Abstract
The introduction of electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI) for the mass spectrometric analysis of peptides and proteins had a dramatic impact on biological science. We now report that a wide variety of compounds, including peptides, proteins, and protein complexes, are transported directly from a solid-state small molecule matrix to gas-phase ions when placed into the vacuum of a mass spectrometer without the use of high voltage, a laser, or added heat. This ionization process produces ions having charge states similar to ESI, making the method applicable for high performance mass spectrometers designed for atmospheric pressure ionization. We demonstrate highly sensitive ionization using intermediate pressure MALDI and modified ESI sources. This matrix and vacuum assisted soft ionization method is suitable for the direct surface analysis of biological materials, including tissue, via mass spectrometry.
The conversion of large and nonvolatile compounds such as proteins into gas-phase ions is of immense fundamental and practical importance. The 2002 Nobel Prize in Chemistry was awarded for the accomplishment of this conversion via electrospray ionization (ESI)1 (1) and matrix-assisted laser desorption/ionization (MALDI) (2) interfaced with mass spectrometry (MS) to obtain the molecular weights of proteins with high accuracy. These methods employ high voltage or a laser to form gaseous analyte ions from a wide variety of compounds in solution or a solid matrix, respectively.
MALDI interfaced with a time-of-flight (TOF) mass spectrometer produces gas-phase analyte ions in vacuum and is the method of choice for the molecular imaging of biological surfaces. Ionization in vacuum provides excellent ion transmission (3), as well as good spatial resolution achieved using a focused laser beam. However, the analysis of protein complexes is very challenging with MALDI, requiring strategies such as first-shot phenomena (4) and chemical crosslinking (5). The necessity of a laser also makes MALDI less soft than ESI and produces background ions, which can hinder the analysis of small molecules (6, 7). MALDI is also of limited utility on high performance mass-to-charge (m/z) analyzers because of mass range issues related to the formation of singly charged ions, which also produce few fragment ions for structural characterization (8).
Multiple charged ions produced directly from solution in ESI bring the m/z ratio within the range of high performance mass spectrometers, allowing the analysis of high-mass compounds. These instruments have advanced features for structural characterization, such as ion mobility spectrometry (IMS) for gas-phase separations (9–11), ultra-high mass resolution and mass accuracy (12–14), and advanced fragmentation such as electron transfer dissociation (ETD) (13, 14). However, ESI is limited for surface characterization, requiring approaches such as desorption-ESI (15) and laser ablation ESI (16), ionization methods that produce multiply charged ions but are not compatible with analyses of larger proteins or fragile complexes.
A softer ionization approach is needed in order to observe fragile molecules and molecular complexes in living organisms at low levels directly from tissue and cell cultures, without extensive sample preparation, while retaining spatial information. Ideally, this approach would be compatible with mass spectrometers having advanced capabilities to aid structural characterization directly from surfaces. The new ionization method described here, in which molecules are transferred from solid-phase to gas-phase ions through the simple exposure of a material of interest in a suitable matrix to vacuum, is an advance toward this goal and is of fundamental interest.
EXPERIMENTAL PROCEDURES
Materials
3-Nitrobenzonitrile (3-NBN), penta-N-acetylchitopentaose, ammonium acetate, ubiquitin (bovine erythrocytes), lysozyme (chicken egg white), and bovine serum albumin (BSA) were purchased from Sigma Aldrich Inc. (St. Louis, MO). Phosphorylated peptide cholecystokinin (10–20) and hirudin (55–65) peptide were obtained from Protea Biosciences (Morgantown, WV). A synthesized acetylated N-terminal fragment peptide 1–16 of myelin basic protein (MBP) was purchased from Anaspec (Fremont, CA), and leucine enkephalin was obtained from Waters Co. (Milford, MA). 2,5-Dihydroxyacetophenone, acetonitrile (ACN), ethanol, and formic acid were purchased from Fisher Scientific Inc. (Pittsburgh, PA), and ammonium hydroxide (NH4OH) from Mallinckrodt Chemicals (Phillipsburg, NJ). Methanol and HPLC-grade water were obtained from EMD Chemicals (Gibbstown, NJ). Gold Seal microslides (Portsmouth, NH) were used as glass plates.
Matrix Assisted Ionization Vacuum Sample Preparation
Stock solutions of the analyte were prepared individually in water. Analyte solutions were further diluted with water to make concentrations ranging from 1 to 5 pmol μl−1, 20 pmol μl−1 for BSA, and 50 fmol μl−1 for ubiquitin. For the noncovalent complex, stock solutions of lysozyme and penta-N-acetylchitopentaose were individually diluted in 25 mMol ammonium acetate buffer with 10% methanol according to published procedures (17). The molar ratio of the protein–ligand mixture was optimized, and a premixed noncovalent complex mixture of 1:285 protein:ligand molar ratio was used. The blood sample was diluted in water at a 1:10 volume ratio. 3-NBN was dissolved in 50 μl of 100% ACN with 0.1% formic acid and 150 μl of 50:50 ACN:water with 0.1% formic acid (positive mode measurements), and 150 μl of 50:50 ACN:water was mixed with with 0.1% NH4OH for peptides and 1% NH4OH for proteins (negative mode measurements). The matrix–analyte solution was prepared at 1:3 and 1:1 volume ratios for positive and negative mode measurements, respectively, and 1 μl was spotted on the target plate. Mouse brain tissue, obtained using a protocol described elsewhere (12, 14, 18, 19), was donated by Professor Ken Mackie (Indiana University). The delipified mouse brain tissue (14) was coated with several 0.5-μl spots of 3-NBN matrix in 50:50 ACN:water with 0.1% formic acid. The matrix deposition on brain tissue sections from mouse brain tissue was miniaturized by spotting, on the same area, a total of 1 μl in five increments of 0.2 μl, covering a ∼400 μm × 150 μm area on the tissue. Blood was extracted from a Band-Aid using water. After solvent evaporation of the analyte–matrix spot, the target plate was introduced into the vacuum of the mass spectrometer.
Matrix Assisted Ionization Vacuum on a Commercial Vacuum MALDI Source
IMS MS SYNAPT G2 (Waters Co., Milford, MA) with a commercial intermediate-pressure MALDI source conventionally operated with an Nd:YAG laser (355 nm) was employed with the laser off. Matrix–analyte sample was prepared in a manner similar to MALDI sample preparation protocols using 3-NBN as matrix. The source pressure when the sample plate was loaded was ∼0.2 mbar. Ions are observed from the matrix and the analyte once the sample plate is positioned so that the matrix spot is near the extraction lens, a process that takes ca. 2 min. Analyte ions were observed with as little as 1 V applied between the sample plate and the extraction lens. Some voltage is necessary to accelerate ions into the hexapole ion guides and to the mass analyzer. In this study, the TOF mass analyzer was operated in positive or negative ion V mode (also referred to as sensitivity mode). “MALDI” (11) or “LSI” (10) settings were used to perform IMS-MS measurements. Acquisition times in MS and MS/MS ranged from 30 s to 2 min for standard samples and up to 4 min for the mouse brain tissue section sample, but single scan acquisitions were sufficient for most analytes. MassLynx version 4.1 and DriftScope version 2.2 were used for data analysis.
Matrix Assisted Ionization Vacuum on Commercial Atmospheric Pressure ESI Sources
Matrix assisted ionization vacuum (MAIV) on the ESI source of the SYNAPT G2 mass spectrometer was performed by modifying the skimmer cone. The outer cone aperture was widened to about ∼4.5 mm to allow a metal ferrule to form a gas-tight seal with the inner skimmer cone. Non-standard inner cones having ∼3 mm inner diameter inlet apertures were employed. The ion source temperature for the experiments described here was 50 °C. The glass plate with the matrix–analyte spot was placed on the ferrule facing the source entrance, and once the vacuum isolation valve was opened, the glass plate was held to the ferrule by the pressure differential between the atmospheric pressure and the instrument vacuum. Ions were observed until the matrix was no longer visible.
The commercial atmospheric pressure source on a Thermo Scientific LTQ Velos mass spectrometer was modified by inserting a ferrule on the outer opening of the inlet tube to hold the glass plate as described above. Nonstandard 750 μm inner diameter inlet capillaries were used. MAIV-ETD fragmentation for sequencing the MBP peptide used conditions similar to those described in a previous report (14). Mass spectra were acquired, summing 2 microscans having a maximum injection time of 50 ms with an inlet capillary tube temperature of 150 °C. ETD acquisition was set at 1 m/z isolation width and 100 ms activation time. Xcalibur 2.1.0 was used for data analysis.
RESULTS AND DISCUSSION
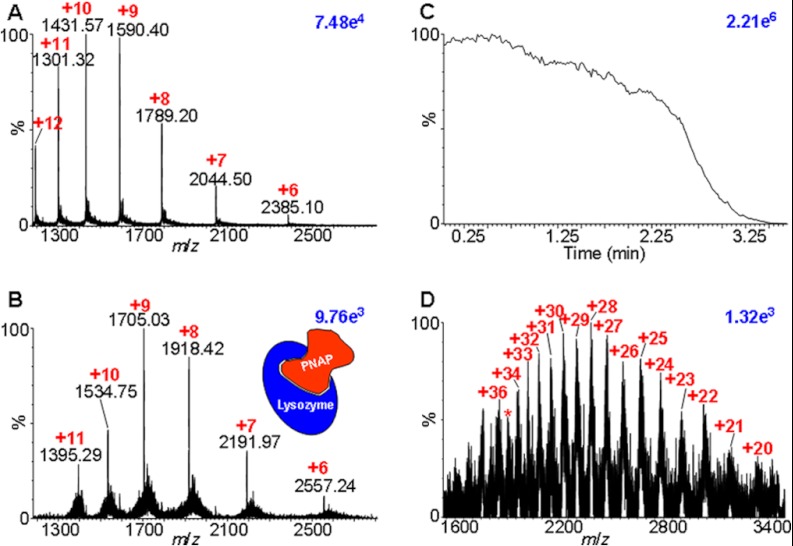
The MAIV method described here, using 3-NBN as matrix, produces ions when placed in vacuum. Similar to ESI (17), multiple charging, even of protein complexes, is observed, but it occurs directly from the solid state. For example, the MAIV mass spectrum of the 14.3 kDa lysozyme protein using the vacuum MALDI source of the SYNAPT G2 is shown in Fig. 1A, and its complex with penta-N-acetylchitopentaose, separated from both the lysozyme and the peptide ions via IMS, is shown in Fig. 1B. A challenge in analyzing molecular complexes via any MS approach is that the sample preparation must keep the complex intact under conditions suitable for ionization. These mass spectra are obtained simply by placing the matrix–analyte sample in vacuum. Ions are continuously formed until the matrix completely sublimes, which, depending on the applied conditions, can take a few seconds to tens of minutes (Fig. 1C). No laser, voltage, or heat is used to initiate the ionization process. The “hot spot phenomenon” (20–22), an intrinsic limitation for quantitation in MALDI, is not an issue because the entire matrix–analyte sample is analyzed by simply summing mass spectra acquired during complete matrix sublimation.
Fig. 1.
MAIV mass spectra. A, lysozyme, 14.3 kDa. B, noncovalent complex of lysozyme/penta-N-acetylchitopentaose (PNAP) extracted from the IMS-MS two-dimensional dataset. C, total ion current of A. D, bovine serum albumin, ∼66 kDa. Red numbers indicate the charge states, and blue numbers in the upper right corner of each spectrum provide relative ion abundances.
3-NBN has no acidic protons needed for ionization, so the protons must be supplied by the solvent, analyte, or acid during sample preparation. Typically, the matrix is dissolved in organic or water–organic solvents, with or without 0.1% formic acid, and the analyte in water–organic solvent or, for complexes, water buffered with ammonium acetate. The matrix and analyte solutions are mixed in the same way as the sample preparation used in MALDI, dried, and placed in vacuum or, for labile compounds, immediately placed in vacuum without drying. Astonishingly, MAIV produces ions from proteins at least as large as BSA (66 kDa) (Fig. 1D).
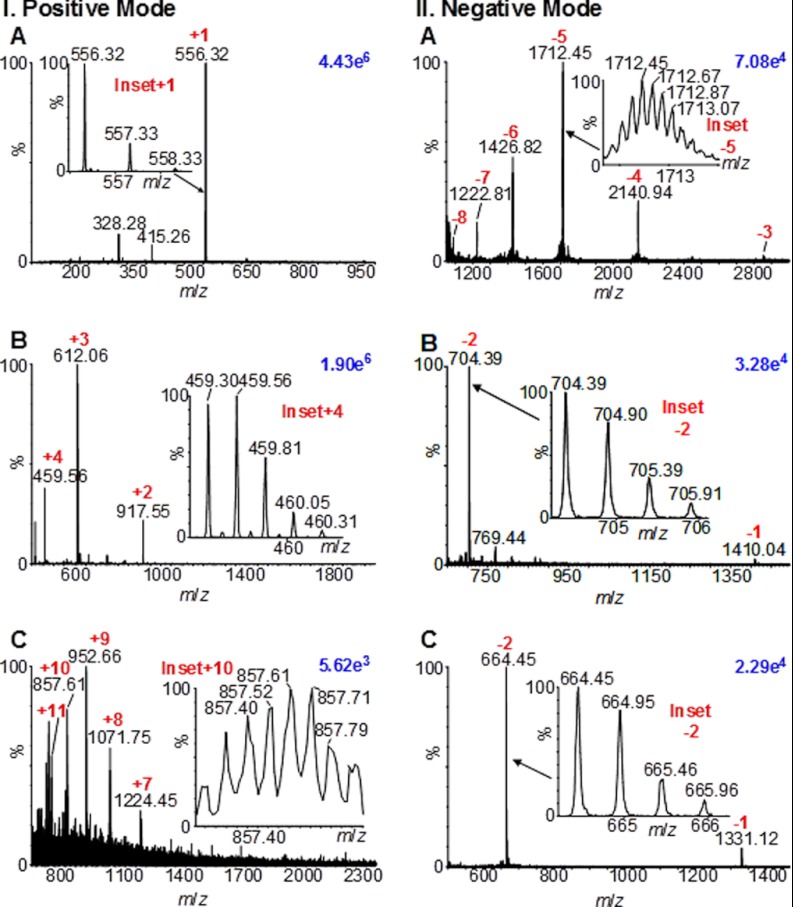
As with ESI, small molecules are detected as singly charged with little matrix-related chemical background (e.g. leucine enkaphelin; Fig. 2IA), and larger peptides (e.g. acetylated N-terminal fragment peptide of myelin basic protein; Fig. 2IB) as multiply charged ions. Mass spectra are obtained using the SYNAPT G2 mass spectrometer vacuum MALDI source, consuming low femtomoles of small proteins such as ubiquitin (50 fmol) (Fig. 2IC). Good negative ion abundances of, for example, ubiquitin (Fig. 2IIA), the hirudin (55–65) peptide (Fig. 2IIB), and the phosphorylated peptide cholecystokinin (10–20) (Fig. 2IIC) are also observed with MAIV.
Fig. 2.
MAIV mass spectra. (I) Positive ion mode: (A) 1 pmol leucine enkephalin (molecular weight (MW) 555 Da), (B) 1 pmol MBP peptide (MW 1833 Da), and (C) 50 fmol ubiquitin (MW 8559 Da). (II) Negative ion mode: (A) ubiquitin, (B) phosphorylated peptide cholecystokinin (10–20) (MW 1332 Da), and (C) hirudin (55–65) peptide (MW 1411 Da).
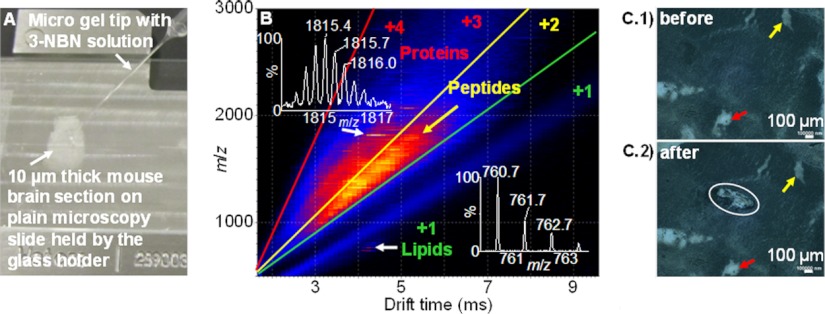
Peptides and proteins are detected by applying 3-NBN in an appropriate solvent onto the surface of a mouse brain tissue section (Fig. 3A) and subjecting the sample to the vacuum of the mass spectrometer (Fig. 3B). Selected areas of interest of a tissue can be exposed to matrix, and even if the entire tissue section is placed in vacuum, only molecular ions from compounds in those specific areas are observed (Fig. 3C). Miniaturizing the matrix deposition area to ca. 150 μm × 400 μm using a micropipette enables enhanced spatial resolution analyses without the need for a laser, as is required with MALDI (23). The high signal-to-noise obtained for this analysis suggests that much smaller spot sizes can be analyzed with appropriate matrix deposition methods and faster introduction of the sample into vacuum. The multiple charging in MAIV aids the separation of the compound classes (proteins, peptides, and lipids) by size, shape, number of charges, and m/z using IMS-MS.
Fig. 3.
MAIV-IMS-MS of a mouse brain tissue section using a commercial vacuum MALDI source with the laser off. A, photograph of application of matrix solution to a section of mouse brain on a glass microscope slide. B, two-dimensional IMS-MS plot of drift time versus m/z of ions showing separation of compound classes by charge, size, and shape of lipids, peptides, and proteins. Insets show isotopic distributions of lipid and protein ions. C, microscopy photograph of the mouse brain tissue before and after ionization. Circled area shows the area on the tissue analyzed in B, and small arrows are included to guide the eye for a better relative comparison of the photographs.
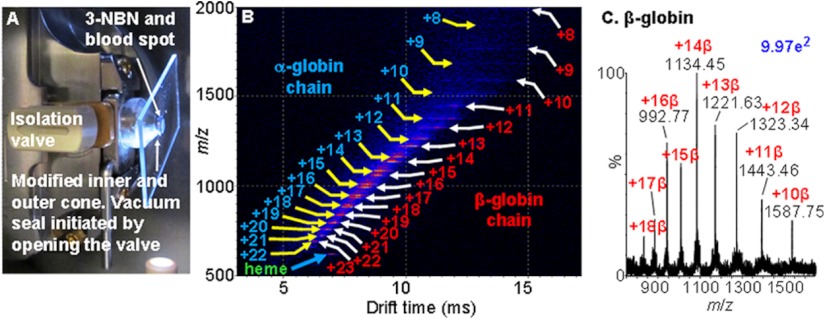
Mass spectrometers with vacuum MALDI sources are not a requirement for MAIV, as ESI sources can be reversibly modified for MAIV operation. Fig. 4A shows a glass microscope slide (details in supplemental Fig. S1) onto which a microliter of blood, diluted 1 part in 10 of water, mixed with 3-NBN matrix in ACN–water, and dried, is held against the inlet of a modified ESI source of the SYNAPT G2 mass spectrometer by the atmospheric-pressure-to-vacuum differential. Unlike with ESI, which detects predominantly the α-subunit of hemoglobin (24), both the α- and β-subunits are detected in good ion abundance from the blood spot using MAIV. Fig. 4B shows the IMS-MS plot of the gas-phase drift-time separated α- and β-subunit multiply charged molecular ions. The extracted mass spectrum of the β-subunit is shown in Fig. 4C. These results correlate well with blood spot results obtained with MAIV using the SYNAPT G2 vacuum MALDI source, indicating the universal utility of this ionization method with vacuum and atmospheric pressure ion sources.
Fig. 4.
MAIV-IMS-MS of a solvent-extracted blood spot from a Band-Aid using a commercial atmospheric pressure ESI source with a modified skimmer cone to provide a larger inlet aperture. A, photograph of the device showing the isolation valve in the open position with a glass microscope slide held by the pressure differential against the skimmer opening, with the matrix–blood spot sample exposed to the vacuum initiating ionization; more details on source modifications are provided in supplemental Fig. S1. B, two-dimensional IMS-MS plot of drift time versus m/z of ions showing separation of compound classes by charge, size, and shape: α-globin (MW 15,126 Da) and β-globin (MW 15,866 Da) chains of hemoglobin. Both α and β chains of hemoglobin are detected with MAIV in good ion abundance (mass spectral details in supplemental Fig. S2), something that has been reported (24) to be problematic with ESI (1). Extraction of the mass spectral information from the two-dimensional display in B provides the individual mass spectra of the proteins. C, extracted full range mass spectrum of the β-globin chain.
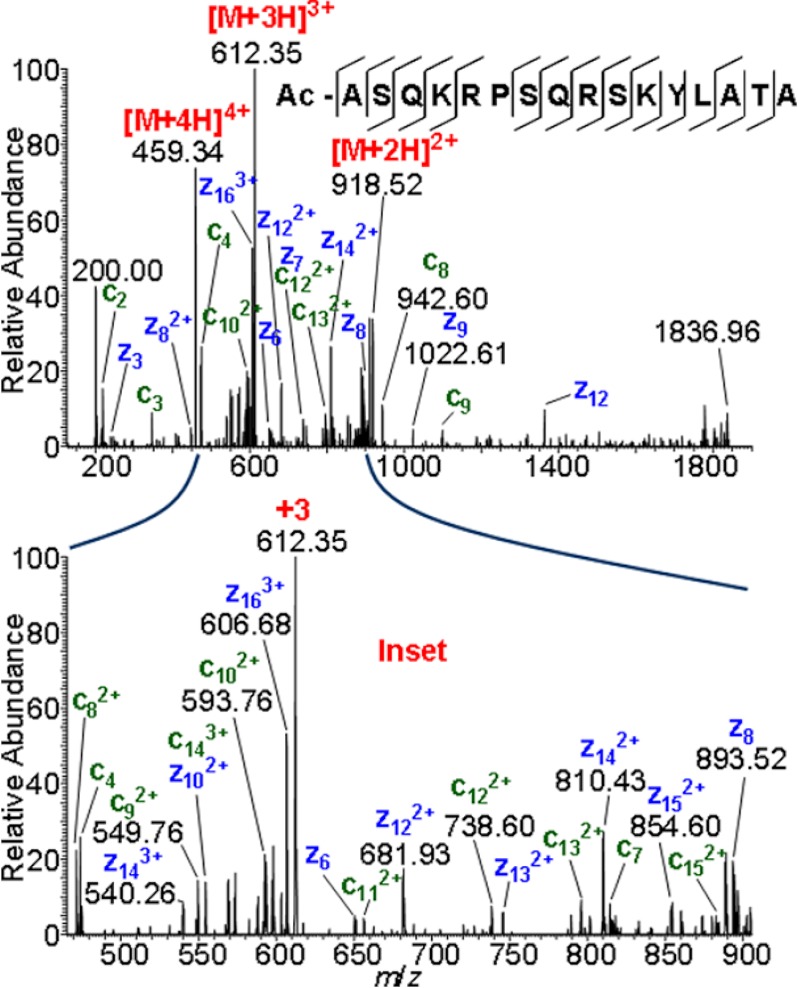
Using MAIV, sufficient ion abundance is obtained with the modified ESI source to perform sequence characterization. ETD fragmentation of the quadruply charged molecular ions of the acetylated N-terminal peptide fragment of myelin basic protein provides complete sequence coverage (Fig. 5) using the modified ESI source, identical to the principle shown in Fig. 4A, of a linear ion trap LTQ Velos. An expansion of the region between m/z 450 and 900 is shown in the inset, graphically demonstrating the degree of fragmentation obtained with this method. ETD is more applicable to the fragmentation of peptides and proteins than collision-induced dissociation, but only with ionization methods that produce abundant multiply charged ions (13, 14).
Fig. 5.
MAIV using the LTQ Velos ESI source to perform ETD fragmentation of the MBP peptide +4 charge state ion (∼m/z 459). Top, total fragment spectrum; bottom, inset m/z 400–900. Sequence coverage is increased relative to previous ETD studies of this peptide (14). Red numbers indicate the charge state, and blue numbers the ion abundance. Sequence coverage is provided (top).
The similarity of the charge states of ions produced via MAIV and ESI suggests similar mechanisms for their formation. A requirement for producing bare ions in ESI is solvent evaporation from highly charged droplets, and MAIV may require matrix sublimation from highly charged particles. A special property of 3-NBN, beside its ability to sublime at ambient temperature when placed in vacuum (25), is triboluminescence (26, 27) producing unusually strong dinitrogen discharge emission under conditions in which crystal fracturing occurs. This discharge is believed to result from a high electric field created between fractured crystal surfaces having opposite charge (26). In vacuum, the fracturing would be driven by sublimation of the matrix or residual solvent, producing charged particles that, upon sublimation of matrix, release analyte ions. 3-NBN matrix particles are observed leaving the surface in vacuum. Other matrices have been found to produce ions via MAIV, but none as well as 3-NBN.
This contribution represents the first example of a new ionization phenomenon demonstrating that small and large molecules are transferred as ions from the solid phase to the gas phase under vacuum conditions, without the application of external energy or fields, apparently via a sublimation-driven process. Multiply charged ions are produced from the solid state with high sensitivity, giving MAIV advantages of both ESI and MALDI. Analyses of surfaces of biological tissue, thin layer chromatography plates, and one- and two-dimensional gels are logical applications. MAIV, requiring only the vacuum necessary for the proper functioning of the mass spectrometer, might be applicable with common electron/chemical ionization mass spectrometers, with the filament used for ionization turned off. The simplicity and wide applicability of MAIV also might prove useful in clinical settings and in field portable instruments.
Supplementary Material
Acknowledgments
E.D.I. and S.T. conceived the experiments and analyzed the data, and S.T. wrote the paper. We are grateful to Prof. Mackie and James Wager-Miller, Indiana University, for providing the mouse brain tissue section (NIH Grant Nos. DA 11322 and DA 21696) and to Dr. Larsen (DuPont), Prof. Pflum (WSU), and Prof. McEwen (USP) for valuable feedback on this manuscript.
Footnotes
* This work was supported by NSF CAREER Award 0955975, an ASMS Research Award (Waters Company), a DuPont Young Professor Award, the Eli Lilly Young Investigator Award in Analytical Chemistry, Waters (COI program), a Schaap Faculty Scholar Award (to S.T.), and a Schaap Graduate Fellowship (to E.D.I.).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- 3-NBN
- 3-nitrobenzonitrile
- ACN
- acetonitrile
- BSA
- bovine serum albumin
- ESI
- electrospray ionization
- ETD
- electron transfer dissociation
- IMS
- ion mobility spectrometry
- MAIV
- matrix assisted ionization vacuum
- MALDI
- matrix-assisted laser desorption/ionization
- MS
- mass spectrometry
- TOF
- time-of-flight.
REFERENCES
- 1. Fenn J. B., Mann M., Meng C. K., Wong S. F., Whitehouse C. M. (1989) Electrospray ionization for mass spectrometry of large biomolecules. Science 246, 64–71 [DOI] [PubMed] [Google Scholar]
- 2. Tanaka K., Waki H., Ido Y., Akita S., Yoshida Y. (1988) Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2, 151–153 [Google Scholar]
- 3. Vestal M. L. (2009) Modern MALDI time-of-flight mass spectrometry. J. Mass Spectrom. 44, 303–317 [DOI] [PubMed] [Google Scholar]
- 4. Rosinke B., Strupat K., Hillenkamp F., Rosenbusch J., Dencher N., Kruger U., Galla H. J. (1995) Matrix-assisted laser desorption/ionization mass-spectrometry (MALDI-MS) of membrane-proteins and noncovalent complexes. J. Mass Spectrom. 30, 1462–1468 [Google Scholar]
- 5. Bich C., Zenobi R. (2009) Mass spectrometry of large complexes. Curr. Opin. Struct. Biol. 19, 632–639 [DOI] [PubMed] [Google Scholar]
- 6. Wei J., Buriak J., Siuzdak G. (1999) Desorption-ionization mass spectrometry on porous silicon. Nature 399, 243–246 [DOI] [PubMed] [Google Scholar]
- 7. Krutchinsky A. N., Chait B. T. (2002) On the nature of the chemical noise in MALDI mass spectra. J. Am. Soc. Mass Spectrom. 13, 129–134 [DOI] [PubMed] [Google Scholar]
- 8. Karas M., Bahr U., Strupat K., Hillenkamp F., Tsarbopoulos A., Pramanik B. N. (1995) Matrix dependence of metastable fragmentation of glycoproteins in MALDI-TOF mass spectrometry. Anal. Chem. 667, 675–679 [Google Scholar]
- 9. Inutan E. D., Trimpin S. (2010) Laserspray ionization-ion mobility spectrometry-mass spectrometry: baseline separation of isomeric amyloids without the use of solvents desorbed and ionized directly from a surface. J. Proteome Res. 9, 6077–6081 [DOI] [PubMed] [Google Scholar]
- 10. Inutan E. D., Wang B., Trimpin S. (2010) Commercial intermediate pressure MALDI ion mobility spectrometry mass spectrometer capable of producing highly charged laserspray ionization ions. Anal. Chem. 83, 678–684 [DOI] [PubMed] [Google Scholar]
- 11. Trimpin S., Wang B., Inutan E. D., Li J., Lietz C. B., Pagnotti V. S., Harron A. F., Sardelis D., McEwen C. N. (2012) A mechanism for ionization of nonvolatile compounds in mass spectrometry: considerations from MALDI and inlet ionization. J. Am. Soc. Mass Spectrom. 23, 1644–1660 [DOI] [PubMed] [Google Scholar]
- 12. Trimpin S., Herath T. N., Inutan E. D., Cernat S. A., Wager-Miller J., Mackie K., Walker J. M. (2009) Field-free transmission geometry atmospheric pressure matrix-assisted laser desorption/ionization for rapid analysis of unadulterated tissue samples. Rapid Commun. Mass Spectrom. 23, 3023–3027 [DOI] [PubMed] [Google Scholar]
- 13. Trimpin S., Inutan E. D., Herath T. N., McEwen C. N. (2010) Laserspray ionization—a new atmospheric pressure MALDI method for producing highly charged gas-phase ions of peptides and proteins directly from solid solutions. Mol. Cell. Proteomics 9, 362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Inutan E. D., Richards A. L., Wager-Miller J., Mackie K., McEwen C. N., Trimpin S. (2011) Laserspray ionization, a new method for protein analysis directly from tissue at atmospheric pressure with ultrahigh mass resolution and electron transfer dissociation. Mol. Cell. Proteomics 10, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takats Z., Wiseman J. M., Gologan B., Cooks R. G. (2004) Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science 306, 471–473 [DOI] [PubMed] [Google Scholar]
- 16. Nemes P., Vertes A. (2007) Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal. Chem. 79, 8098–8106 [DOI] [PubMed] [Google Scholar]
- 17. Hopper J. T. S., Oldham N. J. (2009). Collision induced unfolding of protein ions in the gas phase studied by ion mobility-mass spectrometry: the effect of ligand binding on conformational stability. J. Am. Soc. Mass Spectrom. 20, 1851–1858 [DOI] [PubMed] [Google Scholar]
- 18. Richards A. L., Lietz C. B., Wager-Miller J., Mackie K., Trimpin S. (2011) Imaging mass spectrometry in transmission geometry. Rapid Commun. Mass Spectrom. 25, 815–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Richards A. L., Lietz C. B., Wager-Miller J., Mackie K., Trimpin S. (2012) Laserspray ionization MSn and high spatial resolution tissue imaging of labile gangliosides. J. Lipid Res. 53, 1390–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luxembourg S. L., McDonnell L. A., Duursma M. C., Guo X. H., Heeren R. M. A. (2003). Effect of local matrix crystal variations in matrix-assisted ionization techniques for mass spectrometry. Anal. Chem. 75, 2333–2341 [DOI] [PubMed] [Google Scholar]
- 21. Tholey A., Heinzle E. (2006) Ionic (liquid) matrices for matrix-assisted laser desorption/ionization mass spectrometry—applications and perspectives. Anal. Bioanal. Chem. 386, 24–37 [DOI] [PubMed] [Google Scholar]
- 22. Jaskolla T. W., Lehmann W. D., Karas M. (2008) 4-Chloro-alpha-cyanocinnamic acid is an advanced, rationally designed MALDI matrix. Proc. Natl. Acad. Sci. U.S.A. 105, 12200–12205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tu T., Gross M. L. (2009) Miniaturizing sample spots for matrix-assisted laser desorption/ionization mass spectrometry. Trends Anal. Chem. 28, 833–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuprowski M. C., Boys B. L., Konermann L. (2007) Analysis of protein mixtures by electrospray mass spectrometry: effects of conformation and desolvation behavior on the signal intensities of hemoglobin subunits. J. Am. Soc. Mass Spectrom. 18, 1279–1285 [DOI] [PubMed] [Google Scholar]
- 25. Mitchell J., Jr., Deveraux H. D. (1992) Determination of traces of organic compounds in the atmosphere: role of detectors in gas chromatography. Anal. Chim. Acta 52, 45–52 [Google Scholar]
- 26. Sweeting L. M., Cashel M. L., Rosenblatt M. M. (1992) Triboluminescence spectra of organic crystals are sensitive to conditions of acquisition. J. Lumin. 52, 281–291 [Google Scholar]
- 27. Sweeting L. M. (2001) Triboluminescence with and without air. Chem. Mater. 13, 854–870 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.