Abstract
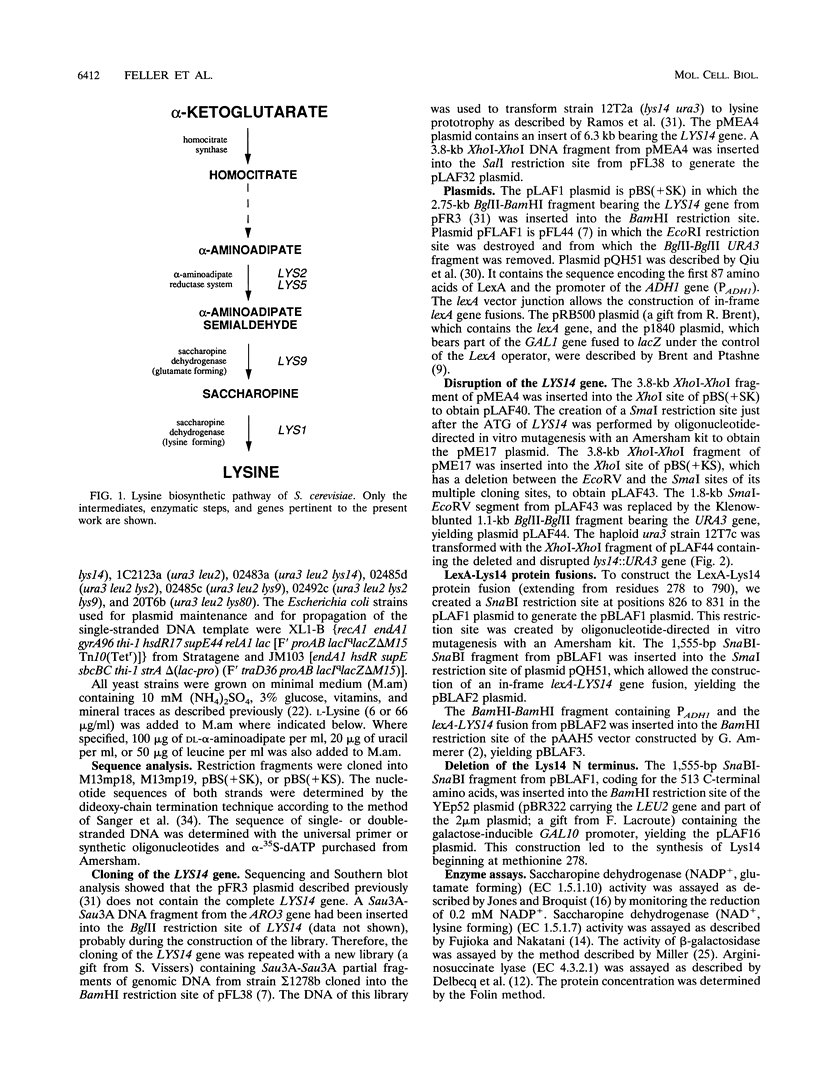
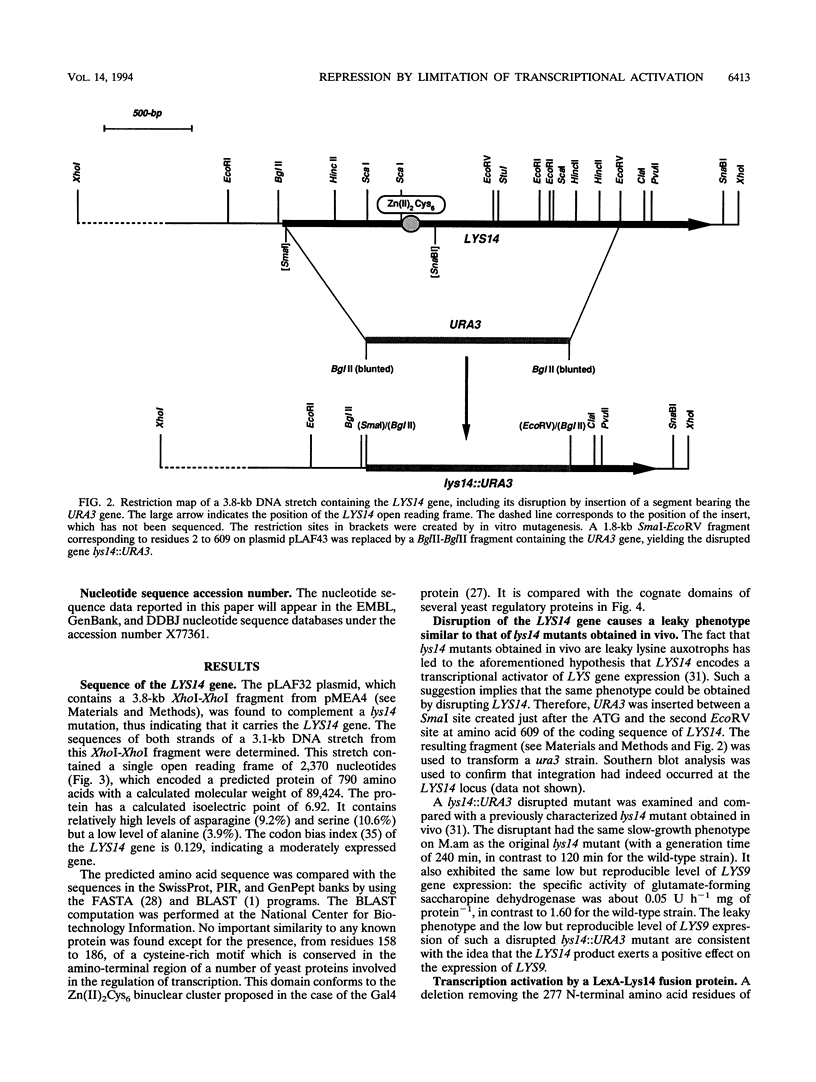
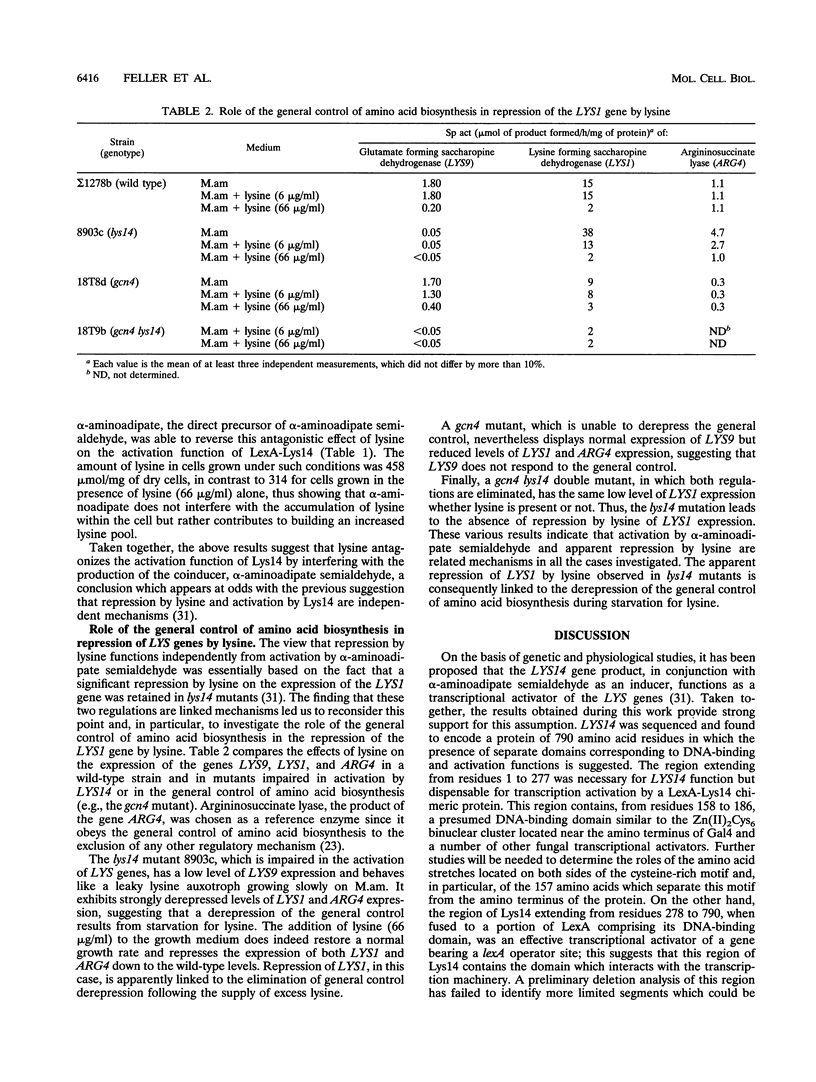
The product of the LYS14 gene of Saccharomyces cerevisiae activates the transcription of at least four genes involved in lysine biosynthesis. Physiological and genetic studies indicate that this activation is dependent on the inducer alpha-aminoadipate semialdehyde, an intermediate of the pathway. The gene LYS14 was sequenced and, from its nucleotide sequence, predicted to encode a 790-amino-acid protein carrying a cysteine-rich DNA-binding motif of the Zn(II)2Cys6 type in its N-terminal portion. Deletion of this N-terminal portion including the cysteine-rich domain resulted in the loss of LYS14 function. To test the function of Lys14 as a transcriptional activator, this protein without its DNA-binding motif was fused to the DNA-binding domain of the Escherichia coli LexA protein. The resulting LexA-Lys14 hybrid protein was capable of activating transcription from a promoter containing a lexA operator, thus confirming the transcriptional activation function of Lys14. Furthermore, evidence that this function, which is dependent on the presence of alpha-aminoadipate semialdehyde, is antagonized by lysine was obtained. Such findings suggest that activation by alpha-aminoadipate semialdehyde and the apparent repression by lysine are related mechanisms. Lysine possibly acts by limiting the supply of the coinducer, alpha-aminoadipate semialdehyde.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ammerer G. Expression of genes in yeast using the ADCI promoter. Methods Enzymol. 1983;101:192–201. doi: 10.1016/0076-6879(83)01014-9. [DOI] [PubMed] [Google Scholar]
- André B. The UGA3 gene regulating the GABA catabolic pathway in Saccharomyces cerevisiae codes for a putative zinc-finger protein acting on RNA amount. Mol Gen Genet. 1990 Jan;220(2):269–276. doi: 10.1007/BF00260493. [DOI] [PubMed] [Google Scholar]
- Balzi E., Chen W., Ulaszewski S., Capieaux E., Goffeau A. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. J Biol Chem. 1987 Dec 15;262(35):16871–16879. [PubMed] [Google Scholar]
- Bechet J., Greenson M., Wiame J. M. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1970 Jan;12(1):31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- Bonneaud N., Ozier-Kalogeropoulos O., Li G. Y., Labouesse M., Minvielle-Sebastia L., Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991 Aug-Sep;7(6):609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- Borell C. W., Urrestarazu L. A., Bhattacharjee J. K. Two unlinked lysine genes (LYS9 and LYS14) are required for the synthesis of saccharopine reductase in Saccharomyces cerevisiae. J Bacteriol. 1984 Jul;159(1):429–432. doi: 10.1128/jb.159.1.429-432.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent R., Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985 Dec;43(3 Pt 2):729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- Brisco P. R., Kohlhaw G. B. Regulation of yeast LEU2. Total deletion of regulatory gene LEU3 unmasks GCN4-dependent basal level expression of LEU2. J Biol Chem. 1990 Jul 15;265(20):11667–11675. [PubMed] [Google Scholar]
- Coornaert D., Vissers S., André B. The pleiotropic UGA35(DURL) regulatory gene of Saccharomyces cerevisiae: cloning, sequence and identity with the DAL81 gene. Gene. 1991 Jan 15;97(2):163–171. doi: 10.1016/0378-1119(91)90048-g. [DOI] [PubMed] [Google Scholar]
- Delbecq P., Werner M., Feller A., Filipkowski R. K., Messenguy F., Piérard A. A segment of mRNA encoding the leader peptide of the CPA1 gene confers repression by arginine on a heterologous yeast gene transcript. Mol Cell Biol. 1994 Apr;14(4):2378–2390. doi: 10.1128/mcb.14.4.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friden P., Schimmel P. LEU3 of Saccharomyces cerevisiae encodes a factor for control of RNA levels of a group of leucine-specific genes. Mol Cell Biol. 1987 Aug;7(8):2708–2717. doi: 10.1128/mcb.7.8.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M., Nakatani Y. A kinetic study of saccharopine dehydrogenase reaction. Eur J Biochem. 1970 Sep;16(1):180–186. doi: 10.1111/j.1432-1033.1970.tb01070.x. [DOI] [PubMed] [Google Scholar]
- JONES E. E., BROQUIST H. P. SACCHAROPINE, AN INTERMEDIATE OF THE AMINOADIPIC ACID PATHWAY OF LYSINE BIOSYNTHESIS. II. STUDIES IN SACCHAROMYCES CEREVISEAE. J Biol Chem. 1965 Jun;240:2531–2536. [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987 Dec;51(4):458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer B., Guyonvarch A., Hubert J. C. Yeast regulatory gene PPR1. I. Nucleotide sequence, restriction map and codon usage. J Mol Biol. 1984 Dec 5;180(2):239–250. doi: 10.1016/s0022-2836(84)80002-9. [DOI] [PubMed] [Google Scholar]
- Kim J., Michels C. A. The MAL63 gene of Saccharomyces encodes a cysteine-zinc finger protein. Curr Genet. 1988 Oct;14(4):319–323. doi: 10.1007/BF00419988. [DOI] [PubMed] [Google Scholar]
- Laughon A., Gesteland R. F. Primary structure of the Saccharomyces cerevisiae GAL4 gene. Mol Cell Biol. 1984 Feb;4(2):260–267. doi: 10.1128/mcb.4.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczak J. E., Brandriss M. C. Analysis of constitutive and noninducible mutations of the PUT3 transcriptional activator. Mol Cell Biol. 1991 May;11(5):2609–2619. doi: 10.1128/mcb.11.5.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F., Dubois E., Descamps F. Nucleotide sequence of the ARGRII regulatory gene and amino acid sequence homologies between ARGRII PPRI and GAL4 regulatory proteins. Eur J Biochem. 1986 May 15;157(1):77–81. doi: 10.1111/j.1432-1033.1986.tb09640.x. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Dubois E. Participation of transcriptional and post-transcriptional regulatory mechanisms in the control of arginine metabolism in yeast. Mol Gen Genet. 1983;189(1):148–156. doi: 10.1007/BF00326068. [DOI] [PubMed] [Google Scholar]
- Messenguy F. Regulation of arginine biosynthesis in Saccharomyces cerevisiae: isolation of a cis-dominant, constitutive mutant for ornithine carbamoyltransferase synthesis. J Bacteriol. 1976 Oct;128(1):49–55. doi: 10.1128/jb.128.1.49-55.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris M. E., Jinks-Robertson S. Nucleotide sequence of the LYS2 gene of Saccharomyces cerevisiae: homology to Bacillus brevis tyrocidine synthetase 1. Gene. 1991 Feb 1;98(1):141–145. doi: 10.1016/0378-1119(91)90117-t. [DOI] [PubMed] [Google Scholar]
- Pan T., Coleman J. E. GAL4 transcription factor is not a "zinc finger" but forms a Zn(II)2Cys6 binuclear cluster. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2077–2081. doi: 10.1073/pnas.87.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer K., Kim K. S., Kogan S., Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989 Jan 27;56(2):291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- Qui H. F., Dubois E., Messenguy F. Dissection of the bifunctional ARGRII protein involved in the regulation of arginine anabolic and catabolic pathways. Mol Cell Biol. 1991 Apr;11(4):2169–2179. doi: 10.1128/mcb.11.4.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos F., Dubois E., Piérard A. Control of enzyme synthesis in the lysine biosynthetic pathway of Saccharomyces cerevisiae. Evidence for a regulatory role of gene LYS14. Eur J Biochem. 1988 Jan 15;171(1-2):171–176. doi: 10.1111/j.1432-1033.1988.tb13773.x. [DOI] [PubMed] [Google Scholar]
- Ramos F., Wiame J. M. Mutation affecting the specific regulatory control of lysine biosynthetic enzymes in Saccharomyces cerevisiae. Mol Gen Genet. 1985;200(2):291–294. doi: 10.1007/BF00425438. [DOI] [PubMed] [Google Scholar]
- Reece R. J., Ptashne M. Determinants of binding-site specificity among yeast C6 zinc cluster proteins. Science. 1993 Aug 13;261(5123):909–911. doi: 10.1126/science.8346441. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze J. Y., Woontner M., Jaehning J. A., Kohlhaw G. B. In vitro transcriptional activation by a metabolic intermediate: activation by Leu3 depends on alpha-isopropylmalate. Science. 1992 Nov 13;258(5085):1143–1145. doi: 10.1126/science.1439822. [DOI] [PubMed] [Google Scholar]
- Tucci A. F., Ceci L. N. Homocitrate synthase from yeast. Arch Biochem Biophys. 1972 Dec;153(2):742–750. doi: 10.1016/0003-9861(72)90393-1. [DOI] [PubMed] [Google Scholar]
- Urrestarazu L. A., Borell C. W., Bhattacharjee J. K. General and specific controls of lysine biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1985;9(5):341–344. doi: 10.1007/BF00421603. [DOI] [PubMed] [Google Scholar]
- Wolfner M., Yep D., Messenguy F., Fink G. R. Integration of amino acid biosynthesis into the cell cycle of Saccharomyces cerevisiae. J Mol Biol. 1975 Aug 5;96(2):273–290. doi: 10.1016/0022-2836(75)90348-4. [DOI] [PubMed] [Google Scholar]
- Zhou K., Brisco P. R., Hinkkanen A. E., Kohlhaw G. B. Structure of yeast regulatory gene LEU3 and evidence that LEU3 itself is under general amino acid control. Nucleic Acids Res. 1987 Jul 10;15(13):5261–5273. doi: 10.1093/nar/15.13.5261. [DOI] [PMC free article] [PubMed] [Google Scholar]



