Abstract
Protein degradation provides an important regulatory mechanism used to control cell cycle progression and many other cellular pathways. To comprehensively analyze the spatial control of protein degradation in U2OS osteosarcoma cells, we have combined drug treatment and SILAC-based quantitative mass spectrometry with subcellular and protein fractionation. The resulting data set analyzed more than 74,000 peptides, corresponding to ∼5000 proteins, from nuclear, cytosolic, membrane, and cytoskeletal compartments. These data identified rapidly degraded proteasome targets, such as PRR11 and highlighted a feedback mechanism resulting in translation inhibition, induced by blocking the proteasome. We show this is mediated by activation of the unfolded protein response. We observed compartment-specific differences in protein degradation, including proteins that would not have been characterized as rapidly degraded through analysis of whole cell lysates. Bioinformatic analysis of the entire data set is presented in the Encyclopedia of Proteome Dynamics, a web-based resource, with proteins annotated for stability and subcellular distribution.
Targeted protein degradation is an important regulatory mechanism that allows co-ordination of cellular pathways in response to environmental and temporal stimuli (1). The control of diverse biochemical pathways, including cell cycle progression and the response to DNA damage, is mediated, at least in part, by dynamic alterations in protein degradation (2). Previous large scale proteomics studies in mammalian cells have shown that the rate of protein degradation can vary from the timescale of minutes, to essentially infinite stability for metastable proteins (3–8).
Most intracellular proteins have similar degradation rates, with a half-life approximating the cell doubling rate. Under 5% of proteins display degradation rates more than threefold faster than the proteome average (3–5, 7). However, degradation rates for individual proteins can change, for example depending on either the cell cycle stage, or signaling events, and can also vary depending on subcellular localization. Disruption of such regulated protein stability underlies the disease mechanisms responsible for forms of cancer, e.g. p53 (9, 10) and the proto-oncogene c-Myc (11).
Detection of rapidly degraded proteins can be difficult because of their low abundance. However, advances in mass spectrometry based proteomics have enabled in-depth quantitative analysis of cellular proteomes (12–14). Stable isotope labeling by amino acids in cell culture (SILAC)1 (15), has been widely used to measure protein properties such as abundance, interactions, modifications, turnover, and subcellular localization under different conditions (16). Subcellular fractionation and protein size separation are also powerful techniques that enhance in-depth analysis of cellular proteomes. Not only do these fractionation techniques increase total proteome coverage, they also provide biological insight regarding how protein behavior differs between subcellular compartments. For example, subcellular fractionation has highlighted differences in the rate of ribosomal protein degradation between the nucleus and cytoplasm, (7, 17). Other studies have also demonstrated the benefit of in-depth subcellular fractionation and created methods for the characterization of how proteomes are localized in organelles (18–20).
In this study we have used SILAC-based quantitative mass spectrometry combined with extensive subcellular and protein-level fractionation to identify rapidly degraded proteins in human U2OS cells. We provide a proteome level characterization of a major feedback mechanism involving inhibition of protein translation when the proteasome is inhibited. We also present the Encyclopedia of Proteome Dynamics, a user-friendly online resource providing access to the entire data set.
EXPERIMENTAL PROCEDURES
Materials
U2OS cells were purchased from the American Type Culture Collection (ATCC, Rockville, MD). Dulbecco's modified Eagle medium (DMEM), fetal calf serum, antibiotics, NuPage gels, LDS sample buffer, MES SDS-PAGE running buffer, nitrocellulose iBlot stacks, SYPRO Ruby, Alexa Fluor 680-conjugated secondary antibodies, and CBQCA assay kit were obtained from Invitrogen (Carlsbad, CA). IrDye 800-conjugated secondary antibodies were obtained from Rockland Immunochemicals (Gilbertsville, PA). HRP-conjugated secondary antibodies were from Cell Signaling Technology (Danvers, MA). bicinchoninic acid (BCA) assay reagents, Coomassie plus (Bradford) reagent, subcellular protein fractionation Kit, detergent removal plates, 16% methanol-free paraformaldehyde, Acclaim Pepmap C18 columns and trapping cartridges, and triscarboxyethylphosphine (TCEP) (Bond-breaker neutral pH solution) were from Thermo Scientific (Waltham MA). Trypsin Gold was from Promega. Sep-Pak tC18 96-well μ-elution plates were from Waters (Milford, MA). Complete protease inhibitor mixture tablets and PhosStop phosphatase inhibitor tablets were from Roche (Basel, Switzerland). All other materials were obtained from Sigma (St. Louis, MO).
Cell Culture
Briefly, U2OS cells were grown in DMEM supplemented with 10% fetal calf serum (FCS), 2 mm l-glutamine, 100 U/l penicillin, and 100 μg/l streptomycin at 37 °C in 10% CO2, and passaged at ∼80% confluence. For SILAC labeling of U2OS cells arginine and lysine free DMEM was used and supplemented with stable isotope labeled arginine and lysine in addition to dialyzed FCS as described previously (21). After splitting by trypsinization, cells in SILAC media were grown to ∼80% confluence over 2–3 days before drug treatment and lysis for fractionation.
Subcellular Fractionation of SILAC-Labeled U2OS Cells
For the SILAC screen U2OS cells were treated with either DMSO, 40 μg/ml cycloheximide or 10 μm MG132 for 6 h and then combined in a 1:1:1 ratio of cells and fractionated by detergent solubility with the subcellular protein fractionation kit (Pierce). These fractions were then chloroform-methanol precipitated (22) and further separated by molecular weight using denaturing size exclusion chromatography before digestion and liquid chromatography-tandem MS (LC-MS/MS) analysis.
Denaturing Size Exclusion Chromatography (SDS-SEC), Trypsin Digestion, and Peptide Clean-up
Using a Dionex Ultimate 3000 HPLC system, fractions resuspended in 4% SDS, 100 mm NaCl, 25 mm TCEP, 50 mm N-ethylmaleimide, 10 mm Na PO4 pH 6.0 were heated to 65 °C for 10 min, then filtered through a 0.45 μm filter. Samples were injected (50 μl per injection - 250 μg protein) onto a mAbPacSEC column (Dionex) equilibrated with 0.2% SDS, 100 mm NaCl, 10 mm Na PO4 pH 6.0. The flow rate was 0.2 ml min−1 and 8 × 200 μl fractions were collected using a low protein binding 96-deep well plate (Eppendorf). Triethylamine bicarbonate (TEAB, 1 m pH 8.0) was added to each fraction to adjust the pH to 8.0, and trypsin diluted in 0.1 m TEAB was added at a ratio of 1:50 with incubation for 18 h at 37 °C. SDS was removed from each fraction using detergent removal resin in 96-well plates as described previously (23). For peptide desalting trifluoroacetic acid (TFA) was added to 1% (v/v) final concentration and peptides were purified using a Sep-Pak tC18 96-well μ-elution plate. Peptides were eluted in 200 μl of 50% (v/v) acetonitrile 0.1% TFA and speedivaced to dryness before resuspension in 5% (v/v) formic acid. Peptide concentrations were determined using the CBQCA assay after 25-fold dilution of peptide samples in 0.1 m borate buffer pH 9.3.
LC-MS/MS and MS Data Analysis
Using a Dionex Ultimate 3000 nanoHPLC system, 4 μg of peptides in 5% (v/v) formic acid were injected onto an Acclaim PepMap C18 nano-trap column (Dionex). After washing with 2% (v/v) acetonitrile 0.1% (v/v) formic acid peptides were resolved on a 50 mm × 75 μm Acclaim PepMap C18 reverse phase analytical column over a 140 min organic gradient with a flow rate of 200 nl min−1. Peptides were ionized by nano-electrospray ionization at 1.2 kV using a fused silica emitter with an internal diameter of 5 μm (New Objective). Tandem mass spectrometry analysis was carried out on a QExactive mass spectrometer (Thermo Scientific). Data were processed, searched, and quantified using the Maxquant software package version 1.2.2.5 as described previously (24, 25), using the default settings and employing the Human Uniprot database (06/07/11) containing 109,824 entries. The settings used for the Maxquant analysis were: two failed cleavages were allowed; fixed modification was N-ethylmaleimide on cysteine; enzymes were Trypsin (K/R not before P); Variable modifications included in the analysis were methionine oxidation, deamidation of glutamine or asparagine, N-terminal pyro-glutamic acid formation, protein N-terminal acetylation. A mass tolerance of 7 ppm was used for precursor ions and a tolerance of 20 ppm was used for fragment ions. Using the default Maxquant settings a maximum false positive rate of 1% was allowed for both peptide and protein identification. This cutoff was used for accepting individual spectra as well as whole proteins in the Maxquant output. This threshold has previously been shown to be a rigorous method for identifying true positive matches (24). For identical protein matches in both Swissprot and Tremble within the same protein group, the match in Swissprot was reported. Isoforms were recorded in the same protein group if they shared the same peptide coverage. All replicates indicated are biological replicates and protein quantitation was derived from at least two of the biological replicates. SILAC ratios for all proteins were normalized using β-Tubulin, Histone H2A, and GAPDH ratios. No outlier data points were removed.
Statistical Analysis of SILAC Data
p values were calculated using a Student's t test comparing the three biological replicate SILAC ratios of each protein, to the three biological replicate SILAC ratios for the control, known stable proteins, i.e. β-tubulin, histone H2A, and GAPDH. For the CHX/DMSO (M/L) and MG132/DMSO (H/L) SILAC ratios a one-tailed and two-tailed t test was used respectively. Values were tested for normality using the Shapiro-Wilk test.
SDS-PAGE and Immunoblotting
For generating total cell lysates, cells were lysed in five times the total volume of cells using 2% SDS in 10 mm HEPES, 250 mm sucrose, and 1 mm EDTA (HES buffer) with 50 mm N-ethylmaleimide, Complete Protease Inhibitors EDTA-free (Roche) and PhosStop (Roche). Lysates were incubated at 65 °C and viscosity because of DNA was removed by passing the lysate through a Qiashredder (Qiagen). Equal amounts of protein were loaded for SDS-PAGE of each sample with 10 μg per lane. SDS-PAGE was performed using 4–12% (w/v) Bis-Tris NuPage gels using MES running buffer according to manufacturer's instructions but with the addition of 25 mm TCEP, in the LDS sample buffer. SYPRO Ruby staining was performed as per manufacturer's instructions (Invitrogen). For Western blotting, separated proteins were electrophoretically transferred to an iBlot nitrocellulose membrane, blocked with 3% nonfat skim milk in 0.1% Tween-20 in TBS (TBST) and incubated with primary antibody in 5% bovine serum albumin (BSA) in TBST overnight at 4 °C. After incubation, membranes were washed three times in TBST and incubated with either HRP-labeled or Alexa fluor 680/IrDye 800 labeled secondary antibodies in 3% nonfat skim milk in TBST. Proteins were visualized using Immobillon chemiluminescent substrate (Millipore) and imaged either with a cooled CCD camera (Fuji) for HRP-labeled secondary antibodies, or a Licor Odyssey CLx imager for Alexa fluor 680/IrDye 800 labeled secondary antibodies.
Immunofluorescence Microscopy
Cells were cultured on glass coverslips as described above. All subsequent steps are at 25 °C. Cells were then fixed with 3% paraformaldehyde in PBS. Fixed cells were washed with PBS, and free aldehyde groups were quenched with 50 mm glycine in PBS. The cells were then permeabilized using 1% Triton X-100 for 10 min followed by washing in PBS. Coverslips were processed for immunolabeling by blocking with 5% BSA in TBST. Primary antibodies were incubated on coverslips for 1 h in 5% BSA in TBST. Coverslips were washed in PBS. Primary antibodies were detected with Alexa Fluor 488 or Alex Fluor 594 conjugated secondary antibodies which were incubated on coverslips for 30 min in 5% BSA in TBST with 2 μg/ml Hoechst 33342. Optical sections were analyzed by confocal microscopy on a Leica SP2 AOBS inverted microscope. Images generated are a single confocal slice from the middle of the cell monolayer.
siRNA Transfection of U2OS Cells
U2OS cells were transfected with 0.45 nmoles of siRNA per condition using RNAiMax (Invitrogen) in Optimem medium (Invitrogen). After 24 h, cells were harvested by trypsinization for analysis either by SDS-PAGE or flow cytometry.
Fluorescence-activated Cell Sorting (FACS)
An asynchronous U2OS cell suspension in DMEM containing 10% FCS was incubated with 25 μg/ml Hoechst 33342 for 5 min at room temperature. Cells were sorted by DNA content into G1, S and G2-phase cell populations using an ACS Vantage DIVA instrument (BD Biosciences). Sorted cells were washed in PBS and lysed in 2% SDS in HES buffer.
RESULTS
Workflow for the Identification of Rapidly Degraded Proteins
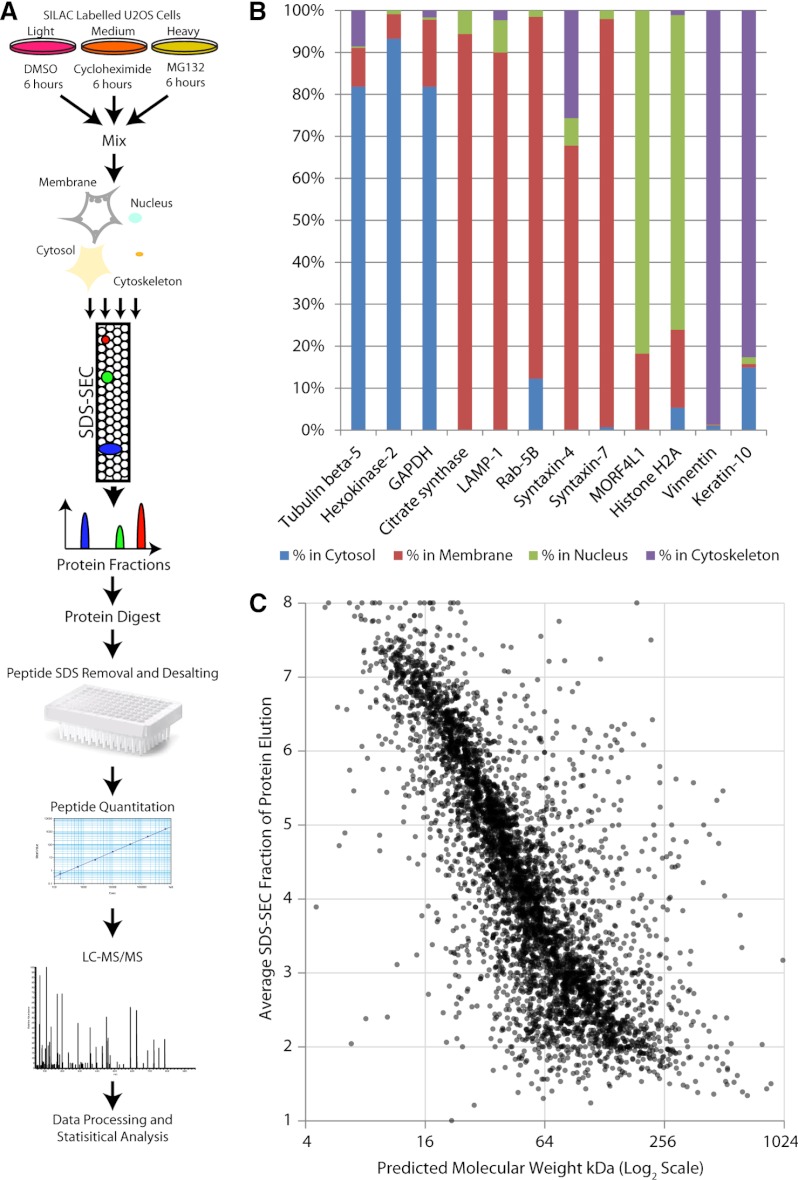
To identify rapidly degraded proteins human U2OS osteosarcoma cells were treated for 6 h with cycloheximide (CHX), which rapidly blocks translation elongation and hence protein synthesis. This method has been used in many studies of protein degradation (26) and is complementary to previous studies using pulse-SILAC to identify rapidly degraded proteins (3, 6, 7). To determine if rapidly degraded proteins are substrates of the ubiquitin-proteasome system a separate population of U2OS cells were treated for 6 h with the proteasome inhibitor MG132. As a control, U2OS cells were treated with DMSO for 6 h. The DMSO control, cycloheximide, and MG132 treated cells were grown, respectively, in either “light,” “medium,” or “heavy” SILAC media (Fig. 1A; see methods). After the 6 h incubation, the three cell populations were mixed in equal amounts and processed for MS analysis as described below (Fig. 1A).
Fig. 1.
Global protein degradation analysis using a comprehensive proteomics workflow. A, Workflow for SILAC-based quantitative mass spectrometry to identify rapidly degraded proteins. B, The subcellular fractionation for the SILAC-screen was assessed using marker proteins of known localization. The markers included GAPDH, β-Tubulin, and hexokinase-2 (cytosolic fraction), citrate synthase, syntaxin-4, syntaxin-7, rab-4b, and LAMP-1 (membrane fraction), histone H2A and MORF4L1 (nuclear fraction), vimentin and keratin-10 (cytoskeletal fraction). The percentage of total protein is indicated on the y axis. The colors indicate percentages in each fraction. C, Analysis of protein SDS-SEC fractionation versus the predicted protein molecular weights based on an in silico translation of the human genome. The average fraction for protein detection in the SILAC-screen is shown on the y axis. The x axis shows the log2 scaled predicted protein molecular weights.
The resulting isotope-encoded proteins isolated from the mixed-cell populations were fractionated both by subcellular compartment and molecular weight (Fig. 1A). Four subcellular fractions were generated, i.e. nucleus, cytosol, cytoskeleton, and membrane. Each subcellular fraction was individually separated by protein molecular weight, using denaturing SDS size exclusion chromatography (SDS-SEC), as previously described (27, 28). Proteins were digested with trypsin and analyzed by LC-MS/MS. Three biological replicates of this experiment were performed.
Peptides were identified and SILAC ratios quantitated using MaxQuant (24, 25). A total of over 74,000 peptides, corresponding to ∼5,000 proteins, were identified and quantified in at least two out of the three biological replicates (supplemental Table S1 and supplemental Table S2). The ∼5000 proteins analyzed in this study are highly representative of the human proteome, based on an analysis of amino acid frequencies and gene ontology (biological process) term frequencies in the total dataset (supplemental Fig. S1A, S1B). Analysis of the subcellular distribution of selected marker proteins known to be enriched in the isolated compartments (Fig. 1B), confirmed >80% enrichment of each fraction. For each subcellular fraction, proteins with masses from >500 kDa down to <10 kDa were efficiently size-separated using SDS-SEC (Fig. 1C).
Characterization of Rapidly Depleted Proteins
The workflow outlined above will detect rapidly depleted proteins (RDPs), whose abundance decreases following cycloheximide treatment, either because of degradation, or secretion. To identify RDPs with high statistical significance, p values were calculated using a t test of the replicate CHX/DMSO (M/L) SILAC ratios of each protein, compared with the CHX/DMSO (M/L) SILAC ratios for control, known stable proteins, i.e. β-Tubulin, Histone H2A, and GAPDH. A significance cutoff value of p < 0.05 (-Log10(p) > 1.3) was used, combined with a requirement of a >50% decrease in CHX/DMSO (M/L) SILAC ratio (Log2 normalized ratio CHX/DMSO < −1), indicating at least 50% depletion in less than 6 h.
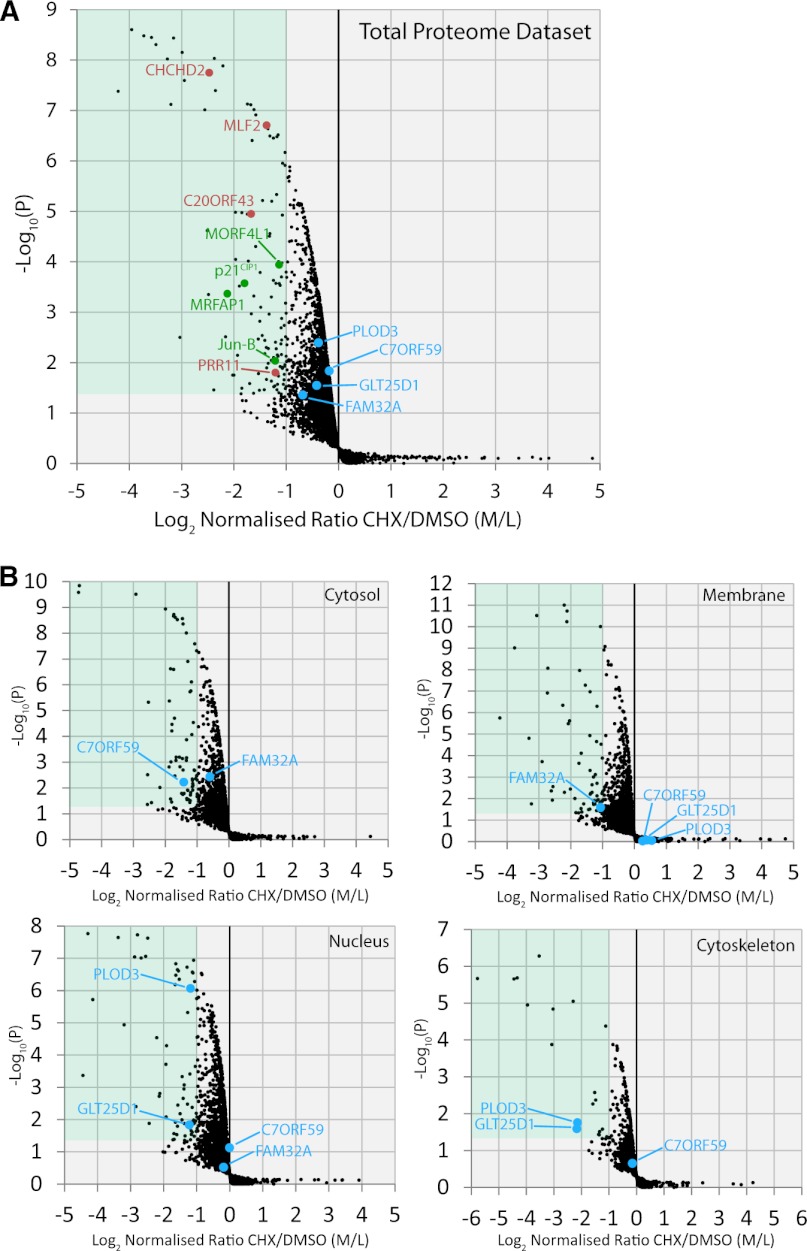
These criteria yielded 110 RDPs out of the total data set of ∼5000 proteins (Fig. 2a green box, and supplemental Table S3). This included proteins previously shown to be rapidly degraded, such as p21 (29), the MORF4L1, and MRFAP1 proteins (28), and Jun-B (30) (Fig. 2A, green text). Some RDPs showed different degradation rates in separate subcellular compartments (supplemental Fig. S2), demonstrating that individual protein stability can vary depending on localization.
Fig. 2.
Identification of rapidly depleted proteins. A, The analysis of ∼5000 proteins is indicated with the mean ratio from three replicates of each protein, represented by a black spot (n = 3). The y axis shows a p value (-log10 transformed) derived from a t test across the three biological replicates for each protein, compared with the biological replicates of negative control proteins, which do not display rapid depletion. The relative fold change is shown on the x axis using the log2 ratio cycloheximide/DMSO (Medium/Light). Proteins with a p < 0.05 and a fold change <-1 are overlaid by the green box and we define these as rapidly depleted proteins (RDPs). Examples of previously uncharacterized proteins highlighted in this study are marked in red and proteins regarded as positive controls are marked in green. B, The analysis of proteins found in each subcellular fraction is shown with the mean ratio from three replicates of each protein represented by a black spot (n = 3). The y axis shows a p value (-log10 transformed) derived from a t test across the three biological replicates for each protein, compared with the biological replicates of negative control proteins, which do not display rapid depletion. The relative fold change is shown on the x axis using the log2 ratio cycloheximide/DMSO (Medium/Light). Proteins with a p < 0.05 and a SILAC ratio <-1 are overlaid by the green box and we define these as fraction-specific rapidly degraded proteins (RDPs).
Rapidly Depleted Proteins Vary Between Subcellular Compartments
The same significance cutoffs (p < 0.05 and >50% decrease in CHX/DMSO M/L SILAC ratio), were used to analyze the data from each subcellular fraction (Fig. 2B). This identified an additional subset of proteins whose abundance rapidly decreased in a specific compartment, but which were not identified as RDPs in the total data set (Fig. 2a,b). C7ORF59, FAM32A, PLOD3 and GLT25D1, provide clear examples of proteins not detected as statistically significant RDPs in the total data set, but where analysis of data from a single subcellular fraction show they behave as compartment-specific RDPs (Fig. 2A, 2B blue points and see supplemental Table S1 for more examples). These examples illustrate that this phenomenon is not confined to any one fraction alone and further show it applies to proteins detected in multiple locations and not only to proteins enriched in a single compartment. For most of the compartment specific RDPs, reciprocal increases in CHX/DMSO (M/L) ratios were not seen in other fractions, indicating that decreased abundance results from degradation and secretion, rather than from intracellular trafficking between compartments (supplemental Table S1).
Comparative Analysis of RDPs
The RDPs identified were analyzed for sequence motifs involved in targeted degradation mechanisms (supplemental Table S3) and compared with data from two recent large scale (>5000 proteins detected), pulse-SILAC analyses of protein turnover in either HeLa cells (7), or mouse NIH-3T3 cells (6). We have also examined the list of RDPs to mark previously known and predicted secreted proteins (supplemental Table S3) (31), which rapidly decrease in relative abundance after cycloheximide treatment because of protein secretion, rather than degradation. Mitochondrial targeting of the RDPs using either motif-based prediction or previous localization data has also been annotated for each protein (supplemental Table S3) (31).
Many of these novel RDPs contain known degradation targeting motifs, such as Anaphase Promoting Complex (APC/C) KEN-boxes (32, 33), APC/C d-boxes (33, 34), and Skp1-Cullin-Fbox (SCF) FBXW7-binding sites (35, 36) (supplemental Table S3). These motifs are found in many proteins whose abundance is regulated during the cell cycle by rapid degradation at specific stages (2). Comparison of all the 110 RDPs identified in this study with data from the previous large scale mammalian protein turnover studies, showed that >34% of our new RDP data were not reported in the earlier studies (supplemental Table S3). Of those RDP proteins previously detected in at least one of the former studies, 70% were also found to be rapidly degraded in the mouse NIH-3T3 data (61 proteins detected in common) and 59% were also rapidly degraded in the HeLa data (37 proteins detected in common). The fact that not all of our RDPs were detected as being rapidly turned over in each of these previous large scale studies may reflect differences in both the cell types and organisms analyzed.
Proteasome Inhibition Triggers the Unfolded Protein Response
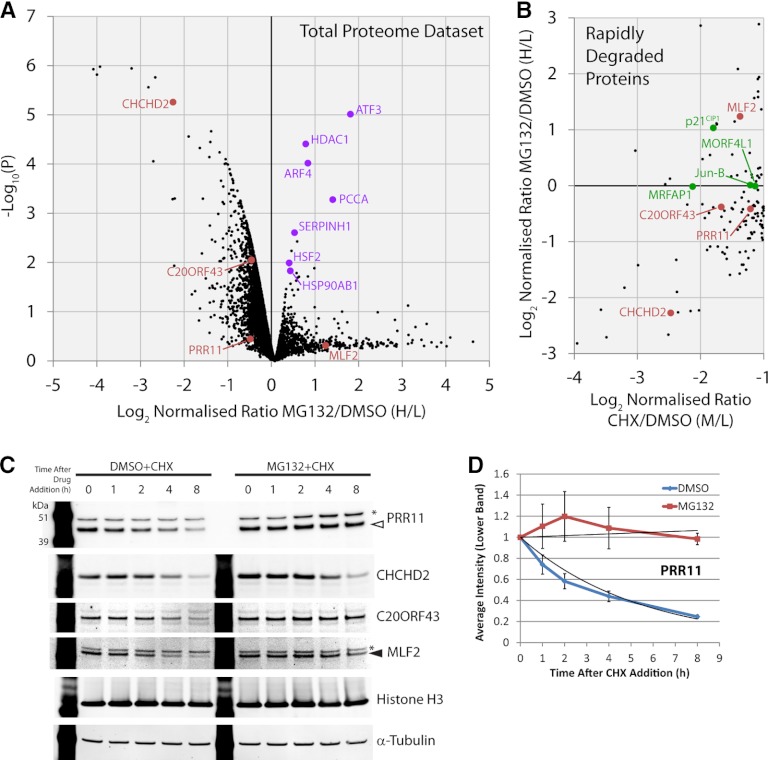
The total proteome data set and the subset of 110 RDPs were examined for their response to the proteasome inhibitor MG132 (Fig. 3A, 3B and supplemental Table S1). Interestingly, in both cases <1% of proteins showed a significant change in abundance after proteasome inhibition, i.e. either increase, or decrease, greater than twofold with p < 0.05, after MG132 treatment. To confirm that the MG132 treatment effectively blocked the proteasome, we analyzed our total data set for the abundance of K48-linked ubiquitin and histone H2A ubiquitin conjugates, which were previously shown to change following proteasome inhibition (37). This showed that MG132 treatment significantly increased the abundance of K48-linked ubiquitin conjugates and significantly decreased the levels of histone H2A-ubiquitin conjugates (supplemental Fig. S3A), confirming proteasome inhibition. Within our total data set, many of the proteins that significantly increase in abundance after MG132 treatment are associated with induction of the unfolded protein response (UPR), including ATF3, SERPINH1, HSF2, and HSP90AB1 (Fig. 3A). To further investigate the MG132-mediated activation of the UPR, we immunoblotted total cell lysates of U2OS cells after 6 h of MG132 treatment to test for phosphorylation of eIF2alpha, which is known to be induced during the UPR (38). This confirmed an increased abundance of phospho-serine51 on eIF2alpha following treatment with MG132, which is inhibitory for translation (supplemental Fig. S3B). Many of the proteins that significantly decrease after MG132 treatment are RDP proteins that are either predicted to be secreted (e.g. collagen COL1A1), or to have mitochondrial (e.g. CHCHD2), or integral membrane localization (e.g. GOLM1). These data therefore indicate which of the RDPs are likely to be proteasome targets, i.e. the subset of RDPs whose abundance does not change significantly after MG132 treatment.
Fig. 3.
Analysis of the proteome-wide response to proteasome inhibition. A, The analysis of ∼5000 proteins is indicated with the mean ratio from three replicates of each protein represented by a black spot (n = 3). The y axis shows a p value (-log10 transformed) derived from a t test across the three biological replicates for each protein, compared with the biological replicates of negative control proteins, which do not display rapid changes. The relative fold change is shown on the x axis using the log2 ratio MG132/DMSO (Heavy/Light). B, Rapidly depleted proteins were analyzed for their response to the proteasome inhibitor MG132. The y axis shows the relative fold change for the RDPs using the log2 ratio MG132/DMSO (Heavy/Light). The x axis shows the relative fold change for the RDPs using the log2 ratio cycloheximide/DMSO (Medium/Light). Examples of previously uncharacterized proteins highlighted in this study are marked in red and proteins regarded as positive controls are marked in green in both scatter plots. C, Immunoblotting of total cell lysates of U2OS cells after a time course of cycloheximide exposure, either in the presence, or absence, of proteasome inhibition to examine protein degradation, (n = 3). Arrowhead (filled white) indicates the PRR11 band, arrowhead (filled black) indicates the MLF2 band, and asterisks mark nonspecific bands. D, Quantitation of the PRR11 band from (C) with error bars indicating standard deviations (n = 3). See also supplemental Fig. S6 for full blot images.
Analysis of Novel RDPs
Four novel RDPs were selected for further characterization, based on the data in supplemental Table S3 and on the availability of antibody reagents i.e. (Proline-rich protein 11, PRR11; Coiled-coil-helix-coiled-coil-helix domain-containing protein 2, CHCHD2; uncharacterized protein, C20ORF43; and Myeloid leukemia factor 2, MLF2) (Fig. 2A and Fig. 3A, 3B red points). Our MS data show that while all four of these proteins decreased in abundance after protein synthesis inhibition, three (PRR11, C20ORF43, and MLF2) showed no significant change after MG132 treatment, consistent with them being proteasome targets, whereas CHCHD2 decreased in abundance (Fig. 3A, 3B). None of these proteins significantly increased in abundance when the proteasome was inhibited. Next, therefore, we analyzed their degradation using a complementary Western blotting method (Fig. 3C).
U2OS total cell lysates, treated either with a time course of cycloheximide alone, or cycloheximide used in combination with MG132, were immunoblotted using specific antibodies for the CHCHD2, PRR11, C20ORF43, and MLF2 proteins (Fig. 3C). These data showed all four proteins decreased in abundance to a similar extent as observed in the SILAC experiments after cycloheximide treatment alone. This independently confirms that all four factors are RDPs. These experiments also showed that PRR11, C20ORF43, and MLF2 were protected from degradation by adding MG132 together with cycloheximide, indicating that they are proteasome targets. However, CHCHD2 was not protected by MG132 and still decreased in abundance with similar kinetics to adding cycloheximide alone. Therefore, CHCHD2 is likely to be degraded by a proteasome-independent mechanism.
PRR11 is Cell Cycle Regulated
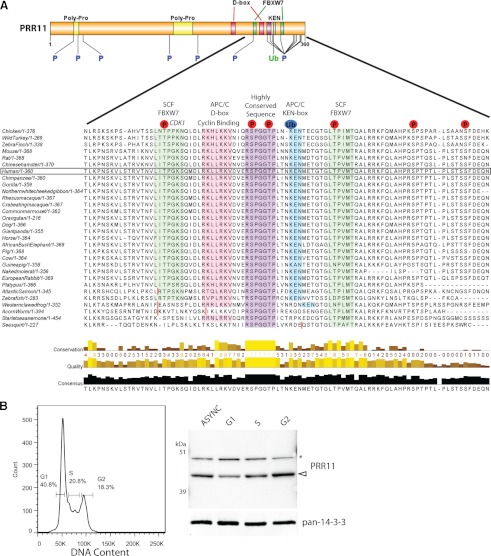
Detection of the PRR11 protein in the immunoblot was validated by siRNA knock-down of PRR11 mRNA (supplemental Fig. S3C). Quantitation of the blotting data showed that PRR11 disappeared exponentially after cycloheximide addition, but was stabilized by simultaneous MG132 treatment of the U2OS cells (Fig. 3D). The PRR11 protein is previously uncharacterised but contains multiple highly conserved sequence motifs associated with cell cycle regulated protein degradation (Fig. 4A and Sup the plemental Table S3). We therefore tested whether the abundance of PRR11 varies during the cell cycle.
Fig. 4.
Analysis of PRR11 cell cycle regulation. A, An overview of the entire human PRR11 protein sequence including features and motifs is shown (top), with the known ubiquitination site and phosphorylation sites indicated. An alignment of the C-terminal region of the PRR11 protein is shown (bottom), with various motifs and relevant known post-translational modifications to the human protein indicated. The conservation of each sequence motif is indicated by a colored overlay. Red vertical bars in the protein sequence represent missing sequence or gaps removed to show homology. The human PRR11 protein is highlighted in the alignment. B, Fluorescence-activated cell sorting of asynchronous U2OS cells stained for DNA content and sorted into G1, S or G2/M-phases of the cell cycle based on the gates indicated, with corresponding percentages of the total population. Immunoblotting of total cell lysates from the starting asynchronous cells and the sorted G1, S and G2/M cells is shown, (n = 2). Arrow head (filled white) indicates the PRR11 band, and an asterisk marks a nonspecific band.
First, the specific PRR11 antibody was used to characterize PRR11 expression and localization in U2OS cells by confocal immunofluorescence microscopy (supplemental Fig. S4). This showed that PRR11 staining was predominantly cytoplasmic and varied in intensity between cells, as compared with vimentin, which showed similar levels of staining in all of the cells. To test whether cell to cell variation in intensity of PRR11 staining reflects cell cycle modulation of its abundance, we used fluorescence-activated cell sorting (FACS) analysis of live U2OS cells, based on DNA content, to yield asynchronous, G1, S, and G2/M phase cell populations. The cell populations were lysed and immunoblotted with the PRR11 antibody (Fig. 4B). This showed that PRR11 levels increased from G1 to G2/M-phase cells. These data demonstrate that the levels of PRR1 protein vary during the cell cycle.
Computational Analysis of the Rapidly Degraded Proteins
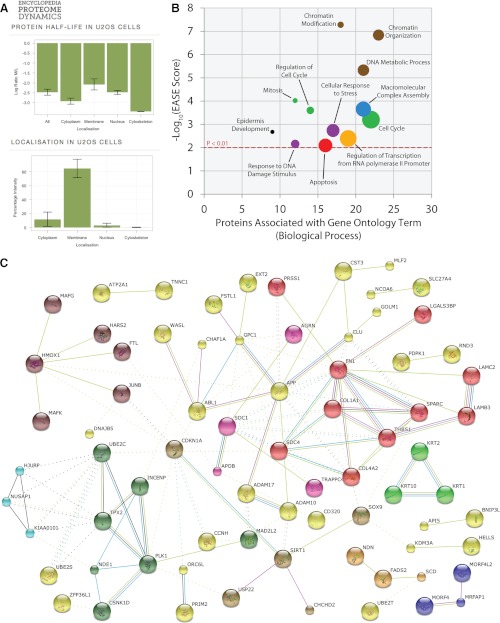
To enable convenient access to the wealth of quantitative proteomics data generated in this study, we have developed the Encyclopedia of Proteome Dynamics (http://www.peptracker.com/encyclopediaInformation/,). This is an online resource that collates all of the protein data from this study, together with data from our other recent proteomics studies, in a publicly available database, (Fig. 5A). Using this database we have analyzed the subset of total proteins identified as RDPs for biological process GO term enrichment, with a significance threshold of p < 0.01 when compared with an in silico translation of the human proteome (Fig. 5B). This showed that GO terms including regulation of cell cycle, mitosis, response to DNA damage stimulus, macromolecular complex assembly, chromatin modification, and epidermis development, were all significantly enriched in the U2OS RDP subset.
Fig. 5.
Computational analysis of the rapidly degraded proteins. A, Data derived from the Encyclopedia of Proteome Dynamics (EPD) is shown. The EPD database, which includes the entire data set presented in this study, is freely accessible at http://www.peptracker.com/encyclopediaInformation/. B, The analysis of enriched biological processes within the RDPs using the DAVID database (48). The y axis indicates the EASE score (-log10 transformed) for each GO term enriched in the RDP data set versus the human proteome, with the p < 0.01 cut-off indicated by a dotted red line. The x axis shows the number of RDPs associated with each GO term (biological process). The size of bubble associated with each GO term increases with increasing frequency of that term in the human proteome. Colors indicate related GO terms. C, The analysis of networks within the RDPs using the STRING database. Each protein is represented by a node and proteins without connections are not shown. Lines between nodes indicate a connection such as experimental interaction (pink), co-expression (black), interaction databases (blue), literature (green), and homology (purple). Kmeans clustering has been used to highlight sub-networks within the RDP data set with common coloring of these nodes.
To search for connections between the RDPs we used the STRING database (39), which analyses multiple parameters including co-expression, protein-protein interactions, and the literature to identify links between proteins (Fig. 5C). Many clusters were identified, of which several interesting networks stand out. This includes the MRFAP1-MORF4L1-MORF4L2 cluster that we identified previously as a network of rapidly degraded interacting proteins involved in chromatin regulation (28), and the PLK1-TPX2-UBE2C cell cycle related cluster that contains components associated with the anaphase promoting complex (Fig. 5C).
A similar clustering analysis was performed on the RDPs within each subcellular fraction (supplemental Fig. S5). This showed several RDP clusters in the cytoplasmic fraction, including a cell cycle related CDC20-UBE2C-UBE2S network and an ABL1-WASL signaling network. As expected, the membrane fraction contains most of the secreted proteins, as well as a cell cycle related p21-CDK4 network. The nuclear fraction highlights the MRFAP1-MORF4L1 network (28), the mediator complex and a cell cycle related signaling network containing Aurora B and PLK1 kinases. Of all the subcellular fractions the cytoskeletal compartment has the fewest RDPs and contains one main cluster comprising cytokeratins 1, 2, and 10. These data show that overall many of the RDPs identified are connected, either as part of multi-protein complexes, or else as components of regulatory pathways.
DISCUSSION
We have used a comprehensive proteomic strategy, combined with subcellular fractionation, to identify rapidly degraded proteins in specific compartments of U2OS osteosarcoma cells. We have characterized the effects of inhibiting either translation, or proteasome-mediated degradation, at the proteome level. Based on analysis of more than 74,000 peptides from over 5000 human proteins, we identified previously uncharacterized, rapidly degraded proteins and proteins whose degradation varied depending on subcellular localization. Using our workflow, the entire data set was annotated for subcellular localization, apparent molecular weight, abundance, and compartment specific degradation. The response to proteasome inhibition was also analyzed at the proteome level and showed a large-scale feedback inhibition on protein synthesis. These data provide a valuable resource detailing the spatial control of proteome degradation and its regulation in the model human U2OS cell line. We have created an Encyclopedia of Proteome Dynamics to provide the community with convenient online access to this, and other, large-scale proteomics data sets that we have generated.
Several features of the proteomics strategy used here enhance the biological value of the resulting data set. For example, because many proteins that are rapidly degraded occur in low abundance, size separation and analysis of subsets of proteins helped to reduce the competition for detection during LC-MS/MS. Subcellular fractionation not only assisted with the detection efficiency, including for example membrane proteins, but also provided insight into protein localization and compartment specific degradation (40). This study provides clear examples of specific proteins that are present in multiple subcellular locations but that behave as RDPs only in a subset of these locations. This is illustrated here both at the level of individual RDPs and at the level of protein interaction networks. Detection of such spatial control relied on the use of subcellular fractionation and was not apparent through the analysis of whole cell lysates. Based on these data, we infer that the spatial control of protein stability may be more common than previously realized. It will therefore be interesting in future to compare this phenomenon in other cell types and model organisms.
We believe that the strategy used here of cycloheximide treatment to subsequently monitor protein degradation, is complementary to studies that use the pulse-SILAC technique. Cycloheximide treatment has the advantage of avoiding possible problems of metabolic labeling, such as delayed incorporation of exogenously added amino acids at short time points and complications with data analysis and interpretation arising from the mixing of different pools of labeled and unlabeled amino acids. On the other hand, blocking translation with drug treatment may result in indirect effects on the proteome that may affect cell physiology and distort proteome dynamics. Therefore we regard both approaches as valid methods to measure protein stability and comparing the resulting data sets is particularly useful, as demonstrated by our comparison of the current cycloheximide-based analysis with previous large scale pulse SILAC studies (see supplemental Table S3).
A particularly striking finding of this study on U2OS cells was that inhibition of the proteasome for 6 h did not cause an increase in the abundance of almost any of the ∼5000 proteins identified. These findings are consistent with two recent reports that showed similar effects in either HCT116, or in HeLa cells, i.e. that most proteins did not substantially change in abundance after proteasome inhibition (37, 41). In our study, even with the subset of RDPs, whose abundance would be expected to increase the most if they are targets for proteasome-mediated degradation, little or no change was observed after treatment with MG132. Our analysis, however, confirmed that the MG132 treatment effectively blocked the proteasome and led to an increased level of K48-linked ubiquitin and decreased levels of histone-ubiquitin conjugates. Although some low abundance proteins that increase after proteasome inhibition may be missing from these data sets, (e.g. p53), nonetheless the data indicate that most proteins do not accumulate when the proteasome is blocked.
This seems paradoxical, because many of the RDPs identified in our study appear to be proteasome targets. We propose that the resolution of this paradox is provided by our current proteomic data showing that proteasome inhibition induces the UPR and thereby causes a global inhibition of translation. In addition, we observed increased abundance of phospho-serine51 on eIF2alpha after MG132 treatment. These observations are summarized in the model shown in Fig. 6. This is consistent with the observations of Jiang and Wek (42), who showed that inhibition of the proteasome leads to phosphorylation of the eukaryotic initiation factor eIF2alpha at ser51 and a reduction in translation. This phosphorylation event occurs in response to multiple forms of cellular stress and results in a broad inhibition of translation by reducing the delivery of Met-tRNAiMet to the ribosome.
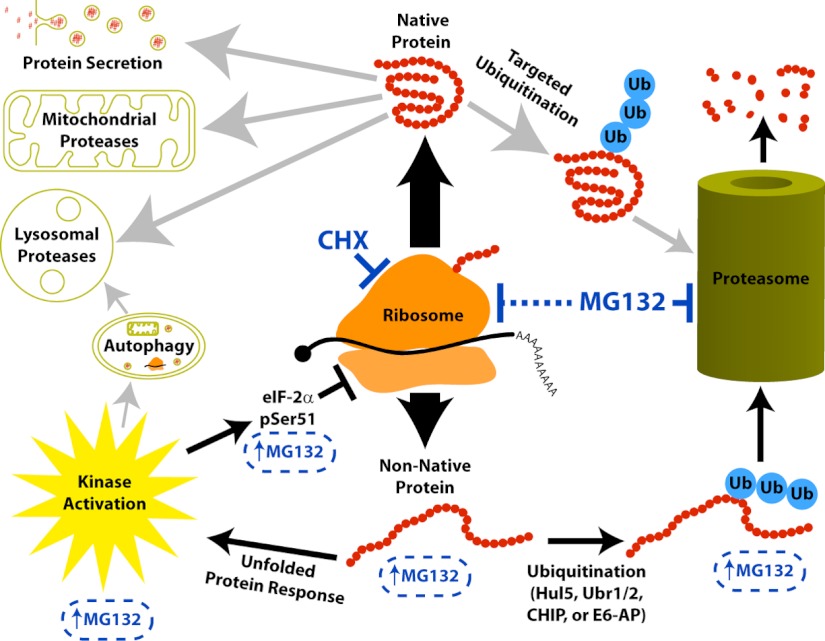
Fig. 6.
Model for proteome-wide responses after translation inhibition or proteasome inhibition. The ribosome produces both native protein and nonnative protein. Native protein may be degraded by mechanisms that may differ depending on subcellular localization (indicated by the gray arrows). Cycloheximide treatment allows us to observe this depletion as a decreased abundance of a protein over time. Under normal conditions the nonnative protein is quickly ubiquitinated and degraded by the proteasome. Proteasome inhibition by MG132 treatment blocks the degradation of both native proteins and nonnative proteins. This induces a build-up of nonnative protein that causes an unfolded protein response. One outcome of this response to MG132 is the inhibition of protein synthesis by eIF-2α pSer51 phosphorylation. Therefore, because MG132 simultaneously blocks the proteasome and protein synthesis, we observe very few proteins increasing in abundance after proteasome inhibition and only the subset of RDPs depleted by nonproteasomal mechanisms decrease in abundance.
What is the source of the unfolded proteins in this model? Previous studies have shown that the ribosome does not display 100% fidelity during translation and suggest up to 30% of translated proteins may be non-native “defective ribosomal products” (DRiPs) (43). This proportion of DRiPs is likely to vary, e.g. under different growth conditions and cell types. Under normal conditions, the DRiPs will be rapidly ubiquitinylated and degraded by the proteasome. In the presence of proteasome inhibitors, when these non-native proteins cannot be degraded by the ubiquitin-proteasome systems normally used for their destruction (44), they will accumulate over time and trigger the UPR. This is consistent with our present data, showing that a variety of proteins indicative of the UPR are up-regulated following inhibition of the proteasome. As illustrated in the model (Fig. 6), inhibition of the proteasome thus causes proteome-wide effects beyond simply blocking substrate degradation, because of the complex feedback inhibition of translation. We note that, as shown in the model, other pathways affecting protein stability and cell physiology, such as autophagy, can also be triggered by proteasome inhibition and the UPR. However, the contribution of other pathways was not addressed directly in this study and will be an interesting topic for future analysis.
Bioinformatics analysis indicated that the RDPs we identified were enriched for specific GO terms (Fig. 5B), indicating that the control of protein stability may be particularly important for regulatory pathways associated with these GO terms. These terms included “mitosis” and “regulation of cell cycle,” processes in which targeted protein degradation is known to be critical. In addition, our analysis showed that APC/C and FBXW7-SCF binding motifs, which are used in the G1-phase of the cell cycle, were associated with many RDPs. This further substantiates the link between protein stability and cell cycle and correlates with the fact that the majority of the asynchronous U2OS cells in our SILAC screen would be in G1 phase. It is interesting that despite the intensive previous analysis of cell cycle regulated proteins, this study, focusing on rapidly degraded proteins, has identified new examples, such as PRR11. Our analysis showed that PRR11 abundance is regulated across the cell cycle. This regulation is likely mediated by the conserved APC/C and FBXW7-SCF targeting motifs in the C terminus of the protein (Fig. 4A). Therefore, in both late mitotic and G1-phase cells, the PRR11 protein may be targeted for degradation by both the APC/C and FBXW7-SCF complexes.
In addition to cell cycle and mitosis, the set of RDPs identified also highlighted GO terms associated with processes in which the role of targeted protein degradation is less well established. This included highly significant enrichment for the GO terms, “chromatin organization” and “chromatin modification,” both of which had multiple protein examples. Other related processes highlighted included “cellular response to stress” and “response to DNA damage stimulus.” The enrichment of the GO term “epidermis development” is also interesting and may reflect the mesenchymal origin of the U2OS cell line. We suggest it will be informative in future to investigate further the potential role of targeted protein degradation mechanisms in these processes.
As well as identifying likely proteasome substrates, our global analysis has also indicated RDPs that are likely to be either degraded by proteasome-independent mechanisms, or else secreted. For example, CHCHD2 is a mitochondrial protein that has been shown previously to regulate oxidative phosphorylation (45). Our data indicate that CHCHD2 was not protected from degradation by proteasome inhibition and therefore must be degraded by other means such as through the mitochondrial degradation pathways of the Clp, Lon, or AAA complexes (46). Proteasome-independent degradation will also be needed for depletion of many membrane proteins and proteins targeted for destruction by autophagy, which uses lysosomal proteases (47). Our RDP data set includes likely examples of secreted proteins, e.g. collagens and proteins dependent on lysosomal degradation, e.g. GOLM1. This supports the view that this data set provides a representative overview of RDPs in human U2OS cells.
In summary, this study provides a resource identifying both global and compartment specific rapidly degraded proteins in U2OS cells, many of which are previously uncharacterized go to sleep. We also document the proteome-wide effects resulting from the negative feedback on translation induced by proteasome inhibition and propose a model for the mechanism responsible. The bioinformatics analysis of RDPs has highlighted biological processes in which the control of protein degradation is likely to play an important role. An interesting avenue for future research would be to examine how protein degradation differs between normal and transformed cells, as many proteins involved in cell proliferation, chromatin regulation, and DNA damage repair may be differentially regulated between these cell states.
Supplementary Material
Footnotes
* This work was supported by grants from the Wellcome Trust (Grant#: 083524/Z/07/Z, 073980/Z/03/Z, 08136/Z/03/Z, and 0909444/Z/09/Z) and the EU FP7 Prospects network (Grant#: HEALTH-F4-2008-201648). A.I.L. is a Wellcome Trust Principal Research Fellow.
 This article contains supplemental material.
This article contains supplemental material.
Author Contributions: M.L. performed all proteomics experiments. M.L. and K.K. performed the immunoblotting and FACS analysis. Y.A. and T.L. performed the statistical analysis. M.L., T.L. and A.I.L. wrote the paper. A.I.L. mentored the project.
1 The abbreviations used are:
- APC
- anaphase-promoting complex
- BCA
- bicinchoninic acid
- CBQCA
- 3-(4-Carboxybenzoyl)quinoline-2-carboxaldehyde
- CHX
- cycloheximide
- DMEM
- Dulbecco's modified eagle medium
- DNA
- deoxyribonucleic acid
- DRiP
- defective ribosomal product
- GO
- gene ontology
- HES
- HEPES, EDTA, sucrose
- HRP
- horseradish peroxidase
- LDS
- lithium dodecyl sulfate
- mRNA
- messenger RNA
- RDP
- rapidly depleted protein
- RT
- room temperature
- SCF
- Skp, Cullin, F-box
- SEC
- size exclusion chromatography
- SILAC
- stable isotope labeling with amino acids in cell culture
- TBST
- TBS tween 20
- TCEP
- triscarboxyethylphosphine
- TEAB
- triethylamine bicarbonate
- UPR
- unfolded protein response.
REFERENCES
- 1. Ciechanover A. (2005) Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Cell Death Differ. 12, 1178–1190 [DOI] [PubMed] [Google Scholar]
- 2. Reed S. I. (2003) Ratchets and clocks: The cell cycle, ubiquitylation and protein turnover. Nat. Rev. Mol. Cell Bio. 4, 855–864 [DOI] [PubMed] [Google Scholar]
- 3. Doherty M. K., Hammond D. E., Clague M. J., Gaskell S. J., Beynon R. J. (2009) Turnover of the human proteome: determination of protein intracellular stability by dynamic SILAC. J. Proteome Res. 8, 104–112 [DOI] [PubMed] [Google Scholar]
- 4. Eden E., Geva-Zatorsky N., Issaeva I., Cohen A., Dekel E., Danon T., Cohen L., Mayo A., Alon U. (2011) Proteome Half-Life Dynamics in Living Human Cells. Science 331, 764–768 [DOI] [PubMed] [Google Scholar]
- 5. Yen H. C., Xu Q., Chou D. M., Zhao Z., Elledge S. J. (2008) Global Protein Stability Profiling in Mammalian Cells. Science 322, 918–923 [DOI] [PubMed] [Google Scholar]
- 6. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 7. Boisvert F. O. M., Ahmad Y., Gierlinski M., Charriere F., Lamont D., Scott M., Barton G., Lamond A. I. (2012) A quantitative spatial proteomics analysis of proteome turnover in human cells. Mol. Cell. Proteomics. 11, M111.011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Savas J. N., Toyama B. H., Xu T., Yates J. R., 3rd, Hetzer M. W. (2012) Extremely long-lived nuclear pore proteins in the rat brain. Science 335, 942–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Levine A. J., Momand J., Finlay C. A. (1991) The P53 Tumor suppressor gene. Nature 351, 453–456 [DOI] [PubMed] [Google Scholar]
- 10. Vojtěsek B., Lane D. P. (1993) Regulation of P53 protein expression in human breast-cancer cell-lines. J. Cell Sci. 105, 607–612 [DOI] [PubMed] [Google Scholar]
- 11. Thomas L. R., Tansey W. P. (2011) Proteolytic Control of the Oncoprotein Transcription Factor Myc. Adv. Cancer Res. 110, 77–106 [DOI] [PubMed] [Google Scholar]
- 12. Cox J., Mann M. (2011) Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 80, 273–299 [DOI] [PubMed] [Google Scholar]
- 13. Bensimon A., Heck A. J., Aebersold R. (2012) Mass spectrometry-based proteomics and network biology. Annu. Rev. Biochem. 81, 379–405 [DOI] [PubMed] [Google Scholar]
- 14. Cravatt B. F., Simon G. M., Yates J. R., 3rd (2007) The biological impact of mass-spectrometry-based proteomics. Nature 450, 991–1000 [DOI] [PubMed] [Google Scholar]
- 15. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 16. Lamond A. I., Uhlen M., Horning S., Makarov A., Robinson C. V., Serrano L., Hartl F. U., Baumeister W., Werenskiold A. K., Andersen J. S., Vorm O., Linial M., Aebersold R., Mann M. (2012) Advancing cell biology through proteomics in space and time (PROSPECTS). Mol. Cell Proteomics. 11, O112.017731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lam Y. W., Lamond A. I., Mann M., Andersen J. S. (2007) Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol. 17, 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gatto L., Vizcaíno J. A., Hermjako H., Huber W., Lilley K. S. (2010) Organelle proteomics experimental designs and analysis. Proteomics. 10, 3957–3969 [DOI] [PubMed] [Google Scholar]
- 19. Dunkley T. P., Watson R., Griffin J. L., Dupree P., Lilley K. S. (2004) Localization of organelle proteins by isotope tagging (LOPIT). Mol. Cell Proteomics. 3, 1128–1134 [DOI] [PubMed] [Google Scholar]
- 20. Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570–574 [DOI] [PubMed] [Google Scholar]
- 21. Boulon S., Pradet-Balade B., Verheggen C., Molle D., Boireau S., Georgieva M., Azzag K., Robert M. C., Ahmad Y., Neel H., Lamond A. I., Bertrand E. (2010) HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol. Cell 39, 912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wessel D., Flügge U. I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 23. Bereman M. S., Egertson J. D., MacCoss M. J. (2011) Comparison between procedures using SDS for shotgun proteomic analyses of complex samples. Proteomics. 11, 2931–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 25. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 26. Zhou P. (2004) Determining protein half-lives. Methods Mol. Biol. 284, 67–77 [DOI] [PubMed] [Google Scholar]
- 27. Larance M., Bailly A. P., Pourkarimi E., Hay R. T., Buchanan G., Coulthurst S., Xirodimas D. P., Gartner A., Lamond A. I. (2011) Stable-isotope labeling with amino acids in nematodes. Nat. Methods. 8, 849–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Larance M., Kirkwood K. J., Xirodimas D. P., Lundberg E., Uhlen M., Lamond A. I. (2012) Characterization of MRFAP1 Turnover and interactions downstream of the NEDD8 pathway. Mol. Cell Proteomics. 11, M111.014407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Starostina N. G., Kipreos E. T. (2012) Multiple degradation pathways regulate versatile CIP/KIP CDK inhibitors. Trends Cell Biol. 22, 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gao M., Labuda T., Xia Y., Gallagher E., Fang D., Liu Y. C., Karin M. (2004) Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase itch. Science 306, 271–275 [DOI] [PubMed] [Google Scholar]
- 31. Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 32. Pfleger C. M., Kirschner M. W. (2000) The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Gene. Dev. 14, 655–665 [PMC free article] [PubMed] [Google Scholar]
- 33. Liu Z. X., Yuan F., Ren J., Cao J., Zhou Y. H., Yang Q., Xue Y. (2012) GPS-ARM: Computational Analysis of the APC/C Recognition Motif by Predicting D-Boxes and KEN-Boxes. Plos One 7, e34370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Glotzer M., Murray A. W., Kirschner M. W. (1991) Cyclin Is degraded by the ubiquitin pathway. Nature 349, 132–138 [DOI] [PubMed] [Google Scholar]
- 35. Welcker M., Clurman B. E. (2008) FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8, 83–93 [DOI] [PubMed] [Google Scholar]
- 36. Nash P., Tang X., Orlicky S., Chen Q., Gertler F. B., Mendenhall M. D., Sicheri F., Pawson T., Tyers M. (2001) Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414, 514–521 [DOI] [PubMed] [Google Scholar]
- 37. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dever T. E. (2002) Gene-specific regulation by general translation factors. Cell 108, 545–556 [DOI] [PubMed] [Google Scholar]
- 39. Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., Jensen L. J., von Mering C. (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ahmad Y., Boisvert F. M., Lundberg E., Uhlen M., Lamond A. I. (2012) Systematic analysis of protein pools, isoforms, and modifications affecting turnover and subcellular localization. Mol. Cell Proteomics. 11, M111.013680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tatham M. H., Matic I., Mann M., Hay R. T. (2011) Comparative Proteomic Analysis Identifies a Role for SUMO in Protein Quality Control. Sci. Signal. 4, rs4. [DOI] [PubMed] [Google Scholar]
- 42. Jiang H. Y., Wek R. C. (2005) Phosphorylation of the alpha-subunit of the eukaryotic initiation factor-2 (eIF2 alpha) reduces protein synthesis and enhances apoptosis in response to proteasome inhibition. J. Biol. Chem. 280, 14189–14202 [DOI] [PubMed] [Google Scholar]
- 43. Schubert U., Anton L. C., Gibbs J., Norbury C. C., Yewdell J. W., Bennink J. R. (2000) Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404, 770–774 [DOI] [PubMed] [Google Scholar]
- 44. Kriegenburg F., Ellgaard L., Hartmann-Petersen R. (2012) Molecular chaperones in targeting misfolded proteins for ubiquitin-dependent degradation. FEBS J. 279, 532–542 [DOI] [PubMed] [Google Scholar]
- 45. Baughman J. M., Nilsson R., Gohil V. M., Arlow D. H., Gauhar Z., Mootha V. K. (2009) A computational screen for regulators of oxidative phosphorylation implicates SLIRP in mitochondrial RNA homeostasis. Plos Genet. 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Baker B. M., Haynes C. M. (2011) Mitochondrial protein quality control during biogenesis and aging. Trends Biochem. Sci. 36, 254–261 [DOI] [PubMed] [Google Scholar]
- 47. Ciechanover A. (2005) Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Bio. 6, 79–87 [DOI] [PubMed] [Google Scholar]
- 48. Huang, da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.