Abstract
Recent genome-wide association studies have revealed an association between variation at the ADAMTS7 locus and susceptibility to coronary artery disease (CAD). Furthermore, in a population-based study cohort, we observed an inverse association between atherosclerosis prevalence and rs3825807, a nonsynonymous SNP (A to G) leading to a Ser-to-Pro substitution in the prodomain of the protease ADAMTS7. In light of these data, we sought a mechanistic explanation for this association. We found that ADAMTS7 accumulated in smooth muscle cells in coronary and carotid atherosclerotic plaques. Vascular smooth muscle cells (VSMCs) of the G/G genotype for rs3825807 had reduced migratory ability, and conditioned media of VSMCs of the G/G genotype contained less of the cleaved form of thrombospondin-5, an ADAMTS7 substrate that had been shown to be produced by VSMCs and inhibit VSMC migration. Furthermore, we found that there was a reduction in the amount of cleaved ADAMTS7 prodomain in media conditioned by VSMCs of the G/G genotype and that the Ser-to-Pro substitution affected ADAMTS7 prodomain cleavage. The results of our study indicate that rs3825807 has an effect on ADAMTS7 maturation, thrombospondin-5 cleavage, and VSMC migration, with the variant associated with protection from atherosclerosis and CAD rendering a reduction in ADAMTS7 function.
Introduction
Recently three genome-wide association studies (GWASs) have revealed that coronary artery disease (CAD) is associated with single nucleotide polymorphisms (SNPs) at the ADAMTS7 (a disintegrin and metalloprotease with thrombospondin motif, 7 [MIM 605009]) locus on chromosome 15q25.1–3 The lead CAD-associated SNP in one of these studies was rs3825807,1 an adenine (A) to guanine (G) polymorphism, resulting in a serine (Ser)-to-proline (Pro) substitution in the prodomain of ADAMTS7. The lead SNPs in the other two studies were rs19940162 and rs43800283 respectively, the former residing in intron 8 of ADAMTS7 and the latter at 7.6 kb upstream of the gene. It has remained unclear as to whether any of these SNPs, or other SNPs in linkage disequilibrium (LD) with them, has a functional effect on ADAMTS7 expression and/or activity and has any effect on the biological processes related to CAD.
ADAMTS7 belongs to the metalloproteinase family. Newly synthesized ADAMTS7 contains a signal peptide, a prodomain, a metalloproteinase domain, a disintegrin-like domain, and a thrombospondin type-1 motif.4 The prodomain is cleaved off during ADAMTS7 maturation and activation.4 Activated ADAMTS7 has proteolytic activity, and its best characterized substrate is thrombospondin-5 (TSP5, also known as cartilage oligomeric matrix protein, COMP),5 an extracellular protein present in such tissues as vascular walls and cartilages.5,6
A recent study in rats demonstrated that ADAMTS7 facilitated vascular smooth muscle cell (VSMC) migration by degrading the extracellular matrix protein TSP5 and thereby promoted neointima formation following vascular mechanical injury.6 Furthermore, in vitro studies have shown that VSMCs produce TSP57 and that TSP5 inhibits VSMC migration.6 Because VSMC migration is an important process in atherogenesis, it is likely that ADAMTS7 can also play a role in the development of atherosclerosis, the pathology underlying the vast majority of CAD.
In this study, we found that ADAMTS7 was present in human atherosclerotic plaques and that variation at the ADAMTS7 locus was associated with atherosclerosis in a population-based, longitudinal study. To seek a mechanistic explanation for this association, we further undertook a series of in vitro experiments. Results of these experiments indicate that the CAD-associated ADAMTS7 genotype has an effect on ADAMTS7 maturation, TSP5 cleavage, and VSMC migration.
Subjects and Methods
The procedures followed were in accordance with the ethical standards of the responsible committees on human experimentation, and appropriate informed consent was obtained.
Bruneck Study Cohort
Subjects of this study were residents of the Bruneck area in Italy, who participated in the Bruneck Study, details of which have been described previously.8–10 DNA samples for genotyping were available for 787 subjects. Ultrasound scanning of the right and left internal carotid and common carotid arteries was performed in 1990 and 1995 by the same experienced sonographer.8–10 Atherosclerotic lesions were defined according to two ultrasound criteria: (1) wall surface (protrusion or roughness of the arterial boundary) and (2) wall texture (echogenicity). The atherosclerosis score, indicative of atherosclerosis severity, was calculated by summing all diameters at 8 well-defined segments of the common and internal carotid arteries. Incident atherosclerosis was defined by the occurrence of atherosclerotic lesions in segments previously free of atherosclerosis or enlargement of nonstenotic lesions by a relative increase in the plaque diameter exceeding twice the measurement error of the method. Intima-media thickness was assessed in plaque-free sections of the common carotid arteries.
Immunohistochemical Analysis
Formaldehyde-fixed paraffin-embedded sections of atherosclerotic coronary arteries or atherosclerotic carotid arteries were deparaffinized, rehydrated, and incubated with sodium citrate for antigen retrieval. The sections were subjected to color or fluorescence immunostaining with an anti-human smooth muscle α-actin (SMA) antibody (Dako, M-0635, or Sigma, A5691 or C6198) and an anti-human ADAMTS7 antibody (Abcam, ab28557). Immunostaining for TSP5 and CD68 respectively in atherosclerotic coronary artery sections was also performed.
Isolation, Culture, and Immunocytochemical Analyses of Primary VSMCs
VSMCs were isolated from umbilical cord arteries. Cultured VSMCs were subjected to immunocytochemical examinations of the VSMC marker SMA, the endothelial cell marker von Willebrand factor (vWF), and the fibroblast marker discoidin domain receptor-2 (DDR2), and verified to be SMA-positive but vWF- and DDR2-negative (data not shown). Cells were also verified for the presence of ADAMTS7 by immunocytochemistry.
Determination of Genotype
Genomic DNA extracted from blood samples of the Bruneck Study subjects, atherosclerotic coronary arteries, and VSMCs was genotyped for rs3825807 with the use of the KASPar (KBiosciences Competitive Allele-Specific PCR SNP genotyping system) method. Accuracy of the genotyping results was verified by sequencing of a random selection of the samples.
Scratch Assay and Cell Migration Assay
Scratch assays were carried out using a previously described method.11 Migration assays were performed with the use of transwells (8 μm pore size; Greiner Bio-One Inc.), following a standard protocol.
Plasmid and Transfection
We utilized a previously constructed plasmid to generate a protein consisting of the ADAMTS7 signal peptide, prodomain (with ADAMTS7-214Ser) and metalloproteinase domain followed by a c-Myc epitope,12 and carried out site-directed mutagenesis (PCR forward and reverse primers: 5′-CAGAGCTGGAGCCTCGACGGGAG-3′ and 5′-CTCCCGTCGAGGCTCCAGCTCTG −3′) to generate a new plasmid for ADAMTS7-214Pro. Cultured HEK293 cells were transfected with the ADAMTS7-214Ser or ADAMTS7-214Pro plasmid.
Immunoblot Analyses
Cell lysates, conditioned media, and cell surface washes (with 0.5 M NaCl4) from VSMCs or transfected HEK293 cells were obtained and subjected to standard immunoblot analysis with an anti-TSP5 antibody (Millipore, MABT36), or an anti-ADAMTS7 prodomain antibody (Abcam, ab45044), or an anti-cMyc antibody (Sigma-Aldrich, M4439).
In Vitro TSP5 Cleavage Assay
Recombinant TSP5 (Abcam, ab104358) was incubated with concentrated media conditioned by VSMCs of the A/A or G/G genotype, or by HEK293 cells transfected with either of the ADAMTS7 plasmids described above, in a digestion buffer5 at 37°C for 8 hr. The digests, along with undigested TSP5 and concentrated conditioned media were subjected to immunoblot analysis with an anti-TSP5 antibody (Millipore, MABT36).
Real-Time RT-PCR
Quantitative RT-PCR analysis of ADAMTS7 was carried out in primary VSMCs from different individuals, as described in the legend of Figure S5 available online.
Allelic Expression Imbalance Analysis
Allelic expression imbalance analysis was performed on VSMCs that were heterozygous for rs3825807, as described in the legend of Figure S6.
Statistical Analysis
In the Bruneck Study, associations of rs3825807 with carotid atherosclerosis were tested by logistic and linear regression analyses. Base models were adjusted for age and sex. Multivariable models were adjusted for variables correlated with atherosclerosis parameters in the Bruneck cohort, including presence or absence of hypertension, smoking status, diabetes mellitus, level of alcohol consumption, levels of high density lipoprotein and low density lipoprotein, ferritin, fibrinogen, anithrombin III, the factor V Leiden mutation, body mass index, waist-to-hip ratio, and loge-transformed concentrations of urinary albumin, high-sensitivity C-reactive protein and lipoprotein(a). Variables with a skewed distribution were normalized by logarithmic transformation. Differences between genotypes in the percentage of ADAMTS7 stain area in atherosclerotic plaque area, the percentage of SMA stain area in plaque area, VSMC migration distance in the migration assays, and band intensity in immunoblot analyses were tested by t tests. ANOVA tests were performed to ascertain differences between genotypes in cell proliferation, senescence, and apoptosis. Real-time RT-PCR results were analyzed by the ΔΔCT method. Allelic expression imbalance analysis results were analyzed by Mann-Whitney test. All p values were two-sided.
Results
Association of Variation in ADAMTS7 with Atherosclerosis
In the Bruneck Study, we observed an inverse association between the ADAMTS7 rs3825807 G/G genotype and atherosclerosis. This genotype was associated with lower prevalence of carotid atherosclerosis (odds ratio [95% CI] = 0.51 [0.31–0.84] and 0.53 [0.31–0.90] in the data from 1990 and 1995 respectively, after adjustment for covariates, Table 1) and similarly lower atherosclerosis scores (β [95% CI] = −0.45 [−0.78, −0.11] and −0.50 [−0.86, −0.14] in the same data, after adjustment for covariates, Table 1). No association was detected for incident atherosclerosis or carotid intima-media thickness (Table 1).
Table 1.
Association between ADAMTS7 rs3825807 and Carotid Atherosclerosis in the Bruneck Study
|
Age/Sex-Adjusted Model |
Multivariable Modelb |
|||
|---|---|---|---|---|
| OR (95% CI)a | p value | OR (95% CI)a | p value | |
| Presence of atherosclerosis in 1990 | 0.58 (0.35–0.96) | 0.035 | 0.51 (0.31–0.84) | 0.008 |
| Presence of atherosclerosis in 1995 | 0.51 (0.32–0.82) | 0.005 | 0.53 (0.31–0.90) | 0.019 |
| Incident atherosclerosis in 1990–1995 | 0.69 (0.43–1.10) | 0.121 | 0.67 (0.40–1.09) | 0.108 |
| β (95% CI)a | p value | β (95% CI)a | p value | |
| Atherosclerosis score in 1990 | −0.43 (−0.77, −0.09) | 0.013 | −0.45 (−0.78, −0.11) | 0.010 |
| Atherosclerosis score in 1995 | −0.53 (−0.89, −0.16) | 0.005 | −0.50 (−0.86, −0.14) | 0.006 |
| Carotid intima-media thickness in 1995 | −0.02 (−0.06–0.02) | 0.427 | −0.02 (−0.06–0.02) | 0.401 |
Odds ratios (OR) and regression coefficients (β) were derived from logistic and linear regression analyses respectively, comparing the G/G genotype versus the A/A and A/G genotypes.
Multivariable analyses were performed with adjustment for variables correlated with atherosclerosis parameters in the Bruneck cohort, as described in Subjects and Methods.
ADAMTS7 in Human Atherosclerotic Plaques
A recent study in rats demonstrated that ADAMTS7 facilitated VSMC migration and thereby promoted neointima formation following vascular mechanical injury.6 In this study, we investigated whether ADAMTS7 is present in and around VSMCs in human atherosclerotic plaques. Double immunostaining for ADAMTS7 and the VSMC marker SMA showed that ADAMTS7 colocalized with a proportion of VSMCs in atherosclerotic plaques of coronary and carotid arteries (Figures 1A–1C). We observed that in the atherosclerotic plaque, VSMCs that accumulated ADAMTS7 were mostly located near the intima-media border and the fibrous cap (Figures 1A and 1B). ADAMTS7 stains were detected both within cells (highlighted by black arrowheads) and in the extracellular spaces (highlighted by green arrowheads) (Figures 1A). In an analysis of coronary atherosclerotic plaques from 44 individuals, no significant association was detected between ADAMTS7 abundance and rs3825807 genotype (Figure S2). There was a trend toward a lower percentage of SMA stain area in atherosclerotic plaque area, in individuals of the G/G genotype (Figure S2). Immunostaining showed that TSP5 was also present in human atherosclerotic coronary arteries (Figure S3).
Figure 1.
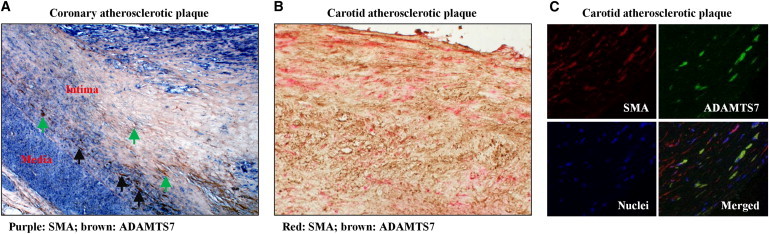
ADAMTS7 Colocalizes with Smooth Muscle Cells in Human Atherosclerotic Plaques
(A) Atherosclerotic coronary artery sections were subjected to double immunostaining of smooth muscle α-actin (SMA) and ADAMTS7. Purple color (NBT/BCIP) indicates SMA staining, and dark brown color with DAB indicates ADAMTS7 staining. Black arrow indicates cell positive for both SMA and ADAMTS7. Green arrow indicates extracellular ADAMTS7 staining. Negative controls are shown in Figure S1.
(B) Atherosclerotic carotid artery sections were subjected to double immunostaining of SMA with Fast red and ADAMTS7 with DAB (brown color). Negative controls are shown in Figure S1.
(C) Atherosclerotic carotid artery sections were subjected to double fluorescent immunostaining for SMA (red) and ADAMTS7 (green) and DAPI fluorescent staining for nuclei (blue), followed by confocal microscopy examination. Upper left panel shows SMA staining; upper right panel shows ADAMTS7 staining; lower left panel shows nuclear staining; lower right panel shows merged image from SMA, ADAMTS7, and nuclear staining.
Influence of ADAMTS7 Genotype on VSMC Migration
To investigate whether the atherosclerosis-associated ADAMTS7 genotype had an influence on VSMC migration, we verified that ADAMTS7 is present in cultured primary VSMCs (Figure 2A) and performed migration assays on primary VSMCs from individuals of different genotypes for rs3825807. The assays showed that VSMCs of the G/G genotype had reduced migratory ability, compared with VSMCs of the A/A genotype (Figures 2B–2D). Replacing the culture media of G/G genotype VSMCs with A/A genotype VSMC conditioned media at the beginning of the migration assay increased the migratory ability of G/G genotype VSMCs (Figures 2C and 2D, the fourth column compared with the second column at 6 hr and at 9 hr respectively), suggesting a migration enhancing effect from A/A genotype VSMC conditioned media, presumably due to a higher concentration of active ADAMTS7.
Figure 2.
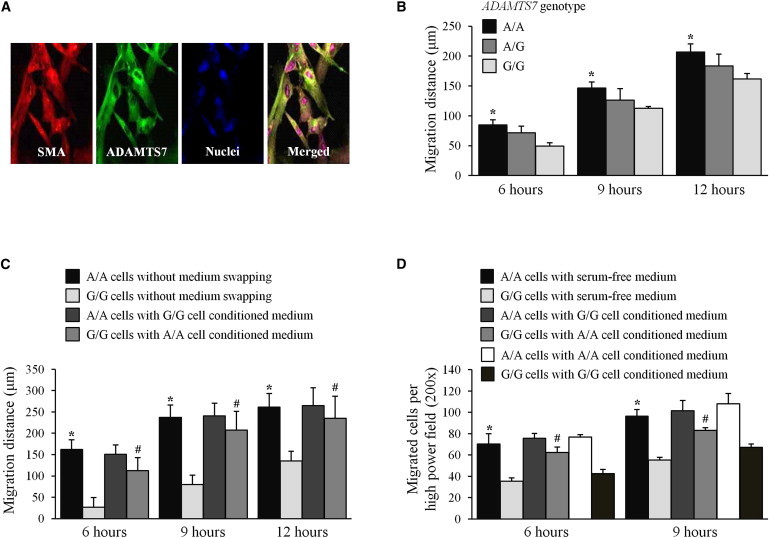
Effect of ADAMTS7 Genotype on VSMC Migration
(A) Primary cultures of VSMCs were subjected to double fluorescent immunostaining for the smooth muscle α-actin (SMA, red) and ADAMTS7 (green) and nucleus staining with propidium iodide (blue). Representative images are shown.
(B) Primary cultures of VSMCs of the A/A, A/G, or G/G genotype for rs3825807 were subjected to scratch assay by a commonly used method.11 Column chart shows migration distances (mean ± SEM) of VSMCs of the A/A, A/G, or G/G genotype for rs3825807 (n = 5 different donor cell preparations for each genotype). ∗p < 0.05 comparing A/A and G/G genotype group.
(C and D) Primary cultures of VSMCs of the A/A or G/G genotype were subjected to scratch (C) and migration (D) assays, without or with medium swapping at the outset of the assay (i.e., at hour 0 of the assay, the culture medium of G/G genotype cells was replaced by A/A genotype cell conditioned medium, and vice versa). Column chart shows migration distances or migrated cell numbers (mean ± SEM) (n = 3 different donor cell preparations for each genotype). ∗p < 0.05 comparing A/A genotype cells without medium swapping and G/G genotype cells without medium swapping; #p < 0.05 comparing G/G genotype cells with A/A genotype cell conditioned medium versus G/G genotype cells without medium swapping.
To ascertain whether ADAMTS7 genotype had an effect on VSMC proliferation and/or apoptosis, we performed proliferation, senescence, and apoptosis assays on primary VSMCs. We detected no difference between the genotypes in proliferation, senescence, or apoptosis (Figure S4).
Influence of ADAMTS7 Genotype on TSP5 Cleavage
It has been shown that VSMCs produce the extracellular matrix protein TSP5, which VSMCs adhere to7 and that ADAMTS7 promotes VSMC migration via degrading TSP5.6 Following the finding that ADAMTS7 genotype influenced VSMC migration, we investigated whether there was a difference in the concentration of cleaved TSP5 in VSMC conditioned media between ADAMTS7 genotypes. We found that media conditioned by VSMCs of the G/G genotype for rs3825807 contained approximately ∼30% less cleaved TSP5, compared with the A/A genotype (Figure 3A).
Figure 3.
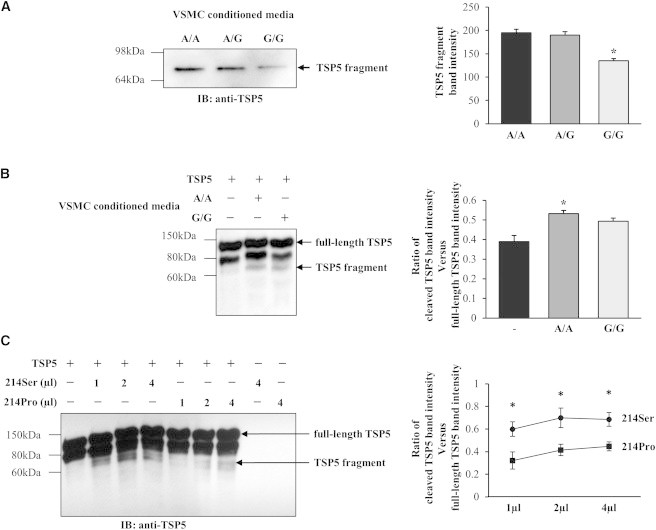
Effect of ADAMTS7 Genotype on Thrombospondin-5 Cleavage
(A) A representative image of immunoblot (IB) analysis of cleaved TSP5 in media conditioned by VSMCs of the A/A, A/G, or G/G genotype for rs3825807. The same amount (20 μg) of proteins for each genotype was loaded. Data shown in column chart are mean (±SEM) values of cleaved TSP5 band intensity in immunoblots (n = 5 different donor cell preparations for each genotype). ∗p < 0.05 comparing A/A and G/G genotypes.
(B) A representative image and quantifications of immunoblot analysis of products from in vitro TSP5 cleavage assays. Recombinant TSP5 (2 μg) was incubated with or without 5 μl concentrated media conditioned by VSMCs of the A/A or G/G genotype, in a digestion buffer at 37°C for 8 hr. The digests, along with undigested TSP5, were subjected to immunoblot analysis with an anti-TSP5 antibody. Data shown in graph are mean (±SEM) values (n = 4 for each genotype). ∗p < 0.05 comparing A/A (second column) and – (first column) or comparing A/A with G/G (third column).
(C) A representative image and quantifications of immunoblot analysis of products from in vitro TSP5 cleavage assays. Recombinant TSP5 (2 μg) was incubated with 1, 2, or 4 μl concentrated media (1 μg total proteins/μl) conditioned by HEK293 cells transfected with a plasmid to produce the ADAMTS7 prodomain-metalloproteinase-domain of either ADAMTS7-214Ser or ADAMTS7-214Pro, in a digestion buffer at 37°C for 8 hr. The digests, along with undigested TSP5 and concentrated conditioned media, were subjected to immunoblot analysis with an anti-TSP5 antibody. Data shown in graph are mean (±SEM) values (n = 3 for each genotype). ∗p < 0.05 comparing ADAMTS7-214Ser and ADAMTS7-214Pro.
Additionally, we carried out in vitro assays of TSP5 cleavage with media conditioned by VSMCs of the A/A or G/G genotype (Figure 3B) or media conditioned by HEK293 cells transfected with a plasmid to produce either ADAMTS7-214Ser or ADAMTS7-214Pro (Figure 3B). The assays showed that the conditioned media were able to cleave TSP5 and that media conditioned by VSMCs of the G/G genotype had lower TSP5 cleavage activity than media conditioned by VSMCs of the A/A genotype (Figure 3B). Similarly, media conditioned by HEK293 cells transfected with the ADAMTS7-214Pro-plasmid had lower TSP5 cleavage activity than media conditioned by ADAMTS7-214Ser-plasmid transfected cells (Figure 3C).
Effect of rs3825807 on ADAMTS7 Prodomain Processing
The Ser-to-Pro substitution resulting from rs3825807 occurs at amino acid residue 214 in the prodomain of the ADAMTS7 protein. Because proline is rigid, the substitution could potentially introduce a conformational change that might affect the secretion of the protein or the cleavage of the prodomain or its interaction with other molecules on the cell surface. Immunoblot analyses showed that VSMC conditioned media contained cleaved ADAMTS7 prodomain (Figure 4A), which was undetectable in cell surface washes (data not shown). Importantly, the analyses showed that the amount of the cleaved ADAMTS7 prodomain in media conditioned by VSMCs of the G/G genotype was ∼5-fold lower than in media conditioned by VSMCs of the A/A genotype (Figure 4A), whereas cell lysates of the two genotypes had similar amounts of full-length ADAMTS7 and ADAMTS7 fragments (Figure 4B). As reported by other researchers,4 the full-length ADAMTS7 band was ∼250 kDa in size (Figure 4B), which is larger than the predicted molecular weight (185 kDa) calculated based on the amino acid sequence, probably due to posttranslational modifications such as glycosylation.4 The ADAMTS7 prodomain band was ∼34 kDa in size (Figure 4A), which is similar to the reported size (∼37 kDa) of the ADAMTS9 [MIM 605421] prodomain with glycosylation.13
Figure 4.
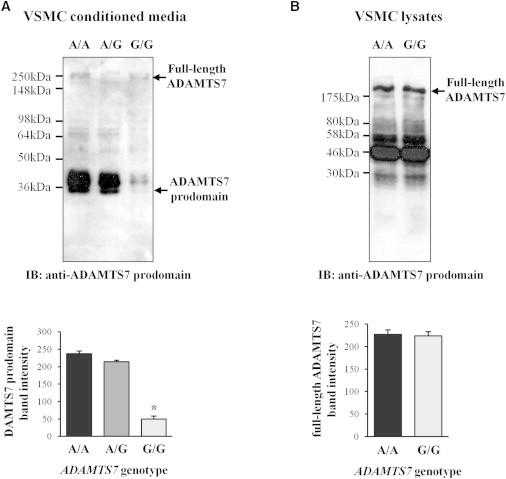
Effect of ADAMTS7 Genotype on the Amount of Cleaved ADAMTS7 Prodomain in VSMC Conditioned Media
(A) A representative image of immunoblot (IB) analysis of cleaved ADAMTS7 prodomain in media conditioned by VSMCs of the A/A, A/G, or G/G genotype for rs3825807. The same amount (20 μg) of proteins for each genotype was loaded. Data shown in column chart are mean (±SEM) values of cleaved ADAMTS7 prodomain band intensity in immunoblots (n = 5 different donor cell preparations for each genotype). ∗p < 0.05 comparing A/A and G/G genotypes.
(B) A representative image of immunoblot analysis of cleaved ADAMTS7 prodomain in lysates of VSMCs of the A/A or G/G genotype for rs3825807. The same amount (20 μg) of proteins for each genotype was loaded. Data shown in column chart are mean (±SEM) values of full-length ADAMTS7 band intensity in immunoblots (n = 3 different donor cell preparations for each genotype).
To further assess effects of the Ser-to-Pro substitution, we transfected cultured HEK293 cells with a plasmid to produce a recombinant protein consisting of the ADAMTS7 single peptide, prodomain (of either ADAMTS7-214Ser or ADAMTS7-214Pro), and metalloproteinase domain followed by a c-Myc epitope tag, and then carried out immunoblot analyses on conditioned media, surface washes (with 0.5M NaCl), and lysates of the transfected cells. Previous studies by other researchers of HEK293 cells transfected with a similar plasmid showed that the recombinant protein underwent prodomain cleavage, leading to an initially processed form (P1), an intermediate form (Pint) and a fully processed form (M)4 (as schematically illustrated in Figure 5A). In our study, immunoblot analyses of conditioned media with an anti-cMyc antibody showed three bands with sizes corresponding to the P1, Pint, and M forms, with the relative intensities of the Pint and M bands (versus the P1 band) being ∼1.7- and ∼4-fold lower, respectively, when comparing media conditioned by cells transfected to produce ADAMTS7-214Pro with media conditioned by cells transfected to produce ADAMTS7-214Ser (Figure 5B). Interestingly, the Pint and P1 forms of ADAMTS7-214Pro had a reduced electrophoretic mobility, compared with the Pint and P1 forms of ADAMTS7-214Ser (Figures 5B), which is in line with reports in the literature that proline-rich proteins tend to migrate more slowly in SDS-PAGE, presumably due to conformational peculiarities that persist even under reducing conditions.14,15 In cell surface washes and lysates, immunoblotting with the anti-cMyc antibody showed only one major band corresponding to the P1 form, with similar intensities for cells transfected with either plasmid (Figures 5E and 5F).
Figure 5.
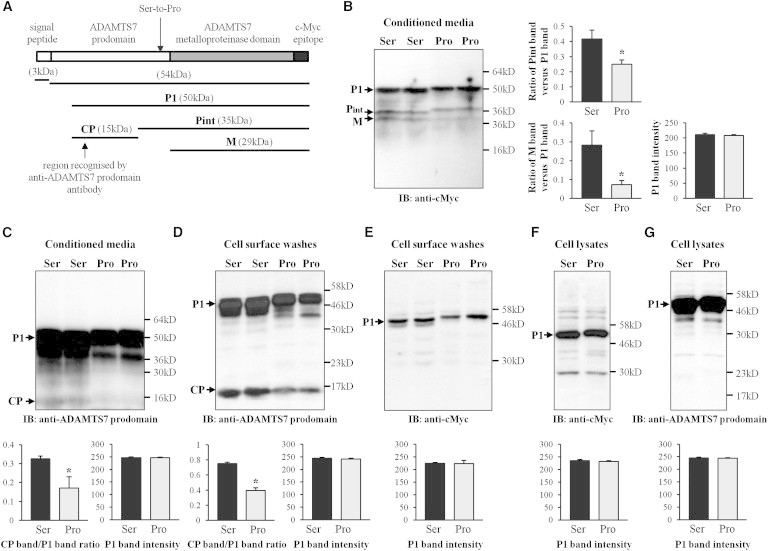
Effect of ADAMTS7 Genotype on ADAMTS7 Prodomain Processing
Cultured HEK293 cells were transfected with a plasmid to produce a recombinant protein containing the ADAMTS7 prodomain and metalloproteinase domain followed by a c-Myc epitope tag, with either serine (Ser) or proline (Pro) at residue 214 in the ADAMTS7 prodomain. Conditioned media, cell surface washes (with 0.5M NaCl), and cell lysates were subjected to immunoblot analyses with an anti-cMyc antibody or an anti-ADAMTS7 prodomain antibody. The same amount (40 μg) of proteins for each allele was loaded.
(A) A schematic diagram for the produced recombinant protein. P1, Pint, and M indicate the previously reported initially processed form, intermediately processing form, and fully processed form, respectively;4 CP indicates a cleaved ADAMTS7 prodomain fragment.
(B to G) Representative images of the immunoblot (IB) analyses and column chart presentations of mean (±SEM) values (n = 4 for each allele). ∗p < 0.05 comparing ADAMTS7-214Ser and ADAMTS7-214Pro.
Immunoblotting with an antibody against the ADAMTS7 prodomain detected a cleaved prodomain fragment (indicated by CP in Figures 5A, 5C, and 5D) in conditioned media (Figure 5C) and cell surface washes (Figure 5D), but not in cell lysates (Figure 5G), of the transfected cells, indicating binding of the cleaved prodomain fragment to the cell surface. Noticeably, the intensity of this band was ∼2-fold lower in conditioned media and cell surface washes, when comparing cells transfected to produce ADAMTS7-214Pro with cells transfected to produce ADAMTS7-214Ser (Figures 5C and 5D).
No Association between rs3825807 and ADAMTS7 Expression in VSMCs
To investigate whether ADAMTS7 genotype had an influence on ADAMTS7 expression in VSMCs, we performed a real-time RT-PCR analysis and an allelic expression imbalance analysis. In the real-time RT-PCR analysis, we detected no association between ADAMTS7 expression level and rs3825807 genotype (Figure S5), consistent with the results of immunoblot analysis that showed that cell lysates of the A/A and G/G genotypes had similar amounts of the ADAMTS7 protein (Figure 4B). In agreement, an allelic expression imbalance analysis showed no difference in ADAMTS7 expression level between the A and G alleles in VSMCs that were heterozygous for rs3825807 (Figure S6).
Discussion
In this study, we found an association between ADAMTS7 variation and atherosclerosis in the Bruneck cohort, with the rs3825807 G/G genotype associating with lower atherosclerosis prevalence and severity (score), which is in line with the finding from a recent GWAS that the G allele is associated to lower susceptibility to CAD.1 In in vitro assays, we found that VSMCs of the G/G genotype had reduced migratory ability and their conditioned media contained less-cleaved product of the ADAMTS7 substrate TSP5 and less-cleaved ADAMTS7 prodomain. Site-directed in vitro mutagenesis assay indicated that the Ser-to-Pro substitution reduces ADAMTS7 prodomain processing. We did not find evidence of between-genotype difference in ADAMTS7 accumulation or secretion, or in VSMC proliferation, senescence, or apoptosis. Taken together, these results indicate that the Ser-to-Pro substitution resulting from rs3825807 has an effect on ADAMTS7 maturation, TSP5 cleavage, and VSMC migration and is associated with atherosclerosis, providing a mechanistic explanation for the recently reported association between the SNP and CAD susceptibility.1
The effect of the Ser-to-Pro substitution on prodomain processing may be due to reduced accessibility to processing proteases such as furin, as a result of a conformational change induced by the rigidity of proline. The possibility of a conformational change is supported by our observation that the processed forms (Pint and M) of ADAMTS7-214Pro had reduced electrophoretic mobility. Although a conformational change might potentially also affect the secretion of the protein, the results of our study indicate that this would not be the case.
Rs3825807 was the lead SNP associated with CAD in one of the recently reported GWASs1 and is in LD with the lead CAD-related SNPs, rs1994016 and rs4380028, in two other recent GWASs2,3(r2 > 0.8 with rs1994016 and > 0.4 with rs4380028, based on data from HapMap and the 1000 Genomes Project). It is unknown whether rs1994016 and/or rs4380028 have a direct functional effect or act as proxy markers for functional SNPs (e.g., SNP rs3825807) due to LD. Rs1994016 resides in intron 8 of ADAMTS7, whereas rs4380028 is located 7.6 kb upstream of the gene. Considering their physical locations in relation to ADAMTS7, they might have an influence on ADAMTS7 expression, although currently there is no reported evidence.
VSMC migration is an important process in the pathogenesis of atherosclerosis.16 Indeed, VSMCs are one of the major constituents of atherosclerotic lesions.17 During atherogenesis, VSMCs migrate from the arterial media into the intima where the VSMCs proliferate and produce extracellular proteins,18 another major constitute of atherosclerotic plaques.17 The finding from our study that in atherosclerotic plaques, VSMCs that accumulate ADAMTS7 are predominantly located near the intima-media border is in line with the notion that ADAMTS7 plays a role in VSMC migration during atherogenesis.
Although a high VSMC content would increase the stability of the atherosclerotic plaque and reduce the risk of plaque rupture to cause such an acute ischemic event as myocardial infarction (MI),19,20 a genetic epidemiological study by Reilly et al. shows that ADAMTS7 variation is associated with coronary atherosclerosis but not with risk of MI.2 The finding of our study that the CAD-related ADAMTS7 genotype enhances VSMC migration is in line with that of the genetic epidemiological study by Reilly et al.2
Apart from the finding of an association between ADAMTS7 variation and CAD in genetic epidemiological studies,1–3 to our knowledge, there has been only one reported study of ADAMTS7 in vascular disease. Results of this latter study indicate that ADAMTS7 plays a role in vascular neointima formation after mechanical injury in a rat model, by inducing VSMC migration resulting from degradation of the extracellular matrix protein TSP5.6 A previous study shows that VSMCs produce TSP5, to which VSMCs adhere.7 Therefore, we focused our study on VSMCs. However, it seems warranted in future studies to investigate whether CAD-related ADAMTS7 variation also affects other cell types such as macrophages and endothelial cells. Our preliminary data, however, did not show an apparent difference in the abundance of CD68 positive cells in atherosclerotic plaques from individuals of rs3825807 A/A or G/G genotype (Figure S7).
Recently over 40 genomic loci have been identified by GWAS to be associated with genetic susceptibility to CAD. However, for many of these loci, the functional mechanisms leading to the genetic effect remain unknown. Functional characterization of these genetic variants can aid the understanding of the underlying biological mechanisms and may facilitate the translation of the genetic discoveries to therapeutic development. The findings of our present study on ADAMTS7 are pertinent in this context.
Acknowledgments
We thank the British Heart Foundation and the William Harvey Research Foundation for support. X.P. and C.F. are recipients of scholarships from the Chinese Scholarship Council. Q.X. is the recipient of a British Heart Foundation Intermediate Basic Science Research Fellowship (FS/09/044/28007). The work forms part of the research themes contributing to the translational research portfolio of Barts Cardiovascular Biomedical Research Unit, which is supported and funded by the National Institute for Health Research.
Supplemental Data
Web Resources
The URL for data presented herein is as follows:
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org/
References
- 1.Schunkert H., König I.R., Kathiresan S., Reilly M.P., Assimes T.L., Holm H., Preuss M., Stewart A.F., Barbalic M., Gieger C., Cardiogenics. CARDIoGRAM Consortium Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reilly M.P., Li M., He J., Ferguson J.F., Stylianou I.M., Mehta N.N., Burnett M.S., Devaney J.M., Knouff C.W., Thompson J.R., Myocardial Infarction Genetics Consortium. Wellcome Trust Case Control Consortium Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet. 2011;377:383–392. doi: 10.1016/S0140-6736(10)61996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coronary Artery Disease (C4D) Genetics Consortium A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat. Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 4.Somerville R.P., Longpré J.M., Apel E.D., Lewis R.M., Wang L.W., Sanes J.R., Leduc R., Apte S.S. ADAMTS7B, the full-length product of the ADAMTS7 gene, is a chondroitin sulfate proteoglycan containing a mucin domain. J. Biol. Chem. 2004;279:35159–35175. doi: 10.1074/jbc.M402380200. [DOI] [PubMed] [Google Scholar]
- 5.Liu C.J., Kong W., Ilalov K., Yu S., Xu K., Prazak L., Fajardo M., Sehgal B., Di Cesare P.E. ADAMTS-7: a metalloproteinase that directly binds to and degrades cartilage oligomeric matrix protein. FASEB J. 2006;20:988–990. doi: 10.1096/fj.05-3877fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L., Zheng J., Bai X., Liu B., Liu C.J., Xu Q., Zhu Y., Wang N., Kong W., Wang X. ADAMTS-7 mediates vascular smooth muscle cell migration and neointima formation in balloon-injured rat arteries. Circ. Res. 2009;104:688–698. doi: 10.1161/CIRCRESAHA.108.188425. [DOI] [PubMed] [Google Scholar]
- 7.Riessen R., Fenchel M., Chen H., Axel D.I., Karsch K.R., Lawler J. Cartilage oligomeric matrix protein (thrombospondin-5) is expressed by human vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 2001;21:47–54. doi: 10.1161/01.atv.21.1.47. [DOI] [PubMed] [Google Scholar]
- 8.Kiechl S., Willeit J. The natural course of atherosclerosis. Part I: incidence and progression. Arterioscler. Thromb. Vasc. Biol. 1999;19:1484–1490. doi: 10.1161/01.atv.19.6.1484. [DOI] [PubMed] [Google Scholar]
- 9.Kiechl S., Willeit J., Bruneck Study Group The natural course of atherosclerosis. Part II: vascular remodeling. Arterioscler. Thromb. Vasc. Biol. 1999;19:1491–1498. doi: 10.1161/01.atv.19.6.1491. [DOI] [PubMed] [Google Scholar]
- 10.Willeit J., Kiechl S., Oberhollenzer F., Rungger G., Egger G., Bonora E., Mitterer M., Muggeo M. Distinct risk profiles of early and advanced atherosclerosis: prospective results from the Bruneck Study. Arterioscler. Thromb. Vasc. Biol. 2000;20:529–537. doi: 10.1161/01.atv.20.2.529. [DOI] [PubMed] [Google Scholar]
- 11.Liang C.C., Park A.Y., Guan J.L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 12.Bai X.H., Wang D.W., Kong L., Zhang Y., Luan Y., Kobayashi T., Kronenberg H.M., Yu X.P., Liu C.J. ADAMTS-7, a direct target of PTHrP, adversely regulates endochondral bone growth by associating with and inactivating GEP growth factor. Mol. Cell. Biol. 2009;29:4201–4219. doi: 10.1128/MCB.00056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koo B.H., Longpré J.M., Somerville R.P., Alexander J.P., Leduc R., Apte S.S. Cell-surface processing of pro-ADAMTS9 by furin. J. Biol. Chem. 2006;281:12485–12494. doi: 10.1074/jbc.M511083200. [DOI] [PubMed] [Google Scholar]
- 14.Hung C.Y., Yu J.J., Seshan K.R., Reichard U., Cole G.T. A parasitic phase-specific adhesin of Coccidioides immitis contributes to the virulence of this respiratory Fungal pathogen. Infect. Immun. 2002;70:3443–3456. doi: 10.1128/IAI.70.7.3443-3456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williamson M.P. The structure and function of proline-rich regions in proteins. Biochem. J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz S.M. Perspectives series: cell adhesion in vascular biology. Smooth muscle migration in atherosclerosis and restenosis. J. Clin. Invest. 1997;99:2814–2816. doi: 10.1172/JCI119472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stary H.C., Chandler A.B., Dinsmore R.E., Fuster V., Glagov S., Insull W., Jr., Rosenfeld M.E., Schwartz C.J., Wagner W.D., Wissler R.W. A definition of advanced types of atherosclerotic lesions and a histological classification of atherosclerosis. A report from the Committee on Vascular Lesions of the Council on Arteriosclerosis, American Heart Association. Circulation. 1995;92:1355–1374. doi: 10.1161/01.cir.92.5.1355. [DOI] [PubMed] [Google Scholar]
- 18.Ross R. The pathogenesis of atherosclerosis: a perspective for the 1990s. Nature. 1993;362:801–809. doi: 10.1038/362801a0. [DOI] [PubMed] [Google Scholar]
- 19.Davies M.J. Stability and instability: two faces of coronary atherosclerosis. The Paul Dudley White Lecture 1995. Circulation. 1996;94:2013–2020. doi: 10.1161/01.cir.94.8.2013. [DOI] [PubMed] [Google Scholar]
- 20.Libby P. Molecular bases of the acute coronary syndromes. Circulation. 1995;91:2844–2850. doi: 10.1161/01.cir.91.11.2844. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


