Abstract
Protein phosphatases undo the post-translational modifications of kinase-signaling networks, but phosphatase activation in cells is difficult to measure and interpret. Here, we report the design of a quantitative and high-throughput assay platform for monitoring cellular phosphatase activity toward specific phosphoprotein targets. Protein substrates of interest are purified recombinantly, phosphorylated in vitro using the upstream kinase, and adsorbed to 96-well plates. Total phosphatase extracts from cells are then added to trigger a solid-phase dephosphorylation reaction. After stopping the reaction, phosphoprotein levels are quantified by ELISA with a phospho-specific antibody, and the loss of phospho-specific immunoreactivity is used as the readout of phosphatase activity. We illustrate the generality of the method by developing specific phosphatase-activity assays for the three canonical mitogen-activated protein phospho-kinases: ERK, JNK, and p38. The assays capture changes in activity with a dynamic range of 25–100-fold and are sensitive to a limit of detection below 25,000 cells. When applied to cytokine-induced signaling, the assays revealed complex and dynamic regulation of phosphatases suggesting cross-communication and a means for cellular memory. Our assay platform should be beneficial for phosphoproteomic surveys and computational-systems models of signaling, where phosphatases are known to be important but their activities are rarely measured.
Phosphatases (PPases)1 reset post-translational modifications by kinases and thus help to sculpt the phosphoproteome (1–3). Once thought of as global attenuators of phosphorylation (2), PPases are now known to recognize specific subsets of phosphoprotein targets (4–7). Cellular PPase activity toward these phosphoprotein subsets is regulated at multiple levels. PPases can be induced transcriptionally (8–10), for example, and their catalytic efficiency is further controlled by diverse post-translational modifications (11–15). Notably, misregulation of PPases has been implicated in various inherited disorders (16, 17) and in diseases such as cancer (18, 19).
Multiple computational studies have indicated that PPases are especially important for the system-level properties of a signaling network (20–23). However, mathematically encoding explicit PPase species is problematic, because many PPases act on multiple substrates (2, 3), and each phosphosite can often be dephosphorylated by multiple PPases (24, 25). Consequently, PPases are often modeled as generic species that are tonically active, although some models include transcriptional regulation in an effort to capture feedback control (21, 23, 26–28). The unfortunate result of this simplification is a model whose generic PPases cannot be constrained by experimental observations. Thus, for network modeling of phosphorylation cascades, there is a need for measurement platforms that capture total PPase activity toward key signaling transducers.
The activity of purified PPases is readily measured with artificial colorimetric substrates (29) or chromogenic indicators of released inorganic phosphate (30, 31). Yet, neither of these detection strategies is compatible with total cellular extracts. Improved selectivity can be achieved with fluorescently labeled peptide substrates (32, 33), but these peptides still lack the structural requirements important for specific recognition by PPases (4–7). One can work around the promiscuity of such substrates by gel electrophoresis of crude extracts and then enzyme renaturation (34, 35), although this focuses on the PPases rather than the phosphosubstrates. Perhaps the clearest way to measure specific PPase activity is with the phosphosubstrate itself. However, previous assays have used radiolabeled substrates that are short-lived and must be precipitated away from the released 32P signal (36, 37), which reduces throughput. More recently, nonradioactive ELISA formats have been explored using broad phospho-motif antibodies (38), but the crossreactivity of such antibodies precludes their use for monitoring specific dephosphorylation events on key signaling proteins. Despite many decades of research on PPases, an assay has not been developed that is quantitative, high-throughput, sensitive, and specific for the conversion of phosphosubstrates.
Here, we report the general design of such an assay and its proof-of-principle application to the PPases deactivating the three canonical mitogen-activated protein kinases (MAPKs): extracellular-regulated kinase (ERK), c-Jun N-terminal kinase (JNK), and p38. MAPK pathways are critical signal-transduction modules that control proliferation, death-survival, differentiation, and stress responses throughout eukaryotes (39, 40). MAPKs are all regulated by phosphorylation of a Thr-X-Tyr (TXY) motif in their activation loop, which is catalyzed by dual-specificity MAPK kinases (MAP2Ks). Complete TXY dephosphorylation is catalyzed by dual-specificity PPases (DUSPs) called MAPK PPases (MKPs) (3, 7). The TXY motif can also be deactivated by the joint action of serine-threonine PPases and tyrosine PPases (41–44). For our assay development and validation, bisphosphorylated MAPKs provide a prototypical phosphosubstrate under complex negative regulation that changes dynamically in response to environmental stimuli (8–14). However, the format described here should generalize to any phosphoprotein that can be prepared in vitro and can be monitored with a high-quality phosphospecific antibody.
EXPERIMENTAL PROCEDURES
Plasmid Construction
Recombinant MAPKs and their upstream constitutively active MAP2Ks were cloned by PCR into pGEX-4T-1 glutathione S-transferase (GST) fusion plasmids containing triple epitope tags for Flag or HA. Rat ERK2 (Addgene, Cambridge, MA; plasmid #8974) (45) was cloned into the BamHI and SalI sites of pGEX-4T-1 (3×Flag) by PCR with the primers gcgcggatccatggcggcggcggcggcggcggg (forward) and gcgcgtcgacttaagatctgtatcctggctgg (reverse) followed by digestion with BamHI and SalI. Human JNK1 (Addgene plasmid #13798) (46) was cloned into the BamHI and SalI sites of pGEX-4T-1 (3×Flag) by PCR with the primers gcgcaggatccatgagcagaagcaagcgtgac (forward) and gcgcgtcgactcactgctgcacctgtgc (reverse) followed by digestion with BamHI and SalI. Murine p38α (Addgene plasmid #20351) (47) was cloned into the BamH1 and Xho1 sites of pGEX-4T-1 (3×Flag) by PCR with the primers gcgcggatccatgtcgcaggagaggccc (forward) and gcgcctcgagtcaggactccatttcttcttgg (reverse) followed by digestion with BamHI and XhoI. Murine MEK1-DD (Addgene plasmid #15268) (48) was cloned into the BamHI and SalI sites of pGEX-4T-1 (3×HA) by PCR with the primers gcgcggatccatgcccaagaagaagccgacg (forward) and gcgcgtcgactcagatgctggcagcgtgg (reverse) followed by digestion with BamHI and SalI. Human MKK4-EE (Addgene plasmid #14813) (49) was cloned into the BamHI and SalI sites of pGEX-4T-1 (3×HA) by PCR with the primers gcgcggatccatgcagggtaaacgcaaagc (forward) and gcgcgtcgactcaatcgacatacatgggagagc (reverse) followed by digestion with BamHI and SalI. Murine MKK7a1-EE (Addgene plasmid #14540) (50) was cloned into the BamHI and SalI sites of pGEX-4T-1 (3×HA) by PCR with the primers gcgcggatccatgctggggctcccatcaac (forward) and gcgcgtcgacctacctgaagaagggcagatg (reverse) followed by digestion with BamHI and SalI. MKK4-EE and MKK7a1-EE were subcloned sequentially into pCDFDuet-1 (Novagen, Madison, WI) for coexpression with pGEX-4T-1 3xFlag-JNK1. Human MKK6-EE (Addgene plasmid #13518) (51) was cloned into the BamHI and SalI sites of pGEX-4T-1 (3×HA) by PCR with the primers gcgcagatctatgtctcagtcgaaaggcaag (forward) and gcgcgtcgacttagtctccaagaatcagttttac (reverse) followed by digestion with BglII and SalI. The ligation products were transformed into electrocompetent E. coli (DH10B; Invitrogen, Grand Island, NY) and ampicillin-resistant clones were screened by endonuclease digestion and sequencing.
Protein Induction and Purification
Sequence-verified clones were transformed into low-copy E. coli (C41 DE3; Avidis, Saint-Beauzire, France) and grown at 37 °C until an optical density of 0.6–0.8 was achieved. The GST-fusion proteins were induced with isopropyl β-d-1-thiogalactopyranoside (IPTG) at 37 °C under the following conditions: GST-3xFlag-ERK2—2 mm IPTG for 5 h, GST-3xFlag-JNK1—0.4 mm IPTG for 4.5 h, GST-3xFlag-p38α—0.4 mm IPTG for 4 h, GST-3xHA-MEK1-DD—2 mm IPTG for 3 h, GST-3xHA-MKK4-EE—0.4 mm IPTG for 5 h, GST-3xHA-MKK7-EE—1 mm IPTG for 5 h, and GST-3xHA-MKK6-EE—0.4 mm IPTG for 4.5 h. Cells were harvested by centrifugation, resuspended in 7.5 ml TNE lysis buffer (50 mm Tris pH 7.4, 150 mm NaCl, 1 mm EDTA, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 1 μg/ml pepstatin) per 250 ml cell culture, and lysed with lysozyme and deoxycholate. Bacterial extracts were clarified by centrifugation and incubated overnight at 4 °C with glutathione-coated agarose beads (Sigma, St. Louis, MO). The bead-bound proteins were washed three times with ice-cold PBS + 0.5% Triton X-100 and then twice with PBS. MAP2Ks were eluted from the beads with 10 mm glutathione in 50 mm Tris pH 8.0 and concentrated with ultrafiltration columns (Millipore, Billerica, MA) before phosphorylation of MAPKs.
In Vitro Phosphorylation of MAPKs
Phosphorylation of the bead-bound MAPKs was accomplished by incubating the beads with their corresponding, concentrated MAP2Ks diluted in kinase assay buffer (10 mm Tris pH 7.5, 1 mm ATP, 15 mm MgCl2, 2.5 mm beta-glycerophosphate, 0.5 mm Na3VO4, 0.5 mm EGTA, 0.2 mm DTT) for 24 h at 37 °C. The phosphorylation reactions were halted by the addition of excess EDTA and the beads were washed three times with PBS. The phosphorylated MAPKs were purified by thrombin digest and quantified by SDS-PAGE with Coomassie staining.
In Vitro Dephosphorylation of pMAPKs
To verify phosphospecificity of the ELISAs, 0.5–1 μg of pMAPK was dephosphorylated with 1250 units lambda PPase (New England Biolabs, Ipswich, MA) in 50 mm HEPES (pH 7.5), 0.1% 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate, 100 mm NaCl, 1 mm MnCl2 for 2 h at 30 °C.
Cell Culture, Lysis, and Stimulation
HT-29 cells (ATCC, Manassas, VA) were cultured according to the manufacturer's specifications. For the cytokine time courses, HT-29 cells were seeded at 50,000 cells/cm2, grown for 24 h, then treated with 200 U/ml interferon gamma (IFNγ) for an additional 24 h (52). IFNγ-sensitized cells were then treated with 100 ng/ml epidermal growth factor (EGF), 100 ng/ml tumor necrosis factor (TNF), or both for the indicated times. Cells were washed once with ice-cold PBS and then lysed in a PPase lysis buffer (50 mm HEPES (pH 7.5), 0.05% saponin, 50 mm 2-mercaptoethanol (βME), 1 mm dithiothreitol (DTT), 2 mm MgCl2, 20 μg/ml aprotinin, 20 μg/ml leupeptin, 1 μg/ml pepstatin). Whole cell lysates were incubated on ice for 15 min then clarified by centrifugation at 16,000 × g for 15 min at 4 °C. Clarified lysates were frozen as single-use aliquots at −80 °C.
For the EGF prestimulation experiment, cells were seeded and sensitized as described above, then treated with 100 ng/ml EGF for 2 h followed by 100 ng/ml TNF for 15 min. Cells were washed once with ice-cold PBS then lysed in radioimmunoprecipitation assay (RIPA) buffer (25 mm Tris-HCl (pH 7.6), 150 mm NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 μg/ml pepstatin). Whole cell lysates were incubated, clarified, and stored as described above.
High-throughput PPase Assay
High protein-binding polystyrene microtiter plates (Corning Costar, Lowell, MA) were coated overnight with the indicated amounts of recombinant phosphorylated MAPK. The plates were then washed three times with PBS + 0.1% Tween-20 (PBS-T), and the dephosphorylation reaction was initiated by the addition of cell lysates (∼27,000 cells/well for pERK2 and pp38 and ∼7,000 cells/well for pJNK). The dephosphorylation reaction was allowed to proceed at 30 °C for the indicated times, and the reaction was terminated by the addition of a concentrated PPase-inhibitor mixture (20 mm NaPP, 60 mm NaF, 400 μm Na3VO4). The plates were then washed three times with PPase-inhibitor wash solution (10 mm NaPP, 30 mm NaF, 200 μm Na3VO4 diluted in PBS-T) before proceeding.
Enzyme-linked Immunosorbent Assay (ELISA)
Inhibitor-washed plates were blocked for 1 h at room temperature with blocking buffer (PBS-T + 5% BSA) before incubation with one of the following primary antibodies diluted in blocking buffer: anti-phospho-p44/p42 (Cell Signaling Technology, Danvers, MA; 1:1000, 1 h), anti-phospho-p38 (Cell Signaling Technology; 1:1000, 1 h), or anti-phospho-SAPK/JNK (Cell Signaling Technology; 1:50, 3 h). The plates were then washed three times with PBS-T and incubated with blocking buffer containing biotinylated goat anti-rabbit secondary antibody (Jackson Immunoresearch, West Grove, PA; 1:10,000) for 1 h at room temperature. After three washes with PBS-T, the plates were incubated with streptavidin-HRP (R&D Systems, Minneapolis, MN; 1:200) for 30 min at room temperature. The plates were again washed three times with PBS-T and incubated with ELISA detection reagents (R&D Systems) for 5–15 min. The ELISA reaction was halted by the addition of 1 m H2SO4 and the signals were measured by spectrophotometry at 450 nm with plate background correction at 540 nm. PPase activation was monitored by the decrease in phosphorylation level of the lysate-treated wells relative to untreated control wells. Relative activity values were calculated by four-parameter logistic regression using a serial dilution of concentrated PPase lysate.
Western Blotting
PPase or RIPA lysates were resolved on 10% or 12% polyacrylamide gels and transferred to 0.45 μm polyvinylidene fluoride (Millipore, Billerica, MA). Membranes were blocked with 0.5× Odyssey blocking buffer (Licor, Lincoln, NB) diluted in PBS and probed with chicken anti-tubulin (Abcam, Cambridge, MA) at 1:5000 dilution and the following antibodies at 1:1000 dilution: rabbit anti-phospho-SAPK/JNK (Cell Signaling Technology), anti-JNK1 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-phospho-p38, anti-IκBα, anti-MKP5 (Cell Signaling Technology), anti-DUSP16/MKP7 (Novus Biologicals, Littleton, CO). The membranes were then probed with 800 CW goat anti-rabbit (Licor) and 680 RD donkey anti-chicken (Licor) secondary antibodies at 1:20,000 dilution and visualized by fluorescence on a Licor Odyssey imager (Licor).
Lentiviral shRNA Knockdown
The following pLKO.1 shRNA constructs were obtained through the RNAi Consortium: shMKP5 (#1: TRCN0000001885 and #2: TRCN0000001887) and shMKP7 (#1: TRCN0000052013 and #2: TRCN0000052017) (Open Biosystems, Lafayette, CO). Lentiviruses were packaged, transduced into HT-29 cells, and selected with 2 μg/ml puromycin as previously described (53).
Statistics
Comparisons between means was performed by Student's t test after correcting for error propagation. Comparisons between control and shMKP knockdown lines were performed by Fisher's method of combining probabilities (54).
RESULTS
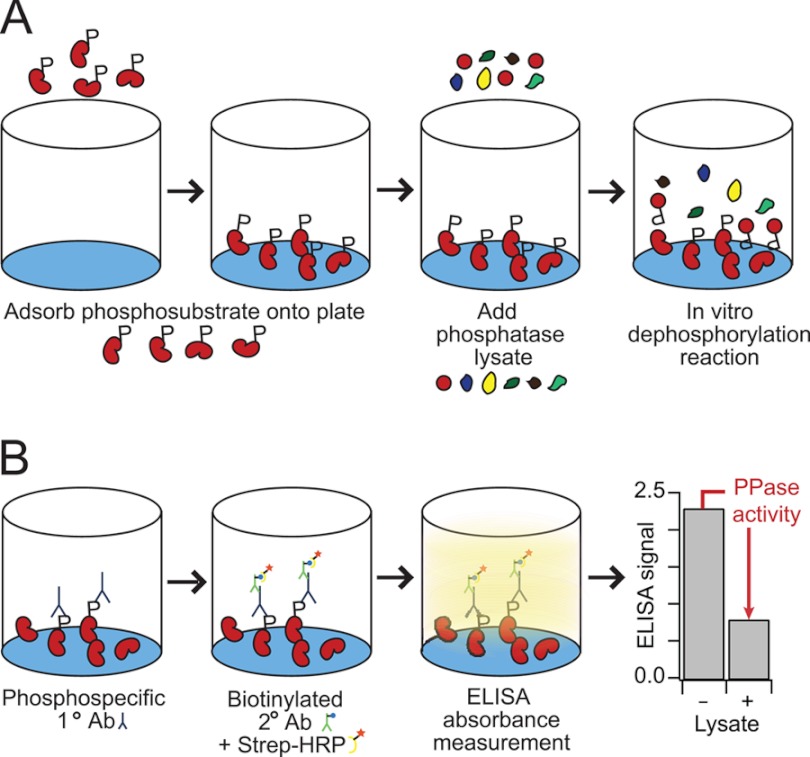
We developed the general assay in a solid-phase format with adsorbed recombinant phosphoprotein as a substrate (Fig. 1A). Phosphoprotein-coated plates are then incubated with cell extracts containing endogenous PPases. The dephosphorylation reaction proceeds for a fixed time before quenching and removal of the lysates. Loss of protein phosphorylation is quantified by an ELISA with high-quality phosphospecific antibodies and used as the measure of phosphoprotein-specific PPase activity (Fig. 1B). This format captures the activity of endogenous PPases toward specific phosphosubstrates in a high-throughput and quantitative manner.
Fig. 1.
A high-throughput phosphatase assay that monitors endogenous activity toward specific phosphoprotein substrates. A, recombinant phosphorylated substrate is adsorbed onto a polystyrene microtiter plate, the immobilized phosphosubstrate are treated with PPase lysate, and the dephosphorylation reaction is halted with the addition of a phosphatase inhibitor mixture. B, the remaining phosphosubstrate is labeled with anti-phosphoprotein primary antibody, biotinylated secondary antibody, and streptavidin-HRP for colorimetric detection. Phosphatase activity is determined by the drop in phosphoprotein signal in lysate-treated samples relative to negative controls.
We chose to develop the assay for PPases targeting the three canonical MAPKs: ERK, JNK, and p38. These proteins are ideal substrates for the assay because their upstream MAP2Ks are known and can be rendered constitutively active with phosphomimetic substitutions in their activation loop. Furthermore, the modest size of the MAPKs allows them to be purified as GST fusion proteins in bacteria. Bead-bound MAPKs from bacterial extracts were phosphorylated with soluble active MAP2Ks, washed extensively, and then eluted by thrombin cleavage of the GST-MAPK linker. We observed no degradation of the MAPKs during the phosphorylation-elution steps (supplemental Fig. S1), and the phosphorylation stoichiometry was equal to or less than that observed when endogenous MAPKs were phosphorylated during cytokine stimulation (supplemental Fig. S2). Phospho-MAPK (pMAPK) yields ranged from ∼50–150 μg, allowing for hundreds of microtiter wells to be coated from a 250 ml bacterial culture.
Quantifying Post-translational Changes in Recombinant MAPKs by Phosphoprotein-specific ELISA
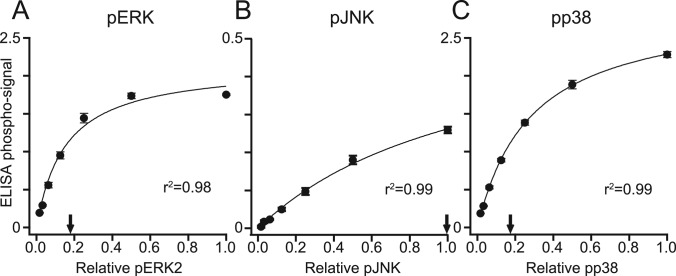
We optimized phosphosubstrate adsorption by immobilizing twofold serial dilutions of pERK, pJNK, or pp38 on polystyrene plates and detecting bound pMAPK by ELISA (Fig. 2A–2C). As expected, ELISA signals for each pMAPK increased hyperbolically, consistent with a standard binding isotherm. Detection of pJNK was notably weaker compared with the other pMAPKs (Fig. 2B), despite extensive optimization of ELISA conditions. We attribute this to the recognized difficulty of preparing high affinity pJNK antibodies that are stringently phosphospecific (55) and the more-complicated preparation involving two different MAP2Ks. For all substrates, we identified the maximum achievable coating below saturation of the ELISA readout and used this coating density in all subsequent experiments (Fig. 2A–2C, arrows).
Fig. 2.
ELISA-based quantification of recombinant pMAPKs adsorbed to 96-well plates. A–C, Twofold dilution series (relative level 1.0 = ∼400 ng) of pERK (A), pJNK (B), and pp38 (C) were adsorbed onto polystyrene microtiter plates and detected by ELISA using phospho-specific antibodies. The detection saturates at ∼75 ng for pERK (A), ∼400 ng for pJNK (B), and ∼75 ng for pp38 (C). Arrows indicate the quantity of pMAPKs used in the PPase assay. Data are shown as the mean ± S.E. of three independent assay replicates.
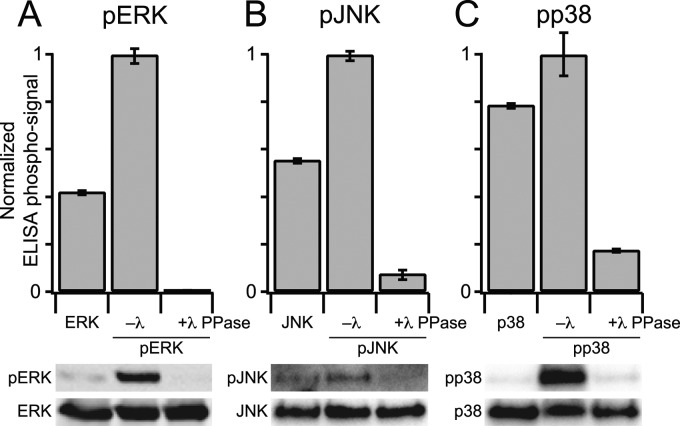
Next, it was important to determine whether the ELISA readout was strongly dependent on MAPK phosphorylation. To investigate the phosphospecificity of the ELISA detection, immobilized pMAPKs were treated with recombinant lambda (λ) PPase. λ PPase is a broad-specificity enzyme with reactivity toward phosphorylated serine, threonine, and tyrosine residues. We found that λ PPase treatment of each pMAPK strongly reduced the measured ELISA signal (Fig. 3A–3C, upper panels). The reduction coincided with the loss of pMAPK immunoreactivity as determined by immunoblotting (Fig. 3A–3C, lower panels). In MAPK purifications that had not been phosphorylated by MAP2Ks, we observed some background immunoreactivity by ELISA and immunoblot, which was stronger than λ PPase-treated pMAPKs. We interpret the background as basal Thr phosphorylation in the bacterial preparations, which is recognized to different extents by the anti-pMAPK antibody of the ELISA (56, 57) (see below). Taken together, we conclude that the phospho-ELISA format accurately measures the change in phosphorylation state of immobilized pMAPKs.
Fig. 3.
The ELISA measurement of pMAPK levels is phospho-specific. A–C (upper panels), immobilized MAPKs or pMAPKs were incubated in the presence (+) or absence (–) of lambda (λ) PPase and the resulting phosphorylation levels of pERK (A), pJNK (B), or pp38 (C) were measured by ELISA. A-C (lower panels), matched λ PPase-treated samples were analyzed by immunoblotting for pERK (A), pJNK (B), or pp38 (C) with Flag or total MAPK used as a loading control. Data are shown as the mean ± S.E. of three independent assay replicates.
Design and Validation of a High-sensitivity MAPK PPase Activity Assay
The success of the solid-phase reaction was contingent upon the extraction of PPases from cells without disrupting their native activity. We optimized a PPase lysis buffer containing saponin (for gentle permeabilization of the plasma membrane), DTT and βME (to maintain stable reducing conditions and avoid inadvertent oxidation of PPase catalytic cysteines) and MgCl2 (to provide Mg2+ cofactor that is critical for PPase catalysis). These conditions were stringent enough to extract cytosolic PPases while minimizing disruptions in PPase activity.
To capture the dephosphorylation activity of cell extracts, we quantitatively mapped the assay readout to changes in the enzymatic properties of the sample. First, we optimized the duration of lysate incubation on the pMAPK-coated plate to ensure that the dephosphorylation reaction was proceeding at a constant reaction velocity. When extracts (∼275,000 cells/well for pERK and pp38, and ∼70,000 cells/well for pJNK) were compared with lysis buffer controls, we observed clear lysate-specific activity that increased linearly for at least 30 min (Fig. 4A–4C). PPase activity plateaued at longer incubation times, likely because of phosphosubstrate depletion. We also observed higher background losses of phosphorylation on pJNK-coated plates (Fig. 4B), consistent with the increased lability of the phosphoprotein in cell extracts (55). For all substrates, we identified the linear regime of dephosphorylation and used this incubation time in all subsequent experiments (Fig. 4A–4C, arrows).
Fig. 4.
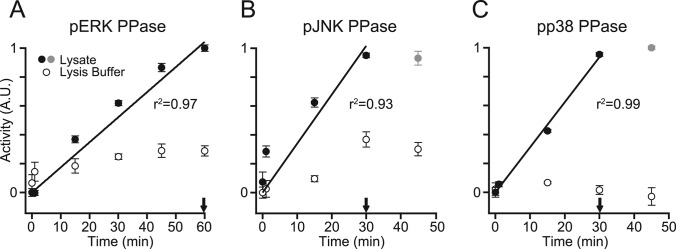
Early reaction kinetics of the MAPK PPase activity assays are linear with time. A–C, immobilized pERK (A), pJNK (B), or pp38 (C) were treated with saturating concentrations of PPase lysate (see Fig. 5) or lysis buffer for the indicated times. Arrows indicate the incubation times used in the optimized assays. Gray markers indicate time points where the kinetics have saturated. Data are shown as the mean ± S.E. of three independent assay replicates.
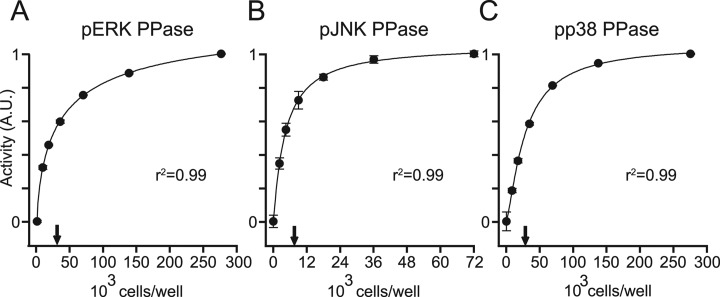
We next explored how measured PPase activity changed with different amounts of cell extract. This titration was designed to characterize the quantitative performance, sensitivity, and dynamic range of the assay. Compared with lysis buffer controls, measured PPase activities increased sharply before saturating with extracts above ∼50,000 cells per well (for pERK and pp38) and ∼12,000 cells per well (for pJNK) (Fig. 5A–5C). The response behavior was accurately captured by a four-parameter logistic (4PL) model of the kind frequently used for ELISA standard curves (58). The steep slopes of activity combined with overall assay precision (2.1% for pERK, 8.8% for pJNK, and 3.6% for pp38) indicate that the absolute limits of detection (59) were less than 30,000 cells/well. By focusing the assay conditions in the middle of the response curve (Fig. 5A-5C, arrows), we were able to capture ∼5–10-fold down-regulation or up-regulation in activity. This dynamic range is sufficient to capture the relative changes in PPase activity reported in cells (60, 61).
Fig. 5.
MAPK PPase activity assays are dose-dependent with respect to the concentration of cell lysate, and measure activity with fewer than 25,000 cells per well. A–C, the PPase assays were performed with 2-fold serial dilutions of concentrated PPase lysate (∼275,000 to ∼9,000 cells/well for pERK (A) and pp38 (C), and ∼70,000 to ∼2,000 cells/well for pJNK (B)). Measured activities were regressed against cell lysate using a four-parameter logistic curve. Arrows indicate the concentration of lysate used in the optimized assays. Data are shown as the mean ± S.E. of three independent assay replicates.
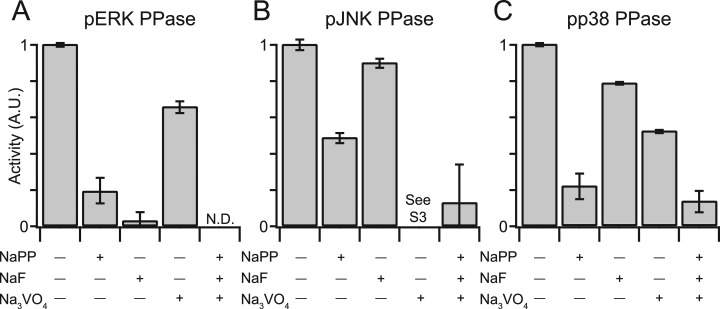
Last, to ensure that the measured activities were indeed catalyzed by PPases, we performed the assays in the presence of various PPase inhibitors. We used sodium pyrophosphate (NaPP) and sodium fluoride (NaF) to inhibit Ser-Thr PPases and sodium orthovanadate (Na3VO4) to inhibit Tyr PPases. Combined treatment with NaPP, NaF, and Na3VO4 abolished PPase activity in all assays (Fig. 6A–6C), whereas individual inhibitors showed differential reductions in PPase activity depending on the pMAPK. In general, NaF and NaPP were more effective at reducing measured PPase activity compared with Na3VO4. These findings agree with previous reports indicating that the anti-pMAPK antibodies used in the ELISAs react with monophosphorylated pTXY in addition to bisphosphorylated pTXpY (56, 57). Furthermore, the results suggest that MKPs, which are inhibited by Tyr PPase inhibitors but not by Ser-Thr PPase inhibitors (8, 62, 63), are only partly responsible for cellular pMAPK dephosphorylation activity (see below). We were unable to test the effect of Tyr PPase inhibition on pJNK PPase activity, because Na3VO4-treated lysates created a false-positive pJNK signal (Fig. 6B and supplemental Fig. S3). This false ELISA signal was not dependent on the immobilized pJNK and was instead caused by the anti-pJNK antibody and the PPase lysate (supplemental Fig. S3A, 3B). The false signal was not caused by the hyperphosphorylation of endogenous JNK, because immunoblot analysis of inhibitor-treated PPase lysates showed no increase in pJNK levels in response to Na3VO4 (supplemental Fig. S3C). Nevertheless, combining Na3VO4 with Ser-Thr PPase inhibitors eliminated the artifact and returned the measured JNK PPase activity to background levels (Fig. 6B). We conclude that the pMAPK PPase assays are accurate, reproducible, sensitive, and specific for endogenous PPase activity in cell extracts.
Fig. 6.
MAPK PPase activity assays specifically report PPase enzymatic activity in cell extracts. A–C, PPase lysates were treated with 10 mm NaPP, 30 mm NaF, 200 μm Na3VO4, or a combination of all three PPase inhibitors, and PPase activity was measured toward pERK (A), pJNK (B), or pp38 (C). See supplemental Fig. S3 for an explanation of the JNK PPase assay results for Na3VO4-treated lysates. Data are shown as the mean ± S.E. of three independent assay replicates.
Application of the MAPK PPase Assays to Cytokine-induced Signaling
To validate the assay in a biological context, we measured the dynamics of PPase activity in cytokine-treated HT-29 colon adenocarcinoma cells (see Experimental Procedures). Cells were sensitized with IFNγ and then stimulated with receptor-saturating concentrations of TNF, EGF, or both, and MAPK PPase activities were monitored for up to 8 h (Fig. 7A–7F). These conditions were chosen to coincide with a large systematic study of signaling in these cells, in which MAPK activities or phosphorylation were measured previously (52, 64–67).
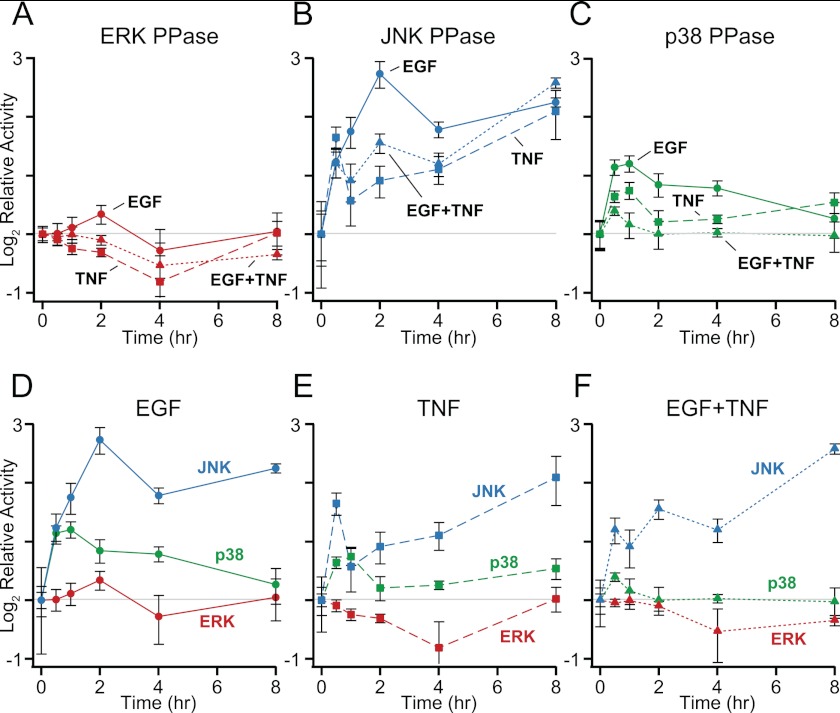
Fig. 7.
Cytokine stimulation evokes quantitatively and temporally distinct MAPK PPase activation profiles. HT-29 cells were seeded at 50,000 cells/cm2, sensitized with 200 U/ml IFNγ, and stimulated with 100 ng/ml EGF, 100 ng/ml TNF, or both for the indicated time points. A–C, cells were lysed and measured for PPase activity against pERK (A), pJNK (B), and pp38 (C) as described in the experimental procedures. D–F, the MAPK PPase activity measurements are replotted with respect to EGF (D), TNF (E), and EGF+TNF (F). Data are shown as the mean ± S.E. of four biological replicates.
We found that all MAPK-specific PPase activities changed in response to cytokine stimulation, but the regulation of each PPase activity was distinct. For example, EGF was consistently the most effective inducer of MAPK PPase activity (Fig. 7A–7C), in agreement with the widespread up-regulation of DUSPs/MKPs triggered by EGF (68). However, EGF caused a sustained activation of JNK PPase while eliciting only a transient activation of p38 PPase and a negligible activation of ERK PPase (Fig. 7D), suggesting pathway specificity in the response. We also observed clear non-additivity in PPase activation when TNF was added together with EGF. TNF costimulation reduced or eliminated MAPK PPase activation triggered by EGF (Fig. 7D–7E), providing an explanation for the higher EGF-induced ERK activity observed when combined with TNF (64). The PPase activity profiles are thus consistent with the literature and quantify the negative regulation of MAPKs in a manner that is more directly interpretable.
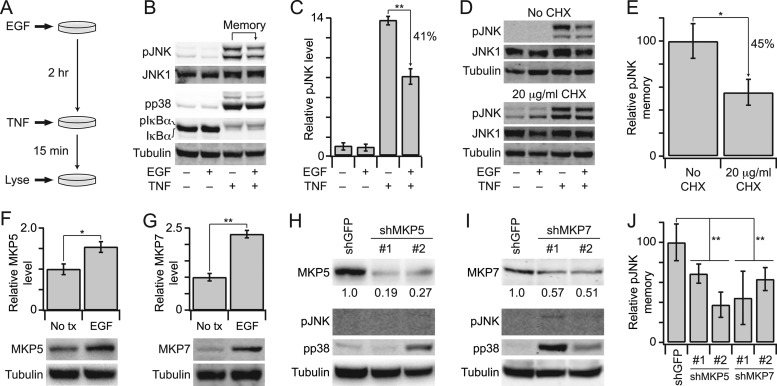
Last, we sought to test a prediction of our in vitro PPase assays in a cellular context. The strongest activation event we observed in our data set was for JNK PPases in cells stimulated with EGF for 2 h (∼eightfold activation, Fig. 7B). This result was surprising, because EGF is a weak activator of JNK signaling (64, 68, 69). We reasoned that the high PPase activity, if true, would create a form of cellular “memory” that would dampen the response of cells to future JNK-activating stimuli, such as TNF (69). To test this prediction, we pretreated cells with EGF for 2 h and then stimulated with TNF for 15 min (Fig. 8A), which maximally activates JNK in HT-29 cells (64). Early TNF-induced JNK signaling is a key point of signal integration with receptor tyrosine kinases and has been shown to be a strong predictor of TNF-induced apoptosis (65, 70). We found that EGF prestimulation led to a significant reduction in early TNF-induced pJNK compared with cells that had not been prestimulated (p < 0.01, Fig. 8B–8C). There was also a slight attenuation of TNF-induced pp38, consistent with the ∼twofold increase in p38 PPase activity caused by EGF prestimulation for 2 h (Figs. 7C–7D, 8B). Conversely, we observed no change in TNF-induced IκBα phosphorylation and degradation, indicating that EGF does not globally upregulate PPase activity (Fig. 8B). These experiments suggested that the measured JNK PPase activity stimulated by EGF could be relevant for JNK activation in vivo.
Fig. 8.
EGF up-regulates MKP5 and MKP7 to prime cells against future JNK-activating stimuli. A, HT-29 cells were seeded at 50,000 cells/cm2, sensitized with 200 U/ml IFN-γ, and stimulated with 100 ng/ml EGF for 2 h to upregulate JNK PPase activity, followed by 100 ng/ml TNF for 15 min. B, cells with or without 100 ng/ml EGF prestimulation or 100 ng/ml TNF stimulation were analyzed for pJNK, pp38, or total IκBα levels with JNK1 and tubulin used as loading controls. pIκBα appears as an upshifted band on the total IκBα immunoblot. C, replicated densitometry of the results shown in (B). D, inhibition of protein synthesis inhibits EGF-induced pJNK memory. Cells were pretreated with 20 μg/ml cycloheximide (CHX) before EGF-TNF stimulation as described in (A). E, replicated densitometry of the results shown in (D). F and G, EGF up-regulates MKP5 and MKP7. Cells were stimulated with 100 ng/ml EGF for 2 h and analyzed for MKP5 or MKP7 by immunoblotting with tubulin used as a loading control. Replicated densitometry is shown in the upper panels. H and I, shRNA-mediated knockdown of MKP5 or MKP7. Cells were transduced with lentiviruses, selected, and analyzed for MKP5 or MKP7, pJNK, or pp38 with tubulin used as a loading control. Relative densitometry for the extent of knockdown is shown beneath the MKP5 and MKP7 immunoblots. J, Knockdown of MKP5 or MKP7 reduces EGF-induced pJNK memory. shMKP5 or shMKP7 cells were stimulated with EGF-TNF as described in (A) and analyzed for pJNK by immunoblotting. Replicated densitometry is shown compared with shGFP control cells. Data are shown as the mean ± S.E. of 3–4 biological replicates. Single asterisk indicates p < 0.05, and double asterisk indicates p < 0.01.
EGF elicits waves of gene expression that feed back on earlier signaling events, and PPases are a major component of this transcriptional signature (68, 71). The gradual kinetics of JNK PPases (Fig. 7D) suggested that changes in activity could be because of EGF-stimulated gene expression. We tested this possibility by inhibiting protein synthesis with cycloheximide (CHX) and found that EGF-induced pJNK memory was significantly reduced in CHX-treated cells (p < 0.05, Fig. 8D-E). To identify specific PPases involved, we took a candidate approach and focused on cytoplasmic MKPs that target pJNK (3) and are regulated by EGF (68).
The JNK/p38-selective PPases, MKP5 and MKP7, were both significantly up-regulated after 2 h of EGF treatment (p < 0.05, Fig. 8F–8G), suggesting that they could contribute to overall JNK PPase activity. To determine whether MKP5 and MKP7 were specifically involved in EGF-induced memory, we stably knocked down each PPase with one of two independent shRNA targeting sequences (Fig. 8H–8I, upper). Despite partial knockdown, shMKP5 and shMKP7 cells showed variable increases in basal p38 phosphorylation, confirming biological efficacy (Fig. 8H–8I, upper). Conversely, there were negligible changes in basal JNK phosphorylation, likely because of minimal upstream pathway activity in the absence of cytokine stimulation. When shRNA-expressing cells were prestimulated with EGF and then challenged with TNF, we found that the paired shMKP5 and shMKP7 lines had reduced memory compared with the shGFP control (p < 0.01, Fig. 8J). Together, we conclude that EGF-induced memory is partly caused by the coordinate up-regulation of MKP5 and MKP7, leading to elevated JNK PPase activity (Fig. 7D). The results overall indicate that the activity assay platform accurately reflect facets of PPase regulation in vivo.
DISCUSSION
In this work, we develop a conceptually different approach for high-throughput monitoring of PPase activity in cell extracts. Rather than striving to measure specific PPase isoforms or holoenzymes, we inverted the assay design to focus on the collective PPase activity toward specific phosphosubstrates. This modification streamlines the assay by eliminating the need for affinity-capture reagents, much like the reverse-phase format that is now widely used for phosphoproteomics (72).
We focused here on PPases that act on the pMAPKs, because these phosphoproteins are negatively regulated at multiple levels and are known to contribute to network function (8–14). However, the overall assay format is general and should apply to many phosphoproteins of interest. The protein targets of most commercial phosphospecific antibodies have been cloned and their upstream kinases identified. In the future, our assay could be adapted to large panels of recombinant proteins that have been phosphorylated in vitro or, perhaps more easily, in vivo using bicistronic vectors (73). Encouragingly, in vivo preparations of phosphoprotein have shown large improvements in yield and ELISA immunoreactivity (supplemental Fig. S4), suggesting a way to streamline expansion of the assay. Thus far, the dephosphorylation reaction conditions for the three MAPK PPases are sufficiently comparable that groups of phosphoproteins could conceivably be combined in a spotted-array format for multiplex activity profiling (74).
With the PPase activity assays reported here, we envision that the most-immediate applications will be toward providing empirical constraints on systems models of MAPK signaling (21, 23, 26–28). Lumped “MAPK PPase” species in such models should conform to the quantitative and temporal changes measured with the assay, thereby reducing the danger of model overfitting. Alternatively, one could exploit the remarkable sensitivity of the assays for high-throughput screening. We routinely detect activity in extracts from ∼25,000 cells, making the assay compatible with 96-well cell-based screens for activators or inhibitors of MAPK PPases. From intracellular networks to small-molecule libraries, we believe that our PPase assay format will prove to be a useful profiling tool for this understudied class of signaling enzymes.
Although simple and effective, the assay has its caveats. The passive adsorption of phosphosubstrate onto the microplate surface (Fig. 1A) could prevent access to docking sites that are important for proper PPase recognition (4, 75). Therefore, the physiological relevance of any measured activity changes should be followed up with independent approaches (Fig. 8). As a biochemical method, our assay also overlooks the role of subcellular localization, which is important for certain PPases (76, 77). In the future, the assay format could be further elaborated to provide additional data on PPase regulation in subcellular compartments. For example, our current PPase lysis buffer uses saponin to gently permeabilize cells and extract cytoplasmic proteins, but the cholesterol-poor inner nuclear envelope is largely unaffected (78). It may be possible to pellet the insoluble material from saponin lysates and extract with a nonionic detergent to isolate nuclear PPase activity in the same cell preparation.
More globally, we see no reason why the overall assay format cannot be adapted to a mass-spectrometry paradigm through triple SILAC labeling (79). Medium- and heavy-labeled arginine could be used to prepare identical phosphosubstrate pools from cells treated with irreversible PPase inhibitors, such as calyculin A (80) and pervanadate (81). After dialyzing away the unreacted inhibitor, one of the pools could be incubated with a PPase lysate as prepared here (containing only light arginine). The second pool would then be added at the end as an internal reference after the PPase reaction has been quenched, with activity read out by the loss of phosphate between the medium and heavy pools. Mass spectrometry has been used to define optimal substrates for specific PPases (82), but the inverted approach we propose has not been described to our knowledge.
Our follow-up experiments with EGF-induced pJNK memory illustrate how activity data obtained with the assay can be further pursued to hone in on specific PPase species. Interestingly, the observed memory is only partially dependent on up-regulated MKP5 and MKP7. This emphasizes the redundancy in the network and raises the possibility that there are other PPases, which may be post-translationally controlled and contribute to the residual memory in CHX-treated cells. The ∼40% reduction in pJNK caused by EGF prestimulation may seem modest considering the ∼eightfold increase in JNK PPase activity under the same conditions (Figs. 7D, 8B–8C). However, this behavior is qualitatively consistent with models of amplified enzymatic signaling cascades, where signal amplitude is much more sensitive to kinase rates than PPase rates (20). The rich information content of the JNK pathway (65, 70) suggests that PPase regulation may play a critical role in fine-tuning an appropriate signaling response.
The overall literature of kinase signaling exceeds that of PPase signaling by a factor of ∼eight. Given the wealth of knowledge and available reagents for phosphoproteomics, it seems wise to leverage these resources for studying the negative regulators of protein phosphorylation. Our method lays the groundwork for such a strategy, which may help to unravel the time-dependent and context-specific control of the enzymatic “brakes” for signal-transduction networks (83).
Supplementary Material
Acknowledgments
We thank David Brautigan and John Lazo for guidance with the PPase lysis conditions and for critically reviewing this manuscript. We also thank Sarah Kinicki for technical assistance.
Footnotes
* This work was supported by the National Institutes of Health Director's New Innovator Award Program (1-DP2-OD006464), the Pew Scholars Program in the Biomedical Sciences, the David and Lucile Packard Foundation, the Joanna M. Nicolay Melanoma Foundation, and a Double ‘Hoo Research Award from the University of Virginia.
 This article contains supplemental Figs. S1 to S4.
This article contains supplemental Figs. S1 to S4.
1 The abbreviations used are:
- PPase
- phosphatase
- MAPK
- mitogen activated protein kinase
- ERK
- extracellular regulated kinase
- JNK
- c-Jun N-terminal kinase
- TXY
- Thr-X-Tyr
- DUSP
- dual-specificity PPase
- GST
- glutathione S-transferase
- EGF
- epidermal growth factor
- TNF
- tumor necrosis factor
- 4PL
- four-parameter logistic
- CHX
- cycloheximide.
REFERENCES
- 1. Tonks N. K. (2006) Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 7, 833–846 [DOI] [PubMed] [Google Scholar]
- 2. Ingebritsen T. S., Cohen P. (1983) Protein phosphatases: properties and role in cellular regulation. Science 221, 331–338 [DOI] [PubMed] [Google Scholar]
- 3. Keyse S. M. (2008) Dual-specificity MAP kinase phosphatases (MKPs) and cancer. Cancer Metastasis Rev. 27, 253–261 [DOI] [PubMed] [Google Scholar]
- 4. Roy J., Cyert M. S. (2009) Cracking the phosphatase code: docking interactions determine substrate specificity. Science signaling 2, re9. [DOI] [PubMed] [Google Scholar]
- 5. Shi Y. (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484 [DOI] [PubMed] [Google Scholar]
- 6. Virshup D. M., Shenolikar S. (2009) From promiscuity to precision: protein phosphatases get a makeover. Mol. Cell 33, 537–545 [DOI] [PubMed] [Google Scholar]
- 7. Farooq A., Zhou M. M. (2004) Structure and regulation of MAPK phosphatases. Cell. Signal. 16, 769–779 [DOI] [PubMed] [Google Scholar]
- 8. Keyse S. M., Emslie E. A. (1992) Oxidative stress and heat shock induce a human gene encoding a protein-tyrosine phosphatase. Nature 359, 644–647 [DOI] [PubMed] [Google Scholar]
- 9. Sun H., Charles C. H., Lau L. F., Tonks N. K. (1993) MKP-1 (3CH134), an immediate early gene product, is a dual specificity phosphatase that dephosphorylates MAP kinase in vivo. Cell 75, 487–493 [DOI] [PubMed] [Google Scholar]
- 10. Brondello J. M., Brunet A., Pouysségur J., McKenzie F. R. (1997) The dual specificity mitogen-activated protein kinase phosphatase-1 and -2 are induced by the p42/p44MAPK cascade. J. Biol. Chem. 272, 1368–1376 [DOI] [PubMed] [Google Scholar]
- 11. Saxena M., Williams S., Taskén K., Mustelin T. (1999) Crosstalk between cAMP-dependent kinase and MAP kinase through a protein tyrosine phosphatase. Nat Cell Biol. 1, 305–311 [DOI] [PubMed] [Google Scholar]
- 12. Blanco-Aparicio C., Torres J., Pulido R. (1999) A novel regulatory mechanism of MAP kinases activation and nuclear translocation mediated by PKA and the PTP-SL tyrosine phosphatase. J. Cell Biol. 147, 1129–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dickinson R. J., Delavaine L., Cejudo-Marin R., Stewart G., Staples C. J., Didmon M. P., Trinidad A. G., Alonso A., Pulido R., Keyse S. M. (2011) Phosphorylation of the kinase interaction motif in mitogen-activated protein (MAP) kinase phosphatase-4 mediates cross-talk between protein kinase A and MAP kinase signaling pathways. J. Biol. Chem. 286, 38018–38026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brondello J. M., Pouysségur J., McKenzie F. R. (1999) Reduced MAP kinase phosphatase-1 degradation after p42/p44MAPK-dependent phosphorylation. Science 286, 2514–2517 [DOI] [PubMed] [Google Scholar]
- 15. Meng T. C., Fukada T., Tonks N. K. (2002) Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol. Cell 9, 387–399 [DOI] [PubMed] [Google Scholar]
- 16. Tartaglia M., Mehler E. L., Goldberg R., Zampino G., Brunner H. G., Kremer H., van der Burgt I., Crosby A. H., Ion A., Jeffery S., Kalidas K., Patton M. A., Kucherlapati R. S., Gelb B. D. (2001) Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet. 29, 465–468 [DOI] [PubMed] [Google Scholar]
- 17. Bottini N., Musumeci L., Alonso A., Rahmouni S., Nika K., Rostamkhani M., MacMurray J., Meloni G. F., Lucarelli P., Pellecchia M., Eisenbarth G. S., Comings D., Mustelin T. (2004) A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat. Genet. 36, 337–338 [DOI] [PubMed] [Google Scholar]
- 18. Bulavin D. V., Demidov O. N., Saito S., Kauraniemi P., Phillips C., Amundson S. A., Ambrosino C., Sauter G., Nebreda A. R., Anderson C. W., Kallioniemi A., Fornace A. J., Jr., Appella E. (2002) Amplification of PPM1D in human tumors abrogates p53 tumor-suppressor activity. Nat. Genet. 31, 210–215 [DOI] [PubMed] [Google Scholar]
- 19. Zhang L., Zhou W., Velculescu V. E., Kern S. E., Hruban R. H., Hamilton S. R., Vogelstein B., Kinzler K. W. (1997) Gene expression profiles in normal and cancer cells. Science 276, 1268–1272 [DOI] [PubMed] [Google Scholar]
- 20. Heinrich R., Neel B. G., Rapoport T. A. (2002) Mathematical models of protein kinase signal transduction. Mol. Cell 9, 957–970 [DOI] [PubMed] [Google Scholar]
- 21. Bhalla U. S., Ram P. T., Iyengar R. (2002) MAP kinase phosphatase as a locus of flexibility in a mitogen-activated protein kinase signaling network. Science 297, 1018–1023 [DOI] [PubMed] [Google Scholar]
- 22. Haugh J. M., Schneider I. C., Lewis J. M. (2004) On the cross-regulation of protein tyrosine phosphatases and receptor tyrosine kinases in intracellular signaling. J. Theor. Biol. 230, 119–132 [DOI] [PubMed] [Google Scholar]
- 23. Blüthgen N., Legewie S., Kielbasa S. M., Schramme A., Tchernitsa O., Keil J., Solf A., Vingron M., Schäfer R., Herzel H., Sers C. (2009) A systems biological approach suggests that transcriptional feedback regulation by dual-specificity phosphatase 6 shapes extracellular signal-related kinase activity in RAS-transformed fibroblasts. FEBS J 276, 1024–1035 [DOI] [PubMed] [Google Scholar]
- 24. Saxena M., Mustelin T. (2000) Extracellular signals and scores of phosphatases: all roads lead to MAP kinase. Semin. Immunol. 12, 387–396 [DOI] [PubMed] [Google Scholar]
- 25. Mustelin T. (2007) A brief introduction to the protein phosphatase families. Methods Mol. Biol. 365, 9–22 [DOI] [PubMed] [Google Scholar]
- 26. Nakakuki T., Birtwistle M. R., Saeki Y., Yumoto N., Ide K., Nagashima T., Brusch L., Ogunnaike B. A., Okada-Hatakeyama M., Kholodenko B. N. (2010) Ligand-specific c-Fos expression emerges from the spatiotemporal control of ErbB network dynamics. Cell 141, 884–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen W. W., Schoeberl B., Jasper P. J., Niepel M., Nielsen U. B., Lauffenburger D. A., Sorger P. K. (2009) Input-output behavior of ErbB signaling pathways as revealed by a mass action model trained against dynamic data. Mol Syst Biol 5, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cirit M., Wang C. C., Haugh J. M. (2010) Systematic quantification of negative feedback mechanisms in the extracellular signal-regulated kinase (ERK) signaling network. J. Biol. Chem. 285, 36736–36744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bessey O. A., Lowry O. H., Brock M. J. (1946) A method for the rapid determination of alkaline phosphates with five cubic millimeters of serum. J. Biol. Chem. 164, 321–329 [PubMed] [Google Scholar]
- 30. Geladopoulos T. P., Sotiroudis T. G., Evangelopoulos A. E. (1991) A malachite green colorimetric assay for protein phosphatase activity. Anal. Biochem. 192, 112–116 [DOI] [PubMed] [Google Scholar]
- 31. Baykov A. A., Evtushenko O. A., Avaeva S. M. (1988) A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal. Biochem. 171, 266–270 [DOI] [PubMed] [Google Scholar]
- 32. Lutz M. P., Pinon D. I., Miller L. J. (1994) A nonradioactive fluorescent gel-shift assay for the analysis of protein phosphatase and kinase activities toward protein-specific peptide substrates. Anal. Biochem. 220, 268–274 [DOI] [PubMed] [Google Scholar]
- 33. Zhang Z. Y., Maclean D., Thieme-Sefler A. M., Roeske R. W., Dixon J. E. (1993) A continuous spectrophotometric and fluorimetric assay for protein tyrosine phosphatase using phosphotyrosine-containing peptides. Anal. Biochem. 211, 7–15 [DOI] [PubMed] [Google Scholar]
- 34. Kameshita I., Baba H., Umeda Y., Sueyoshi N. (2010) In-gel protein phosphatase assay using fluorogenic substrates. Anal. Biochem. 400, 118–122 [DOI] [PubMed] [Google Scholar]
- 35. Burridge K., Nelson A. (1995) An in-gel assay for protein tyrosine phosphatase activity: detection of widespread distribution in cells and tissues. Anal. Biochem. 232, 56–64 [DOI] [PubMed] [Google Scholar]
- 36. Mitsuhashi S., Shima H., Kikuchi K., Igarashi K., Hatsuse R., Maeda K., Yazawa M., Murayama T., Okuma Y., Nomura Y. (2000) Development of an assay method for activities of serine/threonine protein phosphatase type 2B (calcineurin) in crude extracts. Anal. Biochem. 278, 192–197 [DOI] [PubMed] [Google Scholar]
- 37. Cohen P., Alemany S., Hemmings B. A., Resink T. J., Strålfors P., Tung H. Y. (1988) Protein phosphatase-1 and protein phosphatase-2A from rabbit skeletal muscle. Methods Enzymol. 159, 390–408 [DOI] [PubMed] [Google Scholar]
- 38. Chuman Y., Iizuka K., Honda T., Onoue H., Shimohigashi Y., Sakaguchi K. (2011) Phosphatase assay for multi-phosphorylated substrates using phosphatase specific-motif antibody. J Biochem. 150, 319–325 [DOI] [PubMed] [Google Scholar]
- 39. Kyriakis J. M., Avruch J. (2001) Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 81, 807–869 [DOI] [PubMed] [Google Scholar]
- 40. Widmann C., Gibson S., Jarpe M. B., Johnson G. L. (1999) Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 79, 143–180 [DOI] [PubMed] [Google Scholar]
- 41. Barr A. J., Knapp S. (2006) MAPK-specific tyrosine phosphatases: new targets for drug discovery? Trends Pharmacol. Sci. 27, 525–530 [DOI] [PubMed] [Google Scholar]
- 42. Alessi D. R., Gomez N., Moorhead G., Lewis T., Keyse S. M., Cohen P. (1995) Inactivation of p42 MAP kinase by protein phosphatase 2A and a protein tyrosine phosphatase, but not CL100, in various cell lines. Curr. Biol. 5, 283–295 [DOI] [PubMed] [Google Scholar]
- 43. Westermarck J., Li S. P., Kallunki T., Han J., Kähäri V. M. (2001) p38 mitogen-activated protein kinase-dependent activation of protein phosphatases 1 and 2A inhibits MEK1 and MEK2 activity and collagenase 1 (MMP-1) gene expression. Mol. Cell. Biol. 21, 2373–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Takekawa M., Maeda T., Saito H. (1998) Protein phosphatase 2Calpha inhibits the human stress-responsive p38 and JNK MAPK pathways. EMBO J. 17, 4744–4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dimitri C. A., Dowdle W., MacKeigan J. P., Blenis J., Murphy L. O. (2005) Spatially separate docking sites on ERK2 regulate distinct signaling events in vivo. Curr. Biol. 15, 1319–1324 [DOI] [PubMed] [Google Scholar]
- 46. Dérijard B., Hibi M., Wu I. H., Barrett T., Su B., Deng T., Karin M., Davis R. J. (1994) JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76, 1025–1037 [DOI] [PubMed] [Google Scholar]
- 47. Enslen H., Raingeaud J., Davis R. J. (1998) Selective activation of p38 mitogen-activated protein (MAP) kinase isoforms by the MAP kinase kinases MKK3 and MKK6. J. Biol. Chem. 273, 1741–1748 [DOI] [PubMed] [Google Scholar]
- 48. Boehm J. S., Zhao J. J., Yao J., Kim S. Y., Firestein R., Dunn I. F., Sjostrom S. K., Garraway L. A., Weremowicz S., Richardson A. L., Greulich H., Stewart C. J., Mulvey L. A., Shen R. R., Ambrogio L., Hirozane-Kishikawa T., Hill D. E., Vidal M., Meyerson M., Grenier J. K., Hinkle G., Root D. E., Roberts T. M., Lander E. S., Polyak K., Hahn W. C. (2007) Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell 129, 1065–1079 [DOI] [PubMed] [Google Scholar]
- 49. Whitmarsh A. J., Shore P., Sharrocks A. D., Davis R. J. (1995) Integration of MAP kinase signal transduction pathways at the serum response element. Science 269, 403–407 [DOI] [PubMed] [Google Scholar]
- 50. Tournier C., Whitmarsh A. J., Cavanagh J., Barrett T., Davis R. J. (1997) Mitogen-activated protein kinase kinase 7 is an activator of the c-Jun NH2-terminal kinase. Proc. Natl. Acad. Sci. U.S.A. 94, 7337–7342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Raingeaud J., Whitmarsh A. J., Barrett T., Dérijard B., Davis R. J. (1996) MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol. Cell. Biol. 16, 1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Janes K. A., Albeck J. G., Peng L. X., Sorger P. K., Lauffenburger D. A., Yaffe M. B. (2003) A high-throughput quantitative multiplex kinase assay for monitoring information flow in signaling networks: application to sepsis-apoptosis. Mol Cell Proteomics 2, 463–473 [DOI] [PubMed] [Google Scholar]
- 53. Wang L., Brugge J. S., Janes K. A. (2011) Intersection of FOXO- and RUNX1-mediated gene expression programs in single breast epithelial cells during morphogenesis and tumor progression. Proc. Natl. Acad. Sci. U.S.A. 108, E803–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fisher R. A. (1954) Statistical methods for research workers, 12th Ed., Oliver and Boyd, Edinburgh [Google Scholar]
- 55. Liu Z. G., Lewis J., Wang T. H., Cook A. (2001) Role of c-Jun N-terminal kinase in apoptosis. Methods Cell Biol. 66, 187–195 [DOI] [PubMed] [Google Scholar]
- 56. Askari N., Beenstock J., Livnah O., Engelberg D. (2009) p38alpha is active in vitro and in vivo when monophosphorylated at threonine 180. Biochemistry 48, 2497–2504 [DOI] [PubMed] [Google Scholar]
- 57. Zhou B., Zhang Z. Y. (2002) The activity of the extracellular signal-regulated kinase 2 is regulated by differential phosphorylation in the activation loop. J. Biol. Chem. 277, 13889–13899 [DOI] [PubMed] [Google Scholar]
- 58. Healy M. J. (1972) Statistical analysis of radioimmunoassay data. Biochem. J. 130, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Armbruster D. A., Tillman M. D., Hubbs L. M. (1994) Limit of detection (LQD)/limit of quantitation (LOQ): comparison of the empirical and the statistical methods exemplified with GC-MS assays of abused drugs. Clin. Chem. 40, 1233–1238 [PubMed] [Google Scholar]
- 60. Lee S. R., Kwon K. S., Kim S. R., Rhee S. G. (1998) Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J. Biol. Chem. 273, 15366–15372 [DOI] [PubMed] [Google Scholar]
- 61. Zhou X., Ferraris J. D., Dmitrieva N. I., Liu Y., Burg M. B. (2008) MKP-1 inhibits high NaCl-induced activation of p38 but does not inhibit the activation of TonEBP/OREBP: opposite roles of p38alpha and p38delta. Proc. Natl. Acad. Sci. U.S.A. 105, 5620–5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wu L. W., Yoon H. K., Baylink D. J., Graves L. M., Lau K. H. (1997) Fluoride at mitogenic doses induces a sustained activation of p44mapk, but not p42mapk, in human TE85 osteosarcoma cells. J. Clin. Endocrinol. Metab. 82, 1126–1135 [DOI] [PubMed] [Google Scholar]
- 63. Charles C. H., Sun H., Lau L. F., Tonks N. K. (1993) The growth factor-inducible immediate-early gene 3CH134 encodes a protein-tyrosine-phosphatase. Proc. Natl. Acad. Sci. U.S.A. 90, 5292–5296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gaudet S., Janes K. A., Albeck J. G., Pace E. A., Lauffenburger D. A., Sorger P. K. (2005) A Compendium of Signals and Responses Triggered by Prodeath and Prosurvival Cytokines. Mol Cell Proteomics 4, 1569–1590 [DOI] [PubMed] [Google Scholar]
- 65. Janes K. A., Albeck J. G., Gaudet S., Sorger P. K., Lauffenburger D. A., Yaffe M. B. (2005) A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science 310, 1646–1653 [DOI] [PubMed] [Google Scholar]
- 66. Janes K. A., Gaudet S., Albeck J. G., Nielsen U. B., Lauffenburger D. A., Sorger P. K. (2006) The Response of Human Epithelial Cells to TNF Involves an Inducible Autocrine Cascade. Cell 124, 1225–1239 [DOI] [PubMed] [Google Scholar]
- 67. Janes K. A., Reinhardt H. C., Yaffe M. B. (2008) Cytokine-induced signaling networks prioritize dynamic range over signal strength. Cell 135, 343–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Amit I., Citri A., Shay T., Lu Y., Katz M., Zhang F., Tarcic G., Siwak D., Lahad J., Jacob-Hirsch J., Amariglio N., Vaisman N., Segal E., Rechavi G., Alon U., Mills G. B., Domany E., Yarden Y. (2007) A module of negative feedback regulators defines growth factor signaling. Nat. Genet. 39, 503–512 [DOI] [PubMed] [Google Scholar]
- 69. Raingeaud J., Gupta S., Rogers J. S., Dickens M., Han J., Ulevitch R. J., Davis R. J. (1995) Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J. Biol. Chem. 270, 7420–7426 [DOI] [PubMed] [Google Scholar]
- 70. Beyer E. M., MacBeath G. (2012) Cross-talk between receptor tyrosine kinase and tumor necrosis factor-alpha signaling networks regulates apoptosis but not proliferation. Mol Cell Proteomics 11, M111.013292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Avraham R., Yarden Y. (2011) Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat. Rev. Mol. Cell Biol. 12, 104–117 [DOI] [PubMed] [Google Scholar]
- 72. Paweletz C. P., Charboneau L., Bichsel V. E., Simone N. L., Chen T., Gillespie J. W., Emmert-Buck M. R., Roth M. J., Petricoin E. F., Liotta L. A. (2001) Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene 20, 1981–1989 [DOI] [PubMed] [Google Scholar]
- 73. Tolia N. H., Joshua-Tor L. (2006) Strategies for protein coexpression in Escherichia coli. Nat Methods 3, 55–64 [DOI] [PubMed] [Google Scholar]
- 74. MacBeath G., Schreiber S. L. (2000) Printing proteins as microarrays for high-throughput function determination. Science 289, 1760–1763 [DOI] [PubMed] [Google Scholar]
- 75. Zhang Y. Y., Wu J. W., Wang Z. X. (2011) A distinct interaction mode revealed by the crystal structure of the kinase p38alpha with the MAPK binding domain of the phosphatase MKP5. Science signaling 4, ra88. [DOI] [PubMed] [Google Scholar]
- 76. Frangioni J. V., Beahm P. H., Shifrin V., Jost C. A., Neel B. G. (1992) The nontransmembrane tyrosine phosphatase PTP-1B localizes to the endoplasmic reticulum via its 35 amino acid C-terminal sequence. Cell 68, 545–560 [DOI] [PubMed] [Google Scholar]
- 77. Lopez-Girona A., Furnari B., Mondesert O., Russell P. (1999) Nuclear localization of Cdc25 is regulated by DNA damage and a 14–3-3 protein. Nature 397, 172–175 [DOI] [PubMed] [Google Scholar]
- 78. Schulz I. (1990) Permeabilizing cells: some methods and applications for the study of intracellular processes. Methods Enzymol. 192, 280–300 [DOI] [PubMed] [Google Scholar]
- 79. Blagoev B., Ong S. E., Kratchmarova I., Mann M. (2004) Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat. Biotechnol. 22, 1139–1145 [DOI] [PubMed] [Google Scholar]
- 80. Ishihara H., Martin B. L., Brautigan D. L., Karaki H., Ozaki H., Kato Y., Fusetani N., Watabe S., Hashimoto K., Uemura D., et al. (1989) Calyculin A and okadaic acid: inhibitors of protein phosphatase activity. Biochem. Biophys. Res. Commun. 159, 871–877 [DOI] [PubMed] [Google Scholar]
- 81. Huyer G., Liu S., Kelly J., Moffat J., Payette P., Kennedy B., Tsaprailis G., Gresser M. J., Ramachandran C. (1997) Mechanism of inhibition of protein-tyrosine phosphatases by vanadate and pervanadate. J. Biol. Chem. 272, 843–851 [DOI] [PubMed] [Google Scholar]
- 82. Wang P., Fu H., Snavley D. F., Freitas M. A., Pei D. (2002) Screening combinatorial libraries by mass spectrometry. 2. Identification of optimal substrates of protein tyrosine phosphatase SHP-1. Biochemistry 41, 6202–6210 [DOI] [PubMed] [Google Scholar]
- 83. Tonks N. K., Muthuswamy S. K. (2007) A brake becomes an accelerator: PTP1B–a new therapeutic target for breast cancer. Cancer Cell 11, 214–216 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.