Abstract
Chromatin immunoprecipitation experiments followed by sequencing (ChIP-seq) detect protein-DNA binding events and chemical modifications of histone proteins. Challenges in the standard ChIP-seq protocol have motivated recent enhancements in this approach, such as reducing the number of cells required and increasing the resolution. Complementary experimental approaches – for example DNaseI hypersensitive site mapping and analysis of chromatin interactions mediated by particular proteins - provide additional information about DNA-binding proteins and their function. These data are now being used to identify variability in the functions of DNA-binding proteins across genomes and individuals. In this Review, I describe the latest advances in methods to detect and functionally characterize DNA-bound proteins.
DNA-binding proteins play critical roles in many major cellular processes such as DNA transcription, splicing, replication, and repair. These proteins include transcription factors that bind preferentially to certain DNA sequences as well as histone proteins that form the core of nucleosomes, the basic unit of chromatin. Neither genomic locations of bound factors nor of modified histones can be accurately predicted in a particular cell type using DNA sequence features alone, and functional assays are necessary to identify these cellular characteristics. Chromatin immunoprecipitation coupled with microarrays (ChIP-chip) or short-tag sequencing (ChIP-seq) has become the standard technique for identifying locations and biochemical modifications of bound proteins genome-wide1-3. Recent advances in ChIP methodology have overcome some of the limitations of the ‘standard’ ChIP experiment and the development of complementary assays and analyses have expanded the number, types, and resolution of protein-DNA interactions discovered.
In this review, I discuss the current state of ChIP-based experiments including modifications of the standard ChIP protocol and review basic features of ChIP-seq analysis pipelines. I then describe alternatives to ChIP such as open chromatin assays such as DNase-seq4-7, FAIRE-seq8-10, and genome-wide DNaseI footprinting11-14. Finally I discuss approaches to characterizing protein-DNA interactions that are improving understanding of function. These include three-dimensional chromatin assays such as chromatin conformation capture15-17 and ChIA-PET18, 19 that provide evidence for functional targets of DNA-bound proteins, and analyses of sequence-based data from ChIP20, 21 and other experiments22-24 that reveal allele-specific effects on protein-DNA binding.
ChIP-seq experiments
Current ChIP-seq experiments
ChIP is the most direct way to identify binding sites of a single DNA-binding protein or locations of modified histones. The basic steps of the ChIP-seq assay have been reviewed elsewhere25-27 and are depicted in Figure 1A for transcription factors and 1B for histone modifications. The ENCODE Consortium28 has performed hundreds of ChIP-seq experiments and has used this experience to develop a set of working standards and guidelines29 (Box 1). It must be noted that given the diversity of cell types, conditions, factors, and modifications being assayed, it is near impossible to define common guidelines that will be appropriate for all situations. From a technical perspective, the success of a ChIP experiment depends on the development and validation of a highly specific antibody to the bound protein or modification. Antibody quality varies, even between independently prepared lots of the same antibody, as demonstrated in a recent assessment of over 200 human, fly, and worm antibodies within the ENCODE and modENCODE projects30. In this study, 25% failed specificity tests and 20% failed immunoprecipitation experiments. In addition, multiple histone modifications can alter the efficacy of certain antibodies31. Other technical challenges include the requirement for large numbers of cells and prior knowledge of the existence of a DNA-binding protein or histone modification. Possible solutions to these issues are considered below and in later sections.
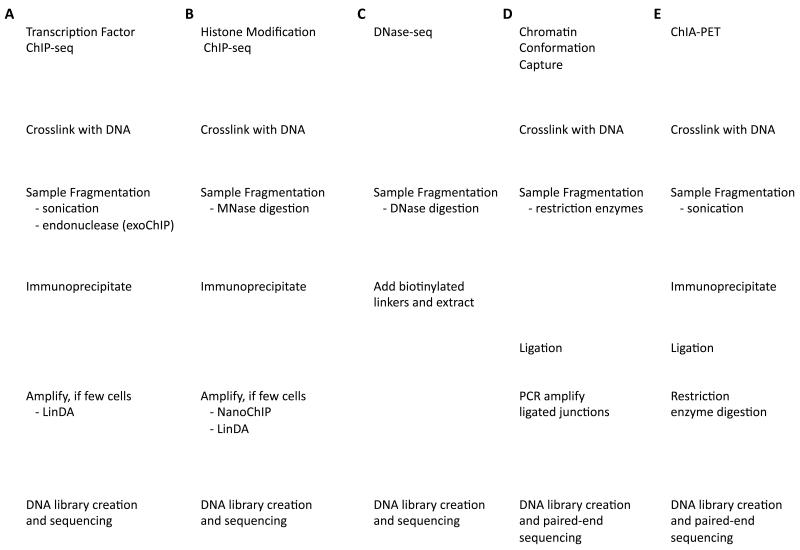
Figure 1. Comparison of experimental protocols.
Experiments to detect different aspects of DNA binding proteins and chromatin conformation share many of the same steps. a | ChIP-seq for DNA binding proteins such as transcription factors. Recent variations on the standard protocol include using endonuclease digestion instead of sonication (ChIP-exo) to increase the resolution of binding site detection and eliminate contaminating DNA, and amplification after ChIP for samples with limited cells. b | ChIP-seq for histone modifications use MNase digestion to fragment DNA and can also now be run on small samples with the additional post-ChIP amplification. c | DNase-seq relies on digestion by the DNaseI nuclease to identify regions of nucleosome-depleted open chromatin where there are binding sites for all types of factors, but it cannot identify what specific factors are bound.
Box 1. Recommended ChIP-seq Standards.
Based on the collective experience of ENCODE and modENCODE labs having performed hundreds of ChIP-seq experiments, a set of standards and guidelines for performing ChIP-seq has been written29. Experiments are classified as point source (highly localized signals, such as for transcription factors), broad source (signal spans large domains, such as for some histone modifications such as H3K36me3) or mixed source (has elements of both, such as RNA PolII). If the type of signal is unknown, multiple peak callers focusing on point source or broad peaks may be applied to determine the best fit to the data. These standards are summarized below.
Antibody validation. Primary characterization of transcription factor antibody using immunoblot or immunofluoresence analysis. Secondary characterization using one of i) factor knockdown by mutation or RNAi; ii) independent ChIP experiments using alternative epitopes or protein members of a complex; iii) immunoprecipitation using epitope-tagged constructs; iv) mass spectrometry; or v) binding site motif analyses. Primary characterization of histone modification antibody using immunoblot analysis. Secondary characterization using one of i) peptide binding tests; ii) mass spectrometry; iii) immunoreactivity analysis in cell lines containing knockdowns of relevant histone modification enzyme or mutants histones; or iv) genome annotation enrichment.
Sequencing depth. 20 million (human) or 8 million (fly/worm) uniquely mapped read sequences for point source, 40 million/10 million for broad source. Increased sequencing depth allows detection of more sites with reduced enrichment. It is noted that setting a minimal signal strength threshold, usually based on a p-value or false discovery rate calculation, to identify peaks does not guarantee discovery of all functional sites. It is also noted that DNA sequencing library complexity, that is the amount of unique DNA molecules, must be sufficient meaning sequencing depths do not exceed complexity. It is suggested that at least 80% of 10 million or more reads be mapped to distinct genomic locations. Low complexity libraries generally indicate a failed experiment where not enough DNA was recovered causing the same PCR amplified products to be sequenced repeatedly and many small peaks to be detected with a high false positive rate.
Experimental replication. Minimum two replicates per experiment, 10 million (human) or 4 million (fly/worm) uniquely mapped reads per replicate for point source, 20 million/5 million for broad source. Each replicate represents an independent cell culture, embryo pool, or tissue sample. For two replicates, either 80% of top 40% of identified targets in one replicate must be among targets in second replicate, or 75% of target lists must be in common between both replicates.
Data quality assessment. No one test is always suitable for all experiments or forms a necessary requirement. Recommended assessments include i) investigating signals at known sites using a genome browser; ii) calculating the fraction of reads in peaks (FRiP), recommended to be greater than 1%; iii) calculating cross correlations, defined as the correlation of the density of sequences aligned to the Watson strand with the density of sequences aligned to the Crick strand after shifting the Watson strand alignments by the average distance between opposite strands reads.
Data and metadata reporting. ChIP results should be submitted to GEO102. Experimental and analyses information provided should include ChIP procedures, antibody validation, DNA sequencing information, identified regions of enrichment and method of identification, and any other analysis.
Limited cells
Typically, large numbers of cells (~10 million) are required for a ChIP experiment limiting both the types of cells that can be assayed as well as the number of ChIP experiments that can be performed on a valuable sample. It can be especially challenging in small model organisms where multiple whole animals may be necessary to achieve these quantities. Two protocols have been recently developed to address this problem through post-ChIP DNA amplification (Figure 1A,B).
Nano-ChIP-seq32 has been successfully performed on as few as 10,000 cells for histone modifications. It is recommended that variable sonication times and antibody concentrations are used, scaled in proportion to the number of starting cells. The small amount of DNA extracted after performing the ChIP experiment is PCR amplified using custom primers that form a hairpin structure at the 5′ end to prevent self-annealing when being added. The primers also contain a BciVI restriction site that allows the direct addition of Illumina sequencing adapters to the resulting DNA, which makes DNA library preparation and sequencing straightforward. The number of cells required is dependent on multiple factors including antibody efficiency and abundance of the target protein. Therefore, while 10,000 cells were sufficient for assaying the H3K4me3 chromatin mark, ChIPs for less abundant histone modifications or transcription factors will likely require more cells and may require further optimization of certain steps such as sonication time.
The second protocol uses single tube linear amplification (LinDA) and has been successfully applied for transcription factor ERα using 5,000 cells and for the histone modification H3K4me3 using 10,000 cells33. The key to this technique is an optimized T7 RNA polymerase linear amplification protocol34. A major concern in any amplification protocol is that technical biases would unevenly amplify the starting material. LinDA was shown to be robust for even amplification of starting material; importantly, it seemed to avoid bias in relation to GC content, which is generally problematic for PCR-based approaches.
Increased precision
Standard ChIP-seq experiments that use sonication to fragment chromatin result in libraries containing DNA molecules that are ~200 bases long, even though each protein typically binds only 6-20 bases. In addition, resulting libraries are often contaminated with DNA not bound by the target factor, which has necessitated the use of the input control experiments and is responsible for some common systematic biases.
ChIP-exo35 uses lambda (λ) exonuclease to digest the 5′ end of protein-bound and formaldehyde cross-linked DNA fragments to a fixed distance from the bound protein (Figure 1A); fixation is a barrier to 5′-3′ digestion. Since DNA fragments are produced from both strands during ChIP, the 5′ ends of sequence-tags align primarily at two genomic locations corresponding the barriers on each strand, the protein being bound to the region inbetween. In addition, the exonuclease largely eliminates contaminating DNA. Experiments in yeast for the Reb1 transcription factor35 showed ChIP-exo could identify binding sites with single basepair precision, a 90-fold greater precision than when using the standard protocol, and with a 40-fold increase in the signal-to-noise ratio indicating lower background (contaminating) signal.
Multiple binding events
DNA bound proteins and histone modifications work together and with other genomic modifications to perform cellular functions. When multiple experiments indicate different proteins or modifications at the same genomic location, it is not clear whether these are simultaneously present or present on different chromosomes in the same cell or in different cells. Sequential ChIP, or re-ChIP or co-immunoprecipitation36, uses antibodies to different proteins in successive experiments to determine genomic locations where both targets are present, but experiments have only been performed at individual loci and not in conjunction with high-throughput sequencing. Recently, assays that perform bisulfite sequencing to identify methylated DNA within immunoprecipitated chromatin fragments have been developed37, 38. These genome-wide experiments showed that DNA methylation and H3K27me3 modified histones can occur simultaneously. More generally, new techniques have been developed to reveal the identities of individual proteins interacting in larger complexes in human and model organisms39-47 providing evidence for combinations of factors that will bind together.
ChIP-seq analysis pipelines
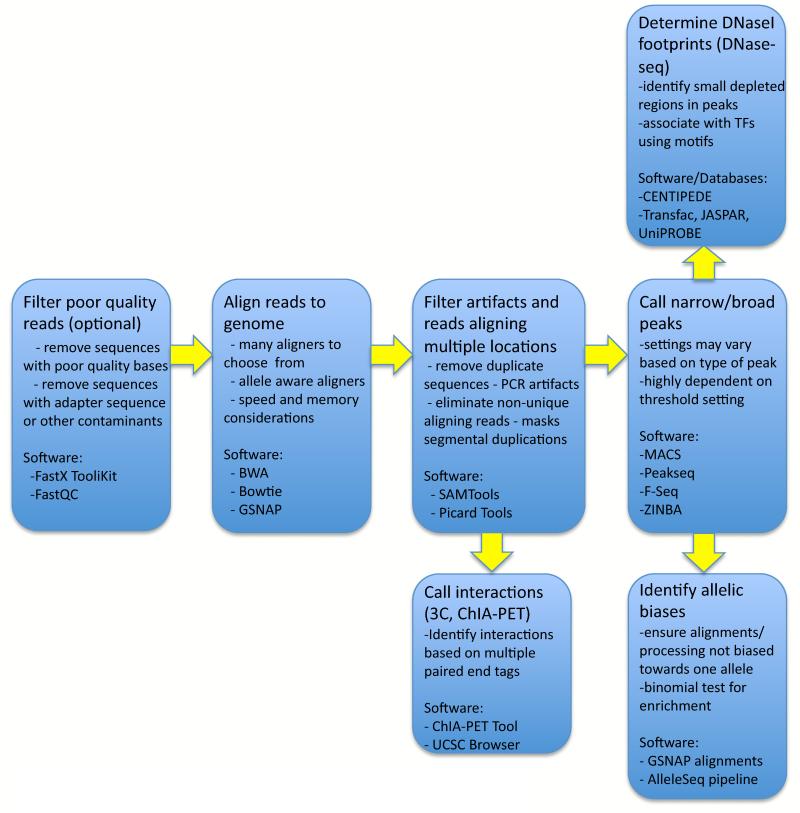
There has also been a large effort to improve analytical tools necessary to interpret the sequence data output from ChIP-seq experiments. Computational processing pipelines are generally implemented to progress from raw sequence reads to usable annotations. Steps common to many pipelines are depicted in Figure 2. Each step has led to the development of specialized software tools, briefly discussed below.
Figure 2. General analysis pipeline for sequence tag experiments.
Experiments using short sequence reads that identify regions with a particular molecular characteristic share many of the same analysis steps. Poor quality reads can be filtered initially, but often the inability to align these reads to the genome sufficiently removes bad sequences. Alignment using one of many possible software programs (Table 1) is followed by filtering artifacts that arose during the PCR amplification step when sequencing, or that appear due to the underrepresentation of certain sequences in the reference genome, such as pericentromeric satellite sequences. Often, reads aligning to more than some number of genomic locations are removed. For experiments identifying independent locations, ‘peak’ calling tools (Table 1) identify genomic regions of signal enrichment indicating a bound protein, histone modification, or open chromatin. In contrast, chromatin interaction experiments use aligned paire-end reads to find evidence of interacting distal genomic regions. DNaseI footprints indicate local protection from DNaseI digestion within a larger DHS region due to a bound protein. The distribution of alleles in sequences spanning heterozygous variants can be analyzed to determine if a bias towards sequences with one of the two alleles exists. This may reflect a functional difference caused by the underlying genotype.
Sequence aligners must be fast and accurate, and several strategies have been developed to achieve these goals (Table 1; see 48 for a recent review). Given a final set of aligned sequences, genomic regions are identified that contain enriched signals, or ‘peaks’, where more sequences are aligned than would be expected by chance, indicating locations of binding sites or histone modifications. Several software programs have been developed to identify these peaks (Table 1; see49-52 for recent comparisons of methods). When available, data from input control experiments are used by most peak callers to represent background levels of signal. Many also control for differences in mappability to regions of the genome. As described in Box 1, peaks can be point source (highly localized signals, such as for transcription factors), broad source (signal spans large domains, such as for some histone modifications such as H3K36me3) or mixed source (has elements of both, such as RNA PolII). Each of these require different detection strategies with some software focused primarily on one type of peak, and others offering different settings that tune the software based on the peak shape.
Table 1. Subset of software tools available for three key steps in the analysis of sequence data.
| Software Tool | Web Address | Notes |
|---|---|---|
|
| ||
| Short Read Aligners | ||
| BWA | http://bio-bwa.sourceforge.net/ | Fast, efficient, based on Burrows- Wheeler transform |
| Bowtie | http://bowtie-bio.sourceforge.net/ | Similar to BWA, part of suite of tools that includes TopHat and CuffLinks for RNA-seq processing |
| GSNAP | http://research-pub.gene.com/gmap/ | Considers set of input variant alleles to better align to heterozygous sites |
| Wikipedia List - Aligners | en.wikipedia.org/wiki/List_of_sequence_alignment_software#Short-Read_Sequence_Alignment | Comprehensive list of available short read aligners with descriptions and links to download software |
| Peak Callers | ||
| MACS | http://liulab.dfci.harvard.edu/MACS/ | Fits data to dynamic Poisson distribution, works with and without control data |
| PeakSeq | http://info.gersteinlab.org/PeakSeq | Takes into account differences in mappability of genomic regions, enrichment based on FDR calculation. |
| ZINBA | http://code.google.com/p/zinba/ | Zero Inflation Negative Binomial Algorithm, can incorporate multiple genomic factors such as mappability, GC content, work with point and broad source peak data |
| Differential Peak Calling | ||
| edgeR | http://www.bioconductor.org/packages/2.9/bioc/html/edgeR.html | Uses negative binomial distribution to model differences in tag counts. Uses replicates to better estimate significant differences. |
| DESeq | http://wwwhuber.embl.de/users/anders/DESeq/ | Also uses negative binomial distribution modelling, but differs in calculation of mean and variance of distribution. |
| baySeq | http://www.bioconductor.org/packages/release/bioc/html/baySeq.html | Uses empirical Bayes approach to identify significant differences. Assumes negative binomial distribution of data. |
| SAMSeq | http://www.stanford.edu/~junli07/research.html#SAM | Based on popular Significance Analysis of Microarrays (SAM) software. Nonparametric method that uses resampling to normalize for differences in sequencing depth. |
It is often desirable to compare data from multiple experiments, for example assaying the same transcription factor in two different cell types or conditions, to investigate common and cell-type specific activity. Simply comparing peaks from each experiment is often used, but this may not identify regions called as peaks in both but with very different strengths of signal, or may incorrectly identify regions that were just above the peak threshold in one but just below in the other. Several software packages, originally developed for RNA-seq data, are now available that can be adapted to identify statistically significant differences based directly on ChIP-seq read count data (Table 1; see 53, 54 for a comparison).
With experimental evidence of factor binding sites, there is an opportunity to improve the characterization of preferred DNA binding motifs for each factor. Several groups have developed software that uses information from ChIP-seq experiments during motif discovery55-60. The more accurate modeling of binding preferences allows for better prediction of significant signals and the precise DNA contact site for factor binding events identified by ChIP-seq.
Sequencing considerations
We are still discovering biases in sequence data due to a combination of genomic characteristics, experimental protocols, specific sequencing technologies, and analytical methods. These have been studied in ChIP-seq data, generated using Illumina’s Genome Analyzer IIx sequencer, to better understand how to uncover true signals61. Findings from this study and others have indicated the need to normalize for chromatin structure and GC content because regions that have open chromatin and higher GC content produced proportionately more sequences. The authors also showed that sequencing paired-ends can nearly double the effective genomic coverage in repeat regions, but with increased sequencing costs. They also assessed the effect of sequencing depth on accuracy and sensitivity and found that some binding sites are missed even at high depths (16.2 million reads in Drosphila, equivalent to approximately 327 million reads in human).
Further analytical challenges
Despite this progress, several challenges remain. As read-length increases, the current short read aligners will likely require further modification48 and alignments to repetitive sequences will remain a challenge62-64. Continued effort is needed to develop or improve methods to identify real events, and to enable a better interpretation. For example, although we would like to think of the assayed binding or modification events as binary, i.e. a protein is or is not bound to a given location, the data is more continuous in nature. Signal strength at a particular location is influenced by the strength of the interaction, which can be modulated by variations in genotype and by the percentage of the population of cells assayed that have the binding or modification event. Signals may reflect not only direct binding events, but also indirect binding where one factor is interacting with another factor that is bound to DNA. Distinguishing between these two events is important but cannot be directly done from ChIP data.
Open Chromatin
Most transcription factors cannot stably interact with their DNA targets if the DNA is nucleosomal. For stable binding to occur, interfering nucleosomes must be displaced or translocated to create a nucleosome-depleted, open chromatin region. Detecting open chromatin complements ChIP-seq data, and can simultaneously identify binding sites for nearly all factors. Two distinct assays, DNase-seq and FAIRE-seq, have been developed to directly detect open chromatin (see 65 for a review of genome accessibility experiments).
DNase-seq and FAIRE-seq
The DNaseI endonuclease non-specifically digests DNA, but in the normal context of chromatin structure it will preferentially digest unbound open chromatin. Since most DNA is wrapped in a nucleosome, DNaseI hypersensitive (DHS) sites largely correspond to nucleosome depleted regions and these are primarily regions that have gene regulatory functions, such as promoters, enhancers, silencers, insulators, and locus control regions66-68. DNase-seq experiments (Figure 1C) combine traditional DHS assays with high-throughput sequencing to simultaneously identify all types of regulatory regions genome-wide4, 7, 69. The 5′ end of a sequence tag generated by DNase-seq indicates the site of a DNaseI digestion event, and regions of enrichment in digestion events are identified as DHS sites, each of which can contain binding sites of multiple factors. Comparisons with ChIP-seq data indicate DNase-seq captures the vast majority of binding sites for most factors4, 6, 7.
The Formaldehyde-Assisted Identification of Regulatory Elements (FAIRE-seq) assay8, 9 starts with formaldehyde cross-linking, similar to ChIP, but then instead of using an antibody to target specific factors, DNA is sonicated, and the extract is subjected to Phenol-chloroform extraction. The nucleosome-depleted fraction of DNA is preferentially segregated to the aqueous phase. FAIRE-enriched DNA has been shown to correspond to regulatory regions8.
Enriched regions from these two assays are highly overlapping but are not identical6. Both show good correspondence to ChIP-seq data for multiple factors with most factor sites found by both methods. However, each method identified a subset of putative regulatory elements not seen in the other. Binding sites of certain factors (FOXA1, FOXA3, GATA1) were better identified by FAIRE-seq while others (ZNF263, CTCF) were more often seen in DNase-seq data. Sites only found in DNase-seq assays were enriched at promoter regions and with promoter-associated H3K4 tri-methylation and H3K9 acetylation histone modifications, while sites specific to FAIRE-seq were more often in internal introns and exons, intergenic, and H3K4 mono-methylated regions6.
The FAIRE-seq assay is fairly easy to perform, though some optimization of cross-linking times may be needed for different cell types or tissues due to variation in fixation efficiency10. DNase-seq can be more difficult at the bench with optimizations of cell lysis procedures and DNaseI concentration required5. The signal-to-noise ratio, i.e. the fraction of sequences in enriched regions vs. non-enriched regions, is higher for DNase-seq than for FAIRE-seq, and these data can additionally be used to identify more precise DNA binding sites, or DNaseI footprints, as described below. Advantages of DNase-seq and FAIRE-seq are that they can identify genomic locations bound by proteins that are uncharacterized or for which antibodies do not exist. However, standard open chromatin analysis does not allow determination of which protein(s) are present in these regions.
Nucleosome positioning experiments such as MNase-seq70, 71 use micrococcal nuclease digestion to determine where nucleosomes are present and, by extension, nucleosome free regions. For large genomes, such as human, this may not be as economically practical since >90% of the genome is nucleosomal. Significantly greater sequencing coverage is required in this case to obtain the same level of resolution as open chromatin assays in these cases.
DNaseI Footprinting
Smaller, more focal areas of DNaseI protection within a larger DHS site, called DNaseI footprints (Figure 3), result from the binding of individual proteins or complexes. Single-site DNaseI footprinting has been used to identify binding sites at individual loci for over 30 years72 and DNase-seq now allows for the discovery of footprints genome-wide7, 11-13.
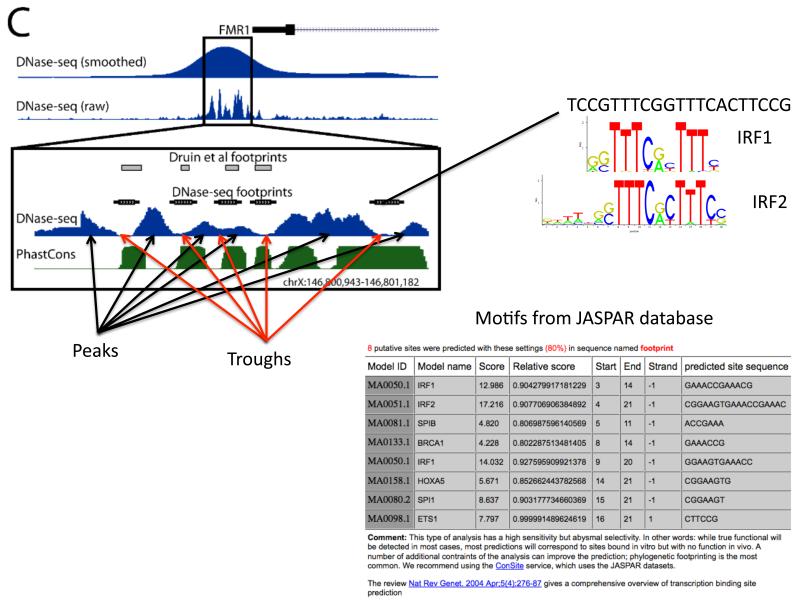
Figure 3. DNaseI footprints correspond to bound proteins.
The distribution of DNaseI digestion sites with DNaseI hypersensitive regions is not uniform with peaks and troughs in the signal, where troughs are due to protection by bound proteins. Transcription factor binding motif databases such as JASPAR74 can be searched using the sequence from each footprint to predict what factor is bound. Shown here is data from the proximal promoter region of the FMR1 gene. DNaseI footprints had been identified previously at this locus103 in lymphoblastoid cells. More recent data from DNase-seq was used to recapitulate these results in a single experiment12.
Note: This figure is taken from 12. It is figure 1C in that manuscript.
Two different basic strategies have been employed for predicting protein binding sites using DNaseI footprints in DNase-seq data. The first tries initially to delineate individual footprints solely based on the distribution of the sequence reads; you would expect a depletion of 5′ends of reads within the footprint compared to the immediately adjacent, non-footprint bases. This strategy has been employed in the yeast and human genomes to identify 8-30bp footprint regions of significantly reduced DNaseI digestion compared to a random background distribution11, 14 and in the human genome using a hidden-Markov model (HMM) to model the characteristic changes in sequence read density in footprints12. To predict what factor may be bound in each identified footprint, transcription factor motif databases such as TRANSFAC73, Jaspar74, and UniPROBE75 can be scanned using the sequence in the footprint. Footprints can also be used to identify novel transcription factor DNA binding motifs. A recent analysis of 41 diverse cell-types showed that approximately 90% of all motifs in TRANSFAC, Jaspar, and UniPROBE could be identified using footprinted sequences, while an additional 289 distinct motifs could be defined14. Comparing ChIP-seq data with motifs in footprints also provides the ability to estimate what sites are being directly vs. indirectly bound by a factor14. As these are predictions, it is recommended that specific binding events are tested experimentally.
An alternative strategy implemented in the CENTIPEDE software tool13 essentially performs the above steps in reverse order. First, the genome is scanned to identify all potential binding sites for a given DNA binding protein based on its motif. CENTIPEDE then employs an unsupervised Bayesian mixture model to predict which of these sites are bound by protein and which are not bound in a particular cell type. This probabilistic model uses evidence based primarily on DNaseI digestion, but can also incorporate evidence from the evolutionary conservation of bases and the presence of histone modifications, if that data is available. A second analysis in this study13 using all 10-mers enriched in DHS sites predicted 49 novel motifs not found in existing motif databases, demonstrating that CENTIPEDE can also find binding sites of undefined factors.
A comparison of the accuracies of the two methods has not been performed. The first method may be more appropriate for a more global annotation of potential binding sites regardless of the existence of a motif, whereas CENTIPEDE provides a more straightforward method to identify footprints for particular factors with known binding site preferences. Both methods are constrained by sequencing depth that can limit their ability to identify footprints in DHS sites with reduced signals in DNase-seq data, and by the lack of knowledge of binding site preferences for factors. Increased sequencing depths will allow for further refinement of footprint models. As DNaseI footprint annotations are generated for more cell types, motif finding algorithms may help predict new factor binding motifs that in turn will help with the annotation of footprints.
Mapping Chromatin Interactions
Identifying protein-DNA binding sites is important, but that by itself does not lead to an understanding of the regulatory programs and other biological processes in cells. ChIP-seq, DNase-seq, and FAIRE-seq do not map each bound protein to the target gene(s) it is helping regulate or to genomic region(s) with which it is interacting to form a higher order chromatin structure. Towards this end, approaches have been developed based on the chromatin conformation capture (3C) method15. This method has been extended to improve scope and/or precision (5C16, Hi-C17), and adapted to identify interactions associated with specific proteins (Chromatin Interaction Analysis with Paired-End Tag (ChIA-PET) sequencing18, 19).
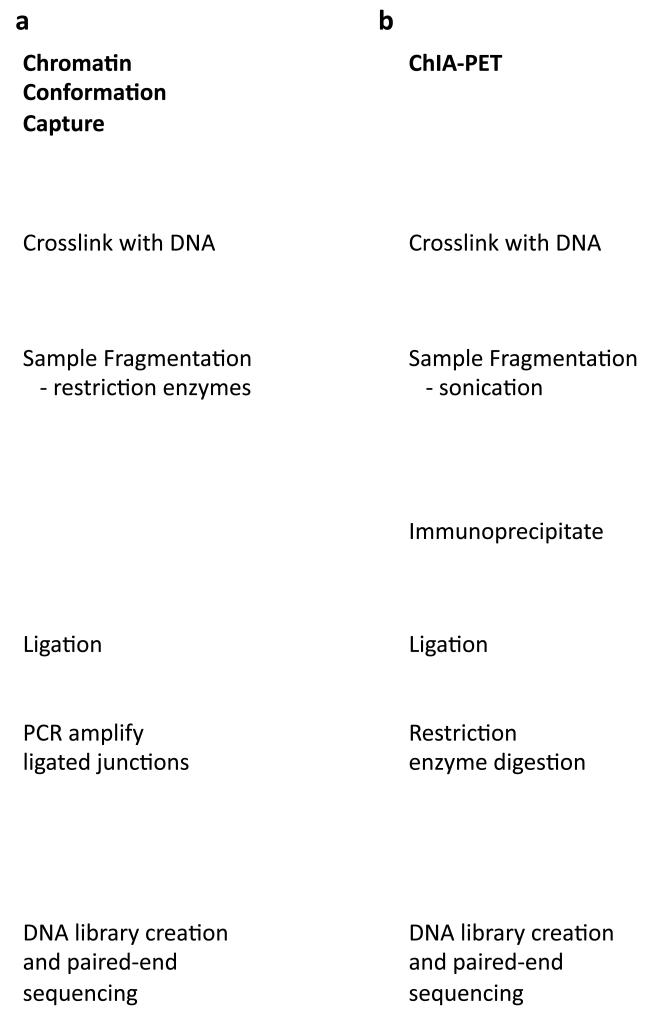
The principle of chromatin conformation capture experiments (Figure 4A) is to cross-link genomic regions that are in close proximity (similar to ChIP-seq), digest the DNA using restriction enzymes to create pairs of DNA cross-linked fragments that originated and identify these pairs of fragments (for example using paired-end sequencing after ligation and amplification of the fragments).
Figure 4. Detecting chromatin interactions.
In three-dimensional space, distal genomic regions on the same or different chromosomes interact mediated by one or more DNA binding proteins. a | Chromatin conformation capture experiments use a ligation step to join distant fragments that are interacting in three-dimensional chromatin space providing information on possible targets for DNA-bound proteins. b | ChIA-PET similarly detects chromatin interactions using a ligation step to pair non-adjacent interaction regions, but using a ChIP step to more specifically identify interactions with a particular bound protein, such as RNA PolII. It should be noted that the DNA that is actually sequenced as part of the paired-end sequencing do not necessarily correspond to the precise region of interaction but are dictated by the presence of restriction enzyme targets. c | The UCSC Genome Browser contains data from both ChIA-PET and 5C experiments. Shown here is a 275Kb region showing interactions near the transcription start site of the ST7 gene. For both experiments, solid blocks represent the sequenced paired ends with lines connecting them when they appear on the same chromosome. Darker shading indicates more confidence in this mapping based on multiple instances of complementary paired end sequences. The ChIA-PET experiment was performed using a RNA PolII antibody, and the corresponding signal from RNA PolII binding is displayed immediately below. Also included beneath these chromatin interaction annotations are signals from DNase-seq experiments in the same cell type. Regions of enriched DNase-seq signal indicate nucleosome-depleted DNA that represents putative regulatory elements. Together, chromatin interaction data provide clues as to the gene targets for these regulatory regions.
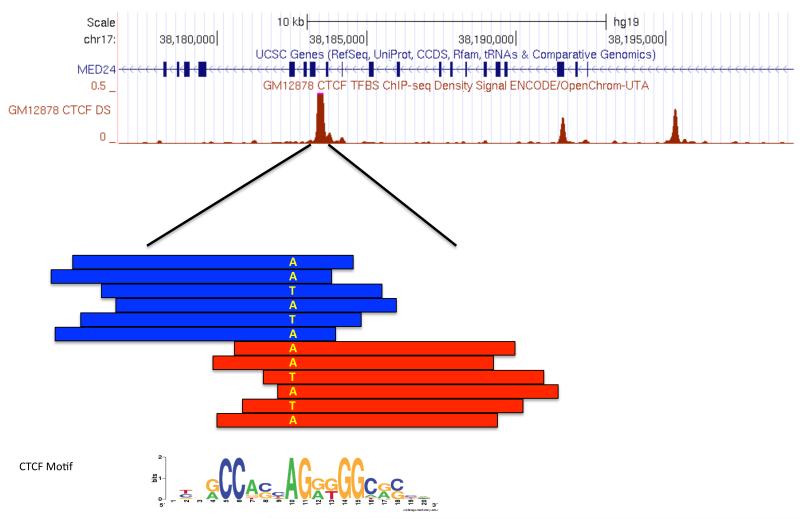
Note: CTCF weblogo from104, figure 2.
3C experiments require PCR primers that are designed for regions of interest and thus are low-throughput. However, designing primers for promoter regions of genes along with regulatory regions identified through ChIP-seq or DNase-seq experiments can identify potential interactions between specific bound proteins and their target genes. 5C experiments simultaneously use thousands of primers in one experiment to detect millions of interactions16. 5C is still limited in the size of the genomic region that can be assayed by the number of primers that are incorporated and sequencing depth to confidently detect interactions. This assay was used to analyze a 400Kb region that included the human β-globin locus and was able to confirm known interactions between regulatory elements and genes in the locus as well as identify new looping interactions. Hi-C does not depend on primers but instead incorporates biotinylated residues after restriction enzyme digestion that allow these fragments to be pulled down using streptavidin beads and the detection of interactions genome-wide. Extremely deep sequencing is required to confidently identify all interactions. While this represents a substantial increase in throughput, the resolution is limited to a megabase scale due to the frequency of restriction sites in the genome76. This limits the ability to confidently associate individual factor binding sites with target genes. A recent study showed that Hi-C was able to correctly separate interaction domains in the HoxA locus that is separated by a known CTCF insulator element76. This information does provide boundaries for potential factor-gene interactions.
ChIA-PET (Figure 4A) also starts with formaldehyde-based cross-linking, but this is followed by fragmentation via sonication and an immunoprecipitation step using a specific antibody, as is done in a ChIP experiment. Ligase is added to create chimeric DNA fragments followed by restriction enzyme digestion and paired-end tag sequencing. ChIP-seq experiments for the factor of interest are also performed to support the interaction data and annotate where the factor is bound.
ChIA-PET provides high-resolution interaction data genome-wide that involves a given DNA-binding protein. An initial study of the estrogen receptor α (ERα) protein revealed ERα binding sites are involved in long range looping interactions with gene promoters and affect transcription rates77. siRNA knockdowns of ERα showed at least some of the interactions disappeared and transcriptional regulation was affected. As with Hi-C, the resolution of ChIA-PET is limited by the frequency and distribution of restriction enzyme digestion sites. Because ChIA-PET relies on an antibody against the factor of interest, as with ChIP an increase in available antibodies will increase the scope of interactions that can be discovered by this method.
Data from both ChIA-PET and 5C experiments are available in the UCSC Genome Browser (Figure 4B), which provides a visual representation of the sequenced paired end tags. Together, the chromatin conformation capture and ChIA-PET technologies offer the ability to generate evidence of what genes are being targeted by DNA bound proteins and regions with specific histone modifications.
Variation in Protein Binding
ChIP-seq, DNase-seq, and chromatin interaction experiments generate complex data sets reflecting the dynamic nature of the biological processes being measured. The results of these experiments provide a snapshot of varying chromatin states and protein binding events across millions of cells that are subject to both genetic and environmental influences. Signals from these data reveal a spectrum of intensities, but the molecular underpinnings of this variation - among loci in the same genome and across multiple individuals – remains unclear. Using data from these experiments, we can begin to understand better both types of variation.
Variation across loci
DNA-binding proteins can generally interact with a range of DNA sequences giving rise to a sequence “motif” to describe their binding preferences. A motif, often more specifically defined as a position weight matrix, describes the nucleotide preferences, most often defined as probabilities, at each position in a binding site. These probabilities are usually based on the frequency at which each nucleotide is present in known binding sites identified across the genome. It is generally thought that the presence of the higher probability nucleotides at a locus indicates an increase in binding affinity and/or specificity. Binding affinity refers to the strength of an interaction and is generally specified in terms of a disassociation constant, whereas binding specificity refers to the preference for binding to specific sequences. Higher affinity or specificity sites may be expected to generate higher signals in protein-binding assays due to increased occupancy and/or stability of the interaction.
Several high-throughput methods are now available to determine binding specificities of proteins in an unbiased manner (see Stormo and Zhao78 for a more detailed review). Protein binding microarrays have been developed that contain all possible 10 base pair sequences79 and have been used, for example, to determine the binding specificities for 104 diverse factors in mouse80. The binding preferences of factors are largely unique and approximately half of the factors show preferences for two motifs. More recently, a similar study was performed in Drosophila using the novel PB-seq method (protein/DNA binding followed by sequencing). In this approach, the protein of interest – in this case, heat shock factor (HSF) - was fused to the 3×FLAG epitope and allowed to bind to fragmented DNA. The HSF-bound DNA was recovered and sequenced81. This study compared the binding preferences of HSF defined by PB-seq in vitro to binding sites defined by ChIP-seq in vivo. Interestingly, in vitro and in vivo binding intensities were not highly correlated when considering all possible binding sites in the genome. A chromatin environment data model was then generated using available DNaseI hypersensitivity data, MNase data, and ChIP-chip data for 21 histone modifications, and was used with the in vitro results to predict binding intensities. This resulted in a high correlation with in vivo data, underscoring the influence of chromatin on protein-DNA binding. In fact, a prior model based solely on DNaseI data produced the highest correlation suggesting that DNA accessibility factors largely into the actual binding of factors in vivo.
Chromatin is dynamic and has substantial, stable differences between phenotypically different cell types and also smaller, more variable differences across a population of similar cells. ChIP-seq and other protein binding experiments provide a snapshot of the occupancy of binding sites, but do not describe the dynamics or function of factor binding. Competition ChIP assays82, 83 have enabled the investigation of binding site turnover in yeast. These studies integrated into a single strain two copies of a factor-encoding gene, each with a different epitope tag with one gene being constitutively expressed and the other inducible. ChIP for each epitope was performed on samples collected at multiple time points after induction of the inducible gene to show the dynamics of factor binding; specifically to show which at sites there is stable binding and at which there is turnover. A study84 of the Rap1 transcription factor showed that sites stably bound by the same factor (resident sites) were associated with efficient transcriptional activation while high-turnover sites (treadmilling sites) were associated lower transcriptional output, even under similar rates of occupancy.
These studies demonstrate that binding sites across a genome are not functionally equivalent and indicate influences on this variation. Complementary information about factor binding, chromatin state, and binding dynamics provide a more complete picture of how protein-DNA interactions at particular loci contribute to cellular processes.
Variation across individuals
The adaptation of ChIP and other experiments to sequencing technologies also provides the opportunity to investigate potential functional effects of the underlying DNA sequence on the presence or absence of a particular event, such as the binding of a protein. Polymorphic bases within regulatory regions can affect the stability of a bound protein or the ability of a region to acquire or propagate chromatin marks. These, in turn, can affect the ability of that locus to regulate the transcription of its target gene.
To identify polymorphic sites associated with functional variation, we can investigate sequences in individual ChIP-seq peaks that align across a heterozygous base in a particular sample; a significant difference in the distribution of sequences containing one allele versus the other indicates a potential allelic effect on protein binding (Figure 5). For example, given ChIP-seq data for transcription factor F, we can investigate each heterozygous site that falls within a called peak (binding site) in that data. For a site with alleles A and B, if the presence of A or B has no effect, we would expect an even distribution of sequences containing A and B at that binding site. If sequences at that site predominantly contain allele A, we could hypothesize that A provides a more favorable binding sequence for that protein, or conversely that B interferes with binding.
Figure 5. Allele-specific bias in a CTCF ChIP-seq experiment.
Sequence-based experiments allow for the investigation of functional differences across individuals due to their underlying genotype. This schematic depicts a region with an enriched number of sequence reads from a ChIP-seq experiment, where each red and blue box indicates an aligned read with blue reads aligned to the forward strand, and red reads to the reverse strand. As is typical of a factor ChIP-seq experiment, forward strand reads accumulate 5′ of the site while reverse strand reads accumulate 3′ of the site. Contained within this locus is a heterozygous polymorphism, denoted by A and T bases. Only one-third of the spanning reads contain the T allele while two-thirds contain the A allele indicating an allelic-imbalance.
Allelic analysis of sequence data requires modifications to the standard analysis pipelines described above (Figure 2). Aligning short read sequences to a single reference sequence creates a bias at heterozygous loci where reads containing the allele present in the reference genome are aligned at a higher rate due to the inherent “mismatch” penalty incurred by the non-reference allele sequences. Ideally, sequences would be aligned to fully defined haplotype genomes, as described in the AlleleSeq computation pipeline21. These are rarely available, but more often the genotype of each individual has been obtained. This can be used to create two reference genomes, each one containing one allele for each heterozygous location, and enable merging of separate alignments of sequences to each of these genome sequences. Alternatively, allele-aware aligners such as GSNAP85 can be used that dynamically consider multiple alleles during alignments. In addition, the alignability of a sequence containing each variant must be considered. The presence of allele A may make a particular sequence unique with respect to the rest of the genome, while the same sequence with allele B is found one or more times elsewhere in the genome. This can be determined by aligning all possible sequences overlapping the site of interest back to the genome and analyzing the uniqueness of these alignments. Overall, a much more careful consideration of non-reference sequence bases is necessary to accurately detect signals at these locations.
Allelic biases have been detected in data from several sequencing based experiments including ChIP-seq20, 86-89 and DNase-seq22, 24. In one study, analysis of ChIP-seq data from 10 human lymphoblastoid cell lines showed that 7.5% of NFkB binding sites and 25% of PolII binding sites differed significantly between individuals, and that 35% and 26% of these corresponded with genetic variations, respectively20. Another study, also using human lymphoblastoid cells, found that 7% of DNaseI HS sites and 11% of CTCF binding sites showed allele specific effects22. Both studies were performed in the context of family trios that showed evidence of the heritability of these allelic functional traits. A more recent study of DNase-seq and expression data from lymphoblastoid cell lines from 70 individuals uncovered just under 9,000 DNaseI Sensitivity Quantitative Trait Loci (dsQTLs) that associated genetic variants with allelic biases in DHS sites with changes in expression of nearby genes24. Many dsQTLs could also be mapped to previously identified DNaseI footprints12, 13 suggesting that the binding of specific factors is altered. Analysis of footprints with predicted binding factors showed enrichment for allelic biases in CTCF, camp-response-element (CRE) and interferon-stimulated response element (ISRE) sites, and depletion in MADS box transcription factor 2 (MEF2) sites.
Perspective
The importance of DNA-binding proteins has motivated the continued development of experimental and analytical methods to better identify and characterize these interactions. While ChIP-seq remains the standard for identifying binding site locations for individual proteins and histone modifications, practical limitations of antibody development, a single factor/modification limit per experiment, the lack of functional annotation, and a static snapshot of a dynamic cell necessitates the use of complementary methods or extensions of ChIP-seq to provide a more complete picture biological processes in the cell, especially transcriptional regulation.
Open chromatin assays like DNase-seq and FAIRE-seq provide a more comprehensive status of all active regulatory elements in a single experiment. Comparing changes in open chromatin profiles across cell-types6, 7, 90, 91, differentiation states92, 93, disease states94-97 and species98 are revealing key changes in factor binding that underlie functional differences across cells. Reduced sequencing costs are enabling deeper coverage of these experiments uncovering more precise positioning of bound proteins in the form of footprints.
Identifying genomic locations of protein-DNA interactions is just the start. Bound proteins interact with other proteins in complexes, create higher order chromatin structures, are involved in specific cellular processes such a the regulation of a particular gene, and vary across time, cell types, and genetic background. Answering these questions requires complementary assays, many of which are presented here. As data from complementary assays accumulate, the challenge will be to integrate these to provide a more complete understanding of transcriptional networks and cellular processes99, 100. Comparisons across cell types will provide new insights into properties of individual and combinations of factors that drive cell-type specific functions. These will require the further development of new analytical and computational modeling techniques as well as focused validation experiments to support model hypotheses.
Results from these studies continue to further our understanding of normal cell biology, but also provide critical information that will benefit efforts to determine the causes and consequences of abnormal cellular states associated with disease. Genome-wide association studies in humans have identified thousands of loci strongly associated with a complex disease or a related trait101, most of which are located in non-coding genomic regions and lack functional annotation. Characterizing the effects of different alleles at single nucleotide polymorphisms (SNPs) on DNA-protein interactions provide potential functional consequences of alleles. These can then be used to suggest testable hypotheses for observed associations of individual SNPs with complex diseases, potentially leading to the development of better diagnoses and treatment options.
Acknowledgements
I gratefully acknowledge support from the National Institutes of Health grants U54-HG004563, R21-DA027040, and U01 CA157703, the Department of Defense grant W81XWH-10-1-0772, and the University Cancer Research Fund from the University of North Carolina at Chapel Hill.
Glossary Terms
- promoters
DNA sequence immediately upstream of transcription start sites at which RNA polymerases and transcription factors bind to initiate gene transcription
- enhancers
DNA sequence at which transcription factors bind that increase the transcription rate of one or more target genes that can be at varying distances to the enhancer
- silencers
DNA sequence at which transcription factors bind that decrease the transcription rate of one or more target genes that can be at varying distances to the silencer
- insulators
DNA sequence that intereferes with enhancer and/or silencer activity
- locus control regions
Regulatory elements that generally control transcription of multiple genes in a single locus
- DNA binding motifs
Degenerate pattern of DNA sequences to which transcription factors prefer to bind, often represented as a probabilistic matrix
- degenerate
In transcription factor binding motifs, most positions in the motif can be more than one base, sometimes with little preference between bases, causing the motif sequence to be degenerate
- allele
Each genomic locus consisting of one or more bases is present in two copies in cells that may not exatly match due to genetic variation. An allele refers to one particular copy
- exonuclease
An enzyme that cleaves a single nucleotide from the end of a DNA molecule
- cross-linked
The strong binding of DNA to interacting proteins via covalent bonds
- sonication
The fragmenting of DNA sequence by exposing it to high frequency sound waves
- mappability
The uniqueness of a stretch of DNA sequence compared to a whole genome sequence. Short sequence reads can be confidently mapped to unique sequence, but less confidently mapped to sequence that occurs multiple times in a genome
- hidden-Markov model (HMM)
A statistical model consisting of states that represent an aspect of a sequence, such as in a footprint, and transitions between states, and are used to label bases in a sequence with the modelled property. HMMs are also used in many gene prediction programs
- Bayesian mixture model
A probabilistic model used to represent the presence of multiple sub-populations, such as a DNaseI footprint, within the whole population, such as the whole DNA sequence. Bayesian mixture models allow for the incorporation of prior knowledge about sub-population frequencies
- biotinylated
A protein or nucleic acid in which a small biotin molecule has been attached. Biotin binds to streptavidin allowing for the isolation of biotinylated molecules
- disassociation constant
Reflects the amount of energy required to separate two interacting molecules, often referred to as Kd
- rule ensembles
A classification model that consists of a linear combination of simple models, or rules, derived from the data
- haplotype
The combintion of alleles on a single chromosome. A genotype then refers to the combination of the two haplotypes in a normal genome
- DNaseI sensitivity quantitative trait loci (dsQTL)
A locus whose sensitivity to DNaseI digestion varies based on the presence of different alleles in that locus. An allelic difference may influence the binding of proteins at this locus causing the variation in digestion
Footnotes
Online links
UCSC Genome Browser: http://genome.ucsc.edu
Gene Expression Omnibus (GEO): http://www.ncbi.nlm.nih.gov/geo/
Picard Sequence Analysis Tools: http://picard.sourceforge.net/
Furey Lab at the University of North Carolina at Chapel Hill: http://fureylab.web.unc.edu/
Access to this interactive links box is free online.
References
- 1.Bhinge AA, Kim J, Euskirchen GM, Snyder M, Iyer VR. Mapping the chromosomal targets of STAT1 by Sequence Tag Analysis of Genomic Enrichment (STAGE) Genome Res. 2007;17:910–916. doi: 10.1101/gr.5574907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valouev A, et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle AP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc. 2010. 2010 doi: 10.1101/pdb.prot5384. pdb prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song L, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012 doi: 10.1038/nature11232. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements) Methods. 2009;48:233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simon JM, Giresi PG, Davis IJ, Lieb JD. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat Protoc. 2012;7:256–267. doi: 10.1038/nprot.2011.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hesselberth JR, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyle AP, et al. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2010;21:456–464. doi: 10.1101/gr.112656.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pique-Regi R, et al. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2010;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neph S, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012 doi: 10.1038/nature11212. This paper describes the identification and anaysis of 8.4 million DNaseI footprints across 41 human cell types corresponding to putative factor binding events and predicting ~300 novel motifs for factor binding.
- 15.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. This paper described the first general approach to characterize interactions between any two genomic loci and provided the first glimpse of the three dimensional structure of chromatin in the nucleus.
- 16.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li G, et al. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol. 2010;11:R22. doi: 10.1186/gb-2010-11-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li G, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasowski M, et al. Variation in transcription factor binding among humans. Science. 2010;328:232–235. doi: 10.1126/science.1183621. This paper demonstrated that functional variation in transcriptioin factor binding due to differences in genotype could be uncovered using data from ChIP-seq experiments.
- 21.Rozowsky J, et al. AlleleSeq: analysis of allele-specific expression and binding in a network framework. Mol Syst Biol. 2011;7:522. doi: 10.1038/msb.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDaniell R, et al. Heritable individual-specific and allele-specific chromatin signatures in humans. Science. 2010;328:235–239. doi: 10.1126/science.1184655. This paper similarly demonstrated that differences in chromatin structure due to genotype variation could be seen using data from DNase-seq data.
- 23.Gertz J, et al. Analysis of DNA methylation in a three-generation family reveals widespread genetic influence on epigenetic regulation. PLoS Genet. 2011;7:e1002228. doi: 10.1371/journal.pgen.1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Degner JF, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farnham PJ. Insights from genomic profiling of transcription factors. Nat Rev Genet. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ku CS, Naidoo N, Wu M, Soong R. Studying the epigenome using next generation sequencing. J Med Genet. 2011;48:721–730. doi: 10.1136/jmedgenet-2011-100242. [DOI] [PubMed] [Google Scholar]
- 28.Consortium TEP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012 doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landt SG, et al. ChIP-seq gudelines and practices used by the ENCODE and modENCODE consortia. Genome Res. 2012 doi: 10.1101/gr.136184.111. Accepted. This paper provides practical guidelines for conducting and analyzing ChIP-seq experiments.
- 30.Egelhofer TA, et al. An assessment of histone-modification antibody quality. Nat Struct Mol Biol. 2011;18:91–93. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD. Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr Biol. 2011;21:53–58. doi: 10.1016/j.cub.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adli M, Bernstein BE. Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP-seq. Nat Protoc. 2011;6:1656–1668. doi: 10.1038/nprot.2011.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shankaranarayanan P, et al. Single-tube linear DNA amplification (LinDA) for robust ChIP-seq. Nat Methods. 2011;8:565–567. doi: 10.1038/nmeth.1626. [DOI] [PubMed] [Google Scholar]
- 34.Liu CL, Schreiber SL, Bernstein BE. Development and validation of a T7 based linear amplification for genomic DNA. BMC Genomics. 2003;4:19. doi: 10.1186/1471-2164-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. This paper describes a modifictation to the tranditional ChIP-seq protocol that allows for greater resolution in identifying binding sites of factors. The key advance is the use of exonuclease to generate more consistent signals of binding locations.
- 36.Markham K, Bai Y, Schmitt-Ulms G. Co-immunoprecipitations revisited: an update on experimental concepts and their implementation for sensitive interactome investigations of endogenous proteins. Anal Bioanal Chem. 2007;389:461–473. doi: 10.1007/s00216-007-1385-x. [DOI] [PubMed] [Google Scholar]
- 37.Brinkman AB, et al. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res. 2012;22:1128–1138. doi: 10.1101/gr.133728.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Statham AL, et al. Bisulfite sequencing of chromatin immunoprecipitated DNA (BisChIP-seq) directly informs methylation status of histone-modified DNA. Genome Res. 2012;22:1120–1127. doi: 10.1101/gr.132076.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Havugimana PC, et al. A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butland G, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 41.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 42.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 43.Guruharsha KG, et al. A protein complex network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 45.Hu P, et al. Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins. PLoS Biol. 2009;7:e96. doi: 10.1371/journal.pbio.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 47.Kuhner S, et al. Proteome organization in a genome-reduced bacterium. Science. 2009;326:1235–1240. doi: 10.1126/science.1176343. [DOI] [PubMed] [Google Scholar]
- 48.Li H, Homer N. A survey of sequence alignment algorithms for next-generation sequencing. Brief Bioinform. 2010;11:473–483. doi: 10.1093/bib/bbq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim H, et al. A short survey of computational analysis methods in analysing ChIP-seq data. Hum Genomics. 2011;5:117–123. doi: 10.1186/1479-7364-5-2-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilbanks EG, Facciotti MT. Evaluation of algorithm performance in ChIP-seq peak detection. PLoS One. 2010;5:e11471. doi: 10.1371/journal.pone.0011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malone BM, Tan F, Bridges SM, Peng Z. Comparison of four ChIP-Seq analytical algorithms using rice endosperm H3K27 trimethylation profiling data. PLoS One. 2011;6:e25260. doi: 10.1371/journal.pone.0025260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laajala TD, et al. A practical comparison of methods for detecting transcription factor binding sites in ChIP-seq experiments. BMC Genomics. 2009;10:618. doi: 10.1186/1471-2164-10-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao D, et al. A survey of statistical software for analysing RNA-seq data. Hum Genomics. 2010;5:56–60. doi: 10.1186/1479-7364-5-1-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kvam VM, Liu P, Si Y. A comparison of statistical methods for detecting differentially expressed genes from RNA-seq data. Am J Bot. 2012;99:248–256. doi: 10.3732/ajb.1100340. [DOI] [PubMed] [Google Scholar]
- 55.Guo Y, Mahony S, Gifford DK. High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Comput Biol. 2012;8:e1002638. doi: 10.1371/journal.pcbi.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boeva V, et al. De novo motif identification improves the accuracy of predicting transcription factor binding sites in ChIP-Seq data analysis. Nucleic Acids Res. 2010;38:e126. doi: 10.1093/nar/gkq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu S, Wang J, Zhao W, Pounds S, Cheng C. ChIP-PaM: an algorithm to identify protein-DNA interaction using ChIP-Seq data. Theor Biol Med Model. 2010;7:18. doi: 10.1186/1742-4682-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu M, Yu J, Taylor JM, Chinnaiyan AM, Qin ZS. On the detection and refinement of transcription factor binding sites using ChIP-Seq data. Nucleic Acids Res. 2010;38:2154–2167. doi: 10.1093/nar/gkp1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kulakovskiy IV, Boeva VA, Favorov AV, Makeev VJ. Deep and wide digging for binding motifs in ChIP-Seq data. Bioinformatics. 2012;26:2622–2623. doi: 10.1093/bioinformatics/btq488. [DOI] [PubMed] [Google Scholar]
- 60.Georgiev S, et al. Evidence-ranked motif identification. Genome Biol. 2010;11:R19. doi: 10.1186/gb-2010-11-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Y, et al. Systematic evaluation of factors influencing ChIP-seq fidelity. Nat Methods. 2012 doi: 10.1038/nmeth.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet. 2011;13:36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J, Huda A, Lunyak VV, Jordan IK. A Gibbs sampling strategy applied to the mapping of ambiguous short-sequence tags. Bioinformatics. 2010;26:2501–2508. doi: 10.1093/bioinformatics/btq460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chung D, et al. Discovering transcription factor binding sites in highly repetitive regions of genomes with multi-read analysis of ChIP-Seq data. PLoS Comput Biol. 2011;7:e1002111. doi: 10.1371/journal.pcbi.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bell O, Tiwari VK, Thoma NH, Schubeler D. Determinants and dynamics of genome accessibility. Nat Rev Genet. 2011;12:554–564. doi: 10.1038/nrg3017. [DOI] [PubMed] [Google Scholar]
- 66.Wu C, Bingham PM, Livak KJ, Holmgren R, Elgin SC. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979;16:797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- 67.Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 68.Cockerill PN. Structure and function of active chromatin and DNase I Hypersensitive Sites. Febs J. 2011;19:1742–4658. doi: 10.1111/j.1742-4658.2011.08128.x. [DOI] [PubMed] [Google Scholar]
- 69.Crawford GE, et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16:123–131. doi: 10.1101/gr.4074106. This paper describes the first DNaseI hypersensitivity experiments that used high-throughput sequencing technology.
- 70.Wei G, Hu G, Cui K, Zhao K. Genome-wide mapping of nucleosome occupancy, histone modifications, and gene expression using next-generation sequencing technology. Methods Enzymol. 2012;513:297–313. doi: 10.1016/B978-0-12-391938-0.00013-6. [DOI] [PubMed] [Google Scholar]
- 71.Wal M, Pugh BF. Genome-Wide Mapping of Nucleosome Positions in Yeast Using High-Resolution MNase ChIP-Seq. Methods Enzymol. 2012;513:233–250. doi: 10.1016/B978-0-12-391938-0.00010-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Galas DJ, Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matys V, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bryne JC, et al. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Newburger DE, Bulyk ML. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2009;37:D77–82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fullwood MJ, Ruan Y. ChIP-based methods for the identification of long-range chromatin interactions. J Cell Biochem. 2009;107:30–39. doi: 10.1002/jcb.22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stormo GD, Zhao Y. Determining the specificity of protein-DNA interactions. Nat Rev Genet. 2010;11:751–760. doi: 10.1038/nrg2845. [DOI] [PubMed] [Google Scholar]
- 79.Berger MF, et al. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Badis G, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guertin MJ, Martins AL, Siepel A, Lis JT. Accurate prediction of inducible transcription factor binding intensities in vivo. PLoS Genet. 2012;8:e1002610. doi: 10.1371/journal.pgen.1002610. This paper described a method that showed the importance of chromatin state dynamics, in addition to sequence preferences, in DNA binding intensities of proteins.
- 82.Dion MF, et al. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 83.van Werven FJ, van Teeffelen HA, Holstege FC, Timmers HT. Distinct promoter dynamics of the basal transcription factor TBP across the yeast genome. Nat Struct Mol Biol. 2009;16:1043–1048. doi: 10.1038/nsmb.1674. [DOI] [PubMed] [Google Scholar]
- 84.Lickwar CR, Mueller F, Hanlon SE, McNally JG, Lieb JD. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature. 2012;484:251–255. doi: 10.1038/nature10985. This paper provides evidence for a model of transcription factor binding in which factors are either stably bound and promote consistent transcription, or are ‘treadmilling’ through bound and unbound states resulting in lower transcription rates.
- 85.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. This paper describes a short-read sequence aligner that can simultaneously align to multiple DNA sequence variants. This removes the bias towards better aligning sequences containing alleles present in a reference genome and penalizing sequences containing non-reference alleles.
- 86.Zheng W, Zhao H, Mancera E, Steinmetz LM, Snyder M. Genetic analysis of variation in transcription factor binding in yeast. Nature. 2010;464:1187–1191. doi: 10.1038/nature08934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marks H, et al. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res. 2009;19:1361–1373. doi: 10.1101/gr.092643.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Motallebipour M, et al. Differential binding and co-binding pattern of FOXA1 and FOXA3 and their relation to H3K4me3 in HepG2 cells revealed by ChIP-seq. Genome Biol. 2009;10:R129. doi: 10.1186/gb-2009-10-11-r129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yildirim E, Sadreyev RI, Pinter SF, Lee JT. X-chromosome hyperactivation in mammals via nonlinear relationships between chromatin states and transcription. Nat Struct Mol Biol. 2011;19:56–61. doi: 10.1038/nsmb.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gaulton KJ, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42:255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bischof JM, et al. A genome-wide analysis of open chromatin in human tracheal epithelial cells reveals novel candidate regulatory elements for lung function. Thorax. 2011;67:385–391. doi: 10.1136/thoraxjnl-2011-200880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Waki H, et al. Global mapping of cell type-specific open chromatin by FAIRE-seq reveals the regulatory role of the NFI family in adipocyte differentiation. PLoS Genet. 2011;7:e1002311. doi: 10.1371/journal.pgen.1002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu W, et al. Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res. 2011 doi: 10.1101/gr.125088.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stitzel ML, et al. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab. 2010;12:443–455. doi: 10.1016/j.cmet.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Magnani L, Ballantyne EB, Zhang X, Lupien M. PBX1 genomic pioneer function drives ERalpha signaling underlying progression in breast cancer. PLoS Genet. 2011;7:e1002368. doi: 10.1371/journal.pgen.1002368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Parker SC, et al. Mutational signatures of de-differentiation in functional non-coding regions of melanoma genomes. PLoS Genet. 2012;8:e1002871. doi: 10.1371/journal.pgen.1002871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.He HH, et al. Differential DNase I hypersensitivity reveals factor-dependent chromatin dynamics. Genome Res. 2012;22:1015–1025. doi: 10.1101/gr.133280.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shibata Y, et al. Extensive Evolutionary Changes in Regulatory Element Activity during Human Origins Are Associated with Altered Gene Expression and Positive Selection. PLoS Genet. 2012;8:e1002789. doi: 10.1371/journal.pgen.1002789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cheng C, et al. Construction and analysis of an integrated regulatory network derived from high-throughput sequencing data. PLoS Comput Biol. 2011;7:e1002190. doi: 10.1371/journal.pcbi.1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Muino JM, Angenent GC, Kaufmann K. Visualizing and characterizing in vivo DNA-binding events and direct target genes of plant transcription factors. Methods Mol Biol. 2011;754:293–305. doi: 10.1007/978-1-61779-154-3_17. [DOI] [PubMed] [Google Scholar]
- 101.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sayers EW, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012;40:D13–25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Drouin R, et al. Structural and functional characterization of the human FMR1 promoter reveals similarities with the hnRNP-A2 promoter region. Hum Mol Genet. 1997;6:2051–2060. doi: 10.1093/hmg/6.12.2051. [DOI] [PubMed] [Google Scholar]
- 104.Essien K, et al. CTCF binding site classes exhibit distinct evolutionary, genomic, epigenomic and transcriptomic features. Genome Biol. 2009;10:R131. doi: 10.1186/gb-2009-10-11-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]