen D., Bian Z., Yan L., Wang Y. X., Lan J., Zhuang G. Q., Ma W. Z., Wang W. (2009) Lysosomal cysteine peptidase cathepsin L protects against cardiac hypertrophy through blocking AKT/GSK3beta signaling. J. Mol. Med. 87, 249–260 [DOI] [PubMed] [Google Scholar]
- 7. Honey K., Nakagawa T., Peters C., Rudensky A. (2002) Cathepsin L regulates CD4+ T cell selection independently of its effect on invariant chain: a role in the generation of positively selecting peptide ligands. J. Exp. Med. 195, 1349–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nakagawa T., Roth W., Wong P., Nelson A., Farr A., Deussing J., Villadangos J. A., Ploegh H., Peters C., Rudensky A. Y. (1998) Cathepsin L: critical role in Ii degradation and CD4 T cell selection in the thymus. Science 280, 450–453 [DOI] [PubMed] [Google Scholar]
- 9. Friedrichs B., Tepel C., Reinheckel T., Deussing J., von Figura K., Herzog V., Peters C., Saftig P., Brix K. (2003) Thyroid functions of mouse cathepsins B, K, and L. J. Clin. Invest. 111, 1733–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Funkelstein L., Toneff T., Mosier C., Hwang S. R., Beuschlein F., Lichtenauer U. D., Reinheckel T., Peters C., Hook V. (2008) Major role of cathepsin L for producing the peptide hormones ACTH, beta-endorphin, and alpha-MSH, illustrated by protease gene knockout and expression. J. Biol. Chem. 283, 35652–35659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gocheva V., Zeng W., Ke D., Klimstra D., Reinheckel T., Peters C., Hanahan D., Joyce J. A. (2006) Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Genes Dev. 20, 543–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gocheva V., Wang H. W., Gadea B. B., Shree T., Hunter K. E., Garfall A. L., Berman T., Joyce J. A. (2010) IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev. 24, 241–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benavides F., Perez C., Blando J., Contreras O., Shen J., Coussens L. M., Fischer S. M., Kusewitt D. F., Digiovanni J., Conti C. J. (2012) Protective role of cathepsin L in mouse skin carcinogenesis. Mol. Carcinog. 51, 352–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dennemarker J., Lohmuller T., Mayerle J., Tacke M., Lerch M. M., Coussens L. M., Peters C., Reinheckel T. (2010) Deficiency for the cysteine protease cathepsin L promotes tumor progression in mouse epidermis. Oncogene 29, 1611–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roth W., Deussing J., Botchkarev V. A., Pauly-Evers M., Saftig P., Hafner A., Schmidt P., Schmahl W., Scherer J., Anton-Lamprecht I., Von Figura K., Paus R., Peters C. (2000) Cathepsin L deficiency as molecular defect of furless: hyperproliferation of keratinocytes and pertubation of hair follicle cycling. FASEB J. 14, 2075–2086 [DOI] [PubMed] [Google Scholar]
- 16. Reinheckel T., Hagemann S., Dollwet-Mack S., Martinez E., Lohmuller T., Zlatkovic G., Tobin D. J., Maas-Szabowski N., Peters C. (2005) The lysosomal cysteine protease cathepsin L regulates keratinocyte proliferation by control of growth factor recycling. J. Cell Sci. 118, 3387–3395 [DOI] [PubMed] [Google Scholar]
- 17. Tobin D. J., Foitzik K., Reinheckel T., Mecklenburg L., Botchkarev V. A., Peters C., Paus R. (2002) The lysosomal protease cathepsin L is an important regulator of keratinocyte and melanocyte differentiation during hair follicle morphogenesis and cycling. Am. J. Pathol. 160, 1807–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deussing J., Roth W., Saftig P., Peters C., Ploegh H. L., Villadangos J. A. (1998) Cathepsins B and D are dispensable for major histocompatibility complex class II-mediated antigen presentation. Proc. Natl. Acad. Sci. U.S.A. 95, 4516–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Halangk W., Lerch M. M., Brandt-Nedelev B., Roth W., Ruthenbuerger M., Reinheckel T., Domschke W., Lippert H., Peters C., Deussing J. (2000) Role of cathepsin B in intracellular trypsinogen activation and the onset of acute pancreatitis. J. Clin. Invest. 106, 773–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vasiljeva O., Reinheckel T., Peters C., Turk D., Turk V., Turk B. (2007) Emerging roles of cysteine cathepsins in disease and their potential as drug targets. Curr. Pharm. Des. 13, 387–403 [DOI] [PubMed] [Google Scholar]
- 21. Guicciardi M. E., Deussing J., Miyoshi H., Bronk S. F., Svingen P. A., Peters C., Kaufmann S. H., Gores G. J. (2000) Cathepsin B contributes to TNF-alpha-mediated hepatocyte apoptosis by promoting mitochondrial release of cytochrome c. J. Clin. Invest. 106, 1127–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vasiljeva O., Korovin M., Gajda M., Brodoefel H., Bojic L., Kruger A., Schurigt U., Sevenich L., Turk B., Peters C., Reinheckel T. (2008) Reduced tumour cell proliferation and delayed development of high-grade mammary carcinomas in cathepsin B-deficient mice. Oncogene 27, 4191–4199 [DOI] [PubMed] [Google Scholar]
- 23. Vasiljeva O., Papazoglou A., Kruger A., Brodoefel H., Korovin M., Deussing J., Augustin N., Nielsen B. S., Almholt K., Bogyo M., Peters C., Reinheckel T. (2006) Tumor cell-derived and macrophage-derived cathepsin B promotes progression and lung metastasis of mammary cancer. Cancer Res. 66, 5242–5250 [DOI] [PubMed] [Google Scholar]
- 24. Sevenich L., Werner F., Gajda M., Schurigt U., Sieber C., Muller S., Follo M., Peters C., Reinheckel T. (2011) Transgenic expression of human cathepsin B promotes progression and metastasis of polyoma-middle-T-induced breast cancer in mice. Oncogene 30, 54–64 [DOI] [PubMed] [Google Scholar]
- 25. Felbor U., Kessler B., Mothes W., Goebel H. H., Ploegh H. L., Bronson R. T., Olsen B. R. (2002) Neuronal loss and brain atrophy in mice lacking cathepsins B and L. Proc. Natl. Acad. Sci. U.S.A. 99, 7883–7888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stahl S., Reinders Y., Asan E., Mothes W., Conzelmann E., Sickmann A., Felbor U. (2007) Proteomic analysis of cathepsin B- and L-deficient mouse brain lysosomes. Biochim. Biophys. Acta 1774, 1237–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boersema P. J., Raijmakers R., Lemeer S., Mohammed S., Heck A. J. (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 [DOI] [PubMed] [Google Scholar]
- 28. Guo K., Ji C., Li L. (2007) Stable-isotope dimethylation labeling combined with LC-ESI MS for quantification of amine-containing metabolites in biological samples. Anal. Chem. 79, 8631–8638 [DOI] [PubMed] [Google Scholar]
- 29. Kleifeld O., Doucet A., auf dem Keller U., Prudova A., Schilling O., Kainthan R. K., Starr A. E., Foster L. J., Kizhakkedathu J. N., Overall C. M. (2010) Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat. Biotechnol. 28, 281–288 [DOI] [PubMed] [Google Scholar]
- 30. Tholen S., Biniossek M. L., Gessler A. L., Muller S., Weisser J., Kizhakkedathu J. N., Reinheckel T., Schilling O. (2011) Contribution of cathepsin L to secretome composition and cleavage pattern of mouse embryonic fibroblasts. Biol. Chem. 392, 961–971 [DOI] [PubMed] [Google Scholar]
- 31. Pedrioli P. G., Eng J. K., Hubley R., Vogelzang M., Deutsch E. W., Raught B., Pratt B., Nilsson E., Angeletti R. H., Apweiler R., Cheung K., Costello C. E., Hermjakob H., Huang S., Julian R. K., Kapp E., McComb M. E., Oliver S. G., Omenn G., Paton N. W., Simpson R., Smith R., Taylor C. F., Zhu W., Aebersold R. (2004) A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 22, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 32. Kessner D., Chambers M., Burke R., Agus D., Mallick P. (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 34. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 35. Cochrane G. R. (2010) The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 38, D142–D148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martens L., Vandekerckhove J., Gevaert K. (2005) DBToolkit: processing protein databases for peptide-centric proteomics. Bioinformatics 21, 3584–3585 [DOI] [PubMed] [Google Scholar]
- 37. Li X. J., Zhang H., Ranish J. A., Aebersold R. (2003) Automated statistical analysis of protein abundance ratios from data generated by stable-isotope dilution and tandem mass spectrometry. Anal. Chem. 75, 6648–6657 [DOI] [PubMed] [Google Scholar]
- 38. Mo F., Mo Q., Chen Y., Goodlett D. R., Hood L., Omenn G. S., Li S., Lin B. (2010) WaveletQuant, an improved quantification software based on wavelet signal threshold de-noising for labeled quantitative proteomic analysis. BMC Bioinformatics 11, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Han D. K., Eng J., Zhou H., Aebersold R. (2001) Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat. Biotechnol. 19, 946–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Soneoka Y., Cannon P. M., Ramsdale E. E., Griffiths J. C., Romano G., Kingsman S. M., Kingsman A. J. (1995) A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23, 628–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kleifeld O., Doucet A., Prudova A., Auf dem Keller U., Gioia M., Kizhakkedathu J., Overall C. M. (2011) System-wide proteomic identification of protease cleavage products by terminal amine isotopic labeling of substrates. Nat. Protoc. 6, 1578–1611 [DOI] [PubMed] [Google Scholar]
- 42. Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 43. Pan C., Kumar C., Bohl S., Klingmueller U., Mann M. (2009) Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol. Cell. Proteomics 8, 443–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Olsen J. V., Ong S. E., Mann M. (2004) Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol. Cell. Proteomics 3, 608–614 [DOI] [PubMed] [Google Scholar]
- 45. Biniossek M. L., Nagler D. K., Becker-Pauly C., Schilling O. (2011) Correction to proteomic identification of protease cleavage sites characterizes prime and non-prime specificity of cysteine cathepsins B, L, and S. J. Proteome Res. 10, 5363–5373 [DOI] [PubMed] [Google Scholar]
- 46. Sounni N. E., Devy L., Hajitou A., Frankenne F., Munaut C., Gilles C., Deroanne C., Thompson E. W., Foidart J. M., Noel A. (2002) MT1-MMP expression promotes tumor growth and angiogenesis through an up-regulation of vascular endothelial growth factor expression. FASEB J. 16, 555–564 [DOI] [PubMed] [Google Scholar]
- 47. Blacher S., Devy L., Burbridge M. F., Roland G., Tucker G., Noel A., Foidart J. M. (2001) Improved quantification of angiogenesis in the rat aortic ring assay. Angiogenesis 4, 133–142 [DOI] [PubMed] [Google Scholar]
- 48. Azzalini A., Capitanio A. (2003) Distributions generated by perturbation of symmetry with emphasis on a multivariate skew t-distribution. J. R. Stat. Soc. B 65, 367–389 [Google Scholar]
- 49. Miles G. P., Samuel M. A., Ellis B. E. (2009) Suppression of MKK5 reduces ozone-induced signal transmission to both MPK3 and MPK6 and confers increased ozone sensitivity in Arabidopsis thaliana. Plant Signal Behav. 4, 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Norris R. A., Damon B., Mironov V., Kasyanov V., Ramamurthi A., Moreno-Rodriguez R., Trusk T., Potts J. D., Goodwin R. L., Davis J., Hoffman S., Wen X., Sugi Y., Kern C. B., Mjaatvedt C. H., Turner D. K., Oka T., Conway S. J., Molkentin J. D., Forgacs G., Markwald R. R. (2007) Periostin regulates collagen fibrillogenesis and the biomechanical properties of connective tissues. J. Cell. Biochem. 101, 695–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Benes P., Vetvicka V., Fusek M. (2008) Cathepsin D—many functions of one aspartic protease. Crit. Rev. Oncol. Hematol. 68, 12–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Egberts F., Heinrich M., Jensen J. M., Winoto-Morbach S., Pfeiffer S., Wickel M., Schunck M., Steude J., Saftig P., Proksch E., Schutze S. (2004) Cathepsin D is involved in the regulation of transglutaminase 1 and epidermal differentiation. J. Cell Sci. 117, 2295–2307 [DOI] [PubMed] [Google Scholar]
- 53. Zeeuwen P. L., van Vlijmen-Willems I. M., Hendriks W., Merkx G. F., Schalkwijk J. (2002) A null mutation in the cystatin M/E gene of ichq mice causes juvenile lethality and defects in epidermal cornification. Hum. Mol. Genet. 11, 2867–2875 [DOI] [PubMed] [Google Scholar]
- 54. Denecker G., Hoste E., Gilbert B., Hochepied T., Ovaere P., Lippens S., Van den Broecke C., Van Damme P., D'Herde K., Hachem J. P., Borgonie G., Presland R. B., Schoonjans L., Libert C., Vandekerckhove J., Gevaert K., Vandenabeele P., Declercq W. (2007) Caspase-14 protects against epidermal UVB photodamage and water loss. Nat. Cell Biol. 9, 666–674 [DOI] [PubMed] [Google Scholar]
- 55. Herron B. J., Rao C., Liu S., Laprade L., Richardson J. A., Olivieri E., Semsarian C., Millar S. E., Stubbs L., Beier D. R. (2005) A mutation in NFkB interacting protein 1 results in cardiomyopathy and abnormal skin development in wa3 mice. Hum. Mol. Genet. 14, 667–677 [DOI] [PubMed] [Google Scholar]
- 56. Kawada A., Hara K., Kominami E., Hiruma M., Akiyama M., Ishibashi A., Abe H., Ichikawa E., Nakamura Y., Watanabe S., Yamamoto T., Umeda T., Nishioka K. (1997) Expression of cathepsin D and B in invasion and metastasis of squamous cell carcinoma. Br. J. Dermatol. 137, 361–366 [PubMed] [Google Scholar]
- 57. Wille A., Gerber A., Heimburg A., Reisenauer A., Peters C., Saftig P., Reinheckel T., Welte T., Buhling F. (2004) Cathepsin L is involved in cathepsin D processing and regulation of apoptosis in A549 human lung epithelial cells. Biol. Chem. 385, 665–670 [DOI] [PubMed] [Google Scholar]
- 58. Zheng X., Chu F., Mirkin B. L., Sudha T., Mousa S. A., Rebbaa A. (2008) Role of the proteolytic hierarchy between cathepsin L, cathepsin D and caspase-3 in regulation of cellular susceptibility to apoptosis and autophagy. Biochim. Biophys. Acta 1783, 2294–2300 [DOI] [PubMed] [Google Scholar]
- 59. Dennemarker J., Lohmuller T., Muller S., Aguilar S. V., Tobin D. J., Peters C., Reinheckel T. (2010) Impaired turnover of autophagolysosomes in cathepsin L deficiency. Biol. Chem. 391, 913–922 [DOI] [PubMed] [Google Scholar]
- 60. Abrahamson M., Alvarez-Fernandez M., Nathanson C. M. (2003) Cystatins. Biochem. Soc. Symp. 70, 179–199 [DOI] [PubMed] [Google Scholar]
- 61. Zeeuwen P. L., Cheng T., Schalkwijk J. (2009) The biology of cystatin M/E and its cognate target proteases. J. Invest. Dermatol. 129, 1327–1338 [DOI] [PubMed] [Google Scholar]
- 62. Cheng T., Hitomi K., van Vlijmen-Willems I. M., de Jongh G. J., Yamamoto K., Nishi K., Watts C., Reinheckel T., Schalkwijk J., Zeeuwen P. L. (2006) Cystatin M/E is a high affinity inhibitor of cathepsin V and cathepsin L by a reactive site that is distinct from the legumain-binding site. A novel clue for the role of cystatin M/E in epidermal cornification. J. Biol. Chem. 281, 15893–15899 [DOI] [PubMed] [Google Scholar]
- 63. Zeeuwen P. L., van Vlijmen-Willems I. M., Cheng T., Rodijk-Olthuis D., Hitomi K., Hara-Nishimura I., John S., Smyth N., Reinheckel T., Hendriks W. J., Schalkwijk J. (2010) The cystatin M/E-cathepsin L balance is essential for tissue homeostasis in epidermis, hair follicles, and cornea. FASEB J. 24, 3744–3755 [DOI] [PubMed] [Google Scholar]
- 64. Dubin G. (2005) Proteinaceous cysteine protease inhibitors. Cell. Mol. Life Sci. 62, 653–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Malanchi I., Santamaria-Martinez A., Susanto E., Peng H., Lehr H. A., Delaloye J. F., Huelsken J. (2011) Interactions between cancer stem cells and their niche govern metastatic colonization. Nature 481, 85–89 [DOI] [PubMed] [Google Scholar]
- 66. Ishidoh K., Kominami E. (1995) Procathepsin L degrades extracellular matrix proteins in the presence of glycosaminoglycans in vitro. Biochem. Biophys. Res. Commun. 217, 624–631 [DOI] [PubMed] [Google Scholar]
- 67. Baril P., Gangeswaran R., Mahon P. C., Caulee K., Kocher H. M., Harada T., Zhu M., Kalthoff H., Crnogorac-Jurcevic T., Lemoine N. R. (2007) Periostin promotes invasiveness and resistance of pancreatic cancer cells to hypoxia-induced cell death: role of the beta4 integrin and the PI3k pathway. Oncogene 26, 2082–2094 [DOI] [PubMed] [Google Scholar]
- 68. Thomas P. D., Kejariwal A., Campbell M. J., Mi H., Diemer K., Guo N., Ladunga I., Ulitsky-Lazareva B., Muruganujan A., Rabkin S., Vandergriff J. A., Doremieux O. (2003) PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 31, 334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Premzl A., Turk V., Kos J. (2006) Intracellular proteolytic activity of cathepsin B is associated with capillary-like tube formation by endothelial cells in vitro. J. Cell. Biochem. 97, 1230–1240 [DOI] [PubMed] [Google Scholar]
- 70. Chang S. H., Kanasaki K., Gocheva V., Blum G., Harper J., Moses M. A., Shih S. C., Nagy J. A., Joyce J., Bogyo M., Kalluri R., Dvorak H. F. (2009) VEGF-A induces angiogenesis by perturbing the cathepsin-cysteine protease inhibitor balance in venules, causing basement membrane degradation and mother vessel formation. Cancer Res. 69, 4537–4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Irving J. A., Pike R. N., Dai W., Bromme D., Worrall D. M., Silverman G. A., Coetzer T. H., Dennison C., Bottomley S. P., Whisstock J. C. (2002) Evidence that serpin architecture intrinsically supports papain-like cysteine protease inhibition: engineering alpha(1)-antitrypsin to inhibit cathepsin proteases. Biochemistry 41, 4998–5004 [DOI] [PubMed] [Google Scholar]
- 72. Schick C., Pemberton P. A., Shi G. P., Kamachi Y., Cataltepe S., Bartuski A. J., Gornstein E. R., Bromme D., Chapman H. A., Silverman G. A. (1998) Cross-class inhibition of the cysteine proteinases cathepsins K, L, and S by the serpin squamous cell carcinoma antigen 1: a kinetic analysis. Biochemistry 37, 5258–5266 [DOI] [PubMed] [Google Scholar]
- 73. Prudova A., auf dem Keller U., Butler G. S., Overall C. M. (2010) Multiplex N-terminome analysis of MMP-2 and MMP-9 substrate degradomes by iTRAQ-TAILS quantitative proteomics. Mol. Cell. Proteomics 9, 894–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schilling O., Overall C. M. (2008) Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat. Biotechnol. 26, 685–694 [DOI] [PubMed] [Google Scholar]
- 75. Schilling O., Huesgen P. F., Barre O., Auf dem Keller U., Overall C. M. (2011) Characterization of the prime and non-prime active site specificities of proteases by proteome-derived peptide libraries and tandem mass spectrometry. Nat. Protoc. 6, 111–120 [DOI] [PubMed] [Google Scholar]
- 76. Schilling O., auf dem Keller U., Overall C. M. (2011) Factor Xa subsite mapping by proteome-derived peptide libraries improved using WebPICS, a resource for proteomic identification of cleavage sites. Biol. Chem. 392, 1031–1037 [DOI] [PubMed] [Google Scholar]
- 77. Schechter I., Berger A. (1968) On the active site of proteases. 3. Mapping the active site of papain; specific peptide inhibitors of papain. Biochem. Biophys. Res. Commun. 32, 898–902 [DOI] [PubMed] [Google Scholar]
- 78. Naso M. F., Liang B., Huang C. C., Song X. Y., Shahied-Arruda L., Belkowski S. M., D'Andrea M. R., Polkovitch D. A., Lawrence D. R., Griswold D. E., Sweet R. W., Amegadzie B. Y. (2007) Dermokine: an extensively differentially spliced gene expressed in epithelial cells. J. Invest. Dermatol. 127, 1622–1631 [DOI] [PubMed] [Google Scholar]
- 79. Tagi T., Matsui T., Kikuchi S., Hoshi S., Ochiai T., Kokuba Y., Kinoshita-Ida Y., Kisumi-Hayashi F., Morimoto K., Imai T., Imoto I., Inazawa J., Otsuji E. (2010) Dermokine as a novel biomarker for early-stage colorectal cancer. J. Gastroenterol. 45, 1201–1211 [DOI] [PubMed] [Google Scholar]
- 80. Lange P. F., Overall C. M. (2011) TopFIND, a knowledgebase linking protein termini with function. Nat. Methods 8, 703–704 [DOI] [PubMed] [Google Scholar]
- 81. Blanpain C., Lowry W. E., Pasolli H. A., Fuchs E. (2006) Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 20, 3022–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lin H. Y., Kao C. H., Lin K. M., Kaartinen V., Yang L. T. (2011) Notch signaling regulates late-stage epidermal differentiation and maintains postnatal hair cycle homeostasis. PLoS One 6, e15842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kasper M., Schnidar H., Neill G. W., Hanneder M., Klingler S., Blaas L., Schmid C., Hauser-Kronberger C., Regl G., Philpott M. P., Aberger F. (2006) Selective modulation of Hedgehog/GLI target gene expression by epidermal growth factor signaling in human keratinocytes. Mol. Cell. Biol. 26, 6283–6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Loeb C. R., Harris J. L., Craik C. S. (2006) Granzyme B proteolyzes receptors important to proliferation and survival, tipping the balance toward apoptosis. J. Biol. Chem. 281, 28326–28335 [DOI] [PubMed] [Google Scholar]
- 85. Kudo A. (2011) Periostin in fibrillogenesis for tissue regeneration: periostin actions inside and outside the cell. Cell. Mol. Life Sci. 68, 3201–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Van Damme P., Arnesen T., Gevaert K. (2011) Protein alpha-N-acetylation studied by N-terminomics. FEBS J. 278, 3822–3834 [DOI] [PubMed] [Google Scholar]
- 87. Goetze S., Qeli E., Mosimann C., Staes A., Gerrits B., Roschitzki B., Mohanty S., Niederer E. M., Laczko E., Timmerman E., Lange V., Hafen E., Aebersold R., Vandekerckhove J., Basler K., Ahrens C. H., Gevaert K., Brunner E. (2009) Identification and functional characterization of N-terminally acetylated proteins in Drosophila melanogaster. PLoS Biol. 7, e1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Helbig A. O., Gauci S., Raijmakers R., van Breukelen B., Slijper M., Mohammed S., Heck A. J. (2010) Profiling of N-acetylated protein termini provides in-depth insights into the N-terminal nature of the proteome. Mol. Cell. Proteomics 9, 928–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Falb M., Aivaliotis M., Garcia-Rizo C., Bisle B., Tebbe A., Klein C., Konstantinidis K., Siedler F., Pfeiffer F., Oesterhelt D. (2006) Archaeal N-terminal protein maturation commonly involves N-terminal acetylation: a large-scale proteomics survey. J. Mol. Biol. 362, 915–924 [DOI] [PubMed] [Google Scholar]
- 90. Aivaliotis M., Gevaert K., Falb M., Tebbe A., Konstantinidis K., Bisle B., Klein C., Martens L., Staes A., Timmerman E., Van Damme J., Siedler F., Pfeiffer F., Vandekerckhove J., Oesterhelt D. (2007) Large-scale identification of N-terminal peptides in the halophilic archaea Halobacterium salinarum and Natronomonas pharaonis. J. Proteome Res. 6, 2195–2204 [DOI] [PubMed] [Google Scholar]
- 91. Arnesen T., Van Damme P., Polevoda B., Helsens K., Evjenth R., Colaert N., Varhaug J. E., Vandekerckhove J., Lillehaug J. R., Sherman F., Gevaert K. (2009) Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. Natl. Acad. Sci. U.S.A. 106, 8157–8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Starheim K. K., Gromyko D., Velde R., Varhaug J. E., Arnesen T. (2009) Composition and biological significance of the human Nalpha-terminal acetyltransferases. BMC Proc. 3 Suppl 6, S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Perrot M., Massoni A., Boucherie H. (2008) Sequence requirements for Nalpha-terminal acetylation of yeast proteins by NatA. Yeast 25, 513–527 [DOI] [PubMed] [Google Scholar]
- 94. Bienvenut W. V., Sumpton D., Martinez A., Lilla S., Espagne C., Meinnel T., Giglione C. (2012) Comparative large-scale characterisation of plant vs. mammal proteins reveals similar and idiosyncratic N-alpha acetylation features. Mol. Cell. Proteomics 11, 10.1074/mcp.M111.015131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Saftig P., Hetman M., Schmahl W., Weber K., Heine L., Mossmann H., Koster A., Hess B., Evers M., von Figura K., Peters C. (1995) Mice deficient for the lysosomal proteinase cathepsin D exhibit progressive atrophy of the intestinal mucosa and profound destruction of lymphoid cells. EMBO J. 14, 3599–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Descargues P., Deraison C., Bonnart C., Kreft M., Kishibe M., Ishida-Yamamoto A., Elias P., Barrandon Y., Zambruno G., Sonnenberg A., Hovnanian A. (2005) Spink5-deficient mice mimic Netherton syndrome through degradation of desmoglein 1 by epidermal protease hyperactivity. Nat. Genet. 37, 56–65 [DOI] [PubMed] [Google Scholar]
- 97. Kersey P. J., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R. (2004) The International Protein Index: an integrated database for proteomics experiments. Proteomics 4, 1985–1988 [DOI] [PubMed] [Google Scholar]
- 98. Colaert N., Helsens K., Martens L., Vandekerckhove J., Gevaert K. (2009) Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6, 786–787 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.