Abstract
The corpus callosum is the principal cerebral commissure connecting the right and left hemispheres. The development of the corpus callosum is under tight genetic control, as demonstrated by abnormalities in its development in more than 1,000 genetic syndromes. We recruited more than 25 families in which members affected with corpus callosum hypoplasia (CCH) lacked syndromic features and had consanguineous parents, suggesting recessive causes. Exome sequence analysis identified C12orf57 mutations at the initiator methionine codon in four different families. C12orf57 is ubiquitously expressed and encodes a poorly annotated 126 amino acid protein of unknown function. This protein is without significant paralogs but has been tightly conserved across evolution. Our data suggest that this conserved gene is required for development of the human corpus callosum.
Main Text
The corpus callosum (CC), the largest commissural white-matter tract in the mammalian brain, is essential for interhemispheric integration of sensory, motor, and higher-order cognitive information. It is traditionally divided into four segments, the rostrum, genu, body, and splenium. Development of the CC occurs during weeks 6–20 of gestation, and developmental defects, observed in 0.3%–0.7% of individuals undergoing brain imaging, are among the most common of brain malformations.1 Abnormalities of the CC are noted in more than 1,000 unique OMIM entries, suggesting that CC development is very sensitive to genetic perturbations. Additionally, twin studies have suggested a high heritability of the CC area.2 However, well-described disorders that specifically affect the CC in the absence of other major abnormalities are quite rare.
We previously described subtypes of CC abnormalities after having conducted detailed structural analysis from brain MRI scans from a cohort of 30 affected individuals from 19 families with a history of parental consanguinity.3 The initial presenting feature was developmental delay or epilepsy in all cases,4 and brain MRIs showed CC abnormalities without other major neuroanatomic defects. We focused on parental consanguinity to enrich the study for cases that were likely to involve recessive inheritance, in which phenotypes were more likely to be fully expressive and fully penetrant. We delineated one major category consisting of CC hypoplasia (CCH) without dysplasia. Most notable was the finding that even within a given CCH-affected family, whose members in all likelihood share a single homozygous-recessive allele, siblings can display a spectrum of severity ranging from almost normal appearance to complete agenesis. The data suggest that there might be wide-ranging phenotypic severity even with presumably identical genetic mutations in siblings and that complete agenesis might be a common end phenotype.
For this study we prioritized families for genetic analysis by focusing on those with first- or second-degree consanguinity and two or more affected members in the setting of CCH; these criteria made it likely that a recessive genetic origin was involved. All family members underwent a routine clinical brain MRI, including midline sagittal and axial views. After worldwide recruitment of individuals identified from 1999–2012 as having neurodevelopmental disease, we focused on those for whom a brain MRI either showed isolated CCH or showed CCH as the major imaging anomaly. Individuals with other major structural brain anomalies or overwhelming evidence of other dysmorphic or syndromic causes were excluded. We genetically evaluated 27 families, four of which were simplex. The remaining familes, including the previously ascertained 19 families, were multiplex with similarly affected siblings. The study was approved by appropriate ethics committees, and the families provided informed consent.
We used QIAGEN reagents to extract blood DNA, in some cases after performing a 5K whole-genome linkage scan on all informative family members by using the Illumina Linkage IVb mapping panel.5 We analyzed the DNA with easyLinkage-Plus software6 to calculate multipoint LOD scores. In each family, we generated whole-exome sequence on two affected individuals if two were available (20 families), one affected individual plus both parents in simplex families (four families), or a single affected individual from either type of family (three families). We used the Agilent SureSelect Human All Exome 50 Mb kit to capture exomes and an Illumina HiSeq2000 instrument to sequence them, resulting in ∼94% recovery at >10× coverage. We used the Genome Analysis Toolkit7 for SNP and INDEL variant identification and SeattleSeq for annotation. We then filtered out variants that were represented with an allele frequency of more than 1% in dbSNP, the Exome Variant Server, or our in-house exome data set of 1400 individuals (dbGaP study ID phs000288). Finally, we prioritized the variants according to scores from prediction programs (PolyPhen, Grantham, Phastcon, or Genomic Evolutionary Rate Profiling).8–11
Two families had mutations in genes already associated with diseases. Family 566 included two affected girls from a first-cousin consanguineous marriage, resulting in four unaffected children. Brain MRIs showed that the affected individuals had CCH, and they also displayed profound developmental delay, visual and motor impairment, and hypogenitalism, and the older child had microcornea; all of this suggested possible Warburg Micro syndrome (MIM 600118). CCH is one of the major findings in Warburg Micro syndrome,12 and not surprisingly, in both affected individuals exome sequencing identified a chr2:135890463A>G homozygous variant at the canonical splice-acceptor site for exon 11 of RAB3GAP (Refseq accession number NM_001172435.1), a gene mutated in this condition.13 Family 1158 included doubly consanguineous parents and three affected children, all three of whom displayed CCH, epilepsy, and delayed milestones. There was evidence of mild progressive symptoms, but metabolic studies were negative. Linkage analysis demonstrated a single linkage peak at chr18q11.2 with maximum LOD score 3.62. Exome sequenceing of two affected individuals identified a shared chr18:21148840G>A homozygous variant, predicting a p.Thr137Met transversion in a conserved threonine in NPC1 (RefSeq accession number NM_000271.4). NPC1 mutations lead to Niemann-Pick type C disease (MIM 257220), a neurovisceral disorder characterized by lysosomal accumulation.14 The major features, including upgaze nystagmus and organomegaly, were not prominent in these individuals, whereas mild phenotypes and CCH are well documented.15–17 These data support the finding of CCH as a part of many described conditions.
Although the remaining families had potentially causative variants, for only one gene were we able to validate the association in two independent CCH-affected families. In these families (542 and 1414; Figure 1), we identified a homozygous variant, chr12:7053285A>G, predicted to result in substitution of the initiator methionine of C12orf57 (c.1A>G, p.Met1?, Refseq accession number NM_138425.1, Entrez ID uc001qrz.3). Family 542, from Kuwait, consisted of first-cousin parents and nine children, three of whom were similarly affected. Family 1414, from the Eastern part of Libya, consisted of nine living children from 11 pregnancies (one with twins); two of these children were similarly affected. Concurrently, we identified two additional consanguineous multiplex families with the same chr12:7053285A>G mutation in C12orf57. Family 1183, clinically reported,18 presented with four healthy individuals and two affected individuals with CCH, optic coloboma, craniofacial, and skeletal abnormalities reminiscent of Temtamy syndrome (MIM 218340). The family was of Palestinian origin and displayed parental consanguinity. Family EZ1, from the United Arab Emirates, consisted of first-cousin parents and six living children, three of whom were similarly affected. One other child had died in adolescence from unrelated causes. Using similar methodology to perform whole-exome sequencing in two affected members per family led to independent identification of this mutation in both of these families. In family 542, we performed SNP analysis of six of the children and both parents by using the Illumina 5K Linkage IVb mapping panel5 to generate LOD scores under a strictly recessive mode of inheritance and identified several linkage peaks with pLOD scores of more than 3.0 on chromosomes 2, 10, and 12 (Figure 1E), which included C12orf57. In family EZ1, we performed SNP analysis by using the Illumina OMNI1 Quad platform on three affected individuals and three unaffected individuals and identified several loci of homozygosity; one of these was at chr12:6488450–23121965 (Figure S1 in the Supplemental Data available with this article online). Thus, the linkage and genotyping data from the two families analyzed with SNP array was consistent with a locus containing C12orf57.
Figure 1.
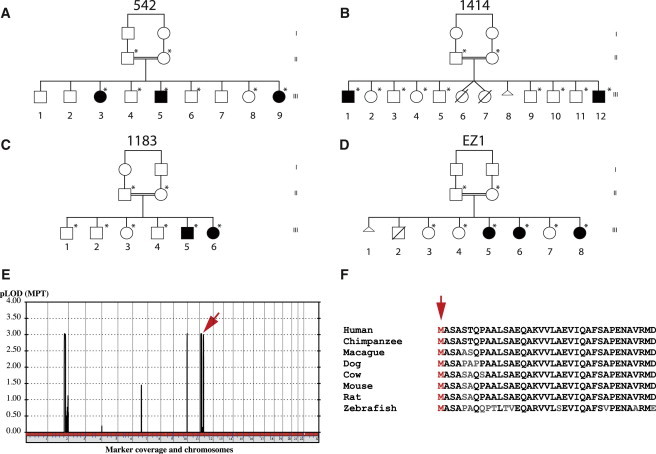
Families 542, 1414, 1183, and EZ1 with a Homozygous C12orf57 Mutation Encoding a p.Met1? Substitution Leading to CCH
(A–D) Pedigrees demonstrating affected and unaffected individuals, all from first-cousin marriages. An asterisk indicates that DNA was ascertained.
(E) Multipoint linkage plot resulting from the analysis of a 5K SNP array performed in all members of 542. Highest pLOD scores were found in chromosomes 3, 10, and 12. A red arrow indicates the location of C12orf57.
(F) Invariant conservation of the Met1 amino acid position of the predicted protein (red arrow). Nonconserved amino acids are in gray.
The clinical phenotype included hypotonia, moderate to severe intellectual disability, and various forms of epilepsy in eight of the ten affected members (Table 1). Four of the ten affected individuals showed evidence of abnormal visual function; such evidence included documented optic atrophy, coloboma, or an abnormal visual evoked potential. All ten individuals displayed autistic features, such as clinical automatisms, absent language, and poor social interactions. The two individuals in family 1183 had lower-extremity spasticity in conjunction with truncal hypotonia. Thus, the clinical phenotype is nonspecific.
Table 1.
Phenotype of C12orf57-Mutated Individuals
| Family ID | 542-III-3 | 542-III-5 | 542-III-9 | 1414-III-1 | 1414-III-12 | 1183-III-5 | 1183-III-6 | EZ1-III-5 | EZ1-III-6 | EZ1-III-8 |
|---|---|---|---|---|---|---|---|---|---|---|
| Family Data | ||||||||||
| Consanguineous | + | + | + | + | + | + | + | + | + | + |
| Multiplex | + | + | + | + | + | + | + | + | + | + |
| Number of affected individuals | 3 | 3 | 3 | 2 | 2 | 2 | 2 | 3 | 3 | 3 |
| Clinical Findings | ||||||||||
| Dysmorphic facies | − | − | − | + | + | + | + | + | + | + |
| Hypotonia | + | + | + | − | + | − | + | + | + | + |
| Microcephaly | − | − | − | − | − | − | + | − | − | − |
| Intellectual disability or developmental delay | + (severe) | + (severe) | + (severe) | + | + | + | + | + | + | + |
| Autistic features (absent language, stereotypies) | + | + | + | + | + | + | + | + | + | + |
| Seizures | + (PC) | + (GTC) | + (PC) | + | − | + (GTC + M) | + | − | + | + |
| Electroencephalogram | NA | multifocal discharges | NA | + generalized spike wave | normal | NA | NA | − | intermittent spike and slow waves, slowing | spike and slow wave discharges |
| Spasticity | − | + | + | + | − | + | + | − | − | − |
| Visual abnormalities | − | + abnormal VEP | + esotropia | − | + optic atrophy | − | left micro-ophthalmia; retinal/iris coloboma | − | − | − |
| Other systemic features | − | ASD | ASD, mild PS, conductive hearing loss | ASD | ASD | distal contractures; elevated BG | diabetes; distal contractures | − | − | − |
| MRI Findings | ||||||||||
| Corpus callosum | hypoplasia | hypoplasia | absent | hypoplasia | absent | hypoplasia | absent with midline colloid cyst | NA | NA | hypoplasia |
| Thalamic hypoplasia | + | + | + | + | + | + | + | NA | NA | + |
| Anterior commissure | − | + (decreased) | − | + (decreased) | − | NA | + (decreased) | NA | NA | + (decreased) |
| Ventriculomegaly | − | + | − | − | + | + | + | NA | NA | + |
| ON abnormalities | − | − | − | − | − | NA | − | NA | NA | − |
| Polymicrogyria | − | − | − | − | − | NA | − | NA | NA | − |
| Septum pellucidum abnormal | + | + | + | + | + | + | − | NA | NA | + |
| Reduced white matter | + | + | + | + | + | + | + | NA | NA | + |
Abbreviations are as follows: +, present; −, absent; NA, not available; ASD, atrial septal defect; BG, blood glucose; GTC, generalized tonic clonic; M, myoclonic seizures; ON, optic nerve; PC, partial complex; PS, pulmonary stenosis; and VEP, visual evoked potentials.
The affected members in the four families displayed clear and specific abnormalities of the corpus callosum (Figure 2); these abnormalities fell within the spectrum of hypoplasia to agenesis. In two individuals from family EZ1, imaging was not available for review. The CC was absent in three of the remaining eight individuals and hypoplastic in five. In family 542, the two older members (III-3 and III-5) displayed CC hypoplasia. Interestingly, the youngest affected member (III-9) displayed complete agenesis of the CC. In family 1414, the older individual (III-1) displayed CC hypoplasia, whereas the younger individual (III-12) displayed complete agenesis. In family 1183, the youngest child had complete callosal agenesis, and the older sibling had partial agenesis of the CC. Individual 1183-III-6 had a midline colloid cyst in the anterior aspect of the third ventricle; this cyst could be incidental or might suggest an underlying midline defect. In each member, notable hypoplasia of the thalamus produced a V-shaped, enlarged third ventricle, best appreciated in coronal images (Figure S2). When we evaluated the remaining 25 families from our CCH cohort, we found no others with this specific third-ventricle defect. Furthermore, enlargement of the third ventricle was apparent in only 2% of the more than 800 brain MRI scans in the UCSF Brain Disorder Research Program database, suggesting a highly specific, possibly pathognomonic radiographic hallmark.
Figure 2.
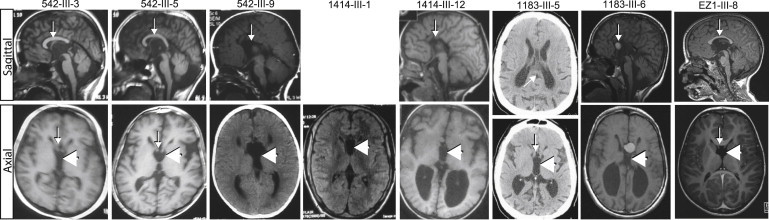
Brain Imaging Demonstrates a Specific CC Abnormality in C12orf57-Mutated Individuals
Upper image: midline sagittal MRI (where available). Lower image: axial MRI Images show CCH (arrows) in 542-III-3, 542-III-5, 1414-III-1, 1183-III-5, and EZ1-III-5 and complete agenesis in 542-III-9, 1414-III-2, and 1183-III-6. The bottom panel shows a prominent fornix (arrow) and unique appearance of the third ventricle (large arrowhead). Individual 1183-III-6 showed a colloid cyst, in addition to complete agenesis of the CC. No sagittal images were available for individual 1414-III-1, only a CT was available for individual 1183-III-5, and no imaging was available for individuals EZ1-III-5 and EZ1-III-6.
We confirmed the mutation on the basis of Sanger sequencing in the affected members in all four families, and we additionally confirmed that the mutation segregated according to a strictly recessive model with full penetrance. All tested parents were heterozygous carriers (one parent in family 1183 was unavailable for testing), whereas the unaffected siblings were either heterozygous or homozygous wild-type. The mutation was not identified in any other individuals from more than 1,400 individuals for whom whole-exome sequencing was available (of these 1,400, ∼1,000 are of matching Arab descent), nor was it evident in any public SNP database. There were no other potentially deleterious variants in this gene in our CCH cohort. We also tested an ethnically matched Kuwaiti and Libyan cohort of 100 control individuals each—Kuwait and Libya are the countries of origin for family 542 and 1414, respectively—and found no carriers for the mutation. Furthermore, other variants identified from exome sequencing were either nonconserved or did not segregate according to a recessive mode of inheritance in family 542 or 1414 (Table S1). For family 1183, only one other variant that we tested from the exome report was found to segregate appropriately, in ANKMY1 (Refseq accession number NM_016552.2), but mutations in this gene were not encountered in any of the other families. The data were consistent with the C12orf57 c.1A>G mutation's being causative in these four families.
Because the four families were of Arab descent, although of different geo-ethnic origin, we considered that c.1A>G might represent a founder mutation. This same mutation was presented as part of a list of 50 candidate genes associated with nonspecific intellectual disability,19 but a haplotype and detailed phenotype were not available. We analyzed the haplotype in families 542, 1414, and 1183 and identified a maximal potential sharing between chr12:7050931 and chr12:7061354, or a distance of about 10 kilobases (Figure 1E), suggesting a common founder mutation. (Family EZ1 sequencing was performed at a clinical referral center and was not available for review.) However, it is possible that the rare exonic variants contributing to this haplotype were relatively recently mutated (and that this is why they were not annotated as common polymorphisms), so we additionally genotyped a series of SNPs surrounding the mutation. However, we could not confirm a shared haplotype greater than this interval (not shown). Thus, although the data suggest that there might be a common founder mutation at this allele, definitive evidence is lacking.
Little is known about C12orf57 function. With 381 bp coding sequence, it predicts a 126 amino acid protein. In order to determine the expression profile of C12orf57, we retrotranscribed mRNA from a collection of human tissue and induced pluripotent stem-cell-derived human neural progenitor cells (NPCs). C12orf57 consensus cDNA was quantified by real-time qPCR. Even though the resulting product was detected in all tissues analyzed, fetal brain was one of the tissues showing the most abundant C12orf57 transcript (Figure 3A). The data suggest ubiquitous expression in all human tissue tested and a relative abundance of expression in fetal brain tissue.
Figure 3.
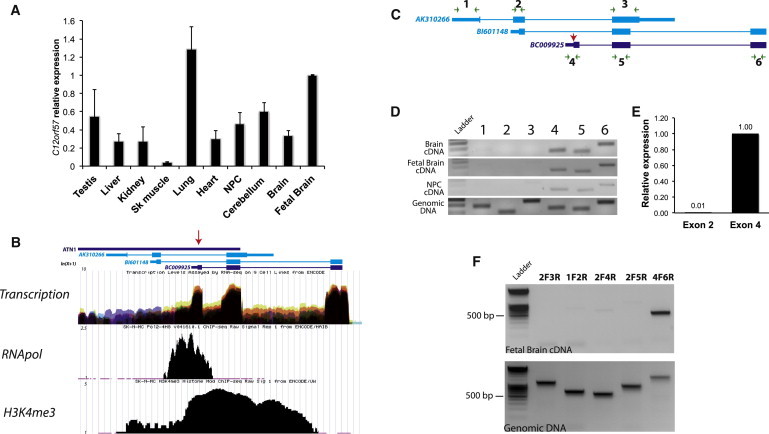
C12orf57 Transcript Expression in Human Tissues and Evidence for a Single Transcript Bearing the p.Met1? Substitution in CCH
(A) qPCR of C12orf57 transcript normalized with GAPDH in adult human testis, liver, kidney, skeletal muscle, lung, heart, NPCs, cerebellum, forebrain, and fetal brain total cDNA. Error bars represent the standard deviation between two independent PCR products for C12orf57 cDNA.
(B) ENCODE project track of UCSC Genome Browser shows transcriptional levels (from RNaseq), RNApol positioning, and H3K4me3 enrichment (from ChIPseq) for three C12orf57 cDNAs (represented by Genebank IDs AK310266, BI601148, and BC0092) with the most relevant alternative start codon, suggesting that the transcript in which Met1 is substituted (red arrow) in CCH is the major active transcript.
(C) Schematic representation of C12orf57 transcripts Primer positioning is indicated by green arrows, and the substituted Met1 is indicated by a red arrow.
(D) PCR of C12orf57 exons in human adult brain, fetal brain, and NPCs from total poly-A reverse-transcribed cDNA compared with total genomic DNA (bottom). Numbers refer to the primer pairs in (C).
(E) qPCR of C12orf57 exon 2, representative of AK310266 and BI601148, and exon 4, representative of BC00925 transcripts, from human fetal brain cDNA.
(F) RT-PCR from human fetal brain cDNA was performed with the indicated combination of primers and is consistent with the idea that the Met1 substituted in CCH derives the major active transcript.
C12orf57 falls within an evolutionarily conserved gene-rich cluster in chr12:p13.31.20 According to the UCSC Genome Browser (see Web Resources), the transcript overlaps with the annotated 3′-UTR of ATN1, encoding Atrophin, which is mutated with a trinucleotide expansion in DentatoRubral-PallidoLuysian Atrophy (DRPLA [MIM 125370]). Despite repeated attempts, we were unable to amplify from cDNA any fragments that would indicate that they are part of a common transcript (not shown). In reviewing evidence for the overlap, we found that it derives from a single spliced cDNA (GenBank AB209345), which is probably a run-on transcript. We conclude that the ATN1 transcript probably ends around chr12:7051500, at least 500 bp prior to the first annotated transcript of C12orf57, and that the two mRNAs are completely separable.
Multiple annotated cDNAs for C12orf57 differ by alternative splicing, different exon boundaries, and truncation of the 5′ end, suggesting three possible alternative promotors (represented by GenBank IDs AK310266, BI601148, and BC009925). This is important because if there are three separate start codons, it is possible that the start codon substitution we identified might be complemented by the other transcripts. However, three independent experimental lines of evidence from the recent Encode project21 suggest that the start codon we identified is the primary start site for the gene. First, RNaseq data from nine cell lines demonstrate active transcription of the BC009925 region. Second, in cell line SK-N-MC, a neuroepithelial cell line derived from a metastatic supra-orbital human brain tumor, the RNA polymerase (RNApol) binds predominantly to the BC009925 transcript start site. Third, histone H3K4 trimethylation (H3K4me3), commonly associated with actively transcribed regions, is enriched at the BC009925 locus start site but not in the upstream AK310266 or BI601148 locus start sites (ENCODE data in the UCSC Genome Browser, year 5 update [see Web Resources]; Figure 3B).22
In order to assess transcription and splicing directly in human brain, we attempted to generate PCR products from healthy human adult brain, fetal brain and NPC cDNA compared with total genomic DNA using primers complementary to the exons predicted by AK310266, BI601148 and BC009925 (Figure 3C). After 30 cycles of amplification, the six primer pairs generated products with similar efficiency from genomic DNA. In the same conditions, however, none of the AK3131266 and BI601148 exon predicted products were detected in human cDNA, whereas all of the BC009925 exon predicted products were well represented. Quantification of the transcripts by real time PCR showed that transcripts represented by BC009925 were at least 100-times more abundant in human fetal brain (Figure 3E). In addition our attempts to identify alternative transcripts extending the mRNA upstream by 5′RACE were unsuccessful and the predicted products for combination of primers that target probable alternative 5′ end transcirpts were not detectable by RT-PCR (Figure 3F). We conclude that BC009925 likely represents the major active transcript from the C12orf57 locus in human neural tissue.
To determine whether the protein is stable upon translation, we cloned the human cDNA upstream of EGFP and transfected COS7 cells. We found stable fluorescently tagged protein signal with generalized cytoplasmic localization (Figure 4A). In order to validate this result in a disease-relevant cells we transduced NPCs with lentivirus encoding EGFP-tagged C12ORF57 in the pINDUCER20 plasmid.23 This system allows dose-dependent doxycycline-inducible expression. After a subsaturating dose of doxycycline for 24 hr, EGFP signal was localized in the cytoplasmic compartment, supporting a cytoplasmic localization in neural cells (Figure 4B). To determine whether the human mutation affects the protein, we introduced the c.1A>G substitution into this lentiviral construct to compare protein levels between the mutant and wild-type. We found that the C12orf57 c.1A>G variant, which results in a GUG start codon, was able to produce some protein, although less efficiently than the wild-type (Figures 4C–4D). In fact, the GUG start codon within an optimal Kozak sequence is capable of initiating protein synthesis, although with less efficiency than AUG.24 Whether this is true for C12orf57 c.1A>G within the genomic context will require further investigtion, but the possibility that the mutation results in reduced protein levels could help explain the phenotypic variability.
Figure 4.
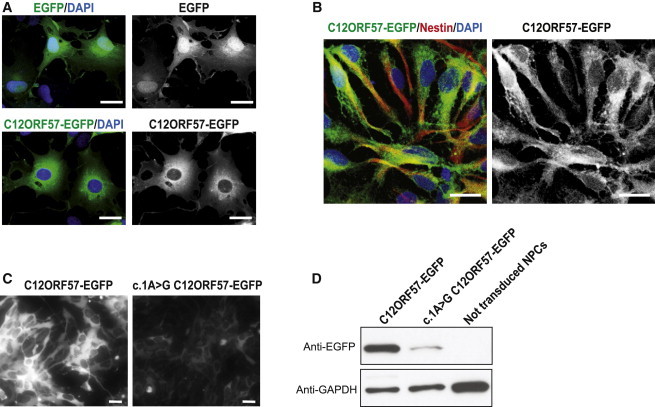
C12ORF57-EGFP Localization Is Cytoplasmic, and c.1A>G Mutation Results in Reduced Translational Efficiency
(A) Cytoplasmic localization of C12ORF57-EGFP fusion protein compared with the diffuse localization of EGFP in transiently transfected COS7 cells.
(B) Cytoplasmic localization of C12ORF57-EGFP in human NPCs. C12orf57-EGFP was subcloned in pINDUCER20 and transduced to NPCs, expression was induced with 30 ng/ml doxycycline for 24 hr, and NPCs were immunostained with Nestin antibody (red).
(C) Comparison between wild-type and c.1A>G C12ORF57-EGFP translated protein levels on transduced NPCs 1 week after positive selection with 200 μg/ml G418 and 24 hr after induction with 30 ng/ml doxycycline. Cells transduced with the mutant construct show notably reduced protein levels.
(D) Immunoblot with EGFP antibody in wild-type and c.1A>G C12ORF57 transduced NPCs treated as in (C). Severely reduced protein levels occur in the presence of the mutation. GAPDH was used as a loading control. The scale bar represents 20 μm.
With the major predicted protein in hand, we attempted to identify both paralogs in humans as well as orthologs in other species. A blastp search of the predicted 126 amino acid protein against the human proteome yielded no significant matches in the human protein database. The closest overlap was with the huntingtin-interacting protein 1-related protein (HIP1R, accession number NP_003950.1, e-value 0.004, 25% maximum identity over a 51 amino acid region), but blastp of HIP1R did not yield C12ORF57, so the relationship is uncertain. More interesting, though, was the phylogenetic analysis of C12ORF57 across evolution; for this analysis, we used a Grishin distance,25 a fast minimum evolution (FME) algorithm,26 and FigTree software. There is evidence that the C12orf57-encoded protein, called C10 (not to be confused with chemokine CC10), is highly conserved as a single copy across evolution. It is clearly traceable to annotated vertebrates, bony fish, most sequenced insects and cephalopods, and even some algae. Half of ten flies treated with RNAi to C10 in a genome-wide screen showed a nonspecific “malformation death” phenotype of the notum or dorsal thorax region of the body,27 but other species have not been subject to genetic perturbation.
In summary, we report a syndrome consisting of CC hypoplasia involving enlargement of the third ventricle, epilepsy, autistic features, and developmental delay. The phenotypic spectrum of CCH, even within a single family with a C12orf57 mutation, supports the idea that complete agenesis is an end phenotype. It also supports the idea that other genetic or environmental modifiers impart an effect on the resultant CC morphology. The C12orf57 locus produces a single major transcript encoding a 126 amino acid protein of unknown function but cytoplasmic localization. Reduced protein production associated with the identified mutation suggests a loss of function as the disease mechanism. The conservation of C12orf57 across evolution, even prior to the emergence of cephalopods, suggests an ancient function, the disruption of which leaves the developing human brain particularly vulnerable.
Acknowledgments
We thank the Center for Inherited Disease Research and UCLA Microarray Core (supported by the National Institutes of Health (NIH) and the NIH Heart, Lung, and Blood Institute) for genotyping support, and the Broad Institute (U54HG003067 to Eric Lander) for sequencing support and analysis. We thank S.J. Elledge for pINDUCER20 vector. We also thank the Wold lab (Caltech), Myers Lab (HudsonAlpha Institute for Biotechnology), and Sandstrom lab (UW ENCODE) for, respectively, RNAseq, RNApol ChIP, and H3K4me3 ChIP data, deposited on the UCSC Genome Browser. This work was supported by NIH grants R01NS048453, R01NS052455, P01HD070494 (to J.G.G.), and R01NS058721 (to E.H.S.), the Simons Foundation Autism Research Initiative, the Howard Hughes Medical Institute (J.G.G.), and a California Institute for Regenerative Medicine training grant (N.A).
Contributor Information
Elliott H. Sherr, Email: sherre@neuropeds.ucsf.edu.
Joseph G. Gleeson, Email: jogleeson@ucsd.edu.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
SeattleSeq Annotation, http://snp.gs.washington.edu/SeattleSeqAnnotation134
Exome Variant Server, http://evs.gs.washington.edu/EVS/
dbSNP Annotation, www.ncbi.nlm.nih.gov/projects/SNP/
UCSC Genome Browser, http://www.genome.ucsc.edu/
References
- 1.Paul L.K. Developmental malformation of the corpus callosum: a review of typical callosal development and examples of developmental disorders with callosal involvement. J. Neurodev. Disord. 2011;3:3–27. doi: 10.1007/s11689-010-9059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scamvougeras A., Kigar D.L., Jones D., Weinberger D.R., Witelson S.F. Size of the human corpus callosum is genetically determined: an MRI study in mono and dizygotic twins. Neurosci. Lett. 2003;338:91–94. doi: 10.1016/s0304-3940(02)01333-2. [DOI] [PubMed] [Google Scholar]
- 3.Hanna R.M., Marsh S.E., Swistun D., Al-Gazali L., Zaki M.S., Abdel-Salam G.M., Al-Tawari A., Bastaki L., Kayserili H., Rajab A. Distinguishing 3 classes of corpus callosal abnormalities in consanguineous families. Neurology. 2011;76:373–382. doi: 10.1212/WNL.0b013e318208f492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sztriha L. Spectrum of corpus callosum agenesis. Pediatr. Neurol. 2005;32:94–101. doi: 10.1016/j.pediatrneurol.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Murray S.S., Oliphant A., Shen R., McBride C., Steeke R.J., Shannon S.G., Rubano T., Kermani B.G., Fan J.B., Chee M.S., Hansen M.S. A highly informative SNP linkage panel for human genetic studies. Nat. Methods. 2004;1:113–117. doi: 10.1038/nmeth712. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann K., Lindner T.H. easyLINKAGE-Plus—automated linkage analyses using large-scale SNP data. Bioinformatics. 2005;21:3565–3567. doi: 10.1093/bioinformatics/bti571. [DOI] [PubMed] [Google Scholar]
- 7.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramensky V., Bork P., Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 10.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper G.M., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A., NISC Comparative Sequencing Program Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derbent M., Agras P.I., Gedik S., Oto S., Alehan F., Saatçi U. Congenital cataract, microphthalmia, hypoplasia of corpus callosum and hypogenitalism: report and review of Micro syndrome. Am. J. Med. Genet. A. 2004;128A:232–234. doi: 10.1002/ajmg.a.30109. [DOI] [PubMed] [Google Scholar]
- 13.Aligianis I.A., Johnson C.A., Gissen P., Chen D., Hampshire D., Hoffmann K., Maina E.N., Morgan N.V., Tee L., Morton J. Mutations of the catalytic subunit of RAB3GAP cause Warburg Micro syndrome. Nat. Genet. 2005;37:221–223. doi: 10.1038/ng1517. [DOI] [PubMed] [Google Scholar]
- 14.Carstea E.D., Morris J.A., Coleman K.G., Loftus S.K., Zhang D., Cummings C., Gu J., Rosenfeld M.A., Pavan W.J., Krizman D.B. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science. 1997;277:228–231. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]
- 15.Walterfang M., Fahey M., Abel L., Fietz M., Wood A., Bowman E., Reutens D., Velakoulis D. Size and shape of the corpus callosum in adult Niemann-Pick type C reflects state and trait illness variables. AJNR Am. J. Neuroradiol. 2011;32:1340–1346. doi: 10.3174/ajnr.A2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trouard T.P., Heidenreich R.A., Seeger J.F., Erickson R.P. Diffusion tensor imaging in Niemann-Pick Type C disease. Pediatr. Neurol. 2005;33:325–330. doi: 10.1016/j.pediatrneurol.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Palmeri S., Battisti C., Federico A., Guazzi G.C. Hypoplasia of the corpus callosum in Niemann-Pick type C disease. Neuroradiology. 1994;36:20–22. doi: 10.1007/BF00599187. [DOI] [PubMed] [Google Scholar]
- 18.Li J., Shivakumar S., Wakahiro M., Mukherjee P., Barkovich A.J., Slavotinek A., Sherr E.H. Agenesis of the corpus callosum, optic coloboma, intractable seizures, craniofacial and skeletal dysmorphisms: an autosomal recessive disorder similar to Temtamy syndrome. Am. J. Med. Genet. A. 2007;143A:1900–1905. doi: 10.1002/ajmg.a.31855. [DOI] [PubMed] [Google Scholar]
- 19.Najmabadi H., Hu H., Garshasbi M., Zemojtel T., Abedini S.S., Chen W., Hosseini M., Behjati F., Haas S., Jamali P. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- 20.Ansari-Lari M.A., Shen Y., Muzny D.M., Lee W., Gibbs R.A. Large-scale sequencing in human chromosome 12p13: experimental and computational gene structure determination. Genome Res. 1997;7:268–280. doi: 10.1101/gr.7.3.268. [DOI] [PubMed] [Google Scholar]
- 21.ENCODE Project Consortium A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenbloom K.R., Sloan C.A., Malladi V.S., Dreszer T.R., Learned K., Kirkup V.M., Wong M.C., Maddren M., Fang R., Heitner S.G. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2012;41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meerbrey K.L., Hu G., Kessler J.D., Roarty K., Li M.Z., Fang J.E., Herschkowitz J.I., Burrows A.E., Ciccia A., Sun T. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 2011;108:3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol. Cell. Biol. 1989;9:5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grishin N.V. A novel approach to phylogeny reconstruction from protein sequences. J. Mol. Evol. 1999;48:264–273. doi: 10.1007/pl00006469. [DOI] [PubMed] [Google Scholar]
- 26.Desper R., Gascuel O. Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. J. Comput. Biol. 2002;9:687–705. doi: 10.1089/106652702761034136. [DOI] [PubMed] [Google Scholar]
- 27.Mummery-Widmer J.L., Yamazaki M., Stoeger T., Novatchkova M., Bhalerao S., Chen D., Dietzl G., Dickson B.J., Knoblich J.A. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987–992. doi: 10.1038/nature07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


