Abstract
A compelling question of how phospholipids interact with their target receptors has been of interest since the first receptor-mediated effects were reported. The recent report of a crystal structure for the S1P1 receptor in complex with an antagonist phospholipid provides interesting perspective on the insights that had previously been gained through structure–activity studies of the phospholipids, as well as modeling and mutagenesis studies of the receptors. This review integrates these varied lines of investigation in the context of their various contributions to our current understanding of phospholipid–receptor interactions. This article is part of a Special Issue entitled Advances in Lysophospholipid Research.
Keywords: S1P, LPA, GPCR structure, Structure–activity relationships
1. Introduction
Twenty years has passed since the late eighties, when the debate began whether lysophospholipids elicit their wide-ranging biological actions through receptors or by detergent-like actions. The first crystal structure of a lipid receptor, the sphingosine-1-phosphate type 1 receptor has now been solved. Twenty years only! Considering that the concept of cholinergic or adrenergic receptors preceded atomic resolution structures of these biomolecules by almost a hundred years, lysophospholipid receptor biology and structural characterization have outpaced the development of any other receptor field. In the early nineties, lysophosphatidic acid (LPA) was humorously called lytic precipitating artifact, while the low cellular level of sphingosine-1-phosphate made some investigators think that it was a trace lipid that could even be a purification artifact. Today, we all know that research has provided solid answers to these early dilemmas. There are five S1P and six LPA plasma membrane receptors that have been firmly established and several others await further characterization. The dilemma whether lysophospholipids work via plasma membrane GPCR or intracellular targets is settled and several cell surface and intracellular targets have been identified.
The interest in lysophospholipids is also fueled by the potential development of drugs that harness the biology of these lipids. The field has received a big impetus from the FDA approval of FTY720/Gilenya as first-line treatment for multiple sclerosis and the granting of orphan drug status to an LPA1 receptor antagonist, AM152 (now BMS-986202), for the treatment of idiopathic pulmonary fibrosis. This is just the beginning. Inhibitors of sphingosine kinases, autotaxin, and receptor antagonists are in the drug development pipeline.
In the present review we provide a selective review of the discovery process that provides the foundation for developing agonist pharmacons targeting lysophospholipid receptors. Our groups started tackling the problem of ligand recognition of lysophospholipid receptors in 1998 and we have been gratified last year when the crystal structure of S1P1 was unveiled confirming the two key residues R3.28 and E3.29 interacting with the polar headgroup of the ligand which we have predicted by computational modeling a decade earlier. This review begins with some of the indirect reflections obtained about the interactions between phospholipids and their receptors through investigations of the structure–activity relationships of phospholipid analogs, proceeds through more direct structural insights, and highlights some of the protein–protein interactions modulated by receptor activation.
1.1. S1P receptor family agonist SAR trends
Several key findings led to an explosive increase in the investigation of structure–activity relationships for members of the S1P receptor family. First among these was the linkage of the immunomodulatory action of FTY720 (fingolimod) with S1P receptor agonism after in vivo phosphorylation [1,2]. Since FTY720 was already in the human clinical trial pipeline [3], its connection to S1P receptors stimulated preparation and testing of a wide variety of phospholipid analogs and subsequently non-lipid and more drug-like molecules. Later discovery that the long-lasting removal of S1P1 from the cell surface occurs after stimulation by FTY720-phosphate (functional antagonism [4]) and is responsible for lymphocyte trafficking effects [5] stimulated consideration of antagonists, as well as agonists. Non-lipid molecules with S1P receptor activity appeared first in the patent literature (reviewed in [6]) and subsequently in peer-reviewed journals. These non-lipid molecules have not often been linked with specific binding sites through mutagenesis studies or confirmation of strictly competitive binding modes. In contrast, the wealth of SAR data generated using S1P analogs and a subset of the non-lipid agonists that have been investigated in the contexts of both wild type and mutated receptors provide valuable indirect information about phospholipid–receptor interactions.
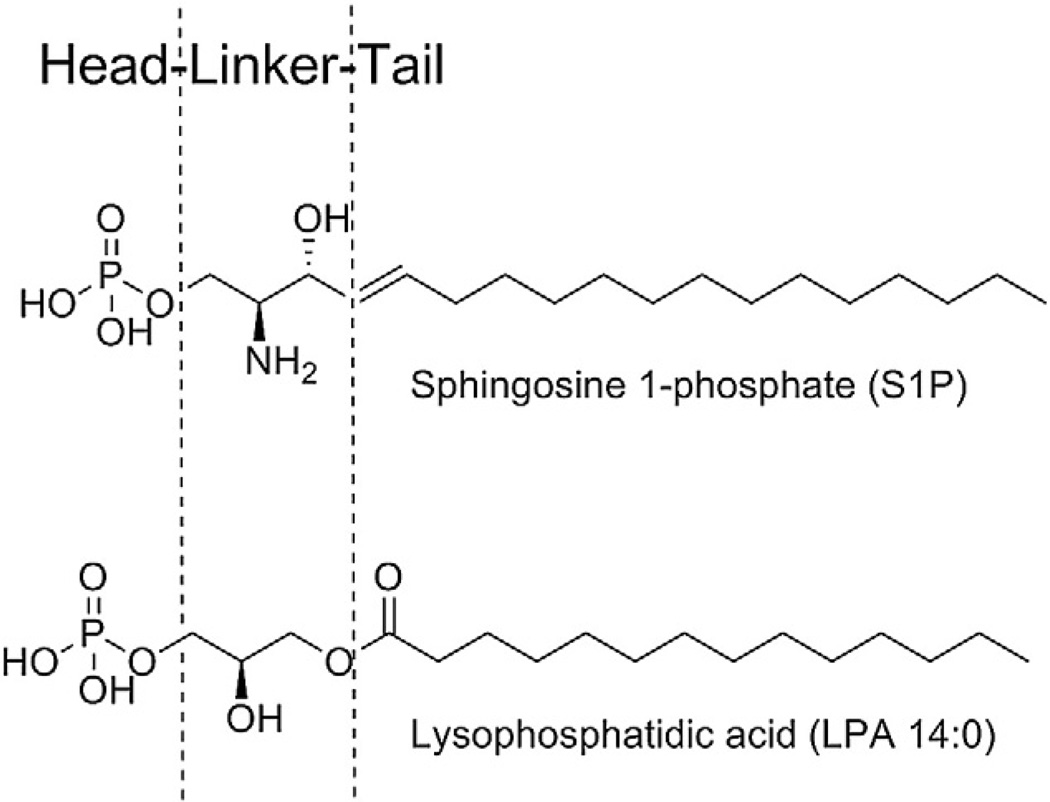
The SAR data for S1P analogs and select non-lipid agonists can be analyzed based on an arbitrary division of these analogs into three pieces, a polar headgroup, a linker, and a tail, as shown for S1P in Fig. 1. Using this framework, S1P has a phosphate headgroup, a 2-aminopropanol linker, and a fifteen carbon tail with a trans double bond near the linker region.
Fig. 1.
Arbitrary division of S1P and LPA phospholipid analogs into headgroup, linker region, and tail for SAR analysis.
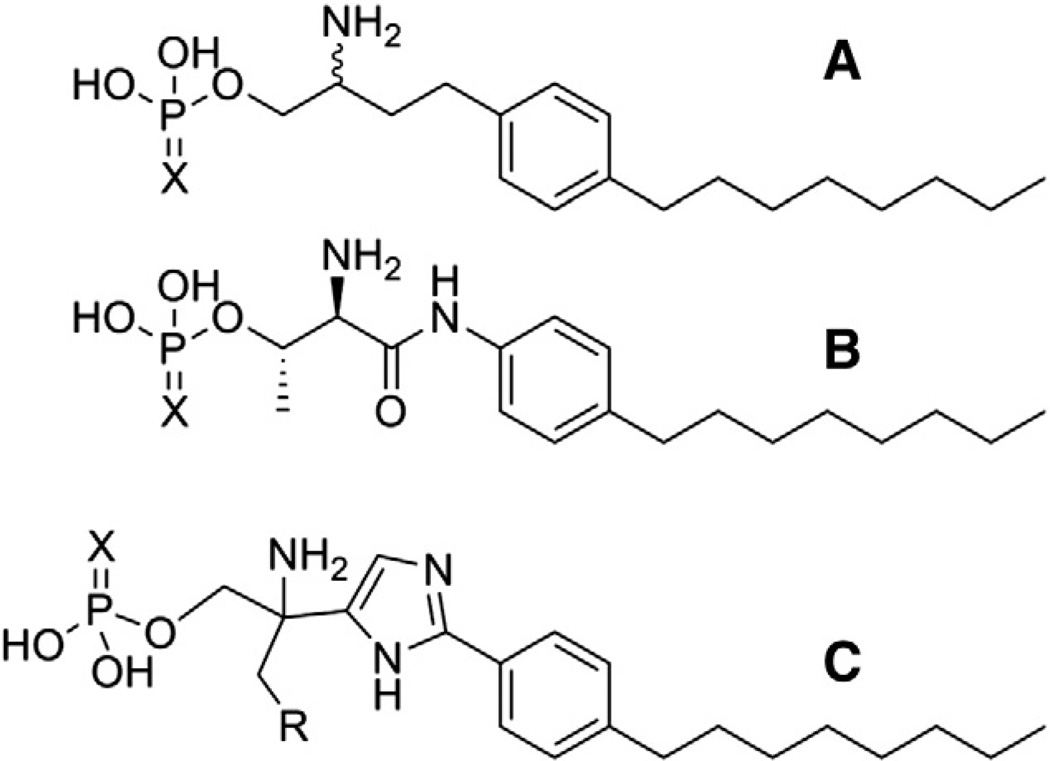
Headgroup changes have been investigated by several research groups in the context of different linker and tail combinations. In one of these studies, 2-amino propane linkers and para-substituted nonyl phenyl methylene tails were used to investigate a range of headgroups including phosphate, phosphonate, alpha-hydroxyphosphonate, sulfonate and carboxylate [7]. Among these headgroups, the carboxylate and sulfonate were poorly tolerated at S1P1, S1P3, S1P4 and S1P5 (S1P2 data not reported) with the smallest potency shift relative to the phosphate headgroup observed for the methylene carboxylate analog at S1P1 (100-fold difference). Replacement of phosphate with methylene phosphonate reduced potency at all receptors tested. Selectivity for S1P1, however, was enhanced due to a more limited 10-fold potency difference at this receptor relative to the 50 to 400-fold potency differences at S1P3–5. The inclusion of a hydroxyl group on the alpha carbon of the phosphonate, in contrast, leads to improved potency at S1P1 relative to the phosphate headgroup, while still showing potency reductions of 5 to 85-fold at the other receptors tested. A second study utilized 2-amino ethane as the linker, and connected the para-substituted nonyl phenyl methylene tail directly to the amino group as the context in which to investigate headgroup preferences [8]. The range of headgroups tested included phosphonate, phosphinate, carboxylate, and both alpha and beta fluorine or hydroxyl-substituted carboxylates. The analog in this series with the methyl phosphonate headgroup showed the best potency at all receptor subtypes. As in the previous series, the methylene carboxylate was best tolerated by the S1P1 receptor (40-fold worse than methylene phosphonate), but all substitutions of hydroxyl groups or fluorine atoms alpha or beta to the carboxylate group further decreased potency [8] in contrast to the increased potency observed for similar substitutions alpha to a phosphonate headgroup [7]. Two different publications investigated S1P receptor preferences between phosphate and thiophosphate head groups using the three linker–tail combinations shown in Fig. 2 [9,10]. S1P1,2,5 showed a preference for phosphate over thiophosphate regardless of linker–tail combination, with the single exception that the two headgroups in the context of scaffold B were equipotent at the S1P1 receptor. The S1P3 receptor also preferred the phosphate headgroup in the context of all scaffolds except B, although only partial agonism at S1P3 was observed for both analogs. The S1P4 receptor also showed a context-sensitive headgroup preference, in which the phosphate headgroup was preferred except for scaffold C when R = OH, although partial agonism was observed for this pair of analogs. The trends in all these studies suggest that anionic headgroups with charges delocalized over three atoms are better able to activate all S1P receptors than anionic headgroups with reduced overall charge, or charge shared only by two atoms as in the carboxylate headgroup. Highly delocalized negative charge is not an absolute requirement, however, as highly potent non-lipid molecules with carboxylate headgroups [11–18] as well as uncharged trifluoromethyl [19,20] and other headgroups [21–25] combined with a variety of aromatic and heteroaromatic linker/tail combinations have been reported. The headgroup pocket also seems to be limited in size, as the sulfur atom in the thiophosphate is substantially larger than the corresponding oxygen atomin a phosphate group. These SAR findings are consistent with prior identification of multiple cationic ion-pairing partners by modeling-driven mutagenesis studies [26,27].
Fig. 2.
Linker–tail combinations used to investigate S1P receptor preferences between phosphate (X = O) and thiophosphate (X = S) headgroups. R = H or OH.
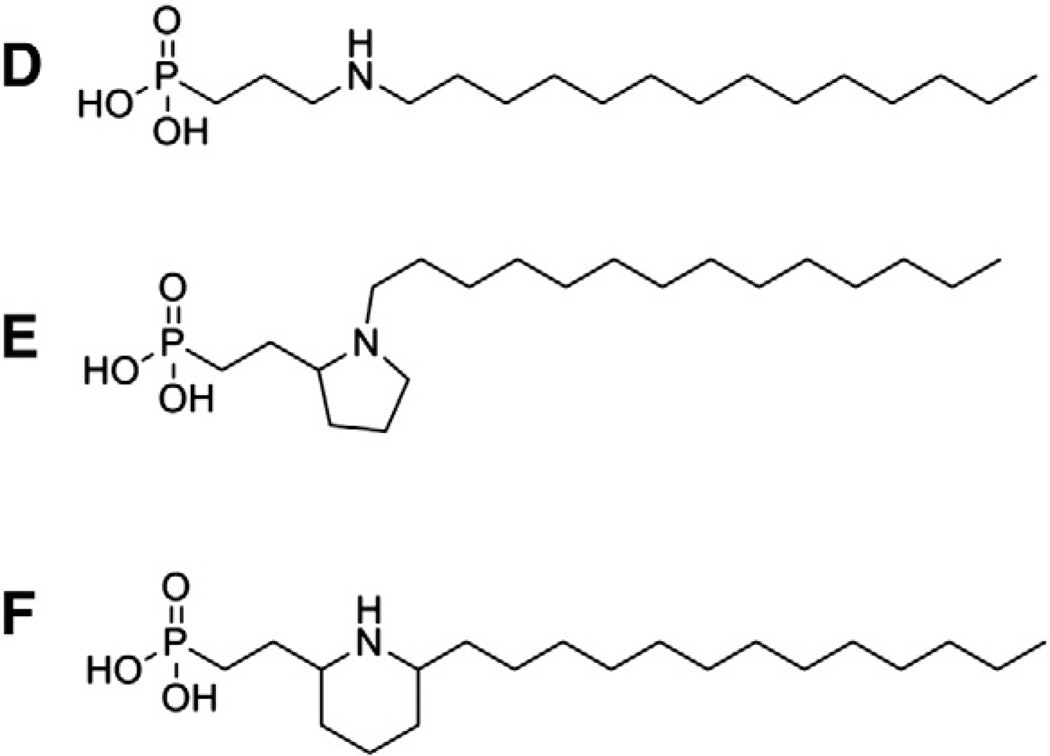
The S1P receptors tolerate a broad selection of linkers, including saturated and unsaturated hydrocarbons with a variety of functional groups [1,7,8,10,28], secondary amines connecting the linker to the tail [8,29], amides [10,30–33], and a selection of aromatic heterocycles [9,34–36]. The single site mutation of the conserved glutamate residue at position 3.29 to glutamine (E3.29Q) is sufficient to confer tolerance of glycerol as a linker [37,38], though the wild type receptors require an ionizable nitrogen in all of these linker types. As with the preferences noted for highly delocalized negative charges in the headgroup, this requirement is also not absolute when combined with substantial changes in the headgroup and tail [39]. In this review, we will focus on one of these SAR studies, as it provides interesting insights into differences in phospholipid recognition in the S1P receptor family. Fig. 3 shows a subset of conformationally-constrained molecules used to define the agonist linker shape preferred by the various S1P receptors [29]. The unconstrained compound, D, showed potencies of 3.7, 580, 3.6, 140, and 13 nM at S1P1–5, respectively. Compound E, with a sharp bend induced by the pyrrolidine ring, showed poorer potencies at all receptors than did compound D. However, the S1P4 receptor showed only a 6.2-fold increase in IC50 in the radioligand binding assay relative to more than 20-fold for S1P1, S1P3, and S1P5. Binding was not detected at S1P2. This finding matches well with comparative modeling of all five S1P receptors which concluded that the S1P4 receptor binding pocket requires ligands to bend sharply near the ammoniumfunctional group [40]. Two isomers of compound F (cis and trans geometries of substituents on the piperidine ring) highlight additional geometry differences in the S1P receptor binding pockets. S1P1 and S1P4 showed several fold better affinity for the trans isomer over the cis isomer. S1P2 and S1P5 showed several fold better affinity for the cis over the trans isomer. Both isomers showed similar affinities for S1P3. The cis isomer can adopt more extended conformations, in contrast to the trans isomer which bends at the ring. As noted previously, the S1P4 receptor showed the most pronounced bend in the binding pocket in modeling studies. These data suggest that S1P1 can also accommodate mildly bent shapes, but that S1P2 and S1P4 prefer more linear ligand geometries. The ability of S1P3 to recognize both bent and linear shapes demonstrates why achieving S1P1-selective agonists that lack the bradycardia side effect mediated by S1P3 action has been achieved by large jumps in structural space from the flexible phospholipid agonist analogs of S1P [11,13,17–19,21,22,25,36,41,42].
Fig. 3.
Examples of conformationally-constrained linkers used to define S1P receptor binding pocket shapes.
SAR studies focused on the hydrophobic tail have defined the preferred length, tolerance for aromatic insertion, and preferred position of aromatic insertions in several headgroup-linker combinations. The linkage of FTY720 with S1P receptors provided the first evidence that the S1P2 receptor does not tolerate aromatic ring insertions immediately adjacent to 2-aminopropanediol linker [1]. This finding extended to most analogs with amide linkers [30–32] as well as secondary amine linkers [8,36]. Interestingly, the aromatic substitution pattern (ortho versus meta versus para) plays an important role in ligand function, with meta-substituted aromatic rings positioned adjacent to the linker showing antagonist activity at S1P1 and S1P3 [31]. This functional impact is discussed further in the context of the S1P1 crystal structure. Aromatic ring position has a tremendous impact on receptor activation profiles, with S1P2 activity evident for analogs in which the aromatic ring is placed at or near the end of the tail region, S1P3/S1P4 potency showing an optimum when the aromatic ring appears near the middle of the hydrophobic tail, and S1P1/S1P5 showing about a ten-fold preference for the aromatic ring to be adjacent to the linker [8]. An atomistic explanation for the anti-S1P2 selectivity of the analogs with aromatic rings adjacent to the linker was difficult to find, as it stems not from any single amino acid substitution, but from differences in the amino acid sequence in the first intracellular loop, located quite distant from the ligand binding pocket [43]. Extensive molecular dynamics simulations indicate that the differences in the intracellular loop sequence impact the mobility of the hydrophobic tail in a manner dependent on the presence or absence of an aromatic ring. The anti-S1P2 selectivity is therefore due to binding entropy differences, rather than differences in binding enthalpy. Chain length preferences have been investigated using saturated linkers [8], amine linkers [8], and amide linkers [32]. These studies all suggest that the majority of S1P receptors are optimized for lengths similar to S1P (18 heavy atoms extending from the phosphate headgroup to the end of the tail) although the S1P5 receptor shows the best retention of potency with shorter chain lengths. A preference for lengths similar to that of S1P was also identified for the S1P4 receptor E3.29Q mutant using LPA species of varying chain lengths, with a preference for the 14-carbon tail and a four-atom glycerol linker [38].
1.2. LPA receptor family agonist SAR trends
Important biological roles for LPA were discovered much earlier than those for S1P, with several hundred publications related to LPA appearing in the literature prior to 1985, and only about ten related to S1P in the same time period. The structure of LPA additionally facilitates SAR explorations due to the natural occurrence of multiple LPA species that differ in the fatty acid linked to the sn-1 position of the glycerol linker (Fig. 2 shows LPA with a 14:0 fatty acid). SAR studies of phospholipid analogs of LPA therefore began earlier and have already been thoroughly reviewed, although most of these studies were limited to examination of the EDG-family LPA receptors, LPA1, LPA2, and LPA3 [44,45]. These reviews note that changes in the phosphate headgroup are, in general, poorly tolerated. Only alpha-hydroxy substituted phosphates and thiophosphates had been incorporated into active EDG LPA receptor agonists. These headgroups have opposing selectivities among LPA receptors, with the alpha-hydroxy substituted phosphonates activating LPA1 and LPA2, and the thiophosphate activating LPA3. A more recently published set of alpha-fluorophosphonate analogs also show selectivity among the EDG LPA receptors [46], but with greater similarity to the LPA3 selectivity of the thiophosphate headgroup; than to the LPA1/LPA2 selectivity of the more structurally similar alpha-hydroxyphosphonates. Although the alpha-fluoro phosphonate and thiophosphate analogs of LPA proved to be selective agonists, alpha-fluoro phosphonate and thiophosphonate analogs of cyclic phosphatidic acid generally showed antagonist activity at LPA1 and LPA3 [47,48]. The unsubstituted phosphonate analogs of cyclic phosphatidic acid (carba-cyclic phosphatidic acids) showed no activity at LPA1–4, but proved to be partial agonists of LPA5 with Emax values of 58 and 71% for the two regioisomers [49]. A final set of phosphonates in which methylene phosphonate is attached to the sn-3 oxygen of glycerol (methylene-extended) proved to be full to partial agonists of LPA1, LPA2 and LPA4, but antagonists of LPA3 [50]. The strict limitations on the headgroup were further validated for the EDG LPA receptors and extended to the non-EDG LPA4 receptor with the finding that two uncharged biomimetics of the phosphate group, trifluoromethanesulfonamide (−NHSO2CF3) and trifluoromethanesulfonamidoxy (−ONHSO2CF3) replacements for phosphate did not produce LPA analogs capable of activating LPA1–4 [51]. These recent additions to the headgroup SAR for the LPA receptors highlight the sensitivity of these receptors to even small changes in headgroup structure, size, and charge.
LPA analog linker SAR has been previously reviewed [44,45]. In summary, LPA receptors have a relatively high tolerance for glycerol with varying numbers and positions of tails and neutral ethanolamines linked into amide functional groups to attach the tail in the linker region. Interestingly, the LPA2 receptor can be activated by a variety of linker-free fatty alcohol phosphates, many of which show weak antagonism of LPA1 and LPA3 [52,53]. The linker, therefore, plays an important functional role in the EDG LPA receptors.
More recently published SAR data has focused largely on the non-EDG LPA receptors, and changes in the hydrophobic tails. The LPA5 receptor is unique in showing a preference for the hydrophobic tails connected to the linker by an ether rather than ester functional group (alkyl-glycerol phosphates, AGP), although AGP is tolerated at the EDG LPA receptors and equipotent with LPA at LPA4 [49,54]. The LPA6 receptor is analogous to the LPA3 receptor in its preference for the hydrophobic tail placement at the central sn-2 position of glycerol, rather than the terminal sn-1 position [55]. This finding is in contrast with results for LPA1 and LPA2, in which little preference between the regioisomers was observed [55,56]. Many short-chain phosphatidic acids with two tails had previously been shown to act as antagonists at members of the EDG LPA receptor subfamily, but display partial agonist activity at LPA5 [54,57].
1.3. Inferred similarities and differences between LPA and S1P recognition
The SAR trends for the S1P and LPA receptor families suggest both similarities and differences in phospholipid recognition. SAR trends all demonstrate a relatively limited tolerance for changes to the phosphate headgroup relative to the natural ligands. Mutagenesis studies done in multiple LPA [49,58,59] and S1P [26,27] receptors as well as the recently published crystallographic structure of the S1P1 receptor [60] allow this limited tolerance to be attributed to the presence of multiple cationic amino acids in close proximity to each other that must be balanced by the distributed negative charge of a headgroup quite similar in size, charge, and charge distribution to a phosphate group. A substantial contribution to phospholipid selectivity between the LPA and S1P receptor families centers on selective recognition of a cationic ammonium group in the linker region of active S1P receptor agonists that is absent in LPA receptor agonists. Mutagenesis studies have demonstrated that S1P receptors select for ammonium-containing phospholipids using an anionic glutamate residue in the third transmembrane domain that is absent in all LPA receptors [37,38].
1.4. An overview of the structural puzzle pieces
1.4.1. Crystal structure of S1P1
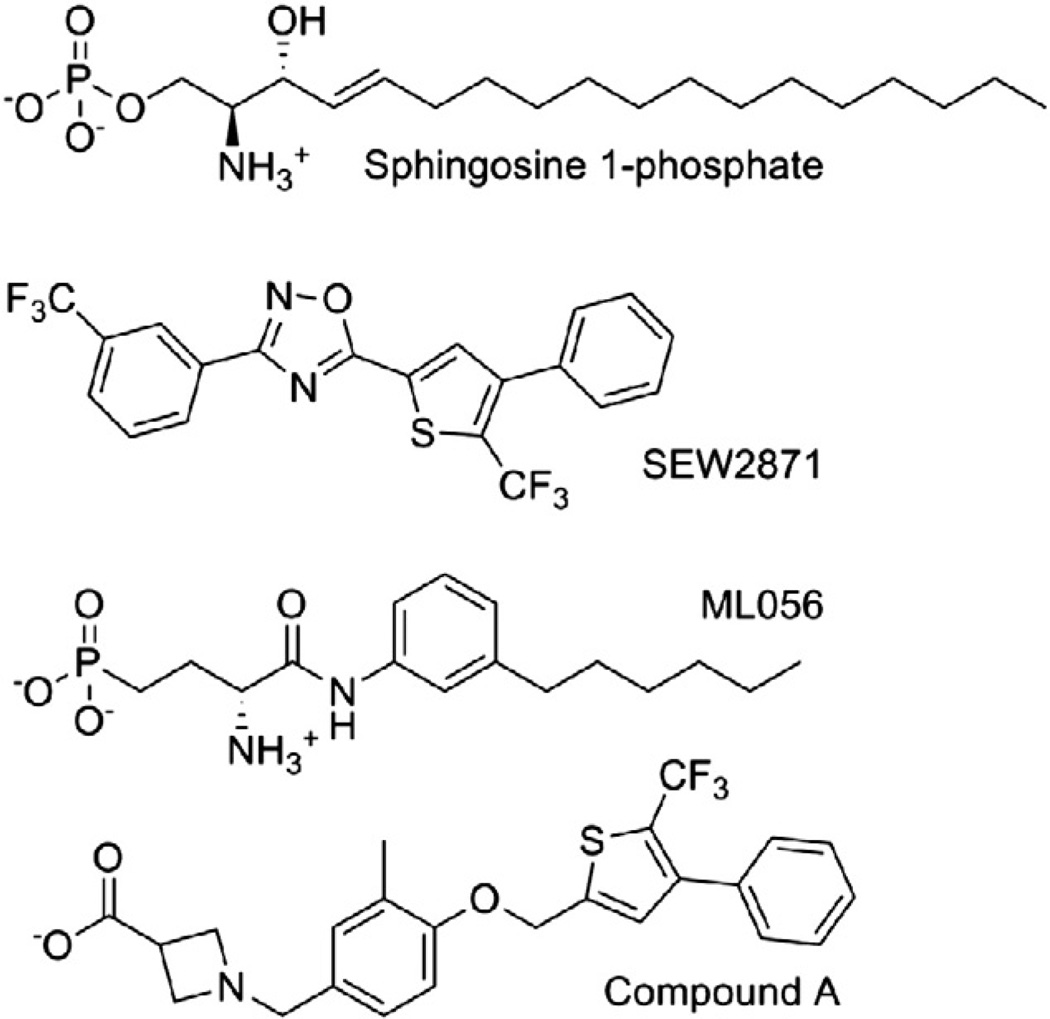
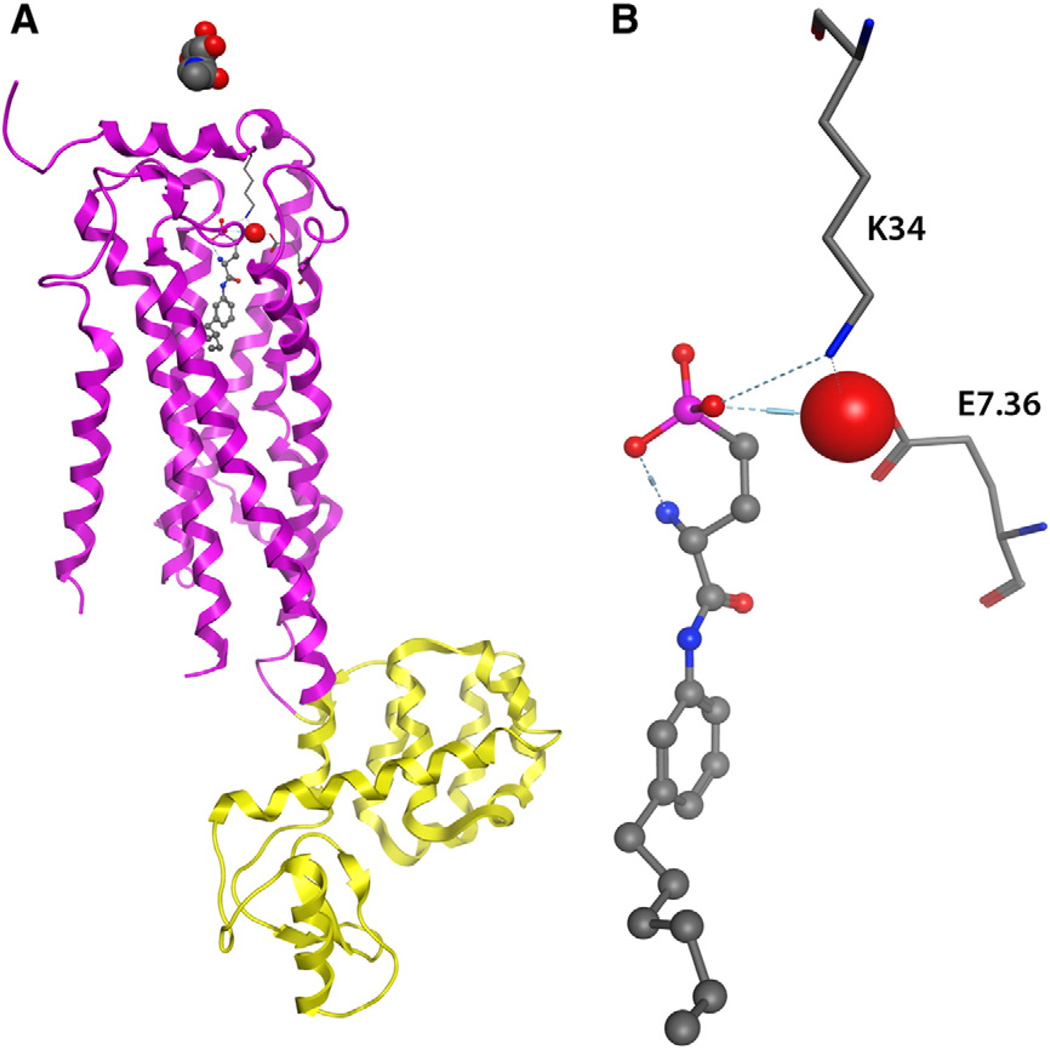
The year 2012 marked the first published and publically available crystal structures of a phospholipid GPCR, the S1P1 receptor in complex with a phospholipid antagonist, ML056 (ML056 structure in Fig. 4) [60]. The crystallized construct is another in a series of GPCR structures that have proven amenable to crystallization after replacement of the third intracellular loop (IL3) with T4 lysozyme, as pioneered in the solution of the structure of the β-2 adrenergic receptor [61]. In the S1P1 receptor, IL3 residues 232–244 were replaced by T4-lysozyme, and therefore are not represented in the structure. Additionally missing due to poorly defined electron density are either 15 or 16 N-terminal amino acids (16 in 3V2W and 15 in 3V2Y), as well as residues 40–46 leading into the first transmembrane segment (3V2W only). The reduced number of missing amino acids in the 3V2Y structure is due to the application of enhanced data assembly methods in processing the same diffraction data. An N-acetylglucosamine molecule is associated with the extracellular surface of the receptor, and one crystallographic water molecule forms bridging interactions between the phosphate headgroup of ML056 and residues in the amino terminus and TM7. Fig. 5 shows these features of the S1P1 crystal structure (PDB ID: 3V2Y [62]).
Fig. 4.
Structures of S1P, SEW2871, ML056, and compound A.
Fig. 5.
S1P1 crystal structure. Panel A: Protein structure rendered in ribbons with residues from S1P1 colored magenta and those from T4 lysozyme colored yellow. N-acetylglucosamine and the water mediating ligand–receptor interactions are shown as spacefilling models. ML056 is shown using ball & stick and select receptor residues are shown using sticks. Panel B: Close up of water-mediated ligand–receptor interactions.
The S1P1 structures provide atomistic experimental confirmation of several phospholipid interactions previously predicted by modeling studies and supported by mutagenesis experiments. In particular, the 3.28R/3.29E motif in the third transmembrane segment conserved in S1P1–5 was observed in the crystal structure to form ion-pairing interactions with the phosphate (R3.28) and ammonium (E3.29) functional groups of ML056. These interactions are directly analogous to the observed interactions with the phosphate and ammonium groups of S1P (structure in Fig. 4) in modeling studies [26,27,37–40,63–66]. These interactions result in phosphate group placement within the transmembrane helical bundle at a depth analogous to the membrane:water interface surrounding the receptor. Additionally, the hydrophobic tail of the bound phospholipid extends deeper into the transmembrane helical bundle, rather than extending upward toward the extracellular loops of the receptor.
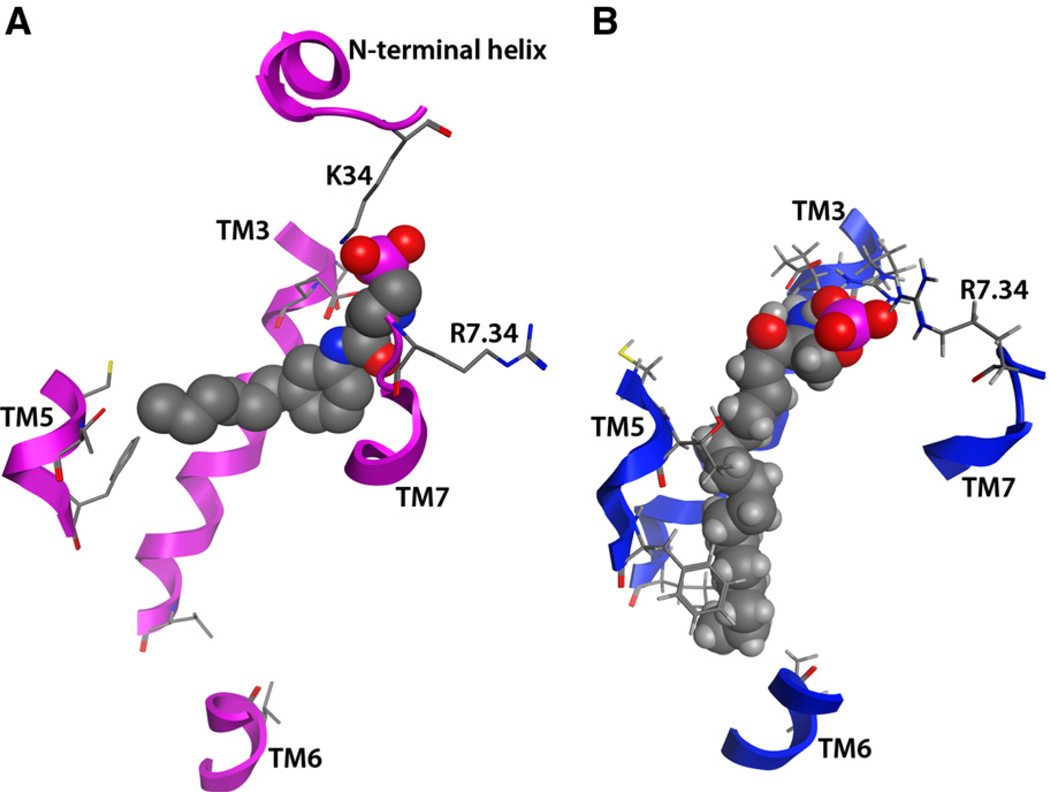
The crystal structure does demonstrate substantial differences in phospholipid:receptor interactions relative to prior modeling studies, which may be attributed either to differences in binding modes between the antagonist crystallized and the agonists investigated in modeling studies, or may indicate errors in the modeling and interpretation of mutagenesis results. These two differences include the hydrophobic tail position and the identity of the second ion-pairing partner for the phosphate head group of the bound phospholipid. Fig. 6 highlights these differences in a side-by-side comparison of the crystallographic complex between S1P1 and ML056 and our modeled complex between S1P1 and S1P.
Fig. 6.
Phospholipid interaction comparison between the S1P1 crystallographic complex with ML056 (Panel A) and an S1P1 modeled complex with S1P (Panel B). Portions of TM3, TM5–7, and the N-terminal helix are shown for reference, and other protein segments are hidden to allow clear depiction of ligand–receptor interactions.
The hydrophobic tail of ML056 in the crystal structure [60] makes a sharp bend toward TM5, passing underneath F125(3.33) and ending near C206(5.44), T207(5.45), F210(5.48) and L276(6.55). In contrast, the hydrophobic tail of S1P in the model complexes reported by ourselves [67] and others [39,65,66] extends in a more linear fashion into the TM bundle from the polar headgroup, ending near V6.40 and L3.43 [67] or near L6.41 and L3.43 [66]. Both mutagenesis [65,67] and structure–activity relationship (SAR) results suggest that differences in the hydrophobic tail position may reflect real differences that differentiate agonists from antagonists. In particular, the C5.44D mutant of S1P1 displayed normal responses to S1P [67],which would not be expected if the hydrophobic tail of S1P occupied the same location as that of ML056 as that would represent an entropically favorable interaction type that should lead to decreased responsiveness to S1P. S1P was also minimally affected by F3.33Y and F3.33A mutations [65], even though F3.33 makes van der Waals contact with five atoms in the hydrophobic tail of ML056, which would be lost upon mutation to alanine and would be expected to showed impaired activity if similar interactions with S1P occur. Interestingly, F3.33A mutants were poorly activated by agonists with aromatic moieties, indicating that this residue differentially contacts ligands of different types [65]. Another residue showing a variable role in ligand binding as a function of ligand structure was identified by the Deng et al. [66]. These studies demonstrated that mutations of L6.55F in S1P1 and F6.55L in S1P3 had little impact on S1P binding, but reversed the S1P1/S1P3 selectivity of a non-lipid agonist, compound A (structure in Fig. 4). In addition to these mutations to residues that do interact with ML056 in the crystallographic complex, mutations at sites that do not interact with ML056 also support different binding modes for S1P and ML056. In particular, the V6.40L mutant of S1P1 showed no response to S1P (although mutations to smaller amino acids were tolerated) [67], suggesting that the increased size of leucine compared to valine blocked a part of the pocket occupied by the hydrophobic tail of S1P. Interestingly, the V6.40L mutant showed reduced response to SEW2871 (Emax at 3 µM = 34.8% and structure in Fig. 4) [67], indicating that the receptor is not grossly misfolded, and a slightly shorter agonist retains some ability to activate the receptor in the presence of the larger sidechain at position 6.40. In contrast, V6.40 is very distant (>17 Å) from any atoms of ML056 in the crystal structure. Finally, SAR data has demonstrated that longer tails on meta-substituted compounds similar in structure to ML056 as well as their para-substituted analogs act as agonists, rather than antagonists [31]. These data in aggregate suggest that the short hydrophobic subpocket angling toward TM5 is likely to allow binding, but does not allow bound ligands to stabilize the active conformation of the receptor. In contrast, the longer and more linearly arranged subpocket that would be occupied by longer analogs and para-substituted analogs must be occupied in order to observe receptor activation. The crystal structure itself clearly includes not only the hydrophobic pocket occupied by the tail of ML056, but also another hydrophobic subpocket similar to the more linear arrangement observed for the S1P hydrophobic tail in modeling studies [68]. Thus the differences in hydrophobic tail location are likely to represent differences that can be used in the rational optimization of agonists versus antagonists.
A second substantial phospholipid interaction difference between the S1P1 crystal structure and previous modeling studies is the identity of the second ion-pairing partner for the phosphate group. ML056 interacts with K34 from the N-terminal helical cap that covers the bound phospholipid in the crystal structure. In contrast, modeling and mutagenesis studies in our group identified R7.34 in TM7 as the second ion-pairing partner for the bound phosphate group of S1P [26,63], although other modeling studies did not suggest any additional ion-pairing partner [39,64–66]. It is worth mentioning that the S1P1 model used in all of these modeling studies did not include N-terminal residues, and therefore could not consider K34 as a potential ion-pairing partner. In support of the our model-derived hypothesis of a direct role for R7.34 in phospholipid binding, the R7.34A mutant of S1P1 showed no ability to bind 1 µM S1P in a radioligand binding assay, and showed no response to 1 µM S1P in a [35S]GTPγS binding assay. Yet the sidechain of R7.34 is located almost 12 Å from the phosphate group of ML056 in the crystal structure. The apparent discrepancy between these data can be attributed either to a ligand-specific and differing role for R7.34, potentially dependent on whether ligands act as agonists or antagonists. An alternative explanation is that R7.34 plays a crucial role in ligand recognition without making direct contact in the final complex. This latter explanation is intriguing due to the location of R7.34 in the crystallographic structure, where the cationic sidechain sits at the top of a gap between TM1 and TM7, suggested to serve as the phospholipid entry and exit pathway between the binding pocket and the surrounding membrane [60]. It is therefore possible that R7.34 is responsible for recruiting phospholipid binding partners from the surrounding membrane, and that its replacement with a non-polar amino acid results in no recruitment event, even if the final complex stability would be unchanged.
Several additional features of published models are interesting to review in the context of the S1P1 crystal structure with ML056. A hydrogen bond between Y98(2.57)was indicated in one modeling study [64], but the role of this residue was suggested to stabilize the polar portion of the binding pocket through a direct interaction with R3.28 in a different modeling study [65]. The latter study experimentally demonstrated that the Y2.57F mutation showed wild-type responses to S1P and other agonists, in contrast to the Y2.57A mutant, which showed impaired responses to all agonists tested with the exception of FTY720-phosphate. This finding seemed to rule out a hydrogen bond between Y2.57 and agonists. The crystal structure [60] demonstrates neither role for Y2.57, which is positioned 9 Å below the sidechain of R3.28 but does form a hydrophobic interaction with V301(7.43) that may contribute to stabilizing the shape of the hydrophobic part of the binding pocket, and makes van der Waals contact with the aromatic ring of ML046 (<4 Å).
1.4.2. NMR structure of first extracellular loop of S1P4
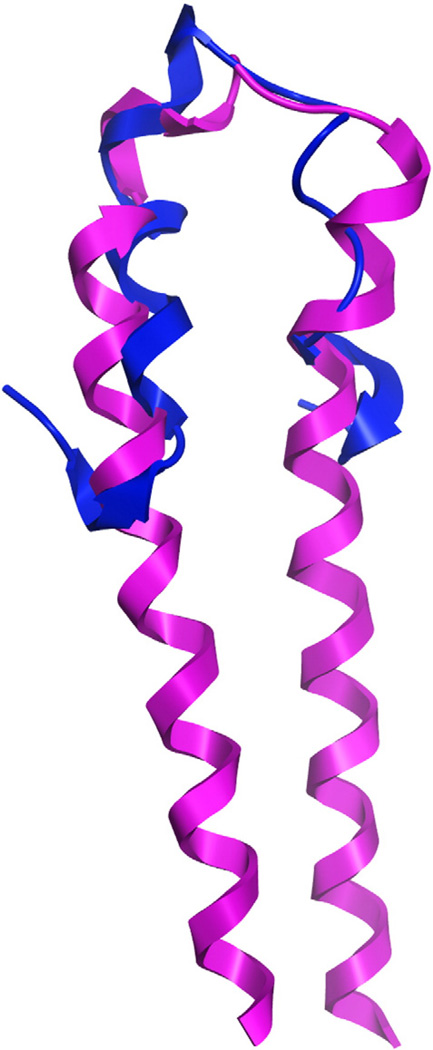
The first atomic-resolution experimental structure that shed light on GPCR–phospholipid interactions was reported five years prior to the report on the S1P1 crystal structure, but was limited to describing the structure of only the first extracellular loop (EL1) of the S1P4 receptor [69]. This study utilized NMR spectroscopy to characterize the structure of a synthetic peptide which included the sequence of the S1P4 EL1 in between two segments of an antiparallel coiled-coil as a folding constraint. The synthetic peptide retained the R3.28/E3.29 motif that had been demonstrated by mutagenesis to be required for phospholipid binding and activation in S1P4 [27]. The structure of S1P4 EL1 includes a short 310 helical segment for residues R109, T110, and F111 at the N-terminal end of the loop. These residues correspond to residues T108, T109, and Y110 in the S1P1 crystal structure, which also form a helical secondary structure (Fig. 7). Notably, ligand titration studies using O-phosphoethanolamine (a mimic of the S1P headgroup) but not N-acylethanolamine (a mimic of the ceramide headgroup) showed chemical shift changes localized to E3.29 and residues in the flexible C-terminal end of the loop (residues 109–116).
Fig. 7.
NMR solution structure of synthetic S1P4 EL1 peptide (PDB ID: 2DCO, blue ribbon) superposed on the crystallographic structure of S1P1 (PDB ID: 3V2Y, magenta ribbon, TM2–3 segment only).
1.5. Extending the structural puzzle pieces to new GPCR
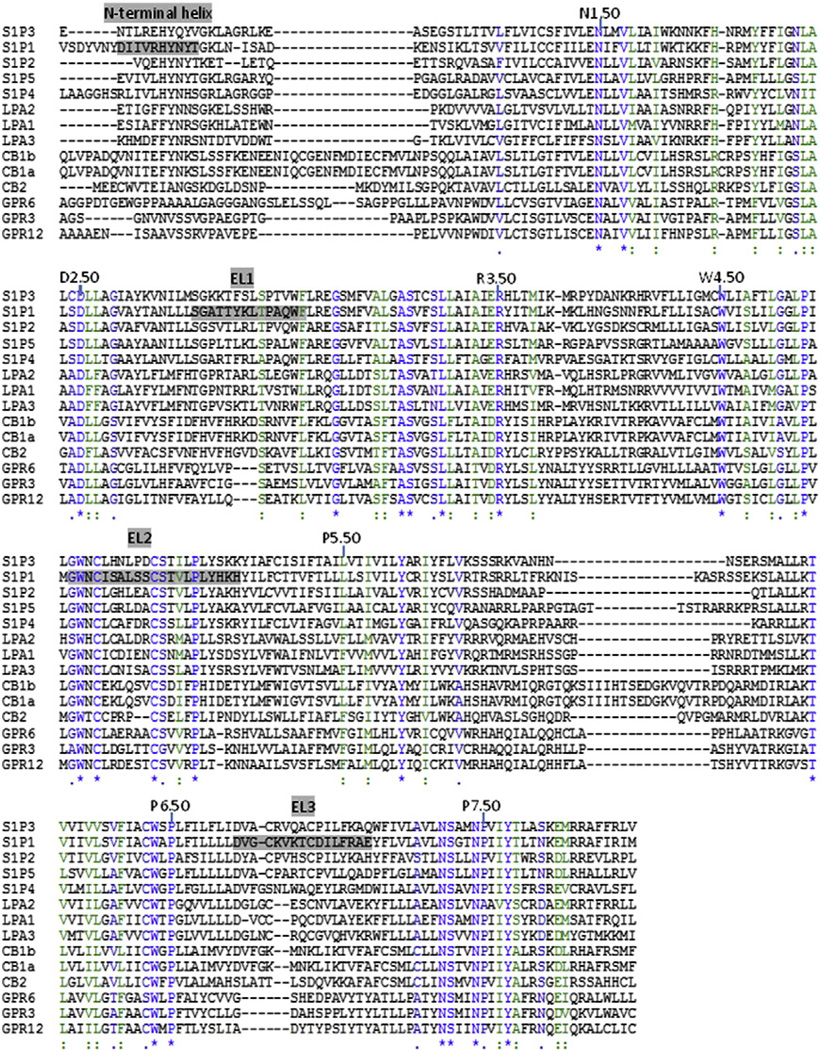
The crystallographic structure of S1P1 represents the first experimental structure of a phospholipid receptor, an important representative of a poorly-characterized segment of GPCR sequence space. S1P1 shares little sequence identity with the more than ten other class A GPCR that have been crystallized (<22%). Several subsets of the class A GPCR that have been documented to respond to a variety of lipid ligands have much higher sequence homology with S1P1 than with any other currently available class A GPCR crystal structure, including the remaining S1P receptors (S1P2–5), the EDG LPA receptors (LPA1–3), the cannabinoid receptors (CB1–2), and the GPR3/6/12 cluster (alignment shown in Fig. 8). Comparison of the NMR structure of a peptide mimetic of EL1 from the S1P4 receptor to the crystallographic structure of S1P1 shows that a short helical segment in this loop (residues GATT in the S1P1 EL1 and GART in the S1P4 EL1) is conserved in two different lipid receptors. It is useful, therefore, to compare sequence features in a larger set of aligned lipid receptors that correspond with interesting structural features in the S1P1 structure in order to suggest which structural features are likely to be observed in other receptors.
Fig. 8.
Alignment of human lipid receptor sequences. Strictly conserved residues are shown in bright blue (marked with * under the alignment). Residues with strong or weak conservation within groups of receptors are colored green (marked with :) and dark blue (marked with .), respectively. Alignment is truncated at both N-terminal and C-terminal ends to exclude residues corresponding to unresolved segments of the S1P1 crystal structure (prior to residue 16 and after residue 326 of S1P1).
1.5.1. Disulfide bonding pattern analogies
The S1P1 receptor crystal structure exhibits a unique disulfide bonding pattern among the currently available class A GPCR crystal structures. This pattern involves two intraloop disulfide bonds, one involving the two cysteine residues in the second extracellular loop (EL2) and the other involving the two cysteine residues in EL3. Fig. 8 shows that the disulfide-bonded cysteine residues in EL2 of S1P1 are strictly conserved in these lipid receptors, although S1P4, LPA1–3, and CB2 have an additional cysteine. Fig. 8 also shows that the disulfide-bonded cysteine residues in EL3 of S1P1 show much more limited conservation only in S1P1–3,5 and LPA1–3. Several of these lipid receptors completely lack cysteine residues in EL3, particularly S1P4, GPR3/6/12, and the cannabinoid receptors. An additional cysteine residue is present in EL3 of LPA1. These comparisons suggest that most of the lipid receptors will share the intraloop disulfide bond found in EL2 of S1P1, with the possible exception of receptors exhibiting an additional cysteine residue in that loop. However, the majority of the lipid receptors cannot be assumed to exhibit the same disulfide bond in EL3 that was observed in S1P1, and further studies will be needed to identify the role disulfide bonds play in determining structural differences among the extracellular regions of these lipid receptors.
1.5.2. N-terminal helical cap analogies
The S1P1 crystal structure exhibits a unique extracellular architecture relative to the other crystallized class A GPCR [60,68]. This architecture includes a helical cap formed from residues 23–32 in the N-terminus. The N-terminal helix includes asparagine 30, which is N-glycosylated in the crystallized construct. Fig. 8 demonstrates that N-glycosylation is not possible in the GPR3/6/12 subset of lipid receptors, due to the absence of asparagine or glutamine as found in the remaining receptors. Tyrosine 29 in the N-terminal helix forms a hydrogen bond with W117 at the C-terminal end of EL1. This pair of residues is conserved in the entire EDG family (S1P1–5 and LPA1–3). Isoleucine 25 of the N-terminal helix sits in a hydrophobic pocket formed by P114 at the C-terminal end of EL1 and L197 near the C-terminal end of EL2. Isoleucine 25 corresponds to branched hydrophobic sidechains in the EDG receptors, with the exception of the linear hydrophobic methionine residue found in LPA3. The EL2 interaction site is highly conserved in the EDG receptors, with the only substitution of isoleucine for leucine appearing in LPA3. The EL1 interaction site is less well-conserved, with the proline residue appearing in the S1P receptors and other hydrophobic residues appearing in the LPA receptors. Given these conserved interaction types, it is likely that the entire set of S1P and LPA receptors may exhibit a similar N-terminal helical cap. It is unlikely that this structural feature generalizes into the cannabinoid or GPR3/6/12 subsets.
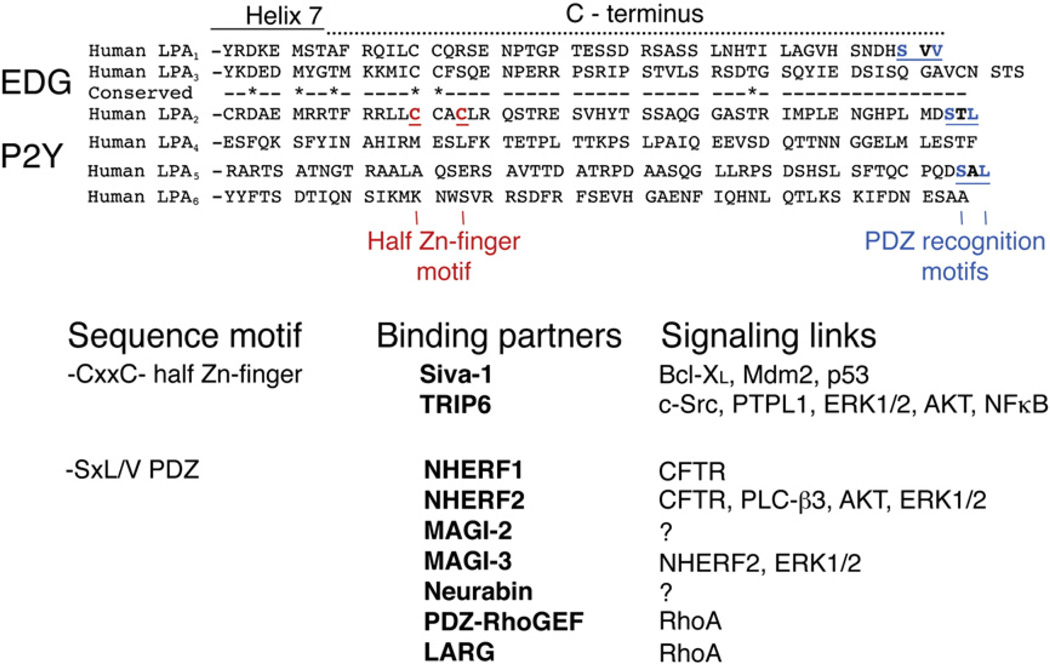
1.6. C-terminal motifs form a signaling hub
Lysophospholipid GPCR engage heterotrimeric G-proteins and activate canonical downstream signal transduction pathways. These pathways have been well characterized in many of the original papers describing these receptors as well as in reviews. One less discussed aspect of signaling by many of the lysophospholipid receptors is not coupled to the activation of G proteins but is mediated by the ligand-dependent assembly of macromolecular signaling complexes via docking sites encoded by distinct sequence motifs in their C-termini (Fig. 9). Thus far, there are two such motifs identified and characterized primarily among LPA-specific lipid receptors. The C-termini of LPA1, LPA2 and LPA5 contain a −SxL/V motif, where x can be any amino acid, which is a docking site for the PSD95/Dlg/ZO-1 (PDZ) family proteins. It was first reported by Yamada and colleagues that PDZ-RhoGEF and leukemia-associated Rho GEF (LARG), two guanine nucleotide exchange factors (GEF) of the small molecular weight G protein RhoA, physically interact with the PDZ motif of LPA1 and LPA2 [70]. PDZ-RhoGEF and LARG contain PDZ domains as well as regulators of G protein signaling (RGS) domains. The PDZ domain mediates their binding to LPA1 and LPA2, whereas the RGS domain binds to the G12 or G13 α subunits. We demonstrated that a PDZ motif-dependent macromolecular complex of the cystic fibrosis transmembrane regulator (CFTR), the Na+/H+ exchange regulatory factor 2 (NHERF2), and the LPA2 receptor subtype is essential for LPA-dependent inhibition of cholera toxin-induced diarrhea. Through this interaction, LPA2-dependent Gi-mediated inhibition of adenylyl cyclase reduces Cl− secretion and attenuates the symptoms of secretory diarrhea in vivo. Zhang et al. [71] screened a library of PDZ proteins for interaction with the LPA2 C-terminus and identified multiple potential interacting proteins including NHERF1, NHERF2, MAGI-2, MAGI-3 and neurabin. Singh et al. provided further functional evidence for the functional regulation of CFTR localization and activity by PDZ-dependent interactions involving LPA2 and either NHERF1 or NHERF2, respectively [72]. MAGI-3 binding to LPA2 regulates LPA-activated ERK1/2 and RhoA activation [71]. Interestingly, if both proteins are expressed in the same cell there is a dynamic competition between MAGI-3 and NHERF2 binding to the PDZ motif of the LPA2 C-terminus, for example in intestinal epithelial cells. In such a case MAGI-3 binding attenuates NF-κB and c-Jun terminal kinase activation mediated by LPA2 [73]. Among the P2Y family LPA receptors, LPA5 has a PDZ motif that has been shown to physically interact with the second PDZ domain of NHERF2 and regulate the Na+/H+ exchanger 3 that mediates fluid absorption in the intestine [74]. Altogether, the type 1 PDZ binding motif in the C-termini of LPA1, LPA2, and LPA5 is a functionally important docking site for several proteins with PDZ domains, which modulate the activation of canonical G protein-coupled signaling.
Fig. 9.
C-terminal docking motifs for signaling complexes formed by LPA receptors. See text for details.
Yeast two hybrid experiments conducted with the C-terminus of LPA2 have identified Siva-1, a non-PDZ protein, and also the thyroid receptor interacting protein 6 (TRIP6) as high-affinity binding partners [75]. Siva-1 is an early response gene regulated by p53 and E2F transcription factors. Siva-1 is rapidly expressed following DNA damage and makes a complex with the antiapoptotic protein Bcl-XL [76,77]. However, upon ligand activation of LPA2, Siva-1 forms a macromolecular complex with this receptor and the complex becomes poly-ubiquitinated and degraded in the proteasome. Siva-1 recognizes the −C311xxC− motif in the LPA2 C-terminus, which is a half zinc finger motif [78]. Double zinc finger structures promote protein–protein interactions [79] and Siva-1 has its own zinc finger motif. TRIP6 belongs to the Lin-11, Isl-1 and Mec-3 (LIM) family of proteins that contain zinc finger motifs. LIM proteins can shuttle between focal adhesions and the nucleus and control gene expression [80,81]. TRIP6 also contains a PDZ motif in its C-terminus and can interact with NHERF2. The zinc finger and PDZ motifs present in TRIP6 are the foundation of a ternary macromolecular complex formed between LPA2–TRIP6–NHERF2 that is required for the antiapoptotic action mediated by this receptor subtype [78]. The physical interaction between LPA2 and TRIP6 is required for LPA-induced activation of NFκB that involves the physical interaction of TRIP6 with the p65 subunit of NFκB [82]. Furthermore, this complex augments the activation of the ERK1/2 kinases. Thus the potent antiapoptotic action of LPA depends on the depletion of the apoptotic mediator Siva-1 as well as activation of the ERK1/2 and NFκB prosurvival pathways all of which involve macromolecular complexes assembled at the C-terminus of LPA2. Although here we focused on macromolecular complexes of the LPA-specific lipid receptors, there is emerging evidence that the S1P1 and S1P2 receptors also interact with the P-Rex1 and LARG PDZ proteins, which are involved in migratory and differentiation responses [83,84].
2. Summary
The coupled progress made over the last twenty years in the areas of phospholipid biology and biomolecular target structural biology has led to an impressive understanding of how phospholipid interactions with their biomolecular targets lead to downstream biological effects. Even in the face of the rapid progress in the field, however, there still remains ample room for future research focused on a wide variety of topics. Compelling unanswered questions remain in areas ranging from the structural differences within receptor subfamilies that can be exploited in the development of receptor-specific therapeutics to detailed understanding of the protein–protein interactions that participate in the complex cascade of interactions that lead to downstream systemic responses. Several groups have effectively applied modeling to discover sources of receptor selectivity. The first of these was our identification of position E/Q3.29 as the source of selectivity between S1P and LPA [37,38]. More applicable to the drug discovery, however, are the findings of Deng et al., and Schurer et al., that position L/F6.55 contributes to S1P1/S1P3 selectivity among the non-lipid agonists [39,66]. However, care must be taken not to ignore the fact that binding free energy differences include both enthalpic differences and entropic differences. Consideration of docked complexes or even crystallographic complex structures and specific interactions within those complexes can provide insights into the enthalpic differences between different complexes, but do not aid in identification of entropic differences that can dramatically influence specificity. We recently undertook an extensive study to define the source of the FTY720-phosphate selectivity against the S1P2 receptor [43]. An extensive set of first site mutations and then S1P1/S1P2 receptor chimeras surprisingly identified intracellular loop 1 as providing this selectivity. Modeling studies that considered the dynamic nature of the receptor ligand complexes demonstrated that entropic penalties due to limited mobility of the hydrophobic tail occurred in the inactive receptor–ligand pairings. Thus modeling studies can provide pathways to understand both the enthalpic and entropic contributions to ligand selectivity.
Acknowledgements
The work was supported by grants to AP from the National Heart, Lung, and Blood Institute, HL084003 and to GT from the National Cancer Institute, CA092160, National Institute of Allergy Medical Countermeasures Radiological and Nuclear Threats Program AI80405 and by the Harriet Van Vleet Endowment for Basic Oncology Research.
Footnotes
This article is part of a Special Issue entitled Advances in Lysophospholipid Research.
References
- 1.Brinkmann V, Davis MD, Heise CE, Albert R, Cottens S, Hof R, Bruns C, Prieschl E, Baumruker T, Hiestand P, Foster CA, Zollinger M, Lynch KR. The immune modulator, FTY720, targets sphingosine 1-phosphate receptors. J. Biol. Chem. 2002;277:21453–21457. doi: 10.1074/jbc.C200176200. [DOI] [PubMed] [Google Scholar]
- 2.Mandala S, Hajdu R, Bergstrom J, Quackenbush E, Xie J, Milligan J, Thornton R, Shei G, Card D, Keohane C, Rosenbach M, Hale J, Lynch CL, Rupprecht K, Parsons W, Rosen H. Alteration of lymphocyte trafficking by sphingosine 1-phosphate receptor agonists. Science. 2002;296:346–349. doi: 10.1126/science.1070238. [DOI] [PubMed] [Google Scholar]
- 3.Napoli KL. The FTY720 story. Ther. Drug Monit. 2000;22:47–51. doi: 10.1097/00007691-200002000-00010. [DOI] [PubMed] [Google Scholar]
- 4.Graler MH, Goetzl EJ. The immunosuppressant FTY720 down-regulates sphingosine 1-phosphate G-protein-coupled receptors. FASEB J. 2004;18:551–553. doi: 10.1096/fj.03-0910fje. [DOI] [PubMed] [Google Scholar]
- 5.Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, Allende ML, Proia RL, Cyster JG. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 6.Buzard DJ, Thatte J, Lerner M, Edwards J, Jones RM. Recent progress in the development of selective S1P1 receptor agonists for the treatment of inflammatory and autoimmune disorders. Expert Opin. Ther. Patents. 2008;18:1141–1159. [Google Scholar]
- 7.Hale JJ, Neway W, Mills SG, Hajdu R, Ann Keohane C, Rosenbach M, Milligan J, Shei GJ, Chrebet G, Bergstrom J, Card D, Koo GC, Koprak SL, Jackson JJ, Rosen H, Mandala S. Potent S1P receptor agonists replicate the pharmacologic actions of the novel immune modulator FTY720. Bioorg. Med. Chem. Lett. 2004;14:3351–3355. doi: 10.1016/j.bmcl.2004.02.106. [DOI] [PubMed] [Google Scholar]
- 8.Hale JJ, Doherty G, Toth L, Li Z, Mills SG, Hajdu R, Ann Keohane C, Rosenbach M, Milligan J, Shei GJ, Chrebet G, Bergstrom J, Card D, Rosen H, Mandala S. The discovery of 3-(N-alkyl)aminopropylphosphonic acids as potent S1P receptor agonists. Bioorg. Med. Chem. Lett. 2004;14:3495–3499. doi: 10.1016/j.bmcl.2004.04.069. [DOI] [PubMed] [Google Scholar]
- 9.Clemens JJ, Davis MD, Lynch KR, Macdonald TL. Synthesis of 4(5)-phenylimidazole-based analogues of sphingosine-1-phosphate and FTY720: discovery of potent S1P1 receptor agonists. Bioorg. Med. Chem. Lett. 2005;15:3568–3572. doi: 10.1016/j.bmcl.2005.05.097. [DOI] [PubMed] [Google Scholar]
- 10.Foss FW, Jr, Clemens JJ, Davis MD, Snyder AH, Zigler MA, Lynch KR, Macdonald TL. Synthesis, stability, and implications of phosphothioate agonists of sphingosine-1-phosphate receptors. Bioorg. Med. Chem. Lett. 2005;15:4470–4474. doi: 10.1016/j.bmcl.2005.07.057. [DOI] [PubMed] [Google Scholar]
- 11.Buzard D, Han SD, Thoresen L, Moody J, Lopez L, Kawasaki A, Schrader T, Sage C, Gao YH, Edwards J, Barden J, Thatte J, Fu LX, Solomon M, Liu L, Al-Shamma H, Gatlin J, Le M, Xing C, Espinola S, Jones RM. Discovery and characterization of potent and selective 4-oxo-4-(5-(5-phenyl-1,2,4-oxadiazol-3-yl)indolin-1-yl)butanoic acids as S1P1 agonists. Bioorg. Med. Chem. Lett. 2011;21:6013–6018. doi: 10.1016/j.bmcl.2011.05.110. [DOI] [PubMed] [Google Scholar]
- 12.Cee VJ, Frohn M, Lanman BA, Golden J, Muller K, Neira S, Pickrell A, Arnett H, Buys J, Gore A, Fiorino M, Horner M, Itano A, Lee MR, McElvain M, Middleton S, Schrag M, Rivenzon-Segal D, Vargas HM, Xu H, Xu Y, Zhang XX, Siu J, Wong M, Burli RW. Discovery of AMG 369, a Thiazolo[5,4-b]pyridine Agonist of S1P(1) and S1P(5) Acs Med. Chem. Lett. 2011;2:107–112. doi: 10.1021/ml100306h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demont EH, Andrews BI, Bit RA, Campbell CA, Cooke JWB, Deeks N, Desai S, Dowell SJ, Gaskin P, Gray JRJ, Haynes A, Holmes DS, Kumar U, Morse MA, Osborne GJ, Panchal T, Patel B, Perboni A, Taylor S, Watson R, Witherington J, Willis R. Discovery of a selective S1P(1) receptor agonist efficacious at low oral dose and devoid of effects on heart rate. Acs Med. Chem. Lett. 2011;2:444–449. doi: 10.1021/ml2000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurata H, Kusumi K, Otsuki K, Suzuki R, Kurono M, Tokuda N, Takada Y, Shioya H, Mizuno H, Komiya T, Ono T, Hagiya H, Minami M, Nakade S, Habashita H. Structure–activity relationship studies of S1P agonists with a dihydronaphthalene scaffold. Bioorg. Med. Chem. Lett. 2012;22:144–148. doi: 10.1016/j.bmcl.2011.11.048. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura T, Asano M, Sekiguchi Y, Mizuno Y, Tamaki K, Kimura T, Nara F, Kawase Y, Shimozato T, Doi H, Kagari T, Tomisato W, Inoue R, Nagasaki M, Yuita H, Oguchi-Oshima K, Kaneko R, Watanabe N, Abe Y, Nishi T. Discovery of CS-2100, a potent, orally active and S1P3-sparing S1P1 agonist. Bioorg. Med. Chem. Lett. 2012;22:1788–1792. doi: 10.1016/j.bmcl.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 16.Saha AK, Yu XA, Lin JA, Lobera M, Sharadendu A, Chereku S, Schutz N, Segal D, Marantz Y, McCauley D, Middleton S, Siu J, Burli RW, Buys J, Horner M, Salyeris K, Schrag M, Vargas HM, Xu Y, McElvain M, Xu H. Benzofuran derivatives as potent, orally active S1P(1) receptor agonists: a preclinical lead molecule for MS. Acs Med. Chem. Lett. 2011;2:97–101. doi: 10.1021/ml100227q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hale JJ, Lynch CL, Neway W, Mills SG, Hajdu R, Keohane CA, Rosenbach MJ, Milligan JA, Shei GJ, Parent SA, Chrebet G, Bergstrom J, Card D, Ferrer M, Hodder P, Strulovici B, Rosen H, Mandala S. A rational utilization of high-throughput screening affords selective, orally bioavailable 1-benzyl-3-carboxyazetidine sphingosine-1-phosphate-1 receptor agonists. J. Med. Chem. 2004;47:6662–6665. doi: 10.1021/jm0492507. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Chen W, Hale JJ, Lynch CL, Mills SG, Hajdu R, Keohane CA, Rosenbach MJ, Milligan JA, Shei GJ, Chrebet G, Parent SA, Bergstrom J, Card D, Forrest M, Quackenbush EJ, Wickham LA, Vargas H, Evans RM, Rosen H, Mandala S. Discovery of potent 3,5-diphenyl-1,2,4-oxadiazole sphingosine-1-phosphate-1 (S1P1) receptor agonists with exceptional selectivity against S1P2 and S1P3. J. Med. Chem. 2005;48:6169–6173. doi: 10.1021/jm0503244. [DOI] [PubMed] [Google Scholar]
- 19.Jo E, Sanna MG, Gonzalez-Cabrera PJ, Thangada S, Tigyi G, Osborne DA, Hla T, Parrill AL, Rosen H. S1P1-selective in vivo-active agonists from high throughput screening: off-the-shelf chemical probes of receptor interactions, signaling and fate. Chem. Biol. 2005;12:703–715. doi: 10.1016/j.chembiol.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 20.Sanna MG, Liao J, Jo E, Alfonso C, Ahn MY, Peterson MS, Webb B, Lefebvre S, Chun J, Gray N, Rosen H. Sphingosine 1-phosphate (S1P) receptor subtypes S1P1 and S1P3, respectively, regulate lymphocyte recirculation and heart rate. J. Biol. Chem. 2004;279:13839–13848. doi: 10.1074/jbc.M311743200. [DOI] [PubMed] [Google Scholar]
- 21.Bolli MH, Abele S, Binkert C, Bravo R, Buchmann S, Bur D, Gatfield J, Hess P, Kohl C, Mangold C, Mathys B, Menyhart K, Muller C, Nayler O, Scherz M, Schmidt G, Sippel V, Steiner B, Strasser D, Treiber A, Weller T. 2-Imino-thiazolidin-4-one derivatives as potent, orally active S1P(1) receptor agonists. J. Med. Chem. 2010;53:4198–4211. doi: 10.1021/jm100181s. [DOI] [PubMed] [Google Scholar]
- 22.Demont EH, Arpino S, Bit RA, Campbell CA, Deeks N, Desai S, Dowell SJ, Gaskin P, Gray JR, Harrison LA, Haynes A, Heightman TD, Holmes DS, Humphreys PG, Kumar U, Morse MA, Osborne GJ, Panchal T, Philpott KL, Taylor S, Watson R, Willis R, Witherington J. Discovery of a brain-penetrant S1P(3)-sparing direct agonist of the S1P(1) and S1P(5) receptors efficacious at low oral dose. J. Med. Chem. 2011;54:6724–6733. doi: 10.1021/jm200609t. [DOI] [PubMed] [Google Scholar]
- 23.Harrington PE, Croghan MD, Fotsch C, Frohn M, Lanman BA, Pennington LD, Pickrell AJ, Reed AB, Sham KKC, Tasker A, Arnett HA, Fiorino M, Lee MR, McElvain M, Morrison HG, Xu H, Xu Y, Zhang XX, Wong M, Cee VJ. Optimization of a potent, orally active S1P(1) agonist containing a quinolinone core. Acs Med. Chem. Lett. 2012;3:74–78. doi: 10.1021/ml200252b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He XH, Wang LL, Zheng ZB, Xiao JH, Zhong W, Li S. Novel immunomodulators based on an oxazolin-2-one-4-carboxamide scaffold. Bioorg. Med. Chem. Lett. 2012;22:553–557. doi: 10.1016/j.bmcl.2011.10.088. [DOI] [PubMed] [Google Scholar]
- 25.Pennington LD, Sham KKC, Pickrell AJ, Harrington PE, Frohn MJ, Lanman BA, Reed AB, Croghan MD, Lee MR, Xu H, McElvain M, Xu Y, Zhang XX, Fiorino M, Horner M, Morrison HG, Arnett HA, Fotsch C, Wong M, Cee VJ. 4-Methoxy-N-[2-(trifluoromethyl)biphenyl-4-ylcarbamoyl]nicotinamide: a potent and selective agonist of S1P(1) Acs Med. Chem. Lett. 2011;2:752–757. doi: 10.1021/ml2001399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parrill AL, Wang D-A, Bautista DL, Van Brocklyn JR, Lorincz Z, Fischer DJ, Baker DL, Liliom K, Spiegel S, Tigyi G. Identification of Edg1 receptor residues that recognize sphingosine 1-phosphate. J. Biol. Chem. 2000;275:39379–39384. doi: 10.1074/jbc.M007680200. [DOI] [PubMed] [Google Scholar]
- 27.Inagaki Y, Pham TT, Fujiwara Y, Kohno T, Osborne DA, Igarashi Y, Tigyi G, Parrill AL. Sphingosine-1-phosphate analog recognition and selectivity at S1P4 within the endothelial differentiation gene family of receptors. Biochem. J. 2005;389:187–195. doi: 10.1042/BJ20050046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valentine WJ, Kiss GN, Liu J, S. E, Gotoh M, Murakami-Murofushi K, Pham TC, Baker DL, Parrill AL, Lu X, Sun C, Bittman R, Pyne NJ, Tigyi G. (S)-FTY720-vinylphosphonate, an analogue of the immunosuppressive agent FTY720, is a pan-antagonist of sphingosine 1-phosphate GPCR signaling and inhibits autotaxin activity. Cell. Signal. 2010;22:1543–1553. doi: 10.1016/j.cellsig.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan L, Hale JJ, Lynch CL, Budhu R, Gentry A, Mills SG, Hajdu R, Keohane CA, Rosenbach MJ, Milligan JA, Shei G, Chrebet G, Bergstrom JD, Card D, Rosen H, Mandala SM. Design and synthesis of conformationally constrained 3-(N-alkylamino) propylphosphonic acids as potent agonists of sphingosine-1-phosphate (S1P) receptors. Bioorg. Med. Chem. Lett. 2004;14:4861–4866. doi: 10.1016/j.bmcl.2004.07.049. [DOI] [PubMed] [Google Scholar]
- 30.Foss FW, Jr, Snyder AH, Davis MD, Rouse M, Okusa MD, Lynch KR, Macdonald TL. Synthesis and biological evaluation of gamma-aminophosphonates as potent, subtype-selective sphingosine 1-phosphate receptor agonists and antagonists. Bioorg. Med. Chem. 2007;15:663–677. doi: 10.1016/j.bmc.2006.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis MD, Clemens JJ, Macdonald TL, Lynch KR. Sphingosine 1-phosphate analogs as receptor antagonists. J. Biol. Chem. 2005;280:9833–9841. doi: 10.1074/jbc.M412356200. [DOI] [PubMed] [Google Scholar]
- 32.Clemens JJ, Davis MD, Lynch KR, Macdonald TL. Synthesis of para-alkyl aryl amide analogues of sphingosine-1-phosphate: discovery of potent S1P receptor agonists. Bioorg. Med. Chem. Lett. 2003;13:3401–3404. doi: 10.1016/s0960-894x(03)00812-6. [DOI] [PubMed] [Google Scholar]
- 33.Im D-S, Clemens J, Macdonald TL, Lynch KR. Characterization of the human and mouse sphingosine 1-phosphate receptor, S1P5 (Edg-8): structure–activity relationship of sphingosine 1-phosphate receptors. Biochemistry. 2001;40:14053–14060. doi: 10.1021/bi011606i. [DOI] [PubMed] [Google Scholar]
- 34.Clemens JJ, Davis MD, Lynch KR, Macdonald TL. Synthesis of benzimidazole based analogues of sphingosine-1-phosphate: discovery of potent, subtype-selective S1P4 receptor agonists. Bioorg. Med. Chem. Lett. 2004;14:4903–4906. doi: 10.1016/j.bmcl.2004.07.030. [DOI] [PubMed] [Google Scholar]
- 35.Foss FW, Jr, Mathews TP, Kharel Y, Kennedy PC, Snyder AH, Davis MD, Lynch KR, Macdonald TL. Synthesis and biological evaluation of sphingosine kinase substrates as sphingosine-1-phosphate receptor prodrugs. Bioorg. Med. Chem. 2009;17:6123–6136. doi: 10.1016/j.bmc.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hale JJ, Doherty G, Toth L, Mills SG, Hajdu R, Keohane CA, Rosenbach M, Milligan J, Shei GJ, Chrebet G, Bergstrom J, Card D, Forrest M, Sun SY, West S, Xie H, Nomura N, Rosen H, Mandala S. Selecting against S1P3 enhances the acute cardiovascular tolerability of 3-(N-benzyl)aminopropylphosphonic acid S1P receptor agonists. Bioorg. Med. Chem. Lett. 2004;14:3501–3505. doi: 10.1016/j.bmcl.2004.04.070. [DOI] [PubMed] [Google Scholar]
- 37.Wang D, Lorincz Z, Bautista DL, Liliom K, Tigyi G, Parrill AL. A single amino acid determines ligand specificity of the S1P1 (EDG1) and LPA1 (EDG2) phospholipid growth factor receptors. J. Biol. Chem. 2001;276:49213–49220. doi: 10.1074/jbc.M107301200. [DOI] [PubMed] [Google Scholar]
- 38.Holdsworth G, Osborne DA, Pham TT, Fells JI, Hutchinson G, Milligan G, Parrill AL. A single amino acid determines preference between phospholipids and reveals length restriction for activation of the S1P4 receptor. BMC Biochem. 2004;5:12. doi: 10.1186/1471-2091-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schurer SC, Brown SJ, Gonzalez-Cabrera PJ, Schaeffer MT, Chapman J, Jo E, Chase P, Spicer T, Hodder P, Rosen H. Ligand-binding pocket shape differences between sphingosine 1-phosphate (S1P) receptors S1P(1) and S1P(3) determine efficiency of chemical probe identification by ultrahigh-throughput screening. ACS Chem. Biol. 2008;3:486–498. doi: 10.1021/cb800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pham TC, Fells JI, Sr, Osborne DA, North EJ, Naor MM, Parrill AL. Molecular recognition in the sphingosine 1-phosphate receptor family. J. Mol. Graph. Model. 2008;26:1189–1201. doi: 10.1016/j.jmgm.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frohn M, Cee VJ, Lanman BA, Pickrell AJ, Golden J, Rivenzon-Segal D, Middleton S, Fiorino M, Xu H, Schrag M, Xu Y, McElvain M, Muller K, Siu J, Burli R. Novel 5- and 6-subtituted benzothiazoles with improved physicochemical properties: potent S1P(1) agonists with in vivo lymphocyte-depleting activity. Bioorg. Med. Chem. Lett. 2012;22:628–633. doi: 10.1016/j.bmcl.2011.10.069. [DOI] [PubMed] [Google Scholar]
- 42.Kurata H, Otsuki K, Kusumi K, Kurono M, Terakado M, Seko T, Mizuno H, Ono T, Hagiya H, Minami M, Nakade S, Habashita H. Structure–activity relationship studies of sphingosine-1-phosphate receptor agonists with N-cinnamyl-beta-alanine moiety. Bioorg. Med. Chem. Lett. 2011;21:1390–1393. doi: 10.1016/j.bmcl.2011.01.029. [DOI] [PubMed] [Google Scholar]
- 43.Valentine WJ, Godwin VI, Osborne DA, Liu J, Fujiwara Y, Van Brocklyn J, Bittman R, Parrill AL, Tigyi G. FTY720 (Gilenya) phosphate selectivity of sphingosine 1-phosphate receptor subtype 1 (S1P1) G protein-coupled receptor requires motifs in intracellular loop 1 and transmembrane domain 2. J. Biol. Chem. 2011;286:30513–30525. doi: 10.1074/jbc.M111.263442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tigyi G, Parrill AL. Molecular mechanisms of lysophosphatidic acid action. Prog. Lipid Res. 2003;42:498–526. doi: 10.1016/s0163-7827(03)00035-3. [DOI] [PubMed] [Google Scholar]
- 45.Parrill AL, Sardar VM, Yuan H. Sphingosine 1-phosphate and lysophosphatidic acid receptors: agonist and antagonist binding and progress toward development of receptor-specific ligands. Semin. Cell Dev. Biol. 2004;15:467–476. doi: 10.1016/j.semcdb.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 46.Xu Y, Aoki J, Shimizu K, Umezu-Goto M, Hama K, Takanezawa Y, Yu SX, Mills GB, Arai H, Qian L, Prestwich GD. Structure–activity relationships of fluorinated lysophosphatidic acid analogues. J. Med. Chem. 2005;48:3319–3327. doi: 10.1021/jm049186t. [DOI] [PubMed] [Google Scholar]
- 47.Jiang G, Xu Y, Fujiwara Y, Tsukahara T, Tsukahara R, Gajewiak J, Tigyi G, Prestwich GD. Alpha-substituted phosphonate analogues of lysophosphatidic acid (LPA) selectively inhibit production and action of LPA. ChemMedChem. 2007;2:679–690. doi: 10.1002/cmdc.200600280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Y, Jiang GW, Tsukahara R, Fujiwara Y, Tigyi G, Prestwich GD. Phosphonothioate and fluoromethylene phosphonate analogues of cyclic phosphatidic acid: novel antagonists of lysophosphatidic acid receptors. J. Med. Chem. 2006;49:5309–5315. doi: 10.1021/jm060351+. [DOI] [PubMed] [Google Scholar]
- 49.Williams JR, Khandoga AL, Goyal P, Fells JI, Perygin DH, Siess W, Parrill AL, Tigyi G, Fujiwara Y. Unique ligand selectivity of the GPR92/LPA(5) lysophosphatidate receptor indicates role in human platelet activation. J. Biol. Chem. 2009;284:17304–17319. doi: 10.1074/jbc.M109.003194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gajewiak J, Tsukahara R, Tsukahara T, Fujiwara Y, Yu S, Lu W, Murph M, Mills GB, Tigyi G, Prestwich GD. Alkoxymethylenephosphonate analogues of (lyso)phosphatidic acid stimulate signaling networks coupled to the LPA(2) receptor. ChemMedChem. 2007;2:1789–1798. doi: 10.1002/cmdc.200700111. [DOI] [PubMed] [Google Scholar]
- 51.Gajewiak J, Prestwich GD. Phosphomimetic sulfonamide and sulfonamidoxy analogues of (lyso)phosphatidic acid. Tetrahedron Lett. 2006;47:7607–7609. [Google Scholar]
- 52.Durgam GG, Virag T, Walker MD, Tsukahara R, Yasuda S, Liliom K, van Meeteren LA, Moolenaar WH, Wilke N, Siess W, Tigyi G, Miller DD. Synthesis, structure–activity relationships, and biological evaluation of fatty alcohol phosphates as lysophosphatidic acid receptor ligands, activators of PPARgamma, and inhibitors of autotaxin. J. Med. Chem. 2005;48:4919–4930. doi: 10.1021/jm049609r. [DOI] [PubMed] [Google Scholar]
- 53.Virag T, Elrod DB, Liliom K, Sardar VM, Parrill AL, Yokoyama K, Durgam G, Deng W, Miller DD, Tigyi G. Fatty alcohol phosphates are subtype-selective agonists and antagonists of LPA receptors. Mol. Pharmacol. 2003;63:1032–1042. doi: 10.1124/mol.63.5.1032. [DOI] [PubMed] [Google Scholar]
- 54.Khandoga AL, Fujiwara Y, Goyal P, Pandey D, Tsukahara R, Bolen A, Guo H, Wilke N, Liu J, Valentine WJ, Durgam GG, Miller DD, Jiang G, Prestwich GD, Tigyi G, Siess W. Lysophosphatidic acid-induced platelet shape change revealed through LPA(1-5) receptor-selective probes and albumin. Platelets. 2008;19:415–427. doi: 10.1080/09537100802220468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yanagida K, Masago K, Nakanishi H, Kihara Y, Hamano F, Tajima Y, Taguchi R, Shimizu T, Ishii S. Identification and characterization of a novel lysophosphatidic acid receptor, p2y5/LPA6. J. Biol. Chem. 2009;284:17731–17741. doi: 10.1074/jbc.M808506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bandoh K, Aoki J, Taira A, Tsujimoto M, Arai H, Inoue K. Lysophosphatidic acid (LPA) receptors of the EDG family are differentially activated by LPA species: structure–activity relationship of cloned LPA receptors. FEBS Lett. 2000;478:159–165. doi: 10.1016/s0014-5793(00)01827-5. [DOI] [PubMed] [Google Scholar]
- 57.Durgam GG, Tsukahara R, Makarova N, Walker MD, Fujiwara Y, Pigg KR, Baker DL, Sardar VM, Parrill AL, Tigyi G, Miller DD. Synthesis and pharmacological evaluation of second-generation phosphatidic acid derivatives as lysophosphatidic acid receptor ligands. Bioorg. Med. Chem. Lett. 2006;16:633–640. doi: 10.1016/j.bmcl.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 58.Fujiwara Y, Sardar V, Tokumura A, Baker D, Murakami-Murofushi K, Parrill A, Tigyi G. Identification of residues responsible for ligand recognition and regioisomeric selectivity of lysophosphatidic acid receptors expressed in mammalian cells. J. Biol. Chem. 2005;280:35038–35050. doi: 10.1074/jbc.M504351200. [DOI] [PubMed] [Google Scholar]
- 59.Valentine WJ, Fells JI, Perygin DH, Mujahid S, Yokoyama K, Fujiwara Y, Tsukahara R, Van Brocklyn JR, Parrill AL, Tigyi G. Subtype-specific residues involved in ligand activation of the endothelial differentiation gene family lysophosphatidic acid receptors. J. Biol. Chem. 2008;283:12175–12187. doi: 10.1074/jbc.M708847200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hanson MA, Roth CB, Jo E, Griffith MT, Scott FL, Reinhart G, Desale H, Clemons B, Cahalan SM, Schuerer SC, Sanna MG, Han GW, Kuhn P, Rosen H, Stevens RC. Crystal structure of a lipid G protein-coupled receptor. Science. 2012;335:851–855. doi: 10.1126/science.1215904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parrill AL, Baker DL, Wang D, Fischer DJ, Bautista DL, van Brocklyn J, Spiegel S, Tigyi G. Structural features of EDG1 receptor–ligand complexes revealed by computational modeling and mutagenesis. In: Goetzl EJ, Lynch KR, editors. Lysophospholipids and Eicosanoids in Biology and Pathophysiology. vol. 905. New York: New York Academy of Sciences; 2000. pp. 330–339. [DOI] [PubMed] [Google Scholar]
- 64.Lim H, Park J, Ko K, Lee M, Chung S. Syntheses of sphingosine-1-phosphate analogues and their interaction with EDG/S1P receptors. Bioorg. Med. Chem. Lett. 2004;14:2499–2503. doi: 10.1016/j.bmcl.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 65.van Loenen PB, de Graaf C, Verzijl D, Leurs R, Rognan D, Peters SL, Alewijnse AE. Agonist-dependent effects of mutations in the sphingosine-1-phosphate type 1 receptor. Eur. J. Pharmacol. 2011;667:105–112. doi: 10.1016/j.ejphar.2011.05.071. [DOI] [PubMed] [Google Scholar]
- 66.Deng QL, Clemas JA, Chrebet G, Fischer P, Hale JJ, Li Z, Mills SG, Bergstrom J, Mandala S, Mosley R, Parent SA. Identification of Leu276 of the S1P(1) receptor and Phe263 of the S1P(3) receptor in interaction with receptor specific agonists by molecular modeling, site-directed mutagenesis, and affinity studies. Mol. Pharmacol. 2007;71:724–735. doi: 10.1124/mol.106.029223. [DOI] [PubMed] [Google Scholar]
- 67.Fujiwara Y, Osborne DA, Walker MD, Wang DA, Bautista DA, Liliom K, Van Brocklyn JR, Parrill AL, Tigyi G. Identification of the hydrophobic ligand binding pocket of the S1P1 receptor. J. Biol. Chem. 2007;282:2374–2385. doi: 10.1074/jbc.M609648200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parrill AL, Lima S, Spiegel S. Structure of the first sphingosine 1-phosphate receptor. Sci. Signal. 2012;5:e23. doi: 10.1126/scisignal.2003160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pham TT, Kriwacki RW, Parrill AL. Peptide design and structural characterization of a GPCR loop mimetic. Biopolymers. 2007;86:298–310. doi: 10.1002/bip.20745. [DOI] [PubMed] [Google Scholar]
- 70.Yamada T, Ohoka Y, Kogo M, Inagaki S. Physical and functional interactions of the lysophosphatidic acid receptors with PDZ domain-containing Rho guanine nucleotide exchange factors (RhoGEFs) J. Biol. Chem. 2005;280:19358–19363. doi: 10.1074/jbc.M414561200. [DOI] [PubMed] [Google Scholar]
- 71.Zhang H, Wang D, Sun H, Hall RA, Yun CC. MAGI-3 regulates LPA-induced activation of Erk and RhoA. Cell. Signal. 2007;19:261–268. doi: 10.1016/j.cellsig.2006.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singh AK, Riederer B, Krabbenhoft A, Rausch B, Bonhagen J, Lehmann U, de Jonge HR, Donowitz M, Yun C, Weinman EJ, Kocher O, Hogema BM, Seidler U. Differential roles of NHERF1, NHERF2, and PDZK1 in regulating CFTR-mediated intestinal anion secretion in mice. J. Clin. Invest. 2009;119:540–550. doi: 10.1172/JCI35541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee SJ, Ritter SL, Zhang H, Shim H, Hall RA, Yun CC. MAGI-3 competes with NHERF-2 to negatively regulate LPA2 receptor signaling in colon cancer cells. Gastroenterology. 2011;140:924–934. doi: 10.1053/j.gastro.2010.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin S, Yeruva S, He P, Singh AK, Zhang H, Chen M, Lamprecht G, de Jonge HR, Tse M, Donowitz M, Hogema BM, Chun J, Seidler U, Yun CC. Lysophosphatidic acid stimulates the intestinal brush border Na(+)/H(+) exchanger 3 and fluid absorption via LPA(5) and NHERF2. Gastroenterology. 2010;138:649–658. doi: 10.1053/j.gastro.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin FT, Lai YJ, Makarova N, Tigyi G, Lin WC. The lysophosphatidic acid 2 receptor mediates down-regulation of Siva-1 to promote cell survival. J. Biol. Chem. 2007;282:37759–37769. doi: 10.1074/jbc.M705025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chu F, Barkinge J, Hawkins S, Gudi R, Salgia R, Kanteti PV. Expression of Siva-1 protein or its putative amphipathic helical region enhances cisplatin-induced apoptosis in breast cancer cells: effect of elevated levels of BCL-2. Cancer Res. 2005;65:5301–5309. doi: 10.1158/0008-5472.CAN-04-3270. [DOI] [PubMed] [Google Scholar]
- 77.Xue L, Chu F, Cheng Y, Sun X, Borthakur A, Ramarao M, Pandey P, Wu M, Schlossman SF, Prasad KV. Siva-1 binds to and inhibits BCL-X(L)-mediated protection against UV radiation-induced apoptosis. Proc. Natl. Acad. Sci. U. S. A. 2002;99:6925–6930. doi: 10.1073/pnas.102182299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shuyu E, Lai YJ, Tsukahara R, Chen CS, Fujiwara Y, Yue J, Yu JH, Guo H, Kihara A, Tigyi G, Lin FT. Lysophosphatidic acid 2 receptor-mediated supramolecular complex formation regulates its antiapoptotic effect. J. Biol. Chem. 2009;284:14558–14571. doi: 10.1074/jbc.M900185200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- 80.Hervy M, Hoffman L, Beckerle MC. From the membrane to the nucleus and back again: bifunctional focal adhesion proteins. Curr. Opin. Cell Biol. 2006;18:524–532. doi: 10.1016/j.ceb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 81.Lin VT, Lin FT. TRIP6: an adaptor protein that regulates cellmotility, antiapoptotic signaling and transcriptional activity. Cell. Signal. 2011;23:1691–1697. doi: 10.1016/j.cellsig.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lai YJ, Lin VT, Zheng Y, Benveniste EN, Lin FT. The adaptor protein TRIP6 antagonizes Fas-induced apoptosis but promotes its effect on cell migration. Mol. Cell. Biol. 2010;30:5582–5596. doi: 10.1128/MCB.00134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ledezma-Sanchez BA, Garcia-Regalado A, Guzman-Hernandez ML, Vazquez-Prado J. Sphingosine-1-phosphate receptor S1P1 is regulated by direct interactions with P-Rex1, a Rac guanine nucleotide exchange factor. Biochem. Biophys. Res. Commun. 2010;391:1647–1652. doi: 10.1016/j.bbrc.2009.12.108. [DOI] [PubMed] [Google Scholar]
- 84.Medlin MD, Staus DP, Dubash AD, Taylor JM, Mack CP. Sphingosine 1-phosphate receptor 2 signals through leukemia-associated RhoGEF (LARG), to promote smooth muscle cell differentiation. Arterioscler. Thromb. Vasc. Biol. 2010;30:1779–1786. doi: 10.1161/ATVBAHA.110.209395. [DOI] [PMC free article] [PubMed] [Google Scholar]