Abstract
Monosodium urate (MSU) is an endogenous danger signal that is crystallized from uric acid released from injured cells. MSU is known to activate inflammatory response in macrophages but the molecular mechanisms involved have remained uncharacterized. Activated macrophages start to secrete proteins to activate immune response and to recruit other immune cells to the site of infection and/or tissue damage. Secretome characterization after activation of innate immune system is essential to unravel the details of early phases of defense responses. Here, we have analyzed the secretome of human primary macrophages stimulated with MSU using quantitative two-dimensional gel electrophoresis based proteomics as well as high-throughput qualitative GeLC-MS/MS approach combining protein separation by SDS-PAGE and protein identification by liquid chromatography-MS/MS. Both methods showed that MSU stimulation induced robust protein secretion from lipopolysaccharide-primed human macrophages. Bioinformatic analysis of the secretome data showed that MSU stimulation strongly activates unconventional, vesicle mediated protein secretion. The unconventionally secreted proteins included pro-inflammatory cytokines like IL-1β and IL-18, interferon-induced proteins, and danger signal proteins. Also active forms of lysosomal proteases cathepsins were secreted on MSU stimulation, and cathepsin activity was essential for MSU-induced unconventional protein secretion. Additionally, proteins associated to phosphorylation events including Src family tyrosine kinases were increased in the secretome of MSU-stimulated cells. Our functional studies demonstrated that Src, Pyk2, and PI3 kinases act upstream of cathepsins to activate the overall protein secretion from macrophages. In conclusion, we provide the first comprehensive characterization of protein secretion pathways activated by MSU in human macrophages, and reveal a novel role for cathepsins and Src, Pyk2, PI3 kinases in the activation of unconventional protein secretion.
The innate immune system is activated in response to microbial infection and tissue damage. Macrophages are the central players of the innate immunity and detect the presence of pathogen-associated molecular patterns (PAMPs)1 and damage-associated molecular patterns (DAMPs) with their pattern recognition receptors. This recognition results in the activation of antimicrobial defense, inflammatory response, tissue regeneration, and recruitment of other inflammatory cells to the site of infection and/or tissue damage (1). Proper innate immune response is essential for the activation of the adaptive immune system. At present it is thought that the activation of innate immunity is most effective when both signals of microbial origin and damage are perceived at the same time (2, 3).
Monosodium urate (MSU) is an endogenous DAMP that is crystallized from uric acid released by injured cells (4). Uric acid is a byproduct of purine degradation, and abnormally high levels of uric acid in serum, or hyperuricemia, is a hallmark of metabolic disorders where balance between intake of purines via food and excretion of uric acid is distorted. A well-known disease associated to hyperuricemia is gouty arthritis, in which deposits of MSU can be found in synovial fluid of peripheral joints, and MSU-induced inflammation is the initial trigger of symptoms (5). Hyperuricemia is also linked to other inflammatory diseases, like metabolic syndrome (6, 7), type 2 diabetes (8), and cardiovascular disease (9). MSU-induced inflammation is driven by the innate immune system. MSU engages antigen-presenting cells, macrophages, and dendritic cells. It is a potent adjuvant, initiating a robust adaptive immune response (4). Recently it has been shown that the adjuvant properties of alum are dependent on release of uric acid in vivo (10).
It is unclear how cells detect the presence of MSU. It has been suggested that MSU activates intracellular signaling pathways in dendritic cells by directly engaging cellular membranes, particularly the cholesterol-rich components of the plasma membrane (11). Recently Uratsuji and coworkers showed that MSU activates inflammatory response in keratinocytes and monocytic THP-1 cells through membrane-associated P2Y6 (12). It is also well-documented that MSU activates the NLRP3 inflammasome in macrophages (13). The NLRP3 inflammasome is a multiprotein complex comprising of NACHT, LRR, and PYD domains-containing protein 3 (NLRP3), Apoptosis-associated speck-like protein containing a CARD (ASC) and cysteine protease Caspase-1. Activation of NLRP3 inflammasome results in the autocleavage of Caspase-1. The activated Caspase-1 then in turn cleaves pro-inflammatory cytokines IL-1β and IL-18 into their biologically active forms, which are then readily secreted (14–17). However, the signaling pathways that are involved in MSU-induced NLRP3 inflammasome activation have remained only partially characterized.
Macrophages respond to activating stimuli by producing inflammatory mediators that are delivered to neighboring cells through multiple protein secretion pathways including both conventional and unconventional protein secretion (18). Conventionally secreted proteins contain an N-terminal signal peptide, which directs their transport to the plasma membrane through the well-characterized endoplasmic reticulum (ER)-Golgi pathway. In contrast, mediators and regulators of unconventional protein secretion pathways are less well understood. At present, different proteomic techniques allow for in-depth analysis of the secretome, the global pattern of secreted proteins. Secretome analysis is important in revealing complex cellular processes that require communication and signaling between the cells, and it has recently been applied to analyze the signaling pathways related to cell differentiation (19, 20), cancer (21, 22), and immune responses (23–25). In the present work we have characterized the secretome of human primary macrophages that have been activated simultaneously by microbial signal lipopolysaccharide (LPS) and endogenous danger signal MSU to get a global view of macrophage response to combined PAMP and DAMP stimulation.
EXPERIMENTAL PROCEDURES
Culturing of Human Macrophages and Stimulations
Human peripheral blood-derived monocytes were isolated from leukocyte-rich buffy coats from healthy blood donors (The Finnish Red Cross Blood Transfusion Service) and differentiated to macrophages for 6 days as previously described (26). The cells used in the experiments express macrophage-specific genes including CD14, CD163, and FcγR1A (27). For proteomic analysis of the secretome, macrophages were primed with 100 ng/ml LPS from Escherichia coli 0111:B4 (Sigma) in Macrophage-SFM (Invitrogen, Carlsbad, CA) supplemented with 10 ng/ml GM-CSF (Biosource International Inc., Camarillo, CA) and antibiotics. After 21 h cell culture media was discarded, cells were washed three times with PBS and RPMI 1640 (supplemented with 10 mm Hepes, l-glutamine, and antibiotics (all from Invitrogen)) was added. Cells were stimulated with 100 μg/ml MSU (Enzo Life Sciences, Farmingdale, NY). After 3 h cell culture supernatants of cells from two to three blood donors were pooled together and part was taken for ELISA or Luminex assay. Cells were collected for Western blotting. The supernatants from same amount of cells in each group were concentrated with Amicon Ultra-15 (Millipore) centrifugal filter devices with a cutoff of 10 kDa NMWL (nominal molecular weight limit). Exosomal fractions were enriched using centrifugal filter units (100 kDa NMWL) and the flow-through was further concentrated with 10 kDa-centrifugal filter units. CA-074 Me (25 μm, Calbiochem, San Diego, CA), PP2 (10 μm, Sigma), PF-431396 (10 μm, Sigma), or LY-294002 (100 μm, Sigma) were added to cells in 1 h before MSU stimulation.
For quantitative real-time PCR analysis, cells were primed with LPS (100 ng/ml) for 21 or 18 h. Stimulation with MSU (100 μg/ml) for 3 or 6 h was performed in fresh Macrophage-SFM without GM-CSF. Cells were collected using Eurozol or EuroGold RNA Pure (EuroClone, Milan, Italy) according to manufacturer's instructions.
2-DE Analysis and SDS-PAGE for GeLC-MS/MS
For two-dimensional gel electrophoresis (2-DE), 100 μl of concentrated cell culture supernatants were purified using 2-D Clean-Up kit (GE Healthcare) according to manufacturer's instructions. Proteins were resuspended in 200 μl of rehydration buffer (7 m urea, 2 m thiourea, 4% CHAPS, 0.6% dithiothreitol, 0.5% IPG-buffer 3–10, 0.004% bromphenol blue) and the sample was loaded onto 11 cm pH 3–10 ReadyStrip IPG strips (Bio-Rad, Hercules, CA). Strips were left to rehydrate for 24 h at room temperature and then isoelectric focusing was performed for a total of 63.3 kVh using Ettan IPGphor II (GE Healthcare). Focused strips were equilibrated at room temperature for 15 min in SDS-equilibration buffer (6 m urea, 50 mm Tris-HCl pH 8.8, 30% glycerol, 2% SDS, 0.004% bromphenolblue) supplemented with 10 mg/ml dithiothreitol, following similar equilibration in SDS-equilibration buffer supplemented with 25 mg/ml iodoacetamide. Then SDS-PAGE was run at 200 V for 1 h using Criterion 10–20% Tris-HCl gels (Bio-Rad). Gels were mass spectrometry compatibly silver stained (28). Spot detection, matching and intensity-based quantitation were performed using ImageMaster 2D Platinum version 6.0 (GE Healtcare). The number of replicate gels analyzed was three in both groups. Statistically significant differences (p < 0.05) in spot intensities based by spot volumes were analyzed by Student's t test.
For GeLC-MS/MS, 20 μl of concentrated cell culture supernatants were loaded onto Criterion 10–20% Tris-HCl gel (Bio-Rad) and proteins were separated at 200 V for 1 h. After that the gel was silver stained (28). Also GeLC-MS/MS analyses were performed on three biological replicate samples.
Protein Identification
For identification, the protein spots were cut out from 2-DE gels, and in GeLC-MS/MS, whole sample lanes were cut into 15 pieces of equal size followed by in-gel digestion with trypsin (29). The resulting peptides were analyzed with Ultimate 3000 nano-LC (Dionex) and a QSTAR Elite hybrid quadrupole time-of-flight mass spectrometer (AB Sciex, Foster City, CA) with nano-ESI ionization. Samples were first loaded on a ProteCol C18 trap column (10 mm × 150 μm, 3 μm, 120 Å) (SGE), and then separated on a PepMap100 C18 analytical column (15 cm × 75 μm, 5 μm, 100 Å) (LC Packings/Dionex) at 200 nl/min. MS data were acquired using Analyst QS 2.0 software (AB Sciex). The information-dependent acquisition method consisted of a 0.5 s TOF-MS survey scan of m/z 400–1400. From every survey scan two most abundant ions with charge states +2 to +4 were selected for product ion scans. Ions selected for MS/MS fragmentation were put to exclusion list for 60 s.
The obtained LC-MS/MS data was searched with in-house Mascot search engine version 2.2 through the ProteinPilot 2.0.1 interface against SwissProt 57.12 (20329 H. sapiens protein entries) for 2-DE data or through ProteinPilot 4.0.8085 interface against the SwissProt database 2011 (20245 H. sapiens protein entries) for GeLC-MS/MS data. The search criteria for Mascot searches were: Homo sapiens as taxonomy; trypsin digestion with one missed cleavage allowed; carbamidomethyl modification of cysteine as a fixed modification and oxidation of methionine as a variable modification. Peptide mass tolerance was 50 ppm and fragment mass tolerance was 0.2 Da. Protein identifications that had probability-based Mascot Mowse scores ≥ 50 and p < 0.05 were considered reliable high-confidence identifications. False discovery rates (FDR) were calculated using a concatenated normal and reversed sequence database, and a previously reported method (30), and were 1.5–4% in the GeLC-MS/MS data sets.
Protein Classification
The identified proteins were classified based on Gene Ontology (GO) (31) terms cellular component and biological process with GoMiner (32) using Uniprot, ENSEMBL, LMP, and PDB as data sources and H. sapiens as organism. GO database version for all queries was 2011–01. The identified proteins were also analyzed with GeneTrail (33) to recognize functional Kyoto Encyclopedia of Genes and Genomes (KEGG, (34)) categories that were over or under-represented among data. Analysis was performed using all genes from H. sapiens as a reference set, significance threshold 0.05, p value adjustment FDR and two as minimum number of genes per pathway. Protein identifications were analyzed with prediction tool SignalP 4.0 to find the proteins having a possible signal peptide (35). The identified proteins were also compared with ExoCarta database version 4.1 containing known exosomal proteins (36), and with Interferome database containing known interferon (IFN) regulated genes (37).
ELISA and Luminex
IL-1β from cell culture supernatants was analyzed with human IL-1β Eli-pair ELISA (Diaclone, Tepnel Research Product & Services) according to manufacturer's instructions. Secretion of CCL2, CCL3, CCL4, CCL5, and CXCL10 from cell culture supernatants was measured with Luminex system Bio-Plex 200 (Bio-Rad) using Bio-Plex Manager 4.1.1 software (Bio-Rad). Custom-made Bio-Plex Pro Assay (Bio-Rad) was used according to manufacturer's instructions.
Western Blotting
For Western blotting, cells were lysed in total cell lysis buffer (10 mm Tris pH 7.4, 150 mm NaCl, 25% ethylene glycol) supplemented with Complement Mini protease inhibitor (Roche Applied Science)) and homogenized by ultrasound sonication. Concentrated cell culture supernatants were further purified with 2-D Clean-Up Kit (GE Healthcare). Ten micrograms protein of cell lysates, or alternatively a protein amount that was equal to 10% of the cell culture supernatants, were loaded on a Mini-Protean TXG precast gel (Bio-Rad) for SDS-PAGE and subsequently transferred to Immobilon-P Transfer Membranes (Millipore). Incubation with primary antibodies was performed overnight at 4 °C. Incubation with appropriate HRP-conjugated secondary antibodies (Dako) was performed at RT for 1 h. Proteins were visualized with Western Lightning Plus ECL system (Perkin Elmer), and images were captured with ImageQuant LAS 4000 mini CCD camera (GE Healthcare). Antibodies against HSP90 (Cell Signaling Technology, Danvers, MA), IFIT-3 (also known as RIG-G) (BD Biosciences), Annexin-1, Cathepsin B (Calbiochem), Cathepsin D, Galectin-3, LAMP-1 (all from Santa Cruz Biotechnology, Santa Cruz, CA), and ASC (Millipore) were purchased. IL-1β, IL-18, and MxA antibodies have been previously described (26, 38, 39). To verify equal loading of cell lysate proteins, blotted membranes were stained with Sypro ruby protein blot stain (Bio-Rad) according to manufacturer's instructions.
RNA Extraction, cDNA Synthesis and Quantitative Real-time PCR
Total cellular RNA was extracted from the cells with Eurozol or EuroGold RNA Pure (EuroClone, Siziano, Italy) according to manufacturer's instructions. High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) was used in cDNA synthesis. Quantitative real-time PCR was performed with ABI Prism 7500 Fast Sequence Detector (Applied Biosystems) controlled with 7500 Fast System SDS Software v1.4 (Applied Biosystems). Reaction was set up in PerfeCta qPCR FastMix (Quanta BioSciences) with predeveloped TaqMan Gene Expression Assay primers and probes (CCL2: Hs00234140_m1, CCL3: Hs00234142_m1, CCL4: Hs99999148_m1, CCL5: Hs00174575_m1, CXCL10: Hs00171042_m1, Applied Biosystems). For each sample PCR amplification of 18S rRNA was determined (4319413E, Applied Biosystems) and it was used as an endogenous control to normalize the amount of cDNA between samples. Relative quantification was performed using a comparative CT method according to manufacturer's instructions (Applied Biosystems). Gene expression values are presented as fold change relative to controls.
RESULTS
MSU Activates Unconventional Protein Secretion From Macrophages
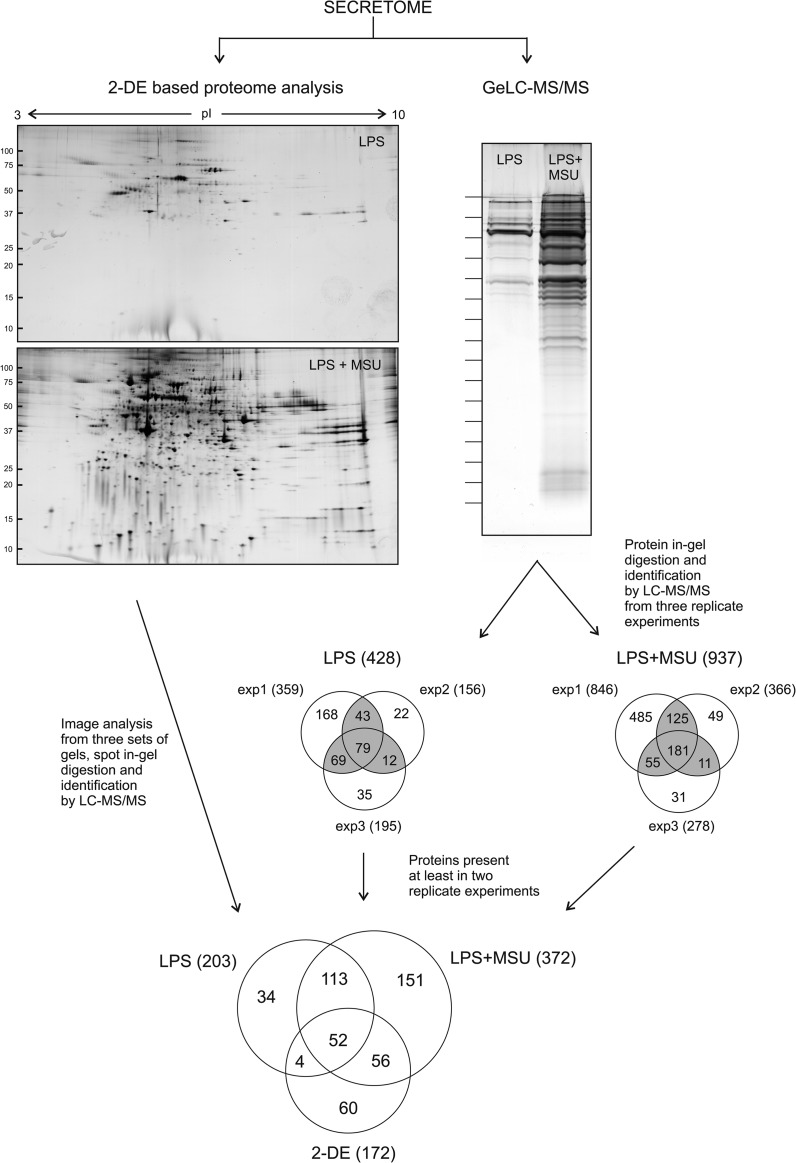
The secretomes from human monocyte-derived macrophages activated with LPS and stimulated with MSU were characterized using quantitative two-dimensional gel electrophoresis (2-DE) based proteomics as well as high-throughput; qualitative GeLC-MS/MS approach combining protein separation by SDS-PAGE and protein identification by LC-MS/MS (Fig. 1). Both methods showed that MSU stimulation induced robust protein secretion from LPS-primed human macrophages. Image analysis of the 2-DE gels revealed that a total of 238 protein spots were more abundant in the secretome of LPS-primed and MSU-stimulated cells compared with the secretome of only LPS-primed cells (Fig. 1). From these 238 spots we could identify proteins from 132 spots. More than one protein was identified from 81 spots, resulting in the identification of a total of 172 distinct proteins (supplemental Fig. S1 and supplemental Table S1). With the GeLC-MS/MS approach we identified altogether 428 proteins from the secretome of LPS-primed macrophages, and 937 proteins from the LPS-primed and MSU-stimulated macrophage secretomes. From these, 203 and 372 proteins from the secretome of LPS-primed, and LPS-primed and MSU-stimulated cells, respectively, were identified in at least two biological replicates (Fig. 1). We used primary cells in our studies, so the starting material for proteomics studies is much less uniform than when using cell lines most likely reflecting also the differential number of proteins identified in the three experiments.
Fig. 1.
Secretome analysis of MSU-stimulated human primary macrophages. Human macrophages were primed with LPS (100 ng/ml) for 21 h, after which they were stimulated with MSU (100 μg/ml) for 3 h. Cell culture supernatants were collected and secreted proteins were analyzed using two different proteomics methods, quantitative 2-DE based proteomics and qualitative, high-throughput GeLC-MS/MS. In 2-DE, the proteins were separated using 11 cm 3–10 IPG-strips in the first dimension followed by second dimension separation with 10–20% Tris-HCl gradient gels. The protein spots were visualized using silver-staining, and after image analysis the differentially secreted proteins were in-gel digested into peptides and identified using LC-MS/MS. In GeLC-MS/MS the proteins were separated using 10–20% Tris-HCl gradient gel and visualized with silver-staining. To identify all the secreted proteins the lanes were cut into 15 pieces, and proteins were identified using a combination of in-gel digestion and LC-MS/MS analysis.
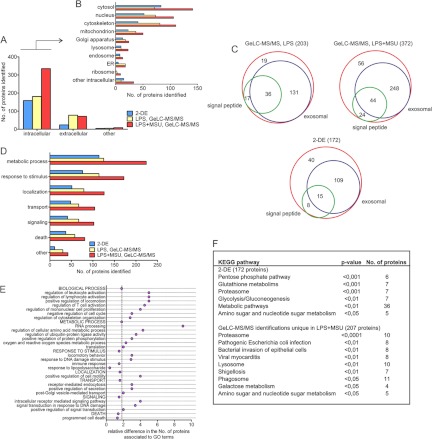
The proteins identified from 2-DE gels and from at least two biological replicates in GeLC-MS/MS were first classified based on their GO term cellular component. This revealed increases mainly in the secretion of intracellular proteins (Fig. 2A). MSU stimulation increased the secretion of proteins associated to all of the main organelles, with the largest relative increase in the number of proteins related to ribosome, lysosome, cytosol, and intracellular proteins that were not associated to any of the main organelles (Fig. 2B). To confirm that the proteins identified from the secretomes are not leaking from the cells because of cell death we performed Western blot analysis with Caspase-3 antibody from total cellular lysates, and measured the amount of apoptotic cells with the Apopersentage assay after MSU stimulation (supplemental Fig. S2). We also measured the lactate dehydrogenase activity in cell culture supernatants. We did not detect Caspase-3 activity in total cellular lysates, apoptotic cells, or increasing activity of lactate dehydrogenase in cell culture supernatants on MSU stimulation (supplemental Fig. S2), indicating that the increased protein secretion is not a result of on-going apoptosis or necrotic rupture of the stimulated cells.
Fig. 2.
Classification of the secretome proteins. A, B, Classification of the secreted proteins based on their GO term cellular component. C, Venn diagrams of the proteins identified having a predicted N-terminal signal peptide and/or being known to be exosomal proteins. D, Classification of the secreted proteins based on the GO term biological process. E, More detailed analysis of the enriched subclasses of biological processes for the proteins identified with GeLC-MS/MS based on comparing the number of proteins with assigned GO terms in MSU-stimulated and LPS-primed to LPS-primed cell secretomes. The vertical line denotes the ratio of proteins in MSU-stimulated and LPS-primed to LPS-primed cell secretomes. F, Over and under-representation analysis of KEGG pathways for the identified proteins.
Proteins are released from cells through various mechanisms including conventional, signal peptide-dependent secretion and unconventional, vesicle-mediated mechanisms (40). We next analyzed the identified proteins using SignalP to determine the presence of signal peptide sequence: 26% of the proteins identified from the secretome of LPS-primed cells had a predicted signal peptide, and only 18% of the proteins identified from the secretomes of LPS-primed and MSU-stimulated macrophages contained signal sequence (Fig. 2C and supplemental Table S2). Exosomes are vesicles known to be secreted in immune reactions (41, 42), so we next compared the identified proteins with an exosomal protein database, ExoCarta (36): 72% of proteins identified from 2-DE gels, and 82 and 78% of proteins identified with GeLC-MS/MS from LPS-primed and LPS-primed and MSU-stimulated cells, respectively, were found in ExoCarta (Fig. 2C and supplemental Table S2). Taken together, our data strongly suggests that MSU activates unconventional, vesicle-mediated protein secretion pathways in macrophages.
The Secretion of Proteins Involved in Inflammatory Response is Induced on MSU Stimulation From LPS-primed Cells
The identified proteins were next classified based on GO term biological process. This showed that the number of proteins involved in a metabolic process increased the most in the secretome on MSU stimulation (Fig. 2D). More detailed classification based on biological processes showed that proteins related to processes such as regulation of leukocytes and lymphocyte activation, regulation of mononuclear cell proliferation, and regulation of locomotion and cytoskeletal organization increased in the secretome on MSU stimulation. Also the secretion of metabolic proteins related to RNA processing, regulation of ubiquitin-protein ligase activity, and protein phosphorylation were clearly increased on MSU stimulation (Fig. 2E). Pathway analysis showed that the mostly enriched KEGG categories identified from 2-DE analysis were pentose phosphate pathway (p < 0.001), glutathione metabolism (p < 0.001), and proteasome (p < 0.001) (Fig. 2F). The enriched pathways from GeLC-MS/MS data included proteasome (p < 0.0001), lysosome (p < 0.01), and phagosome (p < 0.05). The enriched pathways indicate that defense responses like inflammatory processes and phagocytosis were activated on MSU-stimulation.
One major group of inflammatory proteins found in our secretome data was the danger signal proteins. These proteins are preformed endogenous molecules, usually with a well-defined intracellular function, released or exposed following an injury or a stress (43, 44). The identified danger signal proteins included several annexins, galectins, heat shock proteins, and S100 proteins (Table I). Most of these proteins did not contain a signal peptide suggesting that they are released from MSU-stimulated macrophages through an unconventional protein secretion pathway. Instead, most of these danger signal proteins were found in ExoCarta indicating that they are secreted through exosomes. To confirm the exosomal protein secretion of danger signal proteins in MSU-stimulated macrophages, we enriched the fraction containing extracellular vesicles from the growth media. Western blot analysis demonstrated the presence of Annexin-1, Galectin-3, and Heat shock protein HSP 90 (HSP90) in the exosomal fraction (Fig. 3A).
Table I. Proteins related to inflammation in secretomes identified by 2-DE and GeLC-MS/MS (identified at least in one experiment). Human macrophages were primed with LPS (100 ng/ml) for 21 h, after which they were stimulated with MSU (100 μg/ml) for 3 h. Cell culture supernatants were collected and secreted proteins were analyzed using two different proteomics methods, quantitative 2-DE based proteomics and qualitative, high-throughput GeLC-MS/MS. Abbreviations: Y, present in a database; N, not present in a database.
| Protein | Uniprot accession | GeLC-MS/MS |
2-DE | signalP | Exocarta | |
|---|---|---|---|---|---|---|
| LPS | LPS+MSU | |||||
| Danger Signal Proteins | ||||||
| Annexin A4 | P09525 | − | + | − | N | Y |
| Annexin A6 | P08133 | − | + | − | N | Y |
| Galectin-9B | Q3B8N2 | − | + | − | N | N |
| Heat shock protein 105 kDa | Q92598 | − | + | − | N | Y |
| Putative heat shock protein HSP 90-alpha A4 | Q58FG1 | − | + | − | N | Y |
| 60 kDa heat shock protein, mitochondrial | P10809 | − | + | − | N | Y |
| Protein S100-A10 | P60903 | − | + | − | N | Y |
| Thymosin beta-4-like protein 3 | A8MW06 | − | + | − | N | N |
| Parathymosin | P20962 | − | + | − | N | N |
| Hepatoma-derived growth factor | P51858 | − | + | − | N | N |
| Putative annexin A2-like protein | A6NMY6 | − | − | + | N | N |
| Annexin A5 | P08758 | + | + | + | N | Y |
| Annexin A11 | P50995 | + | + | + | N | Y |
| Galectin-1 | P09382 | + | + | + | N | Y |
| Galectin-7 | P47929 | + | + | + | N | Y |
| Heat shock protein beta-1 | P04792 | + | + | + | N | Y |
| Protein S100-A4 | P26447 | + | + | + | N | Y |
| Protein S100-A11 | P31949 | + | + | + | N | Y |
| Endoplasmin | P14625 | + | + | + | Y | Y |
| Annexin A1 | P04083 | + | + | − | N | Y |
| Annexin A2 | P07355 | + | + | − | N | Y |
| Galectin-3 | P17931 | + | + | − | N | Y |
| Galectin-3-binding protein | Q08380 | + | + | − | Y | Y |
| Galectin-9 | O00182 | + | + | − | N | N |
| Heat shock cognate 71 kDa protein | P11142 | + | + | − | N | Y |
| Heat shock protein HSP 90-beta | P08238 | + | + | − | N | Y |
| Heat shock protein HSP 90-alpha | P07900 | + | + | − | N | Y |
| Heat shock 70 kDa protein 1A/1B | P08107 | + | + | − | N | Y |
| Heat shock 70 kDa protein 4 | P34932 | + | + | − | N | Y |
| 78 kDa glucose-regulated protein | P11021 | + | + | − | Y | Y |
| Putative heat shock 70 kDa protein 7 | P48741 | + | + | − | N | Y |
| Heat shock 70 kDa protein 6 | P17066 | + | + | − | N | Y |
| Heat shock-related 70 kDa protein 2 | P54652 | + | + | − | N | Y |
| Protein S100-A8 | P05109 | + | + | − | N | Y |
| Protein S100-A6 | P06703 | + | + | − | N | Y |
| Protein S100-A9 | P06702 | + | + | − | N | Y |
| Protein S100-A7 | P31151 | + | + | − | N | N |
| Prothymosin alpha | P06454 | + | + | − | N | N |
| Cathelicidin antimicrobial peptide | P49913 | + | + | − | N | Y |
| Neutrophil defensin 1 | P59665 | + | + | − | Y | Y |
| Heat shock 70 kDa protein 1-like | P34931 | + | − | − | N | Y |
| Protein S100-A7-like 2 | Q5SY68 | + | − | − | N | Y |
| Chemokines and cytokines | ||||||
| C-C motif chemokine 5 (CCL5) | P13501 | − | + | − | Y | N |
| C-C motif chemokine 22 (CCL22) | O00626 | − | + | − | Y | N |
| Growth-regulated alpha protein (CXCL1) | P09341 | − | + | − | Y | N |
| Macrophage inflammatory protein 2-beta (CXCL3) | P19876 | − | + | − | Y | N |
| Platelet basic protein (CXCL7) | P02775 | − | + | − | Y | N |
| C-X-C motif chemokine 10 (CXCL10) | P02778 | − | + | − | Y | N |
| C-C motif chemokine 24 (CCL24) | O00175 | − | + | − | Y | N |
| Platelet factor 4 (CXCL4) | P02776 | − | + | − | Y | N |
| Macrophage migration inhibitory factor (MIF) | P14174 | − | + | − | N | Y |
| Interleukin-18 | Q14116 | − | + | − | N | N |
| Interleukin-12 subunit beta | P29460 | + | + | + | Y | N |
| Interleukin-1 receptor antagonist protein (IL-1ra) | P18510 | + | + | + | Y | Y |
| C-C motif chemokine 3 (CCL3) | P10147 | + | + | − | Y | N |
| C-C motif chemokine 8 (CCL8) | P80075 | + | + | − | Y | N |
| C-X-C motif chemokine 5 (CXCL5) | P42830 | + | + | − | Y | N |
| C-X-C motif chemokine 6 (CXCL6) | P80162 | + | + | − | Y | N |
| Interleukin-6 | P05231 | + | + | − | Y | N |
| Interleukin-8 | P10145 | + | + | − | Y | Y |
| Interferon-responsive proteinsa | ||||||
| Caspase-1 | P29466 | − | + | + | N | N |
| Interferon-induced GTP-binding protein Mx1 (MxA) | P20591 | − | + | − | N | Y |
| Interferon-induced GTP-binding protein Mx2 (MxB) | P20592 | − | + | − | N | N |
| Interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) | P09914 | − | + | − | N | N |
| Interferon-induced protein with tetratricopeptide repeats 2 (IFIT2) | P09913 | − | + | − | N | N |
| Interferon-induced protein with tetratricopeptide repeats 3 (IFIT3) | O14879 | − | + | − | N | N |
| Interferon-induced guanylate-binding protein 1 (GBP1) | P32455 | − | + | − | N | N |
| Interferon-induced guanylate-binding protein 2 (GBP2) | P32456 | − | + | − | N | N |
| Interferon-induced 17 kDa protein (ISG15) | P05161 | − | + | − | N | N |
| Interferon-induced 35 kDa protein (IFI35) | P80217 | − | + | − | N | N |
| Signal transducer and activator of transcription 1-alpha/beta (STAT1) | P42224 | − | + | − | N | Y |
| C-X-C motif chemokine 10 (CXCL10) | P02778 | − | + | − | N | N |
| Ubiquitin/ISG15-conjugating enzyme E2 L6 | O14933 | − | + | − | N | N |
| Indoleamine 2,3-dioxygenase 1 | P14902 | − | + | − | N | N |
| Galectin-9 | O00182 | − | + | − | N | N |
| Tryptophanyl-tRNA synthetase, cytoplasmic | P23381 | + | + | + | N | Y |
| C-C motif chemokine 8 (CCL8) | P80075 | + | + | − | N | N |
| Plasma protease C1 inhibitor | P05155 | + | + | − | Y | Y |
| Galectin-3-binding protein | Q08380 | + | + | − | N | Y |
| Interleukin-6 | P05231 | + | + | − | N | N |
| Proteasome subunit beta type-8 | P28062 | + | + | − | N | Y |
a Based on Interferome database, minimum of 5 IFN type 1-related occurrences in database was required.
Fig. 3.
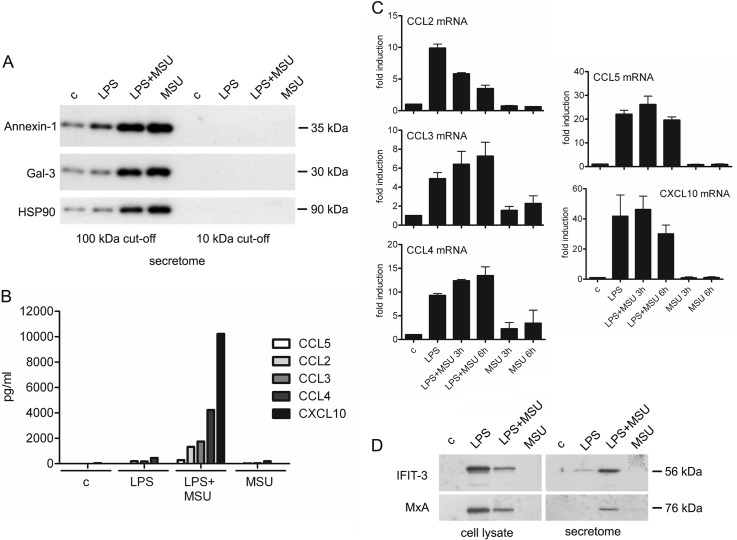
The secretion of danger signal proteins, chemokines and interferon-induced proteins is induced on MSU stimulation from LPS-primed macrophages. A, Human macrophages were left unstimulated or primed with LPS for 21 h. After this macrophages were left untreated or activated with MSU for 3 h and the cell culture supernatants were collected. Exosomal fractions were enriched as depicted in Materials and Methods, and Western blotting analysis of danger signal proteins Annexin-1, Galectin-3 (Gal-3), and HSP90 was performed. B, Human macrophages were stimulated as in (A), and the secretion of chemokines CCL2, CCL3, CCL4, CCL5, and CXCL10 was quantified by Luminex assay. Representative data from one experiment is shown; n = 2 independent experiments. C, Macrophages were left untreated or primed with LPS after which they were stimulated with MSU for 3 and 6 h. Total cellular RNA was isolated, and expression of chemokine genes was analyzed with quantitative RT-PCR as depicted in Materials and Methods. The data are reported as fold induction compared with controls. Mean with S.D. of two independent experiments is shown. D, Western blot analysis of IFIT-3 and MxA expression from total cellular lysates and cell culture supernatants of untreated and LPS-primed macrophages with and without MSU stimulation.
LPS Priming Induces the Gene Expression of Cytokines, Chemokines, and Interferon-induced Proteins, but MSU Stimulation is Needed for Their Robust Secretion
Cytokines and chemokines are small cell signaling proteins that are involved in the activation of inflammatory response. We identified several cytokines and chemokines from the secretomes of LPS-primed and/or from LPS-primed and MSU-stimulated macrophages (Table I). For example IL-6 and IL-8 were identified from both secretomes, and several chemokines like CCL5, CCL24, CXCL1, and CXCL10 were identified only from the secretome of MSU-stimulated cells. To quantify the secreted chemokines we used fluorescent bead-based immunoassay Luminex (Fig. 3B): all the quantified chemokines (CCL2–5 and CXCL10) were clearly more efficiently secreted after LPS priming and MSU stimulation compared with control cells, whereas MSU or LPS stimulation alone had only a modest effect on chemokine secretion. Next, we studied whether stimulation with MSU and/or LPS alone induces also the transcription of CCL2–5 and CXCL10 genes in human macrophages. Stimulation with MSU alone for 3 or 6 h had little effect on the transcription of chemokines in macrophages, whereas LPS priming alone and in combination with MSU stimulation clearly induced the transcription of these genes (Fig. 3C).
Interferon (IFN) signaling is an important defense response in microbial infections. We next compared our identification results with Interferome database, which consists of IFN-regulated gene and protein data collected from public databases (37). We identified more than 200 proteins reported to be IFN-regulated from the LPS-primed and MSU-stimulated macrophage secretomes, and from these 21 proteins have been reported to be regulated by type I IFNs in five or more different studies (Table I). The secretome data was verified using Western blot analysis of two of the proteins, Interferon-induced protein with tetratricopeptide repeats 3 (IFIT-3) and Interferon-induced GTP-binding protein Mx1 (MxA). IFIT-3 and MxA were clearly more abundant in the supernatants of LPS-primed macrophages stimulated with MSU than in the supernatants of LPS-primed cells (Fig. 3D). In contrast, Western blot analysis of total cellular lysates showed that the intracellular amounts of IFIT-3 and MxA were higher in LPS-primed macrophages compared with LPS-primed and MSU-stimulated cells (Fig. 3D). Taken together our results show that LPS priming is able to induce the gene expression, but MSU stimulation is needed to induce robust secretion of cytokines, chemokines, and IFN-induced proteins from human macrophages.
MSU-induced Unconventional Protein Secretion is Dependent on Cathepsin Activity
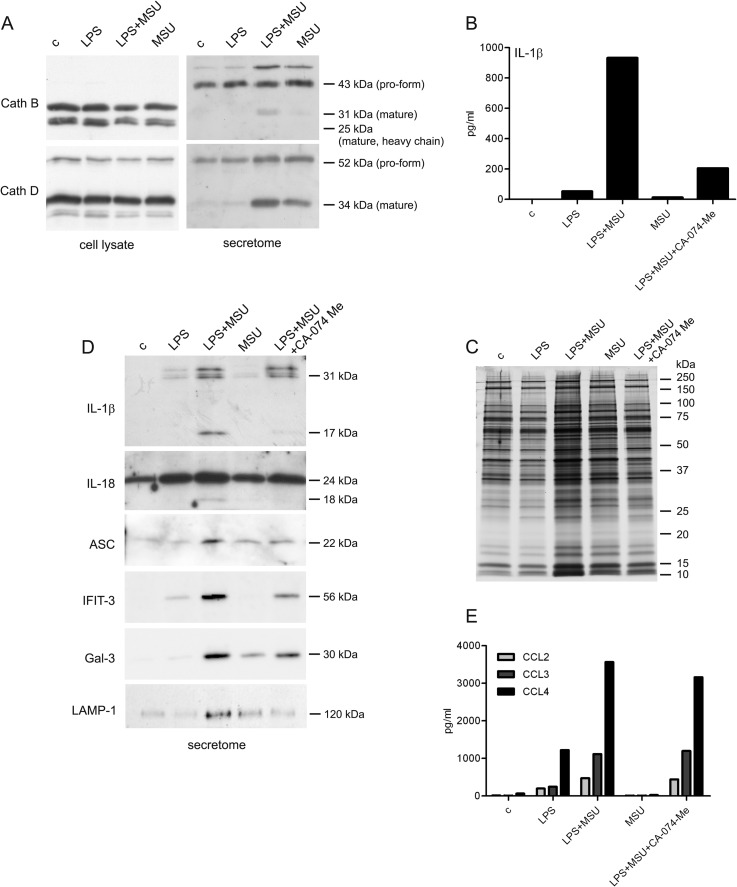
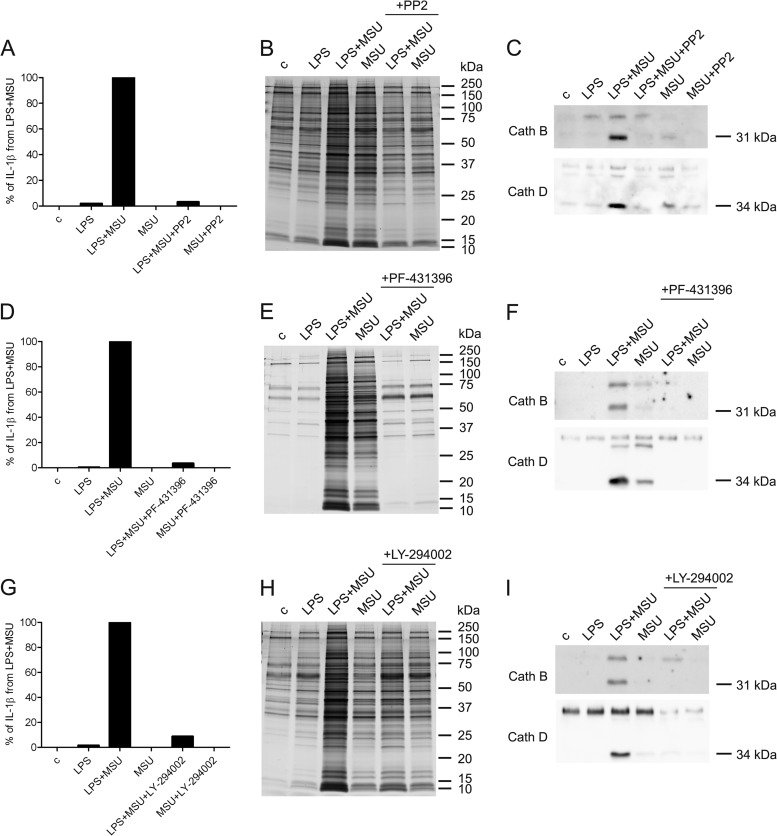
Secretome analysis revealed the release of ASC, Caspase-1, and IL-18 in LPS-primed and MSU-stimulated macrophages demonstrating the activation of NLRP3 inflammasome. Inflammasome activation was verified by measuring enhanced secretion of IL-1β and IL-18 in response to LPS priming and MSU stimulation (supplemental Fig. S3). We also identified lysosomal proteases cathepsins B, D, S, and Z from both LPS-primed and LPS-primed MSU-stimulated cell secretomes, and pro-cathepsin H from the secretome of MSU-stimulated cells using GeLC-MS/MS. Quantitative 2-DE analysis showed that cathepsins D, S, and Z were secreted more efficiently after MSU stimulation compared with LPS priming alone (supplemental Table S1). To study the intracellular levels and secretion of cathepsin B and D in more detail, we performed Western blot analysis. Maturation of cathepsins proceeds through proteolytic processing of pro-enzyme to mature forms (45). Mature forms of cathepsins B (single-chain form of 31 kDa, and heavy-chain form of 25 kDa) and D (34 kDa) were detected in total cellular lysates from control macrophages as well as from LPS-primed and/or MSU-stimulated cells. However, the secretion of mature forms of cathepsins was detected only after LPS priming and MSU stimulation of the cells. In addition, the mature form of Cathepsin D was also detectable after MSU stimulation alone (Fig. 4A).
Fig. 4.
Cathepsin activity is required for unconventional protein secretion on MSU stimulation. A, Macrophages were left untreated or LPS-primed after which they were activated with MSU for 3 h. Cathepsin B and D expression in total cellular lysates and secretion to supernatants was analyzed with Western blotting. B, Untreated and LPS-primed macrophages were activated with MSU for 3 h in the presence and absence of CA-074 Me. After this IL-1β secretion was measured from supernatants with ELISA. Representative data from one experiment is shown; n = 4 independent experiments. C, Untreated and LPS-primed macrophages were activated with MSU for 3 h in the presence and absence of CA-074 Me. Secreted proteins were visualized by silver staining. D, Secretion of IL-1β, IL-18, ASC, IFIT-3, Galectin-3 (Gal-3), and LAMP-1 was analyzed by Western blotting. E, The secretion of CCL2, CCL3 and CCL4 was quantified with Luminex assay. Representative data from one experiment is shown; n = 4 independent experiments.
Cathepsin B-specific inhibitor CA-074 Me has been reported to inhibit the activation of NLRP3 inflammasome (46–49). In accordance with this, CA-074 Me inhibited the inflammasome activation after MSU stimulation as measured by reduced secretion of IL-1β (Fig. 4B). To characterize the effect of cathepsin inhibition on overall protein secretion in more detail, we used silver staining to visualize the secreted proteins from macrophages that had been activated with LPS and MSU in the presence and absence of CA-074 Me (Fig. 4C). Inhibition of cathepsin activity clearly reduced protein secretion in LPS-primed and MSU-stimulated macrophages.
To characterize the effect of cathepsin inhibition on unconventional protein secretion, we performed Western blot analysis with proteins that do not have a signal peptide including inflammasome-related IL-1β, IL-18 and ASC, IFN-induced IFIT-3, danger signal protein Galectin-3, and phagolysosomal lysosome-associated membrane glycoprotein 1 (LAMP-1) (Fig. 4D). The secretion of mature forms of IL-1β and IL-18 was completely inhibited, and the secretion of ASC was markedly reduced in LPS-primed and MSU-stimulated macrophages in response to CA-074 Me treatment. Similarly, CA-074 Me treatment reduced the secretion of IFIT-3, Galectin-3, and LAMP-1 in LPS-primed and MSU-stimulated macrophages.
Next, we studied whether cathepsin activity is needed for MSU-induced conventional protein secretion: we quantified CCL2, CCL3, and CCL4 from the secretome of LPS-primed and MSU-stimulated cells treated with CA-074 Me using Luminex assay. The secretion of these chemokines was unaffected by CA-074 Me treatment (Fig. 4E) demonstrating that cathepsin activity is not essential for MSU-induced conventional protein secretion.
The Activity of Src, Pyk2, and PI3 Kinases is Essential for MSU-induced Protein Secretion
Protein phosphorylation has a major role in signal transduction at various cellular events including innate immune responses. In our experiments, proteins associated with phosphorylation were enriched in the secretome of MSU-stimulated cells (Fig. 2E). The identified kinases include Src family tyrosine kinases Src and Lyn (supplemental Table S3). We have previously shown that the activity of Src kinases is essential for inflammasome activation during influenza A virus infection of human primary macrophages (24). Therefore, we wanted to study whether Src kinase activity is needed for protein secretion and inflammasome activation in response to MSU stimulation. We measured the amount of secreted IL-1β from the cells that were LPS-primed and MSU-stimulated in the presence and absence of PP2. Interestingly, PP2 treatment totally abolished IL-1β secretion from MSU-stimulated cells (Fig. 5A). In addition to this, PP2 totally abolished MSU-induced overall protein secretion as assessed by silver staining of secretomes (Fig. 5B). To characterize whether Src kinases function upstream of cathepsin activity we analyzed secretion of cathepsins B and D in LPS-primed and MSU-stimulated macrophages in the presence and absence of PP2. Strikingly, mature forms of cathepsins B and D were not detectable in cell culture supernatants that were collected from PP2-treated macrophages (Fig. 5C).
Fig. 5.
MSU-induced protein secretion is dependent on the activity of Src, Pyk2, and PI3 kinases. Macrophages were left untreated or primed with LPS after which they were activated with MSU for 3 h in the absence and presence of Src kinase inhibitor PP2 (10 μm). After this the cell culture supernatants were collected. A, The secretion of IL-1β was measured by ELISA. The values obtained in LPS and MSU co-stimulated cells were set at 100%. Representative data from one experiment is shown; n = 3 independent experiments. B, Secreted proteins were visualized with silver staining and (C) the secretion of Cathepsins B and D was analyzed by Western blotting. Macrophages were left untreated or primed with LPS after which they were activated with MSU for 3 h in the absence and presence of Pyk2 kinase inhibitor PF-431396 (10 μm). D, The secretion of IL-1β was measured by ELISA. Representative data from one experiment is shown; n = 2 independent experiments. E, Secreted proteins were visualized with silver staining and (F) the secretion of Cathepsins B and D was analyzed by Western blotting. Macrophages were activated with LPS and MSU as described with or without PI3K inhibitor LY-294002 (100 μm). G, The secretion of IL-1β was measured by ELISA. Representative data from one experiment is shown; n = 2 independent experiments. H, Secreted proteins were visualized with silver staining, and (I) the secretion of cathepsins was analyzed by Western blotting.
Src, Pyk2, and PI3 kinases are part of the integrin signaling cascade both in macrophages and neutrophils (50, 51). Our current data shows that Src kinases are essential for MSU-induced protein secretion in human macrophages and therefore we were interested whether Pyk2 and PI3 kinases function also in MSU signaling. We measured the amount of secreted IL-1β from macrophages that were LPS-primed and MSU-stimulated in the presence and absence of Pyk2 kinase inhibitor PF-431396 and PI3 kinase inhibitor LY-294002. Both inhibitors completely blocked IL-1β secretion in response to MSU stimulation in LPS-primed macrophages (Figs. 5D and 5G). Similarly, LY-294002 and PF-431396 treatment of macrophages completely blocked overall protein secretion in LPS-primed and MSU-activated macrophages (Figs. 5E and 5H). Furthermore, the secretion of mature forms of cathepsins B and D was also totally abolished on MSU stimulation in LPS-primed macrophages. (Figs. 5F and 5I). In conclusion, our data demonstrates an essential role for Src, Pyk2, and PI3 kinases in MSU-induced protein secretion.
DISCUSSION
Inflammatory response is activated on concerted detection of DAMPs and PAMPs through pattern recognition receptors of the innate immune system (3). Activated innate immune cells, including macrophages, start to secrete proteins to activate the immune response and recruit other immune cells to the site of infection and/or tissue damage. Secretome characterization after activation of innate immune system is essential to unravel the details of early phases of the defense responses. Monosodium urate is an endogenous DAMP molecule that activates the innate immune system (4). In this work, we have characterized the secretome of human macrophages stimulated with MSU, when macrophages were already primed with Gram-negative bacterial cell wall component LPS, a well-known PAMP extensively altering the transcription profile of genes in human macrophages (52). Importantly, we used primary human macrophages differentiated from PBMCs rather than commonly used, but much less physiologically relevant, macrophage cell lines in our studies.
Secretome analysis showed robust protein secretion from LPS-primed and MSU-stimulated human macrophages, whereas LPS alone had only a modest effect on protein secretion in our experimental settings. The majority of the proteins secreted by MSU-stimulated cells did not have a signal peptide sequence. We did not detect any sign of cell death, including apoptosis or necrosis, in MSU-stimulated cells demonstrating that the observed protein secretion is an active process involving unconventional protein secretion pathways. Unconventional protein secretion is mainly vesicle-mediated, but can also be mediated through specific channels in plasma membrane (40, 41). Budding of plasma membrane microvesicles, release of exosomes from multivesicular bodies, or exocytosis of secretory lysosomes are different types of vesicle-mediated unconventional protein secretion. Exosomes are 40–100 nm diameter vesicles that are secreted to extracellular space when multivesicular bodies are fused with plasma membrane and release their cargo (53). Secretion of exosomes has been associated to situations requiring communication between cells like antigen presentation, or to pathological conditions like cancer cell signaling (41, 53). According to ExoCarta analysis of our data, exosomal protein secretion was activated in MSU-stimulated human macrophages and is the major secretory pathway activated by MSU in human macrophages. Macrophages are one of the few cell types that have secretory lysosomes (54). In our analysis, KEGG pathway lysosome was enriched in the secretomes of MSU-stimulated macrophages, and also lysosomal proteins like cathepsins, v-type ATPases, and LAMP-1 were shown to be secreted from MSU-stimulated cells (supplemental Table S3). Our data suggests that MSU stimulation also triggers exocytosis of secretory lysosomes resulting in the secretion of lysosomal contents into extracellular space.
Conventional protein secretion machinery mediates trafficking of proteins with N-terminal signal peptide through ER and Golgi apparatus to the cell surface. Chemokines are proteins that are typically released from the cells through the conventional protein secretion pathway. They recruit and attract other immune cells to the site of infection and/or tissue damage. Here, we show that MSU stimulation activated robust secretion of chemokines, including CCL2, CCL3, CCL4, CCL5, and CXCL10, from LPS-primed macrophages. RT-PCR analysis showed that MSU stimulation of macrophages did not induce the transcription of chemokine encoding genes. In contrast, LPS strongly activated the transcription of chemokine genes. Our data demonstrates that MSU stimulation of macrophages enhances protein secretion also through conventional ER-Golgi pathway. In addition, the proteins identified only from the secretomes of LPS-stimulated macrophages included many classically secreted proteins like Calreticulin, Lipocalin-1, and Leukosialin demonstrating that LPS activates classical protein secretion.
IFN-induced proteins are essential for antiviral defense. They are typically induced on virus infection via production of IFNs, but they are also induced in bacterial infections (55). Our results show a robust secretion of IFN-responsive proteins from LPS-primed macrophages on MSU stimulation. We identified the secretion of MxA, MxB, IFIT-(1–3), GBP-1, GBP-2, ISG15, and IFI-35 from MSU-stimulated macrophages. We identified a range of IFN-induced proteins only from one biological replicate, which can likely be explained by higher protein amounts in that set, enabling MS/MS identification of these proteins. The secretion of IFN-induced proteins on MSU-stimulation was further verified by Western blotting of independent secretome samples. LPS binding to TLR4 leads to signaling cascade which activates transcription factor Interferon regulatory factor 3 (IRF-3) (56). IRF-3 in turn activates transcription of IFN-induced genes (57). In accordance with this, LPS induced the expression of IFIT-3 and MxA proteins in human macrophages. However, LPS was not able to activate secretion of these proteins. In contrast, MSU stimulation of macrophages triggered secretion of IFIT-3 and MxA. We have previously shown that IFN-induced proteins are also secreted during virus infection (24). In a recent report, it was shown that IFN-inducible proteins IFIT-1 and IFIT-5 can bind viral double-stranded RNA extracellularly (58) further demonstrating that IFN-inducible proteins may have important extracellular functions. Interestingly, IFN-induced proteins were not found in ExoCarta suggesting nonexosomal release for these proteins. Future studies are needed to identify molecular machinery that mediates secretion of IFN-inducible proteins.
Our secretome data shows that MSU stimulation activates robust secretion of danger signal proteins from LPS-primed macrophages. Danger signal proteins are usually constitutively produced and stored in the cytoplasm and/or nucleus, but on infection-initiated tissue damage, they are actively secreted by innate immune cells through a nonclassical secretory pathway (2). In accordance with this, our data suggest that MSU-stimulated macrophages release danger signal proteins through an exosome-mediated protein secretion mechanism. Once secreted, danger signal proteins act as modulators of the innate immune system by enhancing inflammatory response. In addition to this cytokine-like function, several danger signal proteins have a pattern recognition receptor-like function. For example, galectins bind carbohydrate structures found in fungi and bacteria and facilitate their recognition by membrane-bound pattern recognition receptors (59, 60).
NLRP3 inflammasome assembly activates Caspase-1, and Caspase-1 has been shown to have a regulatory role in unconventional protein secretion of IL-1α and IL-1β (61, 62). Previous studies have shown that NLRP3 inflammasome activation in response to silica (47), cholesterol crystals (49, 63), β-glucans (48), and influenza A virus (24, 64) infection is dependent on cathepsin activity. Similarly, it was recently shown that Cathepsin B is a positive factor in autophagy-driven IL-1β secretion (65). In our experiments, MSU-induced secretion of mature IL-1β and IL-18 was completely blocked by cathepsin specific inhibitor CA-074 Me. Interestingly, inhibition of cathepsin activity clearly decreased total protein secretion in MSU-stimulated macrophages. However, CA-074 Me treatment had no effect on chemokine release, which is mediated by conventional ER-Golgi-mediated pathway, demonstrating that MSU enhances conventional protein secretion through a cathepsin-independent pathway. In contrast, CA-074 Me treatment markedly reduced the secretion of ASC, IFIT-3, Galectin-3, and LAMP-1 in response to MSU stimulation. These proteins do not contain a signal peptide sequence and they are supposed to be released by macrophages through an unconventional protein secretion pathway. Collectively, our data shows an essential role for cathepsins in the activation of unconventional protein secretion.
MSU-induced protein secretion was completely dependent on the activity of Src, Pyk2, and PI3 kinases in human macrophages. The activity of these kinases was essential for the secretion of mature forms of cathepsins in response to MSU stimulation demonstrating that Src, Pyk2, and PI3 kinases act upstream of cathepsins and before the involvement of lysosomes. The function of Src, Pyk2, and PI3 kinases has been linked to phagocytosis (66–68) and it may be that these kinases are activated during the phagocytosis of MSU. Interestingly, Src and PI3K signaling is required for the activation of MSU-induced secretory pathway called degranulation in neutrophils (69). This suggests that macrophages and neutrophils use similar signaling molecules in the activation of protein secretion.
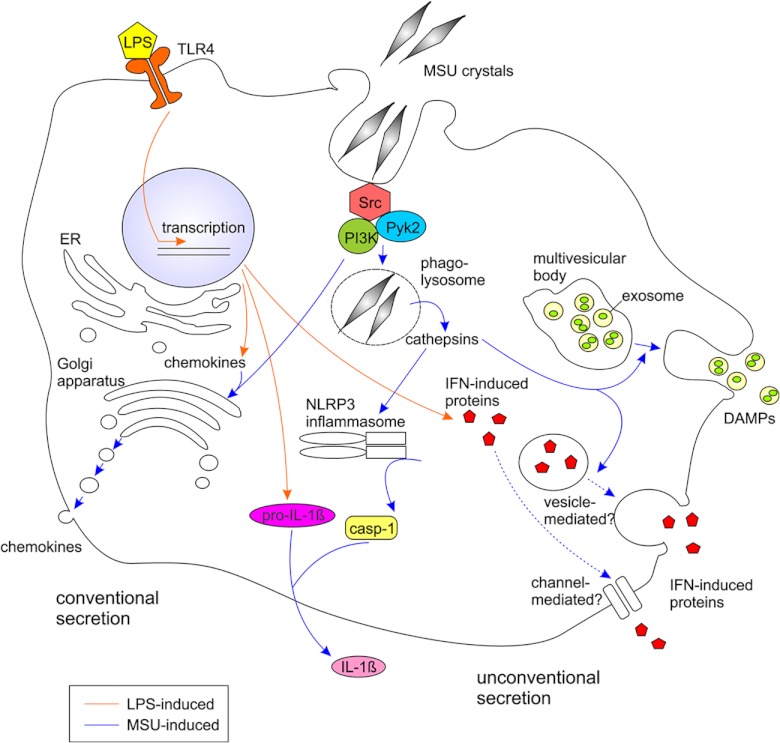
In conclusion, we show, first, that MSU stimulation induced robust protein secretion from LPS-primed human primary macrophages. Both conventional and unconventional protein secretion pathways were activated on MSU stimulation (Fig. 6). Unconventional protein secretion pathways included exosomes and possibly exocytosis of secretory lysosomes. Second, we show that pro-inflammatory proteins including multiple chemokines, IFN-induced proteins, and danger signal proteins were efficiently secreted from LPS-primed macrophages on MSU stimulation. However, MSU did not induce the transcription of pro-inflammatory proteins, and therefore synergistic action of PAMP (LPS) and DAMP (MSU) was required for the activation of a strong inflammatory burst from human macrophages. Third, cathepsin activity was necessary for the MSU-induced unconventional protein secretion, but not for the conventional protein secretion. Finally, we demonstrate that the activity of Src, Pyk2, and PI3 kinases is required for the MSU-activated protein secretion. Our data highlights the importance of secretome analysis to gain in-depth information of global intercellular signaling mechanisms.
Fig. 6.
Schematic model of MSU-induced signaling and protein secretion in human macrophages.
Supplementary Material
Acknowledgments
We thank Niina Ahonen for the assistance with 2-DE analysis.
Footnotes
* This work was supported by grants from the Academy of Finland (grants 135628, 140950, 255842), The Finnish Work Environment Fund, the Sigrid Jusélius Foundation, the Waldemar von Frenckell Foundation, National Doctoral Programme in Informational and Structural Biology (EV), Helsinki Biomedical Graduate School (JJM) and Helsinki Graduate Program in Biotechnology and Molecular Biology (NL).
 This article contains supplemental Figs. S1 to S3 and Tables S1 to S3.
This article contains supplemental Figs. S1 to S3 and Tables S1 to S3.
1 The abbreviations used are:
- PAMP
- pathogen-associated molecular pattern
- 2-DE
- two-dimensional gel electrophoresis
- ASC
- Apoptosis-associated speck-like protein containing a CARD
- DAMP
- damage-associated molecular pattern
- ER
- endoplasmic reticulum
- GO
- Gene Ontology
- HSP90
- Heat shock protein HSP 90
- IFIT-3
- Interferon-induced protein with tetratricopeptide repeats 3
- IFN
- interferon
- IRF-3
- Interferon regulatory factor 3
- KEGG
- Kyoto encyclopedia of genes and genomes
- LAMP-1
- Lysosome-associated membrane glycoprotein 1
- LPS
- lipopolysaccharide
- MSU
- monosodium urate
- MxA
- Interferon-induced GTP-binding protein Mx1
- NLRP3
- NACHT, LRR and PYD domains-containing protein 3
- NMWL
- Nominal molecular weight limit
- PI3K
- Phosphatidyl-inositol 3-kinase
- Pyk2
- Protein-tyrosine kinase 2-beta
- Src
- Proto-oncogene tyrosine-protein kinase Src.
REFERENCES
- 1. Iwasaki A., Medzhitov R. (2010) Regulation of Adaptive Immunity by the Innate Immune System. Science 327, 291–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bianchi M. E. (2007) DAMPs, PAMPs and alarmins: all we need to know about danger. J. Leukocyte Biol. 81, 1–5 [DOI] [PubMed] [Google Scholar]
- 3. Kono H., Rock K. L. (2008) How dying cells alert the immune system to danger. Nat. Rev. Immunol. 8, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shi Y., Evans J. E., Rock K. L. (2003) Molecular identification of a danger signal that alerts the immune system to dying cells. Nature 425, 516–521 [DOI] [PubMed] [Google Scholar]
- 5. Martinon F. (2010) Mechanisms of uric acid crystal-mediated autoinflammation. Immunol. Rev. 233, 218–232 [DOI] [PubMed] [Google Scholar]
- 6. Choi H. K., Ford E. S. (2007) Prevalence of the metabolic syndrome in individuals with hyperuricemia. Am. J. Med. 120, 442–447 [DOI] [PubMed] [Google Scholar]
- 7. See L. C., Kuo C. F., Chuang F. H., Li H. Y., Chen Y. M., Chen H. W., Yu K. H. (2009) Serum uric acid is independently associated with metabolic syndrome in subjects with and without a low estimated glomerular filtration rate. J. Rheumatol. 36, 1691–1698 [DOI] [PubMed] [Google Scholar]
- 8. Dehghan A., van Hoek M., Sijbrands E. J., Hofman A., Witteman J. C. (2008) High serum uric acid as a novel risk factor for type 2 diabetes. Diabetes Care 31, 361–362 [DOI] [PubMed] [Google Scholar]
- 9. Baker J. F., Krishnan E., Chen L., Schumacher H. R. (2005) Serum uric acid and cardiovascular disease: Recent developments, and where do they leave us? Am. J. Med. 118, 816–826 [DOI] [PubMed] [Google Scholar]
- 10. Kool M., Soullié T., van Nimwegen M., Willart M. A., Muskens F., Jung S., Hoogsteden H. C., Hammad H., Lambrecht B. N. (2008) Alum adjuvant boosts adaptive immunity by inducing uric acid and activating inflammatory dendritic cells. J. Exp. Med. 205, 869–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ng G., Sharma K., Ward S. M., Desrosiers M. D., Stephens L. A., Schoel W. M., Li T., Lowell C. A., Ling C. C., Amrein M. W., Shi Y. (2008) Receptor-independent, direct membrane binding leads to cell-surface lipid sorting and Syk kinase activation in dendritic cells. Immunity 29, 807–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Uratsuji H., Tada Y., Kawashima T., Kamata M., Hau C. S., Asano Y., Sugaya M., Kadono T., Asahina A., Sato S., Tamaki K. (2012) P2Y6 receptor signaling pathway mediates inflammatory responses induced by monosodium urate crystals. J. Immunol. 188, 436–444 [DOI] [PubMed] [Google Scholar]
- 13. Martinon F., Pétrilli V., Mayor A., Tardivel A., Tschopp J. (2006) Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440, 237–241 [DOI] [PubMed] [Google Scholar]
- 14. Agostini L., Martinon F., Burns K., McDermott M. F., Hawkins P. N., Tschopp J. (2004) NALP3 Forms an IL-1β-processing inflammasome with increased activity in muckle-wells autoinflammatory disorder. Immunity 20, 319–325 [DOI] [PubMed] [Google Scholar]
- 15. Martinon F., Burns K., Tschopp J. (2002) The Inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 10, 417–426 [DOI] [PubMed] [Google Scholar]
- 16. Martinon F., Mayor A., Tschopp J. r. (2009) The inflammasomes: guardians of the body. Annu. Rev. Immunol. 27, 229–265 [DOI] [PubMed] [Google Scholar]
- 17. Franchi L., Eigenbrod T., Muñoz-Planillo R., Nuñez G. (2009) The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat. Immunol 10, 241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stow J. L., Low P. C., Offenhäuser C., Sangermani D. (2009) Cytokine secretion in macrophages and other cells: Pathways and mediators. Immunobiology 214, 601–612 [DOI] [PubMed] [Google Scholar]
- 19. Sintiprungrat K., Singhto N., Sinchaikul S., Chen S. T., Thongboonkerd V. (2010) Alterations in cellular proteome and secretome upon differentiation from monocyte to macrophage by treatment with phorbol myristate acetate: Insights into biological processes. J Preoteomics 73, 602–618 [DOI] [PubMed] [Google Scholar]
- 20. Chan C. Y. X. A., Masui O., Krakovska O., Belozerov V. E., Voisin S., Ghanny S., Chen J., Moyez D., Zhu P., Evans K. R., McDermott J. C., Siu K. W. M. (2011) Identification of Differentially Regulated Secretome Components During Skeletal Myogenesis. Moll Cell Proteomics 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schiarea S., Solinas G., Allavena P., Scigliuolo G. M., Bagnati R., Fanelli R., Chiabrando C. (2010) Secretome analysis of multiple pancreatic cancer cell lines reveals perturbations of key functional networks. J. Proteome Res. 9, 4376–4392 [DOI] [PubMed] [Google Scholar]
- 22. Chang Y. H., Lee S. H., Liao I. C., Huang S. H., Cheng H. C., Liao P. C. (2012) Secretomic analysis identifies A1AT as a required protein in cancer cell migration, invasion, and pericellular fibronectin assembly for facilitating lung colonization of lung adenocarcinoma cells. Moll Cell Proteomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Villiers C., Chevallet M., Diemer H. L. N., Couderc R., Freitas H., Van Dorsselaer A., Marche P. N., Rabilloud T. (2009) From secretome analysis to immunology. Moll. Cell Proteomics 8, 1252–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lietzén N., Öhman T., Rintahaka J., Julkunen I., Aittokallio T., Matikainen S., Nyman T. A. (2011) Quantitative subcellular proteome and secretome profiling of influenza A virus-infected human primary macrophages. PLoS Pathog 7, e1001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miettinen J. J., Matikainen S., Nyman T. A. (2012) Global secretome characterization of herpes simplex virus 1-infected human primary macrophages. J. Virol. 86, 12770–12778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pirhonen J., Sareneva T., Kurimoto M., Julkunen I., Matikainen S. (1999) Virus infection activates IL-1β and IL-18 production in human macrophages by a caspase-1-dependent pathway. J. Immunol. 162, 7322–7329 [PubMed] [Google Scholar]
- 27. Lehtonen A., Ahlfors H., Veckman V., Miettinen M., Lahesmaa R., Julkunen I. (2007) Gene expression profiling during differentiation of human monocytes to macrophages or dendritic cells. J. Leukocyte Biol. 82, 710–720 [DOI] [PubMed] [Google Scholar]
- 28. O'Connell K. L., Stults J. T. (1997) Identification of mouse liver proteins on two-dimensional electrophoresis gels by matrix-assisted laser desorption/ionization mass spectrometry of in situ enzymatic digests. Electrophoresis 18, 349–359 [DOI] [PubMed] [Google Scholar]
- 29. Öhman T., Lietzén N., Välimäki E., Melchjorsen J., Matikainen S., Nyman T. A. (2010) Cytosolic RNA recognition pathway activates 14–3-3 protein mediated signaling and caspase-dependent disruption of cytokeratin network in human keratinocytes. J Proteome Res. 9, 1549–1564 [DOI] [PubMed] [Google Scholar]
- 30. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Meth 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 31. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zeeberg B. R., Feng W., Wang G., Wang M. D., Fojo A. T., Sunshine M., Narasimhan S., Kane D. W., Reinhold W. C., Lababidi S., Bussey K. J., Riss J., Barrett J. C., Weinstein J. N. (2003) GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biology 4, R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Backes C., Keller A., Kuentzer J., Kneissl B., Comtesse N., Elnakady Y. A., Müller R., Meese E., Lenhof H. P. (2007) GeneTrail-advanced gene set enrichment analysis. Nucleic Acids Res. 35, W186–W192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, D277–D280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Meth 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 36. Mathivanan S., Fahner C. J., Reid G. E., Simpson R. J. (2012) ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 40, D1241–D1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Samarajiwa S. A., Forster S., Auchettl K., Hertzog P. J. (2009) INTERFEROME: the database of interferon regulated genes. Nucleic Acids Res. 37, D852–D857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rintahaka J., Wiik D., Kovanen P. E., Alenius H., Matikainen S. (2008) Cytosolic antiviral RNA recognition pathway activates caspases 1 and 3. J. Immunol. 180, 1749–1757 [DOI] [PubMed] [Google Scholar]
- 39. Ronni T., Melén K., Malygin A., Julkunen I. (1993) Control of IFN-inducible MxA gene expression in human cells. J. Immunol. 150, 1715–1726 [PubMed] [Google Scholar]
- 40. Nickel W. (2010) Pathways of unconventional protein secretion. Curr. Opin. Biotechnol. 21, 621–626 [DOI] [PubMed] [Google Scholar]
- 41. Théry C., Ostrowski M., Segura E. (2009) Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol 9, 581–593 [DOI] [PubMed] [Google Scholar]
- 42. Qu Y., Franchi L., Nunez G., Dubyak G. R. (2007) Nonclassical IL-1β secretion stimulated by P2X7 receptors is dependent on inflammasome activation and correlated with exosome release in murine macrophages. J. Immunol. 179, 1913–1925 [DOI] [PubMed] [Google Scholar]
- 43. Rock K. L., Latz E., Ontiveros F., Kono H. (2010) The sterile inflammatory response. Annu. Rev. Immunol. 28, 321–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen G. Y., Nuñez G. (2010) Sterile inflammation: sensing and reacting to damage. Nat. Rev. Immunol 10, 826–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Turk V., Turk B., Turk D. (2001) Lysosomal cysteine proteases: facts and opportunities. EMBO J. 20, 4629–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Halle A., Hornung V., Petzold G. C., Stewart C. R., Monks B. G., Reinheckel T., Fitzgerald K. A., Latz E., Moore K. J., Golenbock D. T. (2008) The NALP3 inflammasome is involved in the innate immune response to amyloid-β. Nat. Immunol. 9, 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hornung V., Bauernfeind F., Halle A., Samstad E. O., Kono H., Rock K. L., Fitzgerald K. A., Latz E. (2008) Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 9, 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kankkunen P., Teirilä L., Rintahaka J., Alenius H., Wolff H., Matikainen S. (2010) (1,3)-β-Glucans Activate Both Dectin-1 and NLRP3 Inflammasome in human macrophages. J. Immunol. 184, 6335–6342 [DOI] [PubMed] [Google Scholar]
- 49. Rajamäki K., Lappalainen J., Öörni K., Välimäki E., Matikainen S., Kovanen P. T., Eklund K. K. (2010) Cholesterol crystals activate the NLRP3 inflammasome in human macrophages: a novel link between cholesterol metabolism and inflammation. PLoS One 5, e11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fuortes M., Melchior M., Han H., Lyon G. J., Nathan C. (1999) Role of the tyrosine kinase pyk2 in the integrin-dependent activation of human neutrophils by TNF. J. Clin. Investigation 104, 327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Okigaki M., Davis C., Falasca M., Harroch S., Felsenfeld D. P., Sheetz M. P., Schlessinger J. (2003) Pyk2 regulates multiple signaling events crucial for macrophage morphology and migration. Proc. Natl. Acad Sci. 100, 10740–10745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nau G. J., Richmond J. F., Schlesinger A., Jennings E. G., Lander E. S., Young R. A. (2002) Human macrophage activation programs induced by bacterial pathogens. Proc. Natl. Acad. Sci. 99, 1503–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Simpson R. J., Jensen S. S., Lim J. W. E. (2008) Proteomic profiling of exosomes: Current perspectives. Proteomics 8, 4083–4099 [DOI] [PubMed] [Google Scholar]
- 54. Blott E. J., Griffiths G. M. (2002) Secretory lysosomes. Nat. Rev. Mol. Cell Biol. 3, 122–131 [DOI] [PubMed] [Google Scholar]
- 55. Monroe K. M., McWhirter S. M., Vance R. E. (2010) Induction of type I interferons by bacteria. Cell. Microbiol. 12, 881–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Navarro L., David M. (1999) p38-Dependent activation of interferon regulatory factor 3 by lipopolysaccharide. J. Biol. Chem. 274, 35535–35538 [DOI] [PubMed] [Google Scholar]
- 57. Doyle S., Vaidya S., O'Connell R., Dadgostar H., Dempsey P., Wu T. T., Rao G., Sun R., Haberland M., Modlin R., Cheng G. (2002) IRF3 Mediates a TLR3/TLR4-specific antiviral gene program. Immunity 17, 251–263 [DOI] [PubMed] [Google Scholar]
- 58. Pichlmair A., Lassnig C., Eberle C. A., Górna M. W., Baumann C. L., Burkard T. R., Bürckstummer T., Stefanovic A., Krieger S., Bennett K. L., Rulicke T., Weber F., Colinge J., Müller M., Superti-Furga G. (2011) IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol. 12, 624–630 [DOI] [PubMed] [Google Scholar]
- 59. Esteban A., Popp M. W., Vyas V. K., Strijbis K., Ploegh H. L., Fink G. R. (2011) Fungal recognition is mediated by the association of dectin-1 and galectin-3 in macrophages. Proc. Natl. Acad Sci. 108, 14270–14275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vasta G. R., Lambris J. D., Hajishengallis G. (2012) Galectins as pattern recognition receptors: structure, function, and evolution, current topics in innate immunity II. Adv. Exp. Med. Biol. 946, 21–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Keller M., Rüegg A., Werner S., Beer H. D. (2008) Active caspase-1 is a regulator of unconventional protein secretion. Cell 132, 818–831 [DOI] [PubMed] [Google Scholar]
- 62. Gross O., Yazdi, Amir S., Thomas C. J., Masin M., Heinz L. X., Guarda G., Quadroni M., Drexler S. K., Tschopp J. (2012) Inflammasome activators induce interleukin-1α secretion via distinct pathways with differential requirement for the protease function of caspase-1. Immunity 36, 388–400 [DOI] [PubMed] [Google Scholar]
- 63. Duewell P., Kono H., Rayner K. J., Sirois C. M., Vladimer G., Bauernfeind F. G., Abela G. S., Franchi L., Nuñez G., Schnurr M., Espevik T., Lien E., Fitzgerald K. A., Rock K. L., Moore K. J., Wright S. D., Hornung V., Latz E. (2010) NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature 464, 1357–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Allen I. C., Scull M. A., Moore C. B., Holl E. K., McElvania-TeKippe E., Taxman D. J., Guthrie E. H., Pickles R. J., Ting J. P. Y. (2009) The NLRP3 Inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 30, 556–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dupont N., Jiang S., Pilli M., Ornatowski W., Bhattacharya D., Deretic V. (2011) Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J. 30, 4701–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Araki N., Johnson M. T., Swanson J. A. (1996) A role for phosphoinositide 3-kinase in the completion of macropinocytosis and phagocytosis by macrophages. J. Cell Biol. 135, 1249–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Berton G., Mócsai A., Lowell C. A. (2005) Src and Syk kinases: key regulators of phagocytic cell activation. Trends Immunol. 26, 208–214 [DOI] [PubMed] [Google Scholar]
- 68. Owen K. A., Thomas K. S., Bouton A. H. (2007) The differential expression of Yersinia pseudotuberculosis adhesins determines the requirement for FAK and/or Pyk2 during bacterial phagocytosis by macrophages. Cell. Microbiol. 9, 596–609 [DOI] [PubMed] [Google Scholar]
- 69. Popa-Nita O., Rollet-Labelle E., Thibault N., Gilbert C., Bourgoin S. G., Naccache P. H. (2007) Crystal-induced neutrophil activation. IX. Syk-dependent activation of class Ia phosphatidylinositol 3-kinase. J. Leukocyte Biol. 82, 763–773 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.