LETTER
In China, IMP-4 is a major plasmid-mediated carbapenemase among multidrug-resistant Enterobacteriaceae, but little is known about the plasmid scaffolds involved in the dissemination of blaIMP-4 (1, 2). In 2010, we identified a Klebsiella pneumoniae CRE1 strain carrying blaIMP-4 from the penile swab of a 49-year-old man who was repatriated from Huizhou, China, where he had been hospitalized for intracranial hemorrhage. The isolate was resistant to all β-lactams, including imipenem, ertapenem, and meropenem, and multiple non-β-lactam antibiotics (chloramphenicol, co-trimoxazole, nitrofurantoin) (3). Combined disc testing (4) showed that carbapenem resistance can be reversed by EDTA but not boronic acid. Multilocus sequence typing (MLST) (5) identified CRE1 as sequence type (ST) 11, which is a major lineage associated with blaKPC dissemination in China (6). Conjugation was carried out in filters with Escherichia coli J53 as the recipient and transconjugants selected with sodium azide (100 μg/ml) and meropenem (1 μg/ml) (7). The blaIMP-4-carrying plasmid was able to be transferred at a frequency of 1.2 × 10−4 per donor cell.
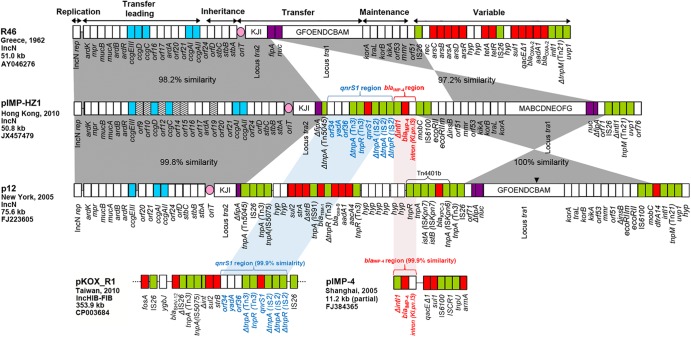
Plasmid DNA from the transconjugant was extracted, amplified, and sequenced using a GS-FLX system (Roche, Mannheim, Germany) as described previously (7). The assembled plasmid pIMP-HZ1 has 50,775 bp (GenBank accession no. JX457479). pIMP-HN1 has a plasmid scaffold typical for the IncN plasmids in general (8, 9). The backbone regions shared by pIMP-HZ1 and the reference IncN plasmid R46 include the replicon repA, the mucA-mucB genes (encoding UV protection), the stbABC operon (for plasmid stability), the ardA-ardB and ardK-ardR genes (with antirestriction function), the ccg genes (ccgEIII-ccgD-ccgC-ccgAI-ccgAII, providing protection from the type I restriction system), and the two transfer gene clusters (locus tra1 and locus tra2, encoding the conjugative apparatus) and genes for plasmid maintenance (Fig. 1). In pIMP-HZ1, there was a genetic rearrangement in the plasmid scaffold leading to inversion of the locus tra1. Despite this, pIMP-HZ1 is conjugative at a high frequency, indicating that functioning of the conjugal apparatus is not affected by the direction of transcription.
Fig 1.
Comparative analysis of linear plasmid maps for three IncN plasmids (R46, pIMP-HZ1, p12), one IncHIB/FIB plasmid (pKOX_R1), and the partially sequenced plasmid fragment pIMP4. The functional blocks for the IncN plasmids are indicated above the linear maps. R46 is used as the IncN reference. White boxes indicate the plasmid scaffold regions. Pink circles indicate the origin of transfer (oriT). The tra genes are indicated in capital letters inside the box. Hatched boxes indicate open reading frames (ORFs) in pIMP-HZ1 with no homologues in R46. Blue boxes indicate genes in the conserved upstream (CUP)-controlled regulon. Purple boxes indicate the fipA and nuc genes. Green boxes indicate the mobile elements. Red boxes indicate the resistance genes. The inverted triangle above p12 represents an 8.7-kb block (tnpA-rec-arsC-arsB-arsA-arsD-arsR) that was inserted into traB. Areas shaded in gray indicate homologies in the plasmid scaffold regions. All genes in the three IncN plasmids are shown, but the genes and regions in the maps are not drawn exactly in proportion to the length of the sequences.
Two resistance genes (qnrS1 and blaIMP-4) were found in the variable region of pIMP-HZ1. In IncN plasmids, the fertility inhibition gene (fipA) includes a hot spot for insertional recombination (9). In pIMP-HZ1, the variable region containing qnrS1 and blaIMP-4 was inserted within the fipA gene. As such, the insertion split the fipA gene into two fragments. The qnrS1-containing region was flanked by relics of Tn3-like tnp genes and a partial IS2. A similar qnrS1-containing region (99.9% nucleotide sequence identity) was found in the IncHIB-FIB plasmid, pKOX_R1 (Fig. 1). In pIMP-HZ1, blaIMP-4 is the first cassette downstream of a truncated integrase gene and is followed by a type II intron (Kl.pn.I3) which disrupted the attC recombination site. The genetic structure (Δintl1-blaIMP-4::Kl.pn.I3) has been given a novel integron number, In823::Kl.pn.I3, by INTEGRALL (http://integrall.bio.ua.pt/). A similar gene array has been described in the related blaIMP-4-carrying structure from plasmid pIMP4 (GenBank accession no. FJ384365), originating from Klebsiella isolates in China (2, 10, 11) (Fig. 1). Finally, part of the variable region (IS6100-ecoRII-ecoRIIm-ΔinsB), the maintenance and transfer genes, was identical to that in the IncN plasmid p12 (GenBank accession no. FJ223605) but lacking the 8.7-kb sequence (tnpA-rec-arsC-arsB-arsA-arsD-arsR) inserted into traB of p12 (Fig. 1). In conclusion, this analysis improves our understanding of the wider genetic context of plasmid-mediated blaIMP-4 in China.
ACKNOWLEDGMENTS
This work was supported by grants from the Research Fund for the Control of Infectious Diseases (RFCID) of the Health and Food Bureau of the Government of the HKSAR.
Footnotes
Published ahead of print 28 December 2012
REFERENCES
- 1. Carattoli A. 2011. Plasmids in Gram negatives: molecular typing of resistance plasmids. Int. J. Med. Microbiol. 301:654–658 [DOI] [PubMed] [Google Scholar]
- 2. Zhao WH, Hu ZQ. 2011. IMP-type metallo-beta-lactamases in Gram-negative bacilli: distribution, phylogeny, and association with integrons. Crit. Rev. Microbiol. 37:214–226 [DOI] [PubMed] [Google Scholar]
- 3. Clinical and Laboratory Standards Institute 2012. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. CLSI M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 4. Giske CG, Gezelius L, Samuelsen O, Warner M, Sundsfjord A, Woodford N. 2011. A sensitive and specific phenotypic assay for detection of metallo-beta-lactamases and KPC in Klebsiella pneumoniae with the use of meropenem disks supplemented with aminophenylboronic acid, dipicolinic acid and cloxacillin. Clin. Microbiol. Infect. 17:552–556 [DOI] [PubMed] [Google Scholar]
- 5. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. 2011. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 66:307–312 [DOI] [PubMed] [Google Scholar]
- 7. Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH, Ang I, Tong AH, Bao JY, Lok S, Lo JY. 2011. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One 6:e17989 doi:10.1371/journal.pone.0017989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brown AM, Willetts NS. 1981. A physical and genetic map of the IncN plasmid R46. Plasmid 5:188–201 [DOI] [PubMed] [Google Scholar]
- 9. Humphrey B, Thomson NR, Thomas CM, Brooks K, Sanders M, Delsol AA, Roe JM, Bennett PM, Enne VI. 2012. Fitness of Escherichia coli strains carrying expressed and partially silent IncN and IncP1 plasmids. BMC Microbiol. 12:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu Y, Zhang B, Cao Q, Huang W, Shen L, Qin X. 2009. Two clinical strains of Klebsiella pneumoniae carrying plasmid-borne blaIMP-4, blaSHV-12, and armA isolated at a pediatric center in Shanghai, China. Antimicrob. Agents Chemother. 53:1642–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yu F, Ying Q, Chen C, Li T, Ding B, Liu Y, Lu Y, Qin Z, Parsons C, Salgado C, Qu D, Pan J, Wang L. 2012. Outbreak of pulmonary infection caused by Klebsiella pneumoniae isolates harbouring blaIMP-4 and blaDHA-1 in a neonatal intensive care unit in China. J. Med. Microbiol. 61:984–989 [DOI] [PubMed] [Google Scholar]